Significance
The human fungal pathogen Candida albicans uses the Target of Rapamycin (TOR) signaling pathway to contend with varying host environments and thereby, regulate cell growth. Seeking unknown components of the C. albicans TOR pathway, we identified a phosphate importer, Pho84, and its molecular link to Target of Rapamycin complex 1 (TORC1). Because phosphorus is a critical element for anabolic processes, like DNA replication, ribosome biogenesis, translation, and membrane biosynthesis, TORC1 monitors its availability in regulating these processes. By depleting the central kinase in the TORC1 pathway, we showed that TORC1 signaling modulates regulation of phosphate acquisition. A Food and Drug Administration-approved small molecule inhibitor of Pho84 inhibits TORC1 signaling and potentiates the activity of the antifungals amphotericin B and micafungin.
Keywords: ribosomal protein S6 phosphorylation, antifungal, amphotericin B, echinocandin, drug potentiation
Abstract
The Target of Rapamycin (TOR) pathway regulates morphogenesis and responses to host cells in the fungal pathogen Candida albicans. Eukaryotic Target of Rapamycin complex 1 (TORC1) induces growth and proliferation in response to nitrogen and carbon source availability. Our unbiased genetic approach seeking unknown components of TORC1 signaling in C. albicans revealed that the phosphate transporter Pho84 is required for normal TORC1 activity. We found that mutants in PHO84 are hypersensitive to rapamycin and in response to phosphate feeding, generate less phosphorylated ribosomal protein S6 (P-S6) than the WT. The small GTPase Gtr1, a component of the TORC1-activating EGO complex, links Pho84 to TORC1. Mutants in Gtr1 but not in another TORC1-activating GTPase, Rhb1, are defective in the P-S6 response to phosphate. Overexpression of Gtr1 and a constitutively active Gtr1Q67L mutant suppresses TORC1-related defects. In Saccharomyces cerevisiae pho84 mutants, constitutively active Gtr1 suppresses a TORC1 signaling defect but does not rescue rapamycin hypersensitivity. Hence, connections from phosphate homeostasis (PHO) to TORC1 may differ between C. albicans and S. cerevisiae. The converse direction of signaling from TORC1 to the PHO regulon previously observed in S. cerevisiae was genetically shown in C. albicans using conditional TOR1 alleles. A small molecule inhibitor of Pho84, a Food and Drug Administration-approved drug, inhibits TORC1 signaling and potentiates the activity of the antifungals amphotericin B and micafungin. Anabolic TORC1-dependent processes require significant amounts of phosphate. Our study shows that phosphate availability is monitored and also controlled by TORC1 and that TORC1 can be indirectly targeted by inhibiting Pho84.
Organisms that fail to maximize growth in response to abundant nutrients can be outcompeted by those that do. Conversely, organisms that fail to cease growth and induce survival programs during stress and starvation lose viability. The Target of Rapamycin (TOR) signaling pathway is conserved in eukaryotes and integrates multiple channels of information regarding the cells’ nutritional and physical environment to induce either growth and proliferation or stress and survival responses (1). In the human fungal pathogen Candida albicans, TOR participates in regulating morphogenesis (2–7) and responses to host cells (8). To control growth, C. albicans TOR also integrates signals of carbon source availability from the PKA pathway with nitrogen source status, its primary nutritional input (9). Similar TOR–PKA intersections have been reported in the model yeast Saccharomyces cerevisiae (10, 11).
In S. cerevisiae, Target of Rapamycin complex 1 (TORC1), which is susceptible to inhibition by rapamycin, is activated by preferred nitrogen sources, such as glutamine and leucine. Leucine activates TORC1 through the Exit from G0 (EGO) complex by inducing GTP loading of one of its subunits, the small GTPase Gtr1 (12). Leucine also promotes TORC1 activity through leucine–tRNA synthase Cdc60, which physically interacts with Gtr1 (13). Many transcriptional regulators responsive to S. cerevisiae and C. albicans TORC1 pathways are conserved (4), but there are important differences between these species as well.
A small Ras-like GTPase upstream of TORC1, Rheb in mammals (14) and Rhb1 in S. cerevisiae (15), also responds to nutritional signals. Rheb is a central activator of mammalian TORC1 and modulated by the TSC1/TSC2 complex (16, 17), whereas in S. cerevisiae, Rhb1 seems to play a minor role in nutritional signaling. In C. albicans, Rhb1 is required for normal tolerance to rapamycin and phosphorylation of ribosomal protein S6, a readout of TORC1 activation (9, 18). C. albicans Rhb1 coregulates expression of genes important in nitrogen source uptake (18) and virulence (19). Unlike S. cerevisiae, C. albicans also has a TSC2 homolog with mutant phenotypes that are consistent with a conserved GTPase-activating activity of C. albicans Tsc2 for Rhb1 (18).
Given the differences between the S. cerevisiae and C. albicans TORC1 pathways, we used a forward genetic approach to find components of C. albicans TORC1 signaling. Using our mariner transposon mutant collection (20), we isolated a rapamycin hypersensitive mutant in a C. albicans homolog of PHO84, the gene encoding the major S. cerevisiae high-affinity phosphate (Pi) transporter.
Having identified a connection between C. albicans Pho84 and the C. albicans TOR pathway in a forward genetic screen, we characterized this link between phosphate homeostasis (PHO) and the cell’s central growth control module. We found that we can indirectly target C. albicans TORC1 using small molecule Pho84 inhibitors, one of which is a Food and Drug Administration (FDA)-approved antiviral drug, and that the antifungals amphotericin B and micafungin are potentiated by Pho84 inhibitors.
Results
A Screen of Haploinsufficient Transposon Mutants for Altered Rapamycin Susceptibility Identified a PHO84 Ortholog.
We screened our heterozygous mutant collection of mariner-transposon insertions marked with our dominant selectable marker NAT1 (20, 21) for altered rapamycin susceptibility. We isolated a transposon mutant hypersensitive to rapamycin, in which the transposon disrupts the promoter of orf19.655 67 bp upstream of the predicted translational start site (SI Appendix, Fig. S1A). This ORF encodes a protein with 66% amino acid identity to S. cerevisiae Pho84 and 55% amino acid homology to the Piriformospora indica PiPT phosphate transporter with a crystal structure that was recently described (22) (SI Appendix, Fig. S1B). According to the Candida Genome Database (CGD) nomenclature, we called this ORF C. albicans PHO84 and used the CGD sequence for additional analysis (23). To confirm that rapamycin hypersensitivity of the transposon mutant was linked to the disrupted PHO84 locus, two independent heterozygous deletion mutants and their homozygous null derivatives were constructed. Mutant phenotypes in these two lineages were the same, and one was chosen for additional characterization (SI Appendix, Fig. S1C). These mutants were also rapamycin hypersensitive (Fig. 1A), confirming that PHO84 is required for normal tolerance of rapamycin.
