Abstract
Panax ginseng C.A. Meyer is a traditional medicinal herb that produces bioactive compounds such as ginsenosides. Here, we investigated the diversity of ginsenosides and related genes among five genetically fixed inbred ginseng cultivars (Chunpoong [CP], Cheongsun [CS], Gopoong [GO], Sunhyang [SH], and Sunun [SU]). To focus on the genetic diversity related to ginsenoside biosynthesis, we utilized in vitro cultured adventitious roots from the five cultivars grown under controlled environmental conditions. PCA loading plots based on secondary metabolite composition classified the five cultivars into three groups. We selected three cultivars (CS, SH, and SU) to represent the three groups and conducted further transcriptome and gas chromatography-mass spectrometry analyses to identify genes and intermediates corresponding to the variation in ginsenosides among cultivars. We quantified ginsenoside contents from the three cultivars. SH had more than 12 times the total ginsenoside content of CS, with especially large differences in the levels of panaxadiol-type ginsenosides. The expression levels of genes encoding squalene epoxidase (SQE) and dammarenediol synthase (DDS) were also significantly lower in CS than SH and SU, which is consistent with the low levels of ginsenoside produced in this cultivar. Methyl jasmonate (MeJA) treatment increased the levels of panaxadiol-type ginsenosides up to 4-, 13-, and 31-fold in SH, SU, and CS, respectively. MeJA treatment also greatly increased the quantity of major intermediates and the expression of the underlying genes in the ginsenoside biosynthesis pathway; these intermediates included squalene, 2,3-oxidosqualene, and dammarenediol II, especially in CS, which had the lowest ginsenoside content under normal culture conditions. We conclude that SQE and DDS are the most important genetic factors for ginsenoside biosynthesis with diversity among ginseng cultivars.
Keywords: Panax ginseng, cultivars, ginsenoside biosynthetic pathway, transcriptome, metabolome
Introduction
Panax ginseng is a medicinal plant that has long been used as a tonic agent throughout Asia, including Korea and China (Jia et al., 2009). Ginseng is rich in useful metabolites, such as polyacetylenes, polysaccharides, phenolic compounds, and terpenes. Triterpene saponins, also known as ginsenosides, are the major compounds in ginseng showing anti-oxidant, anti-inflammatory, and anti-cancer activity (Kim, 2012; Zhang et al., 2013). Dozens of ginseng accessions have been bred through pure line selection from three local landrace populations (Jakyung, Chungkyung, and Hwangsook) and registered as cultivated varieties (cultivars) in Korea (Kim et al., 2012). These cultivars exhibit different morphological and physiological characters (Kim et al., 2012; Lee et al., 2015), as well as different metabolite accumulation patterns (Ahn et al., 2008; Kim et al., 2009; Lee et al., 2011; Cho et al., 2012).
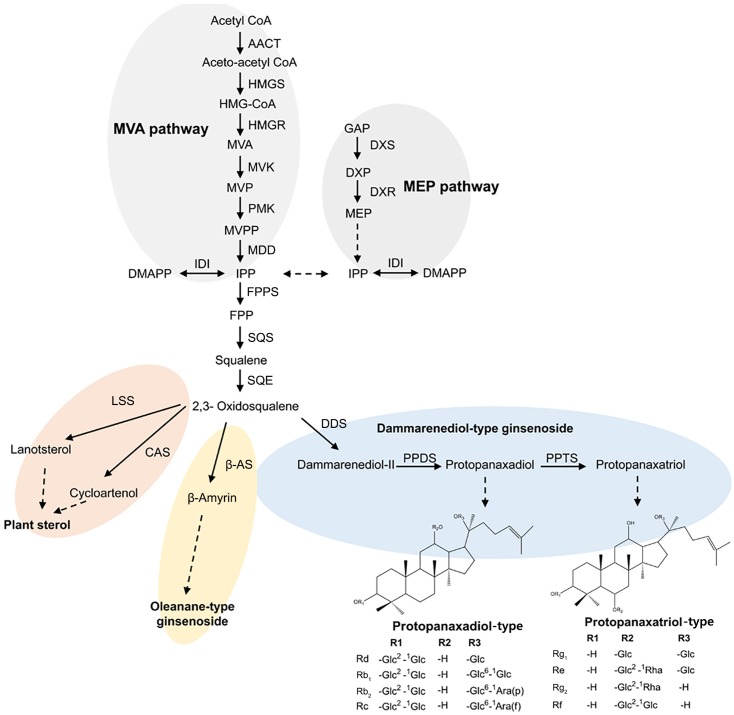
In P. ginseng, ginsenosides are mainly of the dammarane type, which are in turn classified into two types according to the number of hydroxyl groups: protopanaxadiol (PPD)-type ginsenosides with hydroxyl groups at positions C3, C12, and C20 and protopanaxatriol (PPT)-type ginsenosides with hydroxyl groups at positions C3, C6, C12, and C20 (Jung et al., 2014; Kim et al., 2014). A representative biosynthetic pathway for dammarane-type ginsenosides in ginseng is shown in Figure 1. Isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) produced via the mevalonate (MVA) pathway undergo condensation and cyclization reactions catalyzed by farnesyl diphosphate synthase (FPPS), squalene synthase (SQS), and squalene epoxidase (SQE) (Kirby and Keasling, 2009; Thimmappa et al., 2014). Through this process, 2,3-oxidosqualene is generated and cyclized by dammarenediol II synthase (DDS) to produce dammarenediol II (Tansakul et al., 2006). Dammarenediol II is then hydroxylated by protopanaxadiol synthase (PgPPDS, CYP716A47) and converted to PPD (Han et al., 2011), which is further hydroxylated by protopanaxatriol synthase (PgPPTS, CYP716A53v2) to produce PPT (Han et al., 2012). Finally, PPD and PPT are glycosylated to produce various types of ginsenosides (Kim et al., 2014).
FIGURE 1.
Schematic representation of the ginsenoside biosynthetic pathway in Panax ginseng; AACT, acetoacetyl CoA transferase; CAS, cycloartenol synthase; DMAPP, dimethylallyl diphosphate; DDS, dammarenediol synthase; DXP, 1-deoxy-D-xylulose 5-phosphate; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; FPP, farnesyl diphosphate; FPPS, farnesyl diphosphate synthase; GAP, glyceraldehyde 3-phosphate; HMGR, HMG-CoA reductase; HMGS, HMG-CoA synthase; IDI, isopentenyl-diphosphate delta-isomerase; IPP, isopentenyl diphosphate; LSS, lanosterol synthase; MEP, 2-C-methyl-D-erythritol 4-phosphate; MVK, mevalonate kinase; MVA, mevalonic acid; MDD, mevalonate diphosphate decarboxylase; MVP, mevalonic acid 5-phosphate; MVPP, 5-diphosphomevalonate; PMK, phosphomevalonate kinase; PPDS, protopanaxadiol synthase; PPTS, protopanaxatriol synthase; SQE, squalene epoxidase; SQS, squalene synthase; β-AS, β-amyrin synthase.
Candidate genes responsible for ginsenoside biosynthesis in the Panax genus have been identified in P. ginseng (Han et al., 2011, 2012; Jung et al., 2014; Kim et al., 2014; Jayakodi et al., 2015; Wang P. et al., 2015; Wei et al., 2015), P. notoginseng (Liu et al., 2015; Rai et al., 2016), and P. vietnamensis (Zhang et al., 2015). Transcriptome analysis of P. notoginseng identified several candidate unigenes encoding cytochrome P450 and glycosyltransferase and revealed that these genes were conserved across all Panax species (Rai et al., 2016). However, the regulatory mechanism for the ginsenoside biosynthesis pathway still remains unknown. Integrated omics analysis of the transcriptome and metabolome is an efficient tool for obtaining a precise understanding of biosynthetic pathways. This technique allows identification of novel functional genes and characterization of the regulatory mechanism and key factors of biosynthetic pathways (Saito et al., 2008; Mounet et al., 2009). The regulatory genes in the tryptophan (Dubouzet et al., 2007), flavonoid (Cho et al., 2016), flavanol (Yonekura-Sakakibara et al., 2008), and glucosinolate biosynthetic pathways (Hirai et al., 2004, 2007) have been explored using integrated analysis, and potential target genes have been identified.
In this study, we applied an integrated metabolomics and transcriptomics approach to explore the ginsenoside biosynthetic pathway using adventitious roots from five ginseng cultivars. Specifically, we compared the expression levels of genes involved in ginsenoside biosynthesis, the accumulation patterns of intermediates, and the composition and quantity of ginsenosides in adventitious roots with or without methyl jasmonate (MeJA) treatment. This analysis allowed us to uncover the dynamic regulatory mechanism for the ginsenoside biosynthetic pathway controlled by genetic factors and MeJA treatment.
