Significance
The cytoplasm of eukaryotic cells is compartmentalized by intracellular membranes that define subcellular organelles. One of these organelles, the endoplasmic reticulum, forms a continuous network of tubules and cisternae that extends throughout all cell compartments, including neuronal dendrites and axons. This network communicates with most other organelles by vesicular transport, and also by contacts that do not lead to fusion but allow cross-talk between adjacent bilayers. Though these membrane contacts have previously been observed in neurons, their distribution and abundance has not been systematically analyzed. Here, we have carried out such analysis. Our studies reveal new aspects of the internal structure of neurons and provide a critical complement to information about interorganelle communication emerging from functional and biochemical studies.
Keywords: FIB-SEM, thin ER, Stim1, spine apparatus, subsurface cisternae
Abstract
Close appositions between the membrane of the endoplasmic reticulum (ER) and other intracellular membranes have important functions in cell physiology. These include lipid homeostasis, regulation of Ca2+ dynamics, and control of organelle biogenesis and dynamics. Although these membrane contacts have previously been observed in neurons, their distribution and abundance have not been systematically analyzed. Here, we have used focused ion beam-scanning electron microscopy to generate 3D reconstructions of intracellular organelles and their membrane appositions involving the ER (distance ≤30 nm) in different neuronal compartments. ER–plasma membrane (PM) contacts were particularly abundant in cell bodies, with large, flat ER cisternae apposed to the PM, sometimes with a notably narrow lumen (thin ER). Smaller ER–PM contacts occurred throughout dendrites, axons, and in axon terminals. ER contacts with mitochondria were abundant in all compartments, with the ER often forming a network that embraced mitochondria. Small focal contacts were also observed with tubulovesicular structures, likely to be endosomes, and with sparse multivesicular bodies and lysosomes found in our reconstructions. Our study provides an anatomical reference for interpreting information about interorganelle communication in neurons emerging from functional and biochemical studies.
The endoplasmic reticulum (ER) is a network of interconnected tubules and cisternae, which was first identified by electron microscopy in the 1950s (1). Subsequent studies have described its pleomorphic structure, elucidated some of the mechanisms responsible for its shape, and revealed the multiplicity of roles that it plays in cell physiology (2, 3). These roles include synthesis of membrane and secretory proteins, synthesis of most membrane lipids, Ca2+ storage, and metabolite processing.
The ER network extends into all cell compartments, including the most distant regions of cells with long processes. It forms specialized subdomains, such as the nuclear envelope, ribosome-rich areas (rough ER) that can further organize themselves into highly packed stacks, and ribosome-free areas (smooth ER). All these regions are continuous with one another, and fragmentation is rapidly counteracted by homotypic fusions (4, 5). Although this continuous network is physically separate from other intracellular membranes, it is functionally connected to organelles of the secretory and endocytic pathways and to the plasma membrane (PM) via membrane traffic (6). Importantly, cross-talk may also occur between the ER and other organelles via direct appositions of their membranes (membrane contact sites, MCSs) not resulting in membrane fusion (7–12).
MCSs were first observed between the ER and either the PM (13–15) or mitochondria (16) and shown to be implicated in the control of Ca2+ fluxes (17–20). Subsequent studies have demonstrated that the ER interacts via tethering proteins with all other membranous organelles of the cell, and have identified a variety of functions for these contacts (7–12). One major function is exchange of lipids between bilayers via lipid-transfer proteins (11, 12, 21–24). The occurrence of this mode of lipid transfer between membranes, which is independent of vesicular transport and membrane fusion, was first described in the late 1960s (25). However, only recently have studies shown that many of the lipid-binding proteins that mediate such transfer act at MCSs. Membrane fusion-independent lipid transport is likely to be especially important in neurons, where many cell compartments are located very far from the Golgi complex, an obligatory transit station for lipids exiting the ER via vesicular traffic (6, 26, 27).
ER cisternae closely apposed to the PM (subsurface cisternae) in neurons were among the first ER–PM contacts reported (14, 15, 28), and proteins implicated in MCSs are expressed in neurons. However, both the functions of MCSs in neurons and their abundance and distribution in neuronal compartments, have not been systematically explored. This analysis requires the generation of large-volume 3D reconstructions, a challenging task using conventional transmission electron microscopy (TEM) techniques, such as serial sectioning or electron tomography (29, 30), despite the power of these techniques to reveal cellular and subcellular nervous system architecture (29, 31, 32). Recent advances in electron microscopy have greatly facilitated this task (33–37) and these advances are being extensively exploited for “connectomics” studies (https://www.neuroscienceblueprint.nih.gov/connectome/) aimed at mapping neuronal wiring in the brain. Here, we have used one such method, focused ion beam-scanning electron microscopy (FIB-SEM), to map the ER and its connections to other membranes within neurons. This analysis represents an important foundation to better understand organelle cross-talk in neurons.
Results
We performed FIB-SEM on samples of aldehyde-fixed mouse brain tissue. Images were collected at 4 × 4 × 4- or 8 × 8 × 8-nm voxel size sampling in the x, y, and z dimensions. Movie S1 illustrates the excellent resolution at which cellular organelles can be observed using this method. Several neuronal compartments were analyzed at high magnification and 3D models of these compartments were generated by direct visual inspection. MCSs were defined as membrane appositions where the distance between two bilayers was ≤30 nm (8, 38–40). The 3D models shown in the main figures of this paper are from neurons in the nucleus accumbens (Fig. 1; see also Figs. 4–7). However, inspection of FIB-SEM volumes of the cerebral cortex and dorsal striatum, including a 3D reconstruction from the cerebral cortex (Fig. S1), suggested that basic features and subcellular organization are similar in neurons across these cell regions.
Fig. 1.
Organization of the ER in a neuronal cell body. Three-dimensional reconstruction of subcellular organelles from a FIB-SEM image stack of a neuronal cell body (nucleus accumbens). (A) A portion of the peripheral cytoplasm, including all membranous organelles in the region, is shown in a view from the inside of the cell. (B) Plasma membrane areas in contact with the ER are shown in red: bright red for wide ER cisternae and dark red for “thin” ER (i.e., ER cisternae where the two opposite faces of the cisternae are tightly apposed to each other so that lumen of the ER can no longer be appreciated at the resolving power of FIB-SEM). Note the spatial separation of these contact areas from postsynaptic areas (white; one asterisk represents symmetric synapses; two asterisks represent asymmetric synapse). (C and D) ER and mitochondria sites of contact are shown in red in D. (E and F) ER and tubules or vesicles not connected to the ER (endosomes and transport vesicles) with contact sites areas shown in red in F. (G and H) ER and lysosomes/multivesicular bodies; contact sites are shown in red in H. (Scale bars: 400 nm; 80 nm in Inset in J.)
