Significance
Inner organization of eukaryotic cells intimately depends on the active transport of diverse intracellular cargo on the ubiquitous actin and microtubule networks. The underlying mechanisms of such directional transport processes have been of outstanding interest. We studied a motor complex composed of Rab27a, melanophilin, and myosin Va and found, surprisingly, that the adaptor protein melanophilin toggled the binding preference toward actin or microtubules in vitro. Our results offer unexpected mechanistic insights into biasing the directionality of a moving organelle on the cytoskeleton through phospho-targeting the adaptor protein rather than its motor in vivo.
Keywords: melanophilin, myosin Va, intracellular transport, transport regulation
Abstract
Pigment organelles, or melanosomes, are transported by kinesin, dynein, and myosin motors. As such, melanosome transport is an excellent model system to study the functional relationship between the microtubule- and actin-based transport systems. In mammalian melanocytes, it is well known that the Rab27a/melanophilin/myosin Va complex mediates actin-based transport in vivo. However, pathways that regulate the overall directionality of melanosomes on the actin/microtubule networks have not yet been delineated. Here, we investigated the role of PKA-dependent phosphorylation on the activity of the actin-based Rab27a/melanophilin/myosin Va transport complex in vitro. We found that melanophilin, specifically its C-terminal actin-binding domain (ABD), is a target of PKA. Notably, in vitro phosphorylation of the ABD closely recapitulated the previously described in vivo phosphorylation pattern. Unexpectedly, we found that phosphorylation of the ABD affected neither the interaction of the complex with actin nor its movement along actin tracks. Surprisingly, the phosphorylation state of melanophilin was instead important for reversible association with microtubules in vitro. Dephosphorylated melanophilin preferred binding to microtubules even in the presence of actin, whereas phosphorylated melanophilin associated with actin. Indeed, when actin and microtubules were present simultaneously, melanophilin’s phosphorylation state enforced track selection of the Rab27a/melanophilin/myosin Va transport complex. Collectively, our results unmasked the regulatory dominance of the melanophilin adaptor protein over its associated motor and offer an unexpected mechanism by which filaments of the cytoskeletal network compete for the moving organelles to accomplish directional transport on the cytoskeleton in vivo.
The timely and correct positioning of intracellular cargo is a prerequisite to the proper functioning and survival of all eukaryotic cells. Three types of molecular motors, the microtubule-associated kinesin and dynein motors and actin-associated myosin motors, actively drive these transport processes (1). Rather than relying on one type of motor for a given transport process, cells often use multiple types of motors to transport cargo (2, 3). The presence of cooperating or even competing sets of motors on the same cargo raises the question of how cells regulate the intricate navigation of intracellular cargo on the actin and microtubule networks.
One prominent example of cargo transport is found in melanin-producing cells, which are called “melanocytes” in mammals. Melanocytes are responsible for the production and packaging of melanin into pigment organelles termed “melanosomes.” The poorly understood transfer of melanosomes from melanocytes into keratinocytes enables pigmentation in mammals and plays essential roles in protection against UV irradiation of the skin (4). Consequently, any failure of melanosomes to protect the skin is linked to skin melanoma.
Previous work on melanocytes demonstrated that melanosomes move on both the microtubule and actin networks (5–7). However, not much is known about the mechanism of microtubule-based transport in melanocytes, including the contributions of the microtubule-associated motors to the melanosome transport (5, 6, 8–10). In contrast, the actin-based transport of melanosomes has been characterized in greater detail. Actin-based transport is accomplished by a tripartite complex consisting of the Rab27a, melanophilin (Mlph), and myosin Va (MyoVa) subunits. The melanosome-bound small GTPase Rab27a associates in a GTP-dependent manner with the adaptor protein Mlph. Mlph in turn links MyoVa to the melanosome to form a tripartite transport complex in vivo (11–18). Importantly, a functional tripartite transport complex could be reconstituted from the Rab27a, Mlph, and MyoVa in vitro (19).
Of the three subunits forming the tripartite complex, Mlph is by far the most versatile, because it has been shown to associate not only with Rab27a and MyoVa (20, 21) but also with actin and the microtubule plus-end tracking EB1 protein via its C-terminal actin-binding domain (ABD) (22–25). In vivo experiments initially argued for an important contribution of the ABD in the efficient distribution of melanosomes on the actin network (24). Subsequent work, however, reached the opposite conclusion from rescue experiments in a melanocyte cell line lacking Mlph (26). In these experiments, the previously observed deleterious effects caused by the absence of the actin-binding capability of Mlph could not be recapitulated, and melanosomes were distributed on the actin network efficiently in the absence of the ABD (reviewed in ref. 27). MyoVa function, on the other hand, was indispensable for the peripheral distribution of melanosomes on the actin network (5–7, 28, 29). Depolymerization of the actin network or the absence of MyoVa resulted in a microtubule-dependent clustering of melanosomes in the cell center (28, 30, 31). However, the signaling pathway(s) that regulate the directionality of the melanosome transport on the microtubule and actin networks have not yet been delineated.
In contrast to mammalian melanocytes, the signaling pathways that elicit reversible redistribution of melanosomes are better characterized in amphibian melanophores. Here, melanosomes are dynamically and reversibly distributed on the microtubule and actin networks by kinesin-2, dynein-1, and myosin V motors to adapt the skin color rapidly in response to environmental cues (32–35). The minus-end–directed transport mediated by the dynein-1 motor on the microtubule network clusters the melanosomes in the cell center. This process is termed “aggregation.” Kinesin-2 and myosin V motors, on the other hand, transport melanosomes away from the center toward the periphery of the cell on microtubules and actin cables to distribute the melanosomes throughout the cytoplasm. This process is called “dispersion.” Cultured melanophores treated with α-melanocyte–stimulating hormone experience a burst of cAMP levels, which in turn activates PKA and leads to the dispersion of melanosomes on the actin network, a process responsible for the darkening the animal’s skin color (36–38). The opposite process occurs during aggregation, when cAMP levels are reduced and melanosomes are transferred from the actin to the microtubule network to be clustered in the cell center (36, 38, 39). Consistent with these observations, PKA was found to associate with melanosomes and even to form complexes with the respective motor proteins in melanophores (40, 41). Whether any of these motor complexes are direct targets of PKA, however, remains to be elucidated.
Here, we turned to the tripartite Rab27a/Mlph/MyoVa complex from melanocytes, which is essential for the dispersion of melanosomes on the actin network in vivo, and asked whether PKA phosphorylates any of the subunits in vitro. We found that PKA phosphorylated the Mlph adaptor protein, particularly its ABD, but not Rab27a or MyoVa. Notably, the in vitro phosphorylation pattern of the ABD closely matched previously identified phosphosites in cell-wide proteomic analyses. The phosphorylation of the Mlph showed no effects on the MyoVa-mediated transport of this tripartite complex in vitro. Unexpectedly, however, we observed that dephosphorylated Mlph interacted robustly with microtubules. Dephosphorylated Mlph even preferred to bind to microtubules over actin. PKA-dependent phosphorylation of the ABD of Mlph blocked its interaction with microtubules and restored its preference for binding to actin. Indeed, the reversible phosphorylation of Mlph had functional consequences. In transport assays, dephosphorylation of Mlph significantly enhanced the interaction of the Rab27a/Mlph/MyoVa complex with microtubules, with frequent back-and-forth switching of the complex between the actin and microtubule networks. Remarkably, the tripartite complex built with phosphorylated Mlph largely ignored microtubules and displayed uninterrupted directional transport on the actin network. In summary, our results unmasked an unexpected regulatory impact of the adaptor protein on its associated motor. Importantly, these results offer mechanistic insights into how cells may regulate the overall directionality of moving cargo on the microtubule and actin networks.
Results and Discussion
PKA Phosphorylates the Adaptor Protein Mlph of the Tripartite Transport Complex.
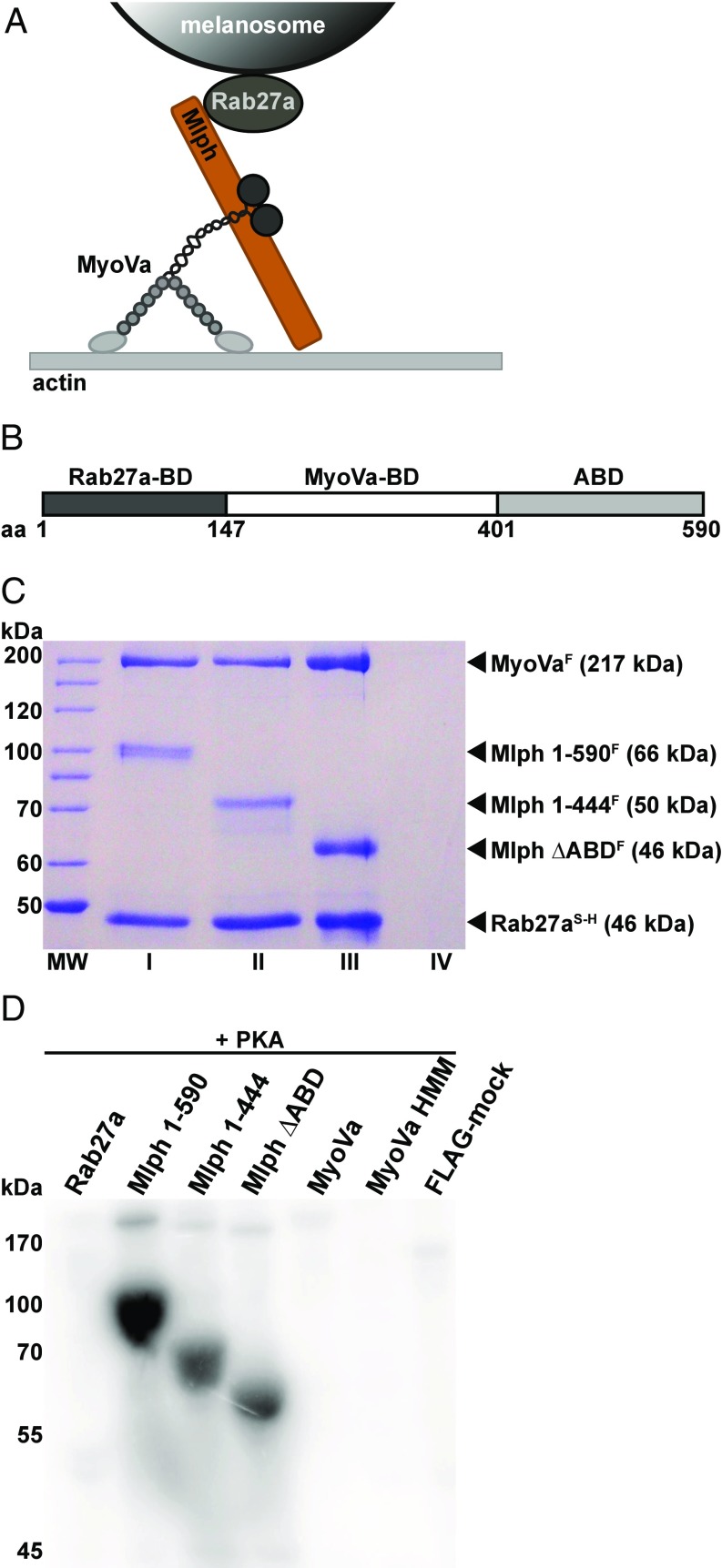
Mlph/MyoVa and Rab27a/Mlph/MyoVa complexes were previously reconstituted by mixing the individually purified components (19, 22). To test the role of PKA-dependent phosphorylation in regulation of the MyoVa-dependent transport, we optimized a strategy to assemble the Rab27a/Mlph/MyoVa complex in vitro (Fig. 1 A and C). To this end, we coexpressed 6×His-SNAP–tagged wild-type Rab27a or Rab27a-Q78L (42) with FLAG-tagged Mlph and tandem-purified the resulting complex via the 6×His and FLAG affinity tags. We then added purified FLAG-tagged MyoVa [the melanocyte-specific isoform (18, 43, 44)] to the preformed Rab27a/Mlph complex that was immobilized on Ni-nitrilotriacetic acid (NTA) affinity resin. The tripartite complex was subsequently eluted from the Ni-NTA resin (Fig. 1C, lane I).
Fig. 1.
The Mlph subunit of the tripartite complex is specifically phosphorylated by PKA. (A) Schematic illustration of the MyoVa-dependent tripartite transport complex on the melanosome surface. The Rab27a GTPase resides in the melanosome membrane and recruits Mlph in a GTP-dependent manner. Mlph in turn recruits the MyoVa motor to form the tripartite transport complex. (B) Domain structure of the adaptor protein Mlph. The Rab27a-binding domain (Rab27a-BD) is located at Mlph’s N terminus (42, 43). MyoVa binds to Mlph’s middle domain with its globular tail domain and the melanocyte-specific alternatively spliced exon F (MyoVa-BD) (15, 65, 66). The C terminus of Mlph harbors an ABD (23, 24). (C) Tripartite complex reconstituted with 6×His-SNAP–tagged Rab27aS-H, FLAG-tagged MyoVaF, and full-length (lane I) or C-terminally truncated (lanes II and III) FLAG-tagged MlphF purified by Ni-NTA affinity purification. As a control for nonspecific binding, FLAG-tagged MyoVaF was also subjected to Ni-NTA affinity purification (lane IV) (Materials and Methods for details). MW, molecular mass marker. (D) The individually expressed full-length subunits of the tripartite complex along with C-terminally truncated Mlph and MyoVa HMM constructs were treated with PKA and radiolabeled ATP. Autoradiography showed specific phosphorylation of Mlph. Deletion of the C terminus of Mlph significantly decreased the phosphorylation levels. A FLAG-mock purification was included to control for unspecific phosphorylation.
