Significance
Recruitment of messenger RNA (mRNA) into the decoding site of the eukaryotic 40S ribosomal subunit is a critical step in the process of mRNA selection for translation. This step is known to require an open conformation of the decoding site to be induced, but the opening mechanism has not yet been determined. We generated a quantitative assay to monitor conformation changes that occur during mRNA recruitment using a reconstituted system. We show that, unexpectedly, mRNA recruitment is dependent on both the eukaryotic initiation factor (eIF) 4A DEAD-box helicase and ATP. This function of eIF4A is completely independent of its unwinding activity and thus functions as a regulator of translation initiation regardless of mRNA secondary structure content.
Keywords: translation initiation, mRNA recruitment, eIF4A, DEAD-box helicase, eIF3j
Abstract
In the scanning model of translation initiation, the decoding site and latch of the 40S subunit must open to allow the recruitment and migration of messenger RNA (mRNA); however, the precise molecular details for how initiation factors regulate mRNA accommodation into the decoding site have not yet been elucidated. Eukaryotic initiation factor (eIF) 3j is a subunit of eIF3 that binds to the mRNA entry channel and A-site of the 40S subunit. Previous studies have shown that a reduced affinity of eIF3j for the 43S preinitiation complex (PIC) occurs on eIF4F-dependent mRNA recruitment. Because eIF3j and mRNA bind anticooperatively to the 43S PIC, reduced eIF3j affinity likely reflects a state of full accommodation of mRNA into the decoding site. Here, we have used a fluorescence-based anisotropy assay to quantitatively determine how initiation components coordinate their activities to reduce the affinity of eIF3j during the recruitment of mRNA to the 43S PIC. Unexpectedly, we show that a full reduction in eIF3j affinity for the 43S PIC requires an ATP-dependent, but unwinding-independent, activity of eIF4A. This result suggests that in addition to its helicase activity, eIF4A uses the free energy of ATP binding and hydrolysis as a regulatory switch to control the conformation of the 43S PIC during mRNA recruitment. Therefore, our results define eIF4A as a universal initiation factor in cap-dependent translation initiation that functions beyond its role in RNA unwinding.
The regulation of messenger RNA (mRNA) recruitment to the ribosome determines which mRNAs from the cellular pool are selected for translation. Before the recruitment of the mRNA, the 40S subunit is bound by eukaryotic initiation factor (eIF) 1, eIF1A, eIF2-GTP-Met-tRNAi, eIF3, and eIF5 to form the 43S preinitiation complex (PIC) (1). To recruit mRNA and enable scanning, the decoding site of the 43S PIC must adopt an “open” conformation (2, 3). eIF1 and eIF1A have been proposed to play critical roles in stabilizing this conformation. However, 40S complexes that include these factors can be captured in both open and closed conformations (4–6), making it unclear as to how the conformation of the decoding site is regulated to enable mRNA recruitment and scanning.
One component missing from all published structures is the eIF4F complex, which is known to promote mRNA recruitment and scanning (1). However, beyond the established function of its cap-binding (eIF4E), helicase (eIF4A), and eIF3-binding (eIF4G) components, very little is known regarding the molecular mechanism by which eIF4F promotes these stages of initiation. Previous work using sucrose density gradient analysis has suggested that the affinity of eIF3j for the 43S PIC is reduced on eIF4F-dependent mRNA recruitment (7, 8). Consistent with this idea, quantitative assays confirmed that eIF3j and mRNA bind in an anticooperative manner to the 40S subunit in the absence of eIF4F in both yeast and humans (9, 10).
Human eIF3j also has been shown to bind with high affinity (∼15 nM) to the mRNA entry channel and A-site of the 43S PIC, where it could be expected to compete with mRNA (11–13). Consistent with this observation, the displacement of eIF3j by mRNA in the entry channel is correlated with an increase in cross-linking between the mRNA and Rps3/uS3 ribosomal protein located at the solvent side of the mRNA entry channel (14). Importantly, that study showed that a short mRNA that does not extend into the entry channel fails to displace eIF3j. A similar observation was also found for initiation mediated by the hepatitis C virus internal ribosome entry site, where an mRNA truncated after the initiation codon failed to displace eIF3j (11). Taken together, these studies suggest a model in which a full accommodation of mRNA in the mRNA entry channel of the 40S subunit corresponds to a reduced affinity of eIF3j for the 40S subunit. This model has allowed us to exploit the change in eIF3j affinity for the 43S PIC to quantitatively monitor the process of mRNA recruitment.
In this study, we demonstrate that the reduction in eIF3j affinity for the 40S subunit is unexpectedly dependent on both the eIF4A DEAD-box helicase and ATP. Importantly, we show that this function of eIF4A is completely independent of its unwinding activity and likely constitutes a new regulated step in mRNA recruitment.
Results
Recruitment of mRNA to the 43S PIC Reduces eIF3j Affinity.