Fig. 4.
Organization of the ER in dendrites. Three-dimensional model of subcellular organelles from a FIB-SEM image stack of a dendritic segment (nucleus accumbens). (A) The model includes all membranous organelles present in this dendrite color-coded as in Fig. 1. Note the continuity of the ER and its penetration into a subset of dendritic spines. (B) The ER, mitochondria, and endosomes and transport vesicles. (C and D) PM and mitochondria, respectively; areas of contacts with the ER shown in red. (E) Endosomes + transport vesicles areas of contact with the ER are shown in red in the Insets. Other contacts with the ER not visible in this view can be seen in Movie S2. (Scale bars: 800 nm.) (F and G) Pie charts showing the percent of spines containing ER in neurons of the nucleus accumbens and of the dorsal striatum, respectively, as assessed by inspection of two FIB-SEM datasets.
Fig. 7.
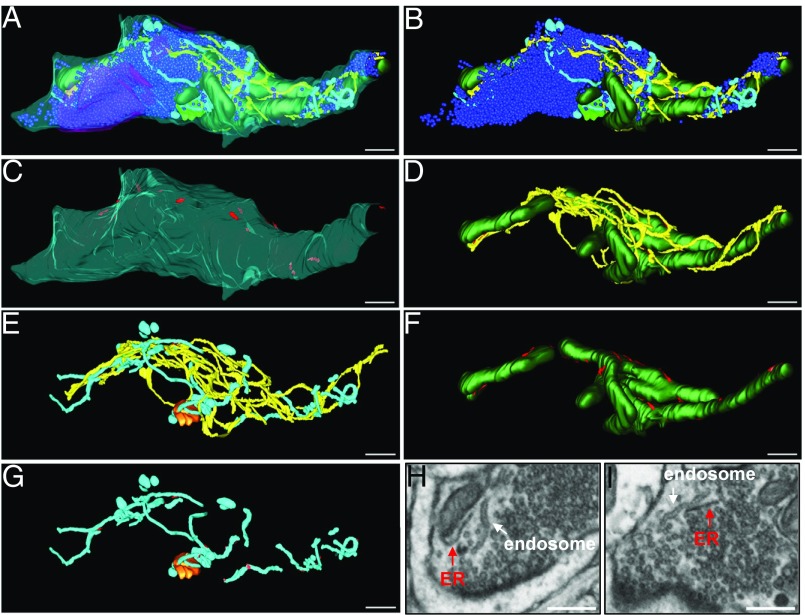
ER and other organelles in a large presynaptic varicosity. (A–G) Three-dimensional model of the nerve terminal (from FIB-SEM image stacks, nucleus accumbens). (A) All membranous organelles and the synaptic interface were reconstructed and color coded as in Fig. 1; synaptic vesicles are in dark blue and the synaptic interface in magenta. (B–G) View of selected organelles with red color indicating contacts with the ER. (H and I) Two images from the FIB-SEM stack used for the 3D reconstruction illustrating the different morphology of two types of tubular elements: the thinner/darker tubules are defined as ER because of their continuity with the ER network of the axon. The larger tubules (shown in light blue in the 3D reconstruction) are disconnected from the ER; they are reminiscent of the tubular endosomal structures observed in some nerve terminals (60, 99). (Scale bars: 400 nm.)
Fig. S1.
Organization of the ER and other organelles in a dendrite and surrounding axon terminals. Cerebral cortex. Three-dimensional model from a FIB-SEM image stack. Color codes are as in Fig. 1. In addition, synaptic vesicles are shown in dark blue and the outer surfaces of the PMs of the dendrite and of the axons are shown in light blue and green, respectively, in A. (A) Outside view. (B) Same as A but with transparent PMs and all organelles shown. (C) PM in contact with the ER shown in red. (D) Mitochondria with areas of contact with the ER shown in red. A long white arrow points to the only mitochondrion present in the dendritic segment, and small white arrows point to shorter mitochondria in axon terminals. (E) Endosomes and vesicles other than synaptic vesicles with contact sites with the ER shown in red. The profiles of the PMs of the dendrite and of axon terminals are shown in dashed light blue and green lines, respectively. (F) Large dendritic spines containing both ER and mitochondria. (G) Thin spine devoid of both ER and mitochondria (ER pictured is presynaptic). In both F and G, synaptic vesicles and the ER of adjacent nerve terminals are also shown. (Scale bars: 800 nm in A–E; 400 nm in F and G.)
Cell Bodies.
Fig. 1A (see also Movie S2) shows a reconstruction of all membranous organelles in the peripheral cytoplasm of a neuronal cell body (the Golgi complex adjacent to this cortical region was not reconstructed). The ER (yellow, irrespective of the presence or absence of ribosomes) is represented by a contiguous network of tubules and cisternae, which are tightly woven throughout the intracellular space and around other organelles. Throughout, these are coded as follows: transparent blue represents the PM, green represents the mitochondria, dark orange represents the multivesicular bodies, gray represents the lysosomes, and light blue represents the vesicles and short tubules not connected into a single network. The latter structures are expected to comprise endosomes and other endocytic vesicles, as well as post-Golgi transport vesicles that carry cargo to various destinations, such as the vesicles that deliver receptors and channels to the plasma membrane. These organelles are collectively referred to below as “endosomes and transport vesicles.” Contacts between ribosome-free areas of the ER and other organelles are shown in red.
All major membranous organelles are involved in ER appositions. In this reconstruction, contacts with the PM are particularly prominent in size (Fig. 1B), cover 12.48% of the PM in the reconstruction (Fig. 2A), and are predominantly mediated by large cisternae (Fig. 1B). These contacts do not colocalize with postsynaptic areas, which are shown in white (Fig. 1B). Contacts with mitochondria cover about 4% of their surface (Fig. 2A) and are primarily mediated by tubules (Figs. 1 C and D and 2E). Similarly, contacts with the few lysosomes and multivesicular bodies observed in the reconstruction (Fig. 2A) are made by tubular ER (Figs. 1 G and H and 2 C and D). Contacts with endosomes and transport vesicles are very small and involve only a few of these structures (Figs. 1 E and F and 2A); most likely they reflect transient interactions.
Fig. 2.
