Significance
Protein disulfide isomerase (PDI) is a ubiquitous enzyme involved in disulfide bond formation during protein folding. It has been related to neurological diseases (Parkinson or Alzheimer’s) because of unfolded protein response phenomena. It also participates in the regulation of the glutathione redox buffer [glutathione/glutathione disulfide (GSH/GSSG)] in the endoplasmic reticulum, also important in protein folding. PDI catalyzes the nucleophilic attack of thiolates to disulfide bonds (thiol-disulfide exchange), enhancing the formation of correct disulfide links that drive protein folding and ensure protein function. This reaction is ubiquitous to disulfide-oxidoreductases across several organisms, and it shows a distinctive chemistry. We present the enzymatic mechanism of PDI in the GSH/GSSG buffer and discuss the chemistry of thiol-disulfide exchange.
Keywords: PDI, GSH/GSSG buffer, thiol-disulfide exchange, protein folding, ONIOM
Abstract
We explore the enzymatic mechanism of the reduction of glutathione disulfide (GSSG) by the reduced a domain of human protein disulfide isomerase (hPDI) with atomistic resolution. We use classical molecular dynamics and hybrid quantum mechanics/molecular mechanics calculations at the mPW1N/6–311+G(2d,2p):FF99SB//mPW1N/6–31G(d):FF99SB level. The reaction proceeds in two stages: (i) a thiol-disulfide exchange through nucleophilic attack of the Cys53-thiolate to the GSSG-disulfide followed by the deprotonation of Cys56-thiol by Glu47-carboxylate and (ii) a second thiol-disulfide exchange between the Cys56-thiolate and the mixed disulfide intermediate formed in the first step. The Gibbs activation energy for the first stage was 18.7 kcal·mol−1, and for the second stage, it was 7.2 kcal·mol−1, in excellent agreement with the experimental barrier (17.6 kcal·mol−1). Our results also suggest that the catalysis by protein disulfide isomerase (PDI) and thiol-disulfide exchange is mostly enthalpy-driven (entropy changes below 2 kcal·mol−1 at all stages of the reaction). Hydrogen bonds formed between the backbone of His55 and Cys56 and the Cys56-thiol result in an increase in the Gibbs energy barrier of the first thiol-disulfide exchange. The solvent plays a key role in stabilizing the leaving glutathione thiolate formed. This role is not exclusively electrostatic, because an explicit inclusion of several water molecules at the density-functional theory level is a requisite to form the mixed disulfide intermediate. In the intramolecular oxidation of PDI, a transition state is only observed if hydrogen bond donors are nearby the mixed disulfide intermediate, which emphasizes that the thermochemistry of thiol-disulfide exchange in PDI is influenced by the presence of hydrogen bond donors.
Protein disulfide isomerase (PDI) is a multifunctional enzyme able to catalyze disulfide bond formation, cleavage, and isomerization in the endoplasmic reticulum (ER) of eukaryotic cells. Despite not being the most effective disulfide redox catalyst, the importance of PDI in the secretory protein pathway, where the formation of native disulfides is a rate-limiting step, has been widely recognized (1–3). In recent years, PDI deletion has been related to diseases involving unfolded protein response, such as Alzheimer’s, Parkinson’s, and type II diabetes (4–6). This unfolded response is mostly a result of the unbalance in the redox buffers in the ER—the glutathione (GSH)/glutathione disulfide (GSSG) and the hydrogen peroxide/molecular oxygen (H2O2/O2) pairs (7, 8). As a result from this misfolding process, these proteins form aggregates through the packing of hydrophobic exposed regions (9, 10). PDI has also been pointed out as a potential target in cancer treatment, because it is frequently overexpressed in cancer cells (11).
Understanding the PDI reaction with the GSH/GSSG buffer is critically important to (i) investigate the role of PDI in the redox balance of the ER; (ii) determine the kinetics and thermochemistry of the reaction catalyzed by PDI, complementing the rather scarce kinetic studies that can be found in the literature (2, 12, 13); (iii) draw mechanistic insights on PDI, because there are no known studies for the reaction cycle with atomistic detail for this enzyme; (iv) provide structural information (transition states) to allow for the rational discovery of therapeutic inhibitors; and (v) provide general insight on the disulfide oxidoreductase family of enzymes, which are responsible for the reduction and isomerization of disulfide bonds, through thiol-disulfide exchange.
Structure and Function of PDI
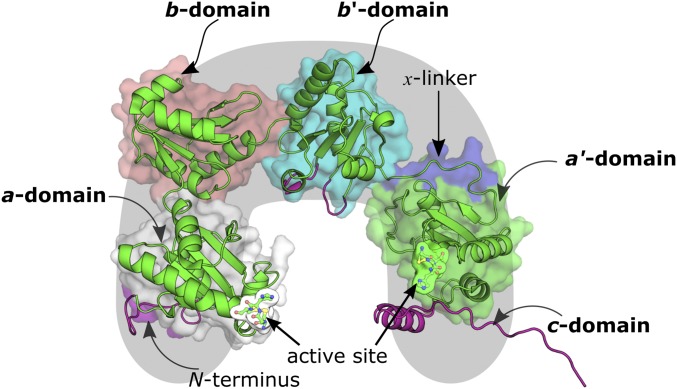
Human protein disulfide isomerase (hPDI) is a U-shaped enzyme with 508 residues. Its tertiary structure is composed of four thioredoxin-like domains (a, b, b′, and a′) and a fifth tail-shaped c domain (Fig. 1) (14, 15). The maximum activity of hPDI is observed when all domains of PDI contribute synergistically to its function (16).
Fig. 1.
Structure of hPDI. The domains are marked by different color surfaces. Cartoon representations in magenta represent regions of the enzyme that have been modeled. Only the a and b domains were used in the calculations of the enzymatic mechanism of hPDI.
Similar to thioredoxin, the a and a′ domains have a catalytic Cys–X–Y–Cys motif (which for hPDI, is Cys53–Gly54–His55–Cys56 and Cys397–Gly398–His399–Cys400) near the N terminus of the α1-helix of the thioredoxin domain (17), which is thought to regulate its (high) redox potential (18). Despite the structural similarity to thioredoxin, the b and b′ domains do not exhibit the usual Cys–X–Y–Cys thioredoxin motif and share a reduced sequence identity of approximately 16.5% (11). Instead, the bb′ region provides a large hydrophobic pocket that binds reversibly to a wide variety of peptides and misfolded proteins (19–23). In particular, the b′ domain has been widely referred to significantly improve the activity of PDI toward misfolded protein substrates (15, 16, 24). Attached to the b′ domain, there is a 19-aa segment (linker x) that connects the b′ and a′ domains. This segment is involved in domain motions that induce changes in the hydrophobic cleft that accommodates the substrates of PDI (22).
The a′ domain does not react with misfolded proteins, unless when at substrate-saturating concentrations (25). Nevertheless, the oxidation of the Cys397/Cys400 pair in the a′ domain has been recently observed to induce domain motions in the b′–x–a′ region, which increase the available hydrophobic surface in the cavity of hPDI, thus enhancing its isomerase and chaperone activities (22). As a result, the oxidation of the Cys397/Cys400 pair should be a critical stage for the isomerase activity of hPDI. Initially, GSSG was thought to play a role in this process; however, currently, it is believed that this oxidation occurs through ER oxidoreductins (26–28), because these enzymes have shown significant interaction with the b′ and a′ domains of hPDI (29). In SI Appendix, Table S1, we summarize the main characteristics and function of the different domains of hPDI.
GSH/GSSG Buffer in Catalysis by hPDI
The main catalytic cycle of PDI as an isomerase can be summed up in two main stages: (i) formation of the mixed disulfide PDI:substrate intermediate through bonding of the Cys53-thiolate to a nonnative disulfide of the misfolded protein and (ii) cleavage of the mixed disulfide intermediate by two possible ways: either formation of an internal protein disulfide between Cys53 and Cys56 or formation of yet another mixed disulfide with another substrate protein Cys or a foreign thiolate species (13, 24, 30).
To support this hypothesis, experimental studies emphasize that Cys53 presents a lower pKa (∼4.4–6.7) than would be expected for equivalent cysteines in thioredoxin-like folds (∼7.1) (24, 31–34) and solution (9.1), whereas Cys56 is more buried within the a domain and presents a much higher pKa (∼12.8) (35). As a result, Cys53 is predominantly deprotonated in the ER and can act as a nucleophile much better than the protonated thiol in the first thiol-disulfide exchange reaction of hPDI. The intriguing pKa and redox properties of Cys53 have been discussed often in the literature (36–38), and three major factors have been pointed out to support these observations: (i) the stabilization of the Cys53-thiolate by nearby positively charged residues; (ii) the effect of several hydrogen bond donors (His55 and Cys56) nearby Cys53 as opposed to Cys56, because thiolates are much better hydrogen bond acceptors than thiols; and (iii) the effect of the macrodipole on a neighboring α-helix. From these aspects, the effect of the hydrogen bond donors in the Cys–X–Y–Cys motif seems to find greater acceptance in academia (39–42). Earlier theoretical studies using cluster models (43–45) suggested that thiol-disulfide exchange is an SN2 reaction, in which a nucleophilic thiolate (Snuc) linearly attacked a disulfide with concomitant dissociation of the latter. In this reaction, the Snuc and the leaving sulfur (Slg) are approximately parallel to each other and orthogonal to the central sulfur (Sox), in such a way that the reactant and product disulfides exhibit a torsion of ∼90° around the S–S bond axis (37, 46–48). As a result, minor conformation rearrangements occur during the SN2 reaction (47). It has been observed also that the near orthogonality of the CSS angles in the disulfide results from a hyperconjugation effect from the interacting p orbitals involved in CS and SS bonds, whereas d orbitals from sulfur mostly do not interact. This effect is proposed to strengthen the disulfide bond (47, 49). For the particular case of the thiol-disulfide exchange between the Cys53-thiolate and the disulfide of GSSG, a second-order turnover rate of 191 M−1·s−1 has been determined (13).
