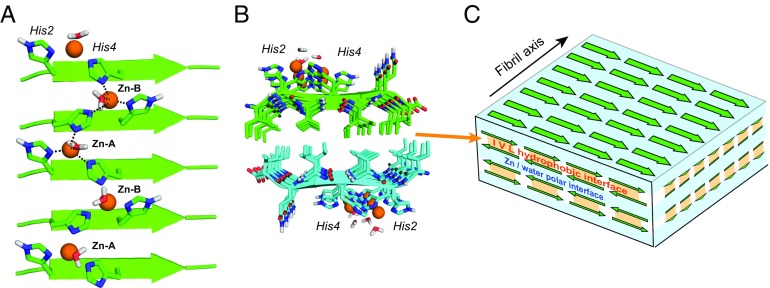
Significance
Functional and pathological amyloid fibrils bind metal ions, but no metal-bound amyloid structures have been determined. Using solid-state NMR and structural bioinformatics, we have determined the oligomeric structure and coordination geometry of a Zn2+-mediated amyloid fibril that catalyzes ester hydrolysis. The peptide assembles into parallel β-sheets in which histidines bridge zinc ions to promote β-strand association in a geometry that mediates water activation for catalysis. The study demonstrates an approach for determining the structures of metalloamyloids. The resulting structure defines how metal ions can stabilize amyloids, lends support to the hypothesis that amyloids can serve as well-structured intermediates between amino acids and proteins during the evolution of life, and provides a framework for potential applications in material science.
Keywords: magic angle spinning, metalloprotein, histidine, metal–peptide framework
Abstract
Throughout biology, amyloids are key structures in both functional proteins and the end product of pathologic protein misfolding. Amyloids might also represent an early precursor in the evolution of life because of their small molecular size and their ability to self-purify and catalyze chemical reactions. They also provide attractive backbones for advanced materials. When β-strands of an amyloid are arranged parallel and in register, side chains from the same position of each chain align, facilitating metal chelation when the residues are good ligands such as histidine. High-resolution structures of metalloamyloids are needed to understand the molecular bases of metal–amyloid interactions. Here we combine solid-state NMR and structural bioinformatics to determine the structure of a zinc-bound metalloamyloid that catalyzes ester hydrolysis. The peptide forms amphiphilic parallel β-sheets that assemble into stacked bilayers with alternating hydrophobic and polar interfaces. The hydrophobic interface is stabilized by apolar side chains from adjacent sheets, whereas the hydrated polar interface houses the Zn2+-binding histidines with binding geometries unusual in proteins. Each Zn2+ has two bis-coordinated histidine ligands, which bridge adjacent strands to form an infinite metal–ligand chain along the fibril axis. A third histidine completes the protein ligand environment, leaving a free site on the Zn2+ for water activation. This structure defines a class of materials, which we call metal–peptide frameworks. The structure reveals a delicate interplay through which metal ions stabilize the amyloid structure, which in turn shapes the ligand geometry and catalytic reactivity of Zn2+.
Metals are essential in enzyme catalysis and protein folding (1). Naturally occurring metalloenzymes possess complex 3D folds to coordinate the metal center with the appropriate geometries for catalysis. A classical example is carbonic anhydrase, where a zinc ion is coordinated by three histidines from two β-strands and a hydroxide ion to catalyze the hydration of carbon dioxide to form bicarbonate (2). It has been hypothesized that such enzyme structure and function might have evolved from short peptides that self-assemble into repeat structures (3–7), in which the metal ions played a significant role by stabilizing the amyloid structure as well as catalyzing reactions. Cu2+ and Zn2+ ions also bind amyloid proteins involved in neurodegenerative disorders at physiological concentrations of these ions (8–14). Structure determination of metal-bound amyloids is thus important for a fundamental understanding of the structural principles of amyloid formation.
NMR spectroscopy has been used to investigate metalloprotein structures by exploiting distance-dependent paramagnetic relaxation enhancement, contact shifts, and pseudocontact shifts of paramagnetic ions such as Cu2+ and Co2+ (15–17). However, this approach cannot be applied to diamagnetic metals such as Zn2+, and direct observation of these quadrupolar nuclei is limited by low sensitivity (18). Zinc, in particular, is abundant and essential in biology (19); thus, it is important to develop a systematic NMR approach for characterizing the inorganic cores of zinc metalloproteins. Solid-state NMR (SSNMR) is the method of choice for structure determination of amyloid fibrils, and high-resolution structures of a number of fibrils have been reported (20–27). However, the metal coordination geometries of amyloid fibrils have not been reported.
Here we present a solid-state NMR investigation of the structure of a designed zinc-binding amyloid fibril that catalyzes ester hydrolysis (5, 28). We have determined the zinc coordination geometry and oligomeric structure of this fibril, which is formed by an amphipathic heptapeptide containing a pair of histidines. Intermolecular distance restraints show that the peptides assemble into hydrogen-bonded parallel in-register β-sheets with alternating dry and wet interfaces between adjacent β-sheets. The hydrophobic interface is stabilized by apolar side chains, whereas the hydrated polar interface houses an array of Zn2+-binding histidines. The 15N and 13C chemical shifts indicate that the two histidines in each peptide adopt singly Nδ1-coordinated and doubly Nδ1, Nε2-coordinated structures at equal populations, whereas measured side chain conformations reveal how the imidazole rings protrude from the β-sheet plane. Combining these solid-state NMR constraints with a structural bioinformatics search, we show that each zinc ion is coordinated by three histidine nitrogens from two adjacent strands, and half of all histidines bridge Zn2+ ions, forming a metal–imidazolate chain that is orthogonal to the direction of the β-strands. This study represents a structure determination of a metalloamyloid and demonstrates a systematic approach for solving the high-resolution structures of diamagnetic metalloproteins from SSNMR data.