Materials and Methods
Chemicals
Nine ginsenosides [Rg1, Re, Rf, Rg2(20S), Rg2(20R), Rb1, Rc, Rb2, and Rd], squalene, 2,3-oxidosqualene, mevalonolactone, and farnesol were purchased from Sigma–Aldrich (St. Louis, MO, United States). Dammarenediol II was kindly provided by Dr. Yong-Eui Choi (Kangwon National University, South Korea). Chemicals for tissue culture were purchased from Duchefa (Haarlem, The Netherlands), and the remaining chemicals were purchased from Sigma–Aldrich (St. Louis, MO, United States).
Plant Materials
Adventitious root formation was induced from five P. ginseng cultivars (Chunpoong [CP], Cheongsun [CS], Gopoong [GO], Sunhyang [SH], and Sunun [SU]), and the roots were cultured as previously described (Jayakodi et al., 2014). Adventitious roots (42 days old) were treated with 200 μM MeJA. For metabolites profiling, the adventitious roots were harvested after 1 week of MeJA treatment, with five biological replicates for both non-treated and MeJA-treated adventitious roots. For quantification of gene expression using RT-PCR, adventitious roots were collected at 0, 12, 24, and 48 h after MeJA treatment, with three biological replicates. For transcriptome analysis, adventitious roots were harvested from plants without MeJA treatment, with three biological replicates. The adventitious roots were immediately stored at -80°C until use.
Metabolome Analysis
Sample Preparation
For UPLC-ESI/MS analysis, 10 mg of lyophilized adventitious root tissue was sonicated with 1 mL of 70% methanol for 30 min at room temperature. The extracts were centrifuged at 13,000 ×g for 5 min, and the supernatants were filtered through a 0.2 μm PTFE syringe filter (Toyo Roshi Kaisha, Japan). For LC-UV/DAD analysis, 10 mg of lyophilized adventitious root tissue was sonicated three times using 2 mL of 70% methanol for 30 min at room temperature. The extracts were dried using N2 gas and resuspended in 200 μL of 80% methanol.
UPLC-ESI/MS Analysis
Metabolite profiling was carried out by ultra-performance liquid chromatography (ACQUITY UPLC system, Waters, United States) coupled with Q-TOF (micrOTOF-QII, Bruker Daltonics, Germany) using an ACQUITY BEH C18 column (2.1 mm × 100 mm, 1.7 μm, Waters, United States). The analysis was performed using a binary mobile phase consisting of A: water + 0.1% formic acid and B: acetonitrile + 0.1% formic acid. Separation was achieved under the following gradient conditions: pre-running with 100% A for 10 min, 100% A at 0 min, 65.5% A at 10 min, 52.5% A at 25 min, 20% A at 35 min, 0% A at 40 min and holding for 10 min. The flow rate was 0.2 mL/min, the column temperature was 40°C, and the injection volume was 5 μL.
The micrOTOF-QII was run in negative mode, and the source conditions were as follows: capillary -4.5 kV, nebulizer pressure at 1.2 bar, dry gas flow rate of 8 L/min, and dry gas temperature at 200°C. The ion transfer and collision stages were set as follows: funnel 1 RF 400 Vpp, funnel 2 RF 400 Vpp, hexapole RF 400 Vpp, quadrupole ion energy 15 eV, collision energy 10 eV, collision RF 400 Vpp, transfer time 100 μs, and pre-pulse storage 5 μs. High-purity nitrogen was used as the nebulizer and dry gas, and argon was used as the collision gas. Lithium formate was used as an internal standard, with a scan range from 50 to 1500 m/z.
LC-UV/DAD Analysis
The nine ginsenosides [Rg1, Re, Rf, Rg2(20S), Rg2(20R), Rb1, Rc, Rb2, and Rd] were quantified using an Agilent 1260 infinity HPLC system (Agilent, United States) with a discovery C18 column (2.1 mm × 100 mm, 5 μm, Sigma–Aldrich, United States). The mobile phase was composed of A: water + 0.1% formic acid and B: acetonitrile + 0.1% formic acid, as used for UPLC-ESI/MS. The flow rate was 0.3 mL/min. The gradient conditions were as follows: 80% A held for 20 min, followed by 70% A for 35 min, 55% A for 45 min, 10% A for 55 min, holding for 5 min, and a post-run with 80% A for 15 min. The UV detector was set at 203 nm, and the injection volume was 5 μL. For quantification, calibration curves of each ginsenoside were obtained, and intra- and interday precision and accuracy parameters were determined for validation (Supplementary Tables S1, S2).
GC-MS Analysis
The squalene, 2,3-oxidosqualene, farnesol, and dammarenediol II contents in CS, SH, and SU were analyzed by GC-MS as described previously (Kim et al., 2011; Lee et al., 2012). Briefly, KOH saponification-based extracts were analyzed using GCMS-QP2010 (Shimadzu, Japan) with a DB-5MS column (30 m × 0.25 mm, 0.25 μm, Agilent technologies, Germany). The carrier gas was helium, the flow rate was 1.0 mL/min, and the injection volume was 1 μL, in split mode (1:2). The mass detector was in electron impact mode of 70 eV, and the ion source temperature was 200°C. The mass detection range was 40–600 m/z. A single peak of each compound was identified using selected single ion monitoring mode and the retention times of authentic compounds. The areas of the compounds were normalized to that of the internal standard.
Data Processing and Multivariate Analysis
Raw data generated from UPLC-ESI/MS were processed using MZmine software version 2.101. The data were subjected to peak deconvolution using a Savitzky-Golay filter and aligned through the RANdom Sample Consensus algorithm. The relative abundance of each peak was centered and scaled to unit variance, with base weight set at 1/standard deviation. The filtered peak lists were imported into SIMCA-P+ (version 12.0, Umetrics, Sweden), and principal component analysis (PCA) and partial least square-discriminant analysis (PLS-DA) were performed. In the PLS-DA model, metabolites with variable influence on the projection (VIP) value above 1 and P-value under 0.05 were selected for the VIP lists.
RNA Isolation and Transcriptome Sequencing Using the Illumina Platform
Total RNA was extracted from adventitious roots of cultivar SU using an RNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacturer’s protocol. The quality and quantity of extracted RNA was checked using an ND-1000 (NanoDrop Technologies Inc., United States) and formaldehyde-agarose gel electrophoresis. An RNA-Seq library with a 300-bp insertion size was then constructed and sequenced on the Illumina sequencing platform (NextSeq 500, LabGenomics Co., South Korea). The three replicated RNA-Seq reads were deposited in the NCBI Sequence Read Archive2 (SRA) under accession numbers SRR2134236, SRR2134277, and SRR2134367. RNA-Seq data for adventitious roots from SH and CS were obtained from a previous study (Lee Y.S. et al., 2016).
Digital Expression Analysis of Genes Related to Ginsenoside Biosynthesis
For expression profiling among cultivars, candidate genes involved in ginsenoside biosynthesis were selected based on a previous report (Jayakodi et al., 2015). The expression levels of genes related to the ginsenoside biosynthetic pathway were determined based on fragments per kilobase of exon per million fragments (FPKM) values from RNA-seq data for CS, SH, and SU using RSEM (Li and Dewey, 2011). Genes showing significantly different expression among the three cultivars were identified by analysis of variance (ANOVA, P-value < 0.05) using R (version 3.1.03) and the R packages mvtnorm (Genz et al., 2008) and multcomp (Hothorn et al., 2008). Mean FPKM values of the genes were converted to log2 values, which were used for visualization using the heatmap package in R. An integrative analysis between expression of the unigenes and the relative quantity of 21 metabolites was conducted with a bidirectional multivariate regression method based on orthogonal projection to latent structures (OPLS) (Bylesjö et al., 2007). The regression method of gene expression profiles (mean-centered, x variables) and metabolite profiles (mean-centered and Pareto-scaled, y variables) was analyzed using O2PLS model implemented in SIMCA tool (version 14.1, trial, Umetrics, Sweden).
RT-PCR Analysis
For RT-PCR analysis, cDNA was produced from total RNA of CS, SH, and SU using a SMART cDNA Synthesis kit (Clontech, Japan) according to the manufacturer’s protocol. The synthesized cDNA was diluted 1/10 and used as template for RT-PCR. The RT-PCR was performed on a LightCycler 480 (Roche, Germany) under the following thermal cycling conditions: 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 58°C for 10 s, and 72°C for 10 s. Primer sets for each gene were designed using Primer3 (Rozen and Skaletsky, 1999), and the specificity of the primer sets was confirmed by sequencing PCR amplicons on an ABI 3730 XL DNA Analyzer (Applied Biosystems, United States). All primer sets used in this study are listed in Supplementary Table S3.
