Significance
Cancer represents a breakdown of molecular mechanisms evolved by multicellular life to impose constraints on cell growth, resulting in more “primitive” proliferative cellular phenotypes. This suggests interpreting the activity of genes in cancer according to their evolutionary origins may provide insights into common mechanisms driving tumorigenesis. We incorporated phylogenetic and interaction data into expression analysis of seven solid tumors, revealing universal strong preferential expression of genes shared with unicellular species in tumors, alongside widespread disruption of links between unicellular and multicellular components of gene regulatory networks. Considering how the constraints imposed on these networks by evolution were altered in tumors identified molecular processes that could be manipulated for therapeutic benefit in cancer and uncovered several promising drug targets.
Keywords: cancer, evolution, transcriptomics, systems biology, atavism
Abstract
Tumors of distinct tissues of origin and genetic makeup display common hallmark cellular phenotypes, including sustained proliferation, suppression of cell death, and altered metabolism. These phenotypic commonalities have been proposed to stem from disruption of conserved regulatory mechanisms evolved during the transition to multicellularity to control fundamental cellular processes such as growth and replication. Dating the evolutionary emergence of human genes through phylostratigraphy uncovered close association between gene age and expression level in RNA sequencing data from The Cancer Genome Atlas for seven solid cancers. Genes conserved with unicellular organisms were strongly up-regulated, whereas genes of metazoan origin were primarily inactivated. These patterns were most consistent for processes known to be important in cancer, implicating both selection and active regulation during malignant transformation. The coordinated expression of strongly interacting multicellularity and unicellularity processes was lost in tumors. This separation of unicellular and multicellular functions appeared to be mediated by 12 highly connected genes, marking them as important general drivers of tumorigenesis. Our findings suggest common principles closely tied to the evolutionary history of genes underlie convergent changes at the cellular process level across a range of solid cancers. We propose altered activity of genes at the interfaces between multicellular and unicellular regions of human gene regulatory networks activate primitive transcriptional programs, driving common hallmark features of cancer. Manipulation of cross-talk between biological processes of different evolutionary origins may thus present powerful and broadly applicable treatment strategies for cancer.
Progression to cancer involves repeated selection for common cellular phenotypes, including sustained proliferation; altered energy metabolism; and abnormal responses to signals controlling cell growth, adhesion, and differentiation. These hallmark features of cancer (1) demonstrate broad alteration of basic cellular processes is a consistent characteristic of tumors regardless of tissue of origin and genetic background. However, the overall principles guiding the convergence to shared molecular properties in cancer remain unclear.
Cancer has been suggested to result from an atavistic process, whereby the activation of primitive, highly conserved programs (2, 3) leads to molecular phenotypes and population dynamics (4) similar to those of unicellular organisms (5). Genes commonly involved in cancer associate with two major evolutionary events: the emergence of self-replicating cellular life and the appearance of simple multicellular organisms (6, 7). The disruption of genes and processes that appeared in early metazoan life to enhance intercellular cooperation is expected to be a recurrent driver of carcinogenesis, as implicated by the widespread occurrence of cancer across the tree of multicellular life (8, 9) and the common dysregulation of pathways that evolved to sustain multicellularity, such as Wnt and integrins (10, 11).
The evidence presented to date to support atavistic transformation in tumors has been primarily observational, with limited comprehensive molecular evidence. It has been reported that disruption of genes tied to multicellularity confers advantages to malignant tumor clones (12), the expression of highly conserved genes is a feature of drug resistance in tumor cells (7), and there is global convergent activation in tumors of transcriptional programs associated with dedifferentiation (12, 13). These findings suggest deeper understanding of the differences in the expression and regulation of ancient unicellular and more recently evolved multicellular gene sets during malignant transformation will be crucial for uncovering the molecular basis of common tumor phenotypes and will provide new targets and strategies for cancer therapy.
To investigate atavistic transformation as a core element of tumorigenesis, we combined phylogenetic and interaction data with RNA-sequencing data from The Cancer Genome Atlas (TCGA) for seven solid tumor types to determine how changes in gene expression patterns in tumors are tied to evolutionary histories of the genes and processes involved.
Results
Genes Originating in Unicellular and Multicellular Ancestors of Humans Show Divergent Expression Patterns in Tumors.