Analysis of contact sites of the ER with the PM and other membranous organelles in different neuronal compartments. (A and B) Morphometric analysis. (A) Percentages of the area of the PM and of the surface of intracellular organelles in close apposition with the ER from the three reconstructions shown in Figs. 1, 3, and 5. Note the greater area of ER–PM contacts in cell bodies. (B) Percentage of PM perimeter of neuronal cell bodies in close apposition with wide and thin ER, respectively, as assessed by TEM micrographs (n = 20 neurons for each region). (C–H) Gallery of micrographs obtained by FIB-SEM at 8 × 8 × 8- (C) and 4 × 4 × 4-nm (D–H) voxel size sampling in the x, y, and z dimensions. (C) ER making contact with a lysosome (red asterisk) in a neuronal cell body. (D–F) Portions of dendrites showing ER making contacts (red arrows) with a multivesicular body (D), a mitochondrion (E), and the PM (F), respectively. Two Insets in F show the ER making contacts (red arrows) with the PM in two small axons. (G and H) ER making contacts (red arrows) with a mitochondrion (E) and with the PM (F) in two axon terminals. All images are from the nucleus accumbens. (Scale bars: 200 nm; 80 nm for Insets in D and F.)
When examined in cross-section, the FIB-SEM images showed that the lumen of ER cisternae in contact with the PM (ER elements in contact with the PM are defined henceforth as “cortical ER”) had variable thickness (Fig. 3 A and B). For the majority of cortical ER, the width of the lumen was greater than 25 nm (mean ± SD = 45.2 ± 9.9 nm; range = 27.7–101.8 nm) (Fig. 3A). However, in all cell bodies of all brain regions examined, the lumen of some cortical ER cisternae narrowed down so much that a space between the two membranes could no longer be perceived at the resolution afforded by FIB-SEM (Fig. 3 B–I). The two membranes, however, could be resolved by conventional TEM (Fig. 3C). These cisternae correspond to “thin ER,” which has been previously described both in neurons (28, 41–46) (https://synapseweb.clm.utexas.edu/112-endoplasmic-reticulum) and in nonneuronal cells (47). In the reconstruction shown in Fig. 1, the thin ER (dark red) and wide ER (bright red) represent 37.3% and 62.7% of the total cortical ER, respectively (see also Fig. 2A). However, inspection of neurons in different brain regions revealed that the abundance of thin versus wide ER at ER–PM contact sites is variable (Fig. 2 A and B). In all cases, cisternae of the cortical ER were continuous with membranes decorated by ribosomes, confirming they are part of the ER (Fig. 3B). Ribosomes were excluded from the thin ER, although they were often present on the face of cisternae opposite to the PM, on wide cortical ER.
Fig. 3.
Thin ER at ER–PM contact sites in neuronal somata. (A and B) ER–PM contacts in FIB-SEM images. Although in A the ER maintains its thickness when making contact with the PM (wide ER), in some ER (B), the lumen virtually disappears (thin ER) upon contact with the PM. Note in B that ribosomes are associated with the dilated portion of the ER (away from the PM), but are absent from the thin ER. (C) TEM micrographs showing the tight apposition of the two opposite faces of thin cortical ER cisternae (red arrows), with near absence of an ER lumen. (D and E) TEM micrographs showing thin cortical ER (thin red arrows) apposed by wide ER with electron-dense accumulations between them (thick short red arrows). (F) Symmetric arrangement of thin cortical ER in two adjacent neurons. The portions enclosed by white rectangles are shown at higher magnification in the two Insets. (G–I) Symmetric cortical ER between two adjacent neurons as seen by FIB-SEM (G) and corresponding 3D reconstruction (H and I) shown in two different orientations. [Scale bars: 200 nm in A–E; 500 nm in F (Inset, 200 nm); 160 nm in G and H.] A and B are from the nucleus accumbens, B–E from the dorsal striatum, and F from the cerebral cortex.
Some thin ER cisternae had additional peculiar features. In some cases, the surface opposite to the PM was apposed to another ER cisterna, which however, had a wide lumen (Fig. 3D and Fig. S2 A and B). In the cytosolic space between these two, ER cisternae electron densities, indicative of protein accumulations, could be observed by TEM (Fig. 3 D and E and Fig. S2 A and B) (48; similar structures are also reported in https://synapseweb.clm.utexas.edu/112-endoplasmic-reticulum). In other cases, the thin cortical ER in one neuron lined up nearly precisely with another thin cortical ER cisterna of an adjacent cell (Fig. 3 E–I and Fig. S2C).
Fig. S2.
Thin ER at ER–PM contact sites. Dorsal striatum. (A and B) TEM micrographs showing thin cortical ER (thin red arrows) flanked by wide ER, with electron dense accumulations between them (thick red arrows). The electron-dense appearance of the thin ER is because of the virtual absence of the lumen, so that the two opposite membranes of the ER cisterns are closely apposed to the each and appear as a single thick membrane at the resolution of FIB-SEM. (C) Thin cortical ER at sites where two adjacent neurons contact each other. The thin ER cisternae (thin red arrows) in the two neurons are in close register with each other. The portion enclosed by white rectangles is shown at higher magnification in the bottom right Inset. Rough ER (RER) in continuity with the smooth thin ER (thin red arrow) is indicated. (Scale bars: 200 nm in A and B and Inset in C; 500 nm in C.)
Dendrites.
Complete reconstructions of all membranous organelles in large dendritic segments are presented in Fig. 4 (nucleus accumbens; see also Movie S3) and Fig. S1 (cerebral cortex). In dendrites the ER is mostly tubular, with occasional small cisternae (Fig. 4 A and B and Fig. S1B) (49–52). Both rough and smooth ER are present. Dendritic ER wraps around mitochondria, which occupy the central portion of the dendrite and also extends, at least partially, into about 50% of the spines (Fig. 4 A, B, F, and G). Other membranous organelles, including endosomes and transport vesicles, are rather sparse in dendrites (Fig. 4E), consistent with a quantitative study of these vesicles in hippocampal neurons (53). Spines with the ER reaching into their head were generally the larger ones, as reported previously (50) (Fig. 4A), including all mushroom spines (Fig. 4 F and G), whereas in other spines (generally smaller ones) the ER only reached into the neck (Fig. 4 F and G). In all cases, the ER of spines was connected to the rest of the network via a thin single tubule that was threaded through the spine neck (Figs. 4 and 5) (50). ER contacts with the PM were also observed in dendrites (Figs. 4C and 5B and Fig. S1C; see also Fig. 2F). However, both the size of these contacts (compare Figs. 1B and 5C) and the fraction of PM involved in such contacts (Fig. 2A) were smaller than in cell bodies. Contacts with mitochondria and with the endosomes and transport vesicle population (Figs. 2A, 4 D and E, and 5C) were similar to those observed in cell bodies.
Fig. 5.