Results and Discussion
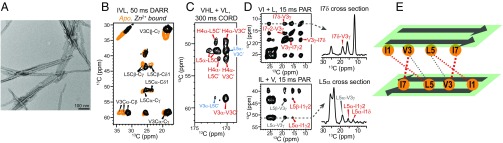
We recently showed that an amphiphilic heptapeptide, Ac-IHIHIQI-CONH2, assembles into micron-length fibrils with high esterase activity (28). The catalytic activity is strictly zinc-dependent, requires histidines at positions 2 and 4, and is enhanced by Gln at position 6 and by β-branched residues at odd-numbered positions. To simplify NMR assignments, we replaced the isoleucines in positions 3 and 5 with Val and Leu, respectively. These substitutions are expected to be well tolerated, given previous work on closely related peptides (5, 28). Indeed, the resulting peptide Ac-IHVHLQI-CONH2 (termed HHQ here) chosen for the SSNMR study forms fibrils that are very similar morphologically and chemically to the ones produced by Ac-IHIHIQI-CONH2. The peptides were fibrilized at pH 8 with varying Zn2+:peptide molar ratios. Transmission electron microscopy (TEM) and thioflavin T fluorescence data confirm that HHQ forms homogeneous fibrils with widths of 20–30 nm, and the Zn2+-bound fibrils catalyze p-nitrophenylacetate hydrolysis with an initial rate that fits to kcat = 0.034 s−1 and KM = 509 µM (Fig. 1A and SI Appendix, Fig. S1), similar to the activity of the parent peptide.
Fig. 1.
Zn2+-bound HHQ peptides form parallel β-strands that stack in antiparallel sheets. (A) TEM image of Zn2+-bound fibrils. (B–D) The 2D 13C–13C correlation spectra of HHQ fibrils. (B) The 50-ms DARR spectra of IVL-labeled fibrils without (orange) and with (black) Zn2+. (C) The 300-ms CORD spectrum of a Zn2+-bound mixed fibril. Most intermolecular backbone cross peaks indicate parallel-in-register strands (red) with a minor component adopting other packing (blue). (D) The 15-ms PAR spectra of Zn2+-bound mixed fibrils, showing V3-I7 and I1-L5 intermolecular cross peaks. (E) Schematic of parallel β-strands in each sheet and two neighboring sheets with opposite strand orientations. Dashed lines indicate intermolecular contacts that have been observed from the 2D correlation spectra, with thicker lines denoting stronger cross peaks or shorter distances.
HHQ Assembles into a Class 1 Steric Zipper with a Parallel Orientation of Adjacent Hydrogen-Bonded Strands.
The conformation and intermolecular packing of HHQ fibrils are determined from 2D 13C–13C correlation spectra (Fig. 1 B–E and SI Appendix, Fig. S2 A–C). With 50-ms 13C spin diffusion based on dipolar-assisted rotational resonance (DARR) mixing (29), intraresidue cross peaks with characteristic β-strand 13C chemical shifts are observed for all labeled residues. Zn2+ binding perturbed the chemical shifts of V3 and L5, suggesting this segment to be the center of the zinc-binding domain (Fig. 1B). With 300-ms combined R2nν-driven (CORD) mixing (30) under echo detection to simplify the spectrum, a mixed labeled fibril (sample 4, SI Appendix, Table S1) shows strong intermolecular cross peaks between V3 Cα and V3 C′ and between L5 Cα and L5 C′ (Fig. 1C), indicating parallel-in-register (PIR) packing of the β-strands. In addition, an intermolecular L5 Cα–V3 C′ cross peak is detected, but only a very weak V3 Cα–L5 C′ cross peak is present. These L5–V3 peaks account for ∼20% of the total intensity and may result from a small amount of parallel-out-of-register strands or antiparallel strands. Such minor structural polymorphism has been observed for various amyloids (31), and we consider only the predominant conformation of the PIR β-strands below. This interpretation is also consistent with the experimentally measured binding stoichiometry, which is 0.75 Zn2+ per peptide, somewhat less than 1.0 Zn2+ per peptide, suggesting the presence of a minor conformer with reduced affinity for Zn2+.
Having established the parallel registry of adjacent strands in the predominant conformer, we examined mixtures of peptides with different 13C, 15N-labeled residues to determine whether interacting sheets were oriented in a parallel or antiparallel fashion across the non–hydrogen-bonded sheet interface. Using samples 9 and 10 (SI Appendix, Table S1), we observed exclusively intermolecular cross peaks between V3 and I7 side chains and between I1 and L5 side chains in 15-ms 2D 13C–13C proton-assisted recoupling (PAR) spectra (32) (Fig. 1D), whereas no L5–I7 side chain contacts were detected. Thus, two adjacent β-sheets stack with the hydrophobic side chains facing each other and with the strands in an antiparallel orientation (Fig. 1E). This cross-β structure, with parallel packing of β-strands within each sheet and antiparallel packing between sheets, has been termed class 1 steric zippers (33). We speculate that it is stabilized by the C2 symmetry along the fibril axis, which allows two adjacent sheets to slide relative to each other to optimize the side chain packing.
His2 and His4 Side Chains of HHQ Each Have Two Chemically Distinct Structures in a 1:1 Molar Ratio.