After the first thiol-disulfide exchange, there are two pathways that compete to complete the catalytic cycle: either the buried Cys56 attacks the mixed disulfide, oxidizing the a domain, or another thiolate (from the substrate or solution) attacks the mixed disulfide intermediate, restoring the reduced state of hPDI. The former step prevents the formation of kinetically trapped intermediates (7, 50, 51). It has been proposed that, before this step occurs, the pKa of Cys56 lowers to make the thiolate form more prevalent and able to attack the mixed disulfide (24). Arg120 has been indicated to contribute to this lowering of pKa (35), and the glutamate of the buried Lys81/Glu47 pair located nearby Cys56 is proposed to be responsible for the deprotonation of the latter (14, 52–55). Because mixed disulfide intermediates were observed to be long-lived (13), the cleavage of the mixed disulfide intermediate has been referred to be the rate-limiting step of the cycle (a turnover of 0.23 s−1, corresponding to a Gibbs energy barrier of 19.2 kcal·mol−1, was determined for the complex between the a domain of hPDI and GSSG) (13).
The thermodynamic and kinetic contours of the isomerization process are still very scarce (30, 56). It is known that the actual isomerization rate of PDI is lower than the rate of thiol-disulfide exchange reactions and that the relative efficiency of each catalytic site in PDI is dependent of the substrate and the synergistic effects in its environment (16, 25, 57). Nevertheless, the kcat of the isomerization by PDI may be up to 103 times larger than that of the uncatalyzed reaction (which exhibits a turnover rate of about 1 min−1) (58).
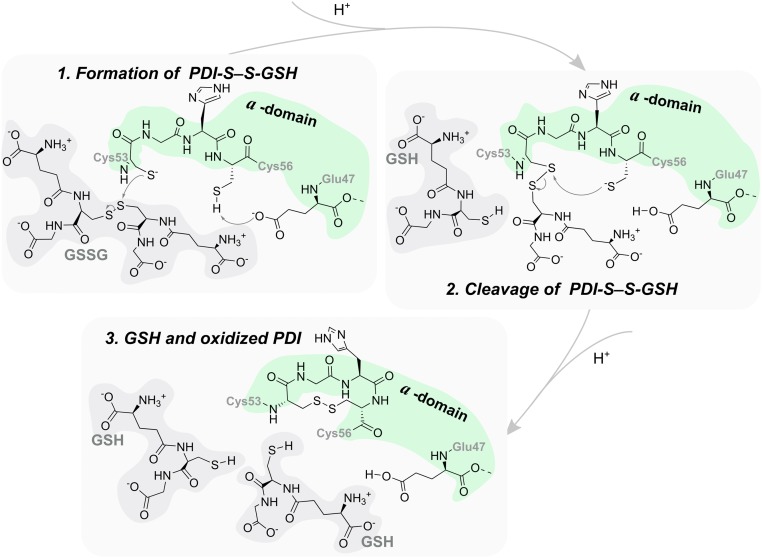
We have studied the reduction of GSSG in the a domain (the postulated mechanism is in Fig. 2). Although GSSG is not a substrate for which PDI exhibits a high affinity, it is one of the smallest and more important substrates of PDI (59). The role of this enzyme in controlling the GSH/GSSG buffer has been a subject of research in the near past (13, 26, 60). It is known that this buffer takes part in disulfide bond redox reactions in the ER, and thus, it can form mixed disulfide intermediates that will also take part in the process of disulfide isomerization of misfolded proteins by PDI (61–63). Finally, the availability of experimental kinetics for this reaction (13) provides important checkpoints to validate our theoretical calculations.
Fig. 2.
Postulated mechanism for the reduction of GSSG by the reduced a domain of hPDI (13, 24, 30).
Methods
Modeling the hPDI:GSSG Complex.
Our molecular model was built from the X-ray crystallographic structure of a human oxidized protein disulfide isomerase (hPDIox) with Protein Data Dank (PDB) ID code 4EL1 (15). It included the four thioredoxin-like domains and the linker sequence of the enzyme (details are in SI Appendix). A total of 237 residues, comprising the a and b domains, were later considered to build the quantum mechanics/molecular mechanics (QM/MM) model. No modeling of any kind was required for these domains.
The GSSG substrate was modeled in the active site of the a domain of hPDIox from the X-ray structure of a homologous enzyme (glutaredoxin) bound to GSSG (PDB ID code 4TR0) (64). Glutaredoxin is a thioredoxin-like enzyme that catalyzes the reduction of GSSG to GSH; like PDI, it also possesses the catalytic Cys–X–Y–Cys motif (Cys12–Pro13–Tyr14–Cys15) (36). We have aligned the heavy atoms of the backbone of the Cys–X–X–Cys motif of glutaredoxin and that of the a domain of hPDIox (rmsd of 0.27 Å) (SI Appendix, Fig. S3) and transposed GSSG and all water molecules within 6 Å of GSSG into oxidized PDI. We have not observed clashes between these waters and hPDI.
Physiological protonation states were attributed to all residues [according to the results from the Propka31.py script (65, 66)], except for Cys53, Cys397, and Cys400, which have protonation states that are extensively described in the literature (15, 52, 67, 68): Cys53 was modeled as a thiolate, and the Cys397 and Cys400 pair was oxidized. All molecular mechanics (MM) parameters for the enzyme were drawn from the FF99SBildn force field (69–71); the parameters for GSSG were determined for the X-ray conformation in glutaredoxin using the Antechamber tool and the PARM99 force field (72, 73) with Merz–Kollman charges (74) derived from a restrained electrostatic potential fitting from a single-point energy calculation at the HF/6–31G(d) level of theory. All parameter and coordinate files are included in Dataset S1.
To minimize the energy of our model at the MM level of theory, we built a rectangular box solvated with transferable intermolecular potential 3-point (TIP3P) waters (75) within a radius of 12 Å from the surface of the enzyme and neutralized the charge of the model (−39 a.u.) with 39 Na+ counterions (details are in SI Appendix). The final minimized model exhibited an rmsd of 1.03 Å relative to the X-ray model (considering only the heavy atoms of the backbone).
We performed a 10-ns classical molecular dynamics (cMD) simulation within an NPT ensemble (temperature of 310 K and pressure of 1 bar) with the solute’s heavy atoms restrained by 20 kcal·mol−1·Å−2 harmonic potentials using the Berendsen barostat (76) and the Langevin thermostat (77) (details on heating are in SI Appendix). We have used the SHAKE algorithm (78) to constrain the motion in H-including bonds to use a 2-fs integration step and the particle–mesh–Ewald summation method (79) to account for the electrostatic interactions beyond the 10-Å cutoff for nonbonded interactions. We have kept the X-ray positions of the solute fixed, because there is evidence that the X-ray structure, being the most common and an average over the enzyme conformations in the crystal form, is the most adequate to be used in single-conformation potential energy surface (PES) studies. We then took the last structure of the cMD run as the representative catalytic conformation of the solvated hPDIox:GSSG complex.
Building the QM/MM Model.
To determine the enzymatic mechanism of hPDI, we used QM/MM calculations with the our own N-layered integrated molecular orbital/molecular mechanics (ONIOM) methodology (80). Nevertheless, several different computational approaches have been widely used to determine enzyme reaction mechanisms (81–90). We have drawn thermochemical and kinetic insight from single-conformation calculations with the X-ray–based model.
The QM/MM model included the a and b domains of hPDIox, GSSG, all water molecules within a 3-Å radius of the included domains, and all water molecules within 6 Å of the active site of the a domain (Cys53–Gly54–His55–Cys56, Glu47, Arg120, and GSSG). These two domains are the only ones that are relevant for the reaction, because the small substrate is far away from the remaining domains. The insights drawn for this study are not intended to represent the chaperone activity of hPDI but solely, the thiol-disulfide exchange reaction.
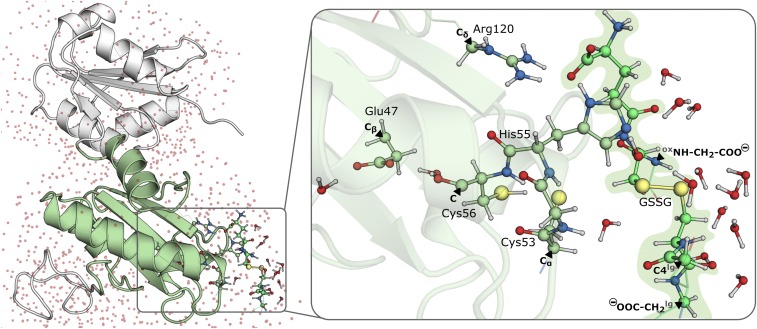
The layer to be studied at the density-functional theory (DFT) level comprised 160 atoms (with an overall charge of −1 and a singlet multiplicity) and included the Cys53–Gly54–His55–Cys56 motif; Glu47; Arg120; a substantial part of GSSG; all water molecules within 6 Å from the sulfur atoms of Cys53, Cys56, and GSSG; and the oxygen atoms of Glu47 (a coordinate file is in Dataset S2). We further used a DFT layer (165 atoms) in another calculation to evaluate the effect of protonating His55 in the active site to the imidazolium form (Dataset S3). We used the link atom approach (80, 91–93) to saturate the valences that resulted from the truncation of bonds across the DFT and MM layers. The MM layer comprised the a and b domains of hPDIox and the remaining water molecules (7,006 atoms). In Fig. 3, we present our QM/MM model, highlighting the DFT and MM layers and the atoms where the DFT layer was cut.
Fig. 3.
