SUMMARY
Organ-specific functions of tissue-resident macrophages in the steady-state heart are unknown. Here we show that cardiac macrophages facilitate electrical conduction through the distal atrioventricular node, where conducting cells densely intersperse with elongated macrophages expressing connexin 43. When coupled to spontaneously beating cardiomyocytes via connexin 43-containing gap junctions, cardiac macrophages have a negative resting membrane potential and depolarize in synchrony with cardiomyocytes. Conversely, macrophages render the resting membrane potential of cardiomyocytes more positive and, according to computational modeling, accelerate their repolarization. Photostimulation of channelrhodopsin 2-expressing macrophages improves atrioventricular conduction, while conditional deletion of connexin 43 in macrophages and congenital lack of macrophages delay atrioventricular conduction. In the Cd11bDTR mouse, macrophage ablation induces progressive atrioventricular block. These observations implicate macrophages in normal and aberrant cardiac conduction.
Keywords: Macrophages, Heart, Atrioventricular node, Electrical conduction, Connexin 43, Gap junctions, Tissue clearing, Single-cell RNA-sequencing, Optogenetics, Computational modeling
Graphical Abstract
INTRODUCTION
Studies in the late 19th century described macrophages as phagocytic cells that defend the organism against pathogens (Metchnikoff, 1892). More recently, it became clear that resident macrophages populate all tissues and pursue organ-specific functions. For instance, macrophages contribute to thermogenesis regulation in adipose tissue (Nguyen et al., 2011), iron recycling in the spleen and liver (Theurl et al., 2016) and synaptic pruning in the brain (Paolicelli et al., 2011). These non-canonical activities highlight macrophages’ functional diversity and emphasize their ability to execute tissue-specific tasks beyond host defense (Davies et al., 2013).
The cardiac conduction system coordinates the heart’s electrical activation. Electrical impulse generation begins in the sinoatrial (SA) node and sequentially propagates activation of the atria, atrioventricular (AV) node, His and Purkinje systems and ventricles. By providing the only electrical connection between the atria and ventricles, the AV node plays an essential role. First described by Tawara in 1906 (Tawara, 1906), the AV node is located at the base of the right atrium and contains cardiomyocytes with distinct action potentials (Billette, 1987). Clinically, AV block delays or abolishes atrial impulse conduction to the ventricles, which can lead to hemodynamic deterioration, syncope and death if not treated with pacemaker implantation (Rubart and Zipes, 2008).
Macrophages are an intrinsic part of the healthy working myocardium, where they appear as spindle-like cells interspersed among myocytes, fibroblasts and endothelial cells (Pinto et al., 2012; Heidt et al., 2014; Epelman et al., 2014). Cardiac healing after injury requires macrophages (Swirski and Nahrendorf, 2013); however, in contrast to other organs, specific functions of cardiac macrophages in the steady-state are unknown. Here we report resident macrophages’ abundance in the distal AV node and show that they contribute to cardiac conduction.
RESULTS
Macrophages Abound in the AV Node
Resident macrophages are present in the left ventricle (LV), but prior work did not report on intra-organ heterogeneity. It therefore remained unclear whether macrophages distribute homogeneously throughout the heart and whether any reside in the conduction system. To investigate macrophages’ presence and spatial distribution, we optically cleared and imaged entire AV nodes of Cx3cr1GFP/+ mice, an extensively validated reporter strain in which green fluorescent protein identifies cardiac macrophages (Pinto et al., 2012; Heidt et al., 2014; Molawi et al., 2014), by confocal microscopy (Figure 1A). We found that HCN4-expressing cardiomyocytes (Biel et al., 2009), particularly in the lower nodal or AV bundle, frequently intersperse with macrophages (Figure 1B). Macrophages assume an elongated, spindle-shaped appearance with far-reaching cytoplasmic projections (Figure 1C) that often approximate stromal AV (Figure S1A) and SA node (Figure S1B) cells. The average cell surface area of an AV node macrophage is 149 ± 24 μm2 (mean ± SEM, n = 17), which is similar to the cell surface area of an LV free wall macrophage (155 ± 15 μm2, n = 15, p = 0.850, Student’s t test). To study the morphological characteristics of AV node macrophages by electron microscopy, we labeled GFP+ macrophages in Cx3cr1GFP/+ mice with diaminobenzidine (DAB). DAB+ macrophages display long cellular processes that closely associate with cardiomyocytes as well as extracellular matrix (Figures 1D and S1C). In the mouse heart, the majority of CD45+ leukocytes are CD11b+ F4/80+ Ly6Clow macrophages. Co-expression of CD64 and CX3CR1 and the lack of CD11c and CD103 expression confirm that these cells are macrophages and not dendritic cells (Figures S1D and S1E). Flow cytometry, quantitative real-time PCR (qPCR) of FACS-purified cardiac cells and immunofluorescence histology (Figures S1E–S1I) concur that CX3CR1+ cells are macrophages and confirm that Cx3cr1GFP/+ mice are an appropriate strain to study macrophages in the AV node; however, comparative studies using other macrophage-specific reporter systems such as Csf1rCreER mice may be valuable.
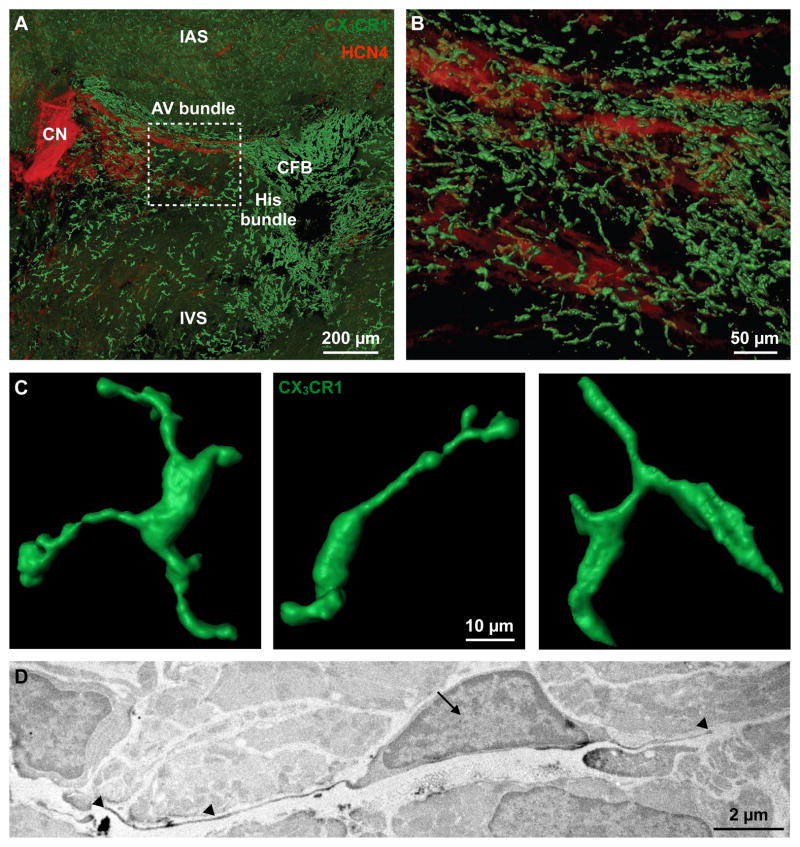
Figure 1. Resident Cardiac Macrophages in the AV Node.
(A) Volumetric reconstruction of confocal microscopy after optical clearing of the atrioventricular (AV) node in a Cx3cr1GFP/+ mouse stained with HCN4 (red). The node is orientated along the AV groove extending from the compact node (CN) into the proximal His bundle. Dashed square indicates the lower nodal or AV bundle. CFB, central fibrous body; IAS and IVS, interatrial and interventricular septum.
(B) Higher magnification of dashed square in (A).
(C) 3D rendering of GFP+ macrophages in the AV bundle.
(D) Electron microscopy of a DAB+ macrophage in the AV node of a Cx3cr1GFP/+ mouse stained with a primary antibody for GFP. Arrow indicates nucleus, arrowheads indicate cellular processes.
See also Figure S1.
To compare macrophage numbers in the AV node and LV myocardium, we investigated microdissected tissue by flow cytometry and histology. The mouse AV node has a higher macrophage density than the LV (Figures 2A and 2B).