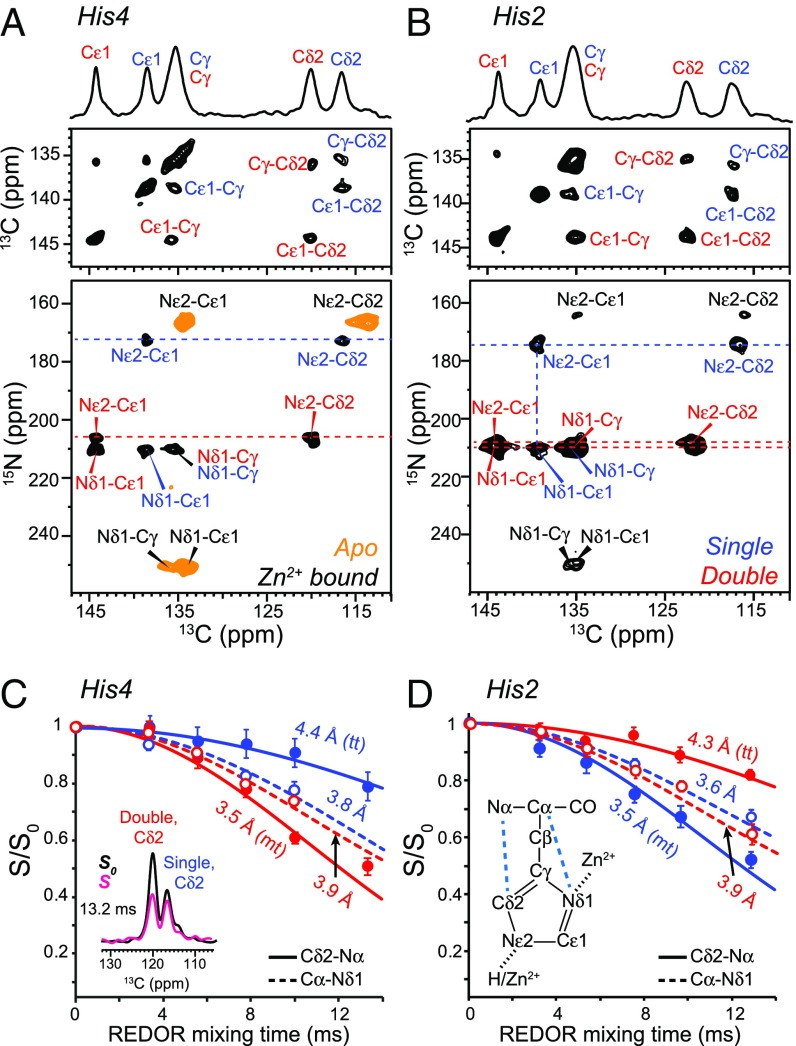
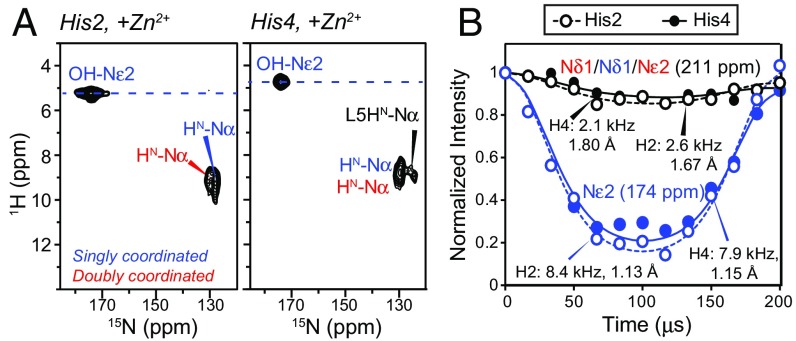
To determine the Zn2+ coordination structure, we measured the chemical shifts and conformation of the two crucial histidines. Without Zn2+, His2 and His4 show 13C and 15N chemical shifts that are diagnostic of a neutral τ-tautomer (34) (Figs. 2 and 3). Zn2+ binding caused pronounced spectral changes, where each His now exhibits two sets of chemical shifts (SI Appendix, Table S2). The 250-ppm 15N peak of unprotonated nitrogen is replaced by two 15N peaks at 207–211 ppm, which are characteristic of Zn2+ coordination (35, 36). One set of signals has a 174-ppm Nε2 chemical shift, indicating Nδ1-only coordination, whereas the other set shows both Nδ1 and Nε2 peaks at ∼210 ppm, indicating double coordination. The two coordination structures have equal intensities, but His2 retains ∼15% unbound signals, which is likely related to the minor conformation described above.
Fig. 2.
(A and B) Histidine structures in HHQ fibrils from chemical shifts and (C and D) distance restraints. (A and B) The 2D 13C–13C and 15N–13C correlation spectra of His4-labeled and His2-labeled HHQ fibrils. The orange spectrum in A is that of the apo sample, whereas the rest correspond to Zn2+-bound fibrils. Singly and doubly coordinated histidine peaks are assigned in blue and red, respectively. (C and D) Cδ2-Nα (solid line) and Cα-Nδ1 (dashed line) REDOR dephasing curves of His4 and His2 to determine the side chain conformation. Representative REDOR spectra and the histidine chemical structure are shown.
Fig. 3.
The 1D 13C and 15N spectra of the HHQ fibrils. (A) The 1D 13C spectra of His2-labeled fibrils without and with zinc. ssb denotes spinning sidebands. Apo, singly coordinated, and doubly coordinated histidine signals are assigned in black, blue, and red, respectively. (B) The 1D 15N spectra of His2-labeled fibrils without and with zinc. (C) The 1D 13C spectra of VHL–VL mixed labeled sample without and with zinc. An 15N–13C dipolar filter was used to select the His4 signals. (D) The 1D 15N spectra of VHL–VL mixed labeled fibrils without and with zinc. Peak assignments are obtained from 2D correlation spectra. Zn2+ binding caused two sets of chemical shifts for His2 and His4.