Results
Secondary Metabolite Profiles in Adventitious Roots of Five Ginseng Cultivars
We performed secondary metabolite profiling in adventitious root tissue from five ginseng cultivars (CP, CS, GO, SH, and SU) using UPLC-ESI/MS to investigate metabolic diversity among ginseng cultivars (Figure 2). A previously reported method using a reverse-phase column (Lee G.J. et al., 2016; Wang et al., 2016) was applied to selectively separate secondary metabolites including ginsenosides from adventitious root extracts. A total of 21 metabolites were identified via comparisons with authentic standards or data found in previous reports (Xie et al., 2008; Qi et al., 2012; MacCrehan and White, 2013). Among these, 16 metabolites with VIP values above 1 and P-value below 0.05 were characterized as biomarkers able to discriminate among adventitious roots from different ginseng cultivars. These biomarkers were PPD/PPT-type ginsenosides, except for four metabolites identified as chikusetsusaponin IVa, gypenoside XVII, calenduloside E, and an isomer of ginsenoside Ro (Table 1). These results indicate that PPD/PPT ginsenosides are the main contributors to the metabolic differences in adventitious roots among these cultivars.
FIGURE 2.
Representative chromatograph of adventitious roots from five ginseng cultivars (CP, CS, GO, SH, and SU) analyzed by UPLC-ESI/MS in negative mode; CP, Chunpoong; CS, Cheongsun; GO, Gopoong; SH, Sunhyang; and SU, Sunun. The identity of each peak is shown in Table 1.
Table 1.
Metabolites in adventitious roots from five ginseng cultivars identified through UPLC-ESI/MS in negative mode (CP, Chunpoong; CS, Cheongsun; GO, Gopong; SH, Sunhyang; and SU, Sunun).
| No. | RT (min) | m/z |
Formula | Error (mDa) | Identified compound | VIP value | P-value | Reference | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Adduct ion | Exact | Measured | ||||||||
| 1 | 6.8 | [M-HCOO]- | 845.49 | 845.48 | C42H72O14 | -6 | Ginsenoside Rg1a | 1.47 | 2.73E-30 | STDb |
| 2 | 6.9 | [M-HCOO]- | 991.55 | 991.54 | C48H82O18 | -5.7 | Ginsenoside Rea | 1.48 | 1.72E-30 | STDb |
| 3 | 7.7 | [M-H]- | 885.48 | 885.48 | C45H74O17 | -2.9 | Malonyl ginsenoside Rg1 | 0.75 | 4.86E-11 | MacCrehan and White, 2013 |
| 4 | 15.8 | [M-H]- | 799.48 | 799.48 | C42H72O14 | -0.7 | Ginsenoside Rfa | 1.29 | 2.78E-13 | STDb |
| 5 | 17.3 | [M-H]- | 1239.64 | 1239.64 | C59H100O27 | 1.3 | Notoginsenoside Fa | 0.83 | 1.98E-25 | Xie et al., 2008 |
| 6 | 17.7 | [M-H]- | 769.47 | 769.47 | C41H70O13 | 0.9 | Notoginsenoside R2a | 1.35 | 1.63E-26 | MacCrehan and White, 2013 |
| 7 | 20.1 | [M-H]- | 783.49 | 783.49 | C42H72O13 | 1.8 | Ginsenoside Rg2 (20S)/(20R) a | 1.45 | 6.58E-21 | STDb |
| 8 | 21.9 | [M-H]- | 1107.59 | 1107.59 | C54H92O23 | -1.1 | Ginsenoside Rb1 a | 1.52 | 2.91E-18 | STDb |
| 9 | 23.3 | [M-H]- | 1193.59 | 1193.59 | C57H94O26 | -4.3 | Malonyl ginsenoside Rb1a | 1.55 | 7.30E-24 | MacCrehan and White, 2013 |
| 10 | 23.9 | [M-H]- | 1077.58 | 1077.58 | C53H90O22 | -2.9 | Ginsenoside Rca | 1.2 | 7.43E-28 | STDb |
| 11 | 25.2 | [M-H]- | 955.49 | 955.49 | C48H76O19 | -2.2 | Ginsenoside Ro isomer | 0.79 | 4.08E-17 | Qi et al., 2012 |
| 12 | 25.4 | [M-H]- | 1163.58 | 1163.58 | C56H92O25 | -5.2 | Malonyl ginsenoside Rb2/Rb3/Rca | 1.39 | 1.45E-25 | Qi et al., 2012 |
| 13 | 26.3 | [M-H]- | 1077.58 | 1077.58 | C53H90O22 | -5.2 | Ginsenoside Rb2a | 1.15 | 8.24E-27 | STDb |
| 14 | 27.3 | [M-H]- | 925.48 | 925.47 | C47H74O18 | -0.6 | Pseudoginsenoside RT1 | 0.83 | 2.77E-11 | Qi et al., 2012 |
| 15 | 27.7 | [M-H]- | 1163.58 | 1163.58 | C56H92O25 | -4.2 | Malonyl ginsenoside Rb2/Rb3/Rca | 1.28 | 1.29E-27 | Qi et al., 2012 |
| 16 | 29.8 | [M-H]- | 793.44 | 793.44 | C42H66O14 | -1.4 | Chikusetsusaponin IVaa | 1.09 | 6.35E-17 | Qi et al., 2012 |
| 17 | 31.3 | [M-H]- | 1163.58 | 1163.59 | C56H92O25 | -0.5 | Malonyl ginsenoside Rb2/Rb3/Rca | 1.31 | 1.02E-21 | Qi et al., 2012 |
| 18 | 31.4 | [M-HCOO]- | 991.55 | 991.55 | C48H82O18 | 5.5 | Ginsenoside Rda | 1.2 | 2.59E-14 | STDb |
| 19 | 31.9 | [M-H]- | 1031.54 | 1031.55 | C51H84O21 | 3.1 | Malonyl ginsenoside Rd isomera | 1.12 | 1.27E-19 | Qi et al., 2012 |
| 20 | 32.8 | [M-HCOO]- | 991.55 | 991.56 | C48H82O18 | 3.1 | Gypenoside XVII | 0.97 | 1.19E-28 | Qi et al., 2012 |
| 21 | 39.2 | [M-H]- | 631.38 | 631.38 | C36H56O9 | -4.2 | Calenduloside Ea | 1.21 | 3.48E-13 | Qi et al., 2012 |
aCharacterized biomarker metabolites (VIP value > 1.0 and P-value < 0.05).
bIdentified metabolites using authentic compounds.
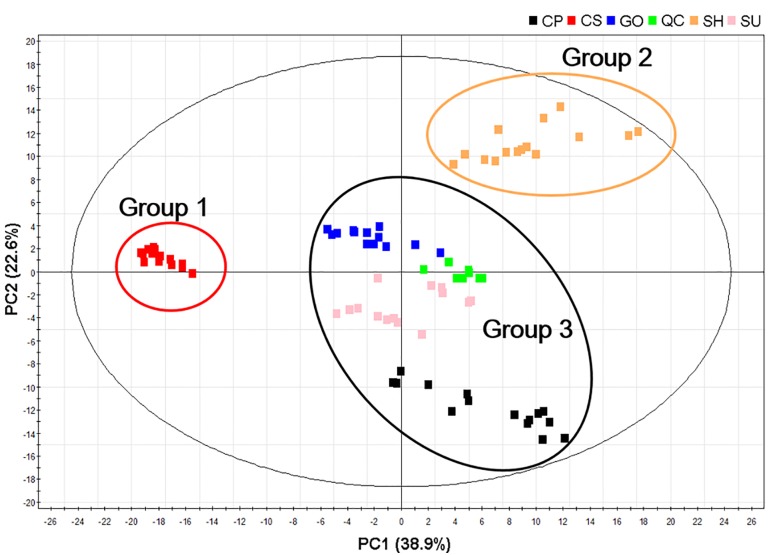
We subjected the UPLC-ESI/MS data to multivariate analysis to visualize the metabolic differences among cultivars. The five ginseng cultivars were divided into three groups according to the PCA loading plots (R2X: 0.809 and d Q2: 0.825): Group 1: CS; Group 2: SH; and Group 3: SU, CP, and GO. This grouping indicated that the accumulation pattern of metabolites varied among cultivars, but that some cultivars shared similar metabolic profiles (Figure 3).
FIGURE 3.
Principal component analysis (PCA) score plots of adventitious roots from five ginseng cultivars (CP, CS, GO, SH, and SU) analyzed by UPLC-ESI/MS in negative mode; CP, Chunpoong; CS, Cheongsun; GO, Gopoong; SH, Sunhyang; and SU, Sunun.
UPLC-ESI/MS analysis also revealed that the relative abundance of representative ginsenosides in adventitious roots differed among the five ginseng cultivars (Figure 4). SH roots were rich in PPD-type ginsenosides such as Rb1, Rb2, Rc, and Rd compared to the other cultivars, although the amount of PPT-type ginsenoside in SH was comparable to that in other cultivars except CS. CS showed the lowest amounts of both PPD and PPT ginsenosides, although it was relatively rich in Rg1. The abundances of PPD-type and PPT-type ginsenosides were not significantly different among cultivars CP, GO, and SU.
