Significance
In plants the hormone jasmonic acid (JA) is synthesized in response to attack by pathogens and herbivores, leading to activation of defense responses. Rapidly following JA accumulation the hormone is metabolized, presumably to prevent inhibitive effects of high JA levels on growth and development. The enzymes that directly inactivate JA were so far unknown. Here, we identify four jasmonate-induced oxygenases (JOXs) in Arabidopsis that hydroxylate jasmonic acid to form inactive 12-OH-JA. A mutant that no longer produces the four enzymes hyperaccumulates JA, exhibits reduced growth, and is highly resistant to attackers that are sensitive to JA-dependent defense. The JOX enzymes thus play an important role in determining the amplitude and duration of JA responses to balance the growth–defense trade-off.
Keywords: jasmonic acid, 2OG oxygenases, 12-OH-JA, plant defense
Abstract
The phytohormone jasmonic acid (JA) is vital in plant defense and development. Although biosynthesis of JA and activation of JA-responsive gene expression by the bioactive form JA-isoleucine have been well-studied, knowledge on JA metabolism is incomplete. In particular, the enzyme that hydroxylates JA to 12-OH-JA, an inactive form of JA that accumulates after wounding and pathogen attack, is unknown. Here, we report the identification of four paralogous 2-oxoglutarate/Fe(II)–dependent oxygenases in Arabidopsis thaliana as JA hydroxylases and show that they down-regulate JA-dependent responses. Because they are induced by JA we named them JASMONATE-INDUCED OXYGENASES (JOXs). Concurrent mutation of the four genes in a quadruple Arabidopsis mutant resulted in increased defense gene expression and increased resistance to the necrotrophic fungus Botrytis cinerea and the caterpillar Mamestra brassicae. In addition, root and shoot growth of the plants was inhibited. Metabolite analysis of leaves showed that loss of function of the four JOX enzymes resulted in overaccumulation of JA and in reduced turnover of JA into 12-OH-JA. Transformation of the quadruple mutant with each JOX gene strongly reduced JA levels, demonstrating that all four JOXs inactivate JA in plants. The in vitro catalysis of 12-OH-JA from JA by recombinant enzyme could be confirmed for three JOXs. The identification of the enzymes responsible for hydroxylation of JA reveals a missing step in JA metabolism, which is important for the inactivation of the hormone and subsequent down-regulation of JA-dependent defenses.
The lipid-derived phytohormone jasmonic acid (JA) is an essential signaling molecule in plant defense. In response to pathogen attack or wounding JA levels accumulate, resulting in activation of a subset of immune genes and the production of defensive secondary metabolites (1). Multiple negative feedback mechanisms control JA levels and JA-responsive gene expression, presumably to minimize inhibition of plant growth that is associated with JA-mediated defense responses (2). In healthy plants, under nonstressed conditions, low levels of JA are present, and the activation of JA-responsive genes is prevented by JAZ repressor proteins that bind transcriptional activators of the JA pathway (3–7). The conjugate of JA with isoleucine (JA-Ile) strongly promotes binding of JAZ repressors to the F-box protein COI1 (4, 8, 9), resulting in the degradation of JAZ proteins and subsequent activation of JA-responsive gene expression (10, 11). At the same time, JAZ gene expression is activated by JA, resulting in the subsequent repression of JA-responsive gene expression (4, 5, 12). Excess JA and JA-Ile are also inactivated by hydroxylation, forming 12-hydroxy-JA (12-OH-JA) and 12-OH-JA-Ile (13–17).
The hydroxylated form of JA is thought to be inactive because it does not trigger degradation of JAZ repressors, and treatment with 12-OH-JA accordingly does not induce JA-responsive gene expression, nor does it inhibit root growth or seed germination (18, 19). 12-OH-JA has been identified in several plant species, including Arabidopsis, maize, potato, tomato, and rice, and accumulates in Arabidopsis after wounding and Botrytis cinerea infection (13, 18, 20–22). Strikingly, hydroxylation of JA to 12-OH-JA by a monooxygenase produced by the blast fungus Magnaporthe oryzae attenuates plant immune responses to this pathogen (19). So far, no plant enzyme that hydroxylates JA has been identified (23). Hydroxylation of JA-Ile, however, has been described in Arabidopsis and is mediated by three cytochrome P450 enzymes, CYP94B3, CYP94B1, and CYP94C1 (Fig. 1A and refs. 14 and 24–26). 12-OH-JA can be formed from 12-OH-JA-Ile by cleavage of the isoleucine group by two amidohydrolases (27, 28). However, an Arabidopsis double mutant that no longer produces these enzymes still accumulates 12-OH-JA, implying that other enzymes catalyze direct hydroxylation of JA to 12-OH-JA.
Fig. 1.
(A) Schematic of metabolism of JA and three JA-derived compounds: JA-Ile, 12-OH-JA, and 12-OH-JA-Ile. Enzymes that catalyze the conversions are indicated: JAR1 conjugates isoleucine to JA to form JA-Ile. CYP94B3 and CYP94C1 hydroxylate JA-Ile to 12-OH-JA-Ile. IAR and ILL6 can hydrolyze the Ile from JA-Ile or from 12-OH-JA-Ile, forming JA or 12-OH-JA, respectively. The enzyme hydroxylating JA to 12-OH-JA is hypothesized to be a 2OG oxygenase. (B) Phylogenetic tree of SA- and MeJA-induced and related 2OG oxygenases of Arabidopsis. The cladogram shows the relatedness of 50 2OG oxygenases selected from the phylogram in SI Appendix, Fig. S1. For each protein model, the name (when available) is supplied, and for each clade, the number assigned by Kawai et al. (29) is indicated. The heat map indicates the log2 fold change of the corresponding genes in Arabidopsis seedlings 3 h after MeJA or SA treatment. Indicated in pink is clade 38, which contains SA-induced S3H/DLO1. The 2OG oxygenases induced by MeJA are in bold. The JOX genes (clade 46) are indicated in blue.