Optimized enzyme–substrate complex. (Left) The full ONIOM model with the a domain in green cartoon, b domain in white cartoon, and water molecules as red dots. (Right) The DFT layer with black triangles pointing to the atoms, labeled in bold, where the DFT layer was capped with H atoms following the link atom approach (the ox and lg in superscript distinguish the two GSH molecules in GSSG).
Determining the Catalytic Mechanism.
All calculations were performed with the ONIOM methodology and the electrostatic embedding scheme (94) as implemented in the Gaussian09 package (95). Throughout the calculations, all water molecules included in the MM layer were kept fixed. The mPW1N/6–31G(d):FF99SB level of theory was used for all geometry optimizations. Several works have shown that the 6–31G(d) basis is adequate for the purpose (85). Furthermore, a previous study has shown that mPW1N (96–101) [along with mPW1K (96–100, 102, 103), BB1K (104–106), and mPWB1K (96–100, 104, 107, 108)] is one of the best density functionals to adequately describe the thermochemistry and kinetics of thiol-disulfide exchange (45).
We performed relaxed geometry optimization calculations for all stationary points along the reaction coordinate. Nuclear vibrational frequencies revealed their nature (one imaginary frequency in the DFT layer for transition states (TS) and no imaginary frequencies in the DFT layer for minima). The 0 K electronic energy was determined with single-point energy calculations at the ONIOM(mPW1N/6–311+G(2d,2p):FF99SB//mPW1N/6–31G(d):FF99SB) level of theory. We have also compared the performance of mPW1N with BB1K. The results state that the energy profiles of the reaction are similar (SI Appendix, Table S3). To build the thermochemical and kinetic profile of the reaction, we determined zero-point energy and thermal and entropic contributions to the Gibbs energy at the mPW1N/6–31G(d):FF99SB level of theory. These corrections were calculated at 310 K and 1 bar using the harmonic oscillator/rigid rotor/particle in a box formalism. In the particular case of vibrational entropy, we used both the harmonic oscillator approximation and the combined free rotor/harmonic oscillator approximation as described elsewhere (109) to evaluate the effect of anharmonicity of low vibrational frequencies in the entropy (SI Appendix). Both approximations showed similar results, with differences in Gibbs energy coming from vibrational entropy and lower than 1 kcal·mol−1.
Results and Discussion
In the optimized QM/MM model, the disulfide of GSSG is linearly aligned (164.5°) with the Cys53-sulfur (SCys53); the backbone NH of His55 and Cys56 are oriented to favor the formation of hydrogen bonds with the Cys53-thiolate; the Glu47-carboxylate, which is postulated to deprotonate the Cys56-thiol, makes two hydrogen bonds with water molecules (1.64 and 1.80 Å); the Arg120-guanidinium makes a salt bridge with the glutamyl-carboxylate of GSSG (35); and His55-imidazole is hydrogen-bonded to the GSSG-glutamyl and a nearby water molecule (2.09 and 2.02 Å). Also in agreement with the literature (37, 46, 48), all angles involving the sulfur atoms from GSSG (Sox and Slg) or Cys53 are close to 90° (88.8° for Cox–Sox···SCys53, 113.0° for Sox···SCys53–C, 104.7° for Clg–Slg–Sox, and 101.2° for Cox–Sox–Slg), and all torsions centered on the S–S axis are also close to 90° (93.3° for CoxSox–SCys53C and −74.4° for ClgSlg–SoxCox).
The GSSG is very solvent-exposed. Our calculations showed that, when only the water molecules from the X-ray crystal were included in the DFT layer (Dataset S4), no TS was observed, and the energy of reaction was much higher than the 30-kcal·mol−1 limit. On inclusion of a considerable number of water molecules nearby GSSG at the DFT level (Dataset S5), we determined a TS with a barrier of ∼17 kcal·mol−1. These results show that, as observed in previous studies (110), the solvent plays a major role in the nucleophilic attack of Cys53 to GSSG (43, 45). A detailed discussion is provided in SI Appendix.
Protonation of His55 in the SN2 Nucleophilic Attack of the Cys53-Thiolate.
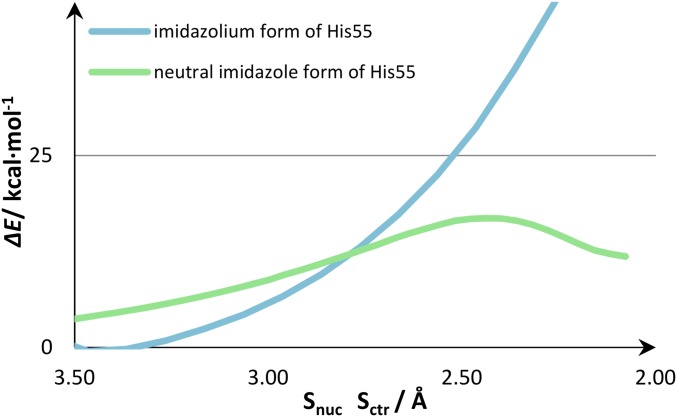
We performed the nucleophilic attack of Cys53 on GSSG with two protonation states for the His55-imidazole in the Cys53–Gly54–His55–Cys56 motif. We built a model complex, in which the His55-imidazole was positively charged (with 165 atoms in the DFT layer) and one with the neutral His55 protonated in the δ-nitrogen (with 160 DFT atoms). Despite that the most accepted hypothesis is that His55 is neutral (36), it has also been suggested that the His55-imidazolium cation could stabilize the Cys53-thiolate (111). Hence, we compared the nucleophilic attack of Cys53 to GSSG on both models (Fig. 4).
Fig. 4.
PES for the SN2 nucleophilic attack of the Cys53-thiolate to the GSSG-disulfide.
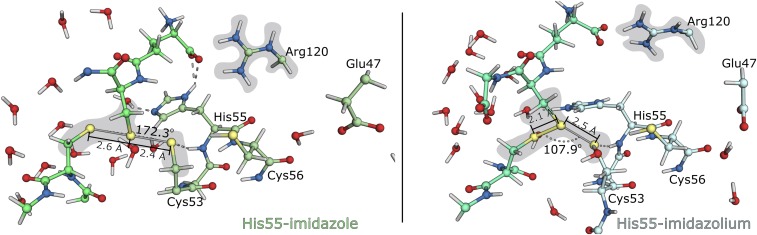
Our calculations showed that, if the His55 was positively charged, the nucleophilic attack of the Cys53-thiolate did not form the linear trisulfide anion arrangement (Fig. 5, Right). The nucleophilic attack in which we modeled an His55-imidazolium exhibited no transition state, the energy of the nucleophilic attack increased throughout the entire linear transit scan (above 60 kcal·mol−1), and there was no formation of any mixed disulfide intermediate. However, the nucleophilic attack in which we had the neutral His55-imidazole (Fig. 5, Left) exhibited a clear transition state, forming a mixed disulfide intermediate.
Fig. 5.
DFT layers of the two ONIOM models in which the His55-imidazole is either (Left) neutral (pale green) or (Right) in the cationic form (blue white). The GSSG substrate is represented in (Left) lime green and (Right) light green. The figure depicts the conformation in which the nucleophilic Cys53-thiolate is at ∼2.4 Å from the GSSG-disulfide (usually the distance for which the linear trisulfide anion is formed). Gray shades indicate the regions where residues occupied substantially different positions in both models.
An analysis of the conformation represented in Fig. 5 indicated that the imidazolium cation was establishing hydrogen bonds with solvent molecules only, whereas the imidazole form was also a hydrogen bond donor to the carboxylate of the glutamyl moiety of GSSG. We observed also that there was a salt interaction between the guanidinium moiety of Arg120 and the glutamyl-carboxylate of GSSG in both models. However, only the neutral imidazole of His55 established hydrogen bonds with the glutamyl-carboxylate of GSSG. In the imidazolium form of His55, the Arg120 and the glutamyl-carboxylate were farther apart. Thus, we believe that the repulsive interaction between the positive Arg120 and His55 weakened the interaction between GSSG and the active site of the a domain. This effect compromised the linear arrangement of the sulfur atoms required for the trisulfide anion to form (the Slg–Sox···SCys53 angle was 172° for the neutral His55 and 108° for the positively charged one). It should also be responsible for the difference in about 40 kcal·mol−1 that was observed in the PES of both nucleophilic attacks (Fig. 4).
At this point, we have asserted that, for the thiol-disulfide exchange reaction to occur, we require that the His55-imidazole in the Cys53–Gly54–His55–Cys56 motif is in the neutral form for the attack of the Cys53-thiolate to the GSSG-disulfide to occur linearly.
SN2 Nucleophilic Attack of the Cys53-Thiolate to the GSSG-Disulfide and Formation of the Mixed Disulfide Intermediate.
The formation of the mixed disulfide intermediate involved a redox reaction to reduce GSSG to the mixed disulfide intermediate and an acid–base reaction to deprotonate the Cys56-thiol via the Glu47-carboxylate. In Figs. 6–8, we show these chemical steps and associated thermochemical quantities (Gibbs energy, enthalpy, and entropy).
Fig. 6.