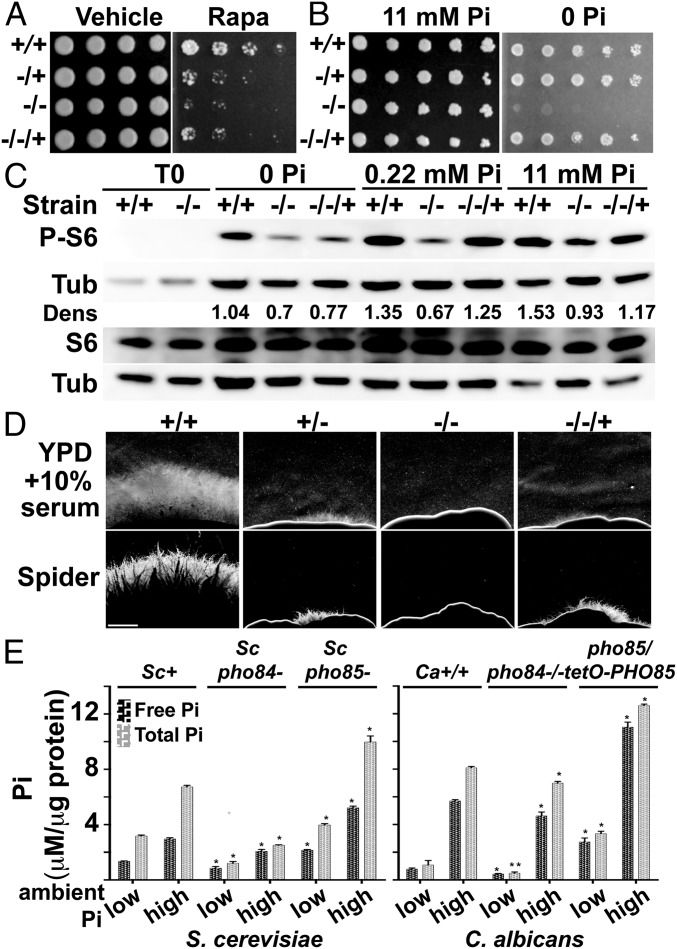
Fig. 1.
C. albicans PHO84 is required for rapamycin tolerance, growth during Pi starvation, normal TORC1 activity and hyphal morphogenesis, and Pi homeostasis. (A) Cell dilutions of WT and a mutant series in PHO84 pinned onto YPD with vehicle or 12 ng/mL rapamycin. PHO84+/+, JKC915; pho84−/+, JKC1583; pho84−/−, JKC1450; pho84−/−/+, JKC1588. (B) Cells as in A pinned onto YNB with 0 or 11 mM Pi. (C) Separate Western blots of the same samples for P-S6, total Rps6, and tubulin of WT (+/+; JKC915), pho84 null (−/−; JKC1450), and PHO84 reintegrant (−/−/+; JKC1588) cells grown in YNB with 0, 0.22, or 11 mM KH2PO4 for 90 min. Dens, densitometric ratio of P-S6 vs. tubulin signal. (D) Strains as in A spotted at equidistant points around agar plates with spot edges imaged. Compare with SI Appendix, Fig. S2A. (Scale bar: 1 mm.) (E) S. cerevisiae and C. albicans WT and pho84 null cells grown in SC medium with 0.22 mM (low Pi) or 11 mM Pi (high Pi) overnight assayed for free and total Pi; pho85 null cells as controls, which hyperaccumulate Pi. Ca+/+ (C. albicans PHO84/PHO84 PHO85/PHO85), JKC915; Capho84−/−, JKC1450; Capho85−/−, CaLC1919 grown in 20 μg/mL doxycycline overnight; Sc+ (S. cerevisiae PHO84 PHO85), BY4741; Scpho84−, EY2960; Scpho85−, from ref. 37. Error bars: SDs of three technical replicates. *P < 0.01; **P < 0.05.
The cytoplasmic membrane protein Pho84 is the major Pi transporter in S. cerevisiae (24–26). PHO84 expression is controlled by the PHO regulon, a homeostatic system that maintains Pi availability for metabolism and growth in fluctuating external Pi conditions (27). C. albicans pho84 mutants, like those in the S. cerevisiae homolog (28), failed to grow on medium without inorganic phosphate (Fig. 1B). Heterozygous and reintegrant cells, apparently haploinsufficient for rapamycin tolerance (Fig. 1A), grew robustly on 0 Pi medium (Fig. 1B), indicating that mechanisms other than haploinsufficiency affect growth during Pi depletion, like the feedback loops between expression of high- and low-affinity Pi transporters characterized in detail in the S. cerevisiae PHO regulon (28). In liquid media with 1 mM Pi, growth of pho84 cells was close to the WT (SI Appendix, Fig. S1D). Expression of the C. albicans PHO84 homolog restored growth on medium lacking Pi to S. cerevisiae pho84 mutants (SI Appendix, Fig. S1E), indicating functional orthology. WT cells secrete acid phosphatase in response to low ambient Pi to mobilize covalently bound Pi from their environment, and this response was used for decades in studies of the S. cerevisiae PHO regulon (29). C. albicans pho84−/− mutants, like those in S. cerevisiae (24), inappropriately derepressed acid phosphatase secretion in high ambient Pi (SI Appendix, Fig. S1F), consistent with a conserved role of C. albicans Pho84 in the PHO regulon.
C. albicans Pho84 Is Required for the Normal TORC1 Response to Pi Availability.
We then examined the relationship between Pho84 and TORC1. To test whether rapamycin hypersensitivity is caused by decreased TORC1 kinase activity in the pho84−/− mutant, we monitored the phosphorylation state of ribosomal protein S6 (P-S6), which we previously showed is controlled by TORC1 signaling (9). Null mutants in PHO84 had a weaker P-S6 signal than the WT during Pi refeeding at every Pi concentration of the media, although they responded to increasing Pi concentrations with an increasing P-S6 signal (Fig. 1C). Pho84, therefore, is required for normal anabolic TORC1 signaling, and TORC1 activity responds to ambient Pi availability.
Heterozygous and Homozygous Deletion Mutants in PHO84 Are Defective in Hyphal Morphogenesis.