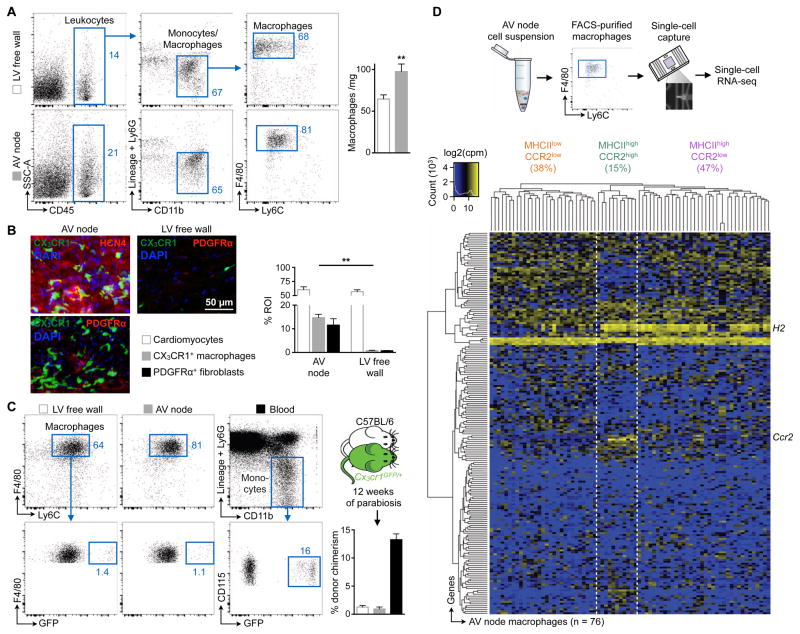
Figure 2. The AV Node Enriches for Macrophages.
(A) Flow cytometric macrophage quantification in microdissected AV node and left ventricular (LV) free wall of C57BL/6 mice. (Left) Representative flow cytometry plots; (right) number of macrophages per mg of heart tissue. Data are mean ± SEM, n = 12 mice from 4 independent experiments, **p < 0.01, Student’s t test.
(B) (Left) Representative immunofluorescence images of the AV node and LV free wall of a Cx3cr1GFP/+ mouse stained for HCN4 (red) and nuclei (blue), or PDGFRα (red) and nuclei (blue). (Right) Percentage of positive staining per region of interest (ROI). Data are mean ± SEM, n = 3–6 mice from 2 independent experiments, **p < 0.01, Kruskal-Wallis test followed by Dunn’s posttest.
(C) Macrophage chimerism in the LV free wall and AV node, and monocyte chimerism in the blood of C57BL/6 mice that had been joined in parabiosis with Cx3cr1GFP/+ mice for 12 weeks (mean ± SEM, n = 3 [AV node] and n = 7 [LV free wall and blood] from 2 independent experiments).
(D) (Top) Workflow; (bottom) heat map of expression levels (cpm, counts per million) among top 200 overdispersed genes from RNA-seq data of 76 AV node macrophages. Unsupervised clustering reflects three macrophage subsets according to expression levels of H2 and Ccr2 (orange, MHCIIlowCCR2low; green, MHCIIhighCCR2high; purple, MHCIIhighCCR2low).
See also Figure S2
Steady-state myocardial tissue-resident macrophages primarily arise from embryonic yolk-sac progenitors and perpetuate independently of monocytes through in situ proliferation (Epelman et al., 2014; Heidt et al., 2014;). Using parabiosis, we determined that circulating cells contributed minimally to AV node macrophages, similar to LV free wall macrophages (Figure 2C).
Single-cell RNA-sequencing (RNA-seq) of mouse AV node macrophages isolated by flow sorting showed cellular subsets that are also present elsewhere in the heart (Epelman et al., 2014) (Figure 2D). These macrophage subsets displayed the characteristic core macrophage gene signature suggested by the Immunological Genome Project (Gautier et al., 2012) (Figure S2A) and separated based on their expression of major histocompatibility complex class II (H2) and chemokine receptor 2 (Ccr2) (Figures S2B–S2D). RNA-seq and qPCR revealed that AV node and LV macrophages express ion channels and exchangers (Figures S2E and S2F), while deposited microarray data (Pinto et al., 2012) show cardiac macrophages’ enrichment of genes associated with conduction (Figure S2G). Thus, murine AV node macrophages have an expression profile similar to that of ventricular resident macrophages, and macrophages from both regions express genes involved in electrical conduction.
We also studied macrophages in six human AV nodes (please see STAR Methods section for clinical information on autopsy cases). Fresh AV nodes were harvested within 24 hours after death and underwent optical clearing after staining with the well-validated human macrophage markers CD68 and CD163 (Murray and Wynn, 2011). Confocal microscopy of 500 μm thick tissue slabs revealed that, in analogy to mice, macrophages distribute heterogeneously and are more abundant in human AV nodes than in working myocardium (Figures 3A–3C). In addition to the AV node, macrophages also enriched in the central fibrous body (Figures 3C and S3A). As in mice, human AV node macrophages exhibit a spindle-shaped appearance with long-reaching protrusions (Figure 3D), which establish close contact with stromal cells (Figure S3B).
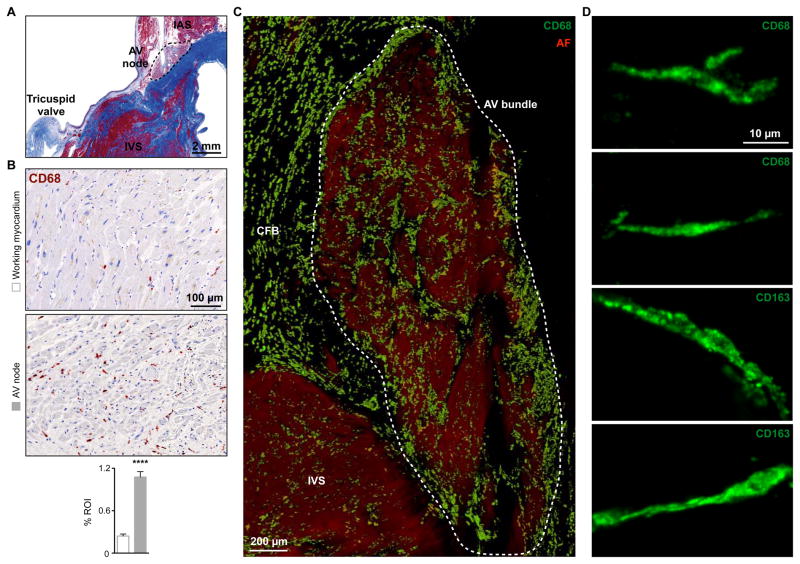
Figure 3. Macrophages in the Human AV Node.
(A) Masson’s trichrome stain of human tissue to identify the AV node. IAS and IVS, interatrial and interventricular septum.
(B) Immunohistochemical stain for CD68 in human working myocardium and AV node. Data are mean ± SEM, n = 20 to 30 high-power fields per section, ****p < 0.0001, Student’s t test.
(C) Volumetric reconstruction of confocal microscopy after optical clearing of a 500 μm section of the human AV bundle stained with CD68 (green). Autofluorescence signal (AF, red) was used for visualization of tissue morphology. Dashed area indicates the AV bundle. CFB, central fibrous body.
(D) Maximum projection images of CD68+ and CD163+ macrophages in the human AV bundle.
See also Figure S3.
Connexin 43 Connects Macrophages with Myocytes
Gap junctions, which are formed by connexin (Cx) proteins, connect the cytoplasm of two adjacent cells to enable their communication (Unger et al., 1999). Most tissues as well as immune cells express Cx43 (Oviedo-Orta and Howard Evans, 2004). Cx43-containing gap junctions electrically couple cardiomyocytes, enable electrical impulse propagation and consequently coordinate synchronous heart muscle contractions (Shibata and Yamamoto, 1977). In addition, Cx43-containing gap junctions couple cardiomyocytes with non-cardiomyocytes, which can then alter the electrophysiological properties of cardiomyocytes (Ongstad and Kohl, 2016).