Apart from cytochrome P450 enzymes, members of the 2-oxoglutarate (2OG) Fe(II)-dependent oxygenase family are involved in oxygenation/hydroxylation reactions in plants (29). Interestingly, several 2OG oxygenases were shown to hydroxylate and inactivate plant hormones, e.g., two different groups of 2OG oxygenases inactivate gibberellic acid (GA) by hydroxylating either bioactive 19-GA or an inactive precursor of GA (30, 31). More recently, the active form of auxin was reported to be hydroxylated and inactivated by the 2OG oxygenase DAO in rice (32) and Arabidopsis (33, 34). Finally, the defense hormone salicylic acid (SA) is hydroxylated by the 2OG oxygenase SA 3-HYDROXYLASE (S3H) (35). Because inactivation of hormones via hydroxylation by 2OG oxygenases is common in plants, we hypothesized that 2OG oxygenases could function as JA-hydroxylases as well. Here, we describe the identification of a clade of four 2OG oxygenases that are transcriptionally induced by JA, which we named JASMONATE-INDUCED OXYGENASES (JOXs). We provide metabolic and biochemical evidence that these enzymes are responsible for hydroxylation of JA to 12-OH-JA. Furthermore, phenotypic studies of mutant and overexpression lines show that the JOXs are involved in down-regulation of JA-dependent responses, thereby affecting plant defense and growth. These results identify a class of enzymes in JA metabolism that perform an essential role in in controlling defense responses to necrotrophs and herbivorous insects.
Results
Four JOXs Group in a Distinct Clade in Arabidopsis.
We set out to investigate whether JA-induced 2OG oxygenases could play a role in hydroxylation of JA to 12-OH-JA (Fig. 1A). First, a phylogenetic tree of 2OG oxygenases was constructed based on 93 Arabidopsis proteins that each contain two conserved 2OG oxygenase Pfam domains: the C-terminal PF03171 [2OG-Fe(II) oxygenase superfamily] and the N-terminal PF14226 (nonhaem dioxygenase in morphine synthesis N-terminal). Phylogenetic clustering of the 93 proteins revealed distinct families (SI Appendix, Fig. S1) that largely overlapped with previously described clades (29), which were based on six plant species ranging from unicellular alga to the flowering plants Arabidopsis and rice. Projection of transcriptome data showed that 2OG oxygenases that are induced in Arabidopsis seedlings by treatment with the defense-related hormones methyl jasmonate (MeJA) or SA were only present in a cluster of 50 proteins that encompass 14 clades as defined by Kawai et al. (29) (Fig. 1B). Clade 38 (in red) contains the S3H/DLO1 and DMR6 genes that are induced by SA (35, 36). The expression of six genes encoding 2OG oxygenases was induced more than twofold by MeJA treatment (indicated in bold, Fig. 1B). Of two weakly induced genes, At5g20550 is a gene of unknown function, whereas LDOX (At4g22880) encodes an enzyme involved in anthocyanin biosynthesis (37). A third MeJA-induced gene is DIN11 (At3g49620), which was described as a senescence-associated gene responsive to viral infection (38). Strikingly, clade 46 (in blue) contains four 2OG-oxygenases, of which three were clearly induced at 3 h after MeJA treatment (Fig. 1B).
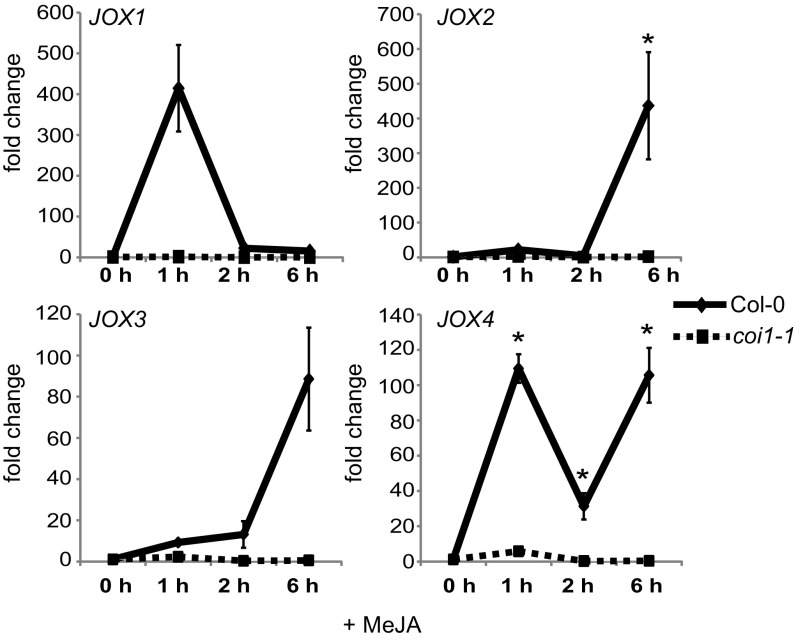
The JA-responsiveness of the four 2OG oxygenase genes of clade 46 was experimentally verified in 5-wk-old plants treated with MeJA. This treatment increased transcript levels of all four genes (Fig. 2), which were, therefore, named JOXs. The expression of JOX1 (At3g11180), which was not MeJA-induced in seedlings according to publically available microarray data (Fig. 1 and ref. 39), was highly induced in adults plants at 1 h after MeJA treatment and was slightly higher than in mock-treated plants at 2–6 h after MeJA treatment (Fig. 2). The expression patterns of JOX2 (At5g05600) and JOX3 (At3g55970) were similar: Induction was low at 1 h and 2 h after treatment but high at 6 h after treatment. Expression of JOX4 (At2g38240) was induced at all time points but showed a different temporal behavior: It was highly induced at 1 h, lower at 2 h, and high again at 6 h after treatment (Fig. 2). In the coi1-1 mutant, which does not have a functional JA receptor, none of the JOX genes was induced by MeJA (Fig. 2). Interestingly, expression of the four JOX genes was strongly induced in plants infected by B. cinerea or infested by the caterpillar Mamestra brassicae, which both induce JA accumulation (SI Appendix, Fig. S2). The fact that the four related JOX oxygenase genes are all activated by JA makes them prime candidates to be involved in JA metabolism, similar to the SA-hydroxylase S3H/DLO1 that is transcriptionally induced by its substrate SA (Fig. 1B). Because 2OG oxygenases are generally involved in hydroxylation and oxygenation reactions, we hypothesized that the JOX enzymes could hydroxylate JA.