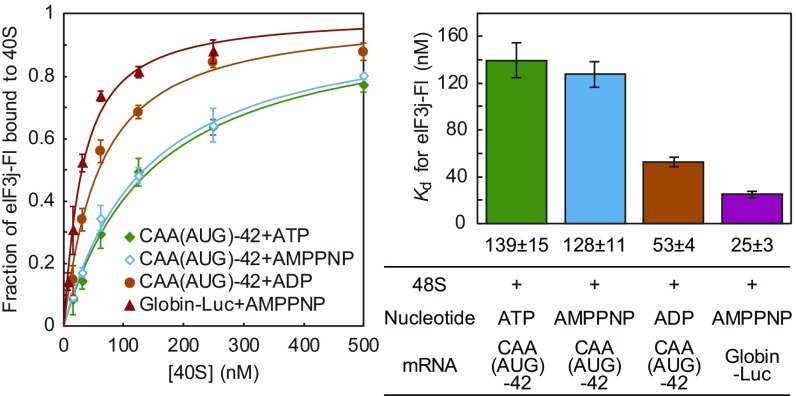
To examine the mechanism of mRNA recruitment to the 43S PIC, we developed an eIF3j affinity assay to monitor conformational changes in the mRNA entry channel during this process (Fig. 1A). Strengths and limitations of this assay are examined in Discussion. To this end, a reconstituted human 43S PIC was incubated with saturating amounts of mRNA, the cap-binding complex eIF4F together with ATP (eIF4F-ATP), and the accessory protein eIF4B. A high-affinity binding of fluorescent-labeled eIF3j (eIF3j-Fl) was confirmed within the 43S PIC, as reported previously (12) (Kd = 13 ± 4 nM; Fig. 1B; anisotropy data are summarized in Table S1). On the addition of a capped Globin-Luc mRNA (encoding luciferase with a globin 5′ UTR), eIF4F-ATP, and eIF4B, we observed a substantial (10-fold) reduction in the apparent equilibrium dissociation constant of eIF3j-Fl for the 43S PIC (Kd = 124 ± 10 nM; 48S PIC in Fig. 1B). This reduction in eIF3j-Fl affinity is dependent on the presence of the cap-binding complex (eIF4F-ATP and eIF4B) and mRNA, because a high-affinity binding of eIF3j-Fl for the 43S PIC is maintained when either of these components is absent (Fig. 1B). The slight weakening of eIF3j-Fl affinity observed when eIF4F-ATP and eIF4B are added in the absence of mRNA reveals the stable recruitment of these components to the 43S PIC (Kd = 38 ± 7 nM; Fig. 1B).
Fig. 1.
Decrease in eIF3j affinity to the 43S PIC on eIF4F-dependent mRNA recruitment. (A) A schematic model for eIF3j affinity change on eIF4F-dependent mRNA recruitment. (B and C) Saturation curves showing the fraction of eIF3j-Fl bound to the initiation complex in each given condition at equilibrium (Left), where B was obtained using endogenous HeLa eIF4G, and C was obtained by using 4GΔN, as described in the text. Solid lines show curve fitting, and the calculated equilibrium dissociation constants are shown as bar graphs in the right panels. Data in B and C represent mean ± SD of three independent experiments.
Table S1.
Summary of fluorescent data and Kd values for eIF3j-Fl
| 43S* | eIF4G | eIF4A/4E/4B | mRNA† | Nucleotide | Kd, nM‡ | rfree§ | rbound¶ |
| + | — | — | — | — | 13 ± 4 | 0.196 ± 0.004 | 0.274 ± 0.006 |
| + | — | — | Globin-Luc | — | 20 ± 3 | 0.184 ± 0.002 | 0.235 ± 0.012 |
| + | HeLa 4G | + | — | ATP | 38 ± 7 | 0.174 ± 0.002 | 0.286 ± 0.003 |
| + | HeLa 4G | + | Globin-Luc | ATP | 124 ± 10 | 0.166 ± 0.003 | 0.266 ± 0.006 |
| + | 4GΔN | + | Globin-Luc | ATP | 125 ± 17 | 0.170 ± 0.001 | 0.262 ± 0.006 |
| + | — | — | CAA(AUG)-42 | — | 24 ± 5 | 0.174 ± 0.002 | 0.239 ± 0.005 |
| + | 4GΔN | + | CAA(AUG)-42 | ATP | 139 ± 15 | 0.174 ± 0.003 | 0.281 ± 0.007 |
| + | 4GΔN | + | CAA(CUC)-42 | ATP | 137 ± 13 | 0.170 ± 0.007 | 0.261 ± 0.005 |
| + | 4GΔN | + | CAA(AUG)-32 | ATP | 57 ± 5 | 0.165 ± 0.006 | 0.269 ± 0.007 |
| + | 4GΔN | + | CAA(CUC)-32 | ATP | 56 ± 5 | 0.175 ± 0.004 | 0.264 ± 0.010 |
| ΔTC | 4GΔN | + | CAA(AUG)-42 | ATP | 24 ± 5 | 0.174 ± 0.010 | 0.271 ± 0.009 |
| + | 4GΔN | + | CAA(AUG)-42 | AMP-PNP | 128 ± 11 | 0.173 ± 0.002 | 0.281 ± 0.006 |
| + | 4GΔN | + | CAA(AUG)-42 | ADP | 53 ± 4 | 0.170 ± 0.002 | 0.274 ± 0.004 |
| + | 4GΔN | + | Globin-Luc | AMP-PNP | 25 ± 3 | 0.174 ± 0.003 | 0.251 ± 0.006 |
| + | 4GΔN | + | Uncap-CAA(AUG)-42 | ATP | 120 ± 12 | 0.178 ± 0.001 | 0.279 ± 0.005 |
| + | — | ++# | CAA(AUG)-42 | ATP | 27 ± 5 | 0.170 ± 0.006 | 0.274 ± 0.011 |
| + | 4GΔN | Δ4E | CAA(AUG)-42 | ATP | 77 ± 5 | 0.187 ± 0.001 | 0.269 ± 0.002 |
| + | 4GΔN | Δ4B | CAA(AUG)-42 | ATP | 42 ± 6 | 0.178 ± 0.003 | 0.259 ± 0.005 |
| Δ1 | 4GΔN | + | CAA(AUG)-42 | ATP | 186 ± 8 | 0.171 ± 0.003 | 0.334 ± 0.006 |
| Δ1A | 4GΔN | + | CAA(AUG)-42 | ATP | 113 ± 11 | 0.169 ± 0.002 | 0.314 ± 0.003 |
| Δ1/Δ1A | 4GΔN | + | CAA(AUG)-42 | ATP | 67 ± 9 | 0.173 ± 0.002 | 0.331 ± 0.001 |
| Δ5 | 4GΔN | + | CAA(AUG)-42 | ATP | 151 ± 7 | 0.170 ± 0.001 | 0.267 ± 0.004 |
Values are mean ± SD of three independent experiments. Δ indicates that the corresponding factor was eliminated from the reaction while all other components indicated in the column were included.