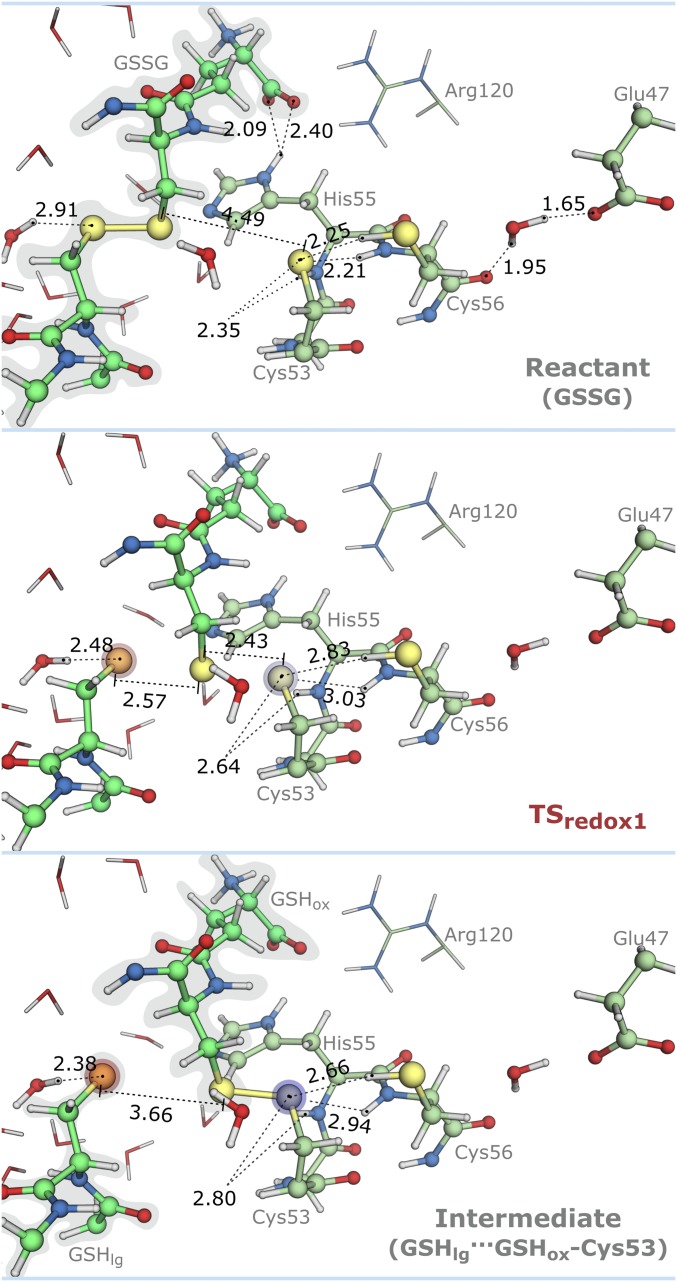
Stationary points for the nucleophilic attack of the Cys53-thiolate to the GSSG-disulfide with distances in angstroms (GSSG, TSredox1, and GSHlg···GSHox–Cys53). Blue to red shades in atoms represent the most relevant changes in atomic charge relative to the GSSG state (from 0.07 to 0.30 a.u.). Blue stands for increase in atomic charge, and red stands for decrease in atomic charge.
Fig. 8.
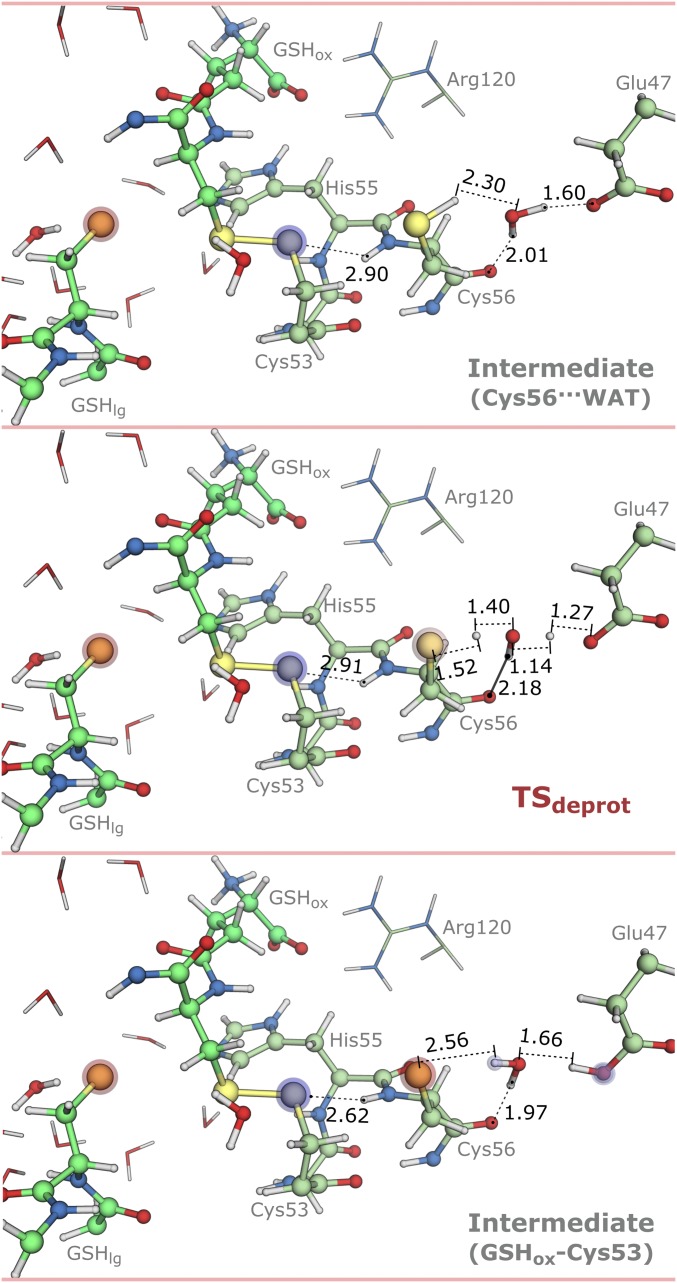
Stationary points for the deprotonation of Cys56 with distances in angstroms (Cys56···WAT, TSdeprot, and GSHox–Cys53). Blue to red shades in atoms represent the most relevant changes in atomic charge relative to the GSSG state (from 0.07 to 0.30 a.u.). Blue stands for increase in atomic charge, and red stands for decrease in atomic charge.
In the reactants (GSSG), the Cys53-sulfur (SCys53) was at 4.49 Å from the GSSG-disulfide and hydrogen-bonded to the backbone NHs of His55 and Cys56 and to the Cys56-thiol. The Glu47-carboxylate was hydrogen-bonded to the backbone carbonyl of Cys56 via a water molecule. The His55-imidazole was hydrogen-bonded to a water molecule and the carboxylate terminus of the GSSG-glutamyl. These hydrogen bonds remain formed throughout the entire catalytic cycle, and as such, these hydrogen bond donors/acceptors were not involved in the chemical step.
In the nucleophilic attack of the Cys53-thiolate (Fig. 6), the hydrogen bonds formed with His55 and Cys56 increased considerably, particularly those involving Cys56. However, an analysis of their associated energy gradient suggested that this increase was not favorable up to the TS. Because the Cys53 only loses its anionic character at the TS (when the linear trisulfide anion is formed), we speculate that the weakening of the hydrogen bonds before the TS could be related to the inability of the backbone NHs of His55 and Cys56 to move along with the Cys53-thiolate during its displacement toward GSSG.
At the transition state (TSredox1), the vibrational mode characterized by an imaginary frequency of 156i cm−1 displayed a linear antisymmetric stretching of the three sulfur atoms. The hydrogen bond established between the His55 peptide amine and the Cys53-thiolate increased the least (2.35–2.64 Å) at the TSredox1. The trisulfide anion (Slg···Sox···SCys53) exhibited an angle of 172° and bond distances of 2.57 Å (Slg–Sox) and 2.43 Å (Sox–SCys53). A water molecule in the close vicinity of the leaving glutathione (GSHlg) approximated substantially to make a shorter hydrogen bond (2.91–2.48 Å) and stabilize the negative charge building on GSHlg. After the S–S bond was formed in the mixed disulfide [GSHlg···oxidized glutathione (GSHox)–Cys53], the hydrogen bonds between Cys53 and Cys56 shortened, contrary to those established with the His55 backbone. There was also an evident charge transfer occurring from the Cys53-thiolate to GSHlg resulting from the cleavage of the GSSG-disulfide bond. The GSHlg was negatively charged and solvated, and the Cys53-sulfur was oxidized and covalently bound to GSHox-sulfur. The thiol-disulfide exchange occurred with little geometry modifications. The ∼90° amplitude for the C–S–Sox/Cox–Sox–S angles (104.7°/101.2° to 107.6°/105.5°) and CS–SoxCox dihedral (−74.4° to 99.0°) was maintained between GSSG and GSHox–Cys53, in agreement with earlier studies that pointed out that hyperconjugation between the SC bond and the SS bond played an important role in the arrangement of SS dihedral angles (47, 49). Moreover, an analysis of the CS and SS natural bond orbitals indicated that the SS bond was a single bond formed by sp hybridization of sulfur valence orbitals (with p orbitals accounting for ∼90% of the interaction); the same occurred for CS bonds (p orbitals accounted for ∼80% of the interaction). The predominant contributions of p orbitals for the binding might also explain why the CS–SC dihedral and the CSS angles were mostly orthogonal (47, 49).
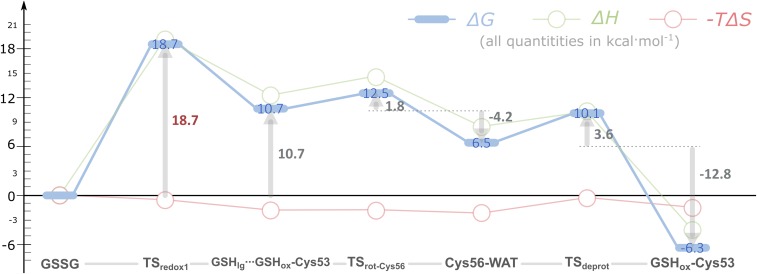
The Gibbs activation energy for the first thiol-disulfide exchange was 18.7 kcal·mol−1 (Fig. 7), a value close to the experimental barrier of 17.6 kcal·mol−1, derived from transition state theory using an experimental turnover of 2.6 s−1. This last value was calculated from the experimental second-order rate constant and the typical concentrations of GSSG and hPDI in the ER (SI Appendix) (13, 112–114).