To determine if AV node macrophages express proteins that give rise to gap junctions, we evaluated six connexins found in leukocytes (Neijssen et al., 2007) in FACS-purified cells harvested from microdissected AV nodes. AV node macrophages mainly express Cx43 (Figure 4A). We next sorted macrophages from the LV and right ventricular (RV) free wall and compared their Cx43 levels with AV node macrophages. In agreement with the RNA-seq data that imply AV node and other cardiac resident macrophages have similar expression profiles (Figure 2D), we found that macrophages from all three cardiac regions express Cx43 (Figure 4B). Because cardiomyocytes express Cx43 at high levels, we ensured the purity and identity of sorted AV node macrophages by measuring different macrophage- (Gautier et al., 2012) and cardiomyocyte-specific markers (Park and Fishman, 2011) in FACS-purified cells. We also included peritoneal macrophages as a negative control population. All macrophage samples display a characteristic macrophage signature (Figure S4A) and lack expression of cardiomyocyte-specific genes (Figure S4B). As reported previously, peritoneal macrophages express Gata6 (Rosas et al., 2014), but AV node macrophages do not (Figure S4B).
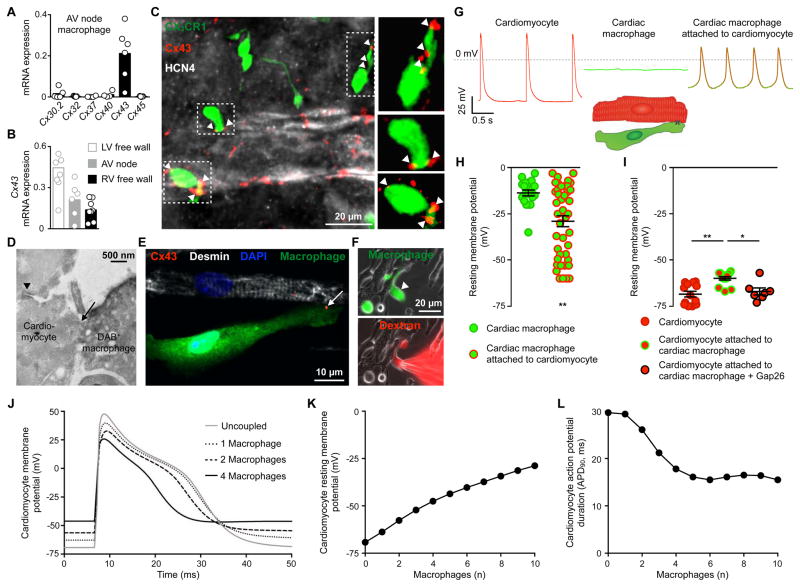
Figure 4. AV Node Macrophages Couple to Conducting Cardiomyocytes and Alter Their Electrophysiological Properties.
(A) Relative connexin (Cx) expression levels in FACS-purified AV node macrophages by qPCR (n = 4 to 6 from 2 independent experiments).
(B) Cx43 levels by qPCR in macrophages FACS-sorted from AV node, and LV and RV free wall. n = 6 to 9 from 2 independent experiments.
(C) Whole-mount immunofluorescence microscopy of AV lower nodal area from a Cx3cr1GFP/+ mouse stained with Cx43 (red) and HCN4 (white). Arrowheads indicate Cx43 colocalization with GFP+ macrophages (green).
(D) Electron microscopy image of a direct membrane contact (arrow) between a DAB+ macrophage and a cardiomyocyte in AV node tissue of a Cx3cr1GFP/+ mouse stained for GFP. The nodal cardiomyocyte is characterized by its typical high mitochondrial content and junctional contact with the neighboring myocyte (arrowhead).
(E) Immunofluorescence image of a co-cultured desmin+ neonatal mouse cardiomyocyte (white) and GFP+ cardiac macrophage (green) stained with Cx43 (red, arrow), illustrating setup for patch clamp experiments (F-I). The cells are grown on cover slips coated with fibronectin in a line pattern.
(F) Immunofluorescence images of dextran diffusion during whole-cell patch clamp with a dextran-loaded pipette. (Top) Arrowhead indicates GFP+ cardiac macrophage (green); (bottom) Texas Red+ dextran (red) diffusion into macrophage.
(G and H) Spontaneous recordings (G) and resting membrane potential (H) of solitary cardiac macrophages (n = 20) and macrophages attached to cardiomyocytes (n = 43) by whole-cell patch clamp. Data are mean ± SEM from 13 independent experiments, **p < 0.01, nonparametric Mann-Whitney test. Rhythmic depolarization was observed in 10/43 macrophages attached to cardiomyocytes.
(I) Resting membrane potential of solitary cardiomyocytes (n = 13) and cardiomyocytes coupled to macrophages before (n = 14) and after (n = 7) addition of the Cx43 inhibitor Gap26. Data are mean ± SEM from 3 independent experiments, *p < 0.05 and **p < 0.01, Kruskal-Wallis test followed by Dunn’s posttest.
(J) Mathematical modeling of ‘single-sided coupling’ between one AV bundle cardiomyocyte and an increasing number of cardiac macrophages. The graph shows the AV bundle cardiomyocyte membrane potential uncoupled or coupled to one, two or four cardiac macrophages at a junctional conductance of 1 nS.
(K and L) Computational modeling of resting membrane potential (K) and action potential duration (L) of an AV bundle cardiomyocyte coupled to an increasing number of cardiac macrophages.
We then analyzed the Cx43 protein expression in AV node macrophages by whole-mount immunofluorescence in the lower AV node, an area in which conducting cells express this connexin (Nikolski et al., 2003; Hucker et al., 2008). Cx43 marked, on average, three punctate contact points between CX3CR1+ macrophages and HCN4+ cardiomyocytes, suggesting gap unction-mediated intercellular communication between these cell types in the distal AV node (Figures 4C and S4C). Likewise, the human AV bundle shows punctate Cx43+ gap junctions between CD163+ macrophages and conducting cardiomyocytes (Figures S4D and S4E). Electron microscopy also visualized direct membrane-membrane contact between AV node macrophages and conducting cardiomyocytes (Figure 4D). Together, these observations indicate the presence of gap junctions between conducting cells and AV node macrophages.
Macrophages Electrically Modulate Myocytes
Since gap junctions electrotonically couple heterotypic cells (Rook et al., 1992), we next tested the hypothesis that macrophages enter electrotonic communication with adjacent cardiomyocytes. When cultured together, FACS-purified GFP+ cardiac macrophages closely interact with neonatal mouse cardiomyocytes (Movie S1). As observed in vivo, Cx43 localized at sites of macrophage-cardiomyocyte interaction (3 ± 0.2 punctate contact points, mean ± SEM, n = 64, 2 independent experiments), thereby indicating gap junction communication between these cell types in culture (Figure 4E). We then used whole-cell patch clamp to investigate the membrane potential of cardiac macrophages attached to cardiomyocytes. Texas Red+ dextran entering GFP+ macrophages from the micropipette (Figure 4F) confirms that the membrane potential we recorded derived from macrophages. Spontaneously-beating cardiomyocytes displayed typical resting membrane and action potentials (Nerbonne et al., 2001) (Figure 4G). The resting membrane potential in solitary cardiac macrophages is depolarized relative to that of cardiomyocytes (Figure 4G). We documented values between −35 and −3 mV that correspond well with data reported for human monocyte-derived and mouse peritoneal macrophages (Gallin and Gallin, 1977; Gallin and Livengood, 1980) (Figure 4H). There was no spontaneous depolarization in solitary cardiac macrophages (Figure 4G). Next, we co-cultured FACS-purified cardiac macrophages with neonatal mouse cardiomyocytes for three days and then recorded the membrane potential in macrophages connected to beating cardiomyocytes. 23% of these macrophages rhythmically depolarized with a distinct action potential-like morphology that was characterized by a slowed upstroke and reduced maximal polarization when compared to cardiomyocytes (Figure 4G). These cardiomyocyte-linked macrophages’ resting membrane potentials were more negative than those of solitary macrophages, thus documenting electrical coupling (Figure 4H). We recorded irregular depolarization in another 23% of co-cultured macrophages and lack of activity in the remaining 54% (Figure S4F). Macrophages with any kind of depolarization, either regular or irregular, had a more negative resting membrane potential than non-depolarizing macrophages (Figure S4G). To simultaneously record action potential-related fluorescence changes in macrophages and cardiomyocytes, we examined cardiomyocyte-driven macrophage depolarization using the ANNINE-6plus voltage-sensitive dye. These data show that macrophage action potentials are synchronous with action potentials of directly coupled cardiomyocytes (Figures S4H and S4I).