43S PIC consisting of the varying amount of 40S subunit and saturating amounts of eIF1, eIF1A, the TC (formed with GMP-PNP), eIF3 (lacking eIF3j), and eIF5.
All mRNAs were 5′-capped unless indicated otherwise.
Equilibrium dissociation constants determined by titration with the 40S subunit under the experimental conditions.
Anisotropy of the fluorescent-labeled component before addition of 40S subunits under the experimental conditions.
Anisotropy of the fluorescent-labeled component in the 40S-bound state under the experimental conditions.
Twofold concentrations of factors (up to 3,200 nM) were included.
These results indicate that the 10-fold reduction in affinity of eIF3j-Fl for the 43S PIC on addition of eIF4F-ATP, eIF4B, and mRNA is consistent with the generation of a 48S PIC (9–14). A recombinant eIF4F complex generated with an N-terminally truncated eIF4GI isoform (4GΔN; residues 557–1,599) was fully active in reducing eIF3j affinity to the 43S PIC (Kd = 125 ± 17 nM; Table S1). This result implies that the N-terminal region of eIF4G that contains the PABP-binding site does not contribute to the stable binding of mRNA to the 43S PIC in our assay. Therefore, we used this eIF4GI truncation in subsequent experiments.
The eIF2-Ternary Complex Is Required for the Reduction in eIF3j Affinity During mRNA Recruitment Independent of AUG Codon Recognition.
We next examined whether AUG initiation codon selection affects eIF3j-Fl affinity during mRNA recruitment. A minimum 42-nt synthetic mRNA consisting of an unstructured CAA repeat sequence and an AUG codon in a strong context [hereinafter denoted as CAA(AUG)-42] generated a reduction in eIF3j-Fl affinity on the addition of eIF4F-ATP and eIF4B (Kd = 139 ± 15 nM for the 48S PIC compared with Kd = 24 ± 5 nM for the 43S PIC; Fig. 1C and Table S1), which is indistinguishable from the globin reporter mRNA. We then replaced the AUG codon with a CUC codon to determine whether our assay is sensitive to start site selection. Remarkably, we found that the AUG codon in the mRNA is completely dispensable for the observed reduction in eIF3j-Fl affinity on mRNA recruitment [hereinafter denoted as CAA(CUC)-42; Fig. 1C]. Thus, we conclude that the reduced eIF3j-Fl affinity state of our complexes does not require a stable tRNA–mRNA interaction at the initiation codon, and likely represents the formation of the 48S PIC with mRNA stably bound to the mRNA entry channel and decoding site of the 40S subunit before start site selection.
Of note, reducing the length of this mRNA to 32 nt resulted in an intermediate eIF3j-Fl affinity [hereinafter denoted as CAA(AUG)-32; Fig. 1C]. The eIF3j-Fl affinity obtained with the 32-nt mRNA is unaffected by the absence of an AUG codon [hereinafter denoted as CAA(CUC)-32; Fig. 1C]. This finding is consistent with the sensitivity of the reduced eIF3j affinity on mRNA binding to the length of the mRNA extending through the mRNA entry channel (11, 14). However, we cannot rule out the possibility that an extension present at the exit channel also may contribute to reduction in eIF3j affinity. This uncertainty is because we do not precisely know in which position the mRNA is bound in the decoding site in the absence of AUG recognition.
The foregoing data suggest that our assay provides a unique opportunity to monitor mRNA recruitment to the 43S PIC before initiation codon selection. Thus, we tested whether the eIF2-GTP-Met-tRNAi ternary complex (TC) contributes to this early step. Our data show that the reduction in eIF3j-Fl affinity on mRNA recruitment is absolutely dependent on the presence of the TC (Fig. 1C). This result implies that the TC functions to stabilize mRNA in the decoding site independent of its role in start site selection, and provides strong evidence to suggest that the TC must be recruited to the 43S PIC before mRNA recruitment (1).
eIF4A Promotes a Reduction in eIF3j Affinity During mRNA Recruitment Independent of Its Helicase Activity.
We next focused on identifying the molecular mechanism by which eIF4F regulates the reduction in eIF3j affinity during mRNA recruitment to the 43S PIC. In the presence of ATP, it is expected that the 43S PIC in our assay translocates along the mRNA until the TC selects the AUG codon. Because we did not observe any AUG-dependent change in eIF3j affinity, we conclude that our assay does not detect the conformational change in the decoding site that represents a closed P-site upon AUG recognition (PIN) (3). In the absence of the AUG codon, we do not know the fate of the scanning 43S PIC when it reaches the 3′ end of the reporter mRNA. Thus, we varied the nucleotide-bound state of the eIF4A component to separate the helicase and mRNA recruitment activities of eIF4F.
To prevent the helicase activity of eIF4F, we first replaced ATP with nonhydrolyzable AMP-PNP. Using the unstructured 42-nt mRNA [CAA(AUG)-42], we found a reduction in eIF3j-Fl affinity to the 43S PIC that is indistinguishable from the ATP-bound state (Fig. 2). This result is consistent with mRNA recruitment not requiring ATP hydrolysis, at least on an unstructured mRNA (15). Along with preventing the unwinding activity of eIF4F, nonhydrolyzable ATP also prevents the process of scanning, adding support for the reduction in eIF3j-Fl affinity occurring during mRNA recruitment. In stark contrast to the ATP-bound state, the affinity of eIF3j-Fl for the 43S PIC remained high on incubation with ADP (Fig. 2). This surprising finding strongly indicates that ATP binding to eIF4A, but not ATP hydrolysis, plays a critical role in recruiting mRNA to the 43S PIC. Importantly, this function of eIF4A is independent of its unwinding activity and provides a plausible molecular basis for why eIF4A is required for translation of all mRNAs regardless of the degree of structure in their 5′ UTR (16, 17). In contrast to the unstructured 42-nt mRNA, we found that recruitment of the Globin-Luc mRNA strictly requires ATP hydrolysis (Fig. 2). This requirement is likely needed by eIF4A for unwinding the secondary structure present in the 5′ UTR of the globin reporter, and strongly suggests that this intermediate in the pathway can serve as a selection step for mRNA recruitment, depending on the secondary structure content.