We determined the point of emergence in evolutionary history of 17,318 human genes by phylostratigraphy (14). Human genes were classified into 16 clades (phylostrata) that represent the major evolutionary innovations, based on the most distant species with a clear ortholog (SI Appendix, Figs. S1 and S2, and Dataset S1). Phylostrata assignments were corroborated by functional enrichment analysis (SI Appendix, Fig. S3), with genes assigned to more primitive phylostrata enriched for basal cellular processes, as opposed to those involved in complex cellular functions assigned to later phylostrata, demonstrating our assignments are not unduly affected by potential biases (15). Human genes assigned to phylostrata 1–3 date back to unicellular ancestors (UC genes), whereas genes assigned to later phylostrata emerged in multicellular ancestors (MC genes).
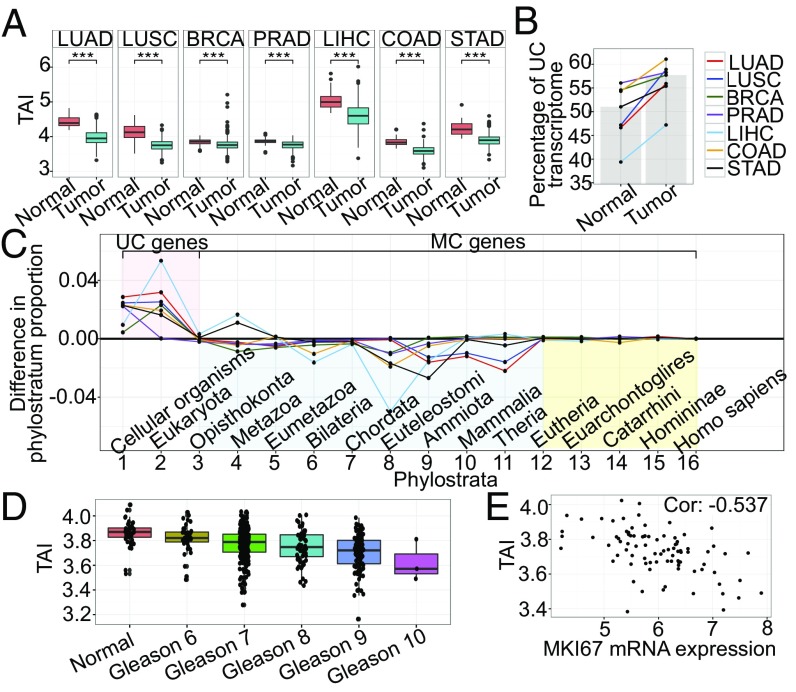
To investigate how the expression of genes in tumors is related to their evolutionary origins, we calculated the transcriptome age index (TAI), using RNAseq gene expression from seven tumor types from TCGA (SI Appendix, Tables S1 and S2): lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), breast (BRCA), prostate (PRAD), liver (LIHC), colon (COAD), and stomach (STAD), with 3,473 tumor samples in total and their respective normal tissues (386 samples). The TAI is a cumulative measure of the expression levels of all genes in a sample weighted by evolutionary age (16) (Eq. 1). Lower values of the TAI are associated with higher expression of ancient genes and represent older transcriptomes. After accounting for cross-contamination (SI Appendix, SI Methods), all tumors had consistently lower TAI values than their normal counterparts (Fig. 1A), with an increased percentage of transcripts coming from unicellular genes with respect to normal tissues from the same organ (Fig. 1B). Thus, tumor transcriptomes shift to stronger expression of more highly conserved genes, through up-regulation of genes originating from primitive unicellular ancestors and broad inactivation of more recently evolved genes. Our results are robust to alternate phylostratum assignments (17), are replicated in microarray data, are not solely driven by replication genes, are consistent across different magnitudes of expression levels, and are unlikely to be a simple consequence of global gene inactivation in tumors (permutation P = 0.035; SI Appendix, Figs. S4–S9 and Table S3). At the level of individual phylostrata, genes with orthologs in bacteria, yeast, and protozoa showed clear and consistently elevated expression in all tumor types (Fig. 1C, pink), whereas genes assigned to metazoan phylostrata predating eutherian (placental) mammals were primarily down-regulated (Fig. 1C, blue). Expression levels of genes, those unique to eutherian mammals, showed little difference between tumor and normal samples (Fig. 1C, yellow).
Fig. 1.
Overexpression of genes that date back to UC ancestors are preferentially expressed in tumors. (A) TAI of tumor and normal samples, by subtype. A lower TAI corresponds to higher expression of genes from earlier phylostrata. Tumors have lower TAI scores (more ancient transcriptomes) than normal samples (Wilcoxon tests: ***P < 0.01). (B) Percentage of transcriptome composed of UC genes increases in all tumor subtypes. Shaded areas: median percentages across samples. (C) Difference in proportion of transcriptome composed of genes from each phylostratum in tumors vs. normal samples, by subtype. (D) TAI decreases as degree of differentiation increases as measured by Gleason score (Jonckheere–Terpstra test P value = 2.79 × 10−16). (E) Negative correlation between the proliferation marker, MKI67, and the TAI (Spearman correlation = −0.537, P value = 1.1 × 10−7) in prostate tumors.
