Significance
The human yolk sac is often considered vestigial. Here, we report RNA-sequencing analysis of the human and murine yolk sacs and compare with that of the chicken. We relate the human RNA-sequencing data to coelomic fluid proteomic data. Conservation of transcripts across the species indicates the human secondary yolk sac likely performs key functions early in development, particularly uptake and processing of macro- and micronutrients, many of which are found in coelomic fluid. More generally, our findings shed light on evolutionary mechanisms giving rise to complex structures such as the placenta. We propose that although a choriovitelline placenta is never established physically in the human, the placental villi, exocoelomic cavity, and secondary yolk sac function together as a physiological equivalent.
Keywords: yolk sac, placenta, evolution
Abstract
The yolk sac is phylogenetically the oldest of the extraembryonic membranes. The human embryo retains a yolk sac, which goes through primary and secondary phases of development, but its importance is controversial. Although it is known to synthesize proteins, its transport functions are widely considered vestigial. Here, we report RNA-sequencing (RNA-seq) data for the human and murine yolk sacs and compare those data with data for the chicken. We also relate the human RNA-seq data to proteomic data for the coelomic fluid bathing the yolk sac. Conservation of transcriptomes across the species indicates that the human secondary yolk sac likely performs key functions early in development, particularly uptake and processing of macro- and micronutrients, many of which are found in coelomic fluid. More generally, our findings shed light on evolutionary mechanisms that give rise to complex structures such as the placenta. We identify genetic modules that are conserved across mammals and birds, suggesting these modules are part of the core amniote genetic repertoire and are the building blocks for both oviparous and viviparous reproductive modes. We propose that although a choriovitelline placenta is never established physically in the human, the placental villi, the exocoelomic cavity, and the secondary yolk sac function together as a physiological equivalent.
The yolk sac is phylogenetically the oldest of the extraembryonic membranes, evolving in anamniotes to absorb nutrients from their lipid-rich megalecithal eggs (1). Although the ova of eutherian mammals are microlecithal, the yolk sac has been recruited to transport maternal nutrients during earliest stages of embryonic development. In the majority of species, it makes contact with the chorion to form a transient choriovitelline placenta. This transient choriovitelline placenta functions during the critical period of organogenesis, at the end of which its functions are generally subsumed by the definitive chorioallantoic placenta. There is, however, considerable species variation, and the most elaborate development is found in rodents and lagomorphs. In these, the yolk sac continues to transport nutrients and immunoglobulins throughout gestation in parallel with the chorioallantoic placenta. For this reason, most of the experimental data on transport have been obtained in the mouse, rat, and guinea pig (2–6), and data on the human yolk sac are limited.
The human yolk sac goes through two developmental phases: A primary yolk sac develops between embryonic days 7 and 9 and is replaced by a secondary yolk sac, which is active until embryonic day 49 (7). The role of the primary yolk sac is unknown. The importance of the secondary yolk sac remains controversial. Although it is known to synthesize proteins, such as alpha fetoprotein, its transport functions are widely considered vestigial, primarily because the secondary yolk sac never makes contact with the chorion to form a choriovitelline placenta. Instead, it floats in the exocoelomic cavity, connected to the embryo by the vitelline duct. Although we, and others, have speculated that the yolk sac plays a critical role during organogenesis (3–5, 8–10), there are limited data to support this claim. Obtaining experimental data for the human is impossible for ethical reasons, and thus we adopted an alternative strategy. Here, we report RNA sequencing (RNA-seq) data derived from human and murine yolk sacs and compare them with published data from the yolk sac of the chicken. We postulate that conservation of transcripts across these species indicates retention of key transport and synthetic functions. We support this hypothesis by comparing the human yolk sac transcriptome with the proteome of the coelomic fluid.
Results and Discussion
We determined the transcript profile for the first-trimester human yolk sac by RNA-seq with a median sequencing depth of 39 million mapped reads per sample (n = 9) (SI Appendix, Table S1). We identified 12,469 transcripts with a mean reads per kilobase per million mapped reads (RPKM) ≥1 (Dataset S1). Similarly, we identified 11,628 transcripts in first-trimester human placental villous samples (n = 11, median sequencing depth 30 million mapped reads) (Dataset S2) and 11,272 transcripts in the mouse yolk sac (n = 8, median sequencing depth 28 million mapped reads) (Dataset S3).
In addition, we investigated the protein composition of the coelomic fluid using gel electrophoresis liquid chromatography (GELC)-MS/MS. We focused on the 165 proteins identified in any four of the five samples after immunoglobulins were excluded (Dataset S4). Proteins were mapped to unique Ensembl gene identifiers, which were used to identify overrepresented gene ontology (GO) terms (Dataset S5).