Fig. 7.
Thermochemical profile for the formation of the mixed disulfide intermediate between the domain a of hPDI and the GSSG substrate. All contributions are represented relative to the initial hPDI:GSSG complex (GSSG).
Our results suggested that the rotation of the Cys56-thiol toward the water molecule nearby (Cys56···WAT) followed quickly. We could not optimize completely the TS for this rotation within the standard convergence criteria of Gaussian09; however, we characterized the highest energy state in the PES and found only one imaginary frequency in the DFT layer (258i cm−1). The Gibbs activation energy for this rotation was 1.8 kcal·mol−1. We also calculated the Gibbs energy profile for this rotation using molecular dynamics (MD) umbrella sampling (115) (SI Appendix, Fig. S11), and the results were similar to those of the QM/MM approach. This result should be expected, because after the first thiol-disulfide exchange, there were no hydrogen bond acceptors/donors for the Cys56-thiol. Hence, its rotation toward the water molecule should be enthalpically favored (−4.2 kcal·mol−1 in Fig. 7), because the latter can function as a hydrogen bond acceptor to Cys56. Other than the rotation of the Cys56-thiol, no changes were observed in the active site (Fig. 8). The Gibbs energy of reaction was +6.5 kcal·mol−1.
In Fig. 7, we see that the Gibbs energy and the enthalpy behave similarly. Entropy contributions were comparatively small, lowering the Gibbs energy of the mixed disulfide in up to 2 kcal·mol−1. They seemed to also favor the positioning of the Cys56-thiol for subsequent deprotonation by Glu47. This increase in entropy was mostly derived from vibrational normal modes in the range 170–350 cm−1 (SI Appendix, Fig. S9), which already significantly involved atoms from both the DFT layer and the MM environment.
The next chemical step was the deprotonation of the Cys56-thiol by Glu47 via a bridging water molecule. The transition state for this reaction (TSdeprot) exhibited one imaginary frequency in the DFT layer (1,059i cm−1) and a very low Gibbs activation energy of 3.6 kcal·mol−1. It was a concerted synchronous proton transfer from the Cys56-thiol to the Glu47-carboxylate via a water molecule (TSdeprot in Fig. 8). This reaction was the most exergonic step of the formation of the mixed disulfide, with a Gibbs energy of reaction of –12.8 kcal·mol−1. However, we did not observe major changes in the active site other than those coming from the rearrangement in hydrogen bonds involving Cys56 and Glu47 (Fig. 8).
The resulting mixed disulfide (GSHox–Cys53) is a relatively long-lived species, because the Gibbs activation energy to return the GSHox–Cys53 to GSSG was ∼25 kcal·mol−1. From the comparison of the enthalpy and the Gibbs energy profiles in Fig. 7, we infer that the reason why this final intermediate was stable, comparatively with GSHlg···GSHox–Cys53, is mainly in the reorganization of the hydrogen bond network in between Cys56 and Glu47 when the nucleophilic character of Cys56 increased. The complete reaction step was characterized by a Gibbs reaction energy of –6.4 kcal·mol−1.
Oxidation of the a Domain: Cleavage of Mixed Disulfide Intermediate.
After the formation of the mixed disulfide, two pathways could take place: either the mixed disulfide intermediate went through a second thiol-disulfide exchange with a nearby thiol group from a protein substrate, or it experienced an intramolecular oxidation through the deprotonated Cys56-thiol. We have studied the intramolecular oxidation through the Cys56-thiolate in the presence of the negatively charged GSHlg. However, the PES along the reaction coordinate exhibited no transition state, with the energy increasing to over 20 kcal·mol−1 from the reactants to the products. This result showed that this intramolecular rearrangement would seldom occur as far as the product of the first stage was still bound to the active site or nearby at least. Because both the products of the first and second steps were negatively charged thiolates, they would repel one another.
Therefore, we protonated GSHlg, which seemed legitimate, because GSHlg is mostly surrounded by water exhibiting a pKa similar to that determined in water (about 8.75) (24). We have not included the change in Gibbs energy for this transformation in Fig. 7. Hence, the first and second thiol-disulfide exchange reactions have to be looked at separately from an energetic point of view. The former optimized state showed that the GSH-thiol was preferably directed to the solvent. However, the conformation in which the GSH-thiol was directed toward the mixed disulfide state was only +1.1 kcal·mol−1 higher than the former. We were not able to optimize the TS for the rotation of the GSH-thiol with standard Gaussian09 convergence criteria, because the PES was quite flat; however, we performed a vibration analysis on the highest energy point in the PES, and we observed only one imaginary frequency (171i cm−1). The estimated Gibbs energy for this transition was +2.5 kcal·mol−1, thus supporting that both conformations should easily interchange. This barrier was also validated through MD simulations of the GSH-thiol rotation with umbrella sampling, which supported that the barriers for the rotation are indeed low (<1 kcal·mol−1) (SI Appendix, Fig. S12) but also indicated that GSH interacts weakly with the active site of PDI (115). Regarding protein substrates, we expect that hydrogen bond donors from the backbone or neighboring side chains may function in a similar way to the solvent, stabilizing the negative leaving Cys.
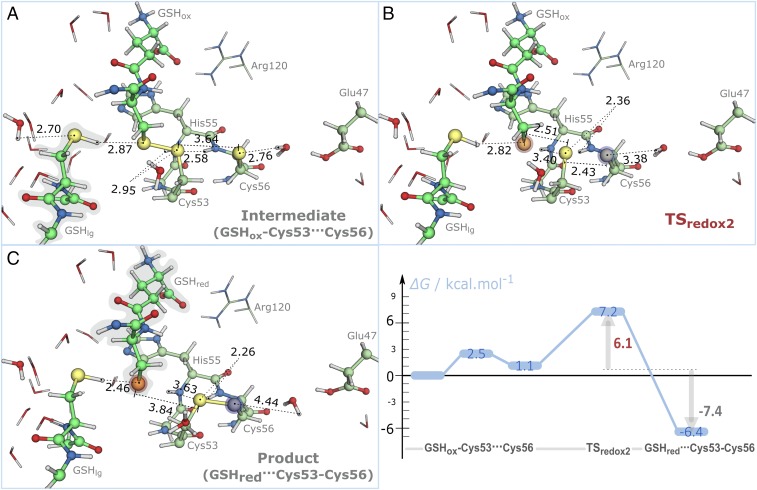
In Fig. 9, we present the stationary points in the PES for the nucleophilic attack of the Cys56-thiolate to the mixed disulfide and the Gibbs energy profile of the reaction. The latter exhibited a transition state (TSredox2) with a low Gibbs activation energy (7.2 kcal·mol−1), and the product (oxidized a domain) was formed with a Gibbs reaction energy of −6.4 kcal·mol−1. This result supports that intramolecular rearrangements will mainly occur to rescue the trapped mixed disulfide intermediate, because intramolecular oxidation is only favorable if there are hydrogen bond donors or positively charged species that are able to stabilize the negative charge of the leaving thiolate.
Fig. 9.
Stationary points for the cleavage of the mixed disulfide intermediate by the Cys56-thiolate: (A) GSH–Cys53···Cys56, (B) TSredox2, and (C) GSHred···Cys53–Cys56. Distances are given in angstroms. Blue to red shades in atoms represent the variation in atomic charge relative to the GSHred–Cys53···Cys56 state (from 0.07 to 0.30 a.u.; blue stands for decrease in atomic charge, and red stands for increase in atomic charge). (C, Right) Gibbs energy profile for the intramolecular oxidation of the a domain. All contributions are represented relative to GSHox–Cys53···Cys56 facing the solvent.
The reaction occurred in an identical way to that pictured in Fig. 6: the Cys56-thiolate formed a hydrogen bond with a nearby water molecule, and it occupied a nearly linear position relative to the mixed disulfide (∼160°). However, contrary to the reactant state (GSSG), the Cys56-thiolate only established one more hydrogen bond with the backbone NH of Ala50. Furthermore, we have proved that this interaction did not compromise our results by studying the same reaction with this interaction described at a DFT level (SI Appendix, Fig. S6).
In the transition state (TSredox2), the hydrogen bond between the Cys56-thiolate and the water molecule stretched (2.76–3.38 Å), whereas the hydrogen bond between the Cys53-sulfur and the backbone NH of Cys56 shortened (2.58–2.36 Å). This latter interaction was not observed in TSredox1, in which there were no hydrogen bonds with the central GSSG-sulfur of the linear trisulfide anion. Together with the fewer number of hydrogen bond donors for the Cys56-thiolate in the reactant state (GSHred–Cys53···Cys56), it may be responsible for the lower Gibbs activation energy that was registered for the step. In fact, because the trisulfide configuration is a minimum energy in vacuum (43, 45), we expected that the Gibbs energy for this reaction depended on the relative stabilization by the environment of the leaving nucleophilic thiolates. TSredox2 was characterized with one imaginary frequency in the DFT layer (111i cm−1). When GSHred formed (GSHred···Cys53–Cys56), it only established a hydrogen bond with the neighboring GSHlg. As for the first thiol-disulfide exchange, the cleavage of the mixed disulfide intermediate was enthalpy-driven, with an entropic contribution for the Gibbs energy lower than 2 kcal·mol−1 (SI Appendix, Table S4).
The intramolecular oxidation of the a domain was not rate-limiting, contrary to experimental suggestions (summarized in SI Appendix, Scheme S1) (13, 116). However, we recall that, in the absence of a polar environment to stabilize the resulting reduced thiolate, this step could be rate-limiting given that no TS was observed for the thiol-disulfide exchange reaction.