ER and other organelles in dendritic spines. (A–D) Three-dimensional models of two adjacent spines emerging from a dendrite (from FIB-SEM image stacks, nucleus accumbens). (A) All membranous organelles were reconstructed (color coded as in Fig. 1). (B and C) PM and mitochondria, respectively; areas of contacts with the ER shown in red. A cluster of endosomes/transport vesicles with two areas of contact with the ER (red) visible in the Inset of C. (D) ER of one of the two spines shown in A. (E–H) Mushroom-shaped dendritic spine in the cerebral cortex containing a typical spine apparatus. (E) Three-dimensional model of the spine showing all organelles (color coded as in Fig. 1). (F) Single FIB-SEM image showing a cross-section of the spine apparatus, which comprises seven cisternae, one of which makes a contact with the PM in the plane of the image (red arrow). (G and H) Three-dimensional reconstruction of the spine apparatus shown in F, demonstrating two contacts with the PM (red) (H). (Scale bars: 400 nm in A–E; 80 nm in Inset in C; 80 nm in F–H.)
In the head of large spines, the ER adopted a variety of shapes and often was in the form of a spine apparatus (Fig. 5) (50, 54) (https://synapseweb.clm.utexas.edu/spine-apparatus). Some small contacts with the plasma membrane could often be observed (Fig. 5 B, F, and H), but we did not observe the ER making direct contact within the postsynaptic membrane in spines. As expected, the cisternae of the spine apparatus were contiguous with one another and with the rest of the ER, as previously observed (55) (Fig. 5 E–H), although discontinuities between cisternae were observed in few cases. Mitochondria were rarely found in spines (Fig. 4D).
Axons.
Smooth ER extended into all axons, where it had a dense appearance with a narrow lumen. In large axons, both myelinated and unmyelinated, the ER formed a system of anastomosed tubules (50, 52, 56), whereas in thin axonal segments a single continuous tubule was observed (Fig. 6 A and B). At synaptic varicosities, the tubules branched or expanded into small cisternae (Fig. 6 A and B and Movie S4) (see also refs. 56 and 57). These ER elements formed a web that surrounded mitochondria, clusters of synaptic vesicles and the few other organelles of nerve terminals. These included vesicles or vacuoles larger than synaptic vesicles [possibly organelles generated by bulk/ultrafast endocytosis (58, 59)] and occasional multivesicular bodies (Fig. 6D). In some varicosities they also included networks of tubules that were physically discontinuous from the ER network (Fig. 7, light blue elements, and Movie S5). These tubules differed in appearance from the ER elements because of a wider lumen and less-dense appearance (Fig. 7 G and H). Most likely, the tubules represent endosomal compartments, as similar tubules connected to clathrin-coated buds have been observed in presynaptic regions (60), although the possibility that some of them may represent ER elements disconnected from the bulk of the ER network cannot be excluded. As in the case of dendrites, the contact area with the PM (both in unmyelinated and in myelinated axons) appeared smaller than the contact area in cell bodies (Figs. 6 C and I and 7C; see also Insets in Fig. 2 F and H). ER appositions with mitochondria (Figs. 6H and 7 D and F; see also Fig. 2G) and endosome-like structures (Figs. 6D and 7 E and G) were present and reminiscent of those observed in cell bodies and dendrites (Fig. 2E). The ER was excluded from active zones of secretion (Figs. 6 A and E and 7A).
Fig. 6.
ER network in an axon. (A–D) Three-dimensional model of a thin axonal segment (from a FIB-SEM image stack, nucleus accumbens). (A) All membranous organelles were reconstructed; they are color-coded as in Fig. 1. In addition, synaptic vesicles are shown in dark blue. A single ER tubule travels along the axon and expands into small tubular networks at presynaptic varicosities. (B) Selective view of the ER, mitochondria, and endosomes and free vesicles excluding synaptic vesicles. (C) PM where areas of contacts with the ER are shown in red. (D) Endosomes and free vesicles excluding synaptic vesicles (synaptic vesicles were defined as the small, homogenously sized small vesicles clustered together at presynaptic sites). Contact sites between the ER and tubulovesicular structures (most likely endosomes) are shown in red in the two insets. (E–H) High-magnification views of selected organelles in the largest presynaptic varicosity of the axonal segment shown in A. The synaptic interface is shown in magenta. Contacts areas between the ER and mitochondria are highlighted in red in field H. (I and J) Three-dimensional model in a myelinated axon showing the PM, the ER, and a membrane tubule not continuous with the ER (endosome or other transport intermediate; nucleus accumbens). Contact between the ER and the PM and with an isolated tubule (endosome or other transport intermediate) are shown in red in I and J, respectively. (K and L) Micrographs showing original FIB-SEM dataset from which the 3D reconstruction of field E was obtained. Active zones of secretion (AZ) are indicated by white stippled rectangles. (M and N) Micrographs from original FIB-SEM data set illustrating cross-sections of the axons shown in A and I, respectively. (Scale bars: 800 nm in A–D; 40 nm in Insets in D; 160 nm in E–H; 400 nm in I–L; 160 nm in M and N.)
Discussion
In recent years, several proteins that function as tethers between intracellular membranes (for example, see refs. 12, 21, and 61–65) have been identified and there is growing appreciation for the importance in cell physiology of MCSs (9, 12, 38, 66). Determining the abundance and extension of MCSs by an ultrastructural analysis is key to place this information into an anatomical context. Here, we used FIB-SEM to analyze the 3D structure of the ER and its relation to other membranous organelles in cell bodies, dendrites, and axons of brain neurons in situ with the goal of identifying the locations of MCSs in the soma, dendrites, and axons of neurons. Only relatively small portions of several selected neurons were reconstructed. Thus, one should be cautious about the generality of our findings, as the abundance of MCSs is expected to vary in different neurons and to be affected by the functional state of the cell. However, as most neurons are likely to share fundamental principles of intracellular organization, the results shown here provide basic information useful to complement biochemical and functional studies of MCSs. The ER populates even the most distant neuronal compartments without any interruptions, although rough ER is only present in the perikaryon and larger dendritic branches (56). As previously noted, remarkably, this continuity persists even in narrow compartments, like the neck of dendritic spines (50, 52) and the thin branches of terminal axons (67), where single thin threads of the ER remain fully connected to the rest of the ER network. The delicate nature of these ER tubules may help explain why loss-of-function mutations in proteins that shape the ER, or mediate homotypic fusion of its membrane, lead to neurodegenerative diseases (68). For example, mutations in atlastin, which is particularly important in counteracting ER fragmentation by promoting homotypic fusion of ER elements (69, 70), have been linked to hereditary spastic paraplegia (68).
The dense packing of membranous organelles within the cytoplasm implies the frequent random occurrence of transient close encounters between organelles. Thus, we cannot conclude that all contacts observed reflect specific direct or indirect interactions between the two participating membranes. Because of the resolution limits of FIB-SEM, the presence at MCSs of protein densities suggestive of protein tethers, a defining feature of stable membrane appositions, could not always be assessed. However, the peculiar morphology of many of the contacts observed (for example, flat ER cisternae closely apposed to the PM) or the extension of such contacts over long distance (for example, ER-mitochondria contacts) strongly suggest that at least a substantial subset of them represent specific interactions.