Fig. 2.
Expression of JOX genes after MeJA treatment. Expression analysis of JOX1, JOX2, JOX3, and JOX4 in response to MeJA in 5-wk-old Col-0 and coi1-1. Shown is the expression (fold change) at 0, 1, 2, or 6 h after MeJA treatment, relative to that in mock-treated plants at the same time. Error bars indicate SE. An asterisk indicates a significant higher expression compared with mock treatment (P ≤ 0.05; two-way ANOVA).
JOX Enzymes Are Negative Regulators of JA Responses.
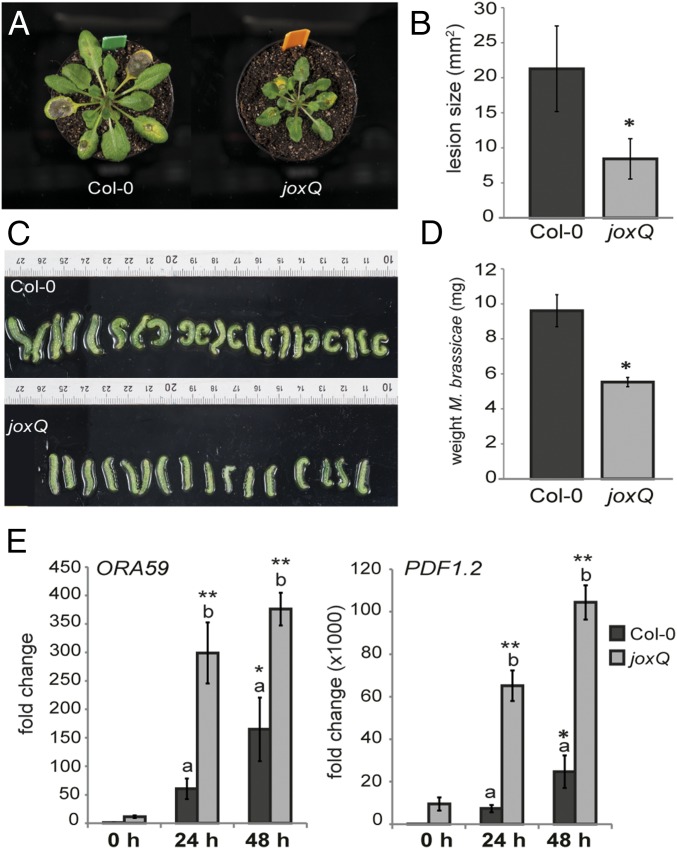
The presumed function of JOX enzymes in JA hydroxylation is expected to influence JA-related biological processes. We therefore set out to test phenotypes affected by JA in mutant plants. Because the four JOX genes could act redundantly, a quadruple mutant jox1 jox2 jox3 jox4, hereafter referred to as joxQ, was generated by crossing four T-DNA insertion lines in each of which one of the four JOX genes was disrupted. JA-related phenotypes (e.g., resistance to necrotrophic pathogens and herbivorous insects) were analyzed in the joxQ mutant. Lesions caused by infection with the necrotrophic fungus B. cinerea were significantly smaller in the joxQ mutant compared with those on wild-type Col-0 leaves (Fig. 3 A and B). In addition, larvae of the generalist caterpillar M. brassicae were smaller and weighed significantly less when fed on joxQ compared with those fed on Col-0 (Fig. 3 C and D). This suggests that defense against both necrotrophic pathogens and herbivorous caterpillars is up-regulated in joxQ plants. To understand the molecular basis of the increased resistance in the joxQ mutant, we measured expression of JA-responsive defense genes before and after infection or infestation. Already before B. cinerea infection expression of the JA/ET-responsive PDF1.2 and ORA59 genes was higher in the joxQ mutant compared with Col-0; the transcription factor gene ORA59 was expressed 15-fold higher, whereas expression of PDF1.2 was increased 9,000-fold (Fig. 3E). During infection with B. cinerea, ORA59 and PDF1.2 levels strongly increased and were significantly higher in the joxQ mutant than in Col-0 (Fig. 3E). Similarly, expression of the JA-responsive transcription factor gene MYC2 was higher in the joxQ mutant than in Col-0 under control conditions. Levels of this gene increased after M. brassicae feeding, and stayed slightly higher in the joxQ mutant (SI Appendix, Fig. S3A).
Fig. 3.
The joxQ quadruple mutant displays phenotypes reminiscent of activated JA signaling. (A) Representative pictures of reduced disease symptoms caused by B. cinerea infection on joxQ compared with Col-0. (B) Area size of necrotic lesions caused by B. cinerea in Col-0 and joxQ. (C and D) Size and weight of M. brassicae caterpillars on Col-0 and joxQ plants after 8 d of feeding. In B and D an asterisk denotes a significant difference between Col-0 and joxQ (t test, P ≤ 0.001). (E) Expression of the JA-responsive genes ORA59 and PDF1.2 before infection (0 h) and after 24 or 48 h of B. cinerea infection of Col-0 or joxQ, relative to Col-0 at 0 h. Different letters indicate significant differences between genotypes. An asterisk indicates a significant higher expression compared with Col-0 plants at 0 h (two-way ANOVA, Tukey post hoc test; *P ≤ 0.05; **P ≤ 0.001).