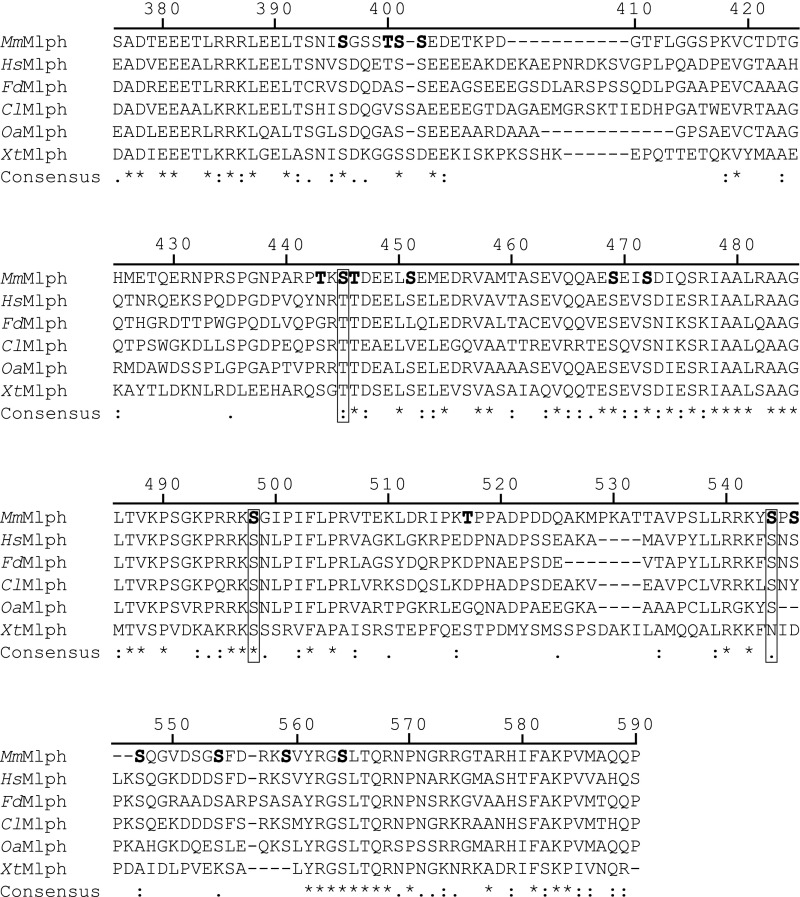
As detailed above, the C-terminal region of Mlph in mouse contains an ABD (Fig. 1B) (23, 24). Of note, several conserved serine/threonine residues that also represent potential cAMP-dependent protein kinase consensus sites in the ABD were found to be phosphorylated in proteomic analyses (Fig. S1) (45, 46). To test whether the ABD is a direct target of PKA and to assess its functional importance in vitro, we deleted the ABD (Mlph 1–400, referred to as “Mlph ΔABD,” and Mlph 1–444). Importantly, ABD-deleted Mlph retained its ability to form the tripartite transport complex, demonstrating the structural integrity of the deletion constructs (Fig. 1C, lanes II and III).
Fig. S1.
Alignment of Mlph ABDs from mouse (Mm), human (Hs), Fukomys (Fd), dog (Cl), sheep (Oa), and Xenopus (Xt). Sequence alignment of selected Mlph ABDs revealed numerous conserved serine/threonine residues that represent potential PKA targets. Serine and threonine residues that were found to be phosphorylated in vivo are in bold. Boxes indicate conserved phosphorylatable serine residues. Numbers represent the residue numbers according to MmMlph. Asterisks, colons, and dots indicate positions that are fully, partially, or weakly conserved, respectively.
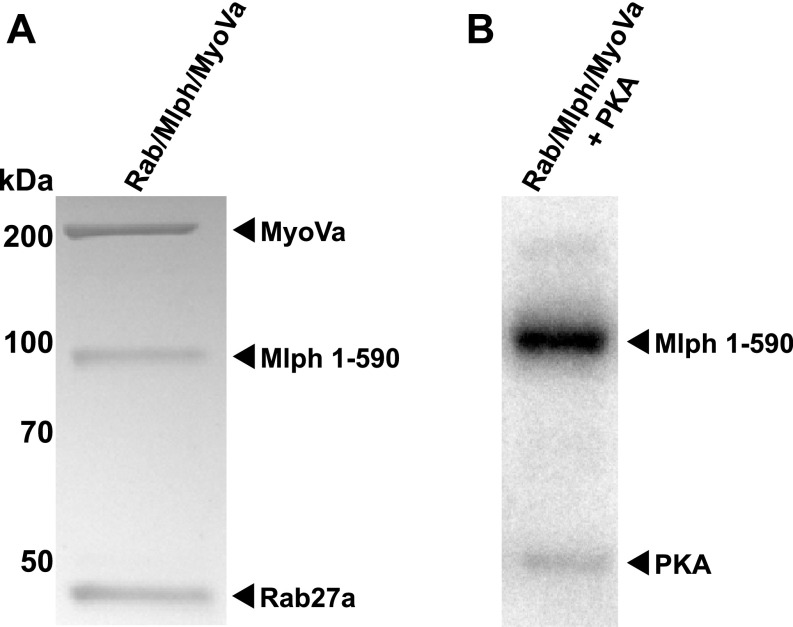
We next assessed whether the PKA directly phosphorylated any of the individually expressed subunits of the transport complex. The full-length MyoVa motor was previously shown to adopt a compact, inactive conformation in which the C-terminal tail domain interacted with the N-terminal head domains (47–50). To exclude the possibility that a folded conformation of MyoVa hampers effective phosphorylation, we additionally phosphorylated a C-terminally truncated MyoVa (MyoVa HMM, residues 1–1,109) and the tripartite Rab27a/Mlph/MyoVa complex in which the MyoVa motor is C-terminally bound to the Mlph subunit and is released from its folded, inactive state (51, 52). Phosphorylation assays with all individual constructs described above and the reconstituted tripartite complex revealed that Mlph was the only target of PKA (Fig. 1D and Fig. S2). Consistent with previous proteomic analyses (45, 46), removal of the ABD substantially decreased the phosphorylation of Mlph in vitro (Fig. 1D). To identify the phosphorylated residues of the ABD, we used quantitative mass spectrometry. Table S1 shows that the great majority of the phosphosites previously identified in proteome-wide studies (45, 46) have indeed been phosphorylated by PKA in our in vitro assays. The conserved and strong PKA-consensus site S498 contributed most to the overall phosphorylation within the ABD (Fig. S1 and Table S1).
Fig. S2.
Mlph is the sole phosphorylation target of the reconstituted tripartite complex. (A) Coomassie-stained SDS/PAGE image of the reconstituted Rab27a/Mlph/MyoVa transport complex that was subjected to in vitro phosphorylation. (B) PKA specifically phosphorylated Mlph in the presence of Rab27a and MyoVa (autoradiograph).
Table S1.
Identification of phosphorylated residues and their quantification in the ABD of wild-type Mlph in vitro
| Modified amino acid | Localization probability | Phospho (STY) probabilities | Score | Occupancy, % |
| 400 | 0.708 | ISGSST(0.708)SSEDE | 183 | 18.1 |
| 401 | 0.949 | SGSSTS(0.949)SEDET | 208 | 37.1 |
| 416 | 0.999 | TFLGGS(0.999)PKVCT | 186 | 51.1 |
| 435 | 1.00 | ERNPRS(1.00)PGNPA | 128 | 60.9 |
| 443 | 0.386 | NPARPT(0.386)KSTDE | 51 | ND |
| 445 | 0.989 | ARPTKS(0.989)TDEEL | 122 | 18.9 |
| 446 | 0.988 | RPTKST(0.988)DEELS | 92 | 8.46 |
| 451 | 0.999 | TDEELS(0.999)EMEDR | 214 | 26.9 |
| 491 | 0.999 | LTVKPS(0.999)GKPRR | 145 | ND |
| 498 | 1.00 | KPRRKS(1.00)GIPIF | 150 | 99.7 |
| 517 | 1.00 | DRIPKT(1.00)PPADP | 137 | 22.2 |
| 537 | 0.999 | TTAVPS(0.999)LLRRK | 49 | ND |
| 544 | 0.999 | LRRKYS(0.999)PSSQG | 185 | 49.2 |
| 546 | 0.836 | RKYSPS(0.836)SQGVD | 126 | ND |
| 554 | 0.994 | GVDSGS(0.994)FDRKS | 143 | ND |
| 559 | 0.972 | SFDRKS(0.972)VYRGS | 100 | ND |
| 564 | 0.982 | SVYRGS(0.982)LTQRN | 89 | ND |
The residues found to be phosphorylated in proteomic analyses are highlighted in bold letters as in Fig. S1. The highly conserved residue S498 that also represents the strongest cAMP-dependent protein kinase consensus site was quantitatively phosphorylated in vitro. ND, not detected.
Interaction with Actin Is Independent of Mlph’s Phosphorylation State.
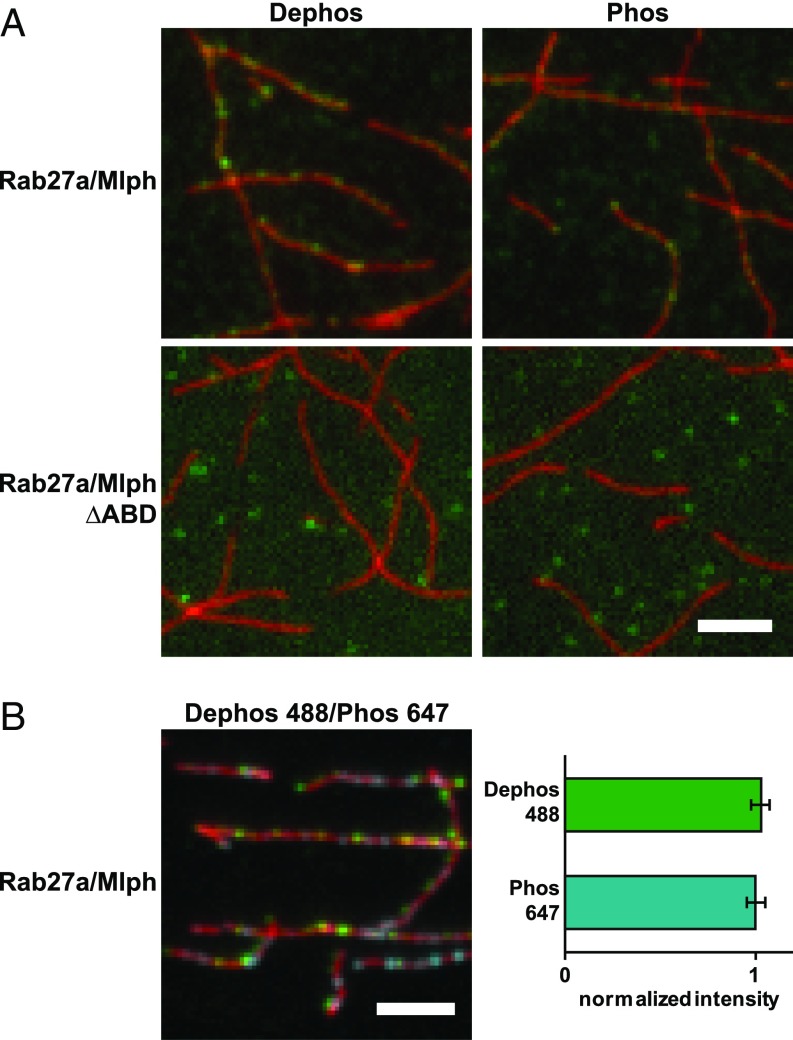
First we assessed whether phosphorylation of Mlph’s ABD affected its association with actin. To this end, we used the Rab27a/Mlph complex that is capable of recruiting the MyoVa motor, as we demonstrated earlier (Fig. 1C). To visualize the interaction, we fluorescently labeled the SNAP-tagged Rab27a subunit of the complex and the actin filaments using two different fluorophores. The phosphatase-treated complex (green channel) efficiently decorated the actin network (red channel), as did the PKA-treated complex, suggesting that the phosphorylation state of Mlph did not significantly affect actin binding (Fig. 2A). Consistent with previous results (19), Mlph lacking its ABD no longer interacted with actin filaments (Fig. 2A).
Fig. 2.
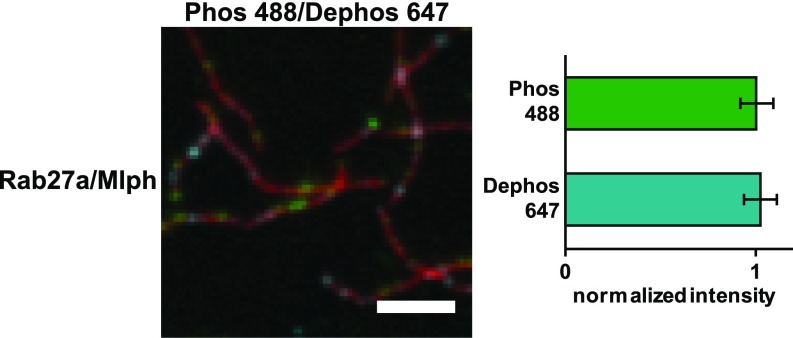
Mlph’s phosphorylation state does not interfere substantially with actin binding. (A) Actin decoration experiments were performed with surface-immobilized and Atto488-labeled actin filaments (red). Filaments were incubated with the complex formed between Mlph and Alexa Fluor 647-labeled Rab27a (green). Dephosphorylated (Dephos; Left) and the phosphorylated (Phos; Right) Mlph decorated actin filaments similarly well. Removal of the C-terminal ABD of Mlph (Rab27a/Mlph ΔABD) abolished this interaction regardless of Mlph’s phosphorylation state. (B) The dephosphorylated, Alexa Fluor 488-labeled Rab27a/Mlph complex was mixed in equal amounts with the phosphorylated, Alexa Fluor 647-labeled Rab27a/Mlph complex and was incubated with surface-attached, Atto565-labeled actin filaments. The quantification of the actin-associated fluorescence signals from the respective PKA- and phosphatase-treated Rab27a/Mlph complexes showed that the phosphorylation state of Mlph did not substantially interfere with actin binding. Error bars represent SD. (Scale bars: 3 µm.)