The experimental barrier for the second-stage reaction was difficult to assign, because two experimental measurements have provided clearly different results in the past: 14 and 19 kcal·mol−1 (13). The reason for the discrepancy between the two experimental values is unknown. The Gibbs activation energy obtained here (7.2 kcal·mol−1) is in disagreement with both of the (contradictory) experimental values. However, the experimental value for the second-order rate constant of the first reduction of the oxidized a domain was measured as 11,200 M−1 s−1. Considering a stable physiologic GSH concentration of 50 mM (117, 118), this rate constant would result in a pseudo first-order rate constant of 560 s−1, which corresponds to a Gibbs activation energy of 14 kcal·mol−1, in excellent agreement with the calculated value of 13.6 kcal·mol−1.
One other aspect is that, in the intramolecular oxidation, the GSHlg was stabilized by hydrogen bonds. Two preliminary MD simulations of an enzyme model in the mixed disulfide state immersed in a water box, having the GSHlg-thiol in the neutral and ionized forms, were carried out (details are in SI Appendix and Dataset S6). These simulations have shown that GSHlg unbinds the active site in less than 10 ns (ionized GSHlg thiol) and 1 ns (neutral GSHlg thiol), which indicate that GSH molecules dissociate in a timescale much smaller than the chemical reaction timescale. Because we have not calculated the Gibbs binding energy of GSH in the periphery of the active site, we could not determine accurately which of the states (bound/unbound GSHlg) will be prevalent. Therefore, we have repeated the intramolecular oxidation in the absence of GSHlg. The reaction was almost thermoneutral and exhibited similar kinetics to that described in Fig. 9 (SI Appendix, Fig. S7).
Conclusions
The catalytic mechanism of the reduction of GSSG by hPDI was studied with hybrid QM/MM using the ONIOM methodology. This study provided mechanistic insights transferable to the catalysis of hPDI in the oxidative protein folding pathway.
The reaction occurred in two stages: (i) nucleophilic attack of a Cys53-thiolate to the GSSG-disulfide with elimination of GSH (GSHlg) followed by a fast rotation (ΔG‡ < 2 kcal·mol−1) and deprotonation of the Cys56-thiol by Glu47 via a bridging water molecule and (ii) protonation, and/or dissociation of GSHlg from the active site followed by Cys56-thiolate nucleophilic attack on the mixed disulfide, forming a second GSH (GSHred) and oxidizing hPDI. We provided atomistic and thermochemical detail for every step along these reactions (Datasets S7–S9 and Movies S1 and S2).
The first thiol-disulfide exchange exhibited a higher Gibbs activation energy than the second (18.7 vs. 7.2 kcal·mol−1). These barriers are in good agreement with the overall experimental turnover (2.6 s−1). We observed also that entropy can lower the Gibbs energy in up to 2 kcal·mol−1 on formation of the mixed disulfide intermediate. During this reaction, the His55-imidazole should be in its neutral state at the beginning of the catalysis. The His55-imidazolium form prevented the linear arrangement of the trisulfide anion (in the transition state), raising the barrier for formation of the mixed disulfide intermediate beyond 60 kcal·mol−1. The hydrogen bonds established between the Cys53-thiolate, the peptidic NHs of His55 and Cys56, and the Cys56-thiol exhibited the largest increase during the SN2 reaction.
The deprotonation of Cys56-thiol exhibited a Gibbs activation energy of 3.6 kcal·mol−1, and it was accompanied by minor changes in the active site. The Gibbs reaction energy determined for the formation of the stable mixed disulfide intermediate was −6.4 kcal·mol−1. Our results are in agreement with experimental data that suggest that the mixed disulfide intermediate is a long-lived species (the Gibbs activation energy to return to the reactant state was 25.0 kcal·mol−1).
The internal disulfide resulted from the cleavage of the mixed disulfide intermediate by the Cys56-thiolate. This second nucleophilic attack occurred quickly if there were polar species that could stabilize the leaving reduced glutathionate. In the presence of negatively charge species, no TS was observed for the same reaction. The Gibbs activation energy that we calculated (7.2 kcal·mol−1) is lower than experimentally derived ones (which are contradictory between themselves). However, our Gibbs activation energy for the first reduction of the oxidized a domain agreed very well with the pseudofirst-order turnovers in the literature (13.6 vs. 14.3 kcal·mol−1).
Overall, our study has provided atomic-level understanding of the reaction mechanism of hPDI. The kinetics of this mechanism may differ in different compartments of the cell, because thiol-disulfide exchange kinetics depends on the ratio GSH/GSSG. However, the structural and thermochemical details provided here will be of use for future studies on regulation/inhibition of the activity of hPDI. Despite that, there are diverse factors that influence its activity other than its catalytic sites; the knowledge on the way hPDI reacts with its substrates will always be helpful to control its physiologic functions.
Supplementary Material
Acknowledgments
This work received financial support from the following institutions: Fundo Europeu de Desenvolvimento Regional (FEDER) Grant POCI/01/0145/FEDER/007728 and Fundação para a Ciência e Tecnologia (FCT) and Ministério da Educação e Ciência Partnership Agreement PT2020, UID/MULTI/04378/2013; NORTE-01-0145-FEDER-000024 supported by Norte Portugal Regional Operational Programme (NORTE 2020) under the PORTUGAL 2020 Partnership Agreement through the European Regional Development Fund and FCT Projects EXCL/QEQ-COM/0394/2012 and EXCL-II/QEQ-COM/0394/2012. R.P.P.N. also thanks the FCT for Grant SFRH/BD/78397/2011.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618985114/-/DCSupplemental.
References
- 1.Narayan M, Welker E, Wedemeyer WJ, Scheraga HA. Oxidative folding of proteins. Acc Chem Res. 2000;33:805–812. doi: 10.1021/ar000063m. [DOI] [PubMed] [Google Scholar]
- 2.Kersteen EA, Barrows SR, Raines RT. Catalysis of protein disulfide bond isomerization in a homogeneous substrate. Biochemistry. 2005;44:12168–12178. doi: 10.1021/bi0507985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sevier CS, Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 4.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galligan JJ, Petersen DR. The human protein disulfide isomerase gene family. Hum Genomics. 2012;6:6. doi: 10.1186/1479-7364-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benham AM. The protein disulfide isomerase family: Key players in health and disease. Antioxid Redox Signal. 2012;16:781–789. doi: 10.1089/ars.2011.4439. [DOI] [PubMed] [Google Scholar]
- 7.Walker KW, Gilbert HF. Effect of redox environment on the in vitro and in vivo folding of RTEM-1 beta-lactamase and Escherichia coli alkaline phosphatase. J Biol Chem. 1994;269:28487–28493. [PubMed] [Google Scholar]
- 8.Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: Mechanisms and consequences. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 10.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu S, Sankar S, Neamati N. Protein disulfide isomerase: A promising target for cancer therapy. Drug Discov Today. 2014;19:222–240. doi: 10.1016/j.drudis.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Darby NJ, Creighton TE. Characterization of the active site cysteine residues of the thioredoxin-like domains of protein disulfide isomerase. Biochemistry. 1995;34:16770–16780. doi: 10.1021/bi00051a027. [DOI] [PubMed] [Google Scholar]
- 13.Lappi AK, Ruddock LW. Reexamination of the role of interplay between glutathione and protein disulfide isomerase. J Mol Biol. 2011;409:238–249. doi: 10.1016/j.jmb.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 14.Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 15.Wang C, et al. Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase. Antioxid Redox Signal. 2013;19:36–45. doi: 10.1089/ars.2012.4630. [DOI] [PubMed] [Google Scholar]
- 16.Darby NJ, Penka E, Vincentelli R. The multi-domain structure of protein disulfide isomerase is essential for high catalytic efficiency. J Mol Biol. 1998;276:239–247. doi: 10.1006/jmbi.1997.1504. [DOI] [PubMed] [Google Scholar]
- 17.Edman JC, Ellis L, Blacher RW, Roth RA, Rutter WJ. Sequence of protein disulphide isomerase and implications of its relationship to thioredoxin. Nature. 1985;317:267–270. doi: 10.1038/317267a0. [DOI] [PubMed] [Google Scholar]
- 18.Dumont E, Loos PF, Laurent AD, Assfeld X. Huge disulfide-linkage’s electron capture variation induced by α-helix orientation. J Chem Theory Comput. 2008;4:1171–1173. doi: 10.1021/ct800161m. [DOI] [PubMed] [Google Scholar]
- 19.Klappa P, Hawkins HC, Freedman RB. Interactions between protein disulphide isomerase and peptides. Eur J Biochem. 1997;248:37–42. doi: 10.1111/j.1432-1033.1997.t01-1-00037.x. [DOI] [PubMed] [Google Scholar]
- 20.Denisov AY, et al. Solution structure of the bb’ domains of human protein disulfide isomerase. FEBS J. 2009;276:1440–1449. doi: 10.1111/j.1742-4658.2009.06884.x. [DOI] [PubMed] [Google Scholar]
- 21.Byrne LJ, et al. Mapping of the ligand-binding site on the b′ domain of human PDI: Interaction with peptide ligands and the x-linker region. Biochem J. 2009;423:209–217. doi: 10.1042/BJ20090565. [DOI] [PubMed] [Google Scholar]