The equal presence of singly and doubly coordinated histidines is unexpected because naturally occurring Zn2+–His complexes in proteins predominantly involve singly coordinated histidines. We determined the histidine rotamers by measuring Cα–Nδ1 and Cδ2–Nα distances using frequency-selective 13C–15N rotational echo double resonance (REDOR) (37). The normalized REDOR intensities (S/S0) as a function of mixing time (Fig. 2 C and D) indicate a Cα–Nδ1 distance of 3.6–3.9 Å for both histidines, which constrains the χ2 angle to trans. However, the Cδ2–Nα distances differ. For His2, the Cδ2–Nα distance is shorter in the singly coordinated form (His2S) than the doubly coordinated form (His2D), indicating an mt rotamer for His2S and a tt rotamer for His2D (SI Appendix, Figs. S3 and S4), whereas the opposite rotamer combination is found for His4.
Determination of the Structure of the Zn2+-Binding Site.
The above NMR data reveal the following structural features of the Zn-bound HHQ fibrils: (i) PIR packing of β-strands in each sheet; (ii) all His residues are coordinated to Zn2+ via Nδ1, and in addition, half of the His residues are also coordinated to Zn2+ via Nε2 (these singly and doubly Zn-coordinated His residues are equally populated); (iii) distinct His rotamers exist at positions 2 and 4 and depend on the coordination number; and (iv) Zn2+ binds in a 1:1 metal ion/peptide ratio. Thus, on average, each Zn2+ is coordinated by three His N ligands (two Nδ1 and one Nε2).
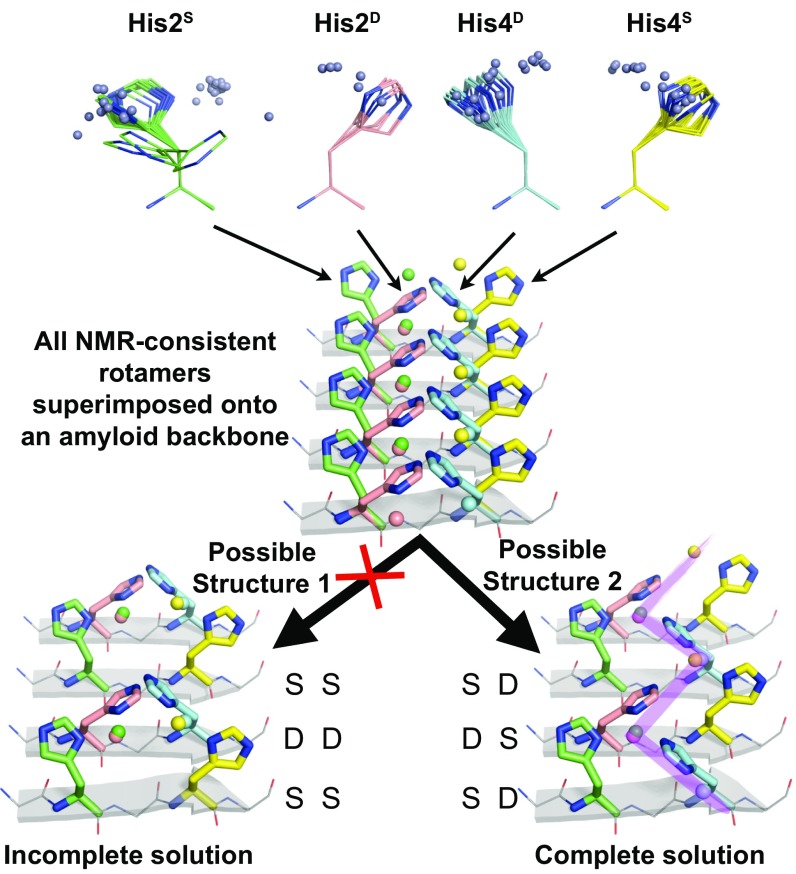
These features allow for two possible Zn coordination structures of the amyloid, with a peptide dimer as the asymmetric unit (Fig. 4). For any two adjacent PIR β-strands j and j + 1, feature ii dictates that His2 of strand j be singly coordinated if His2 of strand j + 1 is doubly coordinated. Likewise, His4 of strand j must be singly coordinated if His4 of strand j + 1 is doubly coordinated. Two possible Zn coordination configurations can arise from the combinatorics: S for His2 and S for His4 in strand j would lead to an SS/DD (j/j + 1) configuration, whereas S for His2 and D for His4 in strand j would lead to an SD/DS (j/j + 1) configuration. We devised a structural bioinformatics approach to eliminate the possibility of one of these two models (SS/DD), as well as to determine the plausibility of the remaining model (SD/DS). Although structural bioinformatics is widely used in protein structure determination and verification (38, 39), it has been much less used for determining metal–protein coordination structure.
Fig. 4.
Determination of the HHQ metalloamyloid structure by structural bioinformatics. His Nδ1-Zn fragments from natural proteins (Top) that are consistent with the SSNMR data of HHQ were docked onto a β-sheet backbone (PDB code: 1YJP). A complete solution to the SSNMR constraints of HHQ is found and produces an infinite zigzag of Zn2+ (purple shade). S (single) and D (double) refer to the coordination number of the His. Zn2+ atoms are shown as spheres.