TORC1 regulates hyphal morphogenesis in C. albicans (2–7, 30), an important virulence determinant. Hyphal morphogenesis was defective in pho84 mutants on yeast extract peptone dextrose (YPD) agar medium with 10% serum, Spider medium, and RPMI 1640 (Fig. 1D and SI Appendix, Fig. S2A), whereas the mutant and the WT grew equally in these media when the hyphal temperature signal was absent (SI Appendix, Fig. S2B). Although many signaling pathways converge on morphogenesis, these findings are formally consistent with defective regulation by TORC1 (2).
Pi Content of Cells Lacking Pho84 Is Diminished.
We asked if C. albicans TORC1 activity may be down-regulated in response to decreased intracellular Pi in pho84 mutants, analogous to the response of S. cerevisiae TORC1 to decreased intracellular amino acids. Using pho85 mutants as controls known to hyperaccumulate intracellular Pi (31), we found that intracellular Pi concentrations were lower in pho84−/− null than WT cells in low- and high-Pi–containing media, although the difference was substantially less than in the homologous S. cerevisiae mutant–WT pair (Fig. 1E). Diminished intracellular Pi concentrations of C. albicans pho84−/− cells may be responsible for the decreased TORC1 activation state, possibly in addition to the lack of a putative TORC1-activating function performed specifically by Pho84.
Gtr1 Links Pho84 to TORC1 in C. albicans.
Seeking a molecular link between Pho84 and TORC1 activity, we considered the possibility that Gtr1 may connect Pho84 to TORC1. GTR1 was first described for its functional and physical proximity to S. cerevisiae PHO84 (32, 33), and its product later was characterized as a component of the TORC1-activating EGO complex (12, 13, 34, 35). We found that the P-S6 response of gtr1−/− cells to phosphate refeeding was blunted (Fig. 2A). To determine whether this is an unspecific effect of decreased upstream TORC1 signaling, mutants in another small TORC1-activating GTPase, RHB1, were tested; rhb1 mutants responded to Pi refeeding like the WT (Fig. 2B), suggesting that a Pi signal to TORC1 is transmitted specifically through Gtr1.
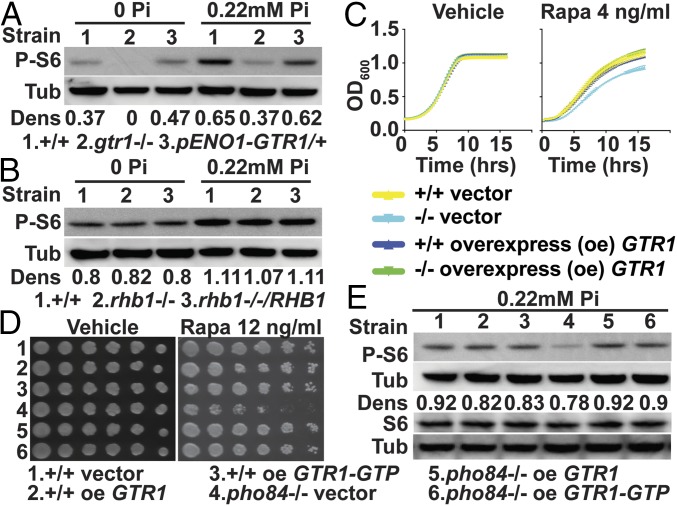
Fig. 2.
Gtr1 links Pho84 to TOR in C. albicans. (A) Western blot of WT (SC5314), gtr1−/− (YGM367), and pENO1-GTR/GTR1 (YGM365) cells grown in YNB with 0 and 0.22 mM KH2PO4 for 90 min. (B) Western blot of WT (SC5314), rhb1−/− (CCT-D1), and rhb1−/−/pADH1-RHB1 (CCT-OE1) cells grown in YNB with 0 or 0.22 mM KH2PO4 for 90 min. (C) Growth in YPD with 4 ng/mL rapamycin or vehicle. OD600 monitored every 15 min. Blue, WT overexpressing GTR1 (JKC1596); cyan, pho84−/− with vector (JKC1598); green, pho84−/− overexpressing GTR1 (JKC1600); yellow, WT with vector (JKC1594). (D) Cell dilutions pinned onto YPD with vehicle or 12 ng/mL rapamycin. Strains WT with vector (JKC1594; 1), WT overexpressing GTR1 (JKC1596; 2), WT overexpressing GTR1-GTP (JKC1619; 3), pho84−/− with vector (JKC1598; 4), pho84−/− overexpressing GTR1 (JKC1600; 5), and pho84−/− overexpressing GTR1-GTP (JKC1616; 6). (E) Western blot of cells grown in YNB with 0.22 mM KH2PO4 for 90 min. Strains 1–6 as in D. Dens, densitrometric ratio of P-S6 vs. tubulin signal intensity.
If Gtr1 acts downstream of Pho84 in activating TORC1, its overexpression may suppress pho84−/− phenotypes. GTR1 was overexpressed from the ACT1 promoter in WT and pho84−/− C. albicans cells. Compared with rapamycin hypersensitive pho84−/− cells transformed with the empty vector, the resulting pho84−/− pACT1-GTR1 cells showed WT tolerance to rapamycin, suggesting recovery of their TORC1 signaling activity (Fig. 2 C and D). To investigate this possibility, TORC1 activity was tested directly by comparing the P-S6 signal of pho84−/− cells transformed with the empty vector with that of pho84−/− pACT1-GTR1 cells. Overexpression of GTR1 recovered Rps6 phosphorylation in pho84−/− cells nearly to WT levels (Fig. 2E). A GTR1 mutant encoding constitutively GTP-bound Gtr1Q67L, homologous to S. cerevisiae Gtr1Q65L (12, 35, 36), was then constructed and overexpressed from the ACT1 promoter. This GTR1-GTP allele suppressed the TORC1 signaling defect of pho84−/− cells apparently equally to the overexpressed WT GTR1 (Fig. 2 D and E). Overexpression of GTR1 or GTR1-GTP did not increase phosphorylation of S6 in WT cells (Fig. 2E). These findings are consistent with the model that, in C. albicans, Gtr1 indirectly or directly conveys a Pi signal to TORC1 and links Pho84 to TORC1 signaling.