Contacts between the ER and the PM were ubiquitous, but covered a larger fraction of the PM in the cell bodies than elsewhere. Possibly, this abundance in cell bodies is related to the lower surface- (PM) to-volume (cytoplasm) ratio, so that functions mediated by ER–PM contact in these compartments must support a greater volume of cytosol and the ER. ER–PM contacts have been proposed to have a multiplicity of functions (see refs. 9, 12, and 66 for recent reviews). These functions include delivery of newly synthesized lipids from the ER to the PM, and return of lipid metabolites from the PM to the ER for metabolic recycling. That lipid exchange between the ER and the PM can occur also in axons is supported by studies of the giant squid axon, indicating the presence of phospholipid-synthesizing enzymes (71) in the core of the axon and the presence of numerous ER–PM contact sites along its PM (72).
Additional housekeeping functions of ER–PM contacts that may operate also in neurons include in trans action of ER-localized enzymes on the PM (73, 74), store-operated Ca2+ entry (SOCE) (18), and coupling between Ca2+ influx from the extracellular medium and Ca2+-release from the ER (75, 76). Both stromal interaction molecule (STIM) and ORAi proteins, key players in SOCE, and ryanodine receptors, ER-localized Ca2+ channels responsible for Ca2+-induced Ca2+ release at the ER–PM junction in muscle (75, 77), are expressed in neurons (78, 79). Similarly, isoforms of junctophilin, another integral protein of the ER membrane that binds the PM in trans and functions as an accessory factor of the ryanodine receptors at muscle triads, are expressed in neurons (80–82).
Interestingly, several of the subsurface cisternae of the cortical ER in cell bodies and large dendrites had an extremely narrow lumen (thin ER), as if the two opposite faces of the cisternae were stapled to one another. Thin cortical ER cisternae (referred to as subsurface cisternae) have been previously observed in neurons (44) (https://synapseweb.clm.utexas.edu/112-endoplasmic-reticulum), in some cases under the postsynaptic membrane (also called hypolemmal cisternae) (28, 41–43, 45, 46), where they have been proposed to be involved in signal transduction. Subsets of thin cortical ER cisternae had additional interesting features, as they were apposed to a wide ER cisterna on the side opposite the PM, or were in precise register with thin ER in an adjacent neuron, suggesting a form of intercellular communication. Similar structures have been observed in cancer cells (83). The peculiar morphology of all these ER structures strongly suggests the occurrence of regional specializations of the cortical ER, although their molecular basis remains unknown. Thin ER expands in cells overexpressing STIM1, leading to the hypothesis that thin ER may be involved in SOCE (47). The predominance of thin ER in cell bodies suggests that SOCE plays a greater role in the soma than in neuronal processes. This finding is in agreement with the dominant role of Ca2+ influx from the PM, rather than from intracellular stores, in the control of Ca2+ dynamics in nerve terminals. However, it remains to be demonstrated whether STIM1 or STIM2 are concentrated in the thin ER of neurons.
Contacts between the ER and mitochondria were abundant in all compartments, consistent with the fact that the two organelles work together in lipid metabolism but lack vesicular transport connecting them (38, 84). Another expected function of ER–mitochondria contacts is the control of direct Ca2+ fluxes between the two organelles without major spillover into the cytosol (19). Our reconstructions revealed a striking tendency of the ER to form a network around mitochondria (see also ref. 85). One might expect this interaction to limit mitochondrial motility. However, at least a fraction of mitochondria are motile in neuronal processes (86), pointing to occurrence of mechanisms to dissociate them or disentangle/fragment them. In fact, contacts with the ER have been implicated in mitochondria fission (87). Interestingly, Gem1 (GTPase EF-hand protein of mitochondria)/miro (mitochondrial rho-like), a GTPase which is found at ER–mitochondria contact sites in yeast (88), has been shown to control mitochondrial motility in neurons (86).
Finally, the identification in all neuronal compartments of contacts between the ER and tubulovesicular structures with the morphology expected for endosomes, complement several recent studies providing evidence for a role of these contacts in lipid transport, control of endosome movement, and endosome fission (12, 89–91). ER appositions were also found with lysosomes and multivesicular bodies (40, 62, 92), as expected. Such appositions were not systematically investigated, given the sparse presence of these organelles in the neuronal volumes that we have reconstructed.
Collectively, our results advance knowledge about the organization of the ER in neurons and of its interaction with other membranes. Mutations affecting ER contacts with various organelles can result in severe neurological diseases, including amyotrophic lateral sclerosis (93), Huntington’s-like chorea (80), and neuroacanthocytosis (64, 94). Abnormal functions of MCSs have also been proposed to have a role in Alzheimer’s disease (95). The information presented in this study, together with further quantitative studies of MCSs in specific neuronal populations and in different activity states, will help to interpret functional studies of MCSs in physiology and pathology in a structural context.
Methods
Animals.
All mice were maintained on a 12-h light/dark cycle with standard mouse chow and water ad libitum. All research and animal care procedures were approved by the Yale University Institutional Animal Care and Use Committee.
Fixation of Mouse Brain Tissue.
Nucleus accumbens for FIB-SEM.
An adult mouse (11-mo-old male C57/BL6J) was deeply anesthetized with sodium pentobarbital (80 mg/kg, i.p.) and intracardially perfused with a mixture of 2% glutaraldehyde/2% depolymerized paraformaldehyde in 0.1 M phosphate buffer at a pH of 6.8. The brain was removed, postfixed overnight at 4 °C, and 50-µm sections were generated on a vibratome. Sections were incubated 30 min in 0.1% CaCl2 dissolved in normal saline (0.9% in water), processed for reduced osmium in 0.1 M cacodylate buffer (0.1 M, pH 7.4) according to the Graham Knott protocol (96, 97), then treated with 2% samarium trichloride and 1% uranyl acetate in maleate buffer pH 6.0, before dehydration and infiltration in Durcupan resin. Sections were sandwiched between two layers of ACLAR plastic and polymerized 48 h at 60 °C. After polymerization, chips from the core of the nucleus accumbens were cut out and glued to custom-made copper specimen holders for FIB-SEM.
Cerebral cortex for FIB-SEM.
Preparation of this material (from a 3-mo-old male C57/BL6 mouse) was described in Hayworth et al. (37).
Dorsal striatum for FIB-SEM and cerebral cortex for TEM.