Cholesterol.
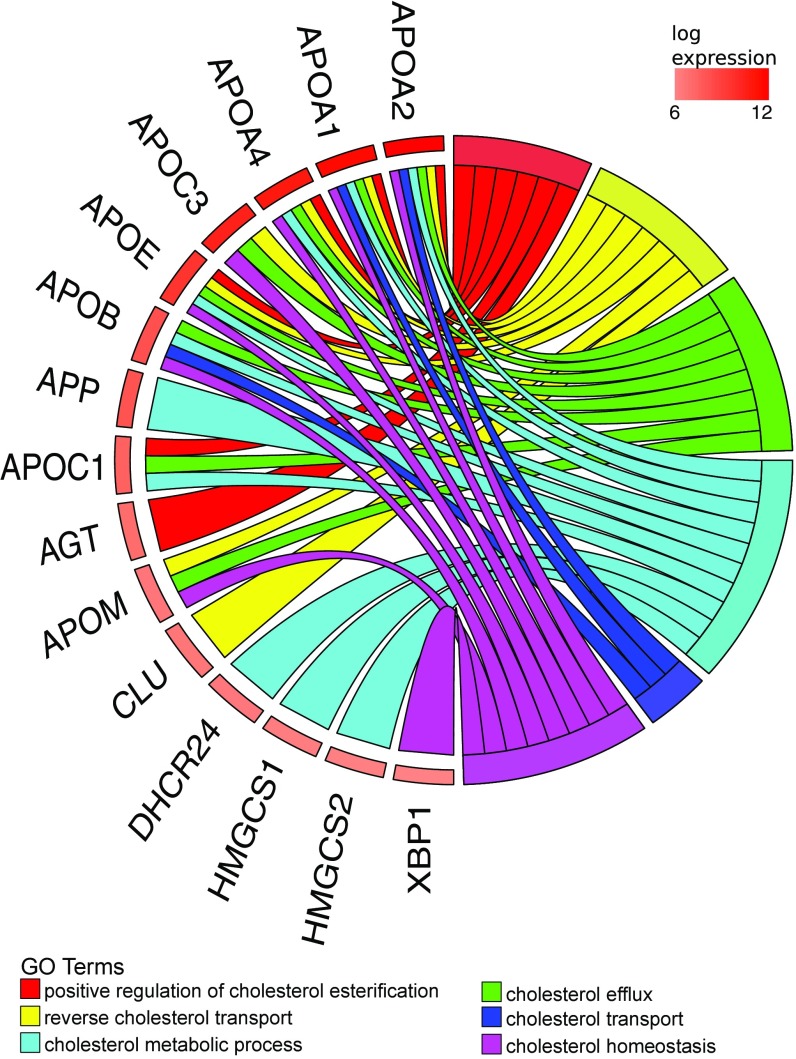
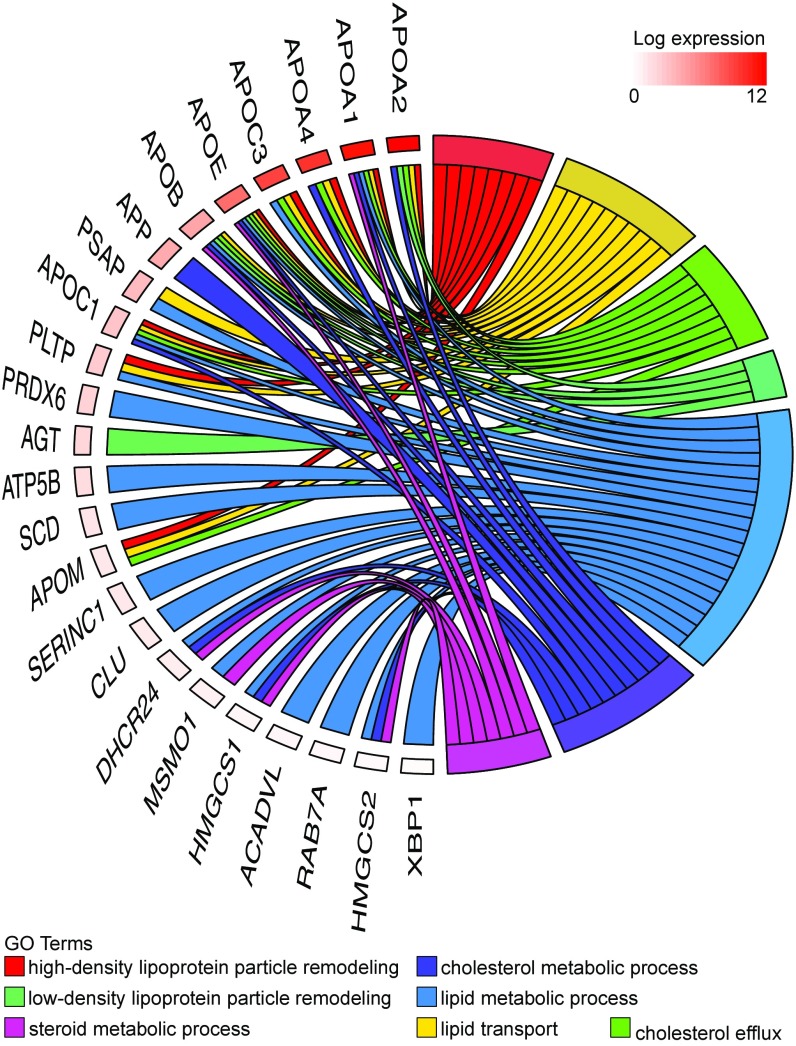
We selected the 400 most abundant human yolk sac transcripts and identified enriched GO terms using Panther (complete reference database with Bonferroni correction) (Dataset S6). Several terms associated with lipid transport were enriched; e.g., the term “very-low-density lipoprotein particle” was enriched 23-fold (P = 4.5 × 10−7). Indeed, the term “cholesterol” featured in many of the enriched biological process terms (Fig. 1). Cholesterol is required for development (3, 5) because it maintains the integrity of cell membranes (11), mediates metabolism through propagation of signaling pathways (12), and is the precursor for steroid hormones. In addition, activity of sonic hedgehog (SHH) proteins, which are responsible for the development of the central nervous system (13–15), is determined by covalent modification with cholesterol and other lipids (16). During organogenesis, the embryo is reliant on maternal sources of cholesterol until its liver is sufficiently mature for synthesis (4, 17). Our data show that the human yolk sac contains abundant mRNAs encoding multiple apolipoproteins, the cholesterol efflux transporter ABCA1, and lipoprotein receptors, including megalin and cubilin (18), albeit at lower levels (Fig. 1). Also present are transcripts encoding all classes of ABC transporters (A–G), which, in addition to transporting cholesterol and lipids, facilitate the excretion of toxins and confer multidrug resistance (Table 1). The high abundance (i.e., top 0.5%) of transcripts encoding apolipoproteins present in lipoprotein particles and chylomicrons (ApoB, ApoA1, ApoA2, and ApoA4) is matched by the high levels of these proteins in the coelomic fluid (Dataset S4). Indeed, most of the proteins found in coelomic fluid are highly ranked in the RNA-seq data (although some were undetectable, i.e., below the threshold of RPKM ≥1). Many of these proteins have functions associated with cholesterol or lipid transport and metabolism (Fig. 2).
Fig. 1.
Chord plot illustrating the GO biological process terms that include “cholesterol” and that are overrepresented in the 400 most abundant yolk sac transcripts (Right) and the genes contributing to that enrichment (Left) arranged in order of their expression level.
Table 1.
Categorization of ABC transporters detected in human yolk sac tissue
| Family | Transporters detected in human yolk sac | Function |
| ABCA | ABCA1*,†, ABCA2, ABCA3*,†, ABCA5, ABCA7* | Responsible for the transportation of cholesterol and lipids |
| ABCB | ABCB1*,†, ABCB10*,†, ABCB6*,†, ABCB7*,†, ABCB8*,† | Several of the B family members are known to confer multidrug resistance in cancer cells. Some are located in the blood–brain barrier, liver, mitochondria, transports, peptides, and bile. |
| ABCC | ABCC1*,†, ABCC10*,†, ABCC2*, ABCC3, ABCC4*,†, ABCC5*,†, ABCC6*,†, ABCC6P1*, ABCC6P2* | Ion-channel and toxin excretion activity and reception on the cell surface; toxin excretion (fungal and bacterial toxins). Includes the CFTR protein, mutations in which cause cystic fibrosis. |
| ABCD | ABCD1*,†, ABCD3*,†, ABCD4*,† | Are all used in peroxisomes. |
| ABCE/F | ABCE1*,†, ABCF1*,†, ABCF2*,†, ABCF3*,† | Not proper transporters; members contain ATP-binding domains without the transmembrane domain. Involved in regulating protein synthesis or expression. |
| ABCG | ABCG1*, ABCG2*,†, ABCG5*,†, ABCG8* | Transports lipids, diverse drug substrates, bile, cholesterol, and other steroids. |
Also present in the mouse yolk sac.
Also present in the chick yolk sac.
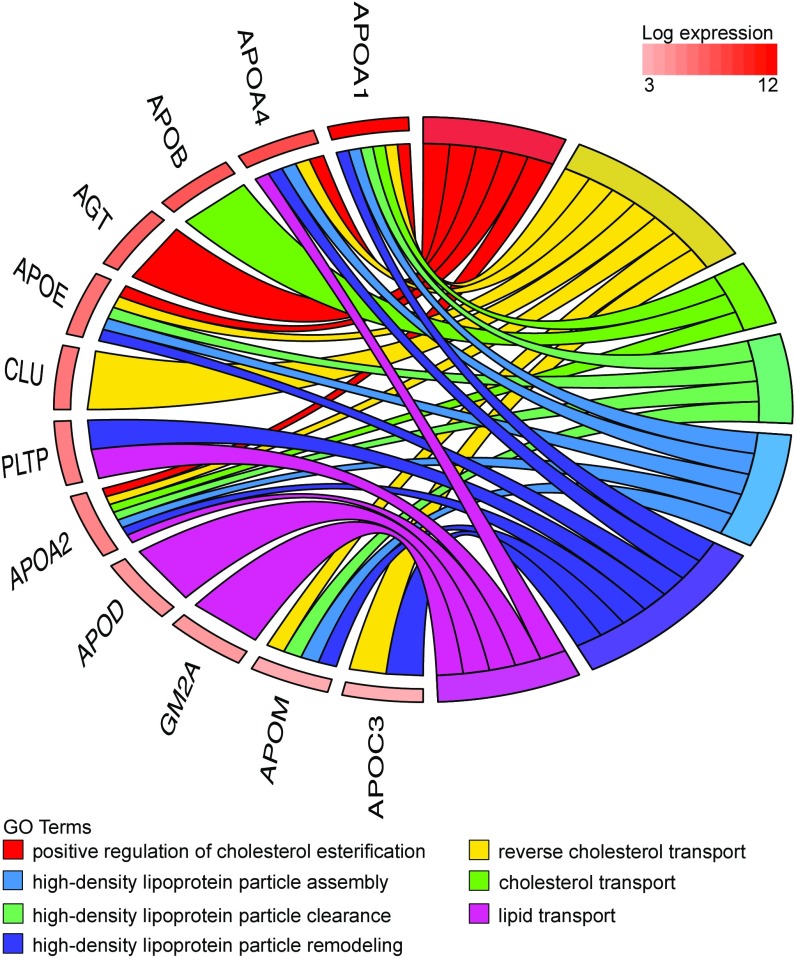
Fig. 2.
Chord plot illustrating proteins present in the coelomic fluid that relate to GO biological process terms involving “cholesterol” and “lipid transport.” The presence of these proteins is consistent with the high level of their transcripts in the human yolk sac illustrated in Fig. 1.
APOB is an essential apolipoprotein found in very-low-density lipoprotein (VLDL) particles and chylomicrons. In the latter, which are assembled in enterocytes, the APOB transcript is edited by a cytidine deaminase (APOBEC1, apolipoprotein B mRNA editing enzyme catalytic subunit 1) to introduce a stop codon leading to the production of a short form of the apolipoprotein (APOB-48). The APOB transcripts in the human secondary yolk sac are all of the unedited hepatic form, and the APOBEC1 transcript was not detected. Transcripts encoding other proteins required for lipid transport (19) are also abundant; for example, the LDL receptor (LDLR) and microsomal triglyceride transfer protein, MTTP, are in the top 10% and 20%, respectively. The presence of these transcripts suggests that the secondary yolk sac plays a role in facilitating the transport of maternal lipids and cholesterol to the fetal compartments before vascularization of the placental villi is fully established.
The fluid of the exocoelom shares many proteins in common with maternal plasma, supplemented by the addition of specific decidual, trophoblastic, and yolk sac proteins. Analogous to maternal serum proteins (20), coelomic fluid proteins can be broadly categorized into common circulating proteins, coagulation and complement factors, blood transport and binding proteins, protease inhibitors, proteases and other enzymes, cytokines and hormones, channel and receptor-derived peptides, and miscellaneous (SI Appendix, Table S2) (21, 22).