Fig. 2.
ATP-dependent reduction in eIF3j affinity during mRNA recruitment. eIF3j-Fl vs. 48S PIC saturation curves (Left) and the calculated equilibrium dissociation constants (Right) as described in Fig. 1B. The 48S PIC was generated in the presence of different adenine nucleotides and mRNA, as indicated.
mRNA Can Bind to the 40S Subunit in a Preaccommodated State.
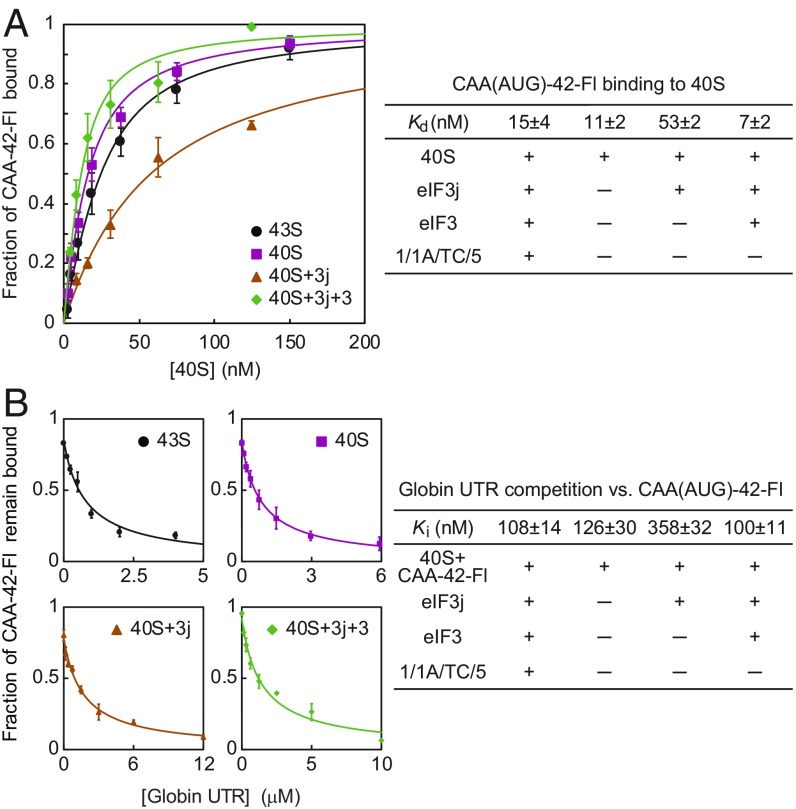
Whereas recruitment of Globin-Luc mRNA requires ATP hydrolysis, we found an appreciable reduction in the eIF3j-Fl anisotropy value in the 48S PIC in the presence of AMP-PNP (0.251 compared with 0.286 in the absence of mRNA; see rbound in Table S1). This would be consistent with the Globin-Luc mRNA binding to the 48S PIC in some kind of preaccommodated state in the absence of duplex unwinding. To characterize this step of mRNA recruitment in more detail, we first measured the affinity of a fluorescent- labeled CAA(AUG)-42 [CAA(AUG)-42-Fl] mRNA to the 43S PIC using anisotropy. We found that this mRNA bound to the 43S PIC with high affinity (Kd = 15 ± 4 nM; Fig. 3A; RNA-binding anisotropy data are provided in Table S2) even though it resides in a preaccommodated state, as demonstrated by a high affinity of eIF3j-Fl in the same complex (Kd = 24 ± 5 nM; Table S1).
Fig. 3.
Binding of the CAA repeat and globin UTR RNAs in the preaccommodated state. (A) The saturation curves showing fractions of CAA(AUG)-42-Fl bound to the 40S subunit as a function of the 40S subunit concentration under the given conditions (Left) and the calculated equilibrium dissociation constants (Right). (B) The inhibition curves showing fractions of CAA(AUG)-42-Fl that remain bound to the 40S subunit as a function of the globin-UTR RNA concentration under the given conditions (Left) and the calculated equilibrium inhibition constants (Right). Data represent mean ± SD of three independent experiments.
Table S2.
Summary of fluorescent data and Kd/Ki values for RNAs
| Complex | Fluorescent RNA | Competitor RNA | Kd, nM* | Ki, nM† | rfree‡ | rbound§ |
| 43S+3j¶ | CAA(AUG)-42-Fl | — | 15 ± 4 | — | 0.214 ± 0.007 | 0.262 ± 0.007 |
| 43S+3j# | CAA(AUG)-42-Fl | Globin UTR | — | 108 ± 14 | — | — |
| 40S¶ | CAA(AUG)-42-Fl | — | 11 ± 2 | — | 0.113 ± 0.007 | 0.233 ± 0.008 |
| 40S# | CAA(AUG)-42-Fl | Globin UTR | — | 126 ± 30 | — | — |
| 40S+3j¶ | CAA(AUG)-42-Fl | — | 53 ± 2 | — | 0.102 ± 0.005 | 0.205 ± 0.004 |
| 40S+3j# | CAA(AUG)-42-Fl | Globin UTR | — | 358 ± 32 | — | — |
| 40S+3+3j¶ | CAA(AUG)-42-Fl | — | 7 ± 2 | — | 0.148 ± 0.008 | 0.224 ± 0.004 |
| 40S+3+3j# | CAA(AUG)-42-Fl | Globin UTR | — | 100 ± 11 | — |
Values are mean ± SD of three independent experiments.
