Significance
Most of the Mycobacterium tuberculosis (Mtb) bacilli in the sputum of most patients with tuberculosis studied to date do not grow on standard agar-based media but rather grow when diluted in liquid media of similar composition. Here, we describe a rigorously standardized and independently replicated method to generate and count these differentially detectable (DD) Mtb in culture. DD Mtb generation required the action of a rifamycin on RNA polymerase after induction of phenotypic tolerance. Generation of these cells in vitro led to identification of one drug that can kill them and should facilitate the discovery of others.
Keywords: Mycobacterium tuberculosis, differentially detectable, phenotypic tolerance, rifampin, thioridazine
Abstract
Mycobacterium tuberculosis (Mtb) encounters stresses during the pathogenesis and treatment of tuberculosis (TB) that can suppress replication of the bacteria and render them phenotypically tolerant to most available drugs. Where studied, the majority of Mtb in the sputum of most untreated subjects with active TB have been found to be nonreplicating by the criterion that they do not grow as colony-forming units (cfus) when plated on agar. However, these cells are viable because they grow when diluted in liquid media. A method for generating such “differentially detectable” (DD) Mtb in vitro would aid studies of the biology and drug susceptibility of this population, but lack of independent confirmation of reported methods has contributed to skepticism about their existence. Here, we identified confounding artifacts that, when avoided, allowed development of a reliable method of producing cultures of ≥90% DD Mtb in starved cells. We then characterized several drugs according to whether they contribute to the generation of DD Mtb or kill them. Of the agents tested, rifamycins led to DD Mtb generation, an effect lacking in a rifampin-resistant strain with a mutation in rpoB, which encodes the canonical rifampin target, the β subunit of RNA polymerase. In contrast, thioridazine did not generate DD Mtb from starved cells but killed those generated by rifampin.
In 2015, an estimated 10.4 million people fell ill with tuberculosis (TB), and 1.8 million died from the disease, making TB one of the top 10 causes of death worldwide (1). An estimated one-third of the world’s population harbors latent TB infection, a massive reservoir of pathogen. If not treated, active TB is contagious and usually lethal. Although treatment can be curative, the currently recommended drug regimens for both latent infection and active disease require at least 6 mo of therapy, have significant side effects and drug–drug interactions (2–5), and are becoming ineffective as drug resistance mounts (1, 6). Limited understanding of clinically latent and drug-tolerant states of Mycobacterium tuberculosis (Mtb) hampers development of effective new drugs.
Heterogeneous subpopulations of Mtb survive stresses in the host such as acidity, exposure to host- or bacteria-derived reactive oxygen and reactive nitrogen species, and limitation in the supply of oxygen, carbon, nitrogen, or iron. In pathologic lesions, these subpopulations can also endure exposure to chemotherapeutics, including within cavities whose bacilli populate infectious respiratory tract secretions and provide most of the material for diagnosis and treatment monitoring. When such stresses are modeled in vitro, they can lead to suppression of Mtb replication and phenotypic tolerance to antimicrobial drugs without impairing the ability of the bacteria to resume growth when the stress is relieved (7–10).
Stresses can also induce a state in which bacteria remain viable but do not necessarily reenter replication in conventional assay conditions upon removal of the stress. Working with Vibrio cholerae and Escherichia coli, Colwell et al. first described bacterial populations unable to grow on media rendered semisolid with agar but able to grow in other formats or at later times and termed them “viable but nonculturable” (VBNC) (11). VBNC states have since been described for over 80 bacterial species (12). This is scientifically and medically problematic, as the gold standard for bacterial enumeration introduced by Koch over 130 y ago involves plating dilutions of a culture suspension on semisolid growth medium and counting colony-forming units (cfus). By this method, VBNC populations would be characterized as dead. In 2010, a study of 25 untreated TB patients in the United Kingdom found that 80–99% of the viable bacteria in the sputum of 80% of the subjects could not be detected as cfus (13). Instead, the initial bacterial number was estimated by the most probable number (MPN) method (13) based on a replicate series of limiting dilutions (LDs), herein termed an MPN–LD assay. In the present report, the mycobacteria estimated by the MPN–LD assay and found to be in numerical excess over those detected as cfus are called “differentially detectable” (DD) Mtb. In 2016, the finding that DD Mtb predominate in the sputum of most treatment-naïve TB patients was confirmed in a study of 110 subjects in South Africa (14).
With the existence of DD Mtb in human subjects now reported by independent groups, it is timely to confront conceptual and technical challenges that DD Mtb poses for the biology, experimental therapeutics, and clinical management of TB (15). A deeper understanding of the phenotype of such cells would greatly aid the development of simpler methods of enumerating them and discovering drugs that kill them. Toward that end we sought a method for generating DD Mtb in vitro that meets three criteria: The MPN–LD assay should yield the same count as the cfu assay in optimal growth conditions; the generation of DD Mtb must be reproducible in the hands of different operators in one laboratory; and the method must be independently confirmed in another laboratory without recourse to exchanges of personnel. Here, we describe a system fulfilling these requirements, involving nutrient starvation followed by exposure to rifampin (RIF), a first-line TB drug. We show that this is a property shared by other rifamycins and that RIF must engage its canonical target to produce DD Mtb. We then demonstrate the utility of this system for identifying compounds that can kill DD Mtb.
Results
Optimization of the MPN–LD Technique.
The method is outlined in Fig. 1 A and B and detailed in Method for Generation of DD Mtb Subpopulation and Movie S1. In brief, after culture of Mtb in standard or experimental conditions, the bacteria are serially diluted in a nutritionally rich medium in a pentuplicate series. An aliquot from each member of each dilution series is pooled for the five samples at a given dilution and plated for detection of cfus on agar solidified in a medium similarly as rich as that used for the dilutions. All wells demonstrating growth across dilutions are recorded as positive until there is no further increase in the number of positive wells over 2 wk, provided that any growth is confirmed to be caused by Mtb as opposed to a contaminant. By this criterion, the reading is usually final by 5 wk but sometimes not until 7 wk. As predicted by the Poisson distribution, not every replicate series reaches its limit at the same dilution. The MPN formula uses the pattern of growth across dilutions and information from replicate series to estimate the concentration of viable bacteria in the original suspension (16). Hereafter in this report, “MPN” denotes the number of bacteria in the original suspension as estimated by this statistical approach. Cfus are calculated by counting colonies at all dilutions that yield ∼10 to ∼150 colonies per plate and multiplying by the relevant dilution factors for those plates. DD Mtb are counted as the excess of MPN over cfus (DD Mtb = MPN – cfus).
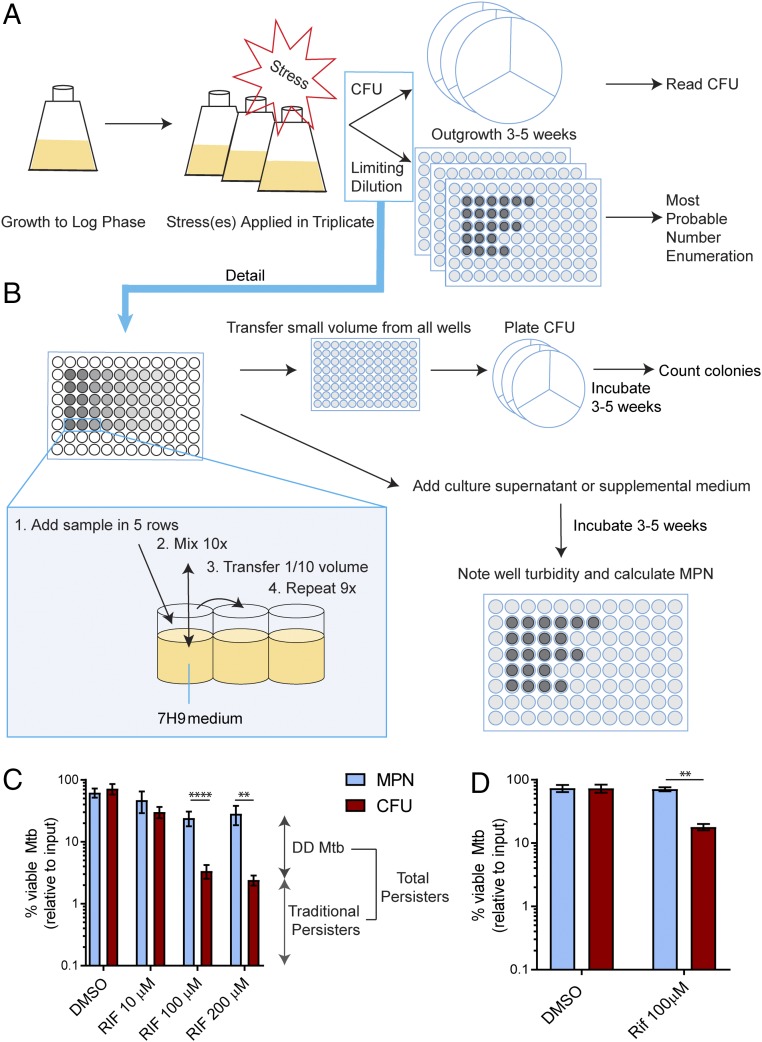
Fig. 1.
Schema for producing and counting DD Mtb. (A) General schema. Mtb is grown to log phase, subjected to stresses, and diluted serially in a rich medium in pentuplicate to the point that no growth is observed at the higher dilutions. Samples from the same dilution series are spread on agar plates made from nutrient-rich medium to count cfus. When there is no further outgrowth in the dilution series, the pattern of growth of Mtb in the dilution series is recorded and is used to determine the MPN. DD Mtb = MPN – cfus. (B) Detailed schema. After vortexing, samples are added to five replicate rows of a 96-well plate filled with 7H9 complete medium. Each well is mixed 10 times before transferring 1/10 of the volume to the next column, using filtered pipet tips and changing tips at each step. This procedure is repeated for a total of nine dilutions. The cfus are determined directly from aliquots of the dilution plates, not from the original suspension. Turbidity of dilution wells is recorded beginning at 3 wk and again as necessary until 2 wk have passed with no turbidity appearing at a higher dilution than at the previous reading. (C) DD Mtb formation. Exposure of H37Rv Mtb to RIF (10 μM, 100 μM, or 200 μM) for 5 d after 2 wk of starvation induces ∼90% DD Mtb. Data are means ± SEM for 21 biological replicates from seven experiments for DMSO and 100 μM RIF and for at least six biological replicates from two experiments for other points. (D) Independent confirmation. Using Method for Generation of DD Mtb Subpopulation as its guide, a second laboratory performed experiments like those in C. Data are means ± SEM for four replicates from two experiments. **P < 0.01; ****P < 0.0001.