We examined the relationship of Pho84 to TORC1 activity in the model yeast S. cerevisiae. A pho84 null mutant in the S288C genetic background (37) was hypersensitive to rapamycin (SI Appendix, Fig. S3A) at an intermediate ambient Pi concentration (1 mM). Of note, the rapamycin phenotype was highly responsive to the Pi concentration of pregrowth media. Rapamycin hypersensitivity was not suppressed, but the S. cerevisiae Sch9 phosphorylation state (36) (SI Appendix, Fig. S3B) and P-S6 signal intensity, which in S. cerevisiae, also responds to TORC1 activation (Fig. 3 and SI Appendix, Fig. S3C), were recovered by constitutively active Gtr1 in pho84 null cells. These findings suggest that Pi homeostasis and TOR signaling are linked in S. cerevisiae as in C. albicans, although specific molecular connections seem to have divergently evolved in these two fungi.
Fig. 3.
A connection between Pho84 and TORC1 is conserved in S. cerevisiae. Western blot of PHO84 with vector (Y1597; 1), pho84 with vector (Y1599; 2), PHO84 with GTR1-GTP GTR2-GDP (Y1596; 3), and pho84 with GTR1-GTP GTR2-GDP (Y1604; 4) cells grown in SC (-His-Ura) containing 0.22 and 11 mM KH2PO4 for 90 min probed for P-S6, S6, and tubulin. Dens, densitrometric ratio of P-S6 vs. tubulin signal intensity.
TORC1 Modulates the PHO Regulon.
Because TORC1 not only responds to nutrient availability but also, directs nutrient uptake (e.g., by regulating expression of amino acid and ammonium transporters), we questioned whether it may play a similar role in phosphate acquisition. Given known discrepancies between rapamycin exposure and physiological TOR modulation (38–41), we examined this potential connection genetically. Repressible tetO was used to control expression of C. albicans TOR1 or a hypomorphic TOR1Δ1–381 encoding a protein lacking the first 381 amino acids which form protein–protein interaction repeat domains. The effect of TOR1 depletion on expression of PHO84 was then examined.
When WT cells were transferred from overnight cultures into fresh rich medium, PHO84 mRNA levels dropped in accordance with the PHO regulon’s response to availability of fresh Pi sources. In cells depleted of either the WT or the N-terminally truncated TOR1 allele, PHO84 expression also decreased but to a significantly lesser extent (SI Appendix, Fig. S4A). Full-length TOR1 permitted greater PHO84 expression than the TOR1Δ1–381 allele, suggesting that structural perturbation of TORC1 by truncation of Tor1 affects its inhibitory as well as activating functions. Active TORC1, signaling nutritional repletion, hence contributes input to the PHO regulon to down-regulate Pi starvation responses, whereas loss of TORC1 activity conveys a starvation signal to dampen these responses (SI Appendix, Fig. S4A). Similarly, overexpression of Gtr1 and Gtr1-GTP blunted up-regulation of secreted acid phosphatase in pho84−/− cells (SI Appendix, Fig. S4B), supporting the model that, in response to Pi, TORC1 signaling down-regulates the PHO regulon to integrate its activity with availability of other nutrients.
Small Molecule Inhibitors of Pho84 Repress TORC1 and Potentiate Antifungal Activity.
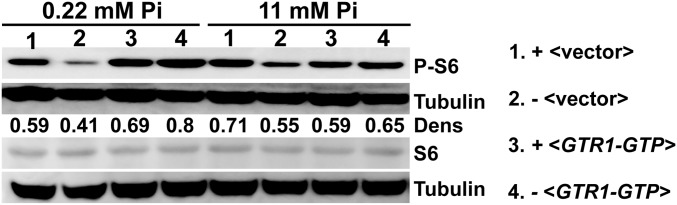
S. cerevisiae Pho84 has been characterized as a Pi transceptor signaling to PKA through identification of point mutations and small molecules that preferentially perturb transport, signaling, or both (42, 43). Direct pharmacological inhibition of C. albicans TORC1 with rapamycin incurs too high a cost on host immune function to be clinically useful (44). We tested whether blocking Pho84 with its known small molecule inhibitors phosphonoformic acid [foscarnet (Fos)] and phosphonoacetic acid (PAA) (42), which we showed inhibit C. albicans growth in dependence on the presence of their target Pho84 (SI Appendix, Fig. S5 A and B), can indirectly inhibit C. albicans TORC1. Exposure of WT cells to the FDA-approved antiviral Fos inhibited Rps6 phosphorylation (P-S6) in a dose-dependent manner (Fig. 4A) at Fos concentrations attained in human plasma during antiviral therapy (45, 46). In heterozygous cells (pho84−/+) with haploinsufficiency phenotypes that likely reflect decreased copies of the drug target, Pho84, P-S6 was hypersensitive to Fos (Fig. 4A). In cells lacking the target Pho84, exposure to Fos did not further decrease the P-S6 signal (Fig. 4A). Pho84 inhibition with small molecules also recapitulated the hyphal growth defect seen in cells genetically depleted of PHO84 (Figs. 1D and 4B and SI Appendix, Fig. S5C).
Fig. 4.
Small molecule inhibition of Pho84 represses TORC1 and hyphal morphogenesis and potentiates amphotericin B and micafungin. (A) Western blot of WT (SC5314) cells with vehicle (1), 100 μM Fos (2), 200 μM Fos (3), and 400 μM Fos (4) and strains PHO84+/+ (JKC915; 5, 7, 9, and 11), pho84−/+ (JKC1583; 6 and 8), and pho84−/− (JKC1450; 10 and 12) grown in standard SC (7.3 mM Pi) for 60 min and probed for P-S6 and tubulin. Dens, densitrometric ratio of P-S6 vs. tubulin signal intensity. (B) WT (SC5314) spotted at equidistant points around RPMI agar (0.22 mM KH2PO4, pH 7) containing vehicle, 8 mM PAA, or 500 μM Fos grown at 37 °C, and spot edges were imaged. Compare with SI Appendix, Fig. S2A. (Scale bar: 1 mm.) (C) WT (SC5314) was exposed to vehicle, 500 μM Fos, 0.2 μg/mL amphotericin B, and amphotericin B plus Fos; OD600 in SC with 0.5 mM KH2PO4 at 30 °C was monitored every 15 min. (D) WT (SC5314) was exposed to vehicle, 500 μM Fos, 25 ng/mL micafungin, and micafungin plus Fos; OD600 in SC with 0.5 mM KH2PO4 at 30 °C was monitored every 15 min. Dens, densitrometric ratio of P-S6 vs. tubulin signal intensity.