Our results reveal a strong global trend of preferential expression of genes of unicellular origin vs. genes of metazoan origin, concordant with an atavistic regression away from multicellularity at the cellular level. Interestingly, the inflection point between up- and down-regulation in tumors coincides with the phylostratum representing genes shared with Opisthokonta, whose life cycles often comprise both free-living unicellular and multicellular colonial stages. Together, these findings suggest a strong association between the evolution of complexity and multicellularity and patterns of gene expression in cancer.
To investigate the association between the preferential expression of unicellular genes in tumors and clinical features, we stratified PRAD samples by Gleason score, a well-defined pathological measure of dedifferentiation that accounts for tumor heterogeneity. The TAI showed a strong decreasing trend with Gleason score, in both the TCGA (Fig. 1E) and an independent dataset (18), and similar results were obtained using the grade of LIHC and STAD tumors (SI Appendix, Figs. S10 and S11). We found negative correlation between the TAI and the proliferation marker MKI67 (19) in prostate and lung (adenocarcinoma and squamous) tumor samples (Fig. 1E and SI Appendix, Fig. S12), indicating that tumor samples with a more ancient transcriptome tend to have higher proliferation rates. The link between the TAI and loss of differentiation and an increased proliferation suggests it mirrors clinically relevant features, and primitive expression phenotypes push tumor cells toward more malignant states and thus provide a signature of potential clinical utility.
Genes of Unicellular Origin Drive Activation of Hallmarks Required for Tumorigenesis.
To examine the functional consequences of the shift toward increased expression of UC genes in tumors, we used the Generic GOslim set from the Gene Ontology (GO) Consortium (20, 21), a comprehensive classification of genes involved in 69 major cellular processes, with low redundancy between sets (SI Appendix, SI Methods and Fig. S13). GOslims were classified as predominantly unicellular (38/69 = 55.07%) or multicellular (19/69 = 27.54%) based on enrichment of gene ages, with GOslims such as cell cycle and metabolic processes dated as unicellular and those related to tissue differentiation or increased organismal complexity labeled multicellular (SI Appendix, Table S4 and Fig. S14).
Differential expression analysis of GOslims revealed a striking level of consistency across all seven tumor types (Fig. 2A, Right and SI Appendix, Fig. S15), suggesting strong convergent evolution at a molecular level. We observed uniform inactivation of cellular processes unique to metazoans, consistent with the widespread reprogramming of the intracellular signaling networks observed during tumorigenesis. In sharp contrast, we observed increased expression of unicellular processes closely tied to enabling hallmarks, including sustaining proliferative signaling, cell death avoidance, and genomic instability (1). This strong bias was consistently found across all seven tumor types, suggesting preferential activation of unicellular genes and concomitant suppression of multicellular processes generate the molecular characteristics essential for tumorigenesis in solid tissues, independent of tissue of origin and etiology. These signatures are driven by processes beyond dedifferentiation, as they are distinct from those observed in stem cells (SI Appendix, SI Note 1 and Figs. S16 and S17).
Fig. 2.
Effect of the expression patterns of UC and MC genes on cellular processes. (A) Expression patterns of cellular processes. (A, Right) LogFC of processes, showing up-regulation of a subset of UC processes and widespread down-regulation of MC processes across tumors. (A, Left) Median logFC of UC and MC components of processes. The logFCs of UC components are more positive or not significantly different from those of MC components, indicating UC genes push processes toward activation. Bars: range in tumors. Triangles: UC component greater (Wilcoxon test P value <0.05) than MC component. (B) Difference in the absolute logFC of UC and MC components of GOslims (y axis) vs. overall logFC for the GOslim in tumors vs. normal samples (x axis). Up-regulated GOslims are driven by UC genes, whereas down-regulated ones are driven by MC genes. Points: median logFC across tumors. Error bars: range in tumors. Linear model P value = 1.9 × 10−10. (C) Response to stress GOterm tree. UC stress-response programs tend to be up-regulated in tumors (55%), whereas recently acquired ones are down-regulated (69%). Node size is proportional to the number of genes annotated with the GOterm.