Two 4-mo-old female mice C57BL6/129 were anesthetized with a ketamine/xylazine anesthetic mixture before perfusion with 2% depolymerized paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer. The brain was then cut into small pieces (less than 1 mm in all dimensions), immersed in perfusion buffer for another 2 h at room temperature, and postfixed in 2% OsO4, 1.5% K4Fe(CN)6 (Sigma-Aldrich), 0.1 M sodium cacodylate buffer 1 h. Subsequently, specimens were stained in block with 2% aqueous uranyl acetate (1 h), dehydrated in increasing concentrations of ethanol, and embedded in Embed 812 (unless otherwise noted, all EM experiment components are from EMS). Ultrathin sections for TEM were imaged in a Philips CM10 microscope at 80 kV and images were taken with a Morada 1 k × 1 k CCD camera (Olympus) and saved in TIFF format. Images were processed with Adobe Photoshop 6.0 software.
FIB-SEM Analysis.
Epon-embedded samples (less than 1 mm in all dimensions) were mounted on sample studs and their tops were trimmed to a 40–100- × 100–300-nm dimension. Samples were then re-embedded in durcupan (EMS, Catalog #14040) by dabbing gently on the trimmed top of the epon-embedded sample with a small volume of uncured durcupan and then exposing them to 60 °C. Next, cured samples were coated with a thin layer of 10-nm gold and 100-nm carbon. For FIB-SEM imaging, a custom built FIB-SEM, with an FEI Magnum FIB column mounted onto a Zeiss Merlin SEM, was used. SEM imaging was performed using a 200-pA electron beam of 700 V landing energy at 200 kHz with 2-nm spacing in the x and y directions. A 7-nA 30-kV gallium ion beam was used to remove 2-nm of material in the z axis after each SEM image to generate a 4- or 8-nm isotropic voxel image stack by an InLens detector (Zeiss electron microscope) in which secondary electrons were filtered out through sample biasing so that only backscattered electrons were detected (98). Volumes sampled were as follows: nuclear accumbens (1,007 μm3 at 8-nm voxel size and 150 μm3 at 4-nm voxel size), cerebral cortex (276 μm3 at 8-nm voxel size) and dorsal striatum (300 μm3 at 4-nm voxel size).
For the generation of the 3D reconstructions, membrane contours were traced semimanually by using 3dmod software. EM images used in the figures are from original TIFF files exported from the 3dmod software via the Movie Montage function. Snap shots of 3D models used in the figures were taken with the Movie Montage function in 3d mod software, with no further processing. For the analysis of the width of the lumen of ER cisternae, measurements were taken at ≤30-nm distance along the length of cross-sectioned cisternae (n = 864 measurements from 207-µm3 stacked images taken at 4 × 4 × 4-nm voxel size).
Supplementary Material
Acknowledgments
P.D.C. was supported by the Howard Hughes Medical Institute, the Kavli Foundation, and NIH Grants R37NS036251, DK45735, and DA018343. C.S.X., K.J.H., and H.F.H. were supported by the Howard Hughes Medical Institute. R.J.W. was supported by the NIH Grant NS 039444.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701078114/-/DCSupplemental.
References
- 1.Porter KR. Observations on a submicroscopic basophilic component of cytoplasm. J Exp Med. 1953;97:727–750. doi: 10.1084/jem.97.5.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shibata Y, Hu J, Kozlov MM, Rapoport TA. Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol. 2009;25:329–354. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- 3.Friedman JR, Voeltz GK. The ER in 3D: A multifunctional dynamic membrane network. Trends Cell Biol. 2011;21:709–717. doi: 10.1016/j.tcb.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNew JA, Sondermann H, Lee T, Stern M, Brandizzi F. GTP-dependent membrane fusion. Annu Rev Cell Dev Biol. 2013;29:529–550. doi: 10.1146/annurev-cellbio-101512-122328. [DOI] [PubMed] [Google Scholar]
- 5.Wang S, Romano FB, Field CM, Mitchison TJ, Rapoport TA. Multiple mechanisms determine ER network morphology during the cell cycle in Xenopus egg extracts. J Cell Biol. 2013;203:801–814. doi: 10.1083/jcb.201308001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 7.Elbaz Y, Schuldiner M. Staying in touch: The molecular era of organelle contact sites. Trends Biochem Sci. 2011;36:616–623. doi: 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefan CJ, Manford AG, Emr SD. ER-PM connections: Sites of information transfer and inter-organelle communication. Curr Opin Cell Biol. 2013;25:434–442. doi: 10.1016/j.ceb.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holthuis JC, Levine TP. Lipid traffic: Floppy drives and a superhighway. Nat Rev Mol Cell Biol. 2005;6:209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- 11.Lahiri S, Toulmay A, Prinz WA. Membrane contact sites, gateways for lipid homeostasis. Curr Opin Cell Biol. 2015;33:82–87. doi: 10.1016/j.ceb.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong R, et al. Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell. 2016;166:408–423. doi: 10.1016/j.cell.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter KR, Palade GE. Studies on the endoplasmic reticulum. III. Its form and distribution in striated muscle cells. J Biophys Biochem Cytol. 1957;3:269–300. doi: 10.1083/jcb.3.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenbluth J. Subsurface cisterns and their relationship to the neuronal plasma membrane. J Cell Biol. 1962;13:405–421. doi: 10.1083/jcb.13.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henkart M, Landis DM, Reese TS. Similarity of junctions between plasma membranes and endoplasmic reticulum in muscle and neurons. J Cell Biol. 1976;70:338–347. doi: 10.1083/jcb.70.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Csordas G, et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis RS. The molecular choreography of a store-operated calcium channel. Nature. 2007;446:284–287. doi: 10.1038/nature05637. [DOI] [PubMed] [Google Scholar]
- 18.Prakriya M, Lewis RS. Store-operated calcium channels. Physiol Rev. 2015;95:1383–1436. doi: 10.1152/physrev.00020.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizzuto R, et al. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 20.Martonosi AN. Mechanisms of Ca2+ release from sarcoplasmic reticulum of skeletal muscle. Physiol Rev. 1984;64:1240–1320. doi: 10.1152/physrev.1984.64.4.1240. [DOI] [PubMed] [Google Scholar]
- 21.Chung J, et al. Intracellular transport. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science. 2015;349:428–432. doi: 10.1126/science.aab1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moser von Filseck J, et al. Intracellular transport. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 2015;349:432–436. doi: 10.1126/science.aab1346. [DOI] [PubMed] [Google Scholar]