FIGURE 4.
Relative abundances of eight ginsenosides in adventitious roots from five ginseng cultivars (CP, CS, GO, SH, and SU) analyzed by UPLC-ESI/MS in negative mode; CP, Chunpoong; CS, Cheongsun; GO, Gopoong; SH, Sunhyang; and SU, Sunun.
Transcriptome Analysis of Adventitious Roots from CS, SH, and SU
UPLC-ESI/MS analysis revealed differences in the accumulation patterns of metabolites among ginseng cultivars, suggesting that the ginsenoside biosynthetic pathway might be differentially regulated in these cultivars (Table 1 and Figure 3). We selected one cultivar to represent each group in the PCA loading plots (CS, SH, and SU; Figure 3) and compared these three cultivars via transcriptome analysis (Table 2). We obtained transcriptome data for adventitious roots from CS and SH from a previous study (Lee Y.S. et al., 2016) and generated transcriptome data for SU in the current study. An average of 15 million (M) reads from three replicates were produced from SU adventitious roots. After trimming, an average of 14 M reads was subsequently used for expression profiling of genes involved in the ginsenoside biosynthetic pathway.
Table 2.
Summary of transcriptome data generated from adventitious roots of Panax ginseng cultivars CS, SH, and SU (CS, Cheongsun; SH, Sunhyang; and SU, Sunun).
| Adventitious root sample (SRA accession number) | Raw data |
QC filtered data |
||
|---|---|---|---|---|
| Total no. of reads | Length (bp) | Total no. of reads | Length (bp) | |
| CS, replicate 1a (SRR2132332) | 16,731,664 | 2,321,621,040 | 15,291,398 | 2,115,447,628 |
| CS, replicate 2a (SRR1688723) | 14,306,820 | 1,977,938,763 | 12,831,592 | 1,766,225,247 |
| CS, replicate 3a (SRR2132333) | 14,801,822 | 2,056,937,756 | 13,407,302 | 1,857,154,376 |
| SH, replicate 1a (SRR1688724) | 17,526,160 | 2,446,652,196 | 15,710,392 | 2,185,318,113 |
| SH, replicate 2a (SRR2132380) | 16,947,000 | 2,354,285,233 | 15,409,700 | 2,134,138,490 |
| SH, replicate 3a (SRR2132382) | 16,426,236 | 2,281,803,413 | 14,810,282 | 2,050,279,385 |
| SU, replicate 1 (SRR2134236) | 15,542,974 | 2,155,072,375 | 14,066,330 | 1,943,820,582 |
| SU, replicate 2 (SRR2134277) | 15,770,684 | 2,187,522,302 | 14,314,738 | 1,979,136,578 |
| SU, replicate 3 (SRR2134367) | 15,461,854 | 2,160,894,730 | 13,846,510 | 1,928,707,399 |
| Total | 186,981,726 | 22,553,682,825 | 172,796,402 | 20,550,270,711 |
aTranscriptomes of CS and SH were obtained from a previous report (Lee Y.S. et al., 2016).
Comparison of Ginsenoside Biosynthesis Pathways in Adventitious Roots from CS, SH, and SU
We previously identified 23 candidate unigenes related to the MVA and triterpene biosynthetic pathways from the P. ginseng root transcriptome database (Jayakodi et al., 2015). To identify genes associated with the differences in ginsenoside contents among cultivars, we analyzed the expression levels of the 23 unigenes based on FPKM values and compared the expression patterns among cultivars (Table 3).
Table 3.
FPKM values of unigenes involved in the ginsenoside biosynthetic pathway in adventitious roots of CS, SH, and SU (CS, Cheongsun; SH, Sunhyang; and SU, Sunun).
| Pathway | Gene name | Candidate unigene | FPKM |
||
|---|---|---|---|---|---|
| CS | SH | SU | |||
| Mevalonate pathway | AACT | Pg_Root114975_c0_seq10 | 15.87 ± 10.88 | 25.5 ± 9.11 | 19.24 ± 7.59 |
| HMGS | Pg_Root104689_c0_seq1 | 48.13 ± 13.29 | 62.2 ± 7.43 | 53.29 ± 9.23 | |
| HMGR | Pg_Root112393_c0_seq27 | 4.57 ± 6.46 | 0 | 0 | |
| HMGR | Pg_Root123699_c0_seq7 | 0.11 ± 0.16 | 0.04 ± 0.06 | 0.17 ± 0.13 | |
| HMGR | Pg_Root123699_c0_seq4 | 4.72 ± 1.44 | 10.55 ± 7.81 | 13.63 ± 0.55 | |
| MVK | Pg_Root113545_c0_seq1 | 15.18 ± 5.56 | 6.47 ± 0.91 | 14.15 ± 3.7 | |
| PMK | Pg_Root127818_c1_seq23 | 27.01 ± 4.2 | 15.8 ± 5.18 | 21.67 ± 7.77 | |
| PMK | Pg_Root127818_c1_seq9 | 3.57 ± 5.05 | 9.79 ± 7.35 | 9.2 ± 13.02 | |
| PMK | Pg_Root101855_c0_seq2 | 0 | 3.15 ± 2.24 | 6.91 ± 6.34 | |
| PMK | Pg_Root101855_c0_seq9 | 2.57 ± 3.64 | 0 | 0.87 ± 1.23 | |
| MDD | Pg_Root126585_c2_seq1 | 21.89 ± 6.6 | 27.93 ± 5.51 | 32.42 ± 12.77 | |
| IDI | Pg_Root110920_c0_seq2 | 74.68 ± 9.86 | 81.49 ± 8.81 | 52.35 ± 18.11 | |
| Triterpene biosynthesis pathway | FPPS | Pg_Root127352_c0_seq7 | 12.96 ± 2.33 | 4.92 ± 4.7 | 12.15 ± 4.85 |
| FPPS | Pg_Root127352_c0_seq34 | 0 | 8.66 ± 12.24 | 0 | |
| FPPS∗ | Pg_Root127352_c0_seq41 | 18.02 ± 3.33ˆa | 10.33 ± 7.4ˆa | 27.13 ± 3.33ˆb | |
| SQS | Pg_Root122824_c0_seq14 | 30.01 ± 5.78 | 37.73 ± 4.06 | 26.69 ± 9.2 | |
| SQE | Pg_Root125745_c1_seq5 | 5.45 ± 2.91 | 4.13 ± 1.47 | 5.97 ± 1.94 | |
| SQE∗ | Pg_Root104873_c0_seq1 | 11.24 ± 7.8ˆa | 39.42 ± 12.87ˆc | 15.07 ± 5.53ˆab | |
| DDS∗ | Pg_Root126438_c1_seq1 | 12.5 ± 2.79ˆa | 82.34 ± 27.84ˆb | 99 ± 4.07ˆb | |
| β-AS | Pg_Root120424_c0_seq1 | 0.63 ± 0.89 | 2.41 ± 1.15 | 2.83 ± 2.57 | |
| β-AS∗ | Pg_Root120424_c0_seq15 | 0.66 ± 0.93ˆa | 2.71 ± 1.34ˆa | 4.04 ± 1.74ˆb | |
| PPDS∗ | Pg_Root123943_c0_seq1 | 40.56 ± 15.36ˆa | 59.18 ± 15.78ˆa | 84.42 ± 8.82ˆb | |
| PPTS | Pg_Root91292_c0_seq1 | 101.35 ± 48.73 | 72.3 ± 10.28 | 117.19 ± 10.06 | |
∗Significantly differentially expressed unigenes among adventitious roots from CS, SH, and SU are marked with an asterisk (P < 0.05), and significant differences are represented by different letters. CS, Cheongsun; SH, Sunhyang; SU, Sunun. The full names of genes are shown in Figure 1.
Among the seven candidate genes in the MVA pathway, unigene Pg_Root110920_c0_seq2 (encoding IDI) showed the highest expression while unigene Pg_Root123699_c0_seq7 (encoding HMGR) showed the lowest expression in adventitious roots from CS, SH, and SU. The expression levels of most unigenes in MVA biosynthesis were not significantly different among cultivars (Table 3). By contrast, the expression levels of unigenes involved in the triterpene biosynthesis pathway differed significantly (Table 3). Unigene Pg_Root127352_c0_seq41 (encoding FPPS) was highly expressed in SU. Unigene Pg_Root104873_c0_seq1 (encoding SQE) and unigene Pg_Root126438_c1_seq1 (encoding DDS) were expressed at extremely low levels in CS compared to SH and SU. The expression level of each gene was correlated with the relative amounts of intermediates and total ginsenosides in adventitious roots of the three cultivars (Figure 4). Unigene Pg_Root123943_c0_seq1 (encoding PPDS) was the most highly expressed in SU whereas unigene Pg_Root91292_c0_seq1 (encoding PPTS) was highly expressed in all cultivars (Table 3). Unigene Pg_Root120424_c0_seq15 (encoding β-amyrin synthase; β-AS) showed different expression among cultivars but its expression level was low compared to that of the DDS unigene (Pg_Root126438_c1_seq1).