Next we assessed quantitatively whether the phosphorylated or dephosphorylated Mlph preferentially associated with actin. To this end, we performed competitive-binding experiments with PKA- and phosphatase-treated Rab27a/Mlph complexes that were fluorescently labeled with Alexa Fluor 647 and Alexa Fluor 488 fluorophores, respectively. To account for the intensity differences in the fluorescence signal, we additionally swapped the fluorophores. The quantification of the actin-associated fluorescence signals from the PKA- and phosphatase-treated Rab27a/Mlph complexes showed no phosphorylation-dependent association of Mlph with actin (Fig. 2B and Fig. S3). We conclude that Mlph’s phosphorylation state does not affect actin binding.
Fig. S3.
Mlph’s phosphorylation state does not interfere substantially with actin binding. Dephosphorylated Alexa Fluor 647-labeled Rab27a/Mlph complex (cyan) was mixed in equal amounts with phosphorylated, Alexa 488-labeled Rab27a/Mlph complex (green) and was incubated with surface-attached, Atto565-labeled actin filaments (red; dye swap control for Fig. 2B). The quantification showed results identical to those demonstrated in Fig. 2B: Phosphorylation of Mlph does not alter the interaction between Mlph and actin filaments. Error bars represent SD. (Scale bar: 3 µm.)
The Role of Mlph’s Phosphorylation in the MyoVa-Dependent Transport of the Tripartite Complex.
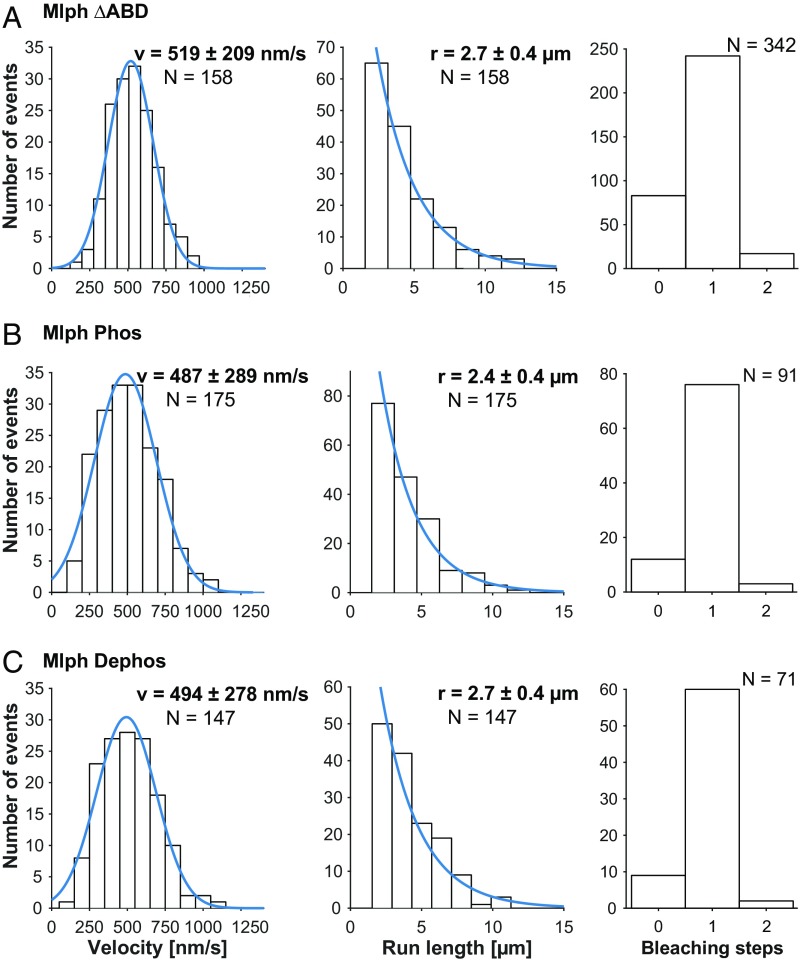
The binding of Mlph to MyoVa was suggested to activate the ATPase of the auto-inhibited motor (51, 52). The presence of ABD was further shown to increase the run frequencies and the run length of MyoVa on the actin filaments (19, 22). Interestingly, however, MyoVa bound to Mlph displayed decreased velocities compared with MyoVa alone in vitro (22). To assess the potential role(s) of Mlph phosphorylation on the MyoVa-dependent transport, we turned to single-molecule total internal reflection fluorescence (TIRF) microscopy assays (53). To this end, we assembled three Rab27a/Mlph/MyoVa tripartite complexes that were all fluorescently labeled via the SNAP-tagged Rab27a subunit. Specifically, we compared the single-molecule transport parameters of the tripartite complexes formed with Mlph and Mlph ΔABD, respectively (Fig. 3 A vs. B and C and Fig. S4). We additionally determined the transport parameters of the phosphorylated and dephosphorylated tripartite complexes to assess the role of Mlph phosphorylation on the MyoVa-dependent transport (Fig. 3 B vs. C). The velocities and run lengths of the respective tripartite complexes were independent of the presence or absence of the ABD (Fig. 3). Similarly, we did not observe any considerable impact of Mlph’s phosphorylation state on the transport properties of the tripartite complex (Fig. 3 B and C). In fact, the velocities of all three tripartite complexes were fully consistent with the previously measured velocities of the full-length MyoVa or the C-terminally truncated MyoVa motors in single-molecule TIRF assays and were not consistent with previously reported effects of Mlph on MyoVa velocity or run length (22, 54).
Fig. 3.
Transport parameters of the tripartite complex on surface-attached actin filaments in single-molecule TIRF assays. The tripartite complex assembled with Mlph that lacked its ABD (A) and the complexes assembled with phosphorylated (B) and dephosphorylated (C) Mlph all moved at consistent velocities. The absence of ABD (A) or the phosphorylation state of the Mlph (B vs. C) did not interfere with the velocities and run lengths of the respective complexes. The majority of the complexes displayed a single-step photobleaching of the SNAP-tagged Rab27a subunit as shown in A–C, Right, demonstrating that the transport parameters are derived from single molecules of Rab27a (also see Fig. S4).
Fig. S4.
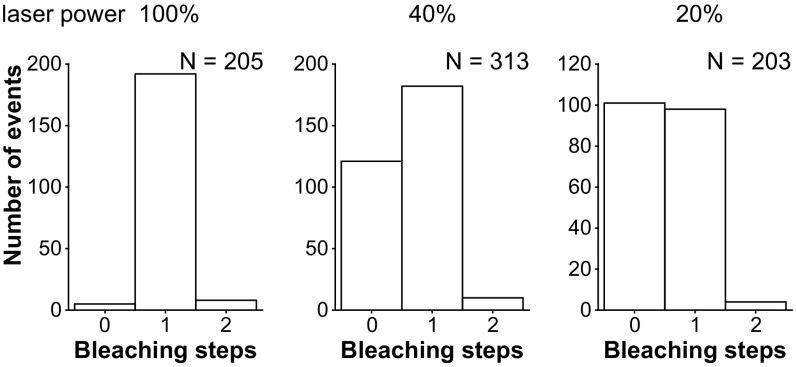
Bleaching step analysis of the Rab27a/Mlph ΔABD complex labeled on the Rab27a subunit dependent on the laser intensity. For each laser intensity, 112 frames with a frame rate of 223 ms were acquired and analyzed according to the procedure described in Materials and Methods. With decreasing laser intensity, the zero-step population increased strongly relative to the one-step population, as is consistent with photobleaching but not dissociation of the complex from the surface.
Taking these findings together, we conclude that PKA-dependent phosphorylation of Mlph, particularly its ABD, does not impact the actin-related processes such as actin binding (Fig. 2) or MyoVa-dependent transport (Fig. 3) in vitro. These results are consistent with the previous finding that ABD deletion did not affect the MyoVa-dependent distribution of the melanosomes on the actin network in vivo (26).
Phosphorylation of PKA Consensus Sites in Mlph’s ABD Regulates Binding to Microtubules.
One of the crucial tasks of the Rab27a/Mlph/MyoVa tripartite complex is to switch from the microtubule to the actin network to enable efficient dispersion of melanosomes in melanocytes (6). We therefore asked whether Mlph associates with microtubules. To this end, we performed filament decoration experiments with microtubules as described above for actin filaments (Fig. 2). Surprisingly, Mlph interacted with microtubules (Fig. 4).
Fig. 4.
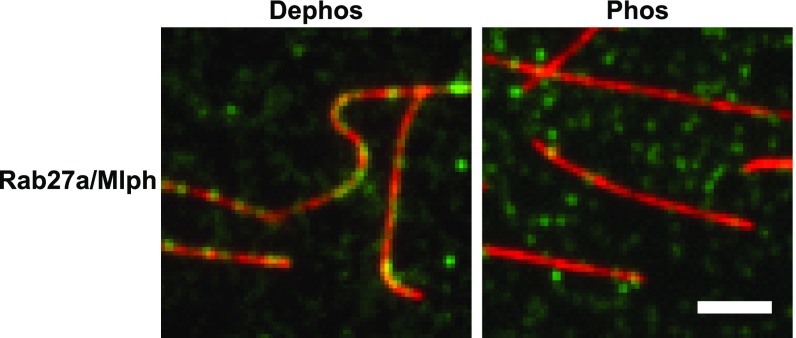
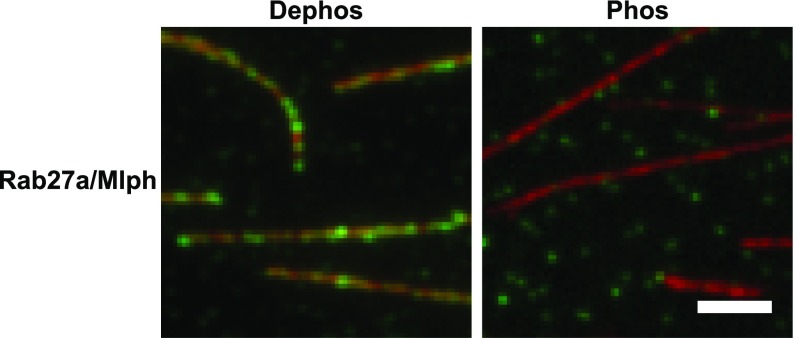
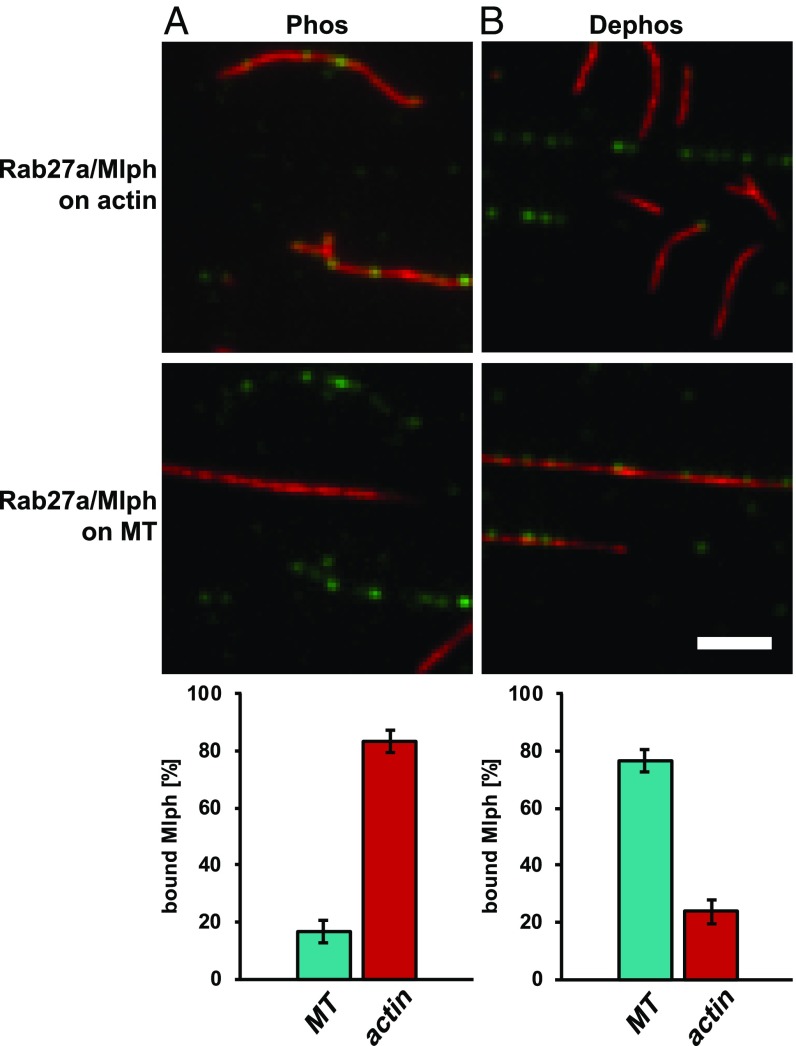
Mlph interacts with microtubules in a phosphorylation-dependent manner. In microtubule decoration experiments Atto488-labeled microtubules (red) were incubated with the Alexa Fluor 647-labeled Rab27a/Mlph complex (green). Decoration of microtubules was strictly dependent on the phosphorylation state of Mlph. The fluorescent background from the phosphorylated and dephosphorylated Rab27a/Mlph complex seen in the green channel was comparable, indicating similar amounts of protein. (Scale bar: 3 µm.)
Unexpectedly, and in stark contrast to actin, the phosphorylation state of Mlph was critical for the interaction with the microtubules. Although phosphatase-treated Mlph decorated microtubules robustly, this association was not detectable with PKA-treated Mlph (Fig. 4). Notably, ABD deletion diminished the interaction between Mlph and microtubules regardless of Mlph’s phosphorylation state (Fig. 5A).
Fig. 5.