Transport.
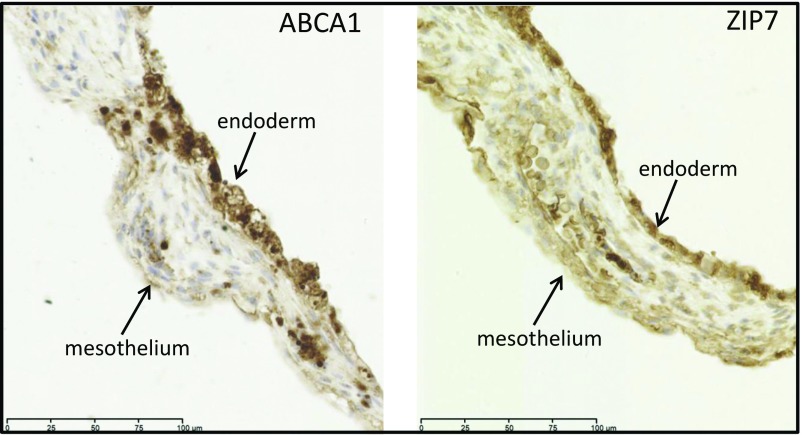
The secondary yolk sac comprises an outer mesothelial epithelium and an inner endodermal layer, separated by dilated capillaries and a small amount of mesoderm (23). Both the mesothelial and endodermal epithelia display ultrastructural features typical of an absorptive epithelium, including numerous microvilli, coated pits, and pinocytotic vesicles. The GO term “transport” (GO:0006810) was overrepresented in the top 400 yolk sac transcripts (2.87 fold, P = 7.16 × 10−47). Many such annotated transcripts were present in the most abundant 20% of transcripts (n = 87) (Dataset S6). Most of these transporter genes are members of the solute carrier (SLC) family of transporters [for example: SLC38A2 (amino acids), SLC4A1 (anions), SLC20A1 (phosphate), and SLC25A37 (iron in the mitochondria)]. Transcripts encoding 259 SLC transporters were identified (the mean RPKM ranged from 134 to 1). We classified these transporters into 11 main groups on the basis of substrate category (24), i.e., amino acids, urea cycle, glucose, nucleoside sugars, metals, vitamins, neurotransmitters, inorganic ions, thyroid, organic ions, and miscellaneous, and matched them to their main substrates that we and others (21, 22, 25–35) have identified in coelomic fluid samples (Table 2). Zinc is the second most abundant trace element and is critical for embryonic development. It plays a role in numerous biological processes, including cell division, growth, and differentiation, and acts as a structural, catalytic, and regulatory component within transcription factors, enzymes, transporters, and receptors (36, 37). Absorbed zinc is mostly bound to albumin and α2-macroglobulin, both of which are abundant in coelomic fluid. In humans, zinc transport is mediated by 14 members of the ZIP family (SLC39A) and 10 members of the ZnT family (SLC30A). We detected mRNAs encoding 12 ZIP proteins and eight ZnT proteins in the secondary yolk sac. Immunostaining for the zinc transporter SLC39A7/ZIP7 was present in both the inner endodermal and outer mesothelial epithelia, suggesting uptake from the coelomic fluid and transport to the fetal circulation (Fig. 3). The outer mesothelial layer also expresses α-tocopherol transport protein to facilitate vitamin E transport (38). Thus, our data suggest the human secondary yolk sac has a role in the transport of multiple nutrients and vitamins, including iron (seven iron-transporting SLC transcripts were identified) (Table 2) (39, 40) and vitamins A, B12, C, E, and folic acid (Table 2).
Table 2.
Categorization of SLC transporters detected in human yolk sac tissue
| Substrate category | SLC transporters detected in human yolk sac | Known substrates | Substrates detected in human coelomic fluid |
| Amino acids | SLC38A2*,†, SLC7A7*, SLC7A2, SLC1A5*, SLC38A3, SLC3A2*, SLC3A1†, SLC25A38*, SLC38A4*, SLC38A10*, SLC25A13*,†, SLC7A8*, SLC7A6*,†, SLC43A1*, SLC7A4, SLC7A9*, SLC1A4*, SLC38A7*,†, SLC7A1*, SLC15A4*,†, SLC38A5*, SLC7A6OS*,†, SLC43A3*,†, SLC38A9*,†, SLC43A2*,†, SLC36A4*, SLC38A6*,†, SLC38A11, SLC6A17, SLC7A10 | Ala, Asn, Cys, Gln, Gly, His, Met, Pro, Ser, Glu, Trp, Asp | Thr, His, Arg, Val, Met, Ile, Leu, Phe, Lys, Ser, Gln, Glu, Tau, Ala, Pro, Tyr, Orn, Trp (21, 22, 25–27) |
| Urea cycle | SLC25A15*,†, SLC2A9† | Lys, Orn, Cit, Asp, Glu, uric acid | Urea (28, 29) |
| Glucose and sugars | SLC2A1*,†, SLC2A3*,†, SLC2A4RG, SLC50A1*, SLC2A2*,†, SLC1A1†, SLC2A10†, SLC2A14, SLC5A9*, SLC45A4*, SLC2A8*, SLC2A12, SLC2A6, SLC2A13†, SLC37A1, SLC2A5†, SLC37A2†, SLC2A7*, SLC45A1, SLC2A11, SLC2A1-AS1 | Glucose, galactose, fructose, mannose, glucosamine | Glucose, galactose, fructose, mannose, glucosamine, galactosamine, erythritol, ribitol, mannitol, inositol, glycerol, sorbitol (22, 30) |
| Nucleoside sugars | SLC35D2*, SLC17A9*, SLC35E2B†, SLC29A1*, SLC35C2*,†, SLC35A4*,†, SLC35A2*, SLC35E1*,†, SLC25A36*,†, SLC35D1*,†, SLC35A5*,†, SLC35F5*,†, SLC37A4*,†, SLC35F6*, SLC35A3*,†, SLC37A3*,†, SLC35B1*,†, SLC35C1*, SLC52A2*, SLC29A2*, SLC29A3*, SLC35B4*,†, SLC35G1*, SLC35E3*,†, SLC29A4, SLC35F2*,†, SLC35F3, SLC25A19*, SLC35G2*, SLC28A1* | UDP-glucuronic acid, UDP-galactose, UDP-N-acetyl-galactosamine, GDP-fructose | |
| Metals | SLC30A5*,†, SLC30A9*,†, SLC40A1*,†, SLC39A7, SLC39A14*,†, SLC25A37*,†, SLC39A6*, SLC39A5*, SLC39A1*, SLC30A1*,†, SLC31A1*,†, SLC25A28*,†, SLC39A13*,†, SLC39A9*,†, SLC39A8*,†, SLC11A2*,†, SLC30A6*,†, SLC30A7*, SLC41A1*, SLC41A3*, SLC39A11*,†, SLC30A10*,†, SLC31A2*, SLC39A10*, SLC41A2†, SLC30A2*, SLC39A4*, SLC11A1, SLC39A3*, SLC30A4*,† | Zinc, iron, magnesium, copper, cadmium, cobalt, manganese, nickel, lead, barium, strontium | Zinc, iron, cadmium, magnesium, copper, manganese, lead, selenium (28, 31, 32) |
| Vitamins | SLC19A2*, SLC46A3*,†, SLC25A32*,†, SLC23A2*, SLC5A6*, SLC46A1*,†, SLC23A1*, SLC19A3*, SLC19A1*, SLC23A3* | Thiamine (vitamin B1), folate, ascorbic acid, biotin, lipoate, pantothenate, thiamine | Vitamins A, E, B12, folate, cobalamin, retinol-binding protein 4, vitamin D-binding protein (21, 27, 29, 33–35) |
| Neurotransmitters | SLC44A2*, SLC44A4*, SLC6A6*,†, SLC44A1*,†, SLC36A1*,†, SLC6A9*,†, SLC1A3*, SLC6A13*, SLC44A3†, SLC17A2, SLC25A22*,†, SLC25A18, SLC25A12*,†, SLC17A4, SLC6A12 | Noradrenaline, serotonin, dopamine, glutamate, glycine, aspartate, choline | Glutamine, glutamic acid (21, 25) |
| Inorganic ions | SLC25A3*,†, SLC9A3R1*, SLC4A1*,†, SLC20A1*, SLC12A7*,†, SLC4A2*,†, SLC9A3R2*, SLC34A2*,†, SLC12A4*,†, SLC26A6*, SLC9A1*, SLC20A2*, SLC9A6*,†, SLC4A1AP*, SLC9A9†, SLC26A11*,†, SLC9B2, SLC26A2*, SLC24A3, SLC9A8*, SLC12A8, SLC8B1*, SLC12A2*,†, SLC24A1, SLC4A7*, SLC12A6*, SLC4A3, SLC26A10, SLC26A1*, SLC5A5, SLC9A7P1, SLC9A5*, SLC8A1, SLC4A4* | Na+, K+, Cl−, HCO3−, Ca2+, phosphate | Na+, K+, Cl−, HCO3−, Ca2+, phosphate (28, 29) |
| Thyroid | SLC16A2†, SLC7A5*,†, SLC16A10*,† | Iodide, iodothyronines | Tyrosine, thyroxine (26) |
| Organic ions | SLCO2B1†, SLC22A7, SLC10A1, SLC10A3*, SLC22A23*,†, SLC51A*, SLC22A9, SLC22A17*, SLCO4C1*, SLCO2A1*, SLC22A31, SLC22A18*,†, SLC22A4*, SLC22A5*,†, SLC10A7*,†, SLC51B, SLCO1B3, SLC22A15, SLC22A18AS, SLCO1B1, SLCO3A1*,†, SLC17A3, SLC22A3* | Estradiol, glucuronide, bilirubin | Bilirubin |