The integration of gene expression with metabolite data by O2PLS model resulted in co-variation of 62% of transcripts with 80% of the metabolites. A pairwise Pearson correlations for those transcripts and metabolites showed a strong correlation (r > 0.7) with more than seven metabolites between unigene Pg_Root104873_c0_seq1 (encoding SQE) and Pg_Root126438_c1_seq1 (encoding DDS) (Supplementary Figure S1).
Ginsenoside Contents Are Altered after MeJA Treatment in Adventitious Roots from CS, SH, and SU
Methyl jasmonate treatment upregulates ginsenoside biosynthesis in P. ginseng (Yendo et al., 2010). We investigated the effects of MeJA treatment on ginsenoside biosynthesis in different ginseng cultivars to understand the underlying genetic factors (Table 4). LC-UV-DAD analysis revealed that the three representative ginseng cultivars had different responses to MeJA treatment. In all three cultivars, the contents of PPD-type ginsenosides under MeJA treatment were higher than those of PPT-type ginsenosides. After 7 days of MeJA treatment, PPD-type ginsenoside levels increased 4.14- (SH), 13.15- (SU), and 31.44- (CS) fold, whereas PPT-type ginsenoside levels increased 1.34- (SH), 4.07- (SU), and 4.00- (CS) fold (Table 4).
Table 4.
Ginsenoside contents in adventitious roots of CS, SH, and SU following 7 days of methyl jasmonate (MeJA) treatment, as analyzed by LC-UV-DAD (CS, Cheongsun; SH, Sunhyang; and SU, Sunun).
| Ginsenosides (μg/mg) | CS |
SH |
SU |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | +MJ | Fold | Control | +MJ | Fold | Control | +MJ | Fold | |
| Rb1 | 0.15 ± 0.06c | 2.58 ± 0.28b | 17.38 | 1.21 ± 0.06a | 4.99 ± 0.73a | 4.13 | 0.43 ± 0.07b | 5.42 ± 0.47a | 12.61 |
| Rb2 | 0.03 ± 0.10c | 1.01 ± 0.10c | 32.92 | 0.90 ± 0.31a | 2.34 ± 0.31a | 2.60 | 0.11 ± 0.16b | 1.71 ± 0.16b | 15.71 |
| Rc | 0.14 ± 0.02c | 4.22 ± 0.52c | 30.43 | 2.23 ± 0.04a | 8.62 ± 0.97a | 3.86 | 0.61 ± 0.05b | 7.17 ± 0.50b | 11.85 |
| Rd | 0.04 ± 0.01c | 3.34 ± 0.51ab | 90.32 | 0.36 ± 0.04a | 3.48 ± 0.63a | 9.78 | 0.13 ± 0.02b | 2.41 ± 0.32b | 18.97 |
| Total panaxadiol | 0.35 ± 0.09c | 11.15 ± 1.33c | 31.44 | 4.7 ± 0.16a | 19.42 ± 2.57a | 4.14 | 1.27 ± 0.12b | 16.71 ± 1.16b | 13.15 |
| Re | 0.24 ± 0.01c | 1.72 ± 0.32c | 7.05 | 3.4 ± 0.20a | 5.60 ± 1.22a | 1.65 | 1.16 ± 0.03b | 7.37 ± 0.80b | 6.36 |
| Rg1 | 0.96 ± 0.07a | 3.08 ± 0.16a | 3.19 | 2.43 ± 0.09b | 2.26 ± 1.05a | 0.93 | 1.24 ± 0.05b | 2.27 ± 0.49a | 1.83 |
| Rg2 (20S)-form | 0.02 ± 0.00c | 0.05 ± 0.00b | 2.50 | 0.10 ± 0.01a | 0.07 ± 0.02ab | 0.69 | 0.03 ± 0.00b | 0.08 ± 0.01a | 2.29 |
| Rg2 (20R)-form | 0.02 ± 0.00c | 0.17 ± 0.02b | 8.24 | 0.52 ± 0.07a | 0.69 ± 0.17a | 1.33 | 0.14 ± 0.01b | 0.88 ± 0.18a | 6.39 |
| Rf | 0.06 ± 0.01c | 0.20 ± 0.01b | 3.30 | 0.23 ± 0.02a | 0.32 ± 0.05a | 1.40 | 0.13 ± 0.01b | 0.39 ± 0.05a | 3.00 |
| Total panaxatriol | 1.31 ± 0.07c | 5.22 ± 0.35b | 4.00 | 6.68 ± 0.28a | 8.94 ± 2.20a | 1.34 | 2.70 ± 0.08b | 10.99 ± 1.41a | 4.07 |
| Total ginsenosides | 1.66 ± 0.19a | 16.37 ± 9.86a | 9.86 | 11.38 ± 0.59c | 28.36 ± 2.49b | 2.49 | 3.97 ± 0.25b | 27.70 ± 6.98b | 6.98 |
∗Significantly differentially expressed unigenes among adventitious roots of CS, SH, and SU are represented by different letters (CS, Cheongsun; SH, Sunhyang; SU, Sunun).
Ginsenoside levels responded most strongly to MeJA treatment in CS, which had the lowest ginsenoside levels of the three cultivars under normal conditions, as well as the lowest expression of ginsenoside biosynthesis genes. In MeJA-treated CS adventitious roots, levels of Rd, a PPD-type ginsenoside, increased 90.32-fold, while the levels of other PPD-type ginsenosides, such as Rb1, Rc, and Rb2, increased 17.38-, 30.43-, and 32.92-fold, respectively. SH, the most ginsenoside-rich cultivar under normal conditions, was less responsive to MeJA treatment compared to CS and SU (Table 4).
Gene Expression and Quantity of Intermediates in Ginsenoside Biosynthesis under MeJA Treatment
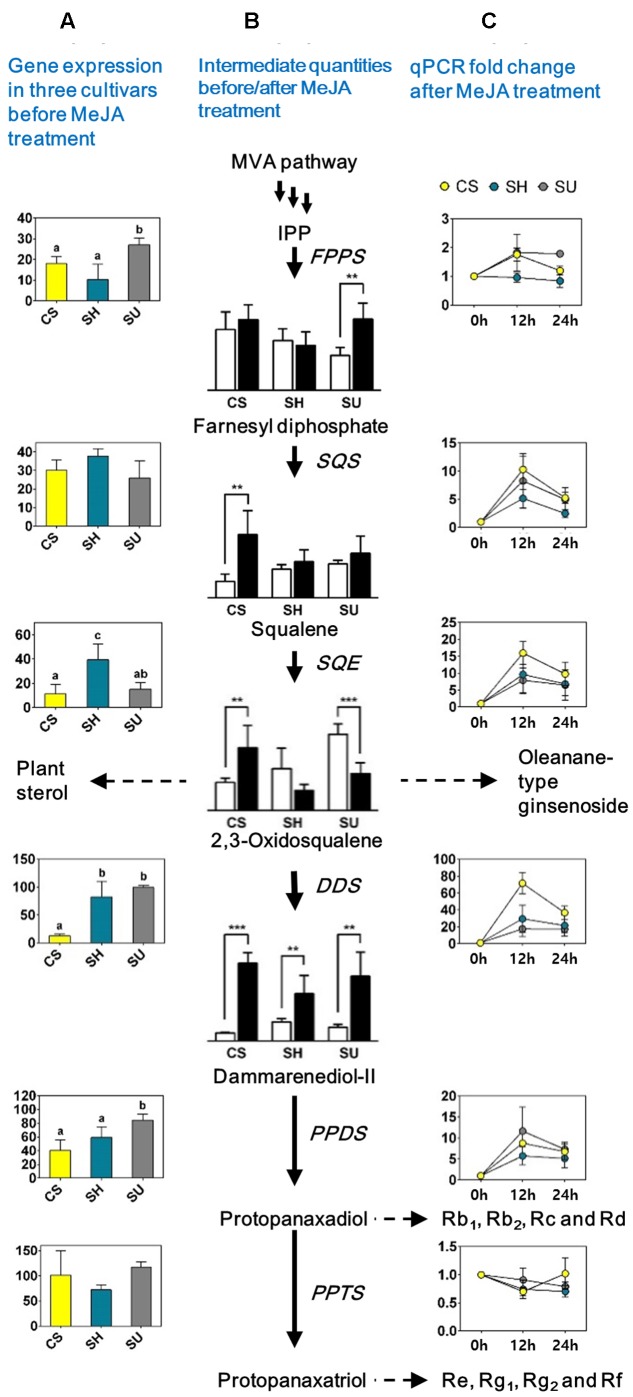
To identify the genes and intermediates associated with changes in ginsenoside contents in the three cultivars following MeJA treatment, we analyzed the expression levels of genes and the contents of intermediates related to the triterpene biosynthetic pathway in MeJA-treated CS, SH, and SU adventitious roots (Figure 5).