Equilibrium dissociation constants determined by titration with the 40S subunit under the experimental conditions.
Equilibrium inhibition constants determined by titration of the unlabeled globin-UTR RNA to the constant amount of 40S subunit in complex with fluorescent CAA(AUG)-42-Fl under the experimental conditions.
Anisotropy of the fluorescent-labeled component before addition of 40S subunits under the experimental conditions.
Anisotropy of the fluorescent-labeled component in the 40S-bound state under the experimental conditions.
Varying amount of the 40S subunit in complex with saturating amounts of additional factors as indicated.
A constant amount of the 40S subunit (65–200 nM; Materials and Methods) in complex with saturating amounts of additional factors as indicated.
We then performed a competition binding assay with the Globin-Luc 5′ UTR sequence to establish its equilibrium dissociation constant to the 43S PIC. Surprisingly, this RNA was found to compete with the CAA repeat RNA with a reasonably high affinity (Ki = 108 ± 14 nM; Fig. 3B). The high-affinity binding of these mRNAs for the 43S PIC in what we term a preaccommodated state implies that mRNA recruitment involves at least two steps: (i) the initial binding step, in which an mRNA can partially bind to the ribosome in a preaccommodated state, and (ii) an accommodation step, in which only single-stranded mRNA binds to the mRNA entry channel of the 40S subunit that likely would be consistent with a scanning-competent conformation. This model implies that the presence of secondary structure in an mRNA could influence the rate and efficiency of the accommodation step and thereby the process of mRNA selection.
To further characterize the preaccommodated state, we performed a similar competition assay but in the absence of initiation factors. Surprisingly, the globin UTR and CAA(AUG)-42-Fl RNAs were found to bind the 40S subunit with an affinity indistinguishable from that determined for the 43S PIC (Fig. 3 A and B). This finding suggests that the mRNA-binding site for the preaccommodated state is composed mainly of the mRNA-binding cleft in the 40S subunit. Consistent with previous data, the addition of saturating eIF3j lowered the affinity of these mRNAs by threefold to fivefold (Fig. 3 A and B). Remarkably, the reduced affinity of both mRNAs was recovered on further addition of the eIF3 complex (Fig. 3 A and B), suggesting that the eIF3 component of the 43S PIC plays an important role in stabilizing mRNA in the preaccommodated state. This is consistent with previous studies showing that eIF3 stabilizes mRNA binding at the entry and exit channels of the 40S subunit during mRNA recruitment (18, 19).
eIF4G, eIF4E, and eIF4B Promote a Reduction in eIF3j Affinity During mRNA Recruitment.
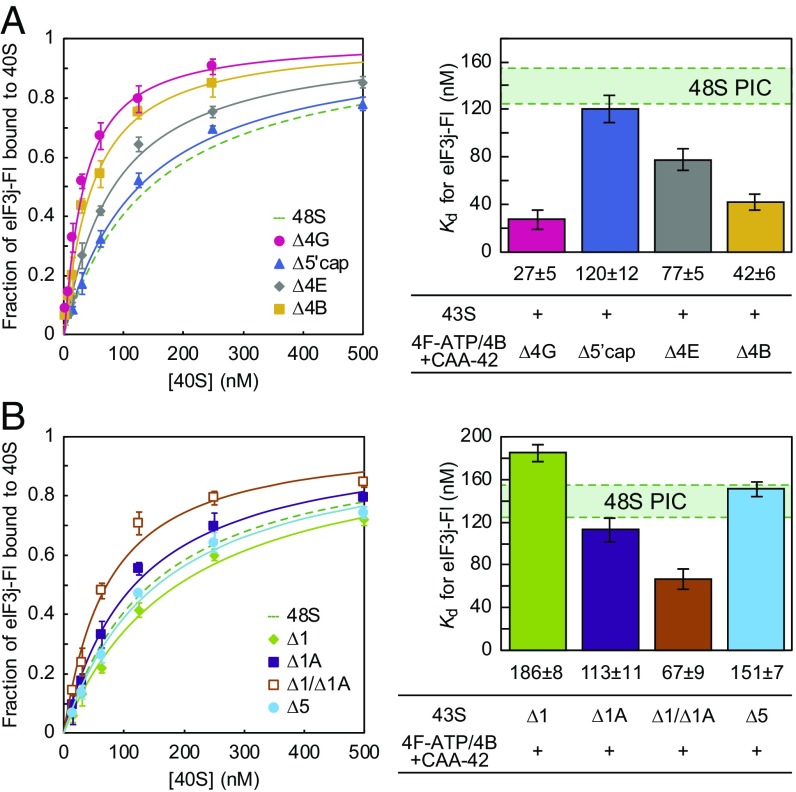
Importantly, eIF4A-ATP must be complexed with eIF4G to reduce the affinity of eIF3j-Fl, given that no such reduction was observed in the absence of eIF4G (Fig. 4A). This holds even when twofold elevated concentrations of eIFs 4A-ATP, 4B, and 4E are present. This finding suggests that eIF4G is essential for tethering eIF4A-ATP to the complex and/or plays an active role in reducing eIF3j affinity. Interestingly, the 5′ m7G cap structure does not appear to play a role in this step, as indicated by equal reduction of eIF3j-Fl affinity for the 48S PIC in its presence or absence (Fig. 4A). However, we note that our equilibrium binding assay was conducted using saturating amounts of components, making it difficult to observe any possible kinetic effect of the m7G cap on recruitment. Unexpectedly, an intermediate eIF3j-Fl affinity resulted in the absence of the cap-binding protein eIF4E (Fig. 4A), implying a function of eIF4E in stabilizing mRNA in the decoding site in addition to its cap-binding and helicase-promoting activities (20, 21).
Fig. 4.