- 22.Wang C, et al. Human protein-disulfide isomerase is a redox-regulated chaperone activated by oxidation of domain a′. J Biol Chem. 2012;287:1139–1149. doi: 10.1074/jbc.M111.303149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irvine AG, et al. Protein disulfide-isomerase interacts with a substrate protein at all stages along its folding pathway. PLoS One. 2014;9:e82511. doi: 10.1371/journal.pone.0082511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatahet F, Ruddock LW. Protein disulfide isomerase: A critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 25.Lyles MM, Gilbert HF. Mutations in the thioredoxin sites of protein disulfide isomerase reveal functional nonequivalence of the N- and C-terminal domains. J Biol Chem. 1994;269:30946–30952. [PubMed] [Google Scholar]
- 26.Cuozzo JW, Kaiser CA. Competition between glutathione and protein thiols for disulphide-bond formation. Nat Cell Biol. 1999;1:130–135. doi: 10.1038/11047. [DOI] [PubMed] [Google Scholar]
- 27.Baker KM, et al. Low reduction potential of Ero1alpha regulatory disulphides ensures tight control of substrate oxidation. EMBO J. 2008;27:2988–2997. doi: 10.1038/emboj.2008.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambers JE, et al. The reduction potential of the active site disulfides of human protein disulfide isomerase limits oxidation of the enzyme by Ero1α. J Biol Chem. 2010;285:29200–29207. doi: 10.1074/jbc.M110.156596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, et al. Reconstitution of human Ero1-Lalpha/protein-disulfide isomerase oxidative folding pathway in vitro. Position-dependent differences in role between the a and a′ domains of protein-disulfide isomerase. J Biol Chem. 2009;284:199–206. doi: 10.1074/jbc.M806645200. [DOI] [PubMed] [Google Scholar]
- 30.Ali Khan H, Mutus B. Protein disulfide isomerase a multifunctional protein with multiple physiological roles. Front Chem. 2014;2:70. doi: 10.3389/fchem.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawkins HC, Freedman RB. The reactivities and ionization properties of the active-site dithiol groups of mammalian protein disulphide-isomerase. Biochem J. 1991;275:335–339. doi: 10.1042/bj2750335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruddock LW, Hirst TR, Freedman RB. pH-dependence of the dithiol-oxidizing activity of DsbA (a periplasmic protein thiol:disulphide oxidoreductase) and protein disulphide-isomerase: Studies with a novel simple peptide substrate. Biochem J. 1996;315:1001–1005. doi: 10.1042/bj3151001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kortemme T, Darby NJ, Creighton TE. Electrostatic interactions in the active site of the N-terminal thioredoxin-like domain of protein disulfide isomerase. Biochemistry. 1996;35:14503–14511. doi: 10.1021/bi9617724. [DOI] [PubMed] [Google Scholar]
- 34.Karala AR, Lappi AK, Ruddock LW. Modulation of an active-site cysteine pKa allows PDI to act as a catalyst of both disulfide bond formation and isomerization. J Mol Biol. 2010;396:883–892. doi: 10.1016/j.jmb.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 35.Lappi AK, et al. A conserved arginine plays a role in the catalytic cycle of the protein disulphide isomerases. J Mol Biol. 2004;335:283–295. doi: 10.1016/j.jmb.2003.10.051. [DOI] [PubMed] [Google Scholar]
- 36.Carvalho AP, Fernandes PA, Ramos MJ. Similarities and differences in the thioredoxin superfamily. Prog Biophys Mol Biol. 2006;91:229–248. doi: 10.1016/j.pbiomolbio.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 37.Dumont E, Loos PF, Assfeld X. Factors governing electron capture by small disulfide loops in two-cysteine peptides. J Phys Chem B. 2008;112:13661–13669. doi: 10.1021/jp806465e. [DOI] [PubMed] [Google Scholar]
- 38.Roos G, Foloppe N, Messens J. Understanding the pK(a) of redox cysteines: The key role of hydrogen bonding. Antioxid Redox Signal. 2013;18:94–127. doi: 10.1089/ars.2012.4521. [DOI] [PubMed] [Google Scholar]
- 39.Rullmann JAC, Bellido MN, van Duijnen PT. The active site of papain. All-atom study of interactions with protein matrix and solvent. J Mol Biol. 1989;206:101–118. doi: 10.1016/0022-2836(89)90527-5. [DOI] [PubMed] [Google Scholar]
- 40.Dillet V, Dyson HJ, Bashford D. Calculations of electrostatic interactions and pKas in the active site of Escherichia coli thioredoxin. Biochemistry. 1998;37:10298–10306. doi: 10.1021/bi980333x. [DOI] [PubMed] [Google Scholar]
- 41.Foloppe N, Nilsson L. The glutaredoxin -C-P-Y-C- motif: Influence of peripheral residues. Structure. 2004;12:289–300. doi: 10.1016/j.str.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 42.Roos G, Loverix S, Geerlings P. Origin of the pKa perturbation of N-terminal cysteine in alpha- and 3(10)-helices: A computational DFT study. J Phys Chem B. 2006;110:557–562. doi: 10.1021/jp0549780. [DOI] [PubMed] [Google Scholar]
- 43.Fernandes PA, Ramos MJ. Theoretical insights into the mechanism for thiol/disulfide exchange. Chemistry. 2004;10:257–266. doi: 10.1002/chem.200305343. [DOI] [PubMed] [Google Scholar]
- 44.Carvalho ATP, et al. Mechanism of thioredoxin-catalyzed disulfide reduction. Activation of the buried thiol and role of the variable active-site residues. J Phys Chem B. 2008;112:2511–2523. doi: 10.1021/jp7104665. [DOI] [PubMed] [Google Scholar]
- 45.Neves RPP, Fernandes PA, Varandas AJC, Ramos MJ. Benchmarking of density functionals for the accurate description of thiol-disulfide exchange. J Chem Theory Comput. 2014;10:4842–4856. doi: 10.1021/ct500840f. [DOI] [PubMed] [Google Scholar]
- 46.Boyd RJ, Perkyns JS, Ramani R. Conformations of simple disulfides and L-cystine. Can J Chem. 1983;61:1082–1085. [Google Scholar]
- 47.Aida M, Nagata C. An ab initio MO study on the disulfide bond: Properties concerning the characteristic S-S dihedral angle. Theor Chim Acta. 1986;70:73–80. [Google Scholar]
- 48.Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta. 2013;1830:3217–3266. doi: 10.1016/j.bbagen.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 49.Pappas JA. Theoretical studies of reactions of sulfur-sulfur bond. 1. General heterolytic mechanisms. J Am Chem Soc. 1977;99:2926–2930. [Google Scholar]
- 50.Walker KW, Gilbert HF. Scanning and escape during protein-disulfide isomerase-assisted protein folding. J Biol Chem. 1997;272:8845–8848. doi: 10.1074/jbc.272.14.8845. [DOI] [PubMed] [Google Scholar]
- 51.Schwaller M, Wilkinson B, Gilbert HF. Reduction-reoxidation cycles contribute to catalysis of disulfide isomerization by protein-disulfide isomerase. J Biol Chem. 2003;278:7154–7159. doi: 10.1074/jbc.M211036200. [DOI] [PubMed] [Google Scholar]
- 52.Kemmink J, Darby NJ, Dijkstra K, Nilges M, Creighton TE. Structure determination of the N-terminal thioredoxin-like domain of protein disulfide isomerase using multidimensional heteronuclear 13C/15N NMR spectroscopy. Biochemistry. 1996;35:7684–7691. doi: 10.1021/bi960335m. [DOI] [PubMed] [Google Scholar]
- 53.Dyson HJ, et al. Effects of buried charged groups on cysteine thiol ionization and reactivity in Escherichia coli thioredoxin: Structural and functional characterization of mutants of Asp 26 and Lys 57. Biochemistry. 1997;36:2622–2636. doi: 10.1021/bi961801a. [DOI] [PubMed] [Google Scholar]
- 54.Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: Substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ. Protein disulfide isomerase: The structure of oxidative folding. Trends Biochem Sci. 2006;31:455–464. doi: 10.1016/j.tibs.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe MM, Laurindo FRM, Fernandes DC. Methods of measuring protein disulfide isomerase activity: A critical overview. Front Chem. 2014;2:73. doi: 10.3389/fchem.2014.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westphal V, Darby NJ, Winther JR. Functional properties of the two redox-active sites in yeast protein disulphide isomerase in vitro and in vivo. J Mol Biol. 1999;286:1229–1239. doi: 10.1006/jmbi.1999.2560. [DOI] [PubMed] [Google Scholar]
- 58.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 59.Westphal V, Spetzler JC, Meldal M, Christensen U, Winther JR. Kinetic analysis of the mechanism and specificity of protein-disulfide isomerase using fluorescence-quenched peptides. J Biol Chem. 1998;273:24992–24999. doi: 10.1074/jbc.273.39.24992. [DOI] [PubMed] [Google Scholar]
- 60.Raturi A, Mutus B. Characterization of redox state and reductase activity of protein disulfide isomerase under different redox environments using a sensitive fluorescent assay. Free Radic Biol Med. 2007;43:62–70. doi: 10.1016/j.freeradbiomed.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 61.Darby NJ, Freedman RB, Creighton TE. Dissecting the mechanism of protein disulfide isomerase: Catalysis of disulfide bond formation in a model peptide. Biochemistry. 1994;33:7937–7947. doi: 10.1021/bi00191a022. [DOI] [PubMed] [Google Scholar]
- 62.Ruoppolo M, Freedman RB, Pucci P, Marino G. Glutathione-dependent pathways of refolding of RNase T1 by oxidation and disulfide isomerization: Catalysis by protein disulfide isomerase. Biochemistry. 1996;35:13636–13646. doi: 10.1021/bi960755b. [DOI] [PubMed] [Google Scholar]
- 63.Bass R, Ruddock LW, Klappa P, Freedman RB. A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein. J Biol Chem. 2004;279:5257–5262. doi: 10.1074/jbc.M304951200. [DOI] [PubMed] [Google Scholar]