FIGURE 5.
Quantification of gene expression levels and levels of intermediates involved in the ginsenoside biosynthetic pathway in adventitious roots with or without methyl jasmonate (MeJA) treatment in three ginseng cultivars; CS, Cheongsun; SH, Sunhyang; and SU, Sunun. (A) Expression levels of the genes among the three ginseng cultivars. The FPKM values (Y-axis) of each gene in Table 3 were compared among cultivars. (B) Ginsenoside biosynthesis pathway and quantity of intermediates among three cultivars before (white bars) and after MeJA treatment (black bars). Relative abundances of intermediates are represented in the bar graphs; significant differences are indicated by asterisks: ∗ P < 0.05; ∗∗ P < 0.01 (t-test). (C) Expression (fold change) of each gene before and after MeJA treatment in adventitious roots of the three ginseng cultivars. Fold changes (Y-axis) in gene expression levels at 12 and 24 h (X-axis) compared to 0 h (analyzed by RT-PCR) are shown in the line graphs.
Gene expression analysis via RT-PCR revealed differences in the expression patterns of genes involved in the MVA pathway and the triterpene biosynthetic pathway (Figure 5 and Supplementary Table S4). Among genes in the MVA pathway, only AACT expression significantly increased after MeJA treatment (Supplementary Table S4). By contrast, most genes corresponding to the triterpene biosynthetic pathway were significantly induced by MeJA treatment, with different levels of increase detected among cultivars, indicating that the triterpene biosynthetic pathway could be responsible for the variations in ginsenoside accumulation among different genotypes of P. ginseng. In particular, the expression of DDS dramatically increased (17.62- to 71.83-fold) at 12 h post-elicitation in all cultivars, and SQE (7.9- to 15.97-fold) and SS (5.16- to 10.32-fold) were also significantly induced. Notably, the changes in expression of all three genes were highest in the cultivar with the lowest ginsenoside content, CS (Figure 5).
GC-MS analysis revealed that the relative abundance of intermediate compounds, from squalene to dammarenediol II, following MeJA treatment increased the most in CS (Figure 5). Comparative analysis of adventitious roots among the three cultivars with or without MeJA treatment suggested that squalene to dammarenediol II might be the major limiting step that affects the variation in ginsenoside content among ginseng cultivars. Among these, DDS likely plays the most critical role in ginsenoside biosynthesis and accumulation.
Discussion
Metabolic Profiles in Ginseng Adventitious Roots
Ginsenoside profiles and accumulation patterns can differ based on genotype (Ahn et al., 2008; Kim et al., 2009) and growth conditions (Matkowski, 2008). In this study, we focused on the genetic component of ginsenoside accumulation by using five genetically fixed inbred cultivars, CP, CS, GO, SH, and SU (Kwon et al., 2001; Ahn et al., 2008; Lee et al., 2008) and isolating ginsenosides from in vitro cultured adventitious roots grown under a controlled environment. We quantified the variation among cultivars in the contents of ginsenosides and intermediates in the ginsenoside biosynthesis pathway as well as the relevant differences in expression of the underlying genes.
We characterized 21 ginsenosides, 15 of which were PPD/PPT-type ginsenosides. Among these PPD/PPT-type ginsenosides, six metabolites were identified as malonyl-ginsenosides (Table 1). Malonylation, a modification process that occurs widely in plants, takes place on the sugar moieties of phytohormones, xenobiotic compounds, and several secondary metabolites (Muth et al., 2008; Kochkin et al., 2013). Malonylation plays a role in improving the solubility of target compounds and facilitating their transport into storage organelles such as vacuoles (Taguchi et al., 2010). In the current study, malonyl-conjugated PPD-type ginsenosides accounted for five of the six identified malonyl-ginsenosides, which is consistent with a previous report that malonylation occurs more often in PPD-type ginsenosides than in PPT-type ginsenosides of Panax (Kochkin et al., 2013). Thus, some PPD-type ginsenosides that accumulate in adventitious roots are malonylated and, therefore, might be stored in organelles such as vacuoles.
The ginsenosides present in adventitious roots had different accumulation patterns in different cultivars. PCA loading plots revealed that that five ginseng cultivars could be divided into three groups according to their secondary metabolite profiles, although the PCA plot based on ginsenoside profile did not correlate precisely with that based on primary metabolite profiles (Lee Y.S. et al., 2016). Notably, however, the CS cultivar exhibited distinct metabolic accumulation in both the PCA loading plots based on primary metabolite profiles (Lee Y.S. et al., 2016) and those based on secondary metabolite profiles (Figure 3). These results indicate that the ginsenoside pathway is likely distinct in CS adventitious roots compared to other ginseng cultivars.
Dammarane-Type Ginsenoside Synthesis Rapidly Increases in Response to MeJA Treatment
Jasmonic acid and MeJA are signaling compounds related to plant defense (Derksen et al., 2013). They also have been used as important plant-derived elicitors for elevated production of secondary metabolites in various plant species (Ali et al., 2006; Han et al., 2006; Lee et al., 2013; Wang J. et al., 2015). Oxidosqualene, a precursor for dammarenediol synthase, is present at a branch point in the triterpene biosynthesis pathway. This compound is cyclized by various oxidosqualene cyclases, such as lanosterol synthase (LSS), cycloartenol synthase (CAS), β-AS, and DDS to produce phytosterols and ginsenosides (Thimmappa et al., 2014). LSS and CAS catalyze the conversion of oxidosqualene to lanosterol and cycloartenol, respectively, which is the first step for entry into phytosterol biosynthesis (Suzuki et al., 2006; Ohyama et al., 2009). The expression levels of LSS and CAS were constant or decreased in response to MeJA treatment in Centella asiatica (Bonfill et al., 2011) and also in P. ginseng (Han et al., 2006), suggesting that the phytosterols in ginseng play a fundamental role in plant growth and development rather than in defense mechanisms involving MeJA. In addition, the expression of β-AS, encoding an enzyme that produces β-amyrin from oxidosqualene during the first committed step of oleanane-type ginsenoside biosynthesis (Suzuki et al., 2006; Tansakul et al., 2006), is not significantly altered under MeJA treatment (Han et al., 2006). By contrast, we found that both the contents of dammarane-type ginsenosides (especially PPD-type) and the expression of DDS significantly increased after MeJA treatment (Figure 5 and Table 4). This result suggests that the biological activities of dammarane-type ginsenosides are more closely involved in defense in ginseng than are those of oleanane-type ginsenosides. The differential regulation of dammarane-type ginsenoside biosynthesis in the ginseng cultivars we examined might influence the properties of these cultivars in terms of plant defense responses.
Dynamic Alteration of Ginsenoside Biosynthesis via MeJA Treatment
In the absence of MeJA treatment, the ginsenoside contents were considerably lower in adventitious roots of CS as compared to the other cultivars (Table 4 and Figure 4), which correlated well with the lower expression in CS of genes that function in the pathway from squalene to dammarenediol II (Table 3). In addition, the ginsenoside contents significantly increased in CS adventitious roots upon MeJA treatment; PPD-type ginsenoside levels were increased 17- to 90-fold, and the expression levels of genes such as SQS, SQE, and DDS, as well as the contents of intermediates such as squalene, 2,3-oxidosqualene, and dammarenediol II, were highly elevated (Figure 5). Remarkably, the expression level of DDS was upregulated 71-fold in CS in response to MeJA treatment and the relative dammarenediol II level in CS adventitious roots became comparable to those in SH and SU (Figure 5). In the case of CS, our data indicate that the low ginsenoside contents in adventitious roots are likely related to the low expression levels of SQE and DDS. The factors that underlie the low expression of these genes in CS are yet not known, but might be directly related to differences in function of the genes, such as unknown transcription factors of DDS.
The dramatic increase in DDS expression under MeJA treatment coincided with the rapid increase in ginsenoside contents, especially in CS adventitious roots. Indeed, the content of dammarane-type ginsenosides is decreased by 84.5% in the roots of transgenic ginseng plants with downregulated DDS expression (Han et al., 2006). These findings suggest that the route from squalene to dammarenediol II is a major factor responsible for the diversity of the flux through the ginsenoside biosynthetic pathway in various ginseng cultivars and that DDS might play a key role in the upregulation of ginsenoside biosynthesis.
Conclusion
We investigated the diversity of the ginsenoside biosynthetic pathway among adventitious roots from five ginseng cultivars using integrated transcriptomic and metabolomic analysis. We elucidated the variation in the ginsenoside biosynthetic pathway by comparing the expression levels of genes, the relative abundance of intermediate compounds, and the ginsenoside contents in adventitious roots of five genetically fixed inbred ginseng cultivars grown in a controlled environment. We also examined the intrinsic variation in ginsenoside contents and gene expression levels in the cultivars before and after MeJA treatment. Major genes in the triterpene biosynthesis pathway, including SQE and DDS, were found to play an important role in regulating ginsenoside biosynthesis, as well as in the variation of ginsenoside contents. The genes responsible for the metabolic diversity of these ginseng cultivars are good candidates to be utilized for the genetic improvement of this important medicinal plant.