Contribution of initiation complex components on mRNA recruitment. eIF3j-Fl vs. 48S PIC saturation curves (Left) and the calculated equilibrium dissociation constants (Right) as described in Fig. 1B using CAA(AUG)-42 mRNA. Shown is eIF3j-Fl affinity in the presence or absence of 4GΔN, m7G cap, eIF4E, and eIF4B (A) or of eIF1, eIF1A, and eIF5 (B). Note that the concentrations of eIF4A, eIF4E, and eIF4B were increased twofold in the absence of eIF4G, to compensate for a possible decrease in affinity of these components. Green dashed lines and green shaded areas represent the control data obtained with the complete set of 48S PIC components and CAA(AUG)-42 mRNA (Fig. 1C). Data represent mean ± SD of three independent experiments.
Recent studies have proposed that eIF4E dissociates from the m7G cap on mRNA recruitment (22), and that the scanning ribosome is not tethered to the m7G cap (23). The identification of two cap-independent roles of eIF4E strongly supports the possibility that eIF4E can regulate initiation if it remains bound to the scanning complex after dissociation from the m7G cap. Surprisingly, we found that the reduction in eIF3j affinity for the 48S PIC was strongly dependent on the eIF4A accessory protein, eIF4B (Fig. 4A). This dependency is apparent even in the absence of secondary structures, suggesting a role of eIF4B in mRNA recruitment beyond its stimulation of eIF4A helicase activity (24). This additional role of eIF4B in mRNA recruitment is consistent with recent data indicating that yeast eIF4B alters the conformation of the mRNA entry channel through its direct binding to the head domain of the 40S subunit (25).
Finally, we investigated whether eIF1, eIF1A, and eIF5 contribute to the reduced eIF3j affinity in the 48S PIC. These initiation factors bind to the intersubunit surface of the 40S subunit, with eIF1 and eIF1A specifically binding to the 43S PIC in an anticooperative manner with eIF3j (12). Interestingly, we found an equal reduction of eIF3j-Fl affinity for the 48S PIC in the absence or presence of any one of these proteins (Fig. 4B). This result implies that these factors do not contribute to mRNA accommodation individually. In contrast, we observed an intermediate eIF3j-Fl affinity for the 48S PIC when both eIF1 and eIF1A were absent (Fig. 4B). Based on these findings, we conclude that the interaction between eIF1 and eIF1A contributes to the reduction of eIF3j affinity and mRNA accommodation into the decoding site of the 40S subunit. This effect is likely associated with their role in promoting the open-latch conformation of the 40S subunit (3, 4).
Discussion
Here, we report the development of an assay that enables the precise detection of intermediates generated during mRNA recruitment to the decoding site of the 43S PIC. The reduction in eIF3j affinity that we observed agrees well with previous studies collectively showing that eIF3j is displaced on eIF4F-dependent accommodation of the mRNA into the 40S subunit entry channel (7–14). Nevertheless, we do note that our observations are limited by the fact that we did not directly monitor the position of mRNA on the 40S subunit during the proposed intermediate steps of accommodation. Thus, future studies will need to use structural approaches together with a detailed kinetic analysis of these events to precisely determine the mechanism of recruitment. Importantly, the reduced eIF3j affinity that we observed is independent of the AUG codon in the mRNA and ATP hydrolysis by eIF4A. This finding strongly indicates that the reduction in eIF3j affinity occurs before the scanning and initiation codon selection steps. Our data show that an mRNA of >32 nt is necessary to promote a complete reduction in eIF3j affinity for the 48S complex. This finding is consistent with accommodation being sensitive to the length of mRNA extending through the mRNA entry channel (11, 14). However, we cannot rule out the possibility that mRNA extending into and beyond the mRNA exit channel is also important for accommodation. Structural models have indicated that ∼25 nt of mRNA is sufficient to extend from the entry channel to the exit channel of the 43S PIC (26). Thus, the requirement of a >32-nt-long mRNA would be entirely consistent with data showing that eIF3 likely binds mRNA protruding from both the entry channel and exit channel of the 40S subunit (18, 19).
Our observation of complete reduction in eIF3j affinity with a 42-nt-long mRNA raises the question of how eIF4A and the 40S subunit can simultaneously interact with such a short mRNA. The crystal structure of eIF4AIII indicates that only six nucleotides are needed to fill the RNA-binding site between the RecA domains (27). Given the 11-nt step size of mRNA duplex unwinding by eIF4A (24), we anticipate that the available nucleotide length for eIF4A binding must be somewhere between these two lengths. As mentioned above, structural models indicate that ∼25 nt of mRNA is needed to span the 40S subunit (26), leaving ∼17 nt of mRNA for eIF4A to interact with (for our 42-nt mRNA). Thus, we believe that it is possible for eIF4A and the 43S PIC to interact simultaneously with only a 42-nt-long mRNA.
Importantly, our data reveal that eIF4A has more than one function in the initiation pathway. It has been generally accepted that the helicase activity of eIF4A plays a critical role in unwinding the mRNA secondary structure to generate a single-stranded region that can be accommodated into the 40S subunit decoding site. This activity of eIF4A requires cycles of binding and hydrolysis of ATP and would be expected to be particularly important for the recruitment of native (structured) mRNAs. Our finding that the reduction in eIF3j affinity with a native mRNA requires ATP hydrolysis is entirely consistent with this function of eIF4A. Unexpectedly, our findings show that a nonhydrolyzable ATP analog (AMP-PNP) is sufficient to promote the reduction in eIF3j affinity with an unstructured mRNA. This result is rather surprising, and the key question is how this helicase-independent function of eIF4A promotes the reduction in eIF3j affinity. A reduction in eIF3j affinity would be consistent with published structures showing an “open” conformation of the 40S subunit latch and decoding site on mRNA binding (3). Thus, it is possible that ATP binding to eIF4A changes the conformation of the 40S subunit to enable full accommodation of mRNA into the entry channel. Importantly, this conformation change could occur indirectly through other components of the 48S PIC, such as eIF4G and/or eIF4B, which also are required for reduced eIF3j affinity. We anticipate that the helicase-independent function of eIF4A likely would be needed for all mRNAs, regardless of the degree of structure in their 5′ UTR. Thus, this activity of eIF4A provides a plausible mechanism to explain why this initiation factor is required for the recruitment and translation of essentially all mRNAs (16, 17). Consequently, we speculate that mRNA recruitment to the 43S PIC might involve conversion of a preaccommodated mRNA-bound state to an accommodated state through an eIF4A-induced change in the conformation of the decoding site (Fig. 5A). Importantly, the accommodation step of this model also would require restructuring of a structured mRNA by the helicase activity of eIF4A to enable binding in the entry channel. As such, this accommodation step could serve as the selection step for mRNAs with stable structure. It will be important in future studies to use structural approaches to rigorously test the predictions of this mRNA recruitment model.