Are cardiac macrophages passive bystanders or do they influence conduction? To address this question, we investigated whether macrophages change the electrical properties of coupled cardiomyocytes. Indeed, macrophages render cardiomyocyte resting membrane potentials more positive, an effect that was reversed by pharmacological Cx43 blockade (Figure 4I). In control experiments, inhibition of Cx43-mediated gap junctions in solitary cardiomyocytes did not change their resting membrane potential (Figure S4J). Voltage dye imaging documented deterioration of macrophage-cardiomyocyte cross talk after gap junction inhibition (Figure S4K).
To explore the consequences of the observed communication between macrophages and cardiomyocytes, we pursued mathematical modeling of electrical interactions between macrophages and AV cardiomyocytes (see STAR Methods section for model parameters). Recapitulating the experimental data (Figure 4I), modeling indicates that the cardiomyocyte resting membrane potential is more depolarized when the cell is coupled to a macrophage, an effect that increases with gap junction conductance (Figure S4L). Modeling further suggests that coupling increasing numbers of macrophages accelerates cardiomyocyte repolarization (Figure 4J). For example, coupling three macrophages to an AV bundle cardiomyocyte, a ratio supported by histology (3 ± 0.3, mean ± SEM, n = 17 in 5 mice; Figures 1, 4C, S1A and S3B), decreases cardiomyocyte action potential duration from 30 ms to 21 ms while depolarizing the resting membrane potential from −69 mV to −52 mV (Figures 4K and 4L), assuming a gap junction conductance of 1 nS. In vivo, a shorter action potential duration would decrease the myocyte’s effective refractory period and increase the frequency at which it can be depolarized. A higher resting membrane potential would facilitate depolarization with less stimulation. Both alterations facilitate AV conduction at higher frequencies. These results correspond well with prior conceptual models of electrotonic interactions between cardiomyocytes and electrically non-excitable cells (Kohl and Gourdie, 2014).
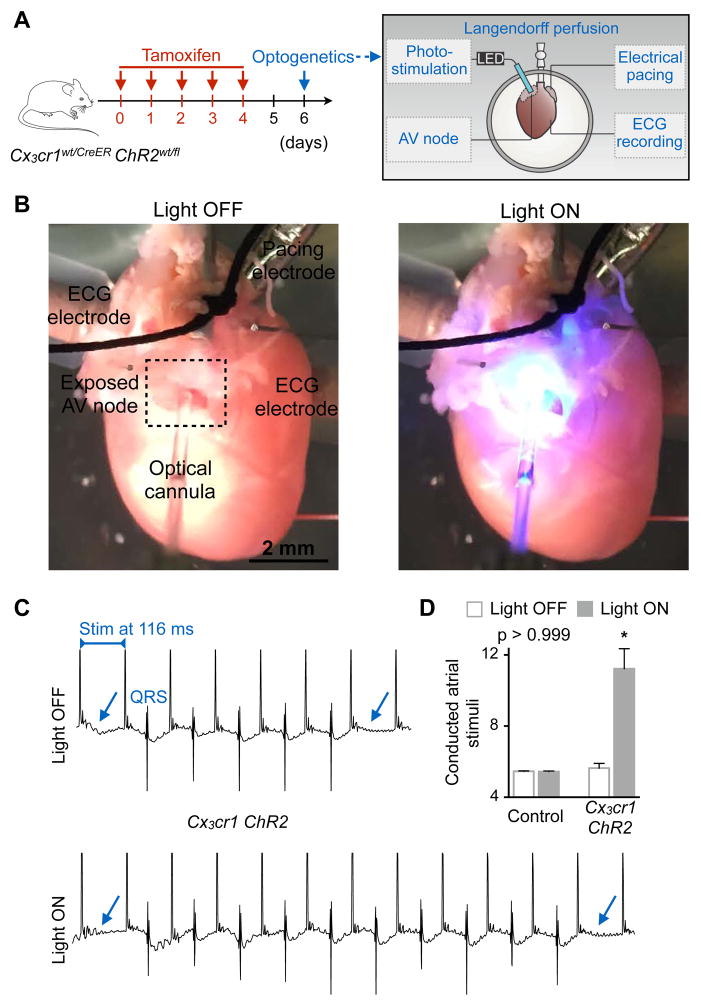
To investigate cell-cell communication directly in the AV node, we expressed the photoactivatable channelrhodopsin 2 (ChR2) (Fenno et al., 2011) in macrophages to control their membrane potential. When illuminated, the cation channel ChR2 undergoes a conformational change that results in an immediate increase in ionic permeability with high conductance for Na+ (Nagel et al., 2003). We posited that light-triggered cation influx into macrophages and their resulting depolarization should alter AV node conduction if the cells are electrotonically coupled to conducting cardiomyocytes. To this end, we bred tamoxifen-inducible Cx3cr1CreER with ChR2fl/fl mice, hereafter denoted Cx3cr1 ChR2, to obtain mice in which tamoxifen treatment triggers ChR2 expression in macrophages. First, we validated macrophage-specific expression of the tamoxifen-inducible Cre recombinase fusion protein (CreER) by measuring YFP fluorescence in heart tissue, as YFP is co-expressed with CreER. We found that YFP signal colocalizes with CX3CR1+ macrophages whereas cardiomyocytes are YFP negative (Figure S5A). In addition, after tamoxifen treatment, AV node macrophages specifically expressed the ChR2 protein, which is fused with YFP (Figure S5B). We then Langendorff-perfused hearts isolated from Cx3cr1 ChR2 mice and inserted a fiber optic cannula into the right atrium to directly illuminate the AV node region (please see Figures 5A and 5B for experimental setup). We assessed AV node conduction by ECG during rapid electrical atrial pacing, comparing continuous 470 nm wavelength illumination with no illumination. To evaluate, with high temporal resolution, the effect of ChR2-induced macrophage depolarization on AV node function, we counted the conducted atrial stimuli between two non-conducted impulses during rapid pacing-induced Wenckebach block. We observed improved AV node conduction during photostimulation of macrophages in hearts harvested from Cx3cr1 ChR2 mice (n = 5). When the light was switched on, the number of conducted atrial stimuli between two non-conducted impulses rose (Figures 5C and 5D). In Cx3cr1wt/CreER control hearts (n = 4), we observed no difference between illuminated and non-illuminated states (Figure 5D). Illumination of the right atrium or LV free wall in Cx3cr1 ChR2 hearts did not alter the number of conducted atrial stimuli (Figure S5C). Thus, opening the cation channel ChR2 in macrophages facilitates AV node conduction during rapid pacing. Modeling indicates that ChR2-induced tonic depolarization of macrophages reduces the minimum heterocellular coupling required to achieve macrophage-mediated passive action potential conduction between otherwise unconnected cardiomyocytes (Figures S5D and S5E). Taken together, these observations suggest that cardiac macrophages can electrically couple to cardiomyocytes via gap junctions containing Cx43. This leads to cyclical macrophage depolarization, modulates cardiomyocytes’ electrophysiological properties and improves AV nodal conduction.
Figure 5. Optogenetics Stimulation of AV Node Macrophages Improves Nodal Conduction.
(A) Experimental outline. Hearts of Cx3cr1wt/CreER (control) or tamoxifen-treated Cx3cr1wt/CreER ChR2wt/fl (Cx3cr1 ChR2) mice were perfused in a Langendorff setup. Recording and pacing electrodes were connected to the heart and illumination with a fiber optic cannula was focused on the AV node.
(B) Images illustrating the optogenetics experimental setup during a light off and on cycle.
(C) Representative ECG recordings from a Cx3cr1 ChR2 heart illustrating the number of conducted atrial stimuli between two non-conducted impulses of a Wenckebach period during light off and on cycles. Arrows indicate failure of conduction leading to missing QRS complexes. Stim, stimulation.
(D) Representative bar graphs of a control and Cx3cr1 ChR2 heart showing the number of conducted atrial stimuli between two non-conducted impulses of a Wenckebach period during light off and on cycles. Data are mean ± SEM, *p < 0.05, Kruskal-Wallis test followed by Dunn’s posttest.
See also Figure S5.