JA is also known to inhibit plant growth and delay flowering time (40, 41). In accordance with this, the joxQ quadruple mutant was consistently smaller than wild-type plants (Fig. 3A) and exhibited reduced root growth on 1/2 MS medium (SI Appendix, Fig. S3B). In addition, in the presence of 50 µM MeJA the length of the main root was more strongly affected in joxQ (main root length 12.5% of untreated) than in Col-0 (20% of untreated) (SI Appendix, Fig. S3B). These phenotypes support the idea that in wild-type plants JOX proteins suppress JA-mediated growth inhibition. Moreover, flowering time was delayed by 6 d in the joxQ mutant compared with Col-0 under short-day conditions and the mutant produced fewer seeds than wild-type Col-0 (SI Appendix, Fig. S3C). The disease resistance and other JA-related phenotypes of the quadruple joxQ mutant are reminiscent of plants with activated JA responses and are possibly caused by high JA levels.
The joxQ Mutant Hyperaccumulates JA.
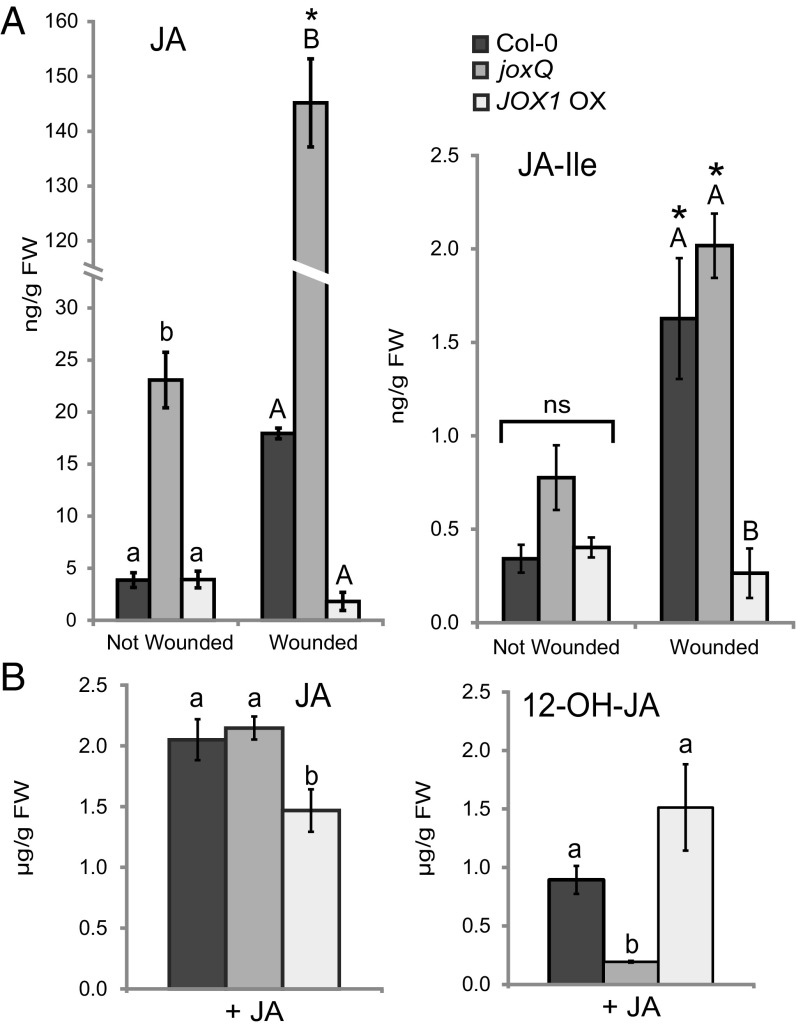
The accumulation of JA could explain the phenotypes of the joxQ quadruple mutant and was therefore measured by liquid chromatography-mass spectrometry (LC-MS) (see SI Appendix, Fig. S4 for standards). We found that JA levels were about five times higher in the joxQ mutant than in Col-0 under nontreated conditions (Fig. 4A). Three hours after wounding, which is known to trigger JA accumulation, JA levels tripled to 18 ng/g fresh weight (FW) in wild-type plants. In the joxQ mutant, JA rose to 146 ng/g FW (Fig. 4A). JA-Ile levels also increased after wounding in Col-0 and the joxQ mutant (Fig. 4A). The increased accumulation of JA in the joxQ mutant suggests that in wild-type plants the JOX proteins negatively affect the accumulation of JA, possibly via hydroxylation. To further study this, JOX1 (At3g11180) was overexpressed in the Col-0 background (JOX1 OX). In accordance with our hypothesis, JA and JA-Ile levels did not increase after wounding in this line (Fig. 4A). The results of both the mutant and overexpression line suggest that after wounding the JOX enzymes act to reduce amounts of JA, possibly via hydroxylation.
Fig. 4.
The joxQ mutant accumulates JA and is impaired in 12-OH-JA production after JA treatment. (A) JA and JA-Ile levels in leaves with or without wounding (3 h after mechanical damage) of Col-0, joxQ, and JOX1 OX plants. Each data point represents the mean of four biological replicates. Error bars indicate SE. JA and JA-Ile levels were calculated by correcting for the internal standard and leaf weight. Different letters indicate statistically significant differences between genotypes at the same treatment, and an asterisk that wounding significantly induced the compound (two-way ANOVA; Tukey post hoc test; P ≤ 0.05). ns, not statistically significant. (B) Accumulation of JA and 12-OH-JA in plants exposed for 3 h to 100 µM JA. Each data point represents the mean of four biological replicates. Error bars indicate SE. JA and 12-OH-JA levels were calculated by correcting for the internal standard and leaf weight. Different letters indicate statistically significant differences between genotypes (two-way ANOVA; Tukey post hoc test; P ≤ 0.05). All measured JA-related metabolites are shown in SI Appendix, Table S2.