However, whether a unicellular process was activated or not was dependent on its functional role, as we observed consistent down-regulation of many unicellular processes, particularly metabolic processes involving complex molecules, reflecting the metabolic reprogramming commonly seen in tumors. These patterns suggest induction of primitive processes in tumors is not merely a side effect of progressive stochastic loss of metazoan gene regulatory mechanisms, but rather the result of coordinated and selective processes targeting specific pathways. Thus, a unifying theme behind the emergence of many of the common hallmarks of cancer could be a controlled transition to more primitive cellular phenotypes.
We hypothesized the transcriptional states of individual cellular processes are functions of the different roles and the relative overall impact of UC and MC genes within each process. Dividing genes within each GOslim into UC and MC components according to the ages of the genes revealed that in 70.4% of GOslims (38/54), the log of the fold change in gene expression (logFC) of the UC component was significantly higher than that of the MC component (Wilcoxon one-sided test adjusted P value <0.05), independent of the age of the cellular process (Fig. 2A, Left). In no case did we find a MC component with a significantly higher logFC than the UC component. Even within suppressed biological processes, the expression of UC genes was often maintained or even enhanced within tumors, in contrast to MC genes, which primarily showed negative logFC. Therefore, the preferential expression of genes of unicellular origin across biological functions underscores their central role in the transition to a tumorigenic state.
UC components also displayed larger absolute logFC than MC components in GOslims up-regulated in tumors (Fig. 2B), demonstrating enhanced expression of UC genes plays a bigger role in driving the changes of up-regulated processes than does suppression of MC genes. In contrast, down-regulated processes show the opposite trend, suggesting that their reduced expression is mostly linked to the inactivation of MC genes. This supports a model whereby genes of a unicellular and multicellular origin modulate the overall expression of cellular processes by acting as two opposing forces, with the resulting activity of the process being the result of a preferential bias toward one of them.
The Activation of Response to Stress Mechanisms in Tumors Is Associated with Their Evolutionary Age.
We investigated the biological processes included under the stress response GO term, a broad and varied category highly relevant to tumor biology thought to be central to the atavistic process in cancer (2, 3, 22). We found 55% of stress responses conserved with unicellular species were up-regulated in tumors, whereas 69% of stress responses exclusive to multicellular organisms were down-regulated (Fig. 2C). This is unlikely due to chance (P = 0.01 for UC; P = 0.02 for MC) and was not seen in stem cells (SI Appendix, SI Methods, SI Note 1, and Fig. S17). Our results suggest tumor survival in response to multiple microenvironmental challenges is dependent on primitive response mechanisms.
This increased expression of processes originating in unicellular ancestors of modern life, such as DNA damage stimulus, is consistent with mechanisms needed to withstand the genetic instability typical of tumors. In contrast, advanced DNA repair processes exploited by multicellular organisms, such as pyrimidine dimer repair and specific double-strand break repair, were down-regulated. An exception was the up-regulation of signal transduction in response to DNA damage, which likely evolved as a support system for the more primitive responses to DNA damage, resulting in strong coregulation.
The transition to a UC state in tumors is supported by preferential activation of stress response mechanisms developed to withstand stresses encountered by unicellular ancestors, many of which (hypoxia, nutrient deprivation, and DNA damage) would be similar to environmental pressures encountered by rapidly expanding tumors. Conversely, the damping down of multicellular functions in tumors appears to extend to stress response processes as well. The resulting phenotypic alterations could significantly impact tumor evolution and response to treatment.
Disruption of the Coexpression Between Unicellular and Multicellular Processes in Tumors Enhances Hallmark Phenotypes.
We hypothesized convergent patterns of expression of cellular processes with respect to evolutionary age were supported by coregulation mechanisms between processes. We first calculated the activity of cellular processes, using single-sample gene set enrichment analysis (ssGSEA) (23), which calculates the degree of coordinated up- and down-regulation of genes in individual samples and can correctly classify samples according to subtype (SI Appendix, Fig. S18). The Spearman correlation between pairs of processes was calculated using ssGSEA scores, constructing a network of correlation of expression between cellular processes.
To distinguish highly coexpressed pairs of processes with high numbers of physical and regulatory interactions from those pairs where coexpression occurred indirectly, we developed a metric, interconnectedness (I), to quantify the degree of interaction between cellular processes (Eq. 3). Our metric normalizes the number of direct physical and genetic interactions between proteins of every pair of processes by the total number of possible interactions, capturing functional dependencies, and the layout of interactions between functional processes obtained during evolution (SI Appendix, Figs. S19–S21, and Dataset S2). The highest interconnectedness occurs between unicellular processes (UC–UC), whereas pairs of multicellular processes (MC–MC) were the least connected, and unicellular and multicellular processes (UC–MC) showed intermediate interconnectedness. We ranked pairs of processes by interconnectedness score and selected the top 10% of each type (UC–UC, UC–MC, and MC–MC) to build a transcriptional network of cellular processes. We used the median correlation across the seven TCGA datasets to represent the typical tumor and normal coexpression states (Fig. 3A). Our network is based on PathwayCommons (24), but similar results are also obtained with other databases (SI Appendix, Figs. S22–S26).