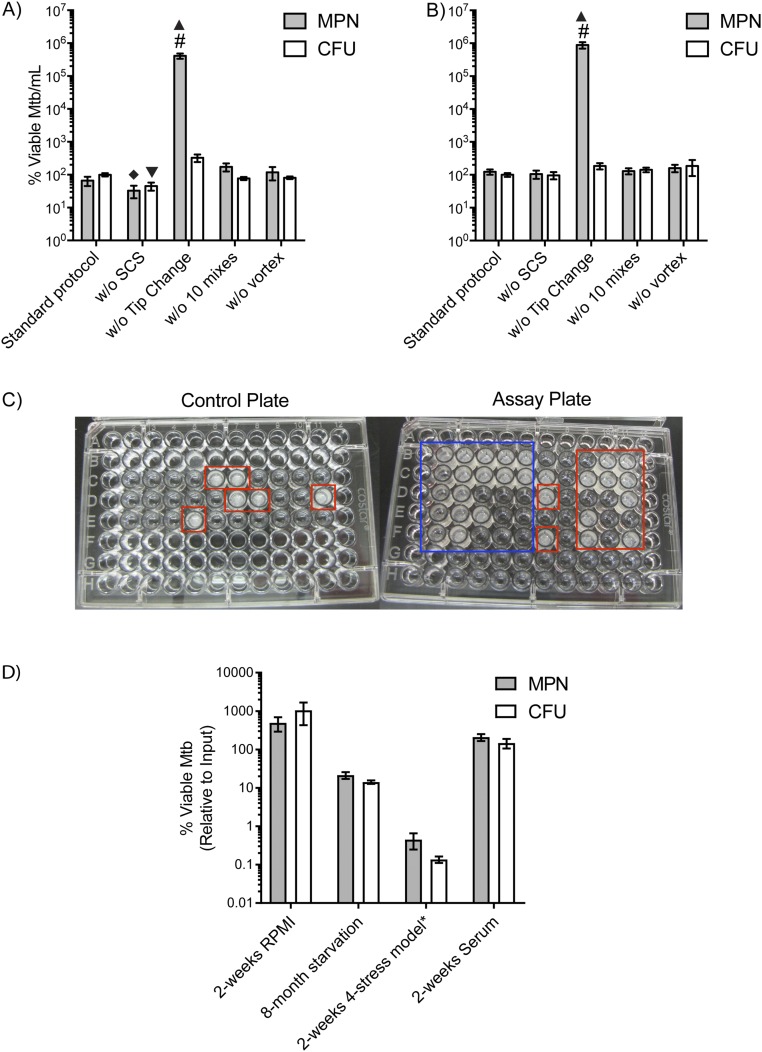
Our experiments before this work often showed apparent DD Mtb in cultures undergoing exponential replication. We considered such results likely to be spurious given the lack of a stress on these bacteria sufficient to induce nonreplication. We took several steps to ensure that MPN = cfu—that is, DD Mtb = 0—in replicating conditions. These included removing large clumps by a low-speed centrifugation at the start of the experiment, vortexing the suspension to break clumps before beginning dilutions, using filter pipet tips for serial dilution, and ensuring that the medium used in agar plates for cfu determination was as rich as and similar to that used for LD. Ten cycles of resuspension of the contents of each well before taking an aliquot for the next dilution increased accuracy compared with three cycles. The cfus had to be calculated from multiple wells in the dilution series used for the MPN assay, as described above, rather than from a separate dilution series derived from the initial suspension. The pipet tips had to be changed between each step in the dilution; failure to change tips led to a >10,000-fold artifactual increase in MPN. These LD and cfu steps were important when using nonstressed replicating bacteria and when using Mtb that was starved for 2 wk and nonreplicating (Fig. S1 A and B). Finally, the addition of Mtb culture filtrate (CF) or any other medium to the serially diluted sample sometimes introduced Mtb or other bacteria as contaminants whose growth exaggerated the MPN (Fig. S1C). To exclude contamination, every experiment included controls lacking intentionally added Mtb but containing all diluting and supplemental media.
Fig. S1.
Variations among different steps in the LD protocol. The standard LD protocol as described in Method for Generation of DD Mtb Subpopulation is compared with MPN and cfu numbers after variables were introduced in several steps with (A) log phase and (B) 2 wk-starved Mtb. Variables include the following: without single-cell suspension (w/o SCS), without changing tips during the dilution (w/o Tip change), with 3 instead of 10 mixes at each dilution (w/o 10 mixes), and without initial vortexing of the sample (w/o vortex). Error bars are SEM. Data are at least three biologic replicates. Two-way ANOVA with Tukey’s multiple comparisons test was performed to determine significance between all MPN results and between all cfu results. #, upper limit of assay; filled diamond, different compared with MPN w/o tip change (P < 0.0001), MPN w/o vortex (P < 0.05), and MPN w/o 10 mixes (P < 0.01); filled triangle, different compared with all other MPNs (P < 0.0001); inverted filled triangle, different compared with cfus w/o tip change (P < 0.0007). (C) Contamination during supernatant addition. Monitoring sterility of culture filtrate after addition to the plates (Left) shows significant growth in this experiment (red boxes), exposing carryover of Mtb through culture filtrate in the LD assay plate (Right). This leads to artifactual increase in MPN results but would not affect cfu results. Red boxes on Right indicate likely contaminant given the pattern is statistically improbable, whereas the blue box indicates an expected dilution series. (D) Screening for DD Mtb using standardized LD protocol. Several of the stresses tested for production of DD Mtb from log-phase culture are shown, including 2 wk of exposure to eukaryotic cell culture medium (RPMI), a four-stress model (*pH 5, 0.5 mM NaNO2, 1% O2, 0.05% butyrate) for 2 wk, prolonged starvation (8-mo starvation), and 2 wk of exposure to serum. None of the stresses create significant differences in number of viable Mtb between MPN enumeration and cfu count by paired t test statistical analysis. Error bars are SEM. Data are of at least six biologic replicates from two experiments.
Once we could reproducibly obtain MPN values that matched cfu values for log phase cultures, we tested diverse stresses for their ability to yield MPN > cfu—that is, to generate DD Mtb. We interpreted it as further evidence of minimal artifact in the MPN–LD method that not all conditions that reduced or blocked the replication of Mtb created DD Mtb but continued to yield MPN = cfu: nutrient deprivation by incubation in PBS for up to 8 mo, incubation in mammalian cell culture medium (RPMI), incubation in human serum, and exposure to a combination of four physiologically relevant stresses that mimic the host environment and have been used in high-throughput screens for agents that can overcome phenotypic tolerance—namely, acidic pH, nitrite as a source of reactive nitrogen intermediates, hypoxia (1% O2), and butyric acid as a carbon source (17, 18). None of these nonreplicating conditions generated DD Mtb (Fig. S1D).
DD Mtb Are Formed with Two Sequential Stresses.
Generation of DD Mtb has been reported in stationary-phase cultures exposed to high doses of RIF (19). We incubated Mtb in PBS to simulate nutrient-limited conditions. Starvation in PBS confers on Mtb a high level of phenotypic tolerance to most TB drugs, including RIF, isoniazid (INH), streptomycin (STREP), moxifloxacin (MOXI), ethambutol (ETH), and bedaquiline (TMC207) (20–22). The reported 90% Loebel-cidal concentration of Mtb nutrient-starved for 14 d was 25 μM for RIF, informing the use of high doses of rifamycins for our experiments (20).
When we incubated Mtb H37Rv for at least 2 wk in PBS and then added a high dose of RIF (100 or 200 μM) for 5 d, we detected two populations of persisters: one that remained cultivable as cfus (traditional persisters), and another detectable only by the MPN–LD assay (DD Mtb). In multiple experiments, DD Mtb produced with this combination of timing and dose constituted about 90% of the viable bacteria (Fig. 1C). As further evidence of the nonartifactual nature of the assay, starvation of Mtb in PBS for 2 wk followed by 5 d of exposure to the drug vehicle, dimethylsulfoxide (DMSO), had minimal impact on survival of Mtb and did not produce any difference between numbers of MPN and cfus (Fig. 1C). Sequential starvation and RIF exposure also generated DD Mtb in experiments carried out independently at the Albert Einstein College of Medicine (Fig. 1D) based only on the material in Method for Generation of DD Mtb Subpopulation.