Author Contributions
T-JY designed the research and organized the manuscript. YSL and H-SP conducted adventitious root culture and transcriptome analysis. D-KL and SWK conducted metabolome analysis. YSL, H-SP, D-KL, MJ, N-HK, HJK, S-CL, and YJK conducted analysis of integrated bioinformatics data. YSL, H-SP, and T-JY wrote and revised the manuscript. All authors approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was carried out with the support of the “Cooperative Research Program for Agriculture Science & Technology Development (PJ01103001),” Rural Development Administration, Republic of Korea.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01048/full#supplementary-material
References
- Ahn I.-O., Lee S.-S., Lee J.-H., Lee M.-J., Jo B.-G. (2008). Comparison of ginsenoside contents and pattern similarity between root parts of new cultivars in Panax ginseng CA Meyer. J. Gingseng Res. 32 15–18. 10.5142/JGR.2008.32.1.015 [DOI] [Google Scholar]
- Ali M. B., Yu K.-W., Hahn E.-J., Paek K.-Y. (2006). Methyl jasmonate and salicylic acid elicitation induces ginsenosides accumulation, enzymatic and non-enzymatic antioxidant in suspension culture Panax ginseng roots in bioreactors. Plant Cell Rep. 25 613–620. 10.1007/s00299-005-0065-6 [DOI] [PubMed] [Google Scholar]
- Bonfill M., Mangas S., Moyano E., Cusido R. M., Palazon J. (2011). Production of centellosides and phytosterols in cell suspension cultures of Centella asiatica. Plant Cell Tissue Organ Cult. 104 61–67. 10.1007/s11240-010-9804-7 [DOI] [Google Scholar]
- Bylesjö M., Eriksson D., Kusano M., Moritz T., Trygg J. (2007). Data integration in plant biology: the O2PLS method for combined modeling of transcript and metabolite data. Plant J. 52 1181–1191. 10.1111/j.1365-313X.2007.03293.x [DOI] [PubMed] [Google Scholar]
- Cho I. H., Lee H. J., Kim Y.-S. (2012). Differences in the volatile compositions of ginseng species (Panax sp.). J. Agric. Food Chem. 60 7616–7622. 10.1021/jf301835v [DOI] [PubMed] [Google Scholar]
- Cho K., Cho K.-S., Sohn H.-B., Ha I. J., Hong S.-Y., Lee H., et al. (2016). Network analysis of the metabolome and transcriptome reveals novel regulation of potato pigmentation. J. Exp. Bot. 67 1519–1533. 10.1093/jxb/erv549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen H., Rampitsch C., Daayf F. (2013). Signaling cross-talk in plant disease resistance. Plant Sci. 207 79–87. 10.3390/ijms140714996 [DOI] [PubMed] [Google Scholar]
- Dubouzet J. G., Ishihara A., Matsuda F., Miyagawa H., Iwata H., Wakasa K. (2007). Integrated metabolomic and transcriptomic analyses of high-tryptophan rice expressing a mutant anthranilate synthase alpha subunit. J. Exp. Bot. 58 3309–3321. 10.1093/jxb/erm179 [DOI] [PubMed] [Google Scholar]
- Genz A., Bretz F., Miwa T., Mi X., Leisch F., Scheipl F., et al. (2008). mvtnorm: Multivariate Normal and t Distributions. R Package Version 0.9-2. Available at: http://CRAN.R-project.org/package=mvtnorm [Google Scholar]
- Han J.-Y., Hwang H.-S., Choi S.-W., Kim H.-J., Choi Y.-E. (2012). Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 53 1535–1545. 10.1093/pcp/pcs106 [DOI] [PubMed] [Google Scholar]
- Han J.-Y., Kim H.-J., Kwon Y.-S., Choi Y.-E. (2011). The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 52 2062–2073. 10.1093/pcp/pcr150 [DOI] [PubMed] [Google Scholar]
- Han J. Y., Kwon Y. S., Yang D. C., Jung Y. R., Choi Y. E. (2006). Expression and RNA interference-induced silencing of the dammarenediol synthase gene in Panax ginseng. Plant Cell Physiol. 47 1653–1662. 10.1093/pcp/pcl032 [DOI] [PubMed] [Google Scholar]
- Hirai M. Y., Sugiyama K., Sawada Y., Tohge T., Obayashi T., Suzuki A., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104 6478–6483. 10.1073/pnas.0611629104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M. Y., Yano M., Goodenowe D. B., Kanaya S., Kimura T., Awazuhara M., et al. (2004). Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101 10205–10210. 10.1073/pnas.0403218101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T., Bretz F., Westfall P. (2008). Simultaneous inference in general parametric models. Biom. J. 50 346–363. 10.1002/bimj.200810425 [DOI] [PubMed] [Google Scholar]
- Jayakodi M., Lee S.-C., Lee Y. S., Park H.-S., Kim N.-H., Jang W., et al. (2015). Comprehensive analysis of Panax ginseng root transcriptomes. BMC Plant Biol. 15:138 10.1186/s12870-015-0527-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakodi M., Lee S.-C., Park H.-S., Jang W., Lee Y. S., Choi B.-S., et al. (2014). Transcriptome profiling and comparative analysis of Panax ginseng adventitious roots. J. Gingseng Res. 38 278–288. 10.1016/j.jgr.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Zhao Y., Liang X.-J. (2009). Current evaluation of the millennium phytomedicine—ginseng (II): collected chemical entities, modern pharmacology, and clinical applications emanated from traditional Chinese medicine. Curr. Med. Chem. 16 2924–2942. 10.2174/092986709788803204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S.-C., Kim W., Park S. C., Jeong J., Park M. K., Lim S., et al. (2014). Two ginseng UDP-glycosyltransferases synthesize ginsenoside Rg3 and Rd. Plant Cell Physiol. 55 2177–2188. 10.1093/pcp/pcu147 [DOI] [PubMed] [Google Scholar]
- Kim J.-H. (2012). Cardiovascular diseases and Panax ginseng: a review on molecular mechanisms and medical applications. J. Gingseng Res. 36 16–26. 10.5142/jgr.2012.36.1.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.-H., Choi H.-I., Ahn I.-O., Yang T.-J. (2012). EST-SSR marker sets for practical authentication of all nine registered ginseng cultivars in Korea. J. Gingseng Res. 36 298–307. 10.5142/jgr.2012.36.3.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-D., Han J.-Y., Huh G. H., Choi Y.-E. (2011). Expression and functional characterization of three squalene synthase genes associated with saponin biosynthesis in Panax ginseng. Plant Cell Physiol. 52 125–137. 10.1093/pcp/pcq179 [DOI] [PubMed] [Google Scholar]
- Kim Y.-J., Lee O. R., Oh J. Y., Jang M.-G., Yang D.-C. (2014). Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme a reductase encoding genes in triterpene saponin-producing ginseng. Plant Physiol. 165 373–387. 10.1104/pp.113.222596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. K., Yoo D. S., Xu H., Park N. I., Kim H. H., Choi J. E., et al. (2009). Ginsenoside content of berries and roots of three typical Korean ginseng (Panax ginseng) cultivars. Nat. Prod. Commun. 4 903–906. [PubMed] [Google Scholar]
- Kirby J., Keasling J. D. (2009). Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu. Rev. Plant Biol. 60 335–355. 10.1146/annurev.arplant.043008.091955 [DOI] [PubMed] [Google Scholar]
- Kochkin D. V., Kachala V. V., Shashkov A. S., Chizhov A. O., Chirva V. Y., Nosov A. M. (2013). Malonyl-ginsenoside content of a cell-suspension culture of Panax japonicus var. repens. Phytochemistry 93 18–26. 10.1016/j.phytochem.2013.03.021 [DOI] [PubMed] [Google Scholar]
- Kwon W. S., Lee M. G., Lee J. H. (2001). Characteristics of flowering and fruiting in new varieties and lines of Panax ginseng CA Meyer. J. Gingseng Res. 25 41–44. [Google Scholar]
- Lee G. J., Shin B.-K., Yu Y.-H., Ahn J., Kwon S. W., Park J. H. (2016). Systematic development of a group quantification method using evaporative light scattering detector for relative quantification of ginsenosides in ginseng products. J. Pharm. Biomed. Anal. 128 158–165. 10.1016/j.jpba.2016.05.030 [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Lee J.-S., Kwon W.-S., Kang J.-Y., Lee D.-Y., In J.-G., et al. (2015). Characteristics of Korean ginseng varieties of Gumpoong, Sunun, Sunpoong, Sunone, Cheongsun, and Sunhyang. J. Gingseng Res. 39 94–104. 10.1016/j.jgr.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-S., Lee S.-S., Lee J.-H., Ahn I.-O. (2008). Effect of seed size and cultivars on the ratio of seed coat dehiscence and seedling performance in Panax ginseng. J. Gingseng Res. 32 257–263. 10.5142/JGR.2008.32.3.257 [DOI] [Google Scholar]