Potentiating existing antifungals is a promising strategy (47–49). Fos at concentrations reached in plasma during antiviral therapy (45, 46) and PAA potentiate activity of the antifungal amphotericin B at concentrations of the latter far below those in serum or tissue during standard dosing (50) (Fig. 4C and SI Appendix, Fig. S5D). Activity of the antifungal micafungin, belonging to the distinct drug class of echinocandins, was also potentiated (Fig. 4D). Because Pho84 is not conserved in mammals, inhibition of Pho84 offers a nonconventional approach to fungal-specific TORC1 inhibition (Fig. 5) and antifungal potentiation as shown in our proof of principle experiments with PAA and Fos.
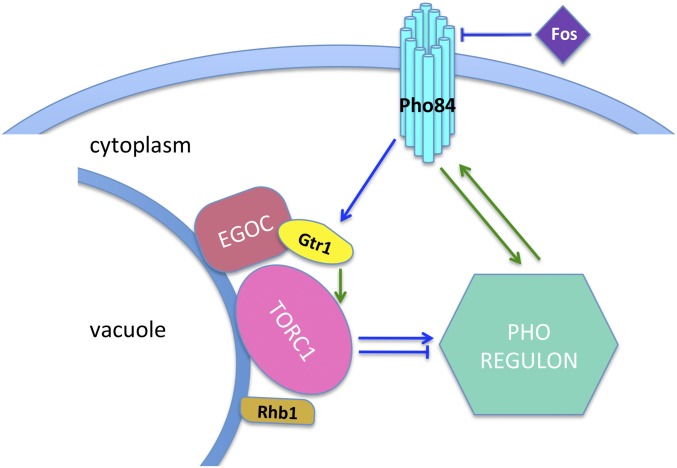
Fig. 5.
Pho84 activates TORC1 via Gtr1, and TORC1, in turn, modulates the PHO regulon. Fos inhibits Pho84 and thereby, indirectly blocks TORC1 activity. Signaling events with known molecular mechanisms in S. cerevisiae are shown as green lines. Blue lines represent predicted activities based on our findings.
Discussion
Eukaryotic cell mass consists to 0.5–1% of phosphorus (51). However, phosphorus constitutes only 0.07% of the Earth’s crust (51), and only ∼15% of soil phosphorus is bioavailable in a soluble form (52). Fungi, like plants and bacteria, have sophisticated regulatory networks to manage Pi starvation. S. cerevisiae cells use the TORC1 and PKA pathways to ensure orderly cessation of growth when nitrogen or carbon sources become limiting (53, 54). C. albicans cells similarly use the TORC1 pathway for calibrated responses to distinct nitrogen sources, with modulating input from PKA according to carbon source type and concentration (9).
Our study shows that C. albicans TORC1 monitors not only nitrogen and carbon source but also, Pi availability (Figs. 1C and 2 A and E). Whether Pho84 activates TORC1 indirectly by an increased intracellular Pi content, directly in a transceptor role, or through a combination of these inputs remains an open question. Our results establish the small GTPase Gtr1, first discovered in S. cerevisiae because of its mutant phenotypes’ resemblance to those of Pho84 (32), as an important element linking C. albicans Pho84 and TORC1 (Fig. 2). How Gtr1, a subunit of the TORC1-associated vacuolar membrane-residing EGO complex (55), receives information from Pho84 remains to be determined. Possible models include an activating signal to the TORC1-activating Seh1-associated complex activating TORC1 (SEACAT) complex (55) through a sensor of cytosolic Pi or vacuolar polyphosphate or a physical interaction between Pho84 and the EGO complex on endosomes while Pho84 is internalized during ambient Pi abundance (56). We cannot exclude alternative explanations of our findings (e.g., a TORC1 response only to intracellular Pi concentrations and up-regulation of low-affinity Pi transporters during GTR1 overexpression in pho84−/− null cells). If Gtr1 is activated by Pho84, this GTPase’s role in the input to TORC1 regarding the cell’s Pi state seems to be specific (Fig. 2), because there was no perturbation of TORC1 activation during Pi refeeding in rhb1−/− cells (Fig. 2B).
In S. cerevisiae, Pho84 signals to PKA as a transceptor (42, 57). A signal from Pho84 to TORC1 has not yet been described in S. cerevisiae. We found that, in the genetic background S288C, loss of Pho84 leads to rapamycin hypersensitivity and TORC1 inactivation as it does in C. albicans (SI Appendix, Fig. S3). Sch9 phosphorylation, like that of Rps6, is known to correspond with the TORC1 activation state (36, 58). Overexpression of constitutively active Gtr1 restores TORC1 activity in S. cerevisiae as assayed by Sch9 and Rps6 phosphorylation (Fig. 3 and SI Appendix, Fig. S3B). However, in contrast to C. albicans, constitutively active Gtr1 did not suppress rapamycin hypersensitivity of S. cerevisiae pho84 null cells, indicating differences in the connections of PHO and TORC1 signaling between these fungi. In S. cerevisiae, control of entry into quiescence (G0) by the Rim15 kinase is coregulated by the PHO pathway cyclin/cyclin-dependent kinase module Pho80/Pho85 as well as TORC1 (59), suggesting multiple levels of cross-talk between phosphate-specific and global nutritional signaling pathways in that organism, which remain to be explored in C. albicans.
In addition to the signal from Pho84 to TORC1, a signal in the opposite direction was seen from TORC1 to expression of PHO84 and the classic readout of the PHO regulon, the acid phosphatase (27) (SI Appendix, Fig. S4A), although clearly, TORC1 input contributes only to a fraction of the PHO regulon responses. TORC1 is well-known to regulate proteins required for acquisition of other nutrients, like amino acids and ammonium, in S. cerevisiae (60, 61). TORC1 input to the PHO regulon may fine-tune the investment of energy and nutrients in Pi acquisition to match the overall state of the cell. In S. cerevisiae, transcriptional regulation favoring Pi acquisition is achieved through the transcription factor Pho4 (31). A C. albicans homolog of Pho4, required for stress resistance (62, 63) and commensalism in a murine model (63), was recently shown to control C. albicans PHO84 expression (62). How C. albicans Pho4 may be coregulated by TORC1 remains to be determined.
Although TORC1 monitors nutrient availability in mammals as in fungi, its relationship with the PHO regulon may have diverged in these phyla. In fungi, H+-Pi symport is the major form of Pi import, because the steep Pi concentration gradient at the plasma membrane imposes a high energetic demand on transport, which is met by the electrochemical proton gradient generated by the P-type H+ ATPase (52). In animals, Na+-Pi pumps predominate (64). Humans largely consume Pi together with amino acids in food consisting of other eukaryotes and their products, so that Pi starvation tends to occur during starvation for protein (65). In contrast, Pi and nitrogen sources must be acquired independently by unicellular organisms, and therefore, the connection that we discovered between the fungal H+-Pi symporter and the TORC1 pathway seems physiologically plausible.