- 64.Lee EH, Kim HY, Hwang KY. The GSH- and GSSG-bound structures of glutaredoxin from Clostridium oremlandii. Arch Biochem Biophys. 2014;564:20–25. doi: 10.1016/j.abb.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 65.Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 66.Søndergaard CR, Olsson MHM, Rostkowski M, Jensen JH. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J Chem Theory Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 67.Kemmink J, et al. The structure in solution of the b domain of protein disulfide isomerase. J Biomol NMR. 1999;13:357–368. doi: 10.1023/a:1008341820489. [DOI] [PubMed] [Google Scholar]
- 68.Dijkstra K, et al. Assignment of 1H, 13C and 15N resonances of the a′ domain of protein disulfide isomerase. J Biomol NMR. 1999;14:195–196. doi: 10.1023/a:1008331225208. [DOI] [PubMed] [Google Scholar]
- 69.Sorin EJ, Pande VS. Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys J. 2005;88:2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cornell WD, et al. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 73.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 74.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges - the resp model. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- 75.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 76.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 77.Langevin P. Sur la théorie du mouvement brownien. C R Acad Sci. 1908;146:530–532. [Google Scholar]
- 78.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 79.Ewald PP. The calculation of optical and electrostatic grid potential. Ann Phys. 1921;64:253–287. [Google Scholar]
- 80.Maseras F, Morokuma K. Imomm - a new integrated ab-initio plus molecular mechanics geometry optimization scheme of equilibrium structures and transition-states. J Comput Chem. 1995;16:1170–1179. [Google Scholar]
- 81.Amata O, Marino T, Russo N, Toscano M. Human insulin-degrading enzyme working mechanism. J Am Chem Soc. 2009;131:14804–14811. doi: 10.1021/ja9037142. [DOI] [PubMed] [Google Scholar]
- 82.Blomberg MRA, Borowski T, Himo F, Liao RZ, Siegbahn PEM. Quantum chemical studies of mechanisms for metalloenzymes. Chem Rev. 2014;114:3601–3658. doi: 10.1021/cr400388t. [DOI] [PubMed] [Google Scholar]
- 83.Senn HM, Thiel W. QM/MM methods for biomolecular systems. Angew Chem Int Ed Engl. 2009;48:1198–1229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- 84.Chung LW, et al. The ONIOM method and its applications. Chem Rev. 2015;115:5678–5796. doi: 10.1021/cr5004419. [DOI] [PubMed] [Google Scholar]
- 85.Sousa SF, et al. Application of quantum mechanics/molecular mechanics methods in the study of enzymatic reaction mechanisms. WIREs Comput Mol Sci. 2017:7:e1281. [Google Scholar]
- 86.Ferrer S, et al. Hybrid schemes based on quantum mechanics/molecular mechanics simulations goals to success, problems, and perspectives. Adv Protein Chem Struct Biol. 2011;85:81–142. doi: 10.1016/B978-0-12-386485-7.00003-X. [DOI] [PubMed] [Google Scholar]
- 87.van der Kamp MW, Mulholland AJ. Combined quantum mechanics/molecular mechanics (QM/MM) methods in computational enzymology. Biochemistry. 2013;52:2708–2728. doi: 10.1021/bi400215w. [DOI] [PubMed] [Google Scholar]
- 88.Dal Peraro M, Ruggerone P, Raugei S, Gervasio FL, Carloni P. Investigating biological systems using first principles Car-Parrinello molecular dynamics simulations. Curr Opin Struct Biol. 2007;17:149–156. doi: 10.1016/j.sbi.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 89.Brunk E, Rothlisberger U. Mixed quantum mechanical/molecular mechanical molecular dynamics simulations of biological systems in ground and electronically excited states. Chem Rev. 2015;115:6217–6263. doi: 10.1021/cr500628b. [DOI] [PubMed] [Google Scholar]
- 90.Kamerlin SCL, Warshel A. The empirical valence bond model: Theory and applications. WIREs Comput Mol Sci. 2011;1:30–45. [Google Scholar]
- 91.Singh UC, Kollman PA. A combined ab initio quantum-mechanical and molecular mechanical method for carrying out simulations on complex molecular-systems - applications to the CH3Cl + Cl- exchange-reaction and gas-phase protonation of polyethers. J Comput Chem. 1986;7:718–730. [Google Scholar]
- 92.Field MJ, Bash PA, Karplus M. A combined quantum-mechanical and molecular mechanical potential for molecular-dynamics simulations. J Comput Chem. 1990;11:700–733. [Google Scholar]
- 93.Das D, et al. Optimization of quantum mechanical molecular mechanical partitioning schemes: Gaussian delocalization of molecular mechanical charges and the double link atom method. J Chem Phys. 2002;117:10534–10547. [Google Scholar]
- 94.Vreven T, et al. Combining quantum mechanics methods with molecular mechanics methods in ONIOM. J Chem Theory Comput. 2006;2:815–826. doi: 10.1021/ct050289g. [DOI] [PubMed] [Google Scholar]
- 95.Frisch MJ, et al. GAUSSIAN 09, Revision E.01. Gaussian, Inc.; Wallingford, CT: 2009. [Google Scholar]
- 96.Burke K, Perdew JP, Wang Y. Electronic Density Functional Theory: Recent Progress and New Directions. Springer Plenum; New York: 1998. [Google Scholar]
- 97.Perdew JP. Electronic Structure of Solids ’91. Akademie Verlag; Berlin: 1991. [Google Scholar]
- 98.Perdew JP, Burke K, Wang Y. Generalized gradient approximation for the exchange-correlation hole of a many-electron system. Phys Rev B Condens Matter. 1996;54:16533–16539. doi: 10.1103/physrevb.54.16533. [DOI] [PubMed] [Google Scholar]
- 99.Perdew JP, et al. Atoms, molecules, solids, and surfaces: Applications of the generalized gradient approximation for exchange and correlation. Phys Rev B. 1992;46:6671–6687, and erratum (1993) 48:4978. doi: 10.1103/physrevb.46.6671. [DOI] [PubMed] [Google Scholar]
- 100.Adamo C, Barone V. Exchange functionals with improved long-range behavior and adiabatic connection methods without adjustable parameters: The mPW and mPW1PW models. J Chem Phys. 1998;108:664–675. [Google Scholar]
- 101.Kormos BL, Cramer CJ. Adiabatic connection method for X− + RX nucleophilic substitution reactions (X = F, Cl) J Phys Org Chem. 2002;15:712–720. [Google Scholar]
- 102.Lynch BJ, Zhao Y, Truhlar DG. Effectiveness of diffuse basis functions for calculating relative energies by density functional theory. J Phys Chem A. 2003;107:1384–1388. [Google Scholar]
- 103.Lynch BJ, Fast PL, Harris M, Truhlar DG. Adiabatic connection for kinetics. J Phys Chem A. 2000;104:4811–4815. [Google Scholar]
- 104.Becke AD. Density-functional thermochemistry. IV. A new dynamical correlation functional and implications for exact-exchange mixing. J Chem Phys. 1996;104:1040–1046. [Google Scholar]
- 105.Becke AD. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A Gen Phys. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 106.Zhao Y, Lynch BJ, Truhlar DG. Development and assessment of a new hybrid density functional model for thermochemical kinetics. J Phys Chem A. 2004;108:2715–2719. [Google Scholar]
- 107.Zhao Y, Truhlar DG. Hybrid meta density functional theory methods for thermochemistry, thermochemical kinetics, and noncovalent interactions: The MPW1B95 and MPWB1K models and comparative assessments for hydrogen bonding and van der Waals interactions. J Phys Chem A. 2004;108:6908–6918. [Google Scholar]
- 108.Boese AD, Martin JML, Handy NC. The role of the basis set: Assessing density functional theory. J Chem Phys. 2003;119:3005–3014. [Google Scholar]
- 109.Grimme S. Supramolecular binding thermodynamics by dispersion-corrected density functional theory. Chemistry. 2012;18:9955–9964. doi: 10.1002/chem.201200497. [DOI] [PubMed] [Google Scholar]
- 110.Antonczak S, Ruizlopez MF, Rivail JL. Ab-initio analysis of water-assisted reaction-mechanisms in amide hydrolysis. J Am Chem Soc. 1994;116:3912–3921. [Google Scholar]
- 111.Hernández G, Anderson JS, LeMaster DM. Electrostatic stabilization and general base catalysis in the active site of the human protein disulfide isomerase a domain monitored by hydrogen exchange. ChemBioChem. 2008;9:768–778. doi: 10.1002/cbic.200700465. [DOI] [PubMed] [Google Scholar]
- 112.Laurindo FRM, Pescatore LA, Fernandes DdeC. Protein disulfide isomerase in redox cell signaling and homeostasis. Free Radic Biol Med. 2012;52:1954–1969. doi: 10.1016/j.freeradbiomed.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 113.Appenzeller-Herzog C, Ellgaard L. In vivo reduction-oxidation state of protein disulfide isomerase: The two active sites independently occur in the reduced and oxidized forms. Antioxid Redox Signal. 2008;10:55–64. doi: 10.1089/ars.2007.1837. [DOI] [PubMed] [Google Scholar]
- 114.Raturi A, Vacratsis PO, Seslija D, Lee L, Mutus B. A direct, continuous, sensitive assay for protein disulphide-isomerase based on fluorescence self-quenching. Biochem J. 2005;391:351–357. doi: 10.1042/BJ20050770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Grossfield A (2013) WHAM: The Weighted Histogram Analysis Method (Department of Biochemistry and Biophysics, University of Rochester Medical Center, Rochester, NY), Version 2.0.9.
- 116.Darby NJ, Creighton TE. Functional properties of the individual thioredoxin-like domains of protein disulfide isomerase. Biochemistry. 1995;34:11725–11735. doi: 10.1021/bi00037a009. [DOI] [PubMed] [Google Scholar]
- 117.Mizunaga T, Katakura Y, Miura T, Maruyama Y. Purification and characterization of yeast protein disulfide isomerase. J Biochem. 1990;108:846–851. doi: 10.1093/oxfordjournals.jbchem.a123291. [DOI] [PubMed] [Google Scholar]
- 118.Gilbert HF. Protein disulfide isomerase. Methods Enzymol. 1998;290:26–50. doi: 10.1016/s0076-6879(98)90005-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.