Our approach begins with identifying structural elements in the Protein Data Bank (PDB) that simultaneously satisfied features i–iv of the HHQ fibril above. Within a nonredundant database of the PDB, we searched for His residues with Nδ1 or Nε2 atoms within 2.5 Å of Zn. To satisfy feature i, we restricted these hits to His residues with β-sheet (ϕ, ψ) angles. To satisfy feature ii, we only considered His–Zn fragments with Nδ1 coordination because all His residues in HHQ coordinate Zn with their Nδ1 nitrogen. From this set of fragments, we only considered His–Zn fragments that satisfy the NMR-derived His rotamer constraints, thus satisfying feature iii. This search process resulted in distinct sets of His–Zn geometries from natural proteins that agree with all NMR constraints for His2S, His2D, His4S, and His4D (four sets total). These His–Zn fragments were superimposed via backbone atoms onto a PIR β-strand amyloid structure from the PDB at positions i and i + 2. This backbone superposition places the accompanying Zn atoms of the His–Zn fragments in space relative to the four His side chains of the dimer asymmetric unit. From this distribution, we sought a Zn coordination structure that is consistent not only with the experimental SSNMR data but also with the observed His–Zn coordination geometries from the PDB (SI Appendix, Figs. S5 and S6). We found that the SD/DS solution yielded excellent overlap between Zn distributions of Nδ1-coordinated His residues that were cross-strand (strands j, j + 1 as well as j, j − 1) and at the same residue position. Furthermore, although Nε2 ligation was not included as an explicit restraint, a single set of rotamers positioned the Nε2 of the third His in an optimal geometry for double coordination. The alternative configuration of SS/DD was eliminated as a possibility because it resulted in a Zn-binding geometry of doubly coordinated His residues that is far from those observed in a large set of nonredundant X-ray crystal structures and that is inconsistent with chemical principles known to determine Zn coordination geometries (40, 41).
The resulting structure (Fig. 5A and SI Appendix, Fig. S7) shows a singly and doubly coordinated histidine in each strand, with the side chains alternately pointing to the N terminus in the SD strand (mt rotamers) and the C terminus (tt rotamers) in the adjacent DS strand. Two β-strands constitute the basic repeat unit, and two different triple-His coordination spheres exist. Zn2+–A is bound to His2S and His4D from one strand and His2D from the neighboring strand, whereas Zn2+–B is chelated by His2D and His4S from one strand and His4D from the adjacent strand. The doubly coordinated histidines bridge the Zn2+ ions in an infinite zigzag along the fibril axis. Although we do not directly measure Zn–N contacts, the computed structure shows N–Zn–N angles near that of a tetrahedron, leaving one free coordination site at each Zn2+ to interact with water or substrates.
Fig. 5.
Coordination structure of Zn2+-bound HHQ fibrils. (A) Energy-minimized structure of one β-sheet. Each Zn2+ (orange) is coordinated by three His nitrogens from two neighboring strands. Half of the histidines bridge two Zn2+ ions. Water molecules are present in the coordination sphere, but their exact positions relative to Zn2+ are unknown. (B) Two β-sheets stack with the hydrophobic residues facing each other and with the strands in the two sheets having antiparallel orientations. (C) Schematic of the 3D assembly of the HHQ fibril. The parallel hydrogen-bonded β-sheets stack into bilayers with alternating hydrophobic and polar interfaces that contain hydrated Zn2+-coordinating histidines.
Hydration of the Zn2+-Binding Site of HHQ.
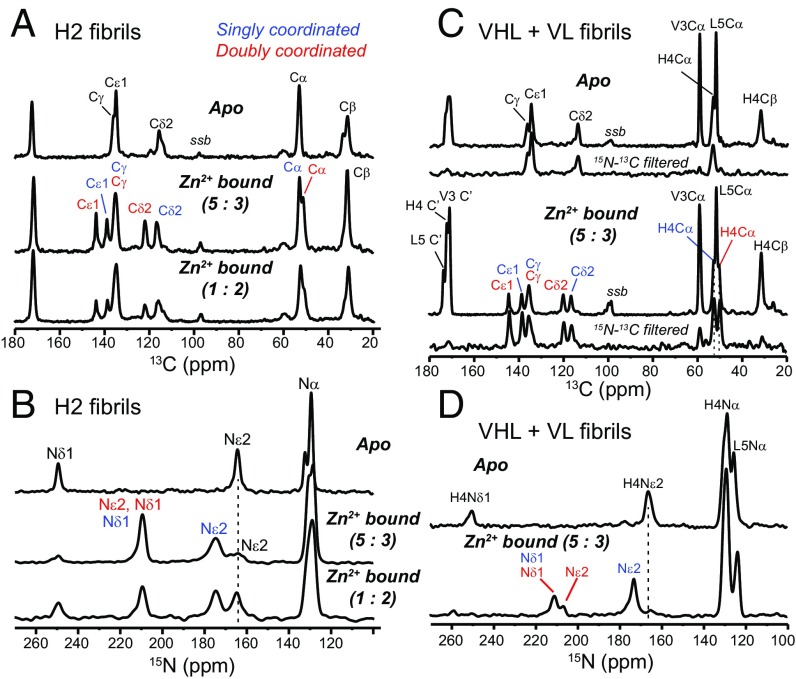
Direct evidence that water hydrates the His side chains and thus lies in the vicinity of Zn2+ is obtained from 2D 1H–15N correlation spectra of the hydrated fibrils (Fig. 6A) (42). The spectra show clear water cross peaks to the 174-ppm Nε2 of His2S and His4S, and the water line widths are narrower than the aromatic 1H line widths (SI Appendix, Fig. S8), indicating that the histidine-associated water exchanges rapidly with bulk water. Under frozen conditions, 15Nε2–1H dipolar couplings are 8.4 kHz for His2S and 7.9 kHz for His4S (Fig. 6B) (43), indicating that the nearest water proton is 1.13 and 1.15 Å away, respectively. In comparison, the 211-ppm nitrogens show much weaker couplings of 2.1 and 2.6 kHz, consistent with the absence of nearby protons for these Zn2+-coordinating nitrogens.
Fig. 6.