- 23.Kornmann B, Walter P. ERMES-mediated ER-mitochondria contacts: Molecular hubs for the regulation of mitochondrial biology. J Cell Sci. 2010;123:1389–1393. doi: 10.1242/jcs.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine T, Loewen C. Inter-organelle membrane contact sites: Through a glass, darkly. Curr Opin Cell Biol. 2006;18:371–378. doi: 10.1016/j.ceb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 25.Wirtz KW, Zilversmit DB. Participation of soluble liver proteins in the exchange of membrane phospholipids. Biochim Biophys Acta. 1969;193:105–116. doi: 10.1016/0005-2736(69)90063-7. [DOI] [PubMed] [Google Scholar]
- 26.Horton AC, et al. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Hanus C, Ehlers MD. Specialization of biosynthetic membrane trafficking for neuronal form and function. Curr Opin Neurobiol. 2016;39:8–16. doi: 10.1016/j.conb.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Smith CA, Sjostrand FS. Structure of the nerve endings on the external hair cells of the guinea pig cochlea as studied by serial sections. J Ultrastruct Res. 1961;5:523–556. doi: 10.1016/s0022-5320(61)80025-7. [DOI] [PubMed] [Google Scholar]
- 29.Harris KM, Weinberg RJ. Ultrastructure of synapses in the mammalian brain. Cold Spring Harb Perspect Biol. 2012;4:a005587. doi: 10.1101/cshperspect.a005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoenger A, McIntosh JR. Probing the macromolecular organization of cells by electron tomography. Curr Opin Cell Biol. 2009;21:89–96. doi: 10.1016/j.ceb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burette AC, et al. Electron tomographic analysis of synaptic ultrastructure. J Comp Neurol. 2012;520:2697–2711. doi: 10.1002/cne.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bushong EA, et al. X-ray microscopy as an approach to increasing accuracy and efficiency of serial block-face imaging for correlated light and electron microscopy of biological specimens. Microsc Microanal. 2015;21:231–238. doi: 10.1017/S1431927614013579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayworth KJ, et al. Imaging ATUM ultrathin section libraries with WaferMapper: A multi-scale approach to EM reconstruction of neural circuits. Front Neural Circuits. 2014;8:68. doi: 10.3389/fncir.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heymann JA, et al. Site-specific 3D imaging of cells and tissues with a dual beam microscope. J Struct Biol. 2006;155:63–73. doi: 10.1016/j.jsb.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayworth KJ, et al. Ultrastructurally smooth thick partitioning and volume stitching for large-scale connectomics. Nat Methods. 2015;12:319–322. doi: 10.1038/nmeth.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: Function of the junction. Nat Rev Mol Cell Biol. 2012;13:607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eden ER. The formation and function of ER-endosome membrane contact sites. Biochim Biophys Acta. 2016;1861:874–879. doi: 10.1016/j.bbalip.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman JR, Dibenedetto JR, West M, Rowland AA, Voeltz GK. Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol Biol Cell. 2013;24:1030–1040. doi: 10.1091/mbc.E12-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boone TB, Aldes LD. Synaptology of the hypoglossal nucleus in the rat. Exp Brain Res. 1984;57:22–32. doi: 10.1007/BF00231128. [DOI] [PubMed] [Google Scholar]
- 42.Davidoff MS, Irintchev AP. Acetylcholinesterase activity and type C synapses in the hypoglossal, facial and spinal-cord motor nuclei of rats. An electron-microscope study. Histochemistry. 1986;84:515–524. doi: 10.1007/BF00482985. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe H. Development of axosomatic synapses of the Xenopus spinal cord with special reference to subsurface cisterns and C-type synapses. J Comp Neurol. 1981;200:323–328. doi: 10.1002/cne.902000304. [DOI] [PubMed] [Google Scholar]
- 44.Spacek J. ‘Free’ postsynaptic-like densities in normal adult brain: Their occurrence, distribution, structure and association with subsurface cisterns. J Neurocytol. 1982;11:693–706. doi: 10.1007/BF01153514. [DOI] [PubMed] [Google Scholar]
- 45.Waxman SG, Pappas GD. An electron microscopic study of synaptic morphology in the oculomotor nuclei of three inframammalian species. J Comp Neurol. 1971;143:41–72. doi: 10.1002/cne.901430105. [DOI] [PubMed] [Google Scholar]
- 46.Fuchs PA, Lehar M, Hiel H. Ultrastructure of cisternal synapses on outer hair cells of the mouse cochlea. J Comp Neurol. 2014;522:717–729. doi: 10.1002/cne.23478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orci L, et al. From the cover: STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc Natl Acad Sci USA. 2009;106:19358–19362. doi: 10.1073/pnas.0911280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siegesmund KA. The fine structure of subsurface cisterns. Anat Rec. 1968;162:187–196. doi: 10.1002/ar.1091620206. [DOI] [PubMed] [Google Scholar]
- 49.Cui-Wang T, et al. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 2012;148:309–321. doi: 10.1016/j.cell.2011.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spacek J, Harris KM. Three-dimensional organization of smooth endoplasmic reticulum in hippocampal CA1 dendrites and dendritic spines of the immature and mature rat. J Neurosci. 1997;17:190–203. doi: 10.1523/JNEUROSCI.17-01-00190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broadwell RD, Cataldo AM. The neuronal endoplasmic reticulum: Its cytochemistry and contribution to the endomembrane system. I. Cell bodies and dendrites. J Histochem Cytochem. 1983;31:1077–1088. doi: 10.1177/31.9.6309951. [DOI] [PubMed] [Google Scholar]
- 52.Martone ME, Zhang Y, Simpliciano VM, Carragher BO, Ellisman MH. Three-dimensional visualization of the smooth endoplasmic reticulum in Purkinje cell dendrites. J Neurosci. 1993;13:4636–4646. doi: 10.1523/JNEUROSCI.13-11-04636.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooney JR, Hurlburt JL, Selig DK, Harris KM, Fiala JC. Endosomal compartments serve multiple hippocampal dendritic spines from a widespread rather than a local store of recycling membrane. J Neurosci. 2002;22:2215–2224. doi: 10.1523/JNEUROSCI.22-06-02215.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gray EG. Electron microscopy of synaptic contacts on dendrite spines of the cerebral cortex. Nature. 1959;183:1592–1593. doi: 10.1038/1831592a0. [DOI] [PubMed] [Google Scholar]
- 55.Spacek J. Three-dimensional analysis of dendritic spines. II. Spine apparatus and other cytoplasmic components. Anat Embryol (Berl) 1985;171:235–243. doi: 10.1007/BF00341418. [DOI] [PubMed] [Google Scholar]
- 56.Lindsey JD, Ellisman MH. The neuronal endomembrane system. III. The origins of the axoplasmic reticulum and discrete axonal cisternae at the axon hillock. J Neurosci. 1985;5:3135–3144. doi: 10.1523/JNEUROSCI.05-12-03135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broadwell RD, Cataldo AM. The neuronal endoplasmic reticulum: Its cytochemistry and contribution to the endomembrane system. II. Axons and terminals. J Comp Neurol. 1984;230:231–248. doi: 10.1002/cne.902300208. [DOI] [PubMed] [Google Scholar]