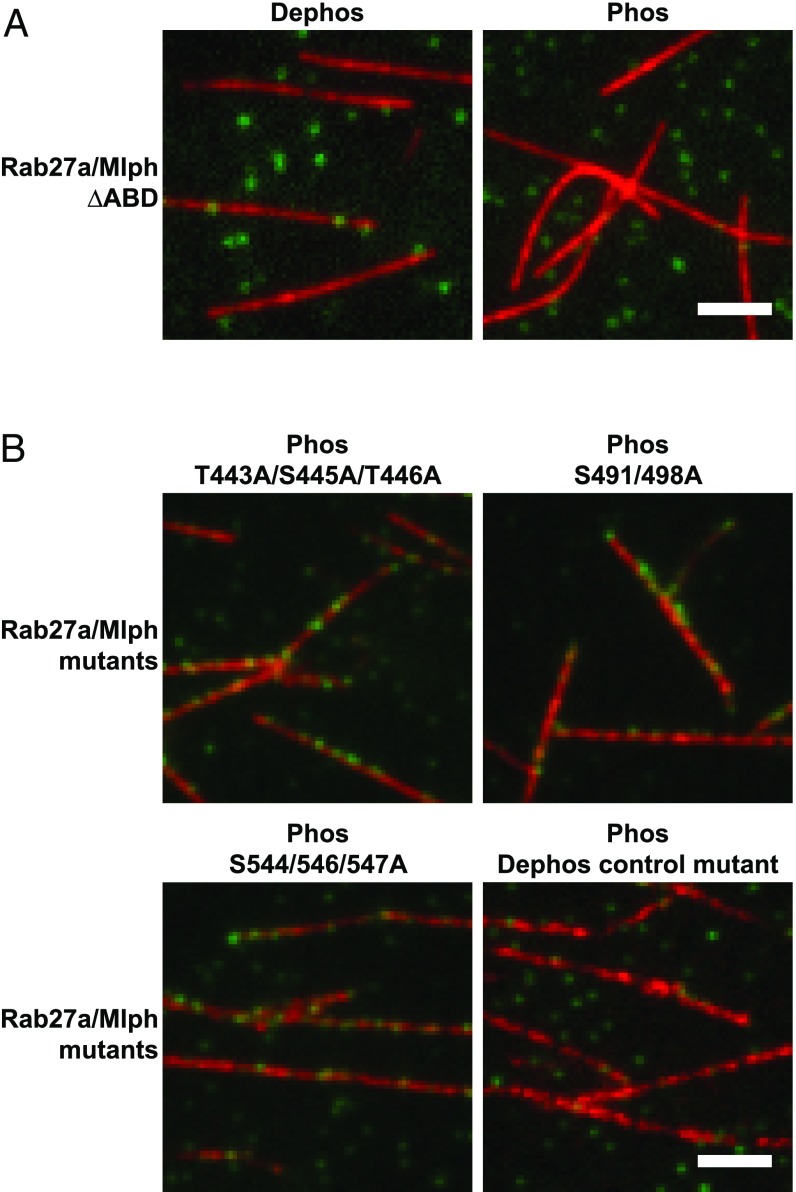
Mlph interacts with microtubules via its ABD, and mutations in candidate phosphorylation sites of Mlph prevent PKA-induced dissociation of Mlph from microtubules. (A) Microtubule decoration experiments were performed as in Fig. 4. Removal of the ABD (Rab27a/Mlph ΔABD) abolished the interaction of Mlph with microtubules. (B) PKA-treated Mlph mutants T443A/S445A/T446A, S491/498A, and S544/546/547A all decorated microtubules, but the Dephos control mutant (T392A/S393/396/398/399A/T400A/S401A) did not. (Scale bars: 3 μm.)
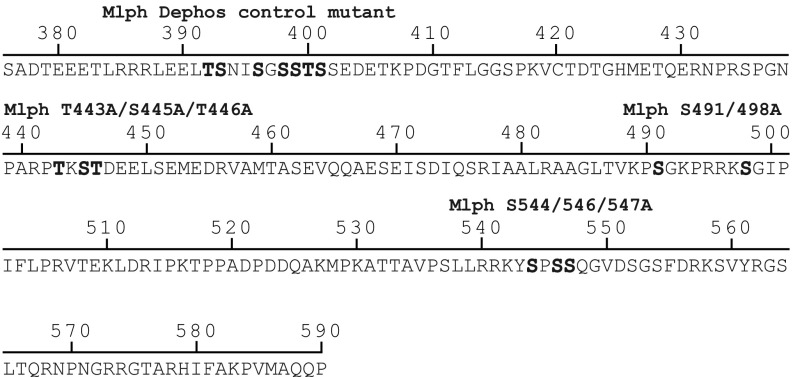
To probe directly whether PKA-dependent phosphorylation of the ABD was indeed responsible for the regulated interaction of Mlph with microtubules, we created three derivatives with mutations in the three phosphorylated clusters within the ABD (Fig. S1 and Table S1): T443A/S445A/T446A, S491/498A, and S544/546/547A (Fig. S5). As a negative control, we mutated an exceptionally serine/threonine-rich stretch (T392A/S393/396/398/399A/T400A/S401A) that bears no resemblance to a PKA consensus site, which we termed the “Dephos control mutant” (Fig. S1 and Fig. S5) (55). If PKA targets any of the respective consensus sites to release Mlph from microtubules, the corresponding mutant will be predicted to decorate microtubules even after PKA treatment.
Fig. S5.
Mutated serine and threonine residues in conserved regions within the ABD of Mlph. Serine and threonine residues in three conserved regions (Fig. S1) were mutated to alanines to mimic the dephosphorylated state. Mutated residues are shown in bold letters. A serine- and threonine-rich stretch outside the ABD was mutated as a negative control.
The mutant proteins and wild-type Mlph were purified side by side, were treated with PKA or phosphatase as needed, and were tested in filament decoration assays. Strikingly, the PKA-treated T443A/S445A/T446A, S491/498A, and S544/546/547A mutants decorated the microtubules (Fig. 5B), but the Dephos control mutant did not (Fig. 5B). As expected from the results shown above (Fig. 4), wild-type Mlph associated strictly with microtubules in its dephosphorylated state (Fig. S6). Collectively, these results demonstrate that the residues mutated in the ABD are necessary for the PKA-dependent release of Mlph from microtubules.
Fig. S6.
Dephosphorylated wild-type Rab27a/Mlph decorates microtubules in vitro, but phosphorylated wild-type Rab27a/Mlph does not. The Rab27a/Mlph complex (green) reconstituted with wild-type Mlph was purified side by side with the Rab27a/Mlph complex containing the point mutations in Mlph’s ABD (see Fig. 5) and used as a control. Consistent with Fig. 4, the dephosphorylated complex decorated microtubules (red), but the phosphorylated Rab27a/Mlph complex failed to decorate microtubules. (Scale bar: 3 μm.)
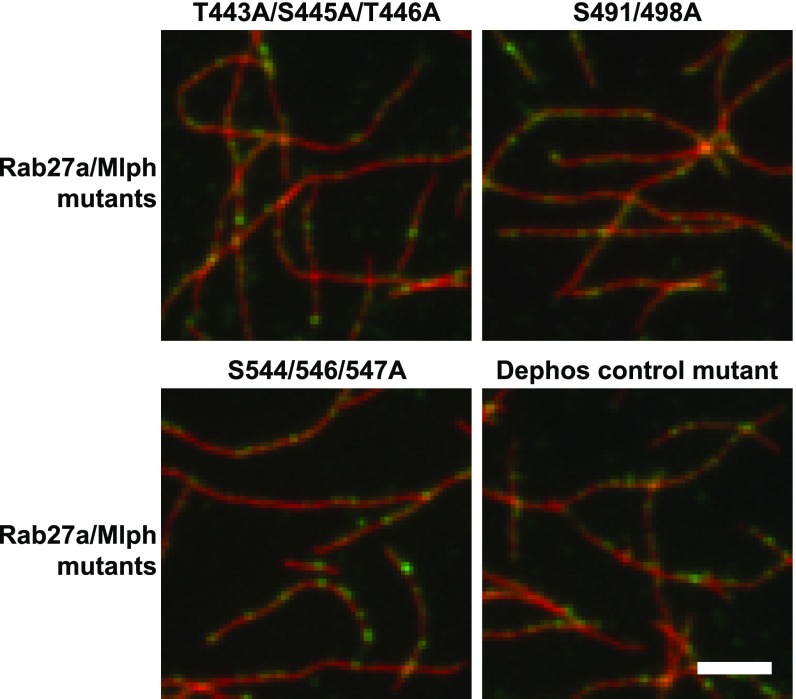
In contrast, all mutants decorated actin filaments similar to the wild-type Mlph (Fig. S7), indicating a nonoverlapping binding of Mlph’s ABD to microtubules and actin, respectively. Indeed, a positively charged stretch (K493, R495, R496, K497) in the ABD of Mlph was previously shown to be responsible for the interaction between Mlph and actin (22, 24). Therefore, the binding of the ABD to actin relies on electrostatic interactions, as described previously, whereas the binding to microtubules takes place in a phosphorylation-dependent manner, as delineated in this work.
Fig. S7.
Mutations in candidate phosphorylation sites of Mlph do not interfere with F-actin binding. Decoration of fluorescently labeled F-actin (red) with the constructs used in Fig. 5 shows that the interaction of all Rab27a/Mlph complexes containing the respective point mutations (green) with F-actin was similar to the interaction of the wild-type complex with F-actin. (Scale bar: 3 µm.)
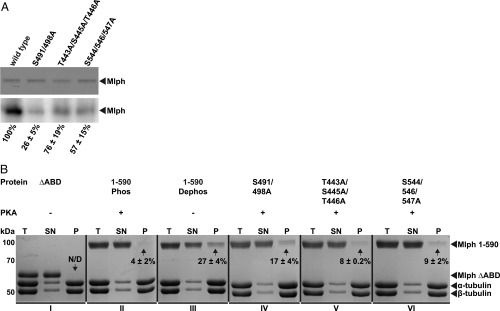
To assess the individual contributions of the respective nonphosphorylatable alanine mutations, we assessed the levels of PKA-dependent phosphorylation of all mutants with respect to wild-type Mlph. Consistent with our previous quantification of the ABD phosphorylation (Table S1), the S491/498A mutant suppressed the phosphorylation of Mlph more robustly than the T443A/S445A/T445A and S544/546/547A mutants (Fig. 6A).
Fig. 6.
Phosphorylation of S491/498A is the main contributor to phosphorylation-dependent binding of Mlph to microtubules. (A) Nonphosphorylatable alanine mutations in three predicted phosphorylation sites reduced PKA-dependent phosphorylation. In vitro phosphorylation assays (Lower: autoradiograph; Upper: corresponding Coomassie-stained SDS/PAGE as loading control) with wild-type Mlph and its mutants showed that the S491/498A mutant suppressed the PKA-dependent phosphorylation of Mlph more efficiently (26%) than the wild-type (100%) or mutants containing weaker and less conserved PKA consensus sites (76% and 57%). (B) Microtubule cosedimentation assays were performed side by side with the truncated Mlph that lacked its ABD (I, ΔABD), phosphorylated wild-type Mlph (II, 1–590 Phos), dephosphorylated wild-type Mlph (III, 1–590 Dephos) along with phosphorylated Mlph that carried the respective nonphosphorylatable alanine mutations (IV–VI). The total reaction (T), supernatant (SN), and pellet (P) were analyzed with SDS/PAGE. As expected from the results shown in Figs. 4 and 5, Mlph that lacked its ABD and the phosphorylated wild-type Mlph failed to display a pronounced interaction with the microtubules (N/D and 4%, I and II, respectively). Also, in line with results from Fig. 5, the dephosphorylated wild-type Mlph and all rescue mutants displayed significant pelleting with microtubules [27%, 17%, 8%, and 9% (III–VI), respectively]. The mutant S491/498A containing the strong and most conserved PKA consensus site S498 also demonstrated the most pronounced effect of rescue (IV). Percentages are obtained from two independent assays ± SD. N/D, not determinable.
The robust suppression of Mph phosphorylation by the S491/498A mutant led us to predict that the S491/498A mutant would display the most pronounced rescue of microtubule binding upon phosphorylation; we next tested this prediction in microtubule cosedimentation assays (Fig. 6B and Fig. S8). As expected from our previous results (compare Figs. 4 and 5A), Mlph lacking its ABD and the phosphorylated Mlph largely failed to interact with microtubules (Fig. 6 B, I and II). In contrast, dephosphorylated wild-type Mlph and all three (phosphorylated) mutants cosedimented with microtubules (Fig. 6B, III–VI and Fig. S8). Of all phosphorylated mutants, the Mlph derivative with the alanine substitution of the highly conserved S498 displayed the most robust microtubule binding (Fig. 6B, IV and Fig. S1). Collectively, both in vitro phosphorylation and microtubule cosedimentation assays highlighted the predominant role of the conserved S498 in mediating the PKA-dependent binding of Mlph to microtubules. We conclude that PKA-dependent phosphorylation regulates the ABD-dependent binding of Mlph to microtubules. In contrast, the phosphorylation state of Mlph does not impact the association with actin, Rab27a, or MyoVa.
Fig. S8.
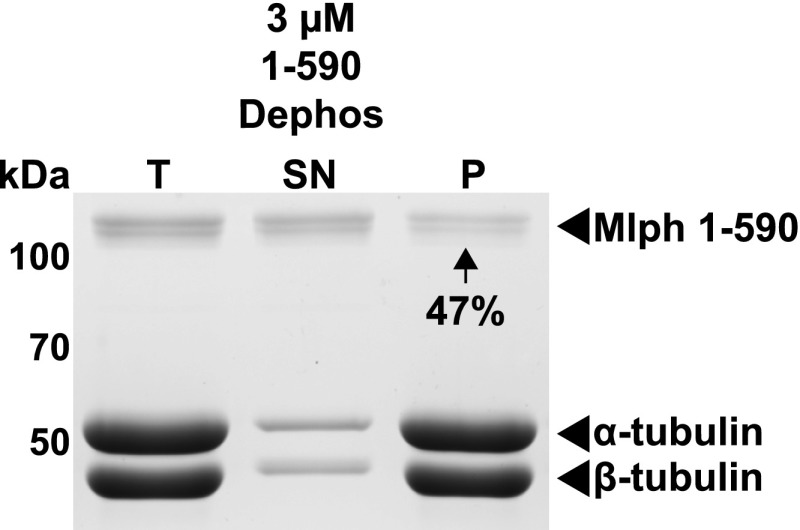
Cosedimentation assay of microtubules (2 µM) with a reduced amount (3 µM) of dephosphorylated wild-type Mlph (1-590 Dephos). Forty-seven percent of the total amount of Mlph protein pelleted with microtubules, noticeably more than in Fig. 6, where excess Mlph (5 µM) was used and only 27% pelleted. These observations suggested that binding sites for Mlph on microtubules become limiting when Mlph exceeds the concentration of microtubules.