| Miscellaneous | SLC25A5*, SLC5A12, SLC25A39*,†, SLC25A1*,†, SLC16A3*, SLC35B2*, SLC25A20*, SLC17A5*,†, SLC16A1*,†, SLC16A4†, SLC13A5, SLC35B3*, SLC6A8*, SLC27A2*,†, SLC25A24*,†, SLC25A23*, SLC25A11*, SLC25A44*, SLC25A25*, SLC45A3*, SLC15A1*, SLC25A46*, SLC25A27, SLC25A42*, SLC27A3*, SLC25A17*,†, SLC25A43*,†, SLC35A1*,†, SLC12A9*,†, SLC16A5, SLC25A29†, SLC18B1*, SLC16A13*,†, SLC25A30*,†, SLC27A4*,†, SLC16A9*,†, SLC48A1*,†, SLC33A1*,†, SLC25A4*, SLC25A14*,†, SLC25A33*, SLC25A16*,†, SLC25A40*, SLC25A34*, SLC27A1*,†, SLC25A26*, SLC47A1*,†, SLC15A3, SLC16A12*,†, SLC25A45*, SLC5A11*, SLC25A51*, SLC13A3*,†, SLC16A14, SLC25A21-AS1, SLC16A7†, SLC16A6*, SLC15A2, SLC27A5*, SLC17A1 | ATP-ADP, carnitine, creatinine, acetyl-CoA, sialic acid, pyruvate, lactate, ketone bodies, bile acids, oxoglutarate, succinate, citrate, ketoglutarate, ornithine, acylcarnitine, melanin, prostaglandin, long-chain and very long-chain fatty acids, haem, ammonia, adenosine, taurocholic acid | Lactate, creatinine (29, 30) |
Also present in the mouse yolk sac.
Also present in the chick yolk sac.
Fig. 3.
Immunolocalization of ABCA1 and SLC39A7/ZIP7 transporter proteins in the human yolk sac at gestational age 11 wk. Sections were immunostained with anti-ABCA1 or anti-ZIP7 antibodies. In both cases, staining was present in the inner endodermal and outer mesothelial layers, although it was stronger in the former.
The total protein concentration is lower in coelomic fluid than in maternal plasma. However, most amino acids are at higher concentrations and must be derived from the villi and/or the yolk sac. This difference suggests that the coelomic cavity is an important route for metabolites required for embryonic development (25). The secondary yolk sac floats within this nutrient-rich milieu. It is therefore possible that uterine secretions supplemented by maternal plasma from spiral arteries are taken up by the trophoblast cells and are passed via the villous stromal channels into the exocoelomic cavity, from which they are taken up by the yolk sac and transferred to the embryonic gut and the fetal circulation via the vitelline duct (38). Thus, there appears to be free interchange between these two compartments of the human gestational sac.
The passage across the trophoblast may require lysosomal digestion of macromolecules, and indeed the GO term “lysosome” (GO:0005764) is enriched fourfold within the most abundant 400 villous transcripts (Bonferroni corrected P = 7.99 x10−9). The efflux amino acid transporters SLC43A2 and SLC7A8 are also highly expressed (above the 95th and 83rd percentiles, respectively). In the rat it has been estimated that ∼95% of the amino acids provided to the fetus in midgestation are obtained by lysosomal digestion of endocytosed maternal proteins (2). In the mouse yolk sac, transcripts encoding five lysosomal cathepsins (Ctsl, Ctsz, Ctsb, Ctsd, and Ctsh) are among the most abundant 400 transcripts, the activity of which would allow the degradation and release of free amino acids to the developing fetus. We have previously shown that the mesothelial layer of the human yolk sac stains for glycodelin, a product of the uterine glands that is present in high concentration in the coelomic fluid (41), indicating exposure to intact maternal proteins and uptake (42). Glycodelin also colocalizes with lysosomal Cathepsin D in human first-trimester villi (43). As in the mouse, several cathepsin transcripts (CTSB, CTSZ, CTSL, and CTSD) are extremely abundant in the human secondary yolk sac. We found that the GO term “lysosome” is enriched in the list of most abundant transcripts from yolk sacs of human, mouse, and chicken (2.79 fold P = 1.35−3; 3.53 fold P = 9.72−5; and 3.6 fold P = 2.13−3, respectively), indicating a similar capacity for digestion of endocytosed macromolecules.
These findings indicate that the exocoelomic cavity is a physiological liquid extension of the early placenta (44) and that the yolk sac is an important route of access for high molecular weight proteins to the embryonic circulation (45).
Hematopoiesis.
In all vertebrates, primitive embryonic and definitive fetal/adult blood cells form successively within the yolk sac, fetal liver, and bone marrow (46). The human secondary yolk sac is the sole site of hematopoiesis for the first 2 wk of pregnancy, and the fetal liver commences blood cell production at week 6 of gestation (47, 48). The term “hemoglobin complex” was significantly enriched among abundant yolk sac transcripts (33 fold, P = 2.95 x10−6). The yolk sac produces predominantly nucleated erythrocytes, which synthesize embryonic hemoglobin (HBZ). There is morphological evidence of the first blood islands in the secondary yolk sac at about day 18 of gestation (49). Yolk sac-derived primitive erythrocytes have been detected in the cardiac cavity as early as the three-somite stage (21 d), indicative of an established functional network between the yolk sac and embryo (50). The human yolk sac, like that of the mouse, also produces macrophage and multipotential hematopoietic progenitors (48).
Transcription Factors.
Within the most abundant 400 transcripts in the human yolk sac, 19 genes are annotated as “regulation of transcription, DNA-templated” (GO:0006355), including several transcription factors (ATF4, FOS, JUN, JUNB, and JUND.) In the mouse dataset, eight genes are similarly annotated (SI Appendix, Table S3). Candidate motifs recognized by these transcription factors (where known) were identified in the regions 1 kb and 5 kb upstream of the transcription start site (TSS) of genes that were highly correlated with the transcription factor transcripts (Datasets S7–S10). The FOS and JUN families and AFT4 are closely related, interact functionally, have multiple target genes, and are widely expressed. There are numerous candidate binding sites in the highly expressed human yolk sac genes (181 genes with sites for the FOS and JUN families and ATF4) (Datasets S7 and S8) and 18 genes in the mouse with candidate Atf4-binding sites (Datasets S9 and S10). The evidence used for the assignment of GO terms varies and for two genes (IGF2 and BHLHE40) depends on a Nontraceable Author Statement (NAS) (SI Appendix, Table S3). Furthermore, binding motifs are not available for all candidate factors even in the most recent JASPAR database.
Yolk Sac and Villi Compared with Adult Lung, Liver, and Kidney.