Fig. 5.
Proposed models for an ATP-driven conformational change of the decoding site during mRNA recruitment and scanning. (A) An mRNA can be recruited from the cellular pool to the 43S PIC in a preaccommodated or accommodated state. The preaccommodated state involves the 43S PIC in a closed latch and decoding site conformation, enabling eIF3j to bind in a high-affinity binding state. In the presence of eIF4F-ATP and the TC, the mRNA is accommodated into the 40S subunit decoding site to form the scanning competent 48S complex. In this state, the latch and decoding site of the 40S subunit adopt the open state, which exhibits a low-eIF3j affinity binding state. In the presence of mRNA secondary structures, the accommodation step requires the generation of single-stranded mRNA through the ATP hydrolysis-dependent helicase activity of eIF4A. (B) In the ATP-bound state, eIF4A adopts a high-RNA affinity closed conformation, and, directly or indirectly, promotes the open conformation of the 40S subunit that allows scanning along the 5′ UTR. In the ADP-bound state, eIF4A adopts a low-RNA affinity open conformation. In this state, the 40S subunit adopts a closed conformation that is likely to be less conducive to scanning. Multiple cycles of ATP hydrolysis by eIF4A during scanning might lead to rounds of opposing opening and closing conformations of eIF4A and the 40S subunit.
The 40S subunit has been proposed to adopt an open conformation during the entire process of scanning until a PIN state is generated on initiation codon recognition (3). Our data raise the interesting possibility that continued cycles of ATP binding and hydrolysis by the eIF4A component of eIF4F might result in coordinated conformational switching of the 40S subunit between open and closed states during scanning (Fig. 5B). Consistent with this idea is the recent finding that scanning 40S subunits have heterogeneous footprint sizes (23). Changes in the conformation of the mRNA entry channel by eIF4A might be important in generating 3′ directional movement of the 40S subunit during scanning (24, 28). Alternatively, eIF4A-dependent changes in the conformation of eIF3j might be part of the mechanism by which eIF3j functions in the fidelity of start site selection (29). Thus, in light of our findings, it appears that unwinding of the mRNA structure, accommodation of mRNA into the 40S subunit decoding site, scanning, and start site selection might be mechanistically coupled through ATP hydrolysis by eIF4A. The ability to precisely monitor the recruitment of mRNA to the 40S subunit provides an exciting opportunity to gain more insight into the mechanism of mRNA selection and how competition between mRNAs regulates the proteome.
Materials and Methods
Sample Preparation.
Fluorescein-labeled human eIF3j, the human 43S PIC components (eIF1, eIF1A, eIF2, eIF3, eIF5, 40S subunit, and initiator Met-tRNAi), and wild-type and truncated eIF4GI, eIF4AI, eIF4B, and eIF4E were prepared as described previously (12, 21). The 32-nt and 42-nt CAA repeat mRNAs [5′-GGACAACAACAACAAACC(AUG/CUC)GAACAACAACAACAACAACAA-3′, where the underlined sequence is specific to the 42-nt mRNAs] were transcribed using T7 RNA polymerase and a synthetic oligo ssDNA template annealed to a T7 promoter strand as described previously (21). The Globin-Luc mRNA and its UTR sequence were transcribed in a similar manner, but using a PCR-amplified DNA as a transcription template. The DNA templates were PCR-amplified from a pUC19 plasmid harboring the T7 promoter, human globin 5′ UTR, followed by a NanoLuc luciferase coding region between the KpnI and HindIII restriction sites, using M13 forward and M13 reverse primers for the Globin-Luc mRNA, or M13 forward and a synthetic reverse primer (5′-GAAATCTTCGAGTGTGAAGACCAT-3′) for the globin UTR sequence. Therefore, the resulting Globin-Luc mRNA included a 160-bp 3′ UTR originating from the sequence between the HindIII and M13 reverse primer sites in the pUC19 backbone. The globin 5′ UTR sequence included the first 24 nt of the NanoLuc coding sequence directly following the globin UTR. Unless indicated otherwise, all mRNAs used in this study were capped with vaccinia capping enzyme (New England BioLabs) according to the manufacturer’s protocol, and then purified with phenol/chloroform extraction, followed by a passage through a Micro Bio-Spin 6 column (Bio-Rad).
For fluorescent labeling, the 3′-end of CAA(AUG)-42 was labeled as described previously (30), using 0.5 mM fluorescein-5-thiosemicarbazide as the reactant. The reaction yielded >95% labeling efficiency as judged by the absorbance spectrum. The labeled RNA was phenol/chloroform-extracted and purified with a Micro Bio-Spin 6 column. Note that the labeling reaction was performed before the capping reaction to eliminate the possibility of the undesired labeling at the 3′ position of m7G cap nucleotide.
Fluorescence Anisotropy Assay.