Deleting Cx43 in Macrophages Delays AV Conduction
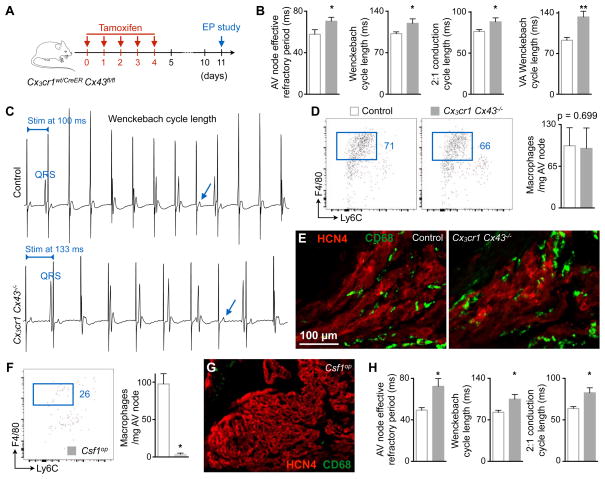
The experiments described above indicate that macrophages present in the AV node may facilitate conduction. To test this hypothesis in loss-of-function experiments, and to directly investigate the importance of Cx43 in macrophages, we bred mice, hereafter denoted Cx3cr1 Cx43−/−, in which tamoxifen treatment deletes Cx43 in CX3CR1-expressing cells. In the AV node, all CX3CR1+ cells are macrophages (Figures S1D-S1I and S4A). All mice underwent analysis seven days after tamoxifen treatment (Figure 6A). Genomic PCR-based examination of the wild-type (Cx43wt), floxed intact (Cx43fl) and recombined (Cx43Δ) alleles of the Cx43 gene in FACS-purified CX3CR1+ cardiac macrophages showed effective Cx43 deletion in cardiac macrophages after tamoxifen treatment (Figure S6A). mRNA analysis supported these findings (Figure S6B). The overall myocardial Cx43 protein level did not change, thereby indicating unaltered Cx43 expression in other cardiac cells (Figure S6C).
Figure 6. Cx43 Deletion in Macrophages and Congenital Lack of Macrophages Delay AV Conduction.
(A) Experimental outline of the electrophysiological (EP) study performed on mice lacking Cx43 in macrophages.
(B) AV node effective refractory period at 120 ms pacing frequency, and pacing cycle lengths at which Wenckebach conduction, 2:1 conduction and ventriculo-atrial (VA) Wenckebach conduction occurred in control (n = 5 to 9) and Cx3cr1 Cx43−/− (n = 6 to 8) mice. Data are mean ± SEM, 2 independent experiments, *p < 0.05 and **p < 0.01, Student’s t test and nonparametric Mann-Whitney test.
(C) Surface ECG from control and Cx3cr1 Cx43−/− mice illustrating the Wenckebach cycle length. Arrows indicate missing QRS complexes. Stim, stimulation.
(D) Flow cytometric quantification of AV node macrophages in control and Cx3cr1 Cx43−/− mice. Data are mean ± SEM, n = 6 mice per group, nonparametric Mann-Whitney test.
(E) Immunofluorescence images of control and Cx3cr1 Cx43−/− AV node stained for CD68 (green) and HCN4 (red).
(F) Quantification of AV node macrophages in control (n = 5) and Csf1op (n = 4) mice by flow cytometry. Data are mean ± SEM, 3 independent experiments, *p < 0.05, nonparametric Mann-Whitney test.
(G) Immunofluorescence image of a Csf1op AV node stained for CD68 (green) and HCN4 (red).
(H) AV node effective refractory period at 120 ms pacing frequency, and pacing cycle lengths at which Wenckebach and 2:1 conduction occurred in control (n = 8) and Csf1op (n = 7) mice. Data are mean ± SEM, 4 independent experiments, *p < 0.05, nonparametric Mann-Whitney test.
To determine how macrophage-specific Cx43 deletion affects AV nodal function, we performed an in vivo electrophysiological (EP) study on Cx3cr1 Cx43−/− mice and littermate controls. The PR interval, AH interval and AV node effective refractory period were prolonged in Cx3cr1 Cx43−/− mice (Figure 6B and Table S1). We examined three additional parameters of AV nodal function including the pacing cycle lengths at which Wenckebach conduction, 2:1 conduction and ventriculo-atrial Wenckebach conduction occur. In Cx3cr1 Cx43−/− mice, each of these parameters was prolonged, a change that indicates impaired AV conduction (Figure 6B). Representative surface ECG tracings of an AV Wenckebach block in control and Cx3cr1 Cx43−/− mice are shown in Figure 6C. There is progressive PR prolongation prior to AV block, which develops at a slower pacing rate in Cx3cr1 Cx43−/− mice compared to controls. The ventricular effective refractory period was also prolonged in Cx3cr1 Cx43−/− mice (Table S1). We did not observe differences in either sinus node function or atrial refractory period (Table S1), and compromised AV conduction in Cx3cr1 Cx43−/− mice was not accompanied by altered AV node macrophage numbers (Figures 6D and 6E). These data indicate that Cx43 is required for macrophage facilitation of AV node conduction.
To explore the effect of congenital macrophage loss on AV node conduction, we performed an EP study in Csf1op mice, which lack Csf1-dependent tissue macrophages in many organs (Cecchini et al., 1994). The absence of AV node macrophages in Csf1op mice (Figures 6F and 6G) prolonged the PR interval, AV node effective refractory period as well as the pacing cycle lengths at which Wenckebach conduction and 2:1 conduction occurred (Figure 6H and Table S1). Interestingly, we also observed an increase in the atrial and ventricular refractory period of Csf1op mice (Table S1).
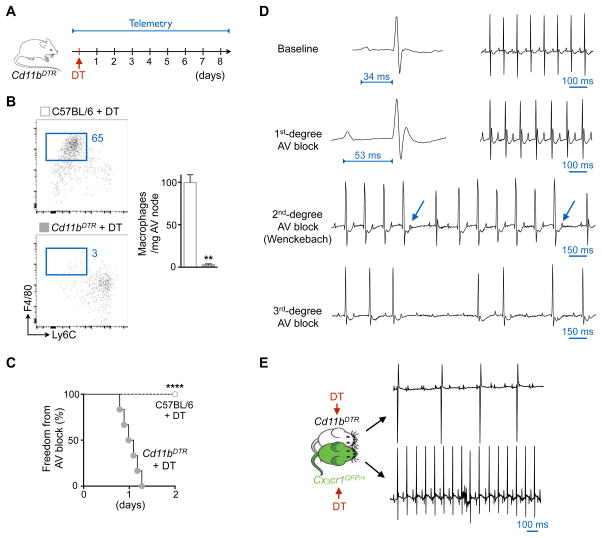
Macrophage Ablation Induces AV Block
Cd11bDTR mice express a diphtheria toxin (DT)-inducible system controlled by the human CD11b promoter that enables efficient depletion of myeloid cells, including resident cardiac macrophages (Heidt et al., 2014). We monitored these mice continuously by implantable ECG telemetry after macrophage ablation (Figure 7A). In the heart, myeloid cell expression of CD11b and CX3CR1 correlates closely, with the exception of neutrophils (Figure S7A). Maximum depletion of AV node macrophages happened three days after a single dose of 25 ng/g body weight DT (Figure 7B). Within one day of DT injection, all mice monitored by telemetry developed first degree AV block (Figure 7C) that progressively evolved into second and third degree AV block (Figure 7D). Complete AV block coincided with the time point of peak AV node macrophage depletion. Lower DT doses led to less pronounced AV node macrophage depletion and a lower incidence of AV block (Figure S7B). AV block after macrophage depletion in Cd11bDTR mice has not been previously reported, including in our own prior work (Heidt et al., 2014), since ECG is not commonly monitored in immunological studies.
Figure 7. Macrophage Ablation Induces AV Block.
(A) Experimental outline. DT, diphtheria toxin.
(B) Flow cytometric quantification of AV node macrophages three days after intraperitoneal injection of DT (25 ng/g) into C57BL/6 and Cd11bDTR mice. Data are mean ± SEM, n = 6 mice per group, **p < 0.01, nonparametric Mann-Whitney test.
(C) Onset of first degree AV block in Cd11bDTR (n = 6) and C57BL/6 (n = 10) animals after DT injection (DT dose: 25 ng/g, 2 independent experiments, ****p < 0.0001, Mantel-Cox test).
(D) Telemetric ECG recordings before and after DT injection (25 ng/g) in Cd11bDTR mice. Arrows indicate non-conducted P waves in second degree AV block.
(E) Surface ECG of Cd11bDTR and Cx3cr1GFP/+ parabionts three days after DT injection (DT dose: 25 ng/g, mean ± SEM, n = 3 parabiosis pairs).