We found that TORC1 is indirectly repressed during Pho84 inhibition with small molecules (Fig. 4 and SI Appendix, Fig. S5). Because TORC1 components are conserved between fungi and humans, whereas Pho84 has no human homologs, this result provides an option for indirect fungal-specific TORC1 inhibition not previously explored in the search for new antifungals. Although TORC1 is only partially inhibited in this manner, other fungal-specific indirect TORC1 activators may provide synergistic targets. More immediately, Pho84 inhibitors potentiate the antifungal effect of micafungin and amphotericin B (Fig. 4 C and D), possibly permitting lower dosing of the latter “gold standard” broad-spectrum agent to obviate its often treatment-limiting toxicities (66). We used two of multiple Pho84 inhibitors previously characterized (42) (Fig. 4 and SI Appendix, Fig. S5D), and more specific, noncompetitive inhibitors with more favorable therapeutic indices than Fos may be found through screening efforts. Because Pho84 homologs are highly conserved among fungi, potentiating amphotericin B activity through their inhibition may prove a viable therapeutic strategy for other fungal species less amenable to other antifungal agents.
Materials and Methods
Strains and Culture Conditions.
The C. albicans and S. cerevisiae strains, plasmids, and primers used are described in SI Appendix, Tables S1 and S2. C. albicans strains were generated using HIS1 and ARG4 markers and strains as described in ref. 67 as well as the CaNAT1 selectable marker as described in refs. 20 and 21. Two independent heterozygotes were used to derive homozygous null and reintegrant mutants of PHO84 as well as tetO-TOR1 mutants. Auxotrophies were complemented, so that only prototrophic strains were compared in an experiment. Introduced mutations were confirmed by PCR spanning the upstream and downstream homologous recombination junctions of transforming constructs and sequencing. Experiments with defined ambient Pi concentrations were performed in yeast nitrogen base (YNB) 0 Pi (ForMedium Ltd) with added KH2PO4 to stated concentrations. Other media were used as in ref. 20.
Screening Transposon Mutants for Altered Rapamycin Susceptibility.
Our heterozygous mutant collection containing a mariner transposon marked with CaNAT1 (20) was used. Mutants were pregrown at room temperature in 96-well plates containing 2× YPD with 8% glucose to minimize hyphal growth. Cells were replicated to YPD agar with vehicle (90% ethanol) or 20 ng/mL rapamycin. Clones showing less growth than the WT (SC5314) were isolated as rapamycin hypersensitive. The transposon insertion site was identified by vectorette PCR (20).
Growth Assays.
For cell dilutions spotted onto agar media as previously described (20), saturated overnight cultures were diluted in fivefold steps from an OD600 of 0.5. For growth curves in liquid media, saturated overnight cultures in YPD were washed once in 0.9% NaCl and diluted to an OD600 of 0.15 in 150 μL medium in flat-bottomed 96-well dishes. For growth assays, including those during drug exposure, OD600 readings were obtained every 15 min in a plate reader, and SDs of three technical replicates were calculated and graphed in Graphpad Prism. Growth during drug exposure was assayed in synthetic complete (SC) medium. Vehicle for Pho84 inhibitors PAA (284270; Sigma) and Fos (SC-253593A; Santa Cruz Biotechnology) was water, and vehicle for amphotericin B (A9528; Sigma) was DMSO. All panels shown represent at least three biological replicates.
Western Blots.
Cell harvesting, lysis, and Western blotting were performed as described in ref. 9. Antibodies are listed in SI Appendix, Table S1. At least three biological replicates were obtained for each experiment shown. For densitometry, ImageJ (https://imagej.net/Downloads) software (open source) was used as in ref. 9.
Hyphal Morphogenesis Assay.
Cells were revived from frozen stocks on solid YPD overnight, washed, and resuspended in 0.9% NaCl to OD600 0.1. Variations between single colonies and colony density effects were minimized by spotting 3-μL cell suspension at four or six equidistant points using a template around the perimeter of an agar medium plate as in ref. 20. For small molecule Pho84 inhibitor effects on hyphal formation, Spider and RPMI were used (R8999-04; TOKU-E), the latter with 0.22 mM KH2PO4 buffered to pH 7 with 50 mM Mops. All panels shown represent at least three biological replicates.
Acid Phosphatase Assays.
As adapted from ref. 25, overnight cultures in SC were diluted to an OD600 of 0.05 into YNB medium buffered to pH 4 with 50 mM sodium citrate containing 0 or 11 mM KH2PO4 and grown overnight. P-Nitrophenyl Phosphate (N4645; Sigma) was added to washed cells to a concentration of 5.62 mg/mL. After 15 min at room temperature, the reaction was stopped with Na2CO3 (pH 11) to a concentration of 0.3 g/mL, and OD420 and OD600 were measured. At least three biological replicates with three technical replicates each were obtained.
Intracellular Pi Assays.
Free and total Pi was measured by colorimetric molybdate assay as described (68). Briefly, cultures were washed with distilled water twice, resuspended in 500 μL 0.1% Triton X-100, and lysed by glass bead homogenization. Lysate protein content was determined using a BioRad Protein Assay kit. Free Pi was measured in unboiled lysate; then, total phosphate was measured after boiling 3–30 μg whole-cell lysate for 10 min in 0.5 M H2SO4. At least three biological replicates with three technical replicates each were obtained.
RT-PCR Expression Analysis.
Cells were grown overnight in YPD medium with 5 ng/mL doxycycline, diluted into YPD with 30 μg/mL doxycycline, and harvested at time 0 and 2 and 4 h. RNA was extracted with the Direct-Zol RNA miniprep kit (R2051; Zymo Research). RT-PCR procedures were performed as indicated (69).
Supplementary Material
Acknowledgments
We thank Gerald R. Fink, Erin K. O’Shea, Bin He, Charles Boone, Françoise Stutz, Kevin Struhl, and Fred Winston for plasmids and S. cerevisiae strains; Leah Cowen, Chung-Yu Lan, and Rajini Rao for C. albicans strains; Gerald R. Fink, Felix Lam, Valmik Vyas, Luke Whitesell, and Bin He for helpful conversations; Brian M. Hoffman, Valeria C. Culotta, and Amit R. Reddi for protocols; and Dennis Wykoff, Bin He, Robert Husson, and Paula Watnick for critical reading of the manuscript. This work was funded by National Institute of Allergy and Infectious Diseases Grants R21AI096054 and R01AI095305. P.R.F. was supported by Science Foundation Ireland Grant 11/RFP.1/GEN/3044. M.E.C. was funded by National Cancer Institute Grant R01 CA154499.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617799114/-/DCSupplemental.