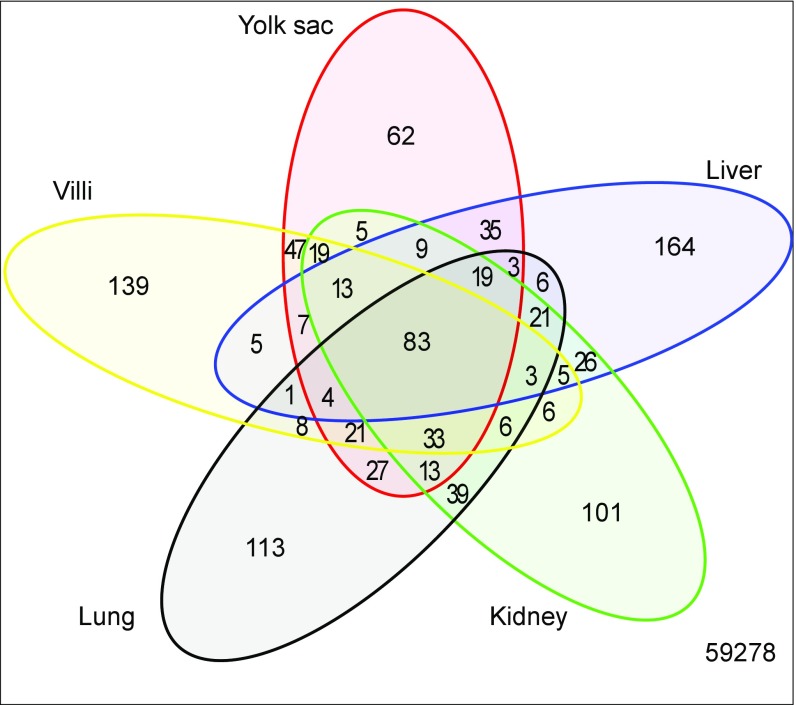
The inaccessibility of the human yolk sac severely constrains any functional investigation of these candidate transcription binding sites. We therefore compared the transcript profile of the human yolk sac with tissues whose function has been better defined experimentally. The placental villi serve functions similar to those of the adult lung, liver, and kidney, and we therefore compared the overlap among the 400 most abundant transcripts from these tissues and the yolk sac (Fig. 4). The transcripts that are unique to each tissue and those shared among these tissues are listed in Dataset S11. As expected, the transcripts shared by all five tissues encoded abundant housekeeping proteins, such as ribosomal proteins, and those involved in mitochondrial energy generation, with GO terms such as “cytosolic small ribosome subunit,” “cytosolic large ribosome subunit,” and “mitochondrial respiratory chain” being significantly overrepresented (P < 8.5 × 10−8 after Bonferroni correction) (Dataset S12). The enriched GO terms associated with the 35 transcripts shared only by the yolk sac and liver include “high-density lipoprotein particle receptor binding,” “cholesterol transporter activity,” and “lipid transporter activity” (all greater than 28-fold enrichment and P < 2 × 10−4 after Bonferroni correction) (Dataset S13).
Fig. 4.
Venn diagram comparing the most abundant 400 transcripts in the human yolk sac with first-trimester placental villi and adult liver, lung, and kidney. Transcripts shared by all five tissues (n = 83) principally encoded housekeeping proteins, whereas those shared uniquely with liver (n = 35) encoded proteins involved in cholesterol and lipid metabolism, suggesting that the yolk sac may perform these functions while the fetal liver develops. By contrast, there are few transcripts shared uniquely with the kidney (n = 5), suggesting that the yolk sac plays little role in excretion.
We also examined the overlap among overrepresented GO terms in the most abundant transcripts in these five tissues (Datasets S6 and S14–S17). For example, the biological process terms “high-density lipoprotein particle remodeling,” “lipid transport,” “cholesterol efflux,” and “cholesterol metabolic process” are all overrepresented and shared between the yolk sac and liver (Fig. 5 and Datasets S18–S20). It is striking that among the shared transcripts many (18 of 35) encode proteins with transport capacity. Besides the genes associated with lipid and cholesterol transport (APOA1, APOA2, APOA4, APOB, APOC1, APOC3, and APOH), the major serum carriers of corticosteroids and progesterone (SERPINA6) are present. In addition, the thyroxine (T4) hormone carrier SERPINA7 (also known as “TBG” and “TTR”) was detected. Maternal thyroxine is essential for fetal brain development before the commencement of fetal thyroid function at the end of the first trimester (26). Furthermore, transcripts encoding several metal-binding or -transporting proteins are present (MT1E, MT1H, TF, HPX, and SEPP1). SEPP1 is a selenoprotein which contains 10 selenocysteine residues per molecule and is postulated to function as a carrier of selenium (51). These data indicate that the yolk sac shares many abundant transcripts with other metabolically active fetal and adult tissues and that it contains transcripts that encode several liver specific functions, including those encoding several essential hormone transporters.
Fig. 5.
Chord plot connecting GO biological process terms associated with “lipid metabolism” and genes encoding transcripts that are shared by the human yolk sac and adult liver. The overlap suggests the yolk sac may perform hepatic functions while the fetal liver differentiates.
Mouse Yolk Sac RNA-Seq Data.
On a comparative basis, the yolk sac provides an important pathway for nutrient uptake in many species during early pregnancy (52, 53), and its role has been well documented in experimental rodents, such as the rat and mouse (54). At the ultrastructural level, the endodermal layers of the human and rodent yolk sacs display many similar morphological characteristics typical of an absorptive epithelium (10, 55). However, their orientation is different, because the human yolk sac floats in the exocoelom with the endodermal cell layer lining the interior of the sac, whereas the rodent yolk sac is inverted, with the endodermal layer facing outwards following degeneration of the parietal layer (56, 57).
Cross-Species Comparison.
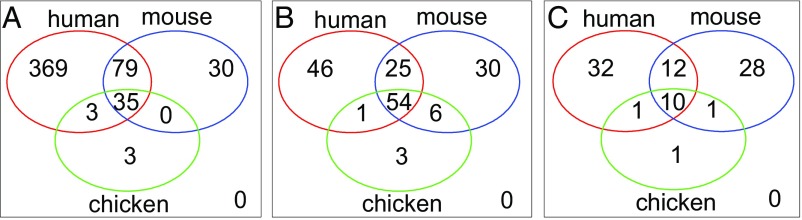
To compare the most abundant transcripts in the human, mouse, and chicken yolk sacs, we identified the homologous genes by mapping the human Ensembl gene identifiers to the corresponding mouse and chicken Ensembl gene IDs using BioMart (release ENSEMBL Genes 85) (Dataset S21). The raw and processed chicken yolk sac data were obtained directly from the authors of ref. 58. GO analysis was carried out as described above, using the 400 most abundant chicken yolk sac transcripts observed at E17 (Dataset S22). The intersection between the overrepresented GO terms in the human, mouse, and chicken yolk sacs was determined as described above (SI Appendix, Table S4). Venn diagrams showing the overlaps are shown in Fig. 6. The P values for all the observed intersections are highly significant (P < 1.70 × 10−22) and are summarized in Dataset S23. The depth of the GO annotation varies among species and is more limited in the chicken, thereby precluding the detection of a high degree of overlap with the human genes. Nonetheless, shared biological process terms include “translation,” “ribosome biogenesis,” “oxidation-reduction process,” and “small molecule metabolic process,” reflecting cellular processes associated with metabolically active tissue. Transcripts reflecting more specialized cellular function, such as lipid and cholesterol transport, which are shared by the human yolk sac and liver (APOA1, APOA4, APOB, RBP4, SEPP1, and TTR) are also present among the 400 most abundant mouse and chicken yolk sac transcripts (P = 2.3 × 10−13).
Fig. 6.
Venn diagrams illustrating the overlap among overrepresented GO terms associated with the 400 most abundant transcripts in each of the human, mouse, and chicken yolk sacs. (A) Biological process terms. (B) Cellular component terms. (C) Molecular function terms. The considerable overlap among the species in all three categories suggests conservation of functions.
Conclusion
Overall, our data show that the human secondary yolk sac appears to be far from vestigial with little or no biological role and that it may perform many key functions in the early weeks of development. In particular, it contains abundant transcripts encoding an array of transporter proteins involved in the uptake of macro- and micronutrients. Our data showing the presence of transporter proteins in both yolk sac epithelia and of their substrates in coelomic fluid support this concept and are consistent with the morphological appearances of absorptive surfaces (23, 59, 60). Our data also confirm significant synthetic activity, especially of apolipoproteins, a function most likely performed in the endodermal cells, given their high content of endoplasmic reticulum and Golgi cisternae (23, 59, 60). The handling of cholesterol, which is essential for synthesis of cell and organelle membranes as well as being a cofactor in signaling pathways involved in axis determination and other fundamental developmental events, appears to be of particular significance. Furthermore, the high level of conservation of transcripts compared with the mouse and the chicken, in which the yolk sac is known to be essential for development, suggests maintenance of function.
The secondary yolk sac thus is likely to be essential for the survival of the embryo during the first weeks of development. Morphological abnormalities of the secondary yolk sac have been reported in 70% of first trimester human miscarriages (61), but separating cause from effect is impossible.
More generally, our findings shed light on the evolutionary mechanisms that give rise to complex structures such as the placenta. The placenta has evolved repeatedly from an oviparous background in mammals, reptiles, and fish, sometimes over very short timescales (62, 63). Development of a yolk sac for direct maternal provisioning of the developing fetus is a common feature of such evolutionary transitions to the extent that the yolk sac may be regarded as a “fundamental vertebrate fetal nutritional system” (64). The yolk sacs studied in this paper constitute a broad, albeit incomplete, sampling of the variation present in the vertebrates. The inverted yolk sac placenta of the mouse is found in several rodents and lagomorphs (1) and also in the distantly related nine-banded armadillo (65). The free-floating secondary yolk sac of the human is found in the other haplorhine primates (Macaca mulatta) (66) but also, surprisingly, in distantly related Afrotherian species. The yolk sac floats freely in the exocoelom in at least one tenrec species, the Nimba otter shrew (Micropotamogale lamottei) (67), in another Afrotherian insectivore, the eastern rock elephant shrew (Elephantulus myurus) (68), and in two species of bat [Myotis lucifugus (little brown bat) (69) and Tadarida brasiliensis (70)]. These observations suggest that mouse and human yolk sacs reflect convergent evolution toward similar forms found elsewhere in the mammalian phylogenetic tree. The yolk sac of the chicken, of course, is characteristic of oviparous species, i.e., all birds and the majority of reptiles.