- 58.Wu Y, et al. A dynamin 1-, dynamin 3- and clathrin-independent pathway of synaptic vesicle recycling mediated by bulk endocytosis. eLife. 2014;3:e01621. doi: 10.7554/eLife.01621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe S, et al. Ultrafast endocytosis at mouse hippocampal synapses. Nature. 2013;504:242–247. doi: 10.1038/nature12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lovas B. Tubular networks in the terminal endings of the visual receptor cells in the human, the monkey, the cat and the dog. Z Zellforsch. 1971;121:341–357. [Google Scholar]
- 61.Giordano F, et al. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 2013;153:1494–1509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henne WM, et al. Mdm1/Snx13 is a novel ER-endolysosomal interorganelle tethering protein. J Cell Biol. 2015;210:541–551. doi: 10.1083/jcb.201503088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stefan CJ, et al. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 64.Lang AB, John Peter AT, Walter P, Kornmann B. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J Cell Biol. 2015;210:883–890. doi: 10.1083/jcb.201502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gatta AT, et al. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife. 2015;4:07253. doi: 10.7554/eLife.07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gallo A, Vannier C, Galli T. Endoplasmic reticulum-plasma membrane associations: Structures and functions. Annu Rev Cell Dev Biol. 2016;32:279–301. doi: 10.1146/annurev-cellbio-111315-125024. [DOI] [PubMed] [Google Scholar]
- 67.Renvoise B, Blackstone C. Emerging themes of ER organization in the development and maintenance of axons. Curr Opin Neurobiol. 2010;20:531–537. doi: 10.1016/j.conb.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blackstone C. Cellular pathways of hereditary spastic paraplegia. Annu Rev Neurosci. 2012;35:25–47. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu J, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 71.Gould RM, Pant H, Gainer H, Tytell M. Phospholipid synthesis in the squid giant axon: Incorporation of lipid precursors. J Neurochem. 1983;40:1293–1299. doi: 10.1111/j.1471-4159.1983.tb13569.x. [DOI] [PubMed] [Google Scholar]
- 72.Metuzals J, Chang D, Hammar K, Reese TS. Organization of the cortical endoplasmic reticulum in the squid giant axon. J Neurocytol. 1997;26:529–539. doi: 10.1023/a:1015482407202. [DOI] [PubMed] [Google Scholar]
- 73.Anderie I, Schulz I, Schmid A. Direct interaction between ER membrane-bound PTP1B and its plasma membrane-anchored targets. Cell Signal. 2007;19:582–592. doi: 10.1016/j.cellsig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 74.Dickson EJ, et al. Dynamic formation of ER-PM junctions presents a lipid phosphatase to regulate phosphoinositides. J Cell Biol. 2016;213:33–48. doi: 10.1083/jcb.201508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Endo M. Calcium-induced calcium release in skeletal muscle. Physiol Rev. 2009;89:1153–1176. doi: 10.1152/physrev.00040.2008. [DOI] [PubMed] [Google Scholar]
- 76.Schneider MF, Chandler WK. Voltage dependent charge movement of skeletal muscle: A possible step in excitation-contraction coupling. Nature. 1973;242:244–246. doi: 10.1038/242244a0. [DOI] [PubMed] [Google Scholar]
- 77.Clarke OB, Hendrickson WA. Structures of the colossal RyR1 calcium release channel. Curr Opin Struct Biol. 2016;39:144–152. doi: 10.1016/j.sbi.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moccia F, et al. Stim and Orai proteins in neuronal Ca(2+) signaling and excitability. Front Cell Neurosci. 2015;9:153. doi: 10.3389/fncel.2015.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Juan-Sanz J, et al. Axonal endoplasmic reticulum Ca2+ content controls release probability in CNS nerve terminals. Neuron. 2017;93:867–881.e6. doi: 10.1016/j.neuron.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Landstrom AP, Beavers DL, Wehrens XH. The junctophilin family of proteins: From bench to bedside. Trends Mol Med. 2014;20:353–362. doi: 10.1016/j.molmed.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: A novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 82.Nishi M, Sakagami H, Komazaki S, Kondo H, Takeshima H. Coexpression of junctophilin type 3 and type 4 in brain. Brain Res Mol Brain Res. 2003;118:102–110. doi: 10.1016/s0169-328x(03)00341-3. [DOI] [PubMed] [Google Scholar]
- 83.Kumegawa M, Cattoni M, Rose GG. Electron microscopy of oral cells in vitro. II. Subsurface and intracytoplasmic confronting cisternae in strain KB cells. J Cell Biol. 1968;36:443–452. doi: 10.1083/jcb.36.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Meer G, de Kroon AI. Lipid map of the mammalian cell. J Cell Sci. 2011;124:5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- 85.Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol. 2010;190:363–375. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang X, Schwarz TL. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009;136:163–174. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kornmann B, Osman C, Walter P. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc Natl Acad Sci USA. 2011;108:14151–14156. doi: 10.1073/pnas.1111314108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Raiborg C, et al. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 2015;520:234–238. doi: 10.1038/nature14359. [DOI] [PubMed] [Google Scholar]
- 90.Alpy F, et al. STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J Cell Sci. 2013;126:5500–5512. doi: 10.1242/jcs.139295. [DOI] [PubMed] [Google Scholar]
- 91.Rowland AA, Chitwood PJ, Phillips MJ, Voeltz GK. ER contact sites define the position and timing of endosome fission. Cell. 2014;159:1027–1041. doi: 10.1016/j.cell.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wijdeven RH, et al. Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat Commun. 2016;7:11808. doi: 10.1038/ncomms11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nishimura AL, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rampoldi L, Danek A, Monaco AP. Clinical features and molecular bases of neuroacanthocytosis. J Mol Med (Berl) 2002;80:475–491. doi: 10.1007/s00109-002-0349-z. [DOI] [PubMed] [Google Scholar]
- 95.Area-Gomez E, et al. Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am J Pathol. 2009;175:1810–1816. doi: 10.2353/ajpath.2009.090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Knott G, Rosset S, Cantoni M. Focussed ion beam milling and scanning electron microscopy of brain tissue. J Vis Exp. 2011;(53):e2588. doi: 10.3791/2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knott G, Marchman H, Wall D, Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J Neurosci. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu CS, et al. Enhanced FIB-SEM systems for large-volume 3D imaging. Elife. May 13, 2017 doi: 10.7554/eLife.25916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hama K, Saito K. Fine structure of the afferent synapse of the hair cells in the saccular macula of the goldfish, with special reference to the anastomosing tubules. J Neurocytol. 1977;6:361–373. doi: 10.1007/BF01178223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.