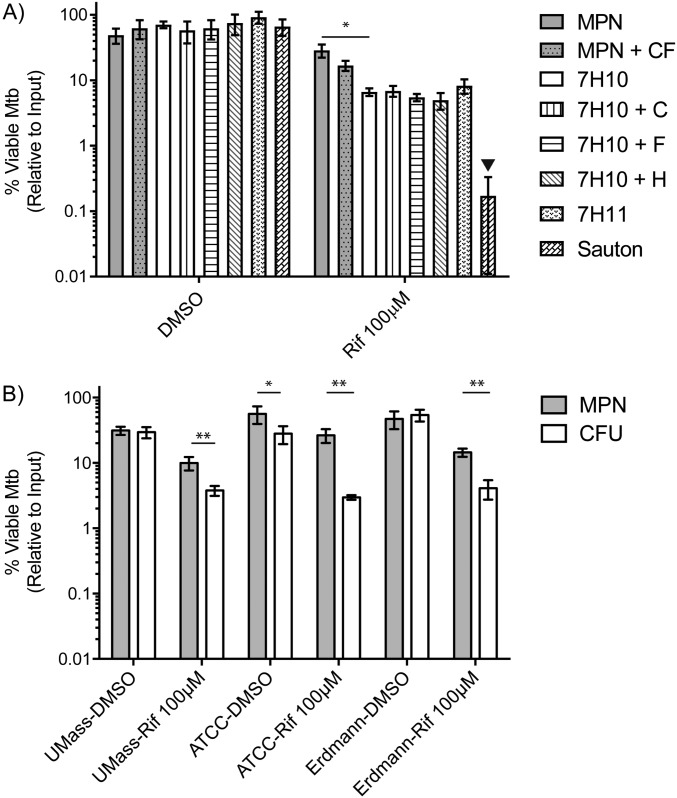
Inclusion of charcoal in the cfu plates to absorb RIF carried over from the original culture (23) did not increase the number of cfus and thereby affect the estimation of DD Mtb, nor did addition of hemin, an alternate source of iron (24), or fumarate, an alternate electron acceptor (25) (Fig. S2A). We wondered if the additional casein or malachite green in 7H11 medium would change cfu counts, but the numbers of cfus were similar on agar plates made with 7H10 or 7H11 medium (7H11 includes a casein digest and contains about fourfold more malachite green than 7H10). Addition of fresh CF from log-phase Mtb, shown in patient sputum to enhance DD Mtb recovery (13, 14), did not change MPN estimates of DD Mtb when supplemented in the MPN plates to a final concentration of 50% (Fig. S2A). Using Sauton’s, a nutrient-poor medium, in agar led to underdetection of cfus compared with cfus plated on 7H10 agar from the same dilutions (Fig. S2A).
Fig. S2.
(A) Variations in outgrowth conditions and recovery of viable Mtb. Two week-starved Mtb exposed for 5 d to either RIF or DMSO are subjected to various outgrowth conditions after washing. Addition of CF from log-phase Mtb does not improve recovery of DD Mtb from RIF-treated cells. The cfu recovery was monitored on 7H10 plates that were supplemented with 4 g/L charcoal (7H10 + C), 20 μM Hemin (7H10 + H), or 10 mM fumarate (7H10 + F). Two different types of agar—namely, 7H11 agar and Sauton’s agar—were also tested. Two-tailed, paired t test was performed to determine significance between MPN and MPN + CF as well as between MPN and cfu on 7H10. Two-way ANOVA with Tukey’s multiple comparisons test was performed to determine significance between all cfu results. Inverted filled triangle, different compared with all other cfus, P < 0.0001. (B) Different strains of Mtb show production of DD Mtb using the protocol. ATCC and UMASS H37Rv strains, as well as the Erdmann Mtb strain, create a DD Mtb population of 0.5–1 log10 after 2 wk of nutrient starvation and 5 d of RIF exposure. Error bars are SEM. Data are at least five biologic replicates from two experiments. *P < 0.05; **P < 0.01.
To ensure results were not laboratory strain-specific, we tested sequential starvation and RIF exposure using Mtb H37Rv obtained from the American Type Culture Collection (ATCC) and the University of Massachusetts (MA) and with the Erdman strain, and all gave similar results (Fig. S2B).
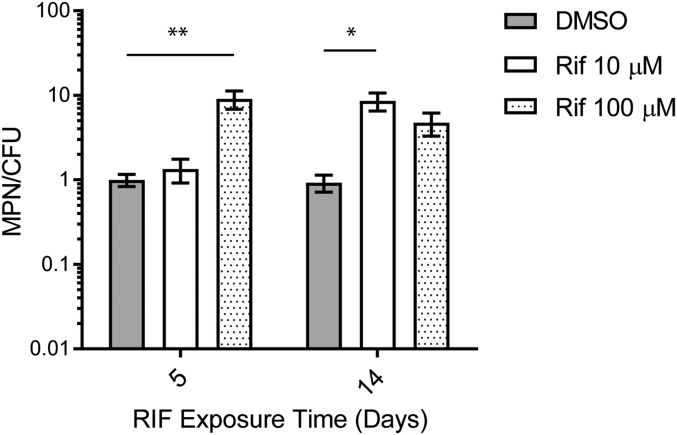
With a longer duration of starvation in PBS before RIF exposure, the overall bactericidal activity of RIF increased, such that only 6–25% of the Mtb before drug addition were recovered as MPN after exposure to 10 μM RIF and 1.5–7% after 100 μM RIF. With longer starvation, a lower dose of RIF (10 μM) sufficed to generate DD Mtb, and the relative size of the DD Mtb population increased to ∼1.5 log10 (∼97% of the surviving population) at certain time points (Fig. 2A). Although 2 wk of starvation followed by 5 d of exposure to 10 μM RIF did not create a significant DD Mtb population, DD Mtb comprised ∼90% of the population remaining when starvation for 2 wk was followed by exposure to 10 μM RIF for 14 d (Fig. S3). Thus, the proportion of DD Mtb increased with the duration of starvation, and longer starvation and longer drug exposure allowed DD Mtb generation with less RIF.
Fig. 2.
Characterization of DD Mtb model. (A) Variation in duration of starvation. Exposure of H37Rv Mtb to RIF (10 or 100 μM) for 5 d after 4, 8, 16, or 32 wk of starvation in PBS. Longer periods of starvation increased the mycobactericidal effect of RIF to 1 log10 but continued to generate ≥90% DD Mtb. Data are means ± SEM for nine biological replicates from three experiments. ****P < 0.0001 (marked associations significant at all time points to Pmax 0.0019). (B) Effect of RIF on log-phase bacteria. Mtb in log phase were exposed to RIF (10 or 100 μM) for 5 or 14 d. Data are means ± SEM from nine biological replicates from three experiments, except for the cfus with charcoal (cfu + C), which are from six biological replicates from two experiments. Points are offset for clarity. (C) Requirement for sequential stresses. Log-phase Mtb exposed simultaneously to starvation and RIF (10 or 100 μM) did not form DD Mtb. Data are means ± SEM of at least six biological replicates from three experiments. Where error bars are not visible, they are shorter than the height of the symbol.
Fig. S3.
10 μM RIF does not produce DD Mtb at 5 d but is comparable to 100 μM RIF at 14 d of exposure. Two week-starved Mtb exposed to 10 μM RIF does not produce a significant DD Mtb population compared with 100 μM RIF. Increasing exposure time to 2 wk with addition of 50% more RIF on day 7 to account for compound degradation creates 90% DD Mtb population for RIF 10 μM. Error bars are SEM. Data are of at least six biologic replicates from two experiments. *P < 0.05; **P < 0.001.
The RIF Tolerance of DD Mtb Is Phenotypic.
In each of two independent experiments in which Mtb was incubated in PBS for 2 wk and exposed to 100 μM RIF for 5 d, we removed 100 μL from each of five positive terminal dilutions (each estimated to contain >5 × 108 viable Mtb per mL, based on the optical density of the suspension) and plated the aliquots on agar containing 100 μM RIF. Only two colonies formed, indicating a frequency of RIF resistance of <2 × 10−9 in the DD Mtb population. Thus, DD Mtb were phenotypically tolerant rather than genetically resistant to RIF.
The Two Stresses That Generate DD Mtb Must Be Sequential.
As noted earlier, starvation of Mtb in PBS did not by itself induce DD Mtb. Neither did exposure of log-phase cultures of Mtb to RIF at 10 or 100 μM for 5 or 14 d, despite significant bactericidal activity (Fig. 2B). The slight difference between MPN and cfus seen at 5 d for RIF at 10 and 100 μM did not persist when agar plates were supplemented with charcoal. Equally ineffective was PBS starvation in the concomitant presence of RIF for 5 or 14 d (Fig. 2C).
RIF Must Engage with Its Canonical Target to Produce DD Mtb.
We applied sequential starvation and RIF exposure to a clinical strain of Mtb that carries an S531L point mutation in the RIF resistance-determining region of RNA polymerase subunit B (Rv0667), the canonical target of RIF, and accordingly displayed high-level resistance to RIF [minimum inhibitory concentration (MIC) > 1 μg/mL]. With either 2 wk or 2 mo of starvation followed by 5 d of 100 μM RIF, there was no loss of MPN or cfus relative to vehicle control in the S531L mutant (Fig. 3A). Thus, off-target effects of relatively high RIF doses were not responsible for DD Mtb generation. Instead, binding of RIF to RpoB appears to be required for formation of DD Mtb in this system. Rifabutin (Rib) and rifapentine (Rfp) also produced DD Mtb in a dose-dependent manner after 4 mo of starvation (Fig. 3B).
Fig. 3.
Role of rifamycins and requirement for engagement of RpoB. (A) Effect on RIF-resistant strain. A clinical RIF-resistant Mtb strain was starved for 2 or 8 wk followed by RIF (100 μM) for 5 d. Data are means ± SEM for six biological replicates from two experiments. (B) Effect of Rfp. H37Rv Mtb was starved for 4 mo and then exposed to Rfp at 1, 10, 30, and 100 μM for 5 d. Data are means ± SEM for three biological replicates from one experiment. (C) Effect of Rib. H37Rv Mtb was starved for 4 mo and then exposed to Rib at 1, 10, 30, and 100 μM for 5 d. Data are means ± SEM for three biological replicates from one experiment. For clarity, only significant differences between MPN and charcoal cfus are noted. *P < 0.05; **P < 0.01; **P < 0.001.
Other Compounds Have Variable Effects on Starved Mtb.