The fluorescence anisotropy assay and data processing were carried out as described previously (12). For the binding assay, the reaction buffer contained 20 mM Tris-acetate (pH 7.5), 70 mM KCl, 2 mM free Mg2+ (supplemented as MgCl2), 0.1 mM spermidine, 1 mM DTT, 10% glycerol, 0.1 mg/mL BSA, 100 μM GMP-PNP-Mg, 0.1 mg/mL creatine phosphokinase, and 0.5 mM adenine nucleotide (ATP-Mg, ADP-Mg, or AMP-PNP-Mg, as indicated). The final concentration of the fluorescent species [eIF3j-Fl or CAA(AUG)-42-Fl] was 10–20 nM. The 40S subunit titration ranges were up to 500 nM depending on the affinity to be measured. The saturating amount of either nonlabeled eIF3j (1,250 nM) or mRNA (800 nM) was included when the corresponding fluorescent species was not included. Unless indicated otherwise, the final concentrations of other components were also saturating according to the available affinities for the 43S PIC (12): 1,250 nM eIF1/eIF1A/eIF5, 800 nM TC/eIF4G/eIF4E/eIF4B, 1,600 nM eIF4A, and 600 nM eIF3 lacking the eIF3j subunit. For the components whose affinity has not been experimentally determined (e.g., eIF4F components, eIF4B, and mRNAs), increasing the concentration by twofold did not change the result for the 48S PIC formation, indicating that these components were saturating at these concentrations. Note that our purified eIF4G (either wild-type or truncated) always included a stoichiometric amount of copurified eIF4A. Thus, the amount of eIF4A represents the sum of copurified and additional eIF4A included in the reaction. The competition assay with the globin 5′ UTR sequence was performed under essentially the same conditions as the CAA(AUG)-42-Fl binding assay but in the presence of a constant amount of the 40S subunit (100 nM for the 43S PIC and 40S+3+3j, 65 nM for the 40S subunit, and 200 nM for 40S+3j) and titration of the globin UTR sequence. All values reported are means of at least three independent experiments, and the errors reported are SD.
Acknowledgments
We thank J. Hershey and J. Lorsch for their critical reading of the manuscript, and E. Baldwin and members of the C.S.F. laboratory for discussions. This research was funded by National Institutes of Health Grant R01 GM092927 (to C.S.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620426114/-/DCSupplemental.
References
- 1.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 2.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llácer JL, et al. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell. 2015;59:399–412. doi: 10.1016/j.molcel.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Passmore LA, et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 6.Lomakin IB, Steitz TA. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013;500:307–311. doi: 10.1038/nature12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benne R, Hershey JWB. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- 8.Unbehaun A, Borukhov SI, Hellen CUT, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser CS, Berry KE, Hershey JWB, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell SF, et al. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol Cell. 2010;39:950–962. doi: 10.1016/j.molcel.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser CS, Hershey JWB, Doudna JA. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat Struct Mol Biol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sokabe M, Fraser CS. Human eukaryotic initiation factor 2 (eIF2)-GTP-Met-tRNAi ternary complex and eIF3 stabilize the 43 S preinitiation complex. J Biol Chem. 2014;289:31827–31836. doi: 10.1074/jbc.M114.602870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aylett CHS, Boehringer D, Erzberger JP, Schaefer T, Ban N. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat Struct Mol Biol. 2015;22:269–271. doi: 10.1038/nsmb.2963. [DOI] [PubMed] [Google Scholar]
- 14.Sharifulin DE, et al. Exploring accessibility of structural elements of the mammalian 40S ribosomal mRNA entry channel at various steps of translation initiation. Biochim Biophys Acta. 2016;1864:1328–1338. doi: 10.1016/j.bbapap.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Kozak M. Role of ATP in binding and migration of 40S ribosomal subunits. Cell. 1980;22:459–467. doi: 10.1016/0092-8674(80)90356-6. [DOI] [PubMed] [Google Scholar]
- 16.Altmann M, et al. Translation initiation factor-dependent extracts from Saccharomyces cerevisiae. Biochim Biophys Acta. 1990;1050:155–159. doi: 10.1016/0167-4781(90)90158-x. [DOI] [PubMed] [Google Scholar]
- 17.Sen ND, Zhou F, Ingolia NT, Hinnebusch AG. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res. 2015;25:1196–1205. doi: 10.1101/gr.191601.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CUT, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aitken CE, et al. Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex. eLife. 2016;5:e20934. doi: 10.7554/eLife.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonenberg N, Rupprecht KM, Hecht SM, Shatkin AJ. Eukaryotic mRNA cap binding protein: Purification by affinity chromatography on sepharose-coupled m7GDP. Proc Natl Acad Sci USA. 1979;76:4345–4349. doi: 10.1073/pnas.76.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci USA. 2013;110:13339–13344. doi: 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar P, Hellen CUT, Pestova TV. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev. 2016;30:1573–1588. doi: 10.1101/gad.282418.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Archer SK, Shirokikh NE, Beilharz TH, Preiss T. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016;535:570–574. doi: 10.1038/nature18647. [DOI] [PubMed] [Google Scholar]
- 24.García-García C, Frieda KL, Feoktistova K, Fraser CS, Block SM. Factor-dependent processivity in human eIF4A DEAD-box helicase. Science. 2015;348:1486–1488. doi: 10.1126/science.aaa5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker SE, et al. Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains. RNA. 2013;19:191–207. doi: 10.1261/rna.035881.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simonetti A, et al. eIF3 peripheral subunits rearrangement after mRNA binding and start-codon recognition. Mol Cell. 2016;63:206–217. doi: 10.1016/j.molcel.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 27.Andersen CBF, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 28.Spirin AS. How does a scanning ribosomal particle move along the 5′-untranslated region of eukaryotic mRNA? Brownian Ratchet model. Biochemistry. 2009;48:10688–10692. [Google Scholar]
- 29.Elantak L, et al. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J Mol Biol. 2010;396:1097–1116. doi: 10.1016/j.jmb.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Ahsen U, Noller HF. Identification of bases in 16S rRNA essential for tRNA binding at the 30S ribosomal P site. Science. 1995;267:234–237. doi: 10.1126/science.7528943. [DOI] [PubMed] [Google Scholar]