References
- 1.Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cutler NS, Pan X, Heitman J, Cardenas ME. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol Biol Cell. 2001;12:4103–4113. doi: 10.1091/mbc.12.12.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastidas RJ, Heitman J, Cardenas ME. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 2009;5:e1000294. doi: 10.1371/journal.ppat.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zacchi LF, Gomez-Raja J, Davis DA. Mds3 regulates morphogenesis in Candida albicans through the TOR pathway. Mol Cell Biol. 2010;30:3695–3710. doi: 10.1128/MCB.01540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Y, Su C, Wang A, Liu H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 2011;9:e1001105. doi: 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Y, Su C, Liu H. Candida albicans hyphal initiation and elongation. Trends Microbiol. 2014;22:707–714. doi: 10.1016/j.tim.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park H, et al. Transcriptional responses of candida albicans to epithelial and endothelial cells. Eukaryot Cell. 2009;8:1498–1510. doi: 10.1128/EC.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhury T, Köhler JR. Ribosomal protein S6 phosphorylation is controlled by TOR and modulated by PKA in Candida albicans. Mol Microbiol. 2015;98:384–402. doi: 10.1111/mmi.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedruzzi I, et al. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell. 2003;12:1607–1613. doi: 10.1016/s1097-2765(03)00485-4. [DOI] [PubMed] [Google Scholar]
- 11.Zurita-Martinez SA, Cardenas ME. Tor and cyclic AMP-protein kinase A: Two parallel pathways regulating expression of genes required for cell growth. Eukaryot Cell. 2005;4:63–71. doi: 10.1128/EC.4.1.63-71.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binda M, et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 13.Bonfils G, et al. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell. 2012;46:105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Yamagata K, et al. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269:16333–16339. [PubMed] [Google Scholar]
- 15.Urano J, Tabancay AP, Yang W, Tamanoi F. The Saccharomyces cerevisiae Rheb G-protein is involved in regulating canavanine resistance and arginine uptake. J Biol Chem. 2000;275:11198–11206. doi: 10.1074/jbc.275.15.11198. [DOI] [PubMed] [Google Scholar]
- 16.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 17.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsao CC, Chen YT, Lan CY. A small G protein Rhb1 and a GTPase-activating protein Tsc2 involved in nitrogen starvation-induced morphogenesis and cell wall integrity of Candida albicans. Fungal Genet Biol. 2009;46:126–136. doi: 10.1016/j.fgb.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Chen YT, et al. Rhb1 regulates the expression of secreted aspartic protease 2 through the TOR signaling pathway in Candida albicans. Eukaryot Cell. 2012;11:168–182. doi: 10.1128/EC.05200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen J, Cowen LE, Griffin AM, Chan L, Köhler JR. The Candida albicans pescadillo homolog is required for normal hypha-to-yeast morphogenesis and yeast proliferation. Proc Natl Acad Sci USA. 2008;105:20918–20923. doi: 10.1073/pnas.0809147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen J, Guo W, Köhler JR. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect Immun. 2005;73:1239–1242. doi: 10.1128/IAI.73.2.1239-1242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedersen BP, et al. Crystal structure of a eukaryotic phosphate transporter. Nature. 2013;496:533–536. doi: 10.1038/nature12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inglis DO, et al. The Candida genome database incorporates multiple Candida species: Multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res. 2012;40:D667–D674. doi: 10.1093/nar/gkr945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bun-Ya M, Nishimura M, Harashima S, Oshima Y. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol. 1991;11:3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wykoff DD, O’Shea EK. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics. 2001;159:1491–1499. doi: 10.1093/genetics/159.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auesukaree C, et al. Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J Biol Chem. 2004;279:17289–17294. doi: 10.1074/jbc.M312202200. [DOI] [PubMed] [Google Scholar]
- 27.Thomas MR, O’Shea EK. An intracellular phosphate buffer filters transient fluctuations in extracellular phosphate levels. Proc Natl Acad Sci USA. 2005;102:9565–9570. doi: 10.1073/pnas.0501122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wykoff DD, Rizvi AH, Raser JM, Margolin B, O’Shea EK. Positive feedback regulates switching of phosphate transporters in S. cerevisiae. Mol Cell. 2007;27:1005–1013. doi: 10.1016/j.molcel.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogel K, Hinnen A. The yeast phosphatase system. Mol Microbiol. 1990;4:2013–2017. doi: 10.1111/j.1365-2958.1990.tb00560.x. [DOI] [PubMed] [Google Scholar]
- 30.Lee CM, Nantel A, Jiang L, Whiteway M, Shen SH. The serine/threonine protein phosphatase SIT4 modulates yeast-to-hypha morphogenesis and virulence in Candida albicans. Mol Microbiol. 2004;51:691–709. doi: 10.1111/j.1365-2958.2003.03879.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee YS, Huang K, Quiocho FA, O’Shea EK. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat Chem Biol. 2008;4:25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bun-Ya M, Harashima S, Oshima Y. Putative GTP-binding protein, Gtr1, associated with the function of the Pho84 inorganic phosphate transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2958–2966. doi: 10.1128/mcb.12.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagerstedt JO, Reeve I, Voss JC, Persson BL. Structure and function of the GTP binding protein Gtr1 and its role in phosphate transport in Saccharomyces cerevisiae. Biochemistry. 2005;44:511–517. doi: 10.1021/bi048659v. [DOI] [PubMed] [Google Scholar]