Microtubules Compete Efficiently for Mlph Binding in the Presence of Actin.
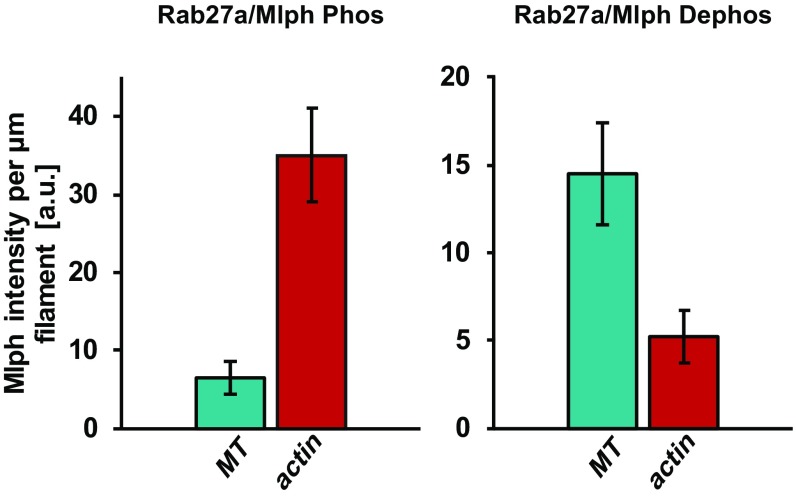
So far, we have shown that the ABD is able to interact not only with actin, as shown previously (22–24), but also with microtubules (Fig. 4). The interaction of Mlph with microtubules—in contrast to its interaction with actin—was strictly phosphorylation dependent (Figs. 2 and 4). To clarify which filament dominated Mlph binding, we performed competitive filament-binding assays using three-color TIRF microscopy. To this end, we fluorescently labeled actin filaments and microtubules with different fluorophores and decorated this mixed network with phosphorylated or dephosphorylated Rab27a/Mlph complex labeled with a third fluorophore. As expected from our previous findings (Fig. 4), the phosphorylated Rab27a/Mlph complex largely ignored the microtubules and associated with the actin filaments, consistent with phosphorylated Mlph having a higher affinity for actin than for microtubules (Fig. 7A and Fig. S9). The dephosphorylated Rab27a/Mlph complex, on the other hand, interacted with both the actin and the microtubules (Figs. 2A and 4). Remarkably, however, dephosphorylated Mlph bound predominantly to microtubules rather than to actin under the competitive conditions (Fig. 7B and Fig. S9). Our in vitro dissection of the Rab27a/Mlph/MyoVa complex thus unmasked an unexpected feature of the Mlph adaptor protein. Instead of regulating actin-dependent processes, such as actin binding or MyoVa-dependent transport, the phosphorylation of Mlph switched the affinity of Mlph toward actin from microtubules. This switch in affinity was sufficient to relocalize the Mlph competitively from the actin to the microtubule network.
Fig. 7.
Dephosphorylation is sufficient to relocate Mlph from actin to microtubules efficiently. Surface-immobilized and Atto565-labeled actin filaments (red; Upper) and Atto488-labeled microtubules (red; Lower) were incubated with phosphorylated (A) and dephosphorylated (B) complex formed between Mlph and Alexa Fluor 647-labeled Rab27a (green). (A) The phosphorylated Rab27a/Mlph complex largely ignored microtubules (MT) and associated with actin filaments (17 ± 4% vs. 83 ± 4%). (B) Upon dephosphorylation, the behavior of the Rab27a/Mlph complex was reversed, and the microtubule binding clearly dominated (76 ± 4%) over the actin binding (24 ± 4%). Error bars represent SD. (Scale bar: 3 µm.)
Fig. S9.
Dephosphorylation is sufficient to relocate Mlph from actin to microtubules efficiently. Alternative analysis of decoration assays with phosphorylated (Left) or dephosphorylated (Right) Rab27a/Mlph complex on both microtubules and actin. The Mlph intensity bound to each filament type was normalized to the total length of this filament type present in the channel. Error bars represent SD. a.u., arbitrary units.
The ABD of Mlph Enforces the Track Selection on the Microtubule and Actin Networks.
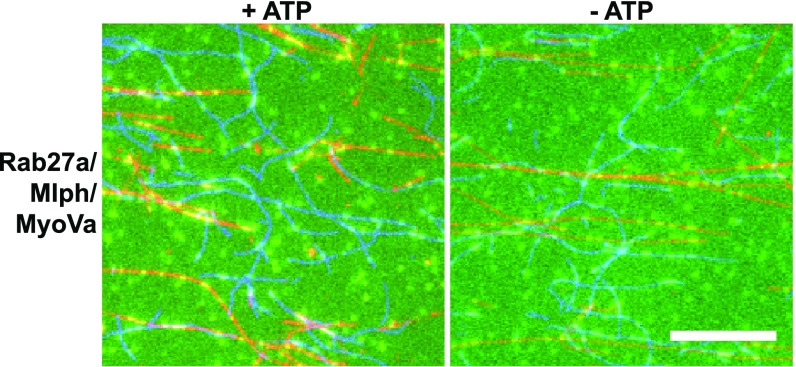
To test the functional consequences of Mlph’s phosphorylation-dependent association with the microtubule on the MyoVa motor, we immobilized the respective networks on a glass surface, as described above, and assessed the behavior of the Rab27a/Mlph/MyoVa tripartite complex built with phosphorylated and dephosphorylated Mlph, respectively. As intuitively expected from the actin-based MyoVa motor, the tripartite complex built with the phosphorylated Mlph largely ignored the microtubules and displayed directional movement on the actin network (Movie S1). Remarkably, however, dephosphorylation of Mlph was sufficient to redirect the complex substantially from the actin to the microtubule network (Fig. S10 and Movie S2).
Fig. S10.
Dephosphorylated Rab27a/Mlph/MyoVa complexes (green) in the presence (Left) or absence (Right) of ATP on surface-attached networks of actin (blue) and microtubules (red) in vitro. (Scale bar: 10 µm.)
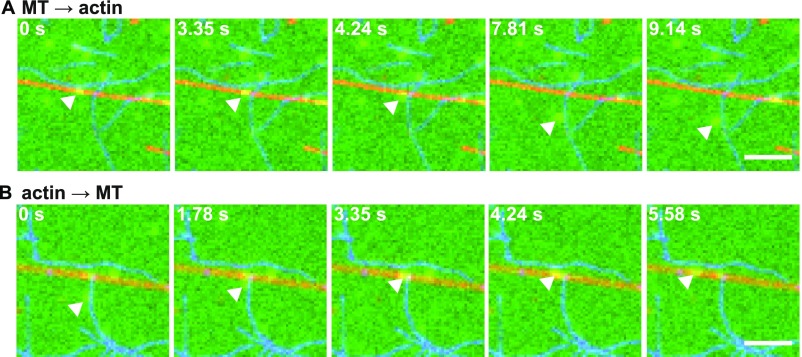
Inspection of the movies showed that some complexes switched the filament type at intersections (Fig. S11 and Movies S3 and S4). Indeed, the switching probabilities of the complexes depended on the phosphorylation state of the Mlph. The tripartite complex containing dephosphorylated Mlph had a pronounced propensity to switch from actin to microtubules (Fig. 8A). Phosphorylation of Mlph completely abolished transfer to the microtubules (Fig. 8B).
Fig. S11.
Examples of switching events at the actin/microtubule intersections. (A) Example of a tripartite complex (green) that diffused on a microtubule (red) and switched to an intersecting actin filament (blue) (Movie S3). (B) Example of a tripartite complex that moved directionally on the actin filament and switched to an intersecting microtubule (Movie S4). (Scale bars: 3 µm.)
Fig. 8.
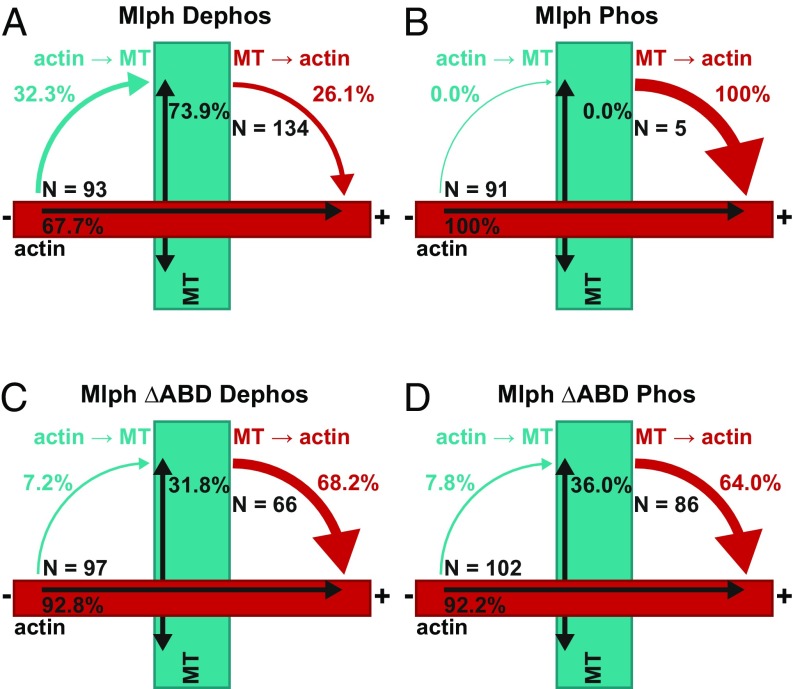
Phosphorylation state of Mlph’s ABD dictates the directionality of switching at the actin–microtubule intersections. The movement of the tripartite complex on actin and microtubules is represented by single- and double-headed black arrows, respectively. Cyan arrows depict switching from actin to microtubules; red arrows indicate switching from microtubules to actin. (A and B) The tripartite complex reconstituted with dephosphorylated Mlph displayed a significantly higher probability of switching from actin to microtubules at the interfilament intersections (A). Although 32.3% of complexes switched from actin to microtubules (67.7% continued directional movement on actin; A), the propensity of switching from actin to microtubules was abolished when the tripartite complex was assembled with phosphorylated Mlph (B). The phosphorylated complex completely ignored the interfilament intersections (0% switching) and continued its directional movement on the actin (100%; B). Conversely, dephosphorylated Mlph significantly suppressed the probability of the complex switching from microtubules to actin (26.1%; A) compared with the complex built with phosphorylated Mlph (100%; B). Indeed, the phosphorylated complex rarely interacted with the microtubules, substantially decreasing the probability of switching events from microtubules to actin (Movie S1). (C and D) In contrast, tripartite complexes assembled with dephosphorylated Mlph ΔABD (C) and phosphorylated Mlph ΔABD (D) displayed similar probabilities of switching between the two filament types, confirming that phosphorylation outside Mlph’s ABD does not interfere with the switching behavior of the tripartite complex. N indicates the number of events for each switching direction.
Because the ABD mediated the interaction with both actin and microtubules (Fig. 2 and 5A), a tripartite complex lacking the ABD should reveal the preference of MyoVa for switching filament type. As expected from an actin-based motor, the ABD-deleted complex switched preferentially from microtubules back to the actin network independent of Mlph’s phosphorylation state (Fig. 8 C and D). Indeed, the propensity of the tripartite complex to switch from actin to the microtubules in the absence of the ABD closely matched the previously reported probability of the C-terminally truncated MyoVa HMM to switch between the respective filaments (56).
Collectively, our results unmasked the surprising regulatory power of an adaptor protein over its associated motor. Although the MyoVa motor in complex with phosphorylated Mlph and the motor in complex with Mlph ΔABD preferred the actin filaments over microtubules, this balance was markedly shifted toward the microtubule network in the presence of a dephosphorylated ABD.
How can such Mlph-mediated regulation benefit directional cargo transport on the microtubule/actin networks in vivo? Previous in vivo tracking of melanosomes from different organisms revealed common features of the motor protein-dependent transport. For example, during aggregation of melanosomes in mouse melanocytes and Xenopus laevis melanophores, the dynein/microtubule system counteracts the MyoVa/actin system, forcing the melanosomes to switch from the actin to the microtubule network so they can be transported toward the cell center (3, 57). In contrast, when melanosomes disperse toward the cell periphery, the MyoVa/actin system wins over the dynein/microtubule system and switches melanosomes to the actin network (3, 38, 58). However, regulatory mechanisms that govern the directionality of the transport between the two cytoskeletal networks remained unclear.
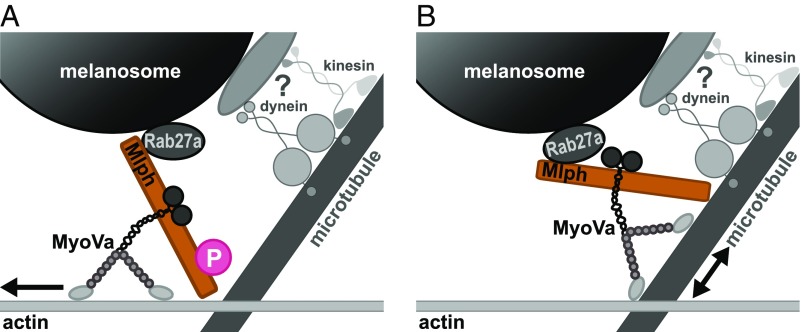
In our in vitro study, we uncovered a surprising phosphorylation-regulated interaction between Mlph and microtubules that was mediated by Mlph’s C-terminal ABD. Importantly, the phosphorylation state of Mlph competitively directed the Rab27a/Mlph as well as the tripartite Rab27a/Mlph/MyoVa complex toward the actin or microtubule network. Our findings thus argue that this regulated competition serves to bias the overall directionality of the moving melanosomes on the actin/microtubule cytoskeleton in vivo (Fig. 9). Taken together, our in vitro dissection of the components involved in the actin-dependent transport of melanosomes in mouse melanocytes delivered unexpected mechanistic insights which deserve further scrutiny in future in vivo studies.