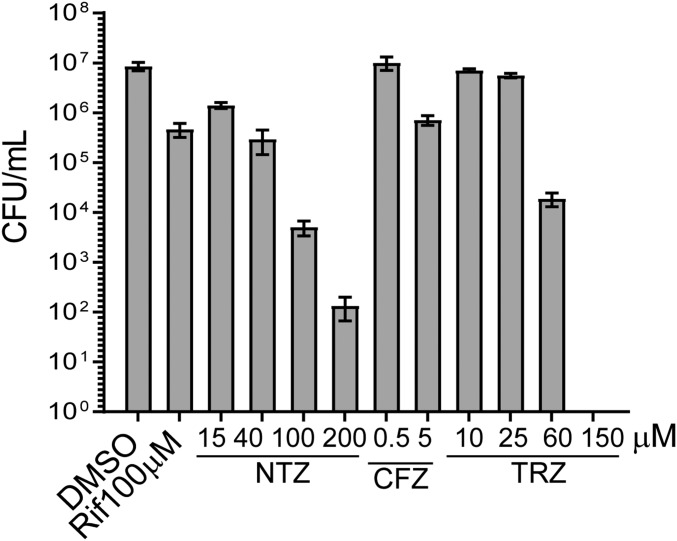
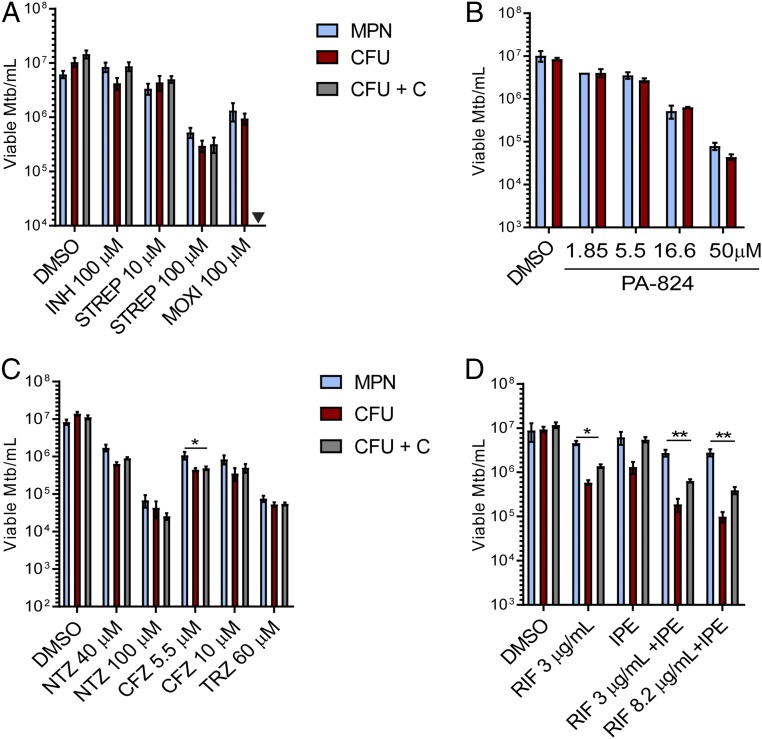
Next, we tested other drugs used to treat TB for their ability to generate DD Mtb. Drugs were tested up to 100 μM; in some cases, the dose was selected based on evidence that it caused a 1–2 log10 drop in cfu count of 2 wk-starved Mtb (Fig. S4). Exposure of 2 wk-starved Mtb for 5 d to INH at 100 μM or STREP at 10 μM showed minimal cidality. However, in contrast to our results with RIF, addition of charcoal to the agar plates was necessary to prevent artifactual reduction of the cfu count after exposure to INH (Fig. 4A). Increasing the concentration of STREP to 100 μM led to ∼1 log10 killing of starved Mtb but without formation of DD Mtb. MOXI at 100 μM after 2 wk of PBS starvation afforded 0.5–1 log10 killing but did not produce DD Mtb (Fig. 4A). PA-824 (pretomanid), a drug showing promise in clinical trials (26, 27), minimally impacted 2 wk-starved Mtb at doses at or below 5.5 μM and did not create DD Mtb under those conditions (Fig. 4B). However, the highest dose tested (50 μM) afforded ∼2 log10 killing compared with vehicle control (Fig. 4B). Clofazimine, previously shown to have potent activity against nonreplicating Mtb in anaerobic conditions (28, 29), led to ∼1 log10 killing at both 5.5 and 10 μM. There was a small (mean, 0.29 log10), statistically significant difference between MPN and cfu enumeration at 5.5 μM of clofazimine. However, this finding was not replicated at 10 μM clofazimine. Thioridazine (TRZ), which has shown activity against both replicating and nonreplicating Mtb (30–33), reduced the viability of 2 wk-starved Mtb by 2 log10 without forming DD Mtb at a dose of 60 μM (Fig. 4C). Similarly, nitazoxanide (NTZ), which is also active against both replicating and nonreplicating Mtb (34, 35), exhibited dose-dependent killing of starved Mtb up to 100 μM but left no significant DD Mtb population (Fig. 4C). As noted earlier in regard to experiments with INH, it was necessary to include charcoal in the agar plates to avoid artifactual results from carryover of drug(s) other than RIF that otherwise impacted the outgrowth of cfus.
Fig. S4.
Dose–response curve of compounds against 2 week-starved Mtb. For the purposes of dose selection in DD Mtb experiments, 2 wk-starved Mtb was exposed for 5 d across several doses for NTZ, CFZ, and TRZ. Data are means ± SEM for three biological replicates from one experiment.
Fig. 4.
Effect of other drugs on starved Mtb. Mtb was starved for 2 wk and treated for 5 d with (A) 100 μM INH, 10 or 100 μM STREP, or 100 μM MOXI; (B) 1.85, 5, 16.6, or 50 μM of PA-824 (H37Rv strain ATCC was used with PA-824); and (C) 40 or 100 μM NTZ, 5.5 or 10 μM clofazimine (CFZ), or 60 μM TRZ. (D) Four week-starved Mtb was exposed to RIF at 3 or 8.2 μg/mL in combination with INH (3 μg/mL), PZA (20 μg/mL) and ETH (2 μg/mL), RIF at 3 μg/mL alone, or the combination of INH, PZA, and ETH without RIF for 5 d. cfu + C refers to cfu counts recovered from 7H10 plates supplemented with 0.4% charcoal. Data are means ± SEM for at least six biological replicates from two experiments, except for IPE alone and RIF 3 μg/mL alone, which are from three biological replicates. For clarity, only significant differences between MPN and charcoal cfus are noted. Inverted filled triangle, not tested. *P < 0.05; **P < 0.01.
Finally, we tested the effect of the standard four-drug combination used to treat drug-sensitive TB, hypothesizing that the therapy given to TB patients might have a role in the formation or treatment of DD Mtb. Doses of RIF, INH, pyrazinamide (PZA), and ETH were selected based on mean peak drug concentrations (Cmax) achieved in patients’ serum (36–38). For these studies, Mtb was starved in PBS for 4 wk, then exposed to the four drugs simultaneously for 5 d, with two different doses of RIF—one based on Cmax data (3 μg/mL), and one already shown to create DD Mtb (10 μM; 8.2 μg/mL). The three drugs—INH (3 μg/mL; 21.8 μM), PZA (20 μg/mL; 162.5 μM), and ETH (2 μg/mL; 9.8 μM)—were tested in combination with and without RIF. With both doses of RIF, 90% of viable Mtb were in the DD Mtb state after 5 d of drug exposure, with minimal cidality when quantified by MPN compared with drug vehicle (Fig. 4D). Addition of the three other first-line drugs to RIF at the two doses did not reduce viability nor alter the size of the DD Mtb subpopulation. Nor did the three-drug combination alone induce DD Mtb.
INH and TRZ Have Different Effects on Preformed DD Mtb.
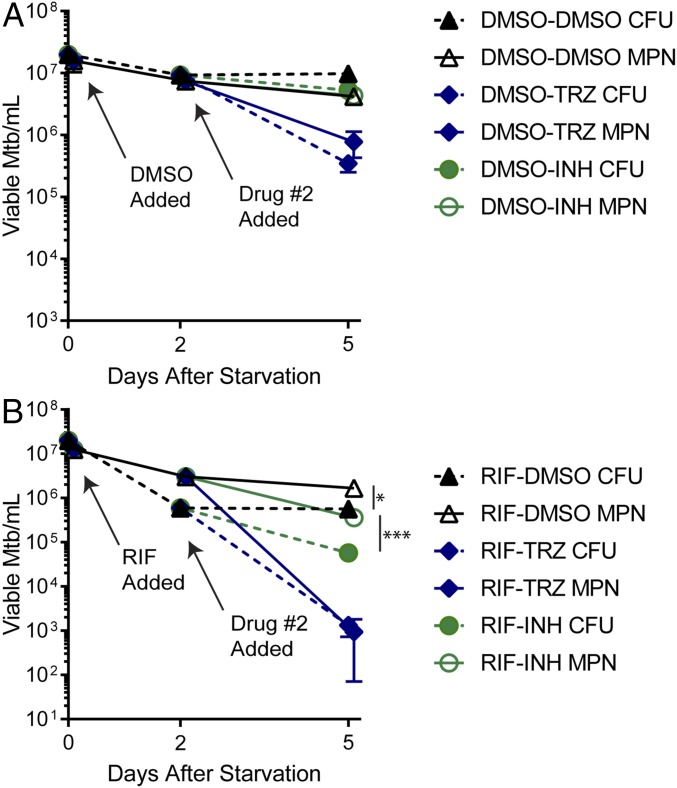
To begin testing compounds for activity against DD Mtb generated in this model, we exposed 2 wk-starved cultures to RIF at 100 μM for 2 d followed by addition of vehicle control or a second drug for 3 further days—TRZ at 60 μM or INH at 100 μM. These compounds were chosen because, as mentioned previously, TRZ has been shown to have activity against nonreplicating Mtb, whereas nonreplicating bacteria become phenotypically tolerant to INH. As a control, starved cells were exposed to DMSO instead of RIF for 2 d followed by the second drug for 3 d. In controls without prior RIF exposure, INH exerted minimal cidality, whereas TRZ resulted in about 1 log10 reduction in both MPN and cfus, but neither TRZ nor INH produced DD Mtb (Fig. 5A). Exposure of aliquots from the same batch of starved cells to 100 μM RIF for 2 d resulted in a culture with 90% DD Mtb, verifying that DD Mtb were formed before addition of the second compound. In the cultures where RIF was added for 2 d and the second drug for 3 d, addition of INH reduced the numbers of viable Mtb by around 0.5 log10 but reduced the DD Mtb and cfu population proportionally. The difference between MPN and cfus remained significant, and the culture remained 90% DD Mtb. In contrast, TRZ led to a 3–4 log10 reduction in both MPN and cfus and eliminated any significant difference in bacterial numbers estimated by MPN and cfus (Fig. 5B). Thus, TRZ addition to RIF killed preformed DD Mtb.