- Lee M.-H., Han J.-Y., Kim H.-J., Kim Y.-S., Huh G. H., Choi Y.-E. (2012). Dammarenediol-II production confers TMV tolerance in transgenic tobacco expressing Panax ginseng dammarenediol-II synthase. Plant Cell Physiol. 53 173–182. 10.1093/pcp/pcr160 [DOI] [PubMed] [Google Scholar]
- Lee M.-J., Choi J.-S., Cha S.-W., Lee K.-S., Lee Z.-W., Hwang G.-S., et al. (2011). Variation in the ginsenoside profiles of cultivated ginseng (Panax ginseng CA Meyer) landraces in Korea. Process Biochem. 46 258–264. 10.1016/j.procbio.2010.08.020 [DOI] [Google Scholar]
- Lee Y. S., Ju H. K., Kim Y. J., Lim T.-G., Uddin M. R., Kim Y. B., et al. (2013). Enhancement of anti-inflammatory activity of Aloe vera adventitious root extracts through the alteration of primary and secondary metabolites via salicylic acid elicitation. PLoS ONE 8:e82479 10.1371/journal.pone.0082479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. S., Park H.-S., Lee D.-K., Jayakodi M., Kim N.-H., Lee S.-C., et al. (2016). Comparative analysis of the transcriptomes and primary metabolite profiles of adventitious roots of five P. ginseng cultivars. J. Gingseng Res. 41 60–68. 10.1016/j.jgr.2015.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.-H., Yang B.-R., Cheung W.-F., Yang K. Y., Zhou H.-F., Kwok J. S., et al. (2015). Transcriptome analysis of leaves, roots and flowers of Panax notoginseng identifies genes involved in ginsenoside and alkaloid biosynthesis. BMC Genomics 16:265 10.1186/s12864-015-1477-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCrehan W. A., White C. M. (2013). Simplified ultrasonically-and microwave-assisted solvent extractions for the determination of ginsenosides in powdered Panax ginseng rhizomes using liquid chromatography with UV absorbance or electrospray mass spectrometric detection. Anal. Bioanal. Chem. 40 4511–4522. 10.1007/s00216-013-6871-8 [DOI] [PubMed] [Google Scholar]
- Matkowski A. (2008). Plant in vitro culture for the production of antioxidants – A review. Biotechnol. Adv. 26 548–560. 10.1016/j.biotechadv.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Mounet F., Moing A., Garcia V., Petit J., Maucourt M., Deborde C., et al. (2009). Gene and metabolite regulatory network analysis of early developing fruit tissues highlights new candidate genes for the control of tomato fruit composition and development. Plant Physiol. 149 1505–1528. 10.1104/pp.108.133967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth D., Marfisden-Edwards E., Kachlicki P., Stobiecki M. (2008). Differentiation of isomeric malonylated flavonoid glyconjugates in plant extracts with UPLC-ESI/MS/MS. Phytochem. Anal. 19 444–452. 10.1002/pca.1073 [DOI] [PubMed] [Google Scholar]
- Ohyama K., Suzuki M., Kikuchi J., Saito K., Muranaka T. (2009). Dual biosynthetic pathways to phytosterol via cycloartenol and lanosterol in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 725–730. 10.1073/pnas.0807675106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L.-W., Wang H.-Y., Zhang H., Wang C.-Z., Li P., Yuan C.-S. (2012). Diagnostic ion filtering to characterize ginseng saponins by rapid liquid chromatography with time-of-flight mass spectrometry. J. Chromatogr. A 1230 93–99. 10.1016/j.chroma.2012.01.079 [DOI] [PubMed] [Google Scholar]
- Rai A., Yamazaki M., Takahashi H., Nakamura M., Kojoma M., Suzuki H., et al. (2016). RNA-seq transcriptome analysis of Panax japonicus, and its comparison with other Panax species to identify potential genes involved in the saponins biosynthesis. Front. Plant Sci. 7:481 10.3389/fpls.2016.00481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (1999). “Primer3 on the WWW for general users and for biologist programmers,” in Bioinformatics Methods and Protocols, eds Misener S., Krawetz S. A. (Totowa, NJ: Humana Press; ), 365–386. 10.1385/1-59259-192-2:365 [DOI] [PubMed] [Google Scholar]
- Saito K., Hirai M. Y., Yonekura-Sakakibara K. (2008). Decoding genes with coexpression networks and metabolomics–‘majority report by precogs’. Trends Plant Sci. 13 36–43. 10.1016/j.tplants.2007.10.006 [DOI] [PubMed] [Google Scholar]
- Suzuki M., Xiang T., Ohyama K., Seki H., Saito K., Muranaka T., et al. (2006). Lanosterol synthase in dicotyledonous plants. Plant Cell Physiol. 47 565–571. 10.1093/pcp/pcj031 [DOI] [PubMed] [Google Scholar]
- Taguchi G., Ubukata T., Nozue H., Kobayashi Y., Takahi M., Yamamoto H., et al. (2010). Malonylation is a key reaction in the metabolism of xenobiotic phenolic glucosides in Arabidopsis and tobacco. Plant J. 63 1031–1041. 10.1111/j.1365-313X.2010.04298.x [DOI] [PubMed] [Google Scholar]
- Tansakul P., Shibuya M., Kushiro T., Ebizuka Y. (2006). Dammarenediol-II synthase, the first dedicated enzyme for ginsenoside biosynthesis, in Panax ginseng. FEBS Lett. 580 5143–5149. 10.1016/j.febslet.2006.08.044 [DOI] [PubMed] [Google Scholar]
- Thimmappa R., Geisler K., Louveau T., O’Maille P., Osbourn A. (2014). Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 65 225–257. 10.1146/annurev-arplant-050312-120229 [DOI] [PubMed] [Google Scholar]
- Wang H.-P., Zhang Y.-B., Yang X.-W., Zhao D.-Q., Wang Y.-P. (2016). Rapid characterization of ginsenosides in the roots and rhizomes of Panax ginseng by UPLC-DAD-QTOF-MS/MS and simultaneous determination of 19 ginsenosides by HPLC-ESI-MS. J. Ginseng Res. 40 382–394. 10.1016/j.jgr.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Qian J., Yao L., Lu Y. (2015). Enhanced production of flavonoids by methyl jasmonate elicitation in cell suspension culture of Hypericum perforatum. Bioresour. Bioprocess. 2:5 10.1186/s40643-014-0033-5 [DOI] [Google Scholar]
- Wang P., Wei Y., Fan Y., Liu Q., Wei W., Yang C., et al. (2015). Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab. Eng. 29 97–105. 10.1016/j.ymben.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Wei W., Wang P., Wei Y., Liu Q., Yang C., Zhao G., et al. (2015). Characterizations of Panax ginseng UDP-glycosyltransferases catalyzing protopanaxatriol and biosyntheses of bioactive ginsenosides F1 and Rh1 in metabolically engineered yeasts. Mol. Plant 8 1412–1424. 10.1016/j.molp.2015.05.010 [DOI] [PubMed] [Google Scholar]
- Xie G. X., Ni Y., Su M. M., Zhang Y. Y., Zhao A. H., Gao X. F., et al. (2008). Application of ultra-performance LC-TOF MS metabolite profiling techniques to the analysis of medicinal Panax herbs. Metabolomics 4 248–260. 10.1007/s11306-008-0115-5 [DOI] [Google Scholar]
- Yendo A. C., de Costa F., Gosmann G., Fett-Neto A. G. (2010). Production of plant bioactive triterpenoid saponins: elicitation strategies and target genes to improve yields. Mol. Biotechnol. 46 94–104. 10.1007/s11306-008-0115-5 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Tohge T., Matsuda F., Nakabayashi R., Takayama H., Niida R., et al. (2008). Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene–metabolite correlations in Arabidopsis. Plant Cell 20 2160–2176. 10.1105/tpc.108.058040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.-H., Ma C.-H., Zhang J.-J., Chen J.-W., Tang Q.-Y., He M.-H., et al. (2015). Transcriptome analysis of Panax vietnamensis var. fuscidicus discovers putative ocotillol-type ginsenosides biosynthesis genes and genetic markers. BMC Genomics 16:159 10.1186/s12864-015-1332-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-J., Huang L.-L., Cai X.-J., Li P., Wang Y.-T., Wan J.-B. (2013). Fatty acid variability in three medicinal herbs of Panax species. Chem. Cent. J. 7:12 10.1186/1752-153X-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.