Water–histidine interactions in HHQ fibrils. (A) The 2D 1H–15N correlation spectra of His2 and His4 in Zn2+-bound HHQ. Nε2 of singly coordinated His exhibits a narrow water cross peak. (B) The 15N–1H dipolar couplings of His2 and His4 at 243 K where peptide motion is frozen. The couplings correspond to Nε2–H distances of 1.13 Å and 1.15 Å for singly coordinated His2 and His4. The Zn2+-ligating nitrogens show much weaker dipolar couplings, as expected.
The Zn2+-coordinated and hydrated histidine structures, together with the intermolecular Ile, Val, and Leu side chain contacts, indicate that the PIR β-strands assemble into bilayers whose interior consists of the hydrophobic Ile, Val, and Leu side chains from two apposing sheets, whereas the exterior is decorated by the polar His2, His4, and water, coordinating Zn2+ (Fig. 5B). Atomic force microscopy data (not shown) indicate that multiple bilayers can stack, stabilized by the hydrated polar interfaces (Fig. 5C). The relative orientation of the strands bracketing the polar interface is not known and may be either parallel or antiparallel. In either case, the hydrated Zn2+ ligand would appear well oriented to interact with substrates diffusing to sites on the surface of the fibril or hydrated sheets within the fibril.
The alternating dry and wet sheet–sheet interfaces resemble the steric zipper structures of crystalline peptide fibrils (44), but the inclusion of an infinite chain of metal–ligand complexes establishes a different class of structures, which we name metal–peptide frameworks by analogy to metal–organic frameworks. This 3D assembly is held together by multiple interactions: hydrogen-bonding and metal–ligand interactions between β-strands, the hydrophobic effect, and polar water-mediated interactions. A similar topology of Cu2+ bridging His residues on adjacent parallel β-strands has been inferred from scanning tunneling microscopy of a Cu2+-bound N-terminal 16-residue peptide of the Alzheimer’s β-amyloid peptide (45).
This bridging-histidine stabilized amyloid fibril structure, heretofore unseen in naturally occurring proteins, may be important in prebiotic molecules for templating enzymatic functions and may also exist in neurodegenerative amyloids (46) to select for pathologically significant 3D folds, conduct redox functions, and regulate metal homeostasis. Determining the metal coordination structures should thus be useful for designing artificial catalysts and materials and might also have implications for the structural stabilities of neurodegenerative amyloids. Although metalloproteins harboring paramagnetic ions such as Cu2+ have been studied with NMR (17), paramagnetic broadening makes the metal center difficult to detect. Thus, diamagnetic Zn2+-containing proteins represent advantageous alternative targets for structure determination of metalloamyloids and metalloproteins. The SSNMR approach of measuring backbone and side chain distances and chemical shifts that are indicative of coordination structures is generally applicable and can be used, as shown here, for the structure determination of other metal–peptide frameworks.
Methods
Peptide Synthesis.
Ac-IHVHLQI-CONH2 was synthesized on a 0.1-mmol scale using Fmoc solid-phase synthesis as described recently (47) and was purified by reverse-phase HPLC to >98% purity. Peptide mass was verified by MALDI-TOF mass spectrometry. A peptide stock solution in 10 mM HCl was prepared for subsequent biochemical and SSNMR experiments.
Transmission Electron Microscopy.
HHQ peptide (3 mg) was dissolved in 8 M urea (450 µL) and incubated at room temperature for 15 min. Fibrilization was initiated by adding 4.43 mL Tris buffer (25 mM Tris, pH 8) containing 1 mM or 0.3 mM Zn2+. After 5 min of incubation, the sample (545 µM peptide) was diluted to 25 µM using the same buffer without Zn. Sample aliquots were adsorbed onto 200-mesh copper grids and then stained with uranyl acetate as previously reported (28). An FEI Tecnai F20 electron microscope at an acceleration voltage of 80 kV was used to obtain the micrographs.
Fibril Preparation for Solid-State NMR.
Ten 13C, 15N-labeled fibril samples with Zn2+:peptide ratios of 0–4:1 were prepared for SSNMR experiments (SI Appendix, Table S1). Unless explicitly stated otherwise, most fibril samples were prepared using excess of zinc (∼1.5-fold) to ensure complete binding of the metal ions to HHQ. From the pH 2 peptide stock, fibrilization was initiated by diluting the stock in pH 8 Tris buffer with or without ZnCl2. The precipitates were collected by centrifugation. Additional fibrilization details are given in SI Appendix, Materials and Methods.
Solid-State NMR Experiments.
The 2D PAR, CORD, and 15N–1H and 13C–1H correlation experiments were measured on a Bruker 800-MHz (18.8-T) spectrometer using a 3.2-mm HCN triple-resonance magic angle spinning (MAS) probe. The 13C–15N REDOR and 2D 15N–1H dipolar-chemical shift correlation (DIPSHIFT) experiments were measured on a 400-MHz (9.4-T) spectrometer using a 4-mm MAS probe. The 2D 15N–13C correlation and 2D DARR experiments were measured on both 400- and 800-MHz spectrometers. Most spectra were measured at 268–298 K, except for the 15N–1H DIPSHIFT data, which were collected at 243 K. Further experimental details and 13C–15N REDOR fitting procedures are given in SI Appendix, Materials and Methods.
Bioinformatics Search and Structural Modeling.