Under a classic Darwinian model of macroevolution, small genetic mutations gradually occur and are accompanied by correspondingly small phenotypic changes until, over time, a high degree of morphological and functional diversification accumulates between species that are distantly related. The results presented here suggest, however, that the genetic systems underlying the function of the yolk sac are robust. We identify a number of genetic modules, including those involved in cholesterol processing, lipid transport, redox processes, and nutrient delivery, which presumably were reorganized and redeployed during evolutionary change while being internally conserved. These findings are in line with an extended evolutionary–developmental model in which it is the gene regulatory networks and underlying transcriptional control elements that change (71). The repeated convergent evolution of yolk sac placentas in all major groups of vertebrates other than birds is characteristic of what has come to be known as “deep homology,” which describes the origin of complex structures through modification and reorganization of preexisting genetic systems (72). Given that the modular conservation of systems active in the yolk sac is shown to extend across mammals and birds, the common ancestor of which was a reptile, it is possible that genetic modules of the yolk sac are part of the core amniote genetic repertoire. That is, conserved genetic systems of yolk sac function, for example cholesterol and lipid metabolism, form part of the common heritage shared by all mammals, reptiles, and birds and are the building blocks for both oviparous and viviparous reproductive modes.
Our findings indicate that extensive high-level morphological diversification of the extraembryonic membranes masks a surprising degree of functional conservation at the molecular genetic level. Evolutionary conservation at the level of nucleotide sequence, gene regulation, and modularity of gene expression is widely regarded as evidence of functional significance in both healthy development and in disease (73–75). Therefore, we propose that, although a choriovitelline placenta is never established physically in the human, the early placental villi, the exocoelomic cavity, and the secondary yolk sac combine to function as a physiological equivalent (Fig. 7).
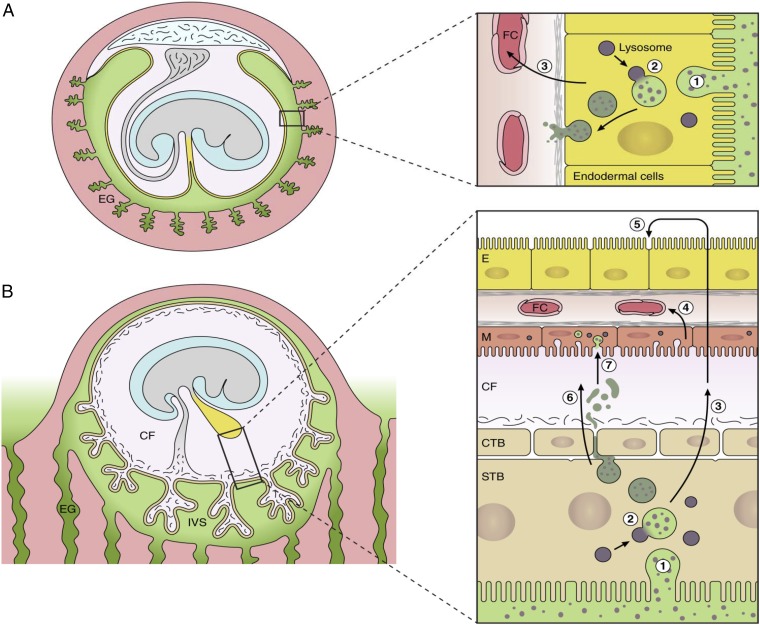
Fig. 7.
Diagrammatic comparison of the nutrient pathway during early pregnancy in the mouse (A) and the speculated pathway in the human (B). In the mouse, histotrophic secretions (green) released from the endometrial glands are phagocytosed (step 1) by the endodermal cells of the visceral layer of the inverted yolk sac. Following fusion with lysosomes (step 2), digestion of maternal proteins leads to release of amino acids that are transported (step 3) to the fetal circulation (FC). In the human, histotrophic secretions are released from the endometrial glands through the developing basal plate of the placenta into the intervillous space (IVS) and are phagocytosed (step 1) by the syncytiotrophoblast (STB) (42). We speculate that following digestion by lysosomal enzymes (step 2), free amino acids are transported (step 3) by efflux transporters to the coelomic fluid (CF), where they accumulate. Nutrients in the CF may be taken up by the mesothelial cells (M) of the yolk sac and transported (step 4) into the fetal circulation (FC). Alternatively, they may diffuse into the cavity of the yolk sac and be taken up by the endodermal cells (step 5). Some intact maternal proteins may also be released into the CF by exocytosis of residual bodies (step 6) and may be engulfed by the mesothelial cells (step 7). CTB, cytotrophoblast cells.
Methods
Human Tissue Collection.
Tissue and fluid samples were collected with informed written patient consent and approval of the Joint University College London/University College London Hospital Committees on the Ethics of Human Research (05/Q0505/82) from 7- to 12-wk uncomplicated pregnancies. Gestational age was confirmed by ultrasound measurement of the crown rump length of the embryo. All samples were collected from patients undergoing surgical pregnancy termination under general anesthesia for psycho-social reasons. Coelomic fluid samples were obtained by transvaginal puncture under sonographic guidance as previously described (27). Villous samples were obtained under transabdominal ultrasound guidance from the central region of the placenta using a chorionic villus sampling (CVS) technique. Intact secondary yolk sacs were obtained by gentle aspiration guided by ultrasound. All samples were snap-frozen in liquid nitrogen and stored at −80 °C until analysis.
Mouse Tissue Collection.
Yolk sacs were collected from time-mated virgin C57BL/6J mice. Experiments were carried out in accordance with the United Kingdom Animals Scientific Procedures Act 1986 which mandates ethical review. A single randomly selected yolk sac was collected from each pregnant female at E9.5 (day of plug = E0.5). Tissue was dissected free from decidua and amnion, snap-frozen, and stored at −80 °C until processing.
RNA Extraction and RNA-Seq.
RNA was extracted from human and mouse yolk sacs and human first-trimester placental villi using the RNeasy Plus Universal Mini Kit (catalog no. 73404; Qiagen). Libraries were made using the Illumina TruSeq Stranded mRNA Library Kit according to the manufacturer’s instructions. Libraries were quantified (kappa qPCR), and equimolar pools were sequenced (single end 50 base reads, SE50) in several lanes of the Illumina HiSeq2500. Additional details are provided in the SI Appendix.
Data Availability.
The datasets generated during the current study are available in the European Nucleotide Archive (ENA www.ebi.ac.uk/ena) under the accession number PRJEB18767 (www.ebi.ac.uk/ena/data/view/PRJEB18767).
Proteomic Analysis of Coelomic Fluid Samples.
Coelomic fluid samples were run on 1D gels, enzymatically digested, and analyzed using an LC-MS/MS (Dionex Ultimate 3000 RSLC nanoUPLC; Thermo Fisher Scientific, Inc.) system and a QExactive Orbitrap mass spectrometer (Thermo Fisher Scientific, Inc.). Detailed methods are supplied in SI Appendix, Methods.
Immunohistochemistry.
Immunohistochemistry was performed as previously described (76) using the following primary antibodies: anti-SLC39A7 (ZIP7; ab117560; Abcam) and anti-ABCA1 (ab7360; Abcam).
Supporting Information.
Additional methods are given in SI Appendix, Methods online.
Supplementary Material
Acknowledgments
We thank Prof. Kathryn Lilley (University of Cambridge) for the proteomic analysis and Dr. Erica Watson (University of Cambridge) for assistance with the mouse yolk sac samples. M.G.E. is the recipient of a Research Fellowship from St. John’s College, University of Cambridge. This study was supported by Medical Research Council Grant MR/L020041/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The datasets generated during the current study are available in the European Nucleotide Archive (ENA) (www.ebi.ac.uk/ena) [accession no. PRJEB18767 (www.ebi.ac.uk/ena/data/view/PRJEB18767)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702560114/-/DCSupplemental.