Fig. 9.
Proposed model for regulating the affinities of the moving organelles on the microtubule and actin networks in vivo. In our mechanistic dissection, we unmasked the regulatory dominance of the adaptor protein Mlph over its associated motor. Even though MyoVa is an actin-associated motor, the dephosphorylation of Mlph’s ABD was sufficient to redirect the MyoVa from directional movement on the actin network to microtubules for diffusive movement. Consequently, the phosphorylation state of Mlph’s ABD regulated the probability of directional switching of MyoVa between the microtubule and actin networks. Based on these findings, we propose that Mlph serves to bias the transport of organelles on the microtubule or actin networks in vivo. (A) Specifically, phosphorylation of Mlph’s ABD promotes MyoVa-dependent motility on the actin network by suppressing the affinity of the tripartite complex toward microtubules. (B) To reverse this process, Mlph is dephosphorylated to increase the affinity of the tripartite complex for the microtubule network.
Materials and Methods
Reagents.
All reagents were the highest purity available and were obtained from Sigma-Aldrich unless stated otherwise.
DNA Constructs.
All constructs were cloned into the vector pFastBacI for subsequent expression in the baculovirus system (Life Technologies). A number of constructs were codon-optimized for expression in insect cells and were synthesized commercially (GenScript).
Rab27a (accession no. NM_023635.4; used for Fig. 1) and the constitutively active Rab27a-Q78L [used for all TIRF assays (42)] with a C-terminal SNAP-tag followed by a 6×His-tag, Mlph (accession no. NM_053015.2) with an N-terminal FLAG-tag, and MyoVa (accession no. NM_010864.1) with an N-terminal FLAG-tag were commercially synthesized (GenScript). The C-terminal truncated Mlph constructs with an N-terminal FLAG-tag were obtained by PCR with the primer pairs 5′-aggggatccctcgagatggactacaaagatgacgacgataaaggcggtggaaaaagattggacc-3′ and 5′-gcctggaggaattgacatccaacatcagcggaagttcgacgtaaggtaccaagcttata-3′ or 5′-cgcaaccctcgttcacccggaaacccagctaggccgacaaagtaaggtaccaagcttata-3′. Four point-mutated and N-terminally FLAG-tagged Mlph constructs mimicking the dephosphorylated state by substituting serine or threonine with alanine residues were synthesized (GenScript). The following point mutants were generated: Mlph T443A/S445A/T446A, Mlph S491/498A, Mlph S544/546/547A, and the Mlph Dephos control mutant T392A/S393/396/398/399A/T400A/S401A. An N-terminal truncation construct of MyoVa (MyoVa HMM, 1–1,109) was generated with the primers 5′-aggggatccctcgagatggattacaaagacgatgatgataaaggcggagcggcttcgg-3′ and 5′-ggaaatgaccttgatgctgaacgtgcctaagcccggctaaggtaccaagcttata-3′.
Protein Expression.
All proteins were expressed using the baculovirus expression system in insect cells (Spodoptera frugiperda, Sf9) according to the manufacturer’s instructions (Life Technologies). Proteins were FLAG- or 6×His-tagged to facilitate purification. Protein purification was carried out as described in SI Materials and Methods and was analyzed by SDS/PAGE.
Reconstitution of the Tripartite Complex.
The 6×His-tagged Rab27a and FLAG-tagged Mlph were coexpressed in Sf9 cells. This complex was purified first via the 6×His-tag, followed by FLAG-tag affinity purification. MyoVa was expressed separately and purified as described above. The Rab27a/Mlph complex was reattached to Ni-NTA beads and was incubated for 1.5 h; then beads were washed before purified MyoVa was added. After 1.5 h of incubation with purified MyoVa, beads were washed with His wash buffer, and the tripartite complex was eluted with the His elution buffer. As a control, purified FLAG-tagged MyoVa was incubated with plain Ni-NTA beads to exclude the possibility of unspecific binding to agarose beads.
In Vitro Phosphorylation Assay with Isotope-Labeled ATP.
Two hundred nanomolar PKA (catalytic subunit from bovine heart) was mixed with 1.2 µM of substrate in kinase buffer [25 mM imidazole (pH 7.5), 25 mM KCl, 10 mM MgCl2, 10 mM DTT]. Fifty nanomolar radiolabeled ATP diluted with 10 µM cold ATP was added to start the reaction and was incubated at 30 °C in a thermo mixer (Eppendorf) for 15 min. The reaction was stopped by adding SDS sample buffer. To determine the stoichiometry of ABD phosphorylation, 0.5 µM of substrate was mixed with 200 nM PKA and was incubated at 30 °C for 20 min. SDS gel electrophoresis was performed, and the gel was placed against a phosphor storage screen overnight. The screen was scanned with a PhosphorImager (Typhoon 9200, Molecular Dynamics), and digital images were used for analysis.
Analysis of the Stoichiometry of Mlph ABD Phosphorylation.
Digital images of scanned phosphor storage screens were analyzed using ImageJ software (59). The mean gray value of each band representing wild-type or point-mutated Mlph was measured using the polygonal selection tool with constant area. For background subtraction, the mean gray values of four different parts of the gel with area identical to that of the Mlph proteins were averaged and subtracted from the mean gray values of the different Mlph proteins. The acquired values were corrected for applied protein amounts determined with the corresponding Coomassie-stained SDS/PAGE. The value obtained for wild-type Mlph was set to 100%, and values of different Mlph point mutants were compared with the wild-type Mlph value.
Fluorescent Labeling of Proteins.
The SNAP-tagged Rab27a was labeled during the protein purification. Before the elution of the protein from the Ni-NTA beads, the wash buffer was supplemented with 20 µM SNAP-tag substrate SNAP-Surface Alexa Fluor 647 or SNAP-Surface Alexa Fluor 488 (New England Biolabs) and was added to the Ni-NTA beads. The labeling reaction was carried out for 30 min at room temperature in the dark on a rotator. The beads were washed extensively with wash buffer to remove excess dye before elution.
Phosphorylation and Dephosphorylation of Recombinant Mlph.
Recombinantly expressed Mlph protein was dephosphorylated or phosphorylated after the purification procedure. To phosphorylate Mlph, 400–900 nM PKA (catalytic subunit from bovine heart) and 10,000 units PKA (catalytic subunit; New England Biolabs) in kinase assay buffer [25 mM imidazole (pH 7.5), 25 mM KCl, 10 mM MgCl2, 10 mM DTT] with 0.25 mM ATP were incubated with purified Mlph for 1 h at room temperature on a rotator. The phosphatase inhibitor mixture PhosSTOP (Roche) was included during phosphorylation to protect Mlph from dephosphorylation. For dephosphorylation, 30–60 units of Antarctic Phosphatase (New England Biolabs) and 30 units of human protein phosphatase 2A (Cayman Chemical) in phosphatase buffer [25 mM imidazole (pH 7.5), 25 mM KCl, 11 mM MgCl2, 10 mM DTT, 50 mM Bis-Tris-Propane-HCl, 0.1 mM ZnCl2] were incubated with purified Mlph protein for 1 h at room temperature on a rotator. Kinase and phosphatases were removed by reattaching Mlph to FLAG-beads and washing the beads four times with the respective wash buffer before elution.
Mlph Binding to Actin Filaments.
Biotinylated G-actin (0.25 µM) and G-actin (4.75 µM) were polymerized with 5 µM Atto488-phalloidin in assay buffer (AB) [25 mM imidazole (pH 7.5), 25 mM KCl, 1 mM EGTA, 4 mM MgCl2, 10 mM DTT] for 30 min at room temperature. Biotinylated BSA was flowed into a 10-µL flow chamber and was incubated for 3 min. The glass surface was blocked with 8 mg/mL BSA in AB. Streptavidin was bound to biotinylated BSA for 3 min followed by a wash step with 8 mg/mL BSA in AB. Biotinylated actin filaments were attached to the Biotin-Streptavidin layer, and unbound filaments were washed out with 8 mg/mL BSA in AB. Alexa Fluor 647-labeled Rab27a/Mlph complex diluted in AB was infused into the flow chamber. The chamber was washed with AB supplemented with 0.4% glucose, 0.18 mg/mL glucose-oxidase, and 0.06 mg/mL catalase, and image acquisition of the sealed chamber was performed at room temperature using a commercially built TIRF microscope (Leica AF6000 Modular Systems; Leica) equipped with an HCX PL Apo 100×/1.47 TIRF objective. Images were acquired with an iXon Ultra X-9360 EMCCD camera (Andor) controlled by the Leica Application Suite Advanced Fluorescence. Acquired images were processed further using ImageJ software.
Quantification of Mlph Binding to Actin Filaments.
For quantification of the Mlph–actin interaction, 400 nM of phosphatase-treated, Alexa Fluor 488-labeled Rab27a/Mlph complex and PKA-treated, Alexa Fluor 647-labeled Rab27a/Mlph complex (or complexes with swapped fluorescent dyes) were perfused into the flow chamber with surface-attached Atto565-labeled actin filaments. The color swap on the Rab27a/Mlph protein controlled for inherent intensity differences of the Alexa Fluor 647 and Alexa Fluor 488 fluorophores. The association of dephosphorylated or phosphorylated Mlph to actin filaments was evaluated using a colocalization analysis script implemented in MATLAB (MathWorks). The filament images were binarized using a common threshold algorithm (60). The available binding area of actin filaments was determined after crossing or overlapping sections of filaments were deleted from the binary images. The background in each Rab27a/Mlph image was calculated as the mean of the image without the actin area and was subtracted from the images. A normalization parameter was introduced to account for differences in the imaging parameters for the two Rab27a/Mlph channels (488 and 647 nm). The images were normalized ignoring a percentage of pixels that was given by the parameter. A check over the parameter space provided a value where both the original and the color swap experiment resulted in the same (inverse) ratio of decoration. This ratio of bound Rab27a/Mlph was calculated from the intensity observed in the actin area for the phosphorylated channel divided by the intensity in the dephosphorylated channel. Data were analyzed from 30 images for the combination of dephosphorylated Alexa Fluor 488–Rab27a/Mlph and phosphorylated Alexa Fluor 647–Rab27a/Mlph complexes and from 44 images for the combination of phosphorylated Alexa Fluor 488–Rab27a/Mlph and dephosphorylated Alexa Fluor 647–Rab27a/Mlph complexes. Errors are given as the SD between ratios for the single images analyzed for each experiment.
Single-Molecule TIRF Assay with Tripartite Complex on Actin Filaments.
Detailed descriptions of the single-molecule TIRF assay with the tripartite complex and subsequent data analysis are provided in SI Materials and Methods.
Photobleaching Assay.
All tripartite complexes used for single-molecule TIRF assays were also subjected to photobleaching assays to ensure that transport parameters were obtained mainly from single tripartite complexes and not from oligomeric complexes. For this purpose, Rab27a/Mlph/MyoVa complexes (Single-Molecule TIRF Assay with Tripartite Complex) were diluted in AB and unspecifically bound to the glass surface of a flow chamber. The time-lapse recordings were performed at 80% laser power and 200-ms exposure time. Spots for analysis were chosen for all constructs by intensity over mean intensity, and the intensity values for a 3 × 3 pixel window were summarized. Observation of the spot intensities over time resulted in bleaching curves. A gliding t-value test with a corresponding threshold identified the bleaching steps (61).
Mlph Binding to Microtubules.
Tubulin was prepared from porcine brain as described previously (62). Biotinylated and Atto488-labeled tubulin was polymerized in BRB80 buffer [80 mM Pipes (pH 6.9), 2 mM MgCl2, 1 mM EGTA, 5 mM DTT] supplemented with 1 mM GTP. To stabilize filaments, 5 µM taxol were added after 30 min of incubation at 35 °C. Microtubules were attached to the surface via the Biotin–Streptavidin bond as described previously. Wash steps were performed with BRB80 buffer supplemented with 8 mg/mL BSA. Alexa Fluor 647-labeled Rab27a/Mlph complex diluted in BRB80 buffer was flowed into the flow chamber, and excess protein was removed by a wash step with BRB80 buffer supplemented with 0.4% glucose, 0.18 mg/mL glucose-oxidase, and 0.06 mg/mL catalase. Sealed chambers were imaged at room temperature as described above. Acquired images were processed further using the ImageJ software.
Microtubule Cosedimentation Assay with Mlph Proteins.
Cosedimentation assays with 3 or 5 µM of the respective Mlph protein and 2 µM microtubules are described in SI Materials and Methods.
Competitive Mlph Binding to Microtubules and Actin Filaments.
Competition experiments with mixed networks (microtubules and actin filaments) were performed with biotinylated, Atto488-labeled microtubules and biotinylated, Atto565-labeled actin filaments. Surface attachment of filaments was performed via the Biotin-Streptavidin layer in BRB80 buffer (Mlph Binding to Microtubules, above). Actin filaments were flowed into the prepared flow chamber first, followed by microtubules. After unbound filaments were removed with a wash step with 8 mg/mL BSA in BRB80 buffer, Alexa Fluor 647-labeled dephosphorylated or phosphorylated Rab27a/Mlph protein was infused into the flow chamber, and image acquisition was performed as described above.
Quantification of Competitive Mlph Binding to Microtubules and Actin Filaments.
Competitive Mlph binding to microtubules and actin filaments was evaluated using a colocalization analysis script implemented in MATLAB as described for Materials and Methods, Quantification of Mlph Binding to Actin Filaments. The available binding area for each filament type was determined as detailed previously. The channel for Mlph-associated fluorescence was also thresholded to avoid the influence of the background intensity on the data. The sum of the previously detected Mlph-associated intensity in the area of filaments was normalized to the total binding area of this filament type, yielding a binding parameter B_filament. The binding proportion for each filament was then calculated as B_filament1/(B_filament1 + B_filament2). Data from 17 images for the dephosphorylated Rab27a/Mlph complex and 29 images for the phosphorylated Rab27a/Mlph complex were analyzed. The actin:microtubules ratio of the binding area for the competition experiments with the dephosphorylated Rab27a/Mlph complex was 4.87 ± 1.58. The actin:microtubules ratio of the binding area for the competition experiments with the phosphorylated Rab27a/Mlph complex was 2.78 ± 1.46. Error bars were calculated from the SD occurring when data from all images were averaged.