See also Figure S7 and Tables S2 and S3.
To determine whether the observed phenotype resulted from DT-related toxicity, we injected C57BL/6 mice with DT and monitored their surface ECG. DT neither altered the number of AV node macrophages in C57BL/6 mice (Figure 7B) nor induced AV block (Figure 7C). Moreover, DT did not induce AV block in Cx3cr1GFP/+ mice joined in parabiosis with Cd11bDTR mice, which did develop AV block while in parabiosis, thereby indicating that circulating factors do not contribute to the observed phenotype (Figure 7E). At the time of complete AV block, we did not observe myocyte death (Figure S7C) or fibrosis (Figure S7D) in AV nodes of Cd11bDTR mice. Because blood electrolyte levels may influence conduction, we measured serum potassium and magnesium levels, which were unchanged in mice with AV block (Figure S7E). Injections of isoproterenol, epinephrine and atropine did not attenuate the AV block (Figure S7F). This suggests the AV block induced by macrophage ablation did not result from imbalanced autonomic nervous control.
When we pursued macrophage depletion with clodronate liposomes (Epelman et al., 2014), flow cytometry of microdissected AV nodes indicated incomplete macrophage depletion (37% decrease in AV node macrophages). We observed no AV node conduction abnormalities via either ECG telemetry or EP study (data not shown). The absence of an AV node phenotype when using clodronate liposomes is likely due to the incomplete depletion of tissue-resident macrophages in the AV node.
Three loss-of-function experiments indicate that macrophages facilitate AV node conduction; however, the observed phenotypes differ in their severity. To better understand the observed differences, we used RNA-seq to compare the whole transcriptome of AV node tissue microdissected from control, Cx3cr1 Cx43−/− and macrophage-depleted Cd11bDTR hearts. The transcriptional profile of Cx3cr1 Cx43−/− AV nodes resembled control nodal tissue with only four genes significantly dysregulated while macrophage depletion led to a distinct expression profile characterized by 1,329 differentially expressed genes (FDR < 0.05; Figure S7G and Tables S2 and S3). Genes associated with cardiac conduction are expressed at lower levels in macrophage-depleted AV nodes than in controls (Figure S7H). Thus, deletion of Cx43 in macrophages had mild effects, while depleting the cells changed the AV node expression profile, and consequently its function, more drastically. These data suggest that AV node macrophages engage in additional, Cx43-independent tasks that may or may not be related to conduction.
DISCUSSION
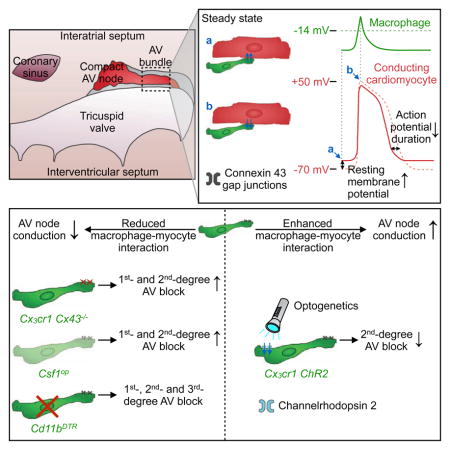
The presence of numerous resident macrophages in the normal myocardium has only recently gained recognition, a development aided by flow cytometry staining of cell surface marker combinations in tissue and macrophage-specific expression of fluorescent proteins (Pinto et al., 2012; Epelman et al., 2014; Heidt et al., 2014). Here, we employed optical clearing in combination with either cell-specific reporter gene expression or surface marker staining to image macrophages in whole AV nodes, thus documenting intra-organ macrophage heterogeneity in mice and humans. Moreover, we show that macrophages couple electrically to cardiomyocytes in the distal AV node via Cx43-containing gap junctions. The presence of Cx43-containing gap junctions in AV node tissue has previously been reported in humans, rabbits and rats (Hucker et al., 2008; Nisbet et al., 2016; Yoo et al., 2006). Cx43 shows relatively little expression within the compact node but is observed in the lower nodal bundle (Temple et al., 2013). This Cx43-expressing region of the distal AV node contains a particularly dense macrophage population in mice and humans. The impaired AV node conduction we observed in macrophage-specific Cx43 knockout mice indicates the functional significance of Cx43 in the mouse AV node. While AV node function was most notably affected, we also observed conduction abnormalities in the atria and ventricles of Cx3cr1 Cx43−/− and Csf1op mice. These data parallel our observations that cardiac macrophages isolated from working myocardium express Cx43, that single AV node macrophage RNA-seq data cluster according to subsets inhabiting the left ventricular myocardium and that normal murine atrial tissues, including the SA node, contain macrophages. We speculate that macrophages may play a role in conduction abnormalities beyond the AV node, perhaps including atrial fibrillation and ischemia-induced ventricular arrhythmias. In addition to these common clinical problems, future studies should investigate macrophages’ role in inherited conduction abnormalities and in the development of the conduction system.
Cardiomyocytes drive rhythmic depolarizations in electrotonically coupled macrophages, which in turn alter the electrophysiological properties of coupled cardiomyocytes. Macrophages’ electrotonic load depolarizes resting cardiomyocytes, reduces their action potential upstroke velocity and overshoot and aids early repolarization. These alternations shorten the cardiomyocyte action potential and ultimately allow for higher rates of conducted beats. While depolarization of the resting membrane potential in working cardiomyocytes can impair excitation and conduction due to sodium channel inactivation, depolarization of AV nodal cells depends chiefly on calcium channels, which have less prominent voltage-dependent inactivation (Catterall, 2011). This phenomenon, also referred to as ‘single-sided coupling’, during which the electrotonic load effects will dominate, is supported by our experimental data. In addition, passive conduction via macrophages, as described for other non-myocytes (Kohl and Gourdie, 2014), may pass on excitatory stimuli between AV node myocytes that are not in direct electrical contact (‘double-sided coupling’). In any case, we found that optogenetic depolarization of AV node macrophages improves AV conduction. Impaired AV node conduction after i) macrophage-specific Cx43 depletion, ii) congenital lack of macrophages and iii) acute macrophage depletion adds further evidence that these cells facilitate nodal conduction.
Clinically, AV block is a common indication for pacemaker implantation, yet up to 60% of AV block cases occur for unknown reasons (Zoob and Smith, 1963). Macrophages change in phenotype and number in response to myocardial infarction (Swirski and Nahrendorf, 2013) and heart failure (Sager et al., 2016), conditions associated with sudden cardiac death as a result of ventricular arrhythmias (Bunch et al., 2007). Moreover, diabetes-induced activation of cardiac macrophages induces arrhythmias through IL-1β production (Monnerat et al., 2016). Other inflammatory diseases of the heart, including Chagas, Lyme, sarcoid and myocarditis cause conduction abnormalities. It will be important to determine if these conditions produce AV block solely by affecting cardiomyocytes and specialized conducting tissues, as is commonly assumed, or if cardiac macrophages, which rapidly adapt their phenotype to inflammatory environments, are involved. Understanding macrophages’ contributions to normal cardiac conduction and to abnormalities in heart rhythm may yield new pathophysiologic insight and suggest novel therapeutic strategies that focus on modulating macrophages.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Matthias Nahrendorf (mnahrendorf@mgh.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Humans
Human AV node tissues were obtained from fully de-identified heart specimens collected during routine autopsy of patients with no known cardiac conduction disease. Patients were 62 ± 5 years old (mean ± SEM, n = 6, 50% male) and their final diagnoses were metastatic lung cancer with multiple pulmonary embolisms; 1 to 2-day-old transmural myocardial infarction with cardiogenic shock; chronic myelomonocytic leukemia, hematopoietic stem cell transplantation with graft-versus-host disease and diffuse alveolar damage; myxoid leiomyosarcoma with pneumonia and sepsis; community acquired pneumonia and diffuse alveolar damage; and leiomyosarcoma and pneumonia. Tissue sampling was approved by the Partners Healthcare Institutional Review Board under protocol #2015P001827. All patients gave written informed consent.
Mice
Genotyping for each strain was performed as described on the Jackson Laboratory website. All experiments were performed with 8- to 40-week-old male and female animals and were carried out using age- and gender-matched groups. One- to 2-day-old male and female C57BL/6 pups were used to isolate neonatal mouse cardiomyocytes. All mice were maintained in a pathogen-free environment of the Massachusetts General Hospital animal facility, and all animal experiments were approved by the Subcommittee on Animal Research Care at Massachusetts General Hospital.