Fig. 5.
Compounds tested for activity against DD Mtb. (A) Control experiments. Two week-starved Mtb was exposed to 1% DMSO (vehicle control) for 2 d followed by 3 d of exposure to 100 μM INH or 60 μM TRZ. (B) Effect of selected drugs on DD Mtb generated by RIF. Two week-starved Mtb was exposed to 100 μM RIF for 2 d followed by 3 d of exposure to 100 μM INH or 60 μM TRZ. MPN and cfu measurements were made on day 0 of drug exposure, 2 d after DMSO or RIF were added, and 3 d after INH or TRZ were added. To maximize recovery of cfus, day 5 cfu plates were supplemented with 0.4% charcoal. Data are means ± SEM for at least six replicates from three experiments. Where error bars are not visible, they are smaller than the symbol. Significant differences where MPN > cfu denoted with *P < 0.05; ***P < 0.0005.
Method for Generation of DD Mtb Subpopulation
Materials.
Reagents.
-
1.
7H9: 7H9 powder (BD; cat. no. 271310) with 0.2% glycerol (Sigma; cat. no. G7893), 10% vol/vol OADC (BD; cat. no. 212351), and 0.02% tyloxapol. The 7H9 broth with the glycerol is autoclaved first. Then OADC and tyloxapol are added and the medium filter-sterilized before storage at 4 °C.
-
2.
PBS (Gibco; cat. no. 14190–144) with 0.02% tyloxapol (PBS-Tx).
-
3.
Tyloxapol (Sigma; cat. no. T8761): 100× stock (2% concentration) is prepared in millipore ddH2O and stored at 4 °C after sterile filtration.
-
4.
DMSO (Sigma; cat. no. D2650).
-
5.
RIF (Sigma; cat. no. R3501): 100× stocks (10 or 1 mM) are prepared in 100% DMSO.
-
6.
7H10 agar (BD; cat. no. 262710): 7H10 agar autoclaved with 0.5% glycerol. OADC (10% vol/vol) supplement is added before dispensing into the triplicates.
-
7.
Activated charcoal (Sigma; cat. no. C5510).
-
8.
Spor-klenz (Steris; cat. no. 6525).
Plasticware.
-
1.
50 mL centrifuge tubes (CORNING; cat. no. 4558).
-
2.
Vented tissue culture-treated flasks (CORNING): 25 cm2 (cat. no. 430639) and 75 cm2 (cat. no. 4430641U).
-
3.
96-well plates (COSTAR; cat. no. 3595).
-
4.
Microcentrifuge tubes for vortexing bacterial samples and for preparing compound (SARSTEDT; cat. no. 72.693.005).
-
5.
Triplates (VWR; cat. no. 25384–306).
Methods.
Initial outgrowth and starvation.
-
1.
Grow MTB to log phase in 7H9 complete media (7H9 + 0.2% glycerol + 10% OADC + 0.02% tyloxapol) to an optical density at 580 nm (OD580) between 0.4 and 0.8.
-
2.
Create single-cell suspension:
-
a.
Wash culture with the same volume of PBS with 0.02% tyloxapol (PBS-Tx) twice; with each wash, spin down culture by spinning at 3,082 × g for 8 min.
-
b.
After the second wash and resuspension, create single-cell suspension by centrifuging at 123 × g for 8 min with NO brake.
-
c.
Transfer the supernatant of this single-cell suspension without touching the pellet (leave 3–5 mL behind) to a new falcon tube.
-
a.
-
3.
Check OD580 of single-cell suspension.
-
4.
Dilute to an OD of 0.1 with PBS-Tx.
-
5.
Plate cfus of this culture 3× (inoculum quantification).
-
6.
Place enough culture for the experiment in triplicate vented flasks. In an experiment comparing RIF 100 μM exposure to DMSO control exposure, each triplicate should have at least 20 mL (75 cm2 vented flasks) of culture, as it will be divided into two 8-mL exposures after the starvation period.
-
7.
Incubate at 37 °C, 20% O2, and 5% CO2, without shaking or agitation for 2 wk or longer.
Drug exposure.
-
8.
Mix the starved cultures (treating each individual replicate separately), being sure to wash down the side of the flask when doing so.
-
9.
Set aside 1 mL of each of the cultures for MPN and cfus (day 0 quantification). You could also measure the OD580, which is normally around 0.05.
-
10.
Divide the remaining volume of each culture into two 8-mL cultures in vented flasks (25 cm2).
-
11.
To the 8-mL cultures, add either DMSO (final 1% by volume) or RIF in DMSO (final 100 μM) in triplicate. For a final concentration of 100 μM RIF, a 10-mM stock is prepared in 100% DMSO, which is then diluted 100-fold (80 μL in 8 mL) to achieve a final DMSO concentration of 1% and a final RIF concentration of 100 μM. Thus, you will have six flasks with cultures; three with DMSO, and three with 100 μM RIF.
-
12.
Incubate the flasks at 37 °C, 20% O2, and 5% CO2, without shaking or agitation for 5 d.
-
13.
Vortex the previously set-aside 1 mL of the original starved cultures 3×, 10 s each.
-
14.
Use the vortexed sample to perform MPN and cfus as described in the last section of this section.
Day 5 after drug exposure.
-
15.
Mix and collect samples into 50-mL falcon tubes.
-
16.
Spin down the samples at 3,082 × g for 8 min.
-
17.
Remove and discard the supernatant, without touching the pellet. We leave behind 0.5–1 mL of the supernatant to ensure this.
-
18.
Wash with 5× the volume of PBS-Tx (40 mL).
-
19.
Spin down again at 3,082 × g for 8 min.
-
20.
Remove the supernatant using a 30-mL pipette first, then a 5-mL pipette to allow for more refined removal of the supernatant. Leave ∼500 μL of liquid at the bottom (or at least less than 1 mL). Note: Do not touch the pellet (or the bottom if no pellet is visualized).
-
21.
Suspend pellet (or mix if no pellet visualized) and transfer the entire volume to a microcentrifuge tube. Record how much volume was transferred.
-
22.
Add enough PBS-Tx to the original 50-mL falcon tube such that transferring this volume to the microcentrifuge tube would add up to 1 mL. Wash the bottom of the falcon tube with this volume and then transfer to the same microcentrifuge tube. Remember for calculations later that this sample is now 8× more concentrated than the original 8-mL sample.
-
23.
Vortex the microcentrifuge tube 3× for 10 s each. This sample is now ready for addition to the MPN assay as described below.
MPN assay.
-
1.
In a 96-well plate, fill all of the wells with 135 μL of 7H9 complete media by using a multichannel pipette with filter tips.
-
2.
Add 15 μL of the 1-mL sample to wells 2B–2F. All samples should be vortexed for 10 s 3×, just before addition to the wells. For technical reasons, when the automated system was used, the volumes were reduced to 12 μL of sample added to 108 μL of 7H9 complete.
-
3.
Mix those wells by pipetting 100 μL up and down 10×.
-
4.
Transfer 15 μL from those five wells using a multichannel pipette into the next column (i.e., 3B–3F) using a multichannel pipette and 20-μL filter microtips; mix twice and then discard tips.
-
5.
Mix by pipetting 100 μL up/down 10×.
-
6.
Repeat this transferring and mixing dilution until column 11.
-
7.
After mixing column 11, discard 15 μL from that column.
-
8.
Next, after the dilutions are complete, transfer 35 μL from columns 2–11 for each row into a new 96-well plate (all of the rows are combined into a single row). This is used for the cfus.
-
9.
Add 100 μL of 7H9 complete from a sterile trough into all wells of the dilution (rows B–F, columns 2–11). If none of the wells are touched, pipette tips do not need to be changed between rows. In a separate 96-well plate, aliquot 100 μL of the 7H9 complete both before addition and after addition to the plates, to ensure contamination has not occurred during addition of 7H9 media.
-
10.
Store plates at 37 °C with 20% O2 and 5% CO2, without shaking. Plates are taped together in stacks of four or less and bagged, with the bag slightly open.
-
11.
Read plates at 3 wk and 5 wk and at continued 1–2-wk intervals until there is no significant change.
-
12.
Input data in the MPN calculator excel sheet, as provided by Jarvis et al. (23).
Cfu plating.
-
1.
Prepare triplates with 7H10 media supplemented with glycerol and OADC, with 9 mL per section.
-
2.
Use the extra volume (35 μL × 5) set aside from each MPN plate and transfer to a separate row in 96-well plates (step 8 in MPN Assay). Mix the dilutions between 10–5 and 100 (note: column B is 10–1, thus 100 will need to be obtained from the tube containing the original sample).
-
3.
Place 10 μL of each dilution between 10–5 and 100, for a total of six dilutions; each replicate will thus require two triplicates.
-
4.
Use a disposable inoculum loop to spread the droplet across the plate.
-
5.
Tape stacks of six plates together.
-
6.
Clean the outside of the plates with a rag soaked in Sporklenz.
-
7.
Bag the plates and clean the outside of the bag with Sporklenz.
-
8.
Place at 37 °C and ambient air.
-
9.
Read plates at 3 wk and 5 wk and at continued 1–2-wk intervals until there is no significant change.
Discussion
With the exclusion of a number of artifacts, we consistently generated Mtb populations in vitro in which 90% or more of the viable bacteria were unable to form colonies on agar while remaining able to grow in liquid medium. Two stresses were required in sequence to elicit this phenomenon: starvation for at least 2 wk in PBS and exposure to RIF for at least 2 d. The proportion of DD Mtb could be increased with longer periods of starvation and longer periods of exposure to RIF. Extended durations of either stress allowed RIF to be used at lower concentrations.