The hydroxylated form of JA, 12-OH-JA, was earlier shown to peak at 3 h after wounding in wild-type Arabidopsis plants (20) and at this time point we measured 21 ng/g FW in Col-0 grown under our conditions (SI Appendix, Fig. S5). If JOXs indeed hydroxylate JA, we would expect low levels of 12-OH-JA in the joxQ mutant. However, 12-OH-JA was about fourfold higher in the joxQ mutant (84 ng/g FW) in response to wounding compared with Col-0 (SI Appendix, Fig. S5 and Table S1). Nevertheless, the 12-OH-JA increase was lower than that of JA that was increased eightfold in joxQ compared with Col-0. Possibly, 12-OH-JA detected in joxQ is generated by cleavage of the conjugated isoleucine of 12-OH-JA-Ile by amidohydrolases ILL6 and IAR3 (Fig. 1A and ref. 27). In JOX1 OX we expected increased 12-OH-JA to accumulate; however, 12-OH-JA could not be detected in this line, not even after wounding (SI Appendix, Fig. S5). We considered two hypotheses for this low 12-OH-JA level: (i) It is further metabolized and therefore not detectable and/or (ii) it is not produced in sufficient amounts due to low levels of the JA substrate in the JOX1 OX line. We did not find evidence for enhanced 12-OH-JA metabolism because we did not detect an increase in the downstream compounds 12-HSO4-JA, 12-OH-JA-Ile and 12-O-glycosyl-JA in JOX1 OX plants (SI Appendix, Table S1 and Fig. S5). In support of the low substrate levels, we find low JA (Fig. 4A) and reduced 12-oxo-phytodienoic acid (OPDA) levels in 5-wk-old JOX1 OX plants (SI Appendix, Table S1). Because a positive feedback loop exists between JA levels and JA biosynthesis, we speculate that this feedback loop generates more JA in Col-0 than in JOX1 OX plants.
To reduce the effect of feedback mechanisms, we added equal amounts of exogenous JA to wild-type joxQ and JOX1 OX plants by immersing the leaves in a 100 µM JA solution and measured jasmonates 3 h later. JA levels in treated leaves of joxQ were similar to those of Col-0 but lower in the JOX1 OX line (Fig. 4B). Strikingly, the level of 12-OH-JA was much lower in the joxQ mutant than in Col-0. In contrast, in JOX1 OX the levels of 12-OH-JA were higher than in Col-0 (Fig. 4B). This indicates that the conversion of JA into 12-OH-JA is reduced in the joxQ mutant and enhanced in the JOX1 OX line. We conclude that the joxQ mutant hyperaccumulates JA under basal and biotic stress conditions and has reduced 12-OH-JA formation after treatment with exogenous JA. In contrast, a line overexpressing JOX1 builds up less JA and accumulates more of the hydroxylated form when treated with JA.
Overexpression of Each Individual JOX Complements joxQ Phenotypes.
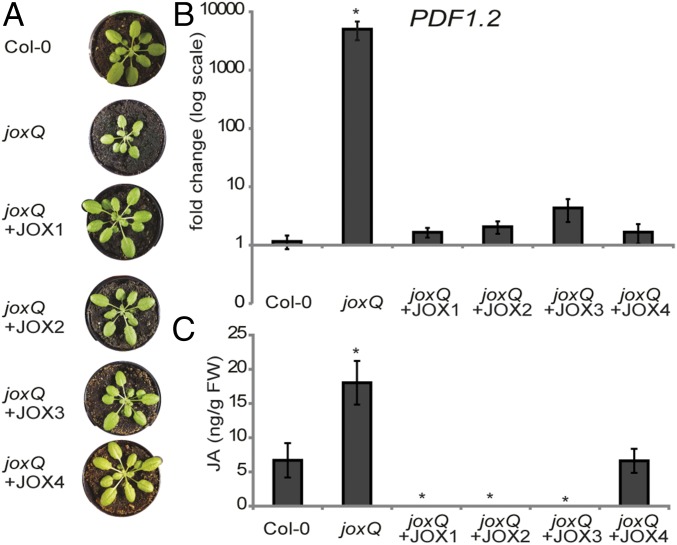
The presumed metabolic function of JOX enzymes is to hydroxylate JA. To determine whether all JOX enzymes have the capability to reduce JA levels and responses, we constitutively expressed each JOX gene from the 35S promoter in the quadruple joxQ mutant. For each JOX, at least two independent homozygous transformants were selected. First, we observed that overexpression of each JOX rescued the growth phenotype of the joxQ mutant. Whereas 5-wk-old joxQ plants were significantly smaller than Col-0, all JOX-transformed plants resembled Col-0 plants (Fig. 5A). Second, the high expression of PDF1.2 that was observed in joxQ plants (Fig. 3E) was reverted to wild-type levels in all lines (Fig. 5B). Finally, we measured levels of JA and JA derivatives in all transformed lines. In untreated adult plants of joxQ, JA levels were about three times higher than in Col-0. Overexpression of JOX1, JOX2, and JOX3 resulted in depletion of JA. Overexpression of JOX4 resulted in JA levels similar to those in Col-0 (Fig. 5C). Taken together, these results show that each individual JOX can reduce JA levels to those in wild-type plants or lower, resulting in complementation of JA-related phenotypes (Fig. 5).