References
- 1.Mossman H. Vertebrate Fetal Membranes: Comparative Ontogeny and Morphology: Evolution; Phylogenetic Significance; Basic Functions, Research Opportunities. Macmillan; London: 1987. [Google Scholar]
- 2.Brent RL, Fawcett LB. Nutritional studies of the embryo during early organogenesis with normal embryos and embryos exhibiting yolk sac dysfunction. J Pediatr. 1998;132:S6–S16. doi: 10.1016/s0022-3476(98)70522-0. [DOI] [PubMed] [Google Scholar]
- 3.Woollett LA. Where does fetal and embryonic cholesterol originate and what does it do? Annu Rev Nutr. 2008;28:97–114. doi: 10.1146/annurev.nutr.26.061505.111311. [DOI] [PubMed] [Google Scholar]
- 4.Baardman ME, et al. The origin of fetal sterols in second-trimester amniotic fluid: Endogenous synthesis or maternal-fetal transport? Am J Obstet Gynecol. 2012;207:202.e19–202.e25. doi: 10.1016/j.ajog.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Baardman ME, et al. The role of maternal-fetal cholesterol transport in early fetal life: Current insights. Biol Reprod. 2013;88:1–9. doi: 10.1095/biolreprod.112.102442. [DOI] [PubMed] [Google Scholar]
- 6.King BF, Enders AC. Protein absorption and transport by the guinea pig visceral yolk sac placenta. Am J Anat. 1970;129:261–287. doi: 10.1002/aja.1001290303. [DOI] [PubMed] [Google Scholar]
- 7.Moore K. The Developing Human Clinically Oriented Embryology. WB Saunders Company, Harcourt Brace Jovanovich, Inc.; Philadelphia, London, Toronto, Montreal, Sydney, Tokyo: 1988. [Google Scholar]
- 8.Zohn IE, Sarkar AA. The visceral yolk sac endoderm provides for absorption of nutrients to the embryo during neurulation. Birth Defects Res A Clin Mol Teratol. 2010;88:593–600. doi: 10.1002/bdra.20705. [DOI] [PubMed] [Google Scholar]
- 9.Jauniaux E, Gulbis B, Burton GJ. The human first trimester gestational sac limits rather than facilitates oxygen transfer to the foetus–a review. Placenta. 2003;24 Suppl A:S86–93. doi: 10.1053/plac.2002.0932. [DOI] [PubMed] [Google Scholar]
- 10.Jones C. In: Embryonic Medicine and Therapy. Jaunaiux E, Barnea E, Edwards R, editors. Oxford Univ Press; Oxford, UK: 1997. [Google Scholar]
- 11.Pörn MI, Ares MP, Slotte JP. Degradation of plasma membrane phosphatidylcholine appears not to affect the cellular cholesterol distribution. J Lipid Res. 1993;34:1385–1392. [PubMed] [Google Scholar]
- 12.Fielding CJ, Fielding PE. Membrane cholesterol and the regulation of signal transduction. Biochem Soc Trans. 2004;32:65–69. doi: 10.1042/bst0320065. [DOI] [PubMed] [Google Scholar]
- 13.Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 14.Cooper MK, et al. A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat Genet. 2003;33:508–513. doi: 10.1038/ng1134. [DOI] [PubMed] [Google Scholar]
- 15.Martí E, Bovolenta P. Sonic hedgehog in CNS development: One signal, multiple outputs. Trends Neurosci. 2002;25:89–96. doi: 10.1016/s0166-2236(02)02062-3. [DOI] [PubMed] [Google Scholar]
- 16.Long J, et al. Identification of a family of fatty-acid-speciated sonic hedgehog proteins, whose members display differential biological properties. Cell Reports. 2015;10:1280–1287. doi: 10.1016/j.celrep.2015.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Witsch-Baumgartner M, et al. Maternal apo E genotype is a modifier of the Smith-Lemli-Opitz syndrome. J Med Genet. 2004;41:577–584. doi: 10.1136/jmg.2004.018085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burke KA, Jauniaux E, Burton GJ, Cindrova-Davies T. Expression and immunolocalisation of the endocytic receptors megalin and cubilin in the human yolk sac and placenta across gestation. Placenta. 2013;34:1105–1109. doi: 10.1016/j.placenta.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao C, Hsieh J, Adeli K, Lewis GF. Gut-liver interaction in triglyceride-rich lipoprotein metabolism. Am J Physiol Endocrinol Metab. 2011;301:E429–E446. doi: 10.1152/ajpendo.00178.2011. [DOI] [PubMed] [Google Scholar]
- 20.Adkins JN, et al. Toward a human blood serum proteome: Analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics. 2002;1:947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 21.Jauniaux E, et al. Relationship between protein concentrations in embryological fluids and maternal serum and yolk sac size during human early pregnancy. Hum Reprod. 1994;9:161–166. doi: 10.1093/oxfordjournals.humrep.a138308. [DOI] [PubMed] [Google Scholar]
- 22.Jauniaux E, Gulbis B. Fluid compartments of the embryonic environment. Hum Reprod Update. 2000;6:268–278. doi: 10.1093/humupd/6.3.268. [DOI] [PubMed] [Google Scholar]
- 23.Jones CJP, Jauniaux E. Ultrastructure of the materno-embryonic interface in the first trimester of pregnancy. Micron. 1995;26:145–173. doi: 10.1016/0968-4328(95)00002-l. [DOI] [PubMed] [Google Scholar]
- 24.Lin L, Yee SW, Kim RB, Giacomini KM. SLC transporters as therapeutic targets: Emerging opportunities. Nat Rev Drug Discov. 2015;14:543–560. doi: 10.1038/nrd4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jauniaux E, Gulbis B, Gerlo E, Rodeck C. Free amino acid distribution inside the first trimester human gestational sac. Early Hum Dev. 1998;51:159–169. doi: 10.1016/s0378-3782(97)00107-2. [DOI] [PubMed] [Google Scholar]
- 26.Contempré B, et al. Detection of thyroid hormones in human embryonic cavities during the first trimester of pregnancy. J Clin Endocrinol Metab. 1993;77:1719–1722. doi: 10.1210/jcem.77.6.8263162. [DOI] [PubMed] [Google Scholar]
- 27.Jauniaux E, Sherwood RA, Jurkovic D, Boa FG, Campbell S. Amino acid concentrations in human embryological fluids. Hum Reprod. 1994;9:1175–1179. doi: 10.1093/oxfordjournals.humrep.a138654. [DOI] [PubMed] [Google Scholar]
- 28.Campbell J, et al. Biochemical composition of amniotic fluid and extraembryonic coelomic fluid in the first trimester of pregnancy. Br J Obstet Gynaecol. 1992;99:563–565. doi: 10.1111/j.1471-0528.1992.tb13821.x. [DOI] [PubMed] [Google Scholar]
- 29.Jauniaux E, et al. Biochemical composition of exocoelomic fluid in early human pregnancy. Obstet Gynecol. 1991;78:1124–1128. [PubMed] [Google Scholar]
- 30.Jauniaux E, Hempstock J, Teng C, Battaglia FC, Burton GJ. Polyol concentrations in the fluid compartments of the human conceptus during the first trimester of pregnancy: Maintenance of redox potential in a low oxygen environment. J Clin Endocrinol Metab. 2005;90:1171–1175. doi: 10.1210/jc.2004-1513. [DOI] [PubMed] [Google Scholar]
- 31.Gulbis B, et al. Distribution of iron and iron-binding proteins in first-trimester human pregnancies. Obstet Gynecol. 1994;84:289–293. [PubMed] [Google Scholar]
- 32.Wathen NC, Delves HT, Campbell DJ, Chard T. The coelomic cavity–a reservoir for metals. Am J Obstet Gynecol. 1995;173:1884–1888. doi: 10.1016/0002-9378(95)90446-8. [DOI] [PubMed] [Google Scholar]
- 33.Campbell J, et al. The coelomic cavity: An important site of materno-fetal nutrient exchange in the first trimester of pregnancy. Br J Obstet Gynaecol. 1993;100:765–767. doi: 10.1111/j.1471-0528.1993.tb14271.x. [DOI] [PubMed] [Google Scholar]
- 34.Campbell J, et al. Concentrations of vitamins A and E in amniotic fluid, extraembryonic coelomic fluid, and maternal serum in the first trimester of pregnancy. Arch Dis Child Fetal Neonatal Ed. 1994;71:F49–F50. doi: 10.1136/fn.71.1.f49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iles RK, et al. Pregnancy-associated plasma protein A levels in maternal serum, extraembryonic coelomic and amniotic fluids in the first trimester. Placenta. 1994;15:693–699. doi: 10.1016/0143-4004(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 36.Vallee BL, Falchuk KH. The biochemical basis of zinc physiology. Physiol Rev. 1993;73:79–118. doi: 10.1152/physrev.1993.73.1.79. [DOI] [PubMed] [Google Scholar]