Our model estimates the number of viable organisms from their growth in replicate LDs. This relies on three assumptions about replicative units: (i) They are randomly distributed in the volume containing them; (ii) they distribute independently of each other or, if not, their mutual association does not change over the course of serial dilution (that is, clump formation and breakage only occur before dilution and have an equivalent effect on the estimation of MPN and cfus); and (iii) one cell in isolation serves as a replicative unit that can produce a visible colony or a turbid culture (16). The first two criteria are difficult to meet with Mtb—it sticks to vessel surfaces, and suspensions are not completely monodisperse even in the presence of detergent. A number of steps become critical: to avoid large clumps before beginning dilution; to take into account any changes in the size of small clumps during dilution by plating the cfus from the dilution series themselves rather than from the original suspension; to use a large number of replicate dilutions; and to change pipet tips with each dilution. If pipet tips are not changed, our results suggest that clumps tend to adhere to them and to break up in subsequent wells in the dilution series, inflating the MPN value by many orders of magnitude. We found that three different strains of Mtb that were passaged in culture for variable periods after being passaged in mice all produced DD Mtb with this method, although with differences in size of the DD population produced. It will be important to test whether recent clinical isolates of Mtb give similar results in vitro. We are in the process of applying the same LD techniques to the sputum of treatment-naïve subjects with TB.
Having standardized the method, we interpret the results to indicate that the stress of prolonged starvation sets Mtb into a state in which exposure to rifamycin can generate DD Mtb. We cannot exclude the possibility that starvation enables Mtb to survive RIF treatment while in liquid media but then RIF proceeds to kill the cells once they have been washed and plated on solid agar with media of similar richness. Arguing against the latter interpretation, results were unaffected by including charcoal in the agar under conditions in which we have documented charcoal’s ability to neutralize the effect of antibiotics that are carried into cfu plates in association with washed bacteria (23). Furthermore, the relative specificity to rifamycins as a class and the fact that the phenomenon does not occur with log-phase bacteria exposed to RIF both suggest that a more complex explanation is required than a delayed drug effect on cfu.
The inability to generate DD Mtb from a clinical isolate with an rpoB mutation conferring resistance to RIF’s activity against replicating Mtb confirms that RIF’s binding to RNA polymerase subunit B is involved. This was in question because DD Mtb generation proceeded most quickly when RIF was used at a concentration (100 µM) far above its minimal bactericidal concentration for replicating bacteria (0.01–0.08 µM) (20, 21). The need for high concentrations of RIF may reflect impaired uptake of RIF by starved Mtb (21) or could be related to the phenotypic RIF resistance attributed to stress-induced protein mistranslation (39). Regardless, the RIF dose requirement did not reflect selection for heritable RIF resistance. Moreover, DD Mtb were generated with RIF at 10-fold lower concentrations (10 µM) when the preceding starvation lasted longer than 2 wk or when the exposure to RIF lasted longer than 5 d.
We speculate that rifamycins, rather than halting transcription altogether in starved Mtb, selectively affect the transcription of different genes in such a way as to produce the DD phenotype. Support for this view comes from our ongoing characterization of the transcriptome of DD Mtb. There is precedent for inhibitors of bacterial macromolecular synthesis exerting differential effects on individual genes or gene products. For example, some ribosome-binding TB drugs “reshape the cellular proteome rather than block global protein synthesis” (40).
Other reported in vitro models for producing DD Mtb include hypoxia and starvation with antibiotic exposure (19), potassium depletion (41), exposure of log-phase cells to antibiotics (42), and gradual acidification (43). It is not clear why we did not succeed in producing DD Mtb with methods like these. Perhaps we failed to reproduce closely enough the conditions pertaining to those studies. Another possibility is that we avoided Sauton’s-based agar, because in our hands it did not support colony formation of Mtb as well as agar based on richer media (7H10 or 7H11) and therefore exaggerated the number of DD Mtb. A variety of methods for DD Mtb generation would be informative, as long as they meet the criteria listed in the Introduction.
Our findings follow a long line of study regarding a population of Mtb that is not detected as cfus. Early evidence for VBNC forms of Mtb came from experiments with human autopsy material from which no Mtb could be grown but which caused TB when inoculated in guinea pigs (44). The first reported experimental generation of VBNC Mtb was in Mtb-infected mice that after combination chemotherapy with INH and PZA yielded no cfus in culture of their organs or even the whole carcass (45–47). This apparent microbiologic cure was proven false when about a third of identically treated mice that were reserved for observation eventually relapsed, and nearly all did so when immunosuppressed with corticosteroids (45–47) or when deficient in inducible nitric oxide synthase (48). Mtb colonies recovered from relapsed mice remained susceptible to INH and PZA (45). Thus, non–colony-forming Mtb were phenotypically tolerant to combination chemotherapy, and their viability was demonstrated by their ability to cause relapse. More recent studies of cfu-negative organ samples from Mtb-infected mice treated with other drug regimens suggested the presence of DD Mtb by detection of Mtb mRNA transcripts (49) or demonstrated the presence of DD Mtb by recovery of Mtb in liquid medium supplemented with CF from log-phase Mtb (50). The ability to use non–agar-based culture methods to grow DD Mtb from mice (22) or human sputum (13, 14) makes the term “nonculturable” inappropriate. Accordingly, Kana and coworkers introduced the term “differentially culturable tubercle bacilli” (DCTB) (14). Because the viability of non–colony-forming Mtb has also been demonstrated by transmission of disease and relapse of disease in the absence of culture, we use a broader term, DD.
Critical phenotypic differences may exist in the DD Mtb populations generated in the four settings discussed above—in human tissue, human sputum, treated mice, or axenic cultures. It is unclear how the regimen of nutrient starvation we used in vitro relates to the physiology of infection, although Mtb is thought to experience nutrient limitation in granulomas (51–53). The requirement of the in vitro model for RIF exposure questions its relevance for latent or untreated active Mtb infection. On the other hand, given that RIF accumulates in necrotic granulomas to 12.15 μM (54), the model may have implications for treatment optimization and monitoring. Recovery of DD Mtb from patients’ sputum often requires the addition of CF (13, 14), whereas CF was immaterial in our in vitro model. This may indicate a fundamental difference between DD Mtb in these two settings, or it may be that CF helps attenuate Mtb growth-inhibitory activities arising from degenerating leukocytes or other factors in tuberculous cavities or respiratory secretions.
The picture becomes further complicated when bacterial populations are classified not by how they are enumerated but by how they respond to antimicrobials. Phenotypic tolerance to bactericidal antibiotics allows the survival of bacterial “persisters”—a population of organisms that remain viable after exposure to an antimicrobial agent at concentrations that kill the vast majority of an isogenic population at the same concentration under standard replicating conditions (7, 12, 55). Class I persisters are a small minority arising stochastically in a replicating bacterial population (7). The upper limit on the size of that minority is set by the precision of the assay used to determine the antibiotic’s MIC. By definition, the MIC is unaffected by the presence of class I persisters and is unchanged when persisters are isolated, expanded, and retested. Class II phenotypic tolerance is displayed by almost all of the bacteria in a population under conditions that prevent net growth during exposure to the antibiotic (7). Bacteria in the state originally called VBNC (12) are class II persisters, in that they do not replicate in some standard growth condition—in our case, solid media normally permissive to growth—and are phenotypically tolerant to diverse antibiotics (56, 57).
We could not generate DD Mtb with a combination of four stresses, each considered physiologic and each capable of inducing phenotypic tolerance to most TB drugs in vitro: mild acidification, hypoxia, a flux of reactive nitrogen intermediates, and a fatty acid as a carbon source (17, 18). This implies that class II phenotypic tolerance encompasses a spectrum of states. Bacteria displaying class IIa phenotypic tolerance in response to certain stresses will grow as cfus once the stress is removed. Those displaying class IIb phenotypic tolerance in response to other stresses will not grow as cfus when the stresses are removed, and yet their viability is demonstrable. Two particular stresses in sequence produced class IIb phenotypic tolerance in vitro. Likewise, a sequence of stresses may lead to the generation of DD Mtb in patients with TB, in contrast to the in vitro model in which we applied four stresses together.
DD Mtb in patient sputum tested ex vivo were found to be phenotypically tolerant to INH and STREP (58). The PBS-starved Mtb studied here (not to be confused with the DD population generated from them) displayed class IIa phenotypic tolerance to RIF, INH, MOXI, SM, PA-824, CFZ, NTZ, TRZ, and the four-drug combination of RIF, INH, PZA and ETH used to treat drug-sensitive TB. DD Mtb generated by sequential PBS starvation and RIF exposure were phenotypically tolerant to INH and to the four-drug combination. High-throughput screens have been developed to find compounds active against Mtb showing class IIa phenotypic tolerance (17, 18). However, it would be exceedingly cumbersome to conduct a screen for Mtb in the class IIb state. TRZ kills Mtb displaying both class IIa phenotypic tolerance and class IIb phenotypic tolerance. Although all compounds active against class IIa phenotypically tolerant Mtb may not be active against class IIb phenotypic tolerance, screening compounds against Mtb under conditions of class IIa phenotypic tolerance may be a viable first step toward finding additional compounds active against DD Mtb.
Using this model as a tool, comparisons of growth characteristics, antibiotic tolerance, and molecular features among DD Mtb populations in various settings can be exploited to better understand the biology and diversity of DD Mtb. It may then become possible to devise easier and faster means of mycobacterial detection, quantification, and susceptibility profiling that are suitable for basic, preclinical, and clinical studies.
Methods
Materials.