Single-Molecule TIRF Assay with the Tripartite Complex on Microtubules and Actin Filaments.
Single-molecule TIRF assays with the Alexa 647-labeled tripartite complex on networks of Atto488-labeled microtubules and Atto565-labeled actin filaments were carried out as described in SI Materials and Methods.
Sequence Alignment.
MmMlph (NP_443748.2), HsMlph (NP_077006.1), FdMlph (XP_010624436.1), ClMlph (NP_001096689.2), OaMlph (NP_001139743.1), and XtMlph (NP_001120194.1 were aligned using ClustalX 2.1 (63). Alignment figures were prepared using the BioEdit Sequence Alignment Editor (64).
LC-MS/MS Analysis.
A complete description of the mass spectrometry analysis is provided in SI Materials and Methods.
SI Materials and Methods
His-Tag Affinity Purification.
Sf9 cells were infected with the respective virus(es) and grown for 72 h. Cells were harvested by centrifugation and resuspended in His lysis buffer [20 mM Tris (pH 8.0), 300 mM NaCl, 10 mM imidazole, 1 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, and Complete EDTA-free protease inhibitor mixture (Roche)]. Cell debris was removed by centrifugation at 40,000 × g for 10 min at 4 °C. The supernatant containing the 6×His-tagged target protein was incubated with Ni-NTA agarose beads (Qiagen) for 1.5 h at 4 °C. Beads were washed six times with His wash buffer [50 mM Tris (pH 8.0), 500 mM NaCl, 40 mM imidazole, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.1% Tween 20]. His-tagged protein was eluted with the His elution buffer [50 mM Tris (pH 7.5), 150 mM NaCl, 500 mM imidazole, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.05% Tween 20].
FLAG-Tag Affinity Purification.
Purification of FLAG-tagged proteins was carried out as described for 6×His-tag affinity purification using a different set of buffers [FLAG lysis buffer: 20 mM Tris (pH 7.5), 300 mM NaCl, 1 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, protease inhibitor mixture; FLAG wash buffer 1: 50 mM Tris (pH 7.5), 500 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.1% Tween 20; FLAG wash buffer 2: 50 mM Tris (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.05% Tween 20; FLAG elution buffer: FLAG wash buffer 2 plus 0.2 mg/mL FLAG peptides]. Anti-FLAG M2 affinity agarose gel was added to the cleared lysate and incubated for 1.5 h at 4 °C. Before elution, beads were first washed three times with FLAG wash buffer 1 and then were washed three times with FLAG wash buffer 2.
FLAG-Tag Affinity Purification of Recombinant MyoVa.
Purification of FLAG-tagged MyoVa was performed as previously described (67) with minor modifications. Untagged calmodulin 1 from Mus musculus was always coexpressed with MyoVa. Briefly, cells were harvested after 72 h of incubation with the viruses. The cell pellet was flash-frozen in liquid nitrogen, resuspended in MyoVa lysis buffer [10 mM 3-(N-morpholino)propanesulfonic acid (Mops) (pH 7.0), 200 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 2 mM ATP, 0.1 mM PMSF, 1 mM DTT, protease inhibitor mixture], and homogenized using a glass homogenizer for 5 min on ice. The suspension was cleared by centrifugation at 47,000 × g for 10 min at 4 °C. The supernatant was incubated with anti-FLAG M2 resin for 1.5 h at 4 °C. Beads were washed on a Poly-Prep chromatography column (Bio-Rad) with 2 mL each, successively, of MyoVa wash buffer 1 [10 mM Mops (pH 7.0), 500 mM NaCl, 0.1 mM EGTA, 1 mM DTT, 0.1 mM PMSF], MyoVa wash buffer 2 [10 mM Mops (pH 7.0), 500 mM NaCl, 0.1 mM EGTA, 1 mM DTT, 0.1 mM PMSF, 1 mM ATP, 5 mM MgCl2], MyoVa wash buffer 1, MyoVa wash buffer 3 [10 mM Mops (pH 7.0), 0.1 mM EGTA, 1 mM DTT, 0.1 mM PMSF, 1 mM EDTA], MyoVa wash buffer 4 [10 mM Mops (pH 7.0), 500 mM NaCl, 0.1 mM EGTA, 1 mM DTT, 0.1 mM PMSF, 1 mM EDTA], MyoVa wash buffer 3, and MyoVa wash buffer 1. MyoVa was eluted with MyoVa elution buffer [10 mM Mops (pH 7.0), 200 mM KCl, 0.1 mM EGTA, 1 mM DTT, 0.1 mM PMSF, 0.3 mg/mL FLAG peptides].
Single-Molecule TIRF Assay with Tripartite Complex on Actin Filaments.
The tripartite MyoVa transport complex Rab27a/Mlph/MyoVa was reconstituted and labeled with Alexa Fluor 488 or 647 on the SNAP-tagged Rab27a subunit as described above. Biotinylated G-actin (0.25 µM) and G-actin (4.75 µM) were polymerized with 5 µM Atto488- or Atto647N-phalloidin in AB for 90–120 min at room temperature and then were stored on ice. Actin filaments were attached to the glass surface via Biotin-Streptavidin (see Materials and Methods, Mlph Binding to Actin Filaments in the main text), and wash steps were carried out with AB supplemented with 1 mg/mL BSA. The glass surface was blocked with 8 mg/mL BSA in AB after incubation with actin filaments. Reconstituted tripartite complex diluted in AB supplemented with 0.4% glucose, 0.18 mg/mL glucose-oxidase, 0.06 mg/mL catalase, 2 mM ATP, and 0.06 mg/mL bovine calmodulin (Calbiochem) was flowed into the chamber. Sealed chambers were imaged at room temperature using the TIRF microscope setup described previously. Acquired images and movies were processed further using ImageJ software.
Data Analysis of the Single-Molecule TIRF Assay with the Tripartite Complex on Actin Filaments.
Movies were recorded at a cycle time of 238 ms and were analyzed using a custom routine implemented in the MATLAB development environment. Spots were selected automatically according to their brightness compared with the mean brightness in each frame. The position of the spots was determined with subpixel accuracy using a radial center approach. A list of spots for each frame was generated, and the distances of spots in each subsequent frame were compared. The connection of each spot to the closest spot in the next frame (with a cutoff maximal distance) resulted in tracking traces for each spot. The distance to the first position of each spot over time was fitted with a linear model. Smooth runs that resulted in a linear fit with an r2 value higher than 97% and that had a length of at least 10 frames were chosen for speed and run length measurements. For run length measurements an additional threshold of 1 µm was applied. The speed was calculated from the slope of the performed fit, and the run length was calculated from the maximal distance of the spot from its original position. Missing run lengths were accounted for in the histogram fit by truncating the single-exponential model.
Microtubule Cosedimentation Assay with Mlph Proteins.
Tubulin was polymerized at 35 °C in BRB80 buffer supplemented with 1 mM GTP. Taxol (5 µM) was added after incubation for 30 min at 35 °C, and polymerized microtubules were kept at 35 °C overnight. Microtubules were centrifuged through a sucrose cushion (40% sucrose in BRB80) and were resuspended in BRB80 buffer supplemented with 5 µM taxol. Cosedimentation assays were carried out in BRB80 buffer with 2 µM microtubules and 3 or 5 µM of the respective Mlph protein. Reactions were mixed thoroughly and incubated at room temperature for 15 min. Microtubule–Mlph mixes were carefully pipetted onto a sucrose cushion and centrifuged at 279,000 × g for 15 min at 22 °C. Supernatant was gently removed, and the sucrose cushion was washed with BRB80. The sucrose cushion was removed, and the pellet was washed twice with BRB80. The pellet was resuspended in BRB80, and equal volumes of total reaction, supernatant, and pellet were prepared and analyzed with SDS/PAGE.
Analysis of the Microtubule Cosedimentation Assay with Mlph Proteins.
Digital images of total, supernatant, and pellet of each reaction on Coomassie-stained SDS gels were analyzed with ImageJ and MATLAB, respectively. To calculate the relative degrees of microtubule-bound protein fraction, a semiautomatic routine was implemented using MATLAB. The area containing the peak of interest was chosen manually from an intensity lane profile of the gel created with ImageJ. By binning the intensity for a histogram and using the upper limit of the heaviest populated bin as a threshold, the peak data were separated from the base line. To account for a linear variation of the base line, a straight line was fitted to the separated base line data. The peak area was calculated integrating the intensity data between the first interceptions of this line with the profile data before and after the peak. Resulting area values were used, and percentages of microtubule-bound protein compared with total protein were calculated. The relative degree of microtubule-bound Mlph ΔABD fraction was not determinable because of the close proximity of protein and tubulin peaks.
Single-Molecule TIRF Assay with Tripartite Complex on Microtubules and Actin Filaments.
Microtubules and actin filaments were prepared as described previously. Briefly, Atto488-labeled microtubules and Atto565-labeled actin filaments, both biotinylated, were used in this experiment. Microtubules were first attached to the surface of a flow chamber, followed by actin filaments. Wash steps were performed using BRB80 buffer. After the actin filaments were perfused into the flow chamber, the surface was blocked with 10 mg/mL BSA in AB. Alexa Fluor 647-labeled dephosphorylated or phosphorylated Rab27a/Mlph/MyoVa complex was diluted in AB supplemented with 0.4% glucose, 0.18 mg/mL glucose-oxidase, 0.06 mg/mL catalase, 2 mM ATP, and 0.06 mg/mL bovine calmodulin and was flowed into the chamber. Time-lapse movies with a cycle time of 223 ms were acquired using the previously described TIRF microscope. To analyze the switching behavior of the MyoVa transport complex on mixed networks, only complexes in close proximity to a microtubule–actin intersection were taken into account. The number of complexes that switched filament at an intersection or passed the intersection without changing the filament was counted manually. The percentage of switching complexes or passing complexes was calculated for dephosphorylated or phosphorylated Rab27a/Mlph/MyoVa complexes with and without Mlph’s ABD.
LC-MS/MS Analysis.
Wild-type Mlph was FLAG-affinity purified and phosphorylated while bound to FLAG-affinity resin. Purified protein was analyzed by the Mass Spectrometry Service of the Max Planck Institute of Biochemistry (Martinsried, Germany) using the following procedure: Protein samples were denatured by adding 4 M guanidium hydrochloride solution (final concentration). The proteins were reduced and alkylated using 10 mM Tris(2-carboxyethyl)phosphine hydrochloride and 40 mM chloroacetamide, respectively. Samples were then diluted to a final concentration of 0.4 M guanidium hydrochloride using a 50 mM ammonium bicarbonate solution. These reduced and alkylated protein samples were digested overnight with trypsin (1 µg trypsin for 50 µg protein). After overnight digestion, peptides were desalted using a C18 StageTip (68) and were used for LC-MS/MS analysis. Peptides were loaded on a 15-cm-long 75-µm i.d. reversed-phase C18 column packed with 1.9-µm beads via the autosampler in the Thermo Easy LC 1000. The peptides were separated over a 120-min linear gradient with 0.1% formic acid (buffer A) as the loading buffer and 0.1% formic acid and 80% acetonitrile (buffer B) as the eluting buffer. Separated peptides were directly electrosprayed into a Q Exactive HF benchtop Orbitrap mass spectrometer (69). The mass spectrometer was operated in data-dependent mode, with up to the top 15 precursors selected for fragmentation. The full scans were acquired at 120,000 resolution (m/z = 200) with up 3E6 charges accumulated in the C-trap, and the higher-energy collision dissociation (HCD) fragmentation spectra were recorded at 15,000 resolution with up to 1E5 charges of the precursor ions. The raw data were processed with MaxQuant computation platform (70) version 1.5.2.22 using the Andromeda (71) search engine. Precursor and fragment ions were searched with 4.5 and 20 ppm mass tolerance, respectively, with the false-discovery rate set to 1% at both the protein and the peptide levels. Carbamidomethylation of cysteine was set as fixed modification, and oxidation of methionine, protein N-terminal acetylation, and phosphorylation on serine, threonine, and tyrosine were used as variation modifications in the database search.
Supplementary Material
Acknowledgments
We thank Günther Woehlke (Technische Universität München), James R. Sellers (National Institutes of Health), and Vladimir I. Gelfand (Northwestern University) for fruitful discussions on the project; Tony Hunter (Salk Institute for Biological Studies) for excellent scientific advice; Edgar Boczek and Florian H. Schopf (Technische Universität München) for providing access to the isotope laboratory and general support with kinase assays; and Nagarjuna Nagaraj (Max Planck Institute for Biochemistry Core Facility) for the mass spectrometry analysis. This work was supported by Deutsche Forschungsgemeinschaft Grant SFB863 and European Research Council Grant 335623 (to Z.Ö.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619473114/-/DCSupplemental.