We downloaded a representative single-chain PDB database from Dunbrack’s PISCES server (48), updated on November 11, 2016: cullpdb_pc50_res2.5_R1.0_d161111_ chains21454. The database contains 21,454 single chains from proteins with X-ray diffraction resolution of ≤2.5 Å and sequence identity ≤50%. We loaded this database into the Python-based bioinformatics program ProDy (49). Within ProDy, we retrieved all histidines (along with i − 1 and i + 1 residues) that coordinate with Zn (His Nδ1 or Nε2 within 2.5 Å of Zn) (Fig. 4). We filtered these by Nδ1–Zn coordination, motivated by the SSNMR data that showed that all His residues within HHQ have their Nδ1 atoms coordinated to Zn. We then filtered these three-residue + Zn fragments by His (ϕ, ψ) angles to select those fragments in β-sheets, as defined by −180° < φ < −45° and 45° < ψ < 225° (50). These fragments were further filtered and binned by the NMR-derived distance constraints (SI Appendix, Fig. S4). The number of fragments found for the singly coordinated His rotamer at position 2 was 35, and the number found for doubly coordinated was 6; the number found for the singly coordinated His rotamer at position 4 was 15, and the number found for doubly coordinated was 18. The His residues along with the coordinating Zn from these fragments were aligned by Cα, C′, N, and O backbone atoms onto positions i and i + 2 of a β-sheet amyloid structure (PDB code: 1YJP) and analyzed for overlapping Zn distributions and simultaneous satisfaction of doubly coordinated His (i.e., His Nε2 is also positioned in a geometry that coordinates Zn). The final model obeys all NMR constraints and yields a Zn:peptide ratio of 1:1. The bioinformatics search results were used to confirm the SSNMR-restrained structure, which was calculated in the CYANA software (51) and refined in Xplor-NIH (52). Further details on the structural modeling are given in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Dr. Paul White for measuring initial SSNMR spectra. This work is partly supported by NIH Grants GM066976 (to M.H.), GM119634 (to I.V.K.), and GM54616 and P01AG002132 (to W.F.D.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The zinc-binding amyloid structure has been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5UGK). The chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank (BMRB ID code 30227).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706179114/-/DCSupplemental.
References
- 1.Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. University Science Books; Mill Valley, CA: 1994. [Google Scholar]
- 2.Christianson DW, Fierke CA. Carbonic anhydrase: Evolution of the zinc binding site by nature and by design. Acc Chem Res. 1996;29:331–339. [Google Scholar]
- 3.Carny O, Gazit E. A model for the role of short self-assembled peptides in the very early stages of the origin of life. FASEB J. 2005;19:1051–1055. doi: 10.1096/fj.04-3256hyp. [DOI] [PubMed] [Google Scholar]
- 4.Greenwald J, Riek R. On the possible amyloid origin of protein folds. J Mol Biol. 2012;421:417–426. doi: 10.1016/j.jmb.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Friedmann MP, et al. Towards prebiotic catalytic amyloids using high throughput screening. PLoS One. 2015;10:e0143948. doi: 10.1371/journal.pone.0143948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero Romero ML, Rabin A, Tawfik DS. Functional proteins from short peptides: Dayhoff’s hypothesis turns 50. Angew Chem Int Ed Engl. 2016;55:15966–15971. doi: 10.1002/anie.201609977. [DOI] [PubMed] [Google Scholar]
- 7.Makhlynets OV, Gosavi PM, Korendovych IV. Short self-assembling peptides are able to bind to copper and activate oxygen. Angew Chem Int Ed Engl. 2016;55:9017–9020. doi: 10.1002/anie.201602480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bush AI, et al. Rapid induction of Alzheimer A beta amyloid formation by zinc. Science. 1994;265:1464–1467. doi: 10.1126/science.8073293. [DOI] [PubMed] [Google Scholar]
- 9.Lovell MA, Robertson JD, Teesdale WJ, Campbell JL, Markesbery WR. Copper, iron and zinc in Alzheimer’s disease senile plaques. J Neurol Sci. 1998;158:47–52. doi: 10.1016/s0022-510x(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 10.Viles JH. Metal ions and amyloid fiber formation in neurodegenerative diseases. Copper, zinc and iron in Alzheimer’s, Parkinson’s and prion diseases. Coord Chem Rev. 2012;256:2271–2284. [Google Scholar]
- 11.Rasia RM, et al. Structural characterization of copper(II) binding to alpha-synuclein: Insights into the bioinorganic chemistry of Parkinson’s disease. Proc Natl Acad Sci USA. 2005;102:4294–4299. doi: 10.1073/pnas.0407881102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtain CC, et al. Alzheimer’s disease amyloid-beta binds copper and zinc to generate an allosterically ordered membrane-penetrating structure containing superoxide dismutase-like subunits. J Biol Chem. 2001;276:20466–20473. doi: 10.1074/jbc.M100175200. [DOI] [PubMed] [Google Scholar]
- 13.Barnham KJ, Bush AI. Metals in Alzheimer’s and Parkinson’s diseases. Curr Opin Chem Biol. 2008;12:222–228. doi: 10.1016/j.cbpa.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Parthasarathy S, et al. Molecular-level examination of Cu2+ binding structure for amyloid fibrils of 40-residue Alzheimer’s β by solid-state NMR spectroscopy. J Am Chem Soc. 2011;133:3390–3400. doi: 10.1021/ja1072178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertini I, Luchinat C, Parigi G, Pierattelli R. Perspectives in paramagnetic NMR of metalloproteins. Dalton Trans. 2008;2008:3782–3790. doi: 10.1039/b719526e. [DOI] [PubMed] [Google Scholar]
- 16.Arnesano F, Banci L, Piccioli M. NMR structures of paramagnetic metalloproteins. Q Rev Biophys. 2005;38:167–219. doi: 10.1017/S0033583506004161. [DOI] [PubMed] [Google Scholar]