The Mtb strains used were as follows: three H37Rv derivatives, Erdman, and the RIF-monoresistant clinical strain (accession no. 2015–3185) obtained from the GHESKIO Center in Port-au-Prince, Haiti. The resistant strain was collected and cultured from sputum in January 2015 and completely de-identified. Mtb were grown in Middlebrook 7H9 medium (BD Biosciences) supplemented with 0.2% glycerol, 10% (vol/vol) Middlebrook OADC (oleic acid, bovine albumin, dextrose, catalase) (BD Biosciences), and 0.02% tyloxapol —hereafter called 7H9 complete—at 37 °C under 5% CO2 and ambient O2. Serial dilutions were performed manually or with an automated pipetting system (BioTek Precision 2000). The cfu enumeration was performed on Middlebrook 7H10 medium supplemented with 0.5% glycerol and 10% (vol/vol) OADC, unless otherwise specified. All reagents were purchased from Sigma-Aldrich unless otherwise specified. PA-824 was obtained from TB Alliance, and NTZ was purchased from Atlantis SciTech group. Optical density was measured in a Genesys 20 visible spectrophotometer (Thermo Scientific). Centrifugation was in an Allegra x-30R centrifuge (Beckman Coulter).
Replicates Within Experiments.
For each stress in each experiment, we used biological triplicates, defined as three cultures prepared and incubated identically but kept separate. This also includes biological triplicates for each secondary stress in sequential stress experiments. In any one experiment, these triplicates are prepared at the same time and are run in parallel. These biological triplicates are distinct from the five technical replicates present within the LD assay and used in the MPN calculations and are not counted as fully distinct experiments.
DD Mtb Generation.
Starvation was performed in PBS (calcium chloride and magnesium chloride-free) (Gibco) with 0.02% tyloxapol at 37 °C under 5% CO2 and ambient O2. A detailed protocol for the generation of DD Mtb is provided in Method for Generation of DD Mtb Subpopulation. To test the effect on the RIF-resistant strain, we starved it for 2 wk in 24-well or 48-well plates and then added 100 μM RIF or 1% DMSO (vehicle control) for 5 d. At the end of 5 d, the protocol in Method for Generation of DD Mtb Subpopulation was followed for washing and outgrowth.
Stress Models.
Testing for all other stresses was performed on cultures grown in 7H9 complete to an OD of 0.4–0.8. To test the effect of RIF on dividing bacteria, single-cell suspension was prepared by removing large clumps by centrifugation (123 × g, 8 min, room temperature), and the suspension was adjusted to an OD of 0.05 in 7H9 complete. For 14-d RIF exposures, 50% of the indicated RIF concentration was added again at day 7 in view of the reported instability of RIF in 7H9 at 37 °C (59). For the four-stress nonreplicating (NR) model, log-phase Mtb was washed with PBS-tyloxapol and suspended in Sauton’s-based media that contained 0.05% KH2PO4, 0.05% MgSO4, 0.005% ferric ammonium citrate, 0.0001% ZnSO4, 0.1% NH4Cl, 0.05% butyrate, 0.5% BSA, 0.085% NaCl, and 0.02% tyloxapol and set to a pH of 5.0 (17). Single-cell suspension was prepared and diluted to an OD of 0.1. Freshly prepared 0.5 mM NaNO2 was added before incubation at 37 °C under 1% O2 and 5% CO2 for 2 wk. Human serum and plasma were collected from fresh whole blood obtained from healthy human volunteers. Whole blood was collected into nonheparinized tubes, allowed to clot for 30 min at room temperature, and centrifuged (1,300 × g, 10 min, 4 °C). The supernatant was collected, and this serum was used the same day. Alternatively for plasma, the blood was collected in heparinized tubes, centrifuged immediately (1,300 × g, 10 min, 4 °C), and the supernatant removed and stored at 4 °C until use on the same day. For serum and RPMI experiments, log-phase Mtb were washed twice in an equal volume of PBS before being suspended into the respective media. A single-cell suspension was then formed by centrifugation (123 × g, 8 min, room temperature), and OD was adjusted to 0.1 when greater than 0.1. OD was often unpredictable after SCS in serum and plasma, and thus an OD between 0.05 and 0.1 was tolerated in those conditions. For RPMI, experiments were adjusted to an OD of 0.005 to simulate conditions in which neutrophils and macrophages might be incubated and infected. The culture was split into triplicates and incubated in 24-well tissue culture plates at 37 °C under 5% CO2 and ambient O2. LD and cfu were performed identically as described in Method for Generation of DD Mtb Subpopulation for DD Mtb generation. For cfu estimations in serum and plasma experiments, 7H11 plates supplemented with 0.5% glycerol and 10% (vol/vol) OADC were used. For RPMI, serum, and four-stress NR models, log-phase CF was supplemented to a final concentration of 50% in MPN plates.
CF Preparation.
CF was collected from log-phase Mtb H37Rv cultures (OD 0.4–0.8) grown in 7H9 complete by centrifugation at 3048 × g for 8 min. Supernatant was filter-sterilized twice using a vacuum-assisted 0.2-μm filter. Samples of the supernatant both before and after use in dilutions were placed in a separate 96-well plate to document the absence of mycobacterial contamination before or during distribution to LD wells.
Variables in Outgrowth Conditions.
For supplementation of agar plates, 10 mM sodium fumarate dibasic or 20 μM hemin was added to 7H10 agar after autoclaving. Hemin was added while the medium was still hot to ensure proper dissolution. For charcoal plates, 4 g/L of activated charcoal were added to standard 7H10 plates before autoclaving. Agar plates based on Sauton’s medium were prepared to contain per liter 6% (vol/vol) glycerol, 15 g agar, 0.5 g potassium phosphate monobasic, 1.4 g magnesium sulfate heptahydrate, 0.5 g ammonium iron III citrate, 4 g l-asparagine, 2 g sodium citrate tribasic dehydrate, 0.0001% ZnSO4·7H2O, and 0.05% Tween80, supplemented with albumin, dextrose, and NaCl as described (60).
MPN Calculation and Statistical Analysis.
Raw data were inputted into a previously published MPN calculator for all MPN calculations (16). For all text figures and legends, comparison of MPN estimates to cfu enumeration was performed using paired two-tailed t tests with a significance cutoff of P < 0.05 after log transformation. All statistical analyses were performed in Graphpad Prism 7.02.
Acknowledgments
We thank Dr. Oksana Ocheretina and the staff of the GHESKIO laboratory in Port-au-Prince, Haiti, for providing the clinical Mtb strain. We are also thankful to Prof. Michael Glickman at Memorial Sloan Kettering Cancer Center in New York for kindly providing the Mtb Erdman strain and Prof. Christopher Sassetti at the University of Massachusetts Medical School, Worcester, for sharing the Mtb H37Rv-UMass strain. We acknowledge Prof. Sabine Ehrt, Dr. Daniel W. Fitzgerald, and their respective teams for helpful input. We thank Ms. Clara Oromendia for statistical advice via the Clinical and Translational Science Center. This work was supported by the Tri-Institutional TB Research Unit (NIH Grant U19 AI111143-01), NIH Grant T32 AI007613 (to K.S.), NIH Grant K08AI108799 (to S.S.-K.), NIH Grant TBRU U19 AI111276 (to W.R.J. and L.K.), NIH Grant R01 AI026170 (to W.R.J. and L.K.), the Human Frontier Science Program Organization (L.K.), and the Milstein Program in Chemical Biology and Translational Medicine. The Department of Microbiology and Immunology is supported by the William Randolph Hearst Trust.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705385114/-/DCSupplemental.