Fig. 3.
Patterns of coexpression between highly interconnected cellular processes are disrupted in tumors. (A) Transcriptional network of cellular processes in tumors, displaying the median correlation (edge brightness) between processes across the seven tumor types. (B) Number of positive and negative interactions between processes. Dots correspond to results from each tumor type. (C) Median correlation in expression between cellular processes in tumors and normal samples. (D) Variance in the correlation of expression between cellular processes, showing decreased variability in tumors with respect to normal samples.
The expression of pairs of UC–UC processes overwhelmingly showed positive correlation in both normal (64/70 pairs) and tumor (66/70 pairs) samples, with correlation being significantly stronger in tumors (Kolmogorov–Smirnov test P value = 0.04; Fig. 3 B and C, pink circle; SI Appendix, Figs. S27 and S28). This suggests promotion of simultaneous activation of UC functions and is consistent with the strong patterns of up-regulation of unicellular processes observed in all tumor types analyzed (Fig. 1). Although pairs of MC processes also showed a consistent positive coexpression in tumors (17/17) and in normal samples (16/17), the distribution of correlations was not significantly different (P value = 0.12), indicating that the coregulation between multicellular regions of the interaction network is mostly unaltered in tumors.
In contrast, UC and MC processes were predominantly negatively correlated, with the number of negatively correlated pairs of processes significantly greater in tumors (57/72, 79.2%) than in normal samples (41/72, 56.9%) (Fisher test P value = 0.0035; Fig. 3 B and C, red circle) or in stem cells (38/72, 52.8%; SI Appendix, SI Note 1). Significant differences were also seen in correlation of expression between UC–MC pairs in tumors and normal samples (KS test P value = 0.0066), and these trends were consistent regardless of the absolute level of interconnectedness between the processes (SI Appendix, Figs. S28 and S29). We propose this to be a form of mutual exclusivity between cellular processes of different evolutionary histories, where the limited integration of unicellular and multicellular processes is exacerbated in tumors, leading to an uncoupling between and increased independence of UC and MC network regions. Although mutual exclusivity has been noted previously for specific pairs of processes (e.g., cell cycle and differentiation; SI Appendix, Table S5), our results demonstrate general and widespread mutual exclusivity in tumors between UC and MC biological functions, indicating its fundamental importance to tumor development.
The strong convergence in these patterns across different tumors is further evidenced by the marked reduction in the variability of coexpression in tumors with respect to normal differentiated tissues (Fig. 3D and SI Appendix, Fig. S30). This consistent loss of diversity of regulation of cellular processes suggests common selective pressures lead to actively regulated changes of coexpression between cellular processes of distinct evolutionary history in cancer. Furthermore, this process is constrained by defined limits, with levels of coexpression in tumors consistently within the ranges of those of normal samples (SI Appendix, Fig. S31), suggesting that system-level constraints limit the viable paths taken by tumor cells.
We found 20 UC–MC interacting pairs switched from positive to negative correlation, whereas only 4 pairs switched in the opposite direction (Fig. 3C, orange square; SI Appendix, Table S6). Nearly half of them involved cell death (11/24; 45.83%), consistent with the view that cell death is tied to many of the major regulatory and signaling changes that occur in tumors. The strong selection for mutually exclusive associations between these specific gene sets across multiple tumor types indicates disruption of the links between them may drive tumor development.
Identifying Genes Modulating the Altered Interactions Between Network Regions of Different Evolutionary Age.
We hypothesized the disruption in coexpression of UC and MC processes was due to altered interactions between genes linking these two parts of the human gene network, forming key vulnerabilities. We focused on a pair of cellular processes whose coexpression was among the most strongly disrupted in tumors, cellular junction organization and chromosome organization, which displayed a pronounced shift from positive correlation in normal tissues (median: 0.21) to strong negative correlation in tumors (median: −0.36). This selection for a drastic change in coexpression suggests mutual exclusivity between these processes is advantageous for tumorigenesis.