- 37.Kambe T, Hashimoto A, Fujimoto S. Current understanding of ZIP and ZnT zinc transporters in human health and diseases. Cell Mol Life Sci. 2014;71:3281–3295. doi: 10.1007/s00018-014-1617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jauniaux E, et al. Distribution and transfer pathways of antioxidant molecules inside the first trimester human gestational sac. J Clin Endocrinol Metab. 2004;89:1452–1458. doi: 10.1210/jc.2003-031332. [DOI] [PubMed] [Google Scholar]
- 39.Gulbis B, Jauniaux E, Cotton F, Stordeur P. Protein and enzyme patterns in the fluid cavities of the first trimester gestational sac: Relevance to the absorptive role of secondary yolk sac. Mol Hum Reprod. 1998;4:857–862. doi: 10.1093/molehr/4.9.857. [DOI] [PubMed] [Google Scholar]
- 40.Evans P, et al. Hepcidin and iron species distribution inside the first-trimester human gestational sac. Mol Hum Reprod. 2011;17:227–232. doi: 10.1093/molehr/gaq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wathen NC, Cass PL, Campbell DJ, Kitau MJ, Chard T. Levels of placental protein 14, human placental lactogen and unconjugated oestriol in extraembryonic coelomic fluid. Placenta. 1992;13:195–197. doi: 10.1016/0143-4004(92)90034-q. [DOI] [PubMed] [Google Scholar]
- 42.Burton GJ, Watson AL, Hempstock J, Skepper JN, Jauniaux E. Uterine glands provide histiotrophic nutrition for the human fetus during the first trimester of pregnancy. J Clin Endocrinol Metab. 2002;87:2954–2959. doi: 10.1210/jcem.87.6.8563. [DOI] [PubMed] [Google Scholar]
- 43.Hempstock J, Cindrova-Davies T, Jauniaux E, Burton GJ. Endometrial glands as a source of nutrients, growth factors and cytokines during the first trimester of human pregnancy: A morphological and immunohistochemical study. Reprod Biol Endocrinol. 2004;2:58. doi: 10.1186/1477-7827-2-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jauniaux E, Gulbis B, Hyett J, Nicolaides KH. Biochemical analyses of mesenchymal fluid in early pregnancy. Am J Obstet Gynecol. 1998;178:765–769. doi: 10.1016/s0002-9378(98)70489-2. [DOI] [PubMed] [Google Scholar]
- 45.Castellucci M, Kaufmann P. A three-dimensional study of the normal human placental villous core: II. Stromal architecture. Placenta. 1982;3:269–285. doi: 10.1016/s0143-4004(82)80004-0. [DOI] [PubMed] [Google Scholar]
- 46.Baron MH, Vacaru A, Nieves J. Erythroid development in the mammalian embryo. Blood Cells Mol Dis. 2013;51:213–219. doi: 10.1016/j.bcmd.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavian M, Péault B. The changing cellular environments of hematopoiesis in human development in utero. Exp Hematol. 2005;33:1062–1069. doi: 10.1016/j.exphem.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 48.Palis J, Yoder MC. Yolk-sac hematopoiesis: The first blood cells of mouse and man. Exp Hematol. 2001;29:927–936. doi: 10.1016/s0301-472x(01)00669-5. [DOI] [PubMed] [Google Scholar]
- 49.Luckett WP. Origin and differentiation of the yolk sac and extraembryonic mesoderm in presomite human and rhesus monkey embryos. Am J Anat. 1978;152:59–97. doi: 10.1002/aja.1001520106. [DOI] [PubMed] [Google Scholar]
- 50.Tavian M, Hallais MF, Péault B. Emergence of intraembryonic hematopoietic precursors in the pre-liver human embryo. Development. 1999;126:793–803. doi: 10.1242/dev.126.4.793. [DOI] [PubMed] [Google Scholar]
- 51.Burk RF, Hill KE. Selenoprotein P: An extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu Rev Nutr. 2005;25:215–235. doi: 10.1146/annurev.nutr.24.012003.132120. [DOI] [PubMed] [Google Scholar]
- 52.Wooding F, Flint A. In: Marshall’s Physiology of Reproduction. Lamming G, editor. Chapman & Hall; London: 1994. [Google Scholar]
- 53.Wooding FP, Burton GJ. Comparative Placentation. Structures, Functions and Evolution. Springer; Berlin: 2008. [Google Scholar]
- 54.Lloyd JB, Beckman DA, Brent RL. Nutritional role of the visceral yolk sac in organogenesis-stage rat embryos. Reprod Toxicol. 1998;12:193–195. doi: 10.1016/s0890-6238(97)00148-2. [DOI] [PubMed] [Google Scholar]
- 55.Carter AM. IFPA Senior Award Lecture: Mammalian fetal membranes. Placenta. 2016;48:S21–S30. doi: 10.1016/j.placenta.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 56.Burton GJ, Hempstock J, Jauniaux E. Nutrition of the human fetus during the first trimester–a review. Placenta. 2001;22 Suppl A:S70–77. doi: 10.1053/plac.2001.0639. [DOI] [PubMed] [Google Scholar]
- 57.Pereda J, Correr S, Motta PM. The structure of the human yolk sac: A scanning and transmission electron microscopic analysis. Arch Histol Cytol. 1994;57:107–117. doi: 10.1679/aohc.57.107. [DOI] [PubMed] [Google Scholar]
- 58.Yadgary L, Wong EA, Uni Z. Temporal transcriptome analysis of the chicken embryo yolk sac. BMC Genomics. 2014;15:690. doi: 10.1186/1471-2164-15-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoyes AD. The human foetal yolk sac. An ultrastructural study offour specimens. Z Zellforsch Mikrosk Anat. 1969;99:469–490. doi: 10.1007/BF00340940. [DOI] [PubMed] [Google Scholar]
- 60.Hesseldahl H, Larsen JF. Ultrastructure of human yolk sac: Endoderm, mesenchyme, tubules and mesothelium. Am J Anat. 1969;126:315–335. doi: 10.1002/aja.1001260306. [DOI] [PubMed] [Google Scholar]
- 61.Nogales F, Beltran E, Gonzales F. The Human Yolk Sac and Yolk Sac Tumors. Springer; Berlin: 1993. [Google Scholar]
- 62.Blackburn DG. Convergent evolution of viviparity, matrotrophy, and specializations for fetal nutrition in reptiles and other vertebrates. Am Zool. 1992;32:313–321. [Google Scholar]
- 63.Dulvy N, Reynolds J. Evolutionary transition among egg-laying, live-bearing and maternal inputs in sharks and rays. Proc Biol Sci. 1997;264:1309–1315. [Google Scholar]
- 64.Stewart J. Yolk sac placentation in reptiles: Structural innovation in a fundamental vertebrate fetal nutritional system. J Exp Zool. 1993;266:431–449. [Google Scholar]
- 65.Enders AC. Development and structure of the villous haemochorial placenta of the nine-banded armadillo (Dasypus novemcinctus) J Anat. 1960;94:34–45. [PMC free article] [PubMed] [Google Scholar]
- 66.King BF, Wilson JM. A fine structural and cytochemical study of the rhesus monkey yolk sac: Endoderm and mesothelium. Anat Rec. 1983;205:143–158. doi: 10.1002/ar.1092050205. [DOI] [PubMed] [Google Scholar]
- 67.Carter AM, Blankenship TN, Enders AC, Vogel P. The fetal membranes of the otter shrews and a synapomorphy for afrotheria. Placenta. 2006;27:258–268. doi: 10.1016/j.placenta.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 68.Oduor-Okelo D, Katema RM, Carter AM. Placenta and fetal membranes of the four-toed elephant shrew, Petrodromus tetradactylus. Placenta. 2004;25:803–809. doi: 10.1016/j.placenta.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 69.Enders AC, Wimsatt WA, King BF. Cytological development of yolk sac endoderm and protein-absorptive mesothelium in the little brown bat, Myotis lucifugus. Am J Anat. 1976;146:1–30. doi: 10.1002/aja.1001460102. [DOI] [PubMed] [Google Scholar]
- 70.Stephens RJ, Cabral LJ. Cytological differentiation of the mesothelial cells of the yolk sac of the bat, Tadarida brasiliensis cynocephala. Anat Rec. 1971;171:293–312. doi: 10.1002/ar.1091710209. [DOI] [PubMed] [Google Scholar]
- 71.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 72.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 73.Boffelli D, Nobrega MA, Rubin EM. Comparative genomics at the vertebrate extremes. Nat Rev Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- 74.Stewart JB, Freyer C, Elson JL, Larsson NG. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat Rev Genet. 2008;9:657–662. doi: 10.1038/nrg2396. [DOI] [PubMed] [Google Scholar]
- 75.Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cindrova-Davies T, et al. Oxidative stress, gene expression, and protein changes induced in the human placenta during labor. Am J Pathol. 2007;171:1168–1179. doi: 10.2353/ajpath.2007.070528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the European Nucleotide Archive (ENA www.ebi.ac.uk/ena) under the accession number PRJEB18767 (www.ebi.ac.uk/ena/data/view/PRJEB18767).