Fig. 5.
Overexpression of each individual JOX complements joxQ phenotypes. (A) Growth phenotype of representative plants of Col-0, joxQ, and overexpression lines of JOX1, JOX2, JOX3, and JOX4 in the joxQ background. (B) Expression of PDF1.2 in untreated Col-0, joxQ, and overexpression lines of JOX1, JOX2, JOX3, and JOX4 in joxQ. Expression is normalized to the reference gene At1g13320 and relative to Col-0 plants. (C) Amount of JA in untreated leaves of Col-0, joxQ, and overexpression lines of JOX1, JOX2, JOX3, and JOX4 in joxQ. JA levels were calculated by correcting for the internal standard and leaf weight. In B and C each data point represents the mean of four to eight biological replicates. Error bars indicate SE. For JOX overexpression lines, average is shown of two or three (JOX2) independent homozygous transformant lines. An asterisk denotes a significant difference between the genotype and Col-0 (t test, P ≤ 0.05).
JOX1, JOX2, and JOX4 Hydroxylate JA in Vitro.
To determine whether the JOX proteins can catalyze the hydroxylation of JA to 12-OH-JA without other plant proteins present, assays were conducted with JA as substrate and recombinant JOX enzymes produced in Escherichia coli. The reaction products were analyzed by LC-MS. Lysates of E. coli expressing JOX1, JOX2, and JOX4 effectively converted JA to 12-OH-JA, resulting in almost complete conversion of the JA substrate and high levels of 12-OH-JA (SI Appendix, Fig. S6A). The lysate of E. coli expressing JOX3 produced low amounts of 12-OH-JA, which were higher than that of the negative control (reaction mixture without lysate) but not higher than the amount of 12-OH-JA produced in the enzymatic assay with E. coli expressing the SA-hydroxylase S3H/DLO1. We thus conclude that recombinant JOX1, JOX2, and JOX4 have JA-12-hydroxylase activity in vitro in the absence of other plant-derived enzymes and compounds.
Discussion
Four JA-induced 2OG oxygenases (JOX) from a single paralogous family in Arabidopsis were identified that strongly contribute to negative regulation of JA-dependent responses. Plants in which the four genes were mutated (joxQ) accumulated high levels of JA and exhibited smaller growth, enhanced expression of defense genes, and resistance to both B. cinerea and M. brassicae. We provide evidence from metabolic measurements and enzymatic assays that JOX enzymes control JA levels by hydroxylation of JA to inactive 12-OH-JA. By keeping JA levels low the JOX enzymes are important in determining the amplitude and duration of JA responses and balance the growth–defense trade-off.
Inactivation of plant hormones by 2OG oxygenase-mediated hydroxylation seems to be a common theme in plants to control their hormone levels. The hormones GA and SA are hydroxylated by 2OG oxygenases that are closely related to the JOXs, that is, GA2OXs that hydroxylate GA (30) (clade 12 in Fig. 1B) and S3H (clade 38) that hydroxylates SA (35). Two more distantly related 2OG oxygenases were recently shown to inactivate the plant hormone auxin (33, 34). Clade 46 of the 2OG oxygenases as defined by Kawai et al. (29) consists of four Arabidopsis members, encoded by the genes At3g11180, At5g05600, At3g55970, and At2g38240, which we named JOX1, JOX2, JOX3, and JOX4, respectively. We show that the members of this clade act by oxidative inactivation of JA and prevent overaccumulation of JA and indirectly its bioactive form JA-Ile.
The observation that JA levels are high in the joxQ mutant already in untreated conditions (Fig. 4A) suggests that JOXs contribute to reduction of JA levels in unstressed growing conditions. Consequently, JA-responsive gene expression is higher in the joxQ plants than in Col-0 (Fig. 3E and SI Appendix, Fig. S3A). JA-Ile is considered the biologically active form of JA because its binding to COI1–JAZ complexes leads to the degradation of JAZ repressor proteins and activation of JA-induced gene expression (4, 5, 9). However, we did not find increased JA-Ile levels in untreated joxQ plants. Possibly, JA-Ile is turned over so quickly that we could not detect it. It has been speculated that other derivatives of JA, or JA itself, can trigger gene activation. Interestingly, wound-induced expression of MYC2 and PDF1.2 was not affected in the jar1 mutant, which has reduced JA-Ile levels (12, 42). We also find increased expression of MYC2 and PDF1.2 in untreated joxQ plants. Further research should determine whether increased JA levels in the joxQ mutant cause these observed responses directly, or whether that occurs via JA-Ile or other JA-related metabolites.
After wounding, JA levels increased dramatically in the joxQ mutant. In wild-type plants JOX enzymes thus could function to reduce accumulation of JA. Because JA biosynthesis genes are JA-responsive, it is likely that the higher JA levels in joxQ lead to increased biosynthesis and thus higher levels of JA. Supporting the idea that JA biosynthesis is up-regulated in joxQ, we detected OPDA levels approximately four times higher in the joxQ mutant than in Col-0. In addition, OPDA and JA levels are low in the JOX1-overexpressing plants (Fig. 4A and SI Appendix, Table S1). The reduced amount of JA substrate is possibly the reason that we do not find increased 12-OH-JA in JOX1 OX plants. After adding exogenous JA to JOX1 OX plants, more 12-OH-JA is generated compared with Col-0 (Fig. 4B), and in an enzymatic assay JOX1 was able to form 12-OH-JA (SI Appendix, Fig. S6). The JA hydroxylase activity of JOX1 is supported by these two experiments.