We reasoned the key genes modulating this change would be highly connected across the two gene sets and have altered coexpression with many of their interaction partners. We developed a “hubness” metric for each gene with annotated interactions between cellular junction organization and chromosome organization, calculated as the sum of the absolute difference of change in expression correlation between its connecting partner genes (Eq. 4). This metric is highly correlated with the number of interactions of a gene (SI Appendix, Fig. S32), a property associated with relative importance in modulating functional responses (25). Ranking by this metric uncovered 12 genes (RCC2, TLN1, VASP, ACTG1, PLEC, CTTN, DSP, ILK, PKN2, CTNNA1, CTNND1, and PKP3) whose hubness was consistently in the top 10% across all seven tumor types, identifying a set of genes that commonly mediate changes in coexpression between cellular junction organization and chromosome organization.
All 12 genes interact with genes belonging to a signature of chromosomal instability associated with poor clinical outcome and metastasis (26) (SI Appendix, Fig. S33), suggesting these genes have roles in regulatory networks linking genomic instability and metastasis during tumor progression. Many are involved in pathways highly relevant to cancer (SI Appendix, Table S8), including 4 within the Rap1 signaling pathway, which has yet to be largely studied in the context of cancer, and experimental evidence suggests they can modulate malignant characteristics in specific tumor types (SI Appendix, Table S7). However, here we associate these genes with seven tumor types, uncovering a wider role as general, pan-cancer modulators of tumor development. CRISPR screen data for these genes and 91 additional genes involved in other pairs of UC–MC processes negatively correlated across tumors revealed knockdown of many of the identified UC genes hinders growth of multiple cancer cell lines (SI Appendix, SI Note 2, Fig. S34, and Tables S9 and S10), indicating their potential as viable drug targets.
The association of key genes promoting mutual exclusivity in tumors with main features of tumorigenesis supports the view that that mutual exclusivity in the coregulation of unicellular and multicellular processes is under positive selection during tumor formation and progression across multiple tumor types. Drugs that target these fundamental points of vulnerability or that abolish this mutual exclusivity would have great potential as effective broad-spectrum treatment strategies.
Discussion
Detailed transcriptome analysis of 3,473 tumor and 386 normal samples from TCGA demonstrates gene expression changes in tumors are closely tied to the evolutionary ages of the genes involved. Our findings suggest convergence of tumors to similar molecular phenotypes is tied to common principles guiding patterns of coexpression of cellular processes according to their evolutionary histories. This is the most comprehensive molecular evidence to date that a widespread shift to preferential expression of genes conserved in primitive, single-celled species is a common feature of tumors. This is concordant with the atavism hypothesis, which states cancer results from a transition to a more “selfish” unicellular mode of life, not merely through a passive stochastic occurrence but as an active, directed process driven by selection (2, 3, 27). Our findings demonstrate the up-regulation of unicellular GO terms is limited to certain processes and pathways, indicative of selection, and loss of coordinated expression between multicellular and unicellular processes across multiple tumor types, implicating altered regulation.
Although convergent evolution in tumors has been described in the context of gene expression (13), here we show convergent evolution is also apparent at the level of coexpression of cellular processes, according to their point of evolutionary emergence. Tumorigenesis reinforces the interdependence between unicellular genes while enhancing segregation between the unicellular and multicellular components of the gene regulatory network. Such mutual exclusivity would promote loss of multicellular features in tumors as activation of unicellular genes occurs in response to selective pressures favoring increased replication or activation of basal cellular processes, leading to an increasingly atavistic malignant phenotype with increased selective advantage.
Gene expression in cancer with respect to evolutionary age is not a simple dichotomy, as multicellular biological processes such as hormone receptors drive several tumor types. However, the highly reproducible nature of our observations and the signs of regulatory control behind them suggests treatment strategies that manipulate the fundamental systems-level rewiring at the interface between more primitive and more advanced components of gene regulatory networks in cancer could have broad therapeutic application and high specificity for tumor cells.
Several compounds already in clinical use target primitive fundamental biological functions, e.g., ref. 28. Our results showing many primitive functions up-regulated together in tumors raise the possibility of going a step beyond, to simultaneously target multiple independent unicellular processes. Another approach involves stressing multicellular systems that are inactive or diminished in cancer, a “target the weakness” approach (29) by altering the intra- or extracellular environment of tumor cells to put cells that have lost or inactivated a particular multicellular pathway at a selective disadvantage. Our analysis shows stress response pathways composed primarily of multicellular genes could also be manipulated for clinical benefit.
Given strong anticorrelation in expression between multicellular and unicellular genes is apparent in many tumors, reestablishing the balance between the activities of unicellular and multicellular processes could push tumors back to a more normal state and/or achieve a form of synthetic lethality. Empirical support for this approach comes from studies showing inhibition of glycolysis increased sensitivity of cancer cells to pharmacologically induced apoptosis (30). Cell death displayed consistently altered correlations with many core metabolic and cell division processes, suggesting manipulation of other biological functions could further prime tumor cells for apoptosis.