References
- 1.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 2.Brown SS. Cooperation between microtubule- and actin-based motor proteins. Annu Rev Cell Dev Biol. 1999;15:63–80. doi: 10.1146/annurev.cellbio.15.1.63. [DOI] [PubMed] [Google Scholar]
- 3.Gross SP, et al. Interactions and regulation of molecular motors in Xenopus melanophores. J Cell Biol. 2002;156:855–865. doi: 10.1083/jcb.200105055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Natarajan VT, Ganju P, Ramkumar A, Grover R, Gokhale RS. Multifaceted pathways protect human skin from UV radiation. Nat Chem Biol. 2014;10:542–551. doi: 10.1038/nchembio.1548. [DOI] [PubMed] [Google Scholar]
- 5.Wei Q, Wu X, Hammer JA., 3rd The predominant defect in dilute melanocytes is in melanosome distribution and not cell shape, supporting a role for myosin V in melanosome transport. J Muscle Res Cell Motil. 1997;18:517–527. doi: 10.1023/a:1018659117569. [DOI] [PubMed] [Google Scholar]
- 6.Wu X, Bowers B, Rao K, Wei Q, Hammer JA., 3rd Visualization of melanosome dynamics within wild-type and dilute melanocytes suggests a paradigm for myosin V function In vivo. J Cell Biol. 1998;143:1899–1918. doi: 10.1083/jcb.143.7.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, Bowers B, Wei Q, Kocher B, Hammer JA., 3rd Myosin V associates with melanosomes in mouse melanocytes: Evidence that myosin V is an organelle motor. J Cell Sci. 1997;110:847–859. doi: 10.1242/jcs.110.7.847. [DOI] [PubMed] [Google Scholar]
- 8.Hume AN, Seabra MC. Melanosomes on the move: A model to understand organelle dynamics. Biochem Soc Trans. 2011;39:1191–1196. doi: 10.1042/BST0391191. [DOI] [PubMed] [Google Scholar]
- 9.Hume AN, Wilson MS, Ushakov DS, Ferenczi MA, Seabra MC. Semi-automated analysis of organelle movement and membrane content: Understanding rab-motor complex transport function. Traffic. 2011;12:1686–1701. doi: 10.1111/j.1600-0854.2011.01283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishida M, Ohbayashi N, Fukuda M. Rab1A regulates anterograde melanosome transport by recruiting kinesin-1 to melanosomes through interaction with SKIP. Sci Rep. 2015;5:8238. doi: 10.1038/srep08238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda M, Kuroda TS, Mikoshiba K. Slac2-a/melanophilin, the missing link between Rab27 and myosin Va: Implications of a tripartite protein complex for melanosome transport. J Biol Chem. 2002;277:12432–12436. doi: 10.1074/jbc.C200005200. [DOI] [PubMed] [Google Scholar]
- 12.Hume AN, et al. The leaden gene product is required with Rab27a to recruit myosin Va to melanosomes in melanocytes. Traffic. 2002;3:193–202. doi: 10.1034/j.1600-0854.2002.030305.x. [DOI] [PubMed] [Google Scholar]
- 13.Hume AN, et al. Rab27a regulates the peripheral distribution of melanosomes in melanocytes. J Cell Biol. 2001;152:795–808. doi: 10.1083/jcb.152.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matesic LE, et al. Mutations in Mlph, encoding a member of the Rab effector family, cause the melanosome transport defects observed in leaden mice. Proc Natl Acad Sci USA. 2001;98:10238–10243. doi: 10.1073/pnas.181336698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagashima K, et al. Melanophilin directly links Rab27a and myosin Va through its distinct coiled-coil regions. FEBS Lett. 2002;517:233–238. doi: 10.1016/s0014-5793(02)02634-0. [DOI] [PubMed] [Google Scholar]
- 16.Provance DW, James TL, Mercer JA. Melanophilin, the product of the leaden locus, is required for targeting of myosin-Va to melanosomes. Traffic. 2002;3:124–132. doi: 10.1034/j.1600-0854.2002.030205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, et al. Rab27a enables myosin Va-dependent melanosome capture by recruiting the myosin to the organelle. J Cell Sci. 2001;114:1091–1100. doi: 10.1242/jcs.114.6.1091. [DOI] [PubMed] [Google Scholar]
- 18.Wu XS, et al. Identification of an organelle receptor for myosin-Va. Nat Cell Biol. 2002;4:271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Sakamoto T, Zhang F, Sellers JR, Hammer JA., 3rd In vitro reconstitution of a transport complex containing Rab27a, melanophilin and myosin Va. FEBS Lett. 2006;580:5863–5868. doi: 10.1016/j.febslet.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 20.Geething NC, Spudich JA. Identification of a minimal myosin Va binding site within an intrinsically unstructured domain of melanophilin. J Biol Chem. 2007;282:21518–21528. doi: 10.1074/jbc.M701932200. [DOI] [PubMed] [Google Scholar]
- 21.Kuroda TS, Fukuda M, Ariga H, Mikoshiba K. The Slp homology domain of synaptotagmin-like proteins 1-4 and Slac2 functions as a novel Rab27A binding domain. J Biol Chem. 2002;277:9212–9218. doi: 10.1074/jbc.M112414200. [DOI] [PubMed] [Google Scholar]
- 22.Sckolnick M, Krementsova EB, Warshaw DM, Trybus KM. More than just a cargo adapter, melanophilin prolongs and slows processive runs of myosin Va. J Biol Chem. 2013;288:29313–29322. doi: 10.1074/jbc.M113.476929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukuda M, Kuroda TS. Slac2-c (synaptotagmin-like protein homologue lacking C2 domains-c), a novel linker protein that interacts with Rab27, myosin Va/VIIa, and actin. J Biol Chem. 2002;277:43096–43103. doi: 10.1074/jbc.M203862200. [DOI] [PubMed] [Google Scholar]
- 24.Kuroda TS, Ariga H, Fukuda M. The actin-binding domain of Slac2-a/melanophilin is required for melanosome distribution in melanocytes. Mol Cell Biol. 2003;23:5245–5255. doi: 10.1128/MCB.23.15.5245-5255.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu XS, Tsan GL, Hammer JA., 3rd Melanophilin and myosin Va track the microtubule plus end on EB1. J Cell Biol. 2005;171:201–207. doi: 10.1083/jcb.200503028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hume AN, Tarafder AK, Ramalho JS, Sviderskaya EV, Seabra MC. A coiled-coil domain of melanophilin is essential for Myosin Va recruitment and melanosome transport in melanocytes. Mol Biol Cell. 2006;17:4720–4735. doi: 10.1091/mbc.E06-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammer J, Wu X. 2012. Function of Rab27a in melanocytes and cytotoxic T lymphocytes. Rab GTPases and Membrane Trafficking, eds Li G, Segev N (Bentham eBooks, Sharjah, United Arab Emirates). pp 77–92.
- 28.Evans RD, et al. Myosin-Va and dynamic actin oppose microtubules to drive long-range organelle transport. Curr Biol. 2014;24:1743–1750. doi: 10.1016/j.cub.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercer JA, Seperack PK, Strobel MC, Copeland NG, Jenkins NA. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature. 1991;349:709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- 30.Koyama YI, Takeuchi T. Differential effect of cytochalasin B on the aggregation of melanosomes in cultured mouse melanoma cells. Anat Rec. 1980;196:449–459. doi: 10.1002/ar.1091960410. [DOI] [PubMed] [Google Scholar]
- 31.Provance DW, Jr, Wei M, Ipe V, Mercer JA. Cultured melanocytes from dilute mutant mice exhibit dendritic morphology and altered melanosome distribution. Proc Natl Acad Sci USA. 1996;93:14554–14558. doi: 10.1073/pnas.93.25.14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nascimento AA, Roland JT, Gelfand VI. Pigment cells: A model for the study of organelle transport. Annu Rev Cell Dev Biol. 2003;19:469–491. doi: 10.1146/annurev.cellbio.19.111401.092937. [DOI] [PubMed] [Google Scholar]
- 33.Tuma MC, Zill A, Le Bot N, Vernos I, Gelfand V. Heterotrimeric kinesin II is the microtubule motor protein responsible for pigment dispersion in Xenopus melanophores. J Cell Biol. 1998;143:1547–1558. doi: 10.1083/jcb.143.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers SL, Gelfand VI. Myosin cooperates with microtubule motors during organelle transport in melanophores. Curr Biol. 1998;8:161–164. doi: 10.1016/s0960-9822(98)70063-6. [DOI] [PubMed] [Google Scholar]
- 35.Rogers SL, Tint IS, Fanapour PC, Gelfand VI. Regulated bidirectional motility of melanophore pigment granules along microtubules in vitro. Proc Natl Acad Sci USA. 1997;94:3720–3725. doi: 10.1073/pnas.94.8.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aspengren S, Hedberg D, Sköld HN, Wallin M. New insights into melanosome transport in vertebrate pigment cells. Int Rev Cell Mol Biol. 2009;272:245–302. doi: 10.1016/S1937-6448(08)01606-7. [DOI] [PubMed] [Google Scholar]
- 37.Daniolos A, Lerner AB, Lerner MR. Action of light on frog pigment cells in culture. Pigment Cell Res. 1990;3:38–43. doi: 10.1111/j.1600-0749.1990.tb00260.x. [DOI] [PubMed] [Google Scholar]
- 38.Reilein AR, Tint IS, Peunova NI, Enikolopov GN, Gelfand VI. Regulation of organelle movement in melanophores by protein kinase A (PKA), protein kinase C (PKC), and protein phosphatase 2A (PP2A) J Cell Biol. 1998;142:803–813. doi: 10.1083/jcb.142.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlsson AM, Lerner MR, Unett D, Lundström I, Svensson SP. Melatonin-induced organelle movement in melanophores is coupled to tyrosine phosphorylation of a high molecular weight protein. Cell Signal. 2000;12:469–474. doi: 10.1016/s0898-6568(00)00089-9. [DOI] [PubMed] [Google Scholar]
- 40.Kashina AS, et al. Protein kinase A, which regulates intracellular transport, forms complexes with molecular motors on organelles. Curr Biol. 2004;14:1877–1881. doi: 10.1016/j.cub.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Park M, Serpinskaya AS, Papalopulu N, Gelfand VI. Rab32 regulates melanosome transport in Xenopus melanophores by protein kinase a recruitment. Curr Biol. 2007;17:2030–2034. doi: 10.1016/j.cub.2007.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuda M. Synaptotagmin-like protein (Slp) homology domain 1 of Slac2-a/melanophilin is a critical determinant of GTP-dependent specific binding to Rab27A. J Biol Chem. 2002;277:40118–40124. doi: 10.1074/jbc.M205765200. [DOI] [PubMed] [Google Scholar]
- 43.Wu X, Wang F, Rao K, Sellers JR, Hammer JA., 3rd Rab27a is an essential component of melanosome receptor for myosin Va. Mol Biol Cell. 2002;13:1735–1749. doi: 10.1091/mbc.01-12-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Au JS, Huang JD. A tissue-specific exon of myosin Va is responsible for selective cargo binding in melanocytes. Cell Motil Cytoskeleton. 2002;53:89–102. doi: 10.1002/cm.10061. [DOI] [PubMed] [Google Scholar]
- 45.Hornbeck PV, et al. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zanivan S, et al. Solid tumor proteome and phosphoproteome analysis by high resolution mass spectrometry. J Proteome Res. 2008;7:5314–5326. doi: 10.1021/pr800599n. [DOI] [PubMed] [Google Scholar]
- 47.Krementsov DN, Krementsova EB, Trybus KM. Myosin V: Regulation by calcium, calmodulin, and the tail domain. J Cell Biol. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li XD, Mabuchi K, Ikebe R, Ikebe M. Ca2+-induced activation of ATPase activity of myosin Va is accompanied with a large conformational change. Biochem Biophys Res Commun. 2004;315:538–545. doi: 10.1016/j.bbrc.2004.01.084. [DOI] [PubMed] [Google Scholar]
- 49.Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 50.Thirumurugan K, Sakamoto T, Hammer JA, 3rd, Sellers JR, Knight PJ. The cargo-binding domain regulates structure and activity of myosin 5. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li XD, Ikebe R, Ikebe M. Activation of myosin Va function by melanophilin, a specific docking partner of myosin Va. J Biol Chem. 2005;280:17815–17822. doi: 10.1074/jbc.M413295200. [DOI] [PubMed] [Google Scholar]
- 52.Yao LL, et al. Melanophilin stimulates myosin-5a motor function by allosterically inhibiting the interaction between the head and tail of myosin-5a. Sci Rep. 2015;5:10874. doi: 10.1038/srep10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toepfer C, Sellers JR. Use of Fluorescent Techniques to Study the In Vitro Movement of Myosins. In: Tosseland CP, Fili N, editors. Fluorescent Methods for Molecular Motors. Springer; Basel: 2014. pp. 193–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Armstrong JM, et al. Full-length myosin Va exhibits altered gating during processive movement on actin. Proc Natl Acad Sci USA. 2012;109:E218–E224. doi: 10.1073/pnas.1109709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 56.Ali MY, et al. Myosin Va maneuvers through actin intersections and diffuses along microtubules. Proc Natl Acad Sci USA. 2007;104:4332–4336. doi: 10.1073/pnas.0611471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slepchenko BM, Semenova I, Zaliapin I, Rodionov V. Switching of membrane organelles between cytoskeletal transport systems is determined by regulation of the microtubule-based transport. J Cell Biol. 2007;179:635–641. doi: 10.1083/jcb.200705146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodionov V, Yi J, Kashina A, Oladipo A, Gross SP. Switching between microtubule- and actin-based transport systems in melanophores is controlled by cAMP levels. Curr Biol. 2003;13:1837–1847. doi: 10.1016/j.cub.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 59.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Otsu N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern. 1979;9:62–66. [Google Scholar]
- 61.Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 62.Mandelkow EM, Herrmann M, Rühl U. Tubulin domains probed by limited proteolysis and subunit-specific antibodies. J Mol Biol. 1985;185:311–327. doi: 10.1016/0022-2836(85)90406-1. [DOI] [PubMed] [Google Scholar]
- 63.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 64.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;(41):95–98. [Google Scholar]
- 65.Fukuda M, Kuroda TS. Missense mutations in the globular tail of myosin-Va in dilute mice partially impair binding of Slac2-a/melanophilin. J Cell Sci. 2004;117:583–591. doi: 10.1242/jcs.00891. [DOI] [PubMed] [Google Scholar]
- 66.Strom M, Hume AN, Tarafder AK, Barkagianni E, Seabra MC. A family of Rab27-binding proteins. Melanophilin links Rab27a and myosin Va function in melanosome transport. J Biol Chem. 2002;277:25423–25430. doi: 10.1074/jbc.M202574200. [DOI] [PubMed] [Google Scholar]
- 67.Wang F, et al. Effect of ADP and ionic strength on the kinetic and motile properties of recombinant mouse myosin V. J Biol Chem. 2000;275:4329–4335. doi: 10.1074/jbc.275.6.4329. [DOI] [PubMed] [Google Scholar]
- 68.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 69.Nagaraj N, et al. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol Cell Proteomics. 2012;11:M111.013722. doi: 10.1074/mcp.M111.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 71.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.