References
- 1.WHO . Global Tuberculosis Report 2016. World Health Organization; Geneva: 2016. [Google Scholar]
- 2.Nahid P, et al. Official American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America clinical practice guidelines: Treatment of drug-susceptible tuberculosis. Clin Infect Dis. 2016;63:e147–e195. doi: 10.1093/cid/ciw376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC . Managing Drug Interactions in the Treatment of HIV-Related Tuberculosis. CDC; Atlanta: 2013. [Google Scholar]
- 4.Yee D, et al. Incidence of serious side effects from first-line antituberculosis drugs among patients treated for active tuberculosis. Am J Respir Crit Care Med. 2003;167:1472–1477. doi: 10.1164/rccm.200206-626OC. [DOI] [PubMed] [Google Scholar]
- 5.Araújo-Mariz C, et al. Hepatotoxicity during treatment for tuberculosis in people living with HIV/AIDS. PLoS One. 2016;11:e0157725. doi: 10.1371/journal.pone.0157725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention (CDC) Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs––Worldwide, 2000-2004. MMWR Morb Mortal Wkly Rep. 2006;55:301–305. [PubMed] [Google Scholar]
- 7.Nathan C. Fresh approaches to anti-infective therapies. Sci Transl Med. 2012;4:140sr2. doi: 10.1126/scitranslmed.3003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manina G, Dhar N, McKinney JD. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe. 2015;17:32–46. doi: 10.1016/j.chom.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Lin PL, et al. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat Med. 2014;20:75–79. doi: 10.1038/nm.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garton NJ, et al. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 2008;5:e75. doi: 10.1371/journal.pmed.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colwell R, et al. Viable but non-culturable Vibrio cholerae and related pathogens in the environment: Implications for release of genetically engineered microorganisms. Nat Biotechnol. 1985;3:817–820. [Google Scholar]
- 12.Li L, Mendis N, Trigui H, Oliver JD, Faucher SP. The importance of the viable but non-culturable state in human bacterial pathogens. Front Microbiol. 2014;5:258. doi: 10.3389/fmicb.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR. Resuscitation-promoting factors reveal an occult population of tubercle Bacilli in Sputum. Am J Respir Crit Care Med. 2010;181:174–180. doi: 10.1164/rccm.200905-0661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chengalroyen MD, et al. Detection and quantification of differentially culturable tubercle bacteria in sputum from patients with tuberculosis. Am J Respir Crit Care Med. 2016;194:1532–1540. doi: 10.1164/rccm.201604-0769OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dartois V, Saito K, Warrier T, Nathan C. New evidence for the complexity of the population structure of Mycobacterium tuberculosis increases the diagnostic and biologic challenges. Am J Respir Crit Care Med. 2016;194:1448–1451. doi: 10.1164/rccm.201607-1431ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarvis B, Wilrich C, Wilrich PT. Reconsideration of the derivation of Most Probable Numbers, their standard deviations, confidence bounds and rarity values. J Appl Microbiol. 2010;109:1660–1667. doi: 10.1111/j.1365-2672.2010.04792.x. [DOI] [PubMed] [Google Scholar]
- 17.Gold B, Warrier T, Nathan C. A multi-stress model for high throughput screening against non-replicating Mycobacterium tuberculosis. In: Parish T, Roberts DM, editors. Mycobacteria Protocols. Springer New York; New York: 2015. pp. 293–315. [DOI] [PubMed] [Google Scholar]
- 18.Gold B, et al. Nonsteroidal anti-inflammatory drug sensitizes Mycobacterium tuberculosis to endogenous and exogenous antimicrobials. Proc Natl Acad Sci USA. 2012;109:16004–16011. doi: 10.1073/pnas.1214188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, et al. High-dose rifampicin kills persisters, shortens treatment duration, and reduces relapse rate in vitro and in vivo. Front Microbiol. 2015;6:641. doi: 10.3389/fmicb.2015.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gengenbacher M, Rao SP, Pethe K, Dick T. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology. 2010;156:81–87. doi: 10.1099/mic.0.033084-0. [DOI] [PubMed] [Google Scholar]
- 21.Sarathy J, Dartois V, Dick T, Gengenbacher M. Reduced drug uptake in phenotypically resistant nutrient-starved nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57:1648–1653. doi: 10.1128/AAC.02202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Z, Siddiqi N, Rubin EJ. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2005;49:4778–4780. doi: 10.1128/AAC.49.11.4778-4780.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gold B, et al. Rapid, semiquantitative assay to discriminate among compounds with activity against replicating or nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2015;59:6521–6538. doi: 10.1128/AAC.00803-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones CM, Niederweis M. Mycobacterium tuberculosis can utilize heme as an iron source. J Bacteriol. 2011;193:1767–1770. doi: 10.1128/JB.01312-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe S, et al. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002287. doi: 10.1371/journal.ppat.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawson R, et al. Efficiency and safety of the combination of moxifloxacin, pretomanid (PA-824), and pyrazinamide during the first 8 weeks of antituberculosis treatment: A phase 2b, open-label, partly randomised trial in patients with drug-susceptible or drug-resistant pulmonary tuberculosis. Lancet. 2015;385:1738–1747. doi: 10.1016/S0140-6736(14)62002-X. [DOI] [PubMed] [Google Scholar]
- 27.Stover CK, et al. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature. 2000;405:962–966. doi: 10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- 28.Cholo MC, Mothiba MT, Fourie B, Anderson R. Mechanisms of action and therapeutic efficacies of the lipophilic antimycobacterial agents clofazimine and bedaquiline. J Antimicrob Chemother. 2017;72:338–353. doi: 10.1093/jac/dkw426. [DOI] [PubMed] [Google Scholar]
- 29.Cho SH, et al. Low-oxygen-recovery assay for high-throughput screening of compounds against nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2007;51:1380–1385. doi: 10.1128/AAC.00055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Soolingen D, et al. The antipsychotic thioridazine shows promising therapeutic activity in a mouse model of multidrug-resistant tuberculosis. PLoS One. 2010;5:e12640. doi: 10.1371/journal.pone.0012640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutta NK, Pinn ML, Karakousis PC. Sterilizing activity of thioridazine in combination with the first-line regimen against acute murine tuberculosis. Antimicrob Agents Chemother. 2014;58:5567–5569. doi: 10.1128/AAC.03408-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sohaskey CD. Nitrate enhances the survival of Mycobacterium tuberculosis during inhibition of respiration. J Bacteriol. 2008;190:2981–2986. doi: 10.1128/JB.01857-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amaral L, Viveiros M. Thioridazine: A non-antibiotic drug highly effective, in combination with first line anti-tuberculosis drugs, against any form of antibiotic resistance of Mycobacterium tuberculosis due to its multi-mechanisms of action. Antibiotics (Basel) 2017;6:E3. doi: 10.3390/antibiotics6010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shigyo K, et al. Efficacy of nitazoxanide against clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57:2834–2837. doi: 10.1128/AAC.02542-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Carvalho LP, Lin G, Jiang X, Nathan C. Nitazoxanide kills replicating and nonreplicating Mycobacterium tuberculosis and evades resistance. J Med Chem. 2009;52:5789–5792. doi: 10.1021/jm9010719. [DOI] [PubMed] [Google Scholar]
- 36.Alsultan A, Peloquin CA. Therapeutic drug monitoring in the treatment of tuberculosis: An update. Drugs. 2014;74:839–854. doi: 10.1007/s40265-014-0222-8. [DOI] [PubMed] [Google Scholar]
- 37.Verbeeck RK, Günther G, Kibuule D, Hunter C, Rennie TW. Optimizing treatment outcome of first-line anti-tuberculosis drugs: The role of therapeutic drug monitoring. Eur J Clin Pharmacol. 2016;72:905–916. doi: 10.1007/s00228-016-2083-4. [DOI] [PubMed] [Google Scholar]
- 38.Chideya S, et al. Isoniazid, rifampin, ethambutol, and pyrazinamide pharmacokinetics and treatment outcomes among a predominantly HIV-infected cohort of adults with tuberculosis from Botswana. Clin Infect Dis. 2009;48:1685–1694. doi: 10.1086/599040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su H-W, et al. The essential mycobacterial amidotransferase GatCAB is a modulator of specific translational fidelity. Nat Microbiol. 2016;1:16147. doi: 10.1038/nmicrobiol.2016.147. [DOI] [PubMed] [Google Scholar]
- 40.Kannan K, Vázquez-Laslop N, Mankin AS. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell. 2012;151:508–520. doi: 10.1016/j.cell.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 41.Salina EG, et al. Potassium availability triggers Mycobacterium tuberculosis transition to, and resuscitation from, non-culturable (dormant) states. Open Biol. 2014;4:140106. doi: 10.1098/rsob.140106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loraine J, Pu F, Turapov O, Mukamolova GV. Development of an in vitro assay for detection of drug induced resuscitation-promoting factor dependent mycobacteria. Antimicrob Agents Chemother. 2016;60:6227–6233. doi: 10.1128/AAC.00518-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shleeva MO, et al. Dormant ovoid cells of Mycobacterium tuberculosis are formed in response to gradual external acidification. Tuberculosis (Edinb) 2011;91:146–154. doi: 10.1016/j.tube.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 44.Opie EL, Aronson J. Tubercle bacilli in latent tuberculous lesions and in lung tissue without tuberculous lesions. Arch Pathol Lab Med. 1927;4:1–21. [Google Scholar]
- 45.McCune RM, Jr, McDermott W, Tompsett R. The fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. II. The conversion of tuberculous infection to the latent state by the administration of pyrazinamide and a companion drug. J Exp Med. 1956;104:763–802. doi: 10.1084/jem.104.5.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCune RM, Feldmann FM, Lambert HP, McDermott W. Microbial persistence. I. The capacity of tubercle bacilli to survive sterilization in mouse tissues. J Exp Med. 1966;123:445–468. doi: 10.1084/jem.123.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCune RM, Feldmann FM, McDermott W. Microbial persistence. II. Characteristics of the sterile state of tubercle bacilli. J Exp Med. 1966;123:469–486. doi: 10.1084/jem.123.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nathan C. Taming tuberculosis: A challenge for science and society. Cell Host Microbe. 2009;5:220–224. doi: 10.1016/j.chom.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 49.Hu Y, et al. Detection of mRNA transcripts and active transcription in persistent Mycobacterium tuberculosis induced by exposure to rifampin or pyrazinamide. J Bacteriol. 2000;182:6358–6365. doi: 10.1128/jb.182.22.6358-6365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Y, et al. Investigation of elimination rate, persistent subpopulation removal, and relapse rates of Mycobacterium tuberculosis by using combinations of first-line drugs in a modified Cornell mouse model. Antimicrob Agents Chemother. 2016;60:4778–4785. doi: 10.1128/AAC.02548-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dietrich J, et al. Differential influence of nutrient-starved Mycobacterium tuberculosis on adaptive immunity results in progressive tuberculosis disease and pathology. Infect Immun. 2015;83:4731–4739. doi: 10.1128/IAI.01055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulrichs T, Kaufmann SH. New insights into the function of granulomas in human tuberculosis. J Pathol. 2006;208:261–269. doi: 10.1002/path.1906. [DOI] [PubMed] [Google Scholar]
- 53.Sosa Belaustegui A, et al. [Immune reconstitution syndrome in an HIV-infected patient and Pneumocystis jirovecii pneumonia] Medicina (B Aires) 2014;74:130–132. [PubMed] [Google Scholar]
- 54.Prideaux B, et al. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat Med. 2015;21:1223–1227. doi: 10.1038/nm.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harms A, Maisonneuve E, Gerdes K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science. 2016;354:aaf4268. doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 56.Ayrapetyan M, Williams TC, Baxter R, Oliver JD. Viable but nonculturable and persister cells coexist stochastically and are induced by human serum. Infect Immun. 2015;83:4194–4203. doi: 10.1128/IAI.00404-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nowakowska J, Oliver JD. Resistance to environmental stresses by Vibrio vulnificus in the viable but nonculturable state. FEMS Microbiol Ecol. 2013;84:213–222. doi: 10.1111/1574-6941.12052. [DOI] [PubMed] [Google Scholar]
- 58.Turapov O, et al. Phenotypically adapted Mycobacterium tuberculosis populations from sputum are tolerant to first-line drugs. Antimicrob Agents Chemother. 2016;60:2476–2483. doi: 10.1128/AAC.01380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu X, et al. Rifampin stability in 7H9 broth and Löwenstein-Jensen medium. J Clin Microbiol. 2011;49:784–789. doi: 10.1128/JCM.01951-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ignatov DV, et al. Dormant non-culturable Mycobacterium tuberculosis retains stable low-abundant mRNA. BMC Genomics. 2015;16:954. doi: 10.1186/s12864-015-2197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]