Our approach uncovered previously unappreciated association between biological processes exploited by tumors. We could narrow down 12 key genes bridging the cell junction organization and chromosome organization processes, which are biomarkers or regulators of malignancy in vitro for at least one cancer, validating the approach, but our analysis also implicates them as potential common drivers of a number of cancers. To our knowledge, no studies have yet published potential therapeutic agents for these genes, making them attractive targets for prioritization in future drug screens.
Our study applies a detailed molecular framework to view cancer as a failure of the systems supporting increased organismal complexity (31, 32). This is an important step toward understanding how macroevolutionary processes left vulnerabilities that lead to cancer and how they may be exploited in practical terms to improve treatment outcomes, facilitating the application of “Darwinian medicine” to oncology (33).
Methods
Phylostratigraphy of Human Genes.
A total of 17,318 human genes were mapped to a phylogenetic tree (Dataset S1), consisting of 16 clades (phylostrata), ranging from including all cellular organisms (phylostratum 1) to Homo sapiens (phylostratum 16) (SI Appendix, Fig. S1). The most ancient phylostratum represented in a group of orthologs from the OrthoMCL database version 5 (34) was considered as the point of emergence of the human protein.
Transcriptomic Analysis Incorporating Gene Ages.
RNAseq expression data were downloaded from TCGA (https://portal.gdc.cancer.gov/) (SI Appendix, Table S2). The transcriptome ages of tumor and normal samples were calculated with the TAI method (16),
| [1] |
where psi is the phylostratum of each gene i and ei is the gene expression value of gene i.
The proportion of the total library size represented by each phylostratum in each sample was calculated using Eq. 2,
| [2] |
where Ppsi is the proportion of expression abundance corresponding to genes of phylostratum i, eij is the expression value of gene j in the phylostratum i, mi is the total number of genes in phylostratum i, and n is the total number of phylostrata. The Ppsi values were averaged across all samples for each normal and tumor type, and the tumor vs. normal difference in proportions was determined by subtraction.
Functional Analyses.
GOslims were obtained from Gene Ontology (geneontology.org/), and their ages were calculated by permutation of the ages of the genes annotated with each GOslim (SI Appendix, Table S4). Tumor vs. normal differential expression analysis of GOslims and UC and MC components was conducted using QuSAGE (35). For the response to stress tree, the average logFC obtained by QuSAGE for each term was calculated across tumors, regardless of the significance of individual false discovery rate. Statistical significance of trends was assessed using permutation tests (SI Appendix, SI Methods).
Construction of Transcriptional Coexpression Networks.
The degree of interaction I (Dataset S2) was calculated for each pair of GOslims (i and j),
| [3] |
with Eij being the number of edges joining genes of GOslim i with GOslim j, and Ei∩ij and Ej∩ij are the number of edges in GOslim i or j that are also found in Eij. Gi and Gj are the numbers of genes of pathways i and j, respectively, and Gi∩j is the number of genes shared by GOslims i and j.
Gene Hubness Score.
Edges connecting genes of cell junction organization and chromosome organization processes were weighted by the Spearman correlation of expression in each tumor and normal type. We defined
| [4] |
with i being a gene in the bipartite graph and j an edge linking genes in separate processes.
Code is available at https://github.com/cancer-evolution/Evolutionary-analysis-of-cancer-transcriptomes.