The induction of the JOX genes by MeJA treatment, by M. brassicae feeding, and by B. cinerea infection, the first of which was shown to be dependent on JA-coreceptor COI1, shows that this JA-inactivating mechanism is controlled by the JA pathway itself. This is reminiscent of mechanisms of oxidative inactivation of hormones as discussed above and in particular of the mechanism of JA-Ile hydroxylation (14, 25). Enzymes from the cytochrome P450 family inactivate JA-Ile: CYP94B1 and CYP94B3 hydroxylate JA-Ile to 12-OH-JA-Ile (24–26) and CYP94C1 further oxygenates this compound to 12-COOH-JA-Ile (14). Our identification of four 2OG oxygenases that convert JA into the inactive 12-OH-JA further elucidates JA metabolism. From 12-OH-JA-Ile, 12-OH-JA can be produced by the amidohydrolases IAR3 and ILL6, which cleave the isoleucine group of JA-Ile and 12-OH-JA-Ile (Fig. 1A and ref. 27). However, the JOX enzymes can hydroxylate JA directly and majorly contribute to the removal of JA in plants in undisturbed growth conditions and in response to wounding or pathogen attack. The quadruple mutant of these enzymes shows similar phenotypes as the JA-Ile hydroxylase mutants (e.g., enhanced expression of JA-responsive genes and increased sensitivity to JA-dependent inhibition of root growth) (Fig. 3 and ref. 14). This suggests that hydroxylation of JA by the JOX enzymes contributes to inactivation of the active JA signal to a similar extent as hydroxylation of JA-Ile. The levels of bioactive JA-Ile are likely directly influenced by the amount of JA, because we show that levels of JA and JA-Ile are in equilibrium after wounding (Fig. 4A).
The importance of hydroxylation of JA is emphasized by the apparent evolution of four different enzymes with the same function. In an enzyme assay, recombinant JOX1, JOX2, and JOX4 were shown to be able to produce 12-OH-JA from JA in the absence of other plant proteins. JOX3 was unable to hydroxylate JA in this in vitro assay. However, in our complementation studies all four enzymes, including JOX3, were shown to be able to complement all phenotypes of the joxQ mutant, indicating that each JOX enzyme can lower JA levels in vivo. Possibly, JOX3 requires activation by another plant protein or compound before it is active as a JA-hydroxylase, or folding of the recombinant JOX3 protein is not correct. JOX proteins are found in a broad taxonomic range of multicellular plant species (29) but the number of JOX orthologs differs per species. Interestingly, the monocot and dicot JOX orthologs group in separate phylogenetic branches, suggesting that ancestral flowering plants had a single JOX gene, whereas extant species have two to four paralogs. It is possible that each paralogous enzyme functions in a different process in the plant or in distinct plant tissues. Preliminary evidence for this comes from the timing and amplitude of the expression induced by MeJA, B. cinerea, and M. brassicae, which was different between the JOX genes (Fig. 2 and SI Appendix, Fig. S2). So far, we have only tested expression in leaf tissue, but spatial expression of each JOX gene within the leaf or within the plant could be different. Similarly, the SA-hydroxylase gene DLO1 and its paralog DMR6 have similar but distinct activities due to their pathogen-induced expression in different parts of downy mildew-infected leaves (36). Experiments to localize the expression of JOXs and complementation assays under their own promoter could further elucidate the different functionalities of the four JOX genes.
In plants, JA is converted to derivatives that are biologically active, reduced active, or inactive (16). The 12-OH-JA has been characterized as an inactive form of JA, because it is not capable of degrading JAZ9 and does not induce expression of JA-responsive genes or inhibition of root growth (15, 18, 19). The compound has been suggested to have tuber-inducing capabilities (22), but what the role of this compound is in non-tuber-forming plants such as Arabidopsis is not clear. It is likely that hydroxylation of JA is a quick mechanism to inactivate JA. Following production of JA, expression of JOX genes would quickly be induced, after which the accumulation of JA, and subsequently the expression JA-responsive genes, is dampened. This is yet another negative feedback system that would be active in the JA pathway, in addition to, for example, the activation of JAZ repressor genes by JA (4, 5, 12). Moreover, similar hydroxylation mechanisms control levels of other plant hormones (25, 33, 35). The identification of JOX enzymes elucidates another major step in plant hormone metabolism. Our data show that it is imperative that plants balance JA levels by controlling JA metabolism, because inhibitive effects on growth are evident in the joxQ mutant. As expression of the JOX genes is induced by JA plants can quickly shut off JA-dependent responses after their activation to prevent negative effect of high JA levels on growth and development. The JOX enzymes thus contribute to balance the growth/defense trade-off.
Materials and Methods
Arabidopsis genes encoding 2OG-oxygenases were selected from Biomart, aligned, processed for phylogenetic analysis, and plotted with gene expression data as described in SI Appendix, Supplemental Material and Methods. The generation of the quadruple jox mutant, JOX OX lines, their phenotypic characterization, and chemical profiles is detailed in SI Appendix, Supplemental Material and Methods, as well as the enzymatic assays on recombinant JOX enzymes produced in E. coli.
Supplementary Material
Acknowledgments
We thank Léon Westerd for rearing of M. brassicae, Hans van Pelt for taking photographs of plants and caterpillars, Tom Raaymakers for help with the figures, and Enza Zaden B.V. for supporting the initial research on the JOX genes. This work was partly supported by the Netherlands Organization for Scientific Research through Dutch Technology Foundation VIDI Grant 11281 (to S.C.M.V.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701101114/-/DCSupplemental.