METHOD DETAILS
In vivo Interventions
Mice were put into parabiosis using either C57BL/6 and Cx3cr1GFP/+ or Cd11bDTR and Cx3cr1GFP/+ mice as described previously (Heidt et al., 2014). Tamoxifen was given as a solution in corn oil (Sigma-Aldrich) to Cx3cr1wt/CreER ChR2wt/fl or Cx3cr1wt/CreER Cx43fl/fl mice by intraperitoneal injection. Animals received 5 doses of 2 mg of tamoxifen with a separation of 24 hours between doses. Cx3cr1wt/CreER ChR2wt/fl and Cx3cr1wt/CreER Cx43fl/fl mice were analyzed 2 and 7 days post-tamoxifen treatment, respectively. Macrophage depletion was achieved by a single intraperitoneal injection of diphtheria toxin (DT; 5, 15 or 25 ng/g body weight) in Cd11bDTR mice (Heidt et al., 2014). C57BL/6 mice injected with DT were used as controls. Clodronate liposomes were kindly provided by Dr. Kory J. Lavine and contained 18 mg of clodronate per mL of liposomes. Depletion studies were performed by intraperitoneal injection of 100 μL / 30 g mouse (Epelman et al., 2014).
EP Study
EP studies were performed under general anaesthesia induced by administering 5% isoflurane driven by an oxygen source into an induction chamber. Anaesthesia was subsequently maintained with 1–2% isoflurane in 95% O2. For EP study, an octapolar catheter (EPR-800) was inserted into the right jugular vein and positioned in the right atrium and ventricle. Programmed electrical stimulation was performed using a standard protocol with 120 ms and 100 ms drive trains and single extrastimuli to measure function of the AV node and the conduction properties of atrial and ventricular tissue. The Wenckebach cycle length was measured by progressively faster atrial pacing rates. Retrograde (VA) conduction cycle length was measured by progressively slower ventricular pacing rates. Sinus node function was determined by measuring the sinus node recovery time (SNRT) following 30 seconds of pacing at three cycle lengths (120, 100 and 80 ms). SNRT was divided by the basic cycle length to adjust for the intrinsic heart rate.
Ambulatory ECG Telemetry
Continuous ambulatory ECG telemetry was performed by implanting an ETA-F10 transmitter during general anaesthesia with isoflurane. The transmitter was implanted in the abdomen and the leads were tunneled subcutaneously to the upper right and lower left chest resulting in a lead II position. Telemetry data was recorded continuously via a receiver placed under the mouse cage. Data analysis was performed using LabChart Pro software.
Surface ECG
Mice were anesthetized as described above and surface ECG was recorded using subcutaneous electrodes connected to the Animal Bio amplifier and PowerLab station (AD Instruments). The ECG channel was filtered between 0.3 and 1000 Hz and analyzed using LabChart Pro software. Atropine (1 mg/kg), epinephrine (2 mg/kg) or isoproterenol (20 mg/kg) were administered intravenously, and changes were examined before and after injection.
Optogenetics
Two days after tamoxifen treatment, Cx3cr1wt/CreER (control) and Cx3cr1wt/CreER ChR2wt/fl (Cx3cr1 ChR2) mice were euthanized and the hearts were perfused in a custom-built, horizontal perfusion bath in Langendorff mode with oxygenized Krebs-Henseleit solution containing (in mM): 118 NaCl, 4.7 KCl, 1.6 CaCl2, 1.2 MgSO4, 24.9 NaHCO3, 1.2 KH2PO4 and 11.1 Dextrose, pH 7.4 (all Sigma-Aldrich). Recording and electrical pacing electrodes were connected to the heart, and the endocardial surface overlying the AV node was exposed by carefully opening the right atrial free wall above the AV groove. Mean perfusion pressure was maintained at between 60–80 mmHg throughout the experiment and adequacy of the preparation was determined by robust return of sinus rhythm in the perfused heart and visual evidence of vigorous contraction. The location of the AV node was identified grossly under a dissecting microscope. The Wenckebach cycle length was first determined without illumination by determining the electrical stimulation atrial pacing rate at which progressive PR interval prolongation occurred, culminating in a non-conducted atrial impulse due to AV block. The heart was subsequently electrically paced at the determined Wenckebach cycle length and the AV node was subjected to alternating 10-second cycles with and without continuous AV node illumination. Continuous illumination of the exposed AV node, right atrium or LV free wall was performed using a 400 μm core fiber optic cannula coupled to a 470 nm LED (ThorLabs) at light intensities of 55.7 mW/mm2. The recorded ECG tracings were analyzed using LabChart Pro software. The average number of conducted atrial stimuli between two non-conducted impulses during rapid pacing-induced Wenckebach block was determined for each light off and on cycle.
Tissue Processing
Peripheral blood for flow cytometric analysis was collected by retro-orbital bleeding using heparinized capillary tubes (BD Diagnostic Systems) and red blood cells were lysed with 1x red blood cell lysis buffer (BioLegend). To determine electrolyte levels, blood was collected by cardiac puncture and electrolytes were measured on serum with EasyLyte PLUS analyzer (Medica). For organ harvest, mice were perfused through the LV with 10 mL of ice-cold PBS. Hearts were excised and processed as whole or subjected to AV node microdissection as described previously (Moskowitz et al., 2007). Briefly, the triangle of Koch, which contains the AV node, was excised by using the following landmarks: ostium of the coronary sinus, tendon of Todaro and septal leaflet of the tricuspid valve. The presence of the AV node was confirmed with HCN4 and acetylcholinesterase staining (see below). After harvest, cardiac tissues were minced into small pieces and subjected to enzymatic digestion with 450 U/mL collagenase I, 125 U/mL collagenase XI, 60 U/mL DNase I, and 60 U/mL hyaluronidase (all Sigma-Aldrich) for 20 minutes (microdissected AV node, LV free wall and RV free wall) or 1 hour (whole heart) at 37°C under agitation. Tissues were then triturated and cells filtered through a 40 μm nylon mesh (Falcon), washed and centrifuged to obtain single-cell suspensions. Peritoneal cells were recovered by lavage with 5 mL of ice-cold PBS supplemented with 3% fetal bovine serum and 2 mM EDTA.
Flow Cytometry
Isolated cells were first stained at 4°C in FACS bu ffer (PBS supplemented with 0.5% bovine serum albumin) with mouse hematopoietic lineage markers including phycoerythrin (PE)- or biotin-conjugated anti-mouse antibodies directed against B220 (1:600), CD49b (1:1200), CD90.2 (1:3000), Ly6G (1:600), NK1.1 (1:600) and Ter119 (1:600). This was followed by a second staining for CX3CR1 (1:600), CD11b (1:600), CD11c (1:600), CD31 (1:400), CD45 (1:600), CD64 (1:600), CD103 (1:600), CD115 (1:600), F4/80 (1:600), Ly6C (1:600), MEFSK4 (1:20) and/or Pacific Orange-conjugated streptavidin (1:600). Monocytes were identified as (B220/CD49b/CD90.2/Ly6G/NK1.1/Ter119)low CD11bhigh CD115high Ly6Clow/high. Cardiac macrophages were identified as (B220/CD49b/CD90.2/CD103/Ly6G/NK1.1/Ter119)low CD45high CD11bhigh Ly6Clow/int F4/80high and cardiac fibroblasts as CD31low CD45low MEFSK4high. Data were acquired on an LSRII (BD Biosciences) and analyzed with FlowJo software.
Cell Sorting
To isolate peritoneal macrophages, depletion of undesired cells including lymphocytes was performed using MACS depletion columns according to the manufacturer’s instructions (Miltenyi Biotec). Briefly, single cell suspensions after peritoneal lavage were stained using a cocktail of PE-conjugated antibodies directed against B220, CD49b, CD90.2, NK1.1 and Ter119, followed by incubation with anti-PE microbeads. The enrichment of peritoneal macrophages was evaluated by flow cytometry. To purify macrophages from AV node, LV free wall or RV free wall tissues, digested samples were stained with hematopoietic lineage markers, CD11b, CD45, F4/80 and Ly6C, and macrophages were FACS-sorted using a FACSAria II cell sorter (BD Biosciences). DAPI was used as a cell viability marker. To isolate cardiac macrophages and fibroblasts from whole heart for qPCR analysis, digested tissue samples were stained with hematopoietic lineage markers, CD11b, CD31, CD45, DAPI, F4/80, Ly6C and MEFSK4, and FACS-sorted using a FACSAria II cell sorter. To isolate cardiac macrophages from UbcGFP hearts for in vitro experiments, digested tissue samples were first enriched for CD11b+ cells using CD11b microbeads and MACS columns according to the manufacturer’s instructions. Next, cells were stained with hematopoietic lineage markers, CD45.2, F4/80 and Ly6C, and FACS-sorted using a FACSAria II cell sorter.