Supplementary Material
Acknowledgments
We thank David Bowtell, Patrick Humbert, David Thomas, Arcadi Cipponi, Ismael Vergara, and Jason Li for helpful comments. This work was supported by a Melbourne International Engagement Award and a Melbourne International Fee Remission Scholarship (to A.S.T.), a National Health & Medical Research Council of Australia (NHMRC) Peter Doherty Early Career Fellowship (to D.L.G.) (APP1052904), and NHMRC Program Grant 1053792 (to R.B.P.), as well as by NHMRC Senior Research Fellowships (to R.B.P. and A.T.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 6160.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617743114/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Davies PC, Lineweaver CH. Cancer tumors as Metazoa 1.0: Tapping genes of ancient ancestors. Phys Biol. 2011;8:015001. doi: 10.1088/1478-3975/8/1/015001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent M. Cancer: A de-repression of a default survival program common to all cells?: A life-history perspective on the nature of cancer. BioEssays. 2012;34:72–82. doi: 10.1002/bies.201100049. [DOI] [PubMed] [Google Scholar]
- 4.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 5.Lambert G, et al. An analogy between the evolution of drug resistance in bacterial communities and malignant tissues. Nat Rev Cancer. 2011;11:375–382. doi: 10.1038/nrc3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domazet-Loso T, Tautz D. Phylostratigraphic tracking of cancer genes suggests a link to the emergence of multicellularity in metazoa. BMC Biol. 2010;8:66. doi: 10.1186/1741-7007-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu A, et al. Ancient hot and cold genes and chemotherapy resistance emergence. Proc Natl Acad Sci USA. 2015;112:10467–10472. doi: 10.1073/pnas.1512396112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aktipis CA, et al. Cancer across the tree of life: Cooperation and cheating in multicellularity. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140219. doi: 10.1098/rstb.2014.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domazet-Lošo T, et al. Naturally occurring tumours in the basal metazoan Hydra. Nat Commun. 2014;5:4222. doi: 10.1038/ncomms5222. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz-Trillo I, Nedelcu A. Evolutionary transitions to multicellular life. In: Ruiz-Trillo I, Nedelcu A, editors. Principles and Mechanisms. Springer, Dordrecht; The Netherlands: 2015. pp. 47–78. [Google Scholar]
- 11.Engler AJ, Humbert PO, Wehrle-Haller B, Weaver VM. Multiscale modeling of form and function. Science. 2009;324:208–212. doi: 10.1126/science.1170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Lin F, Xing K, He X. The reverse evolution from multicellularity to unicellularity during carcinogenesis. Nat Commun. 2015;6:6367. doi: 10.1038/ncomms7367. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, He X. The convergent cancer evolution toward a single cellular destination. Mol Biol Evol. 2016;33:4–12. doi: 10.1093/molbev/msv212. [DOI] [PubMed] [Google Scholar]
- 14.Domazet-Loso T, Brajković J, Tautz D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007;23:533–539. doi: 10.1016/j.tig.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Moyers BA, Zhang J. Phylostratigraphic bias creates spurious patterns of genome evolution. Mol Biol Evol. 2015;32:258–267. doi: 10.1093/molbev/msu286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domazet-Lošo T, Tautz D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature. 2010;468:815–818. doi: 10.1038/nature09632. [DOI] [PubMed] [Google Scholar]
- 17.Domazet-Loso T, Tautz D. An ancient evolutionary origin of genes associated with human genetic diseases. Mol Biol Evol. 2008;25:2699–2707. doi: 10.1093/molbev/msn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erho N, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andor N, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gene Ontology Consortium Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cipponi A, Thomas DM. Stress-induced cellular adaptive strategies: Ancient evolutionarily conserved programs as new anticancer therapeutic targets. BioEssays. 2014;36:552–560. doi: 10.1002/bies.201300170. [DOI] [PubMed] [Google Scholar]
- 23.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerami EG, et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 26.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 27.Greaves M. Evolutionary determinants of cancer. Cancer Discov. 2015;5:806–820. doi: 10.1158/2159-8290.CD-15-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devlin JR, et al. Combination therapy targeting ribosome biogenesis and mRNA translation synergistically extends survival in MYC-driven lymphoma. Cancer Discov. 2016;6:59–70. doi: 10.1158/2159-8290.CD-14-0673. [DOI] [PubMed] [Google Scholar]
- 29.Lineweaver CH, Davies PC, Vincent MD. Targeting cancer’s weaknesses (not its strengths): Therapeutic strategies suggested by the atavistic model. BioEssays. 2014;36:827–835. doi: 10.1002/bies.201400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meynet O, et al. Glycolysis inhibition targets Mcl-1 to restore sensitivity of lymphoma cells to ABT-737-induced apoptosis. Leukemia. 2012;26:1145–1147. doi: 10.1038/leu.2011.327. [DOI] [PubMed] [Google Scholar]
- 31.Aktipis CA, Nesse RM. Evolutionary foundations for cancer biology. Evol Appl. 2013;6:144–159. doi: 10.1111/eva.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aktipis CA, Boddy AM, Gatenby RA, Brown JS, Maley CC. Life history trade-offs in cancer evolution. Nat Rev Cancer. 2013;13:883–892. doi: 10.1038/nrc3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greaves M. Darwinian medicine: A case for cancer. Nat Rev Cancer. 2007;7:213–221. doi: 10.1038/nrc2071. [DOI] [PubMed] [Google Scholar]
- 34.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yaari G, Bolen CR, Thakar J, Kleinstein SH. Quantitative set analysis for gene expression: A method to quantify gene set differential expression including gene-gene correlations. Nucleic Acids Res. 2013;41:e170. doi: 10.1093/nar/gkt660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.