References
- 1.Campos ML, Kang JH, Howe GA. Jasmonate-triggered plant immunity. J Chem Ecol. 2014;40:657–675. doi: 10.1007/s10886-014-0468-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huot B, Yao J, Montgomery BL, He SY. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant. 2014;7:1267–1287. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Z, et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 5.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 6.Fernández-Calvo P, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23:701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niu Y, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot. 2011;62:2143–2154. doi: 10.1093/jxb/erq408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheard LB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fonseca S, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5:344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 10.Devoto A, et al. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 2002;32:457–466. doi: 10.1046/j.1365-313x.2002.01432.x. [DOI] [PubMed] [Google Scholar]
- 11.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung HS, et al. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008;146:952–964. doi: 10.1104/pp.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seto Y, et al. Purification and cDNA cloning of a wound inducible glucosyltransferase active toward 12-hydroxy jasmonic acid. Phytochemistry. 2009;70:370–379. doi: 10.1016/j.phytochem.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Heitz T, et al. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone Jasmonoyl-isoleucine for catabolic turnover. J Biol Chem. 2012;287:6296–6306. doi: 10.1074/jbc.M111.316364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gidda SK, et al. Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J Biol Chem. 2003;278:17895–17900. doi: 10.1074/jbc.M211943200. [DOI] [PubMed] [Google Scholar]
- 16.Wasternack C, Strnad M. Jasmonate signaling in plant stress responses and development - active and inactive compounds. N Biotechnol. 2016;33:604–613. doi: 10.1016/j.nbt.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Guranowski A, Miersch O, Staswick PE, Suza W, Wasternack C. Substrate specificity and products of side-reactions catalyzed by jasmonate:amino acid synthetase (JAR1) FEBS Lett. 2007;581:815–820. doi: 10.1016/j.febslet.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 18.Miersch O, Neumerkel J, Dippe M, Stenzel I, Wasternack C. Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 2008;177:114–127. doi: 10.1111/j.1469-8137.2007.02252.x. [DOI] [PubMed] [Google Scholar]
- 19.Patkar RN, et al. A fungal monooxygenase-derived jasmonate attenuates host innate immunity. Nat Chem Biol. 2015;11:733–740. doi: 10.1038/nchembio.1885. [DOI] [PubMed] [Google Scholar]
- 20.Glauser G, et al. Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J Biol Chem. 2008;283:16400–16407. doi: 10.1074/jbc.M801760200. [DOI] [PubMed] [Google Scholar]
- 21.Aubert Y, Widemann E, Miesch L, Pinot F, Heitz T. CYP94-mediated jasmonoyl-isoleucine hormone oxidation shapes jasmonate profiles and attenuates defence responses to Botrytis cinerea infection. J Exp Bot. 2015;66:3879–3892. doi: 10.1093/jxb/erv190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshihara T, et al. Structure of a tuber-inducing stimulus from potato leaves (Solanum tuberosum L.) Agric Biol Chem. 1989;53:2835–2837. [Google Scholar]
- 23.Koo AJ, Howe GA. Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front Plant Sci. 2012;3:19. doi: 10.3389/fpls.2012.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koo AJ, et al. Endoplasmic reticulum-associated inactivation of the hormone jasmonoyl-L-isoleucine by multiple members of the cytochrome P450 94 family in Arabidopsis. J Biol Chem. 2014;289:29728–29738. doi: 10.1074/jbc.M114.603084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koo AJ, Cooke TF, Howe GA. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc Natl Acad Sci USA. 2011;108:9298–9303. doi: 10.1073/pnas.1103542108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitaoka N, et al. Arabidopsis CYP94B3 encodes jasmonyl-L-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 2011;52:1757–1765. doi: 10.1093/pcp/pcr110. [DOI] [PubMed] [Google Scholar]
- 27.Widemann E, et al. The amidohydrolases IAR3 and ILL6 contribute to jasmonoyl-isoleucine hormone turnover and generate 12-hydroxyjasmonic acid upon wounding in Arabidopsis leaves. J Biol Chem. 2013;288:31701–31714. doi: 10.1074/jbc.M113.499228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhosale R, et al. Predicting gene function from uncontrolled expression variation among individual wild-type Arabidopsis plants. Plant Cell. 2013;25:2865–2877. doi: 10.1105/tpc.113.112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawai Y, Ono E, Mizutani M. Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 2014;78:328–343. doi: 10.1111/tpj.12479. [DOI] [PubMed] [Google Scholar]
- 30.Rieu I, et al. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell. 2008;20:2420–2436. doi: 10.1105/tpc.108.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JA, Amasino RM. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell. 2003;15:151–163. doi: 10.1105/tpc.005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Z, et al. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell. 2013;27:113–122. doi: 10.1016/j.devcel.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, et al. DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2016;113:11010–11015. doi: 10.1073/pnas.1604769113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porco S, et al. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc Natl Acad Sci USA. 2016;113:11016–11021. doi: 10.1073/pnas.1604375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang K, Halitschke R, Yin C, Liu CJ, Gan SS. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc Natl Acad Sci USA. 2013;110:14807–14812. doi: 10.1073/pnas.1302702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeilmaker T, et al. DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 2015;81:210–222. doi: 10.1111/tpj.12719. [DOI] [PubMed] [Google Scholar]
- 37.Shan X, Zhang Y, Peng W, Wang Z, Xie D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot. 2009;60:3849–3860. doi: 10.1093/jxb/erp223. [DOI] [PubMed] [Google Scholar]
- 38.Fernández-Calvino L, et al. Activation of senescence-associated Dark-inducible (DIN) genes during infection contributes to enhanced susceptibility to plant viruses. Mol Plant Pathol. 2016;17:3–15. doi: 10.1111/mpp.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goda H, et al. The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55:526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhai Q, et al. Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell. 2015;27:2814–2828. doi: 10.1105/tpc.15.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Turner JG. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One. 2008;3:e3699. doi: 10.1371/journal.pone.0003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suza WP, Staswick PE. The role of JAR1 in Jasmonoyl-l-isoleucine production during Arabidopsis wound response. Planta. 2008;227:1221–1232. doi: 10.1007/s00425-008-0694-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.