- 17.Knight MJ, et al. Structure and backbone dynamics of a microcrystalline metalloprotein by solid-state NMR. Proc Natl Acad Sci USA. 2012;109:11095–11100. doi: 10.1073/pnas.1204515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipton AS, Heck RW, Ellis PD. Zinc solid-state NMR spectroscopy of human carbonic anhydrase: Implications for the enzymatic mechanism. J Am Chem Soc. 2004;126:4735–4739. doi: 10.1021/ja0305609. [DOI] [PubMed] [Google Scholar]
- 19.Frederickson CJ, Koh JY, Bush AI. The neurobiology of zinc in health and disease. Nat Rev Neurosci. 2005;6:449–462. doi: 10.1038/nrn1671. [DOI] [PubMed] [Google Scholar]
- 20.Tuttle MD, et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat Struct Mol Biol. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wälti MA, et al. Atomic-resolution structure of a disease-relevant Aβ(1-42) amyloid fibril. Proc Natl Acad Sci USA. 2016;113:E4976–E4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao Y, et al. Aβ(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colvin MT, et al. Atomic resolution structure of monomorphic Aβ42 amyloid fibrils. J Am Chem Soc. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitzpatrick AW, et al. Atomic structure and hierarchical assembly of a cross-β amyloid fibril. Proc Natl Acad Sci USA. 2013;110:5468–5473. doi: 10.1073/pnas.1219476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wasmer C, et al. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 26.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy-Smith K, Moore E, Schneider J, Tycko R. Molecular structure of monomorphic peptide fibrils within a kinetically trapped hydrogel network. Proc Natl Acad Sci USA. 2015;112:9816–9821. doi: 10.1073/pnas.1509313112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rufo CM, et al. Short peptides self-assemble to produce catalytic amyloids. Nat Chem. 2014;6:303–309. doi: 10.1038/nchem.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 30.Hou G, Yan S, Trébosc J, Amoureux JP, Polenova T. Broadband homonuclear correlation spectroscopy driven by combined R2(n)(v) sequences under fast magic angle spinning for NMR structural analysis of organic and biological solids. J Magn Reson. 2013;232:18–30. doi: 10.1016/j.jmr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tycko R. Physical and structural basis for polymorphism in amyloid fibrils. Protein Sci. 2014;23:1528–1539. doi: 10.1002/pro.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Paëpe G, Lewandowski JR, Loquet A, Böckmann A, Griffin RG. Proton assisted recoupling and protein structure determination. J Chem Phys. 2008;129:245101. doi: 10.1063/1.3036928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 34.Hu F, Schmidt-Rohr K, Hong M. NMR detection of pH-dependent histidine-water proton exchange reveals the conduction mechanism of a transmembrane proton channel. J Am Chem Soc. 2012;134:3703–3713. doi: 10.1021/ja2081185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S, Hong M. Protonation, tautomerization, and rotameric structure of histidine: a comprehensive study by magic-angle-spinning solid-state NMR. J Am Chem Soc. 2011;133:1534–1544. doi: 10.1021/ja108943n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou L, et al. Interaction between histidine and Zn(II) metal ions over a wide pH as revealed by solid-state NMR spectroscopy and DFT calculations. J Phys Chem B. 2013;117:8954–8965. doi: 10.1021/jp4041937. [DOI] [PubMed] [Google Scholar]
- 37.Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. Frequency selective heteronuclear dipolar recoupling in rotating solids: Accurate (13)C-(15)N distance measurements in uniformly (13)C,(15)N-labeled peptides. J Am Chem Soc. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- 38.Rieping W, et al. ARIA2: Automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 39.Shen Y, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amin EA, Truhlar DG. Zn coordination chemistry: Development of benchmark suites for geometries, dipole moments, and bond dissociation energies and their use to test and validate density functionals and molecular orbital theory. J Chem Theory Comput. 2008;4:75–85. doi: 10.1021/ct700205n. [DOI] [PubMed] [Google Scholar]
- 41.Rulísek L, Vondrásek J. Coordination geometries of selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in metalloproteins. J Inorg Biochem. 1998;71:115–127. doi: 10.1016/s0162-0134(98)10042-9. [DOI] [PubMed] [Google Scholar]
- 42.Williams JK, Hong M. Probing membrane protein structure using water polarization transfer solid-state NMR. J Magn Reson. 2014;247:118–127. doi: 10.1016/j.jmr.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong M, et al. Coupling amplification in 2D MAS NMR and its application to torsion angle determination in peptides. J Magn Reson. 1997;129:85–92. doi: 10.1006/jmre.1997.1242. [DOI] [PubMed] [Google Scholar]
- 44.Riek R, Eisenberg DS. The activities of amyloids from a structural perspective. Nature. 2016;539:227–235. doi: 10.1038/nature20416. [DOI] [PubMed] [Google Scholar]
- 45.Yugay D, et al. Copper ion binding site in β-amyloid peptide. Nano Lett. 2016;16:6282–6289. doi: 10.1021/acs.nanolett.6b02590. [DOI] [PubMed] [Google Scholar]
- 46.Dong J, Shokes JE, Scott RA, Lynn DG. Modulating amyloid self-assembly and fibril morphology with Zn(II) J Am Chem Soc. 2006;128:3540–3542. doi: 10.1021/ja055973j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elkins MR, et al. Structural polymorphism of Alzheimer’s β-amyloid fibrils as controlled by an E22 switch: A solid-state NMR study. J Am Chem Soc. 2016;138:9840–9852. doi: 10.1021/jacs.6b03715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang G, Dunbrack RL., Jr PISCES: A protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 49.Bakan A, Meireles LM, Bahar I. ProDy: Protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27:1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hovmöller S, Zhou T, Ohlson T. Conformations of amino acids in proteins. Acta Crystallogr D Biol Crystallogr. 2002;58:768–776. doi: 10.1107/s0907444902003359. [DOI] [PubMed] [Google Scholar]
- 51.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 52.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.