- 34.Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152:853–867. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kingsbury JM, Sen ND, Cardenas ME. Branched-chain aminotransferases control TORC1 signaling in Saccharomyces cerevisiae. PLoS Genet. 2015;11:e1005714. doi: 10.1371/journal.pgen.1005714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kingsbury JM, Sen ND, Maeda T, Heitman J, Cardenas ME. Endolysosomal membrane trafficking complexes drive nutrient-dependent TORC1 signaling to control cell growth in Saccharomyces cerevisiae. Genetics. 2014;196:1077–1089. doi: 10.1534/genetics.114.161646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 38.Cox KH, Kulkarni A, Tate JJ, Cooper TG. Gln3 phosphorylation and intracellular localization in nutrient limitation and starvation differ from those generated by rapamycin inhibition of Tor1/2 in Saccharomyces cerevisiae. J Biol Chem. 2004;279:10270–10278. doi: 10.1074/jbc.M312023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puria R, Zurita-Martinez SA, Cardenas ME. Nuclear translocation of Gln3 in response to nutrient signals requires Golgi-to-endosome trafficking in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2008;105:7194–7199. doi: 10.1073/pnas.0801087105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puria R, Cardenas ME. Rapamycin bypasses vesicle-mediated signaling events to activate Gln3 in Saccharomyces cerevisiae. Commun Integr Biol. 2008;1:23–25. doi: 10.4161/cib.1.1.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kingsbury JM, Cardenas ME. Vesicular trafficking systems impact TORC1-controlled transcriptional programs in Saccharomyces cerevisiae. G3 (Bethesda) 2016;6:641–652. doi: 10.1534/g3.115.023911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popova Y, Thayumanavan P, Lonati E, Agrochão M, Thevelein JM. Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. Proc Natl Acad Sci USA. 2010;107:2890–2895. doi: 10.1073/pnas.0906546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samyn DR, et al. Mutational analysis of putative phosphate- and proton-binding sites in the Saccharomyces cerevisiae Pho84 phosphate:H(+) transceptor and its effect on signalling to the PKA and PHO pathways. Biochem J. 2012;445:413–422. doi: 10.1042/BJ20112086. [DOI] [PubMed] [Google Scholar]
- 44.Cruz MC, et al. Rapamycin and less immunosuppressive analogs are toxic to Candida albicans and Cryptococcus neoformans via FKBP12-dependent inhibition of TOR. Antimicrob Agents Chemother. 2001;45:3162–3170. doi: 10.1128/AAC.45.11.3162-3170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chrisp P, Clissold SP. Foscarnet. A review of its antiviral activity, pharmacokinetic properties and therapeutic use in immunocompromised patients with cytomegalovirus retinitis. Drugs. 1991;41:104–129. doi: 10.2165/00003495-199141010-00009. [DOI] [PubMed] [Google Scholar]
- 46.Minor JR, Baltz JK. Foscarnet sodium. DICP. 1991;25:41–47. doi: 10.1177/106002809102500109. [DOI] [PubMed] [Google Scholar]
- 47.Robbins N, et al. An antifungal combination matrix identifies a rich pool of adjuvant molecules that enhance drug activity against diverse fungal pathogens. Cell Reports. 2015;13:1481–1492. doi: 10.1016/j.celrep.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butts A, et al. A repurposing approach identifies off-patent drugs with fungicidal cryptococcal activity, a common structural chemotype, and pharmacological properties relevant to the treatment of cryptococcosis. Eukaryot Cell. 2013;12:278–287. doi: 10.1128/EC.00314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butts A, Palmer GE, Rogers PD. Antifungal adjuvants: Preserving and extending the antifungal arsenal. Virulence. 2017;8:198–210. doi: 10.1080/21505594.2016.1216283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamill RJ. Amphotericin B formulations: A comparative review of efficacy and toxicity. Drugs. 2013;73:919–934. doi: 10.1007/s40265-013-0069-4. [DOI] [PubMed] [Google Scholar]
- 51.Audesirk T, Audesirk G, Byers BE. Biology: Life on Earth. 5th Ed Prentice-Hall; Upper Saddle River, NJ: 1999. [Google Scholar]
- 52.Johri AK, et al. Fungal association and utilization of phosphate by plants: Success, limitations, and future prospects. Front Microbiol. 2015;6:984. doi: 10.3389/fmicb.2015.00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barbet NC, et al. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Broach JR. Nutritional control of growth and development in yeast. Genetics. 2012;192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Panchaud N, Péli-Gulli MP, De Virgilio C. SEACing the GAP that nEGOCiates TORC1 activation: Evolutionary conservation of Rag GTPase regulation. Cell Cycle. 2013;12:2948–2952. doi: 10.4161/cc.26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lagerstedt JO, et al. Mutagenic and functional analysis of the C-terminus of Saccharomyces cerevisiae Pho84 phosphate transporter. FEBS Lett. 2002;526:31–37. doi: 10.1016/s0014-5793(02)03109-5. [DOI] [PubMed] [Google Scholar]
- 57.Giots F, Donaton MC, Thevelein JM. Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 2003;47:1163–1181. doi: 10.1046/j.1365-2958.2003.03365.x. [DOI] [PubMed] [Google Scholar]
- 58.Urban J, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 59.Wanke V, Pedruzzi I, Cameroni E, Dubouloz F, De Virgilio C. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J. 2005;24:4271–4278. doi: 10.1038/sj.emboj.7600889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crespo JL, Powers T, Fowler B, Hall MN. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci USA. 2002;99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ikeh MA, et al. Pho4 mediates phosphate acquisition in Candida albicans and is vital for stress resistance and metal homeostasis. Mol Biol Cell. 2016;27:2784–2801. doi: 10.1091/mbc.E16-05-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Urrialde V, Prieto D, Pla J, Alonso-Monge R. The Candida albicans Pho4 transcription factor mediates susceptibility to stress and influences fitness in a mouse commensalism model. Front Microbiol. 2016;7:1062. doi: 10.3389/fmicb.2016.01062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Escoubet B, Djabali K, Amiel C. Adaptation to Pi deprivation of cell Na-dependent Pi uptake: A widespread process. Am J Physiol. 1989;256:C322–C328. doi: 10.1152/ajpcell.1989.256.2.C322. [DOI] [PubMed] [Google Scholar]
- 65.Hearing SD. Refeeding syndrome. BMJ. 2004;328:908–909. doi: 10.1136/bmj.328.7445.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mora-Duarte J, et al. Caspofungin Invasive Candidiasis Study Group Comparison of caspofungin and amphotericin B for invasive candidiasis. N Engl J Med. 2002;347:2020–2029. doi: 10.1056/NEJMoa021585. [DOI] [PubMed] [Google Scholar]
- 67.Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McNaughton RL, et al. Probing in vivo Mn2+ speciation and oxidative stress resistance in yeast cells with electron-nuclear double resonance spectroscopy. Proc Natl Acad Sci USA. 2010;107:15335–15339. doi: 10.1073/pnas.1009648107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh AK, et al. Investigating essential gene function in Mycobacterium tuberculosis using an efficient CRISPR interference system. Nucleic Acids Res. 2016;44:e143. doi: 10.1093/nar/gkw625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.