Isolation and Culture of Neonatal Mouse Cardiomyocytes
Neonatal mouse cardiomyocytes were isolated by use of enzymatic dissociation. One- to 2-day-old pups were sacrificed, the hearts removed and the ventricles harvested. The tissue was dissociated in HBSS containing 0.1% trypsin (Sigma-Aldrich) overnight at 4°C under agitation, followed by three consecutive digestion steps in HBSS containing 335 U/mL collagenase II (Worthington Biochemical Corporation) for 2 minutes at 37°C with gentle agitation. The digest was filtered through a 40 μm nylon mesh, washed and resuspended in mouse culture medium which consisted of DMEM supplemented with 14% FBS and 2% penicillin/streptomycin. Cell suspensions were preplated into 100 mm cell tissue culture dishes and incubated at 37°C for 45 minutes to allow preferential attachment of non-myocyte cell populations and enrichment of the cardiomyocyte population. For whole-cell patch clamp and voltage dye imaging, cardiac cells remaining in suspension were collected and seeded at a density of 0.2–1x105 cells/cm2 on 5 or 8 mm cover slips (Warner Instruments) stamped with fibronectin in a line pattern and pre-seeded with 5x104 FACS-purified GFP+ cardiac macrophages. For real-time acquisition microscopy, cardiomyocytes were seeded at a density of 1x104 cells/cm2 on 35 mm glass-bottom dishes (MatTek Corporation) stamped with fibronectin in a line-pattern and pre-seeded with 7x104 FACS-purified GFP+ cardiac macrophages. Medium exchanges were performed on the first day after seeding and every other day thereafter with mouse culture medium supplemented with 1 μM cytosine β-D-arabinofuranoside hydrochloride (Sigma-Aldrich). Experiments were performed on day 3.
Whole-cell Patch Clamp
Membrane potentials were recorded with whole-cell patch clamp technique in tight-seal current-clamp mode at 37°C. Borosilicate-glass electrodes f illed with pipette solution had 4 to 6 MΩ tip resistance, and were connected with an Axopatch 200B amplifier and a Digidata 1440A A/D converter. Data were analyzed with Clampfit. The bath solution contained (in mM): 136 NaCl, 5.4 KCl, 1.8 CaCl2, 1 MgCl2, 0.33 NaH2PO4, 10 Dextrose and 5 HEPES, pH 7.4; and the pipette solution contained (in mM): 110 K-aspartate, 20 KCl, 1 MgCl2, 5 MgATP, 0.1 GTP, 10 HEPES, 5 Na-Phosphocreatine and 0.05 EGTA, pH 7.3 (all Sigma-Aldrich). To identify the patched cell, the pipette was additionally loaded with 0.2 mg/mL Texas Red+ dextran (MW 3000). To block Cx43-mediated gap junction communication, 200 μM of the Cx43-mimetic peptide Gap26 was added to the bath solution during patch clamp recording.
Voltage Dye Imaging
Cardiomyocyte-macrophage co-cultures were loaded with 4 μM of ANNINE-6plus for 5 minutes in Tyrode’s solution containing (in mM): 140 NaCl, 5.4 KCl, 1.8 CaCl2, 1 MgCl2, 10 glucose and 10 HEPES, pH 7.4 (all Sigma-Aldrich). After washing, cover slips were transferred to Tyrode’s solution containing 20 μM of blebbistatin to uncouple the excitation-contraction process in cardiomyocytes. To optically detect action potentials, line scans were obtained from the surface membranes of cardiomyocytes and attached macrophages using an Olympus IV100 microscope. The acquired line scans were filtered with a collaborative filter to increase the signal-to-noise ratio and analyzed in Matlab as previously described (Bu et al., 2009). In detail, the average signal intensity of each successive line in the line scan image corresponding to the membrane of the cell of interest was calculated to obtain the time course of the averaged fluorescence [F(t)]. The time course of normalized fractional fluorescence changes [ΔF/F0(t)], where ΔF is F(t) – F0(t) and F0(t) is the baseline trace, was subsequently determined for the cardiomyocyte and attached macrophage. To block Cx43-mediated gap junction communication, 200 μM of the Cx43-mimetic peptide Gap26 was added to the Tyrode’s solution during line scan imaging.
Real-Time Acquisition Microscopy
Cardiomyocyte-macrophage co-cultures were imaged using an Olympus VivaView FL incubator microscope in stream acquisition mode. The acquired images were processed using MetaMorph software.
Histology
Immunofluorescence Staining
To eliminate blood contamination, hearts were perfused with 10 mL of ice-cold PBS. Hearts from Cx3cr1GFP/+ mice were fixed using periodate-lysine-paraformaldehyde (PLP) for 1 hour at room temperature, washed in PBS and embedded in OCT compound. Hearts from Cx3cr1 ChR2, Cx3cr1 Cx43−/−, Csf1op and Cd11bDTR mice were directly embedded in OCT. Serial frozen 6 to 25 μm sections were prepared and acetylcholinesterase staining was carried out to identify the AV node. The selected sections from Cx3cr1GFP/+ hearts were fixed with 4% PFA for 5 minutes, washed in PBS and then blocked with 4% normal goat or rabbit serum in PBS for 30 minutes at room temperature. For F4/80 staining, sections were additionally treated with 0.3% Triton X-100 in PBS for 30 minutes at room temperature. After blocking, sections were incubated with a rat anti-mouse CD64, rat anti-mouse CD68, rat anti-mouse MerTK, rat anti-mouse F4/80, rabbit anti-mouse collagen I, rabbit anti-mouse DDR2, rabbit anti-mouse periostin or guinea pig anti-mouse vimentin antibody overnight at 4°C, followed by a biotinylated rabbit anti-rat, goat anti-rabbit or goat anti-guinea pig IgG antibody for 45 minutes and DyLight 649-streptavidin for 30 minutes at room temperature. The sections were additionally incubated with a chicken anti-GFP antibody overnight at 4°C. Alexa F luor 488 goat anti-chicken IgY antibody was used as a secondary antibody. The selected sections from Cx3cr1 ChR2, Cx3cr1 Cx43−/−, Csf1op and Cd11bDTR hearts were fixed with 10% formalin for 5 minutes, washed and permeabilized with 0.1% Triton X-100 in PBS for 30 minutes. The tissue sections were then blocked with 4% normal goat serum in PBS for 30 minutes at room temperature. After blocking, sections were incubated with a rabbit anti-mouse HCN4 antibody (Alomone Labs) overnight at 4°C, followed by a biotinylated goat anti-rabbit IgG antibody for 45 minutes and DyLight 649-streptavidin for 30 minutes at room temperature. The sections from Cx3cr1 ChR2 hearts were additionally incubated with a chicken anti-GFP antibody overnight at 4°C. Alexa Fluor 568 goat anti-chicken IgY antibody was used as a secondary antibody. The sections from Cx3cr1 Cx43−/− and Csf1op hearts were additionally incubated with a rat anti-mouse CD68 antibody for 2 hours at room temperature. Alexa Fluor 568 goat anti-rat IgG antibody was used as a secondary antibody. TUNEL staining was performed using DeadEnd Fluorometric TUNEL system according to the manufacturer’s protocol and DAPI was applied for nuclear counterstaining. Cover slips seeded with cardiomyocytes and GFP+ FACS-purified cardiac macrophages were fixed with 4% PFA for 10 minutes at room temperature. After washing, cells were permeabilized with 0.1% Triton X-100 in PBS for 10 minutes at room temperature, washed and blocked in blocking solution (PBS containing 10% goat serum, 0.1% Tween-20 and 0.3 M glycine) for 1 hour at room temperature. Cells were then stained with rabbit anti-mouse Cx43 antibody in blocking solution for 1 hour at room temperature, followed by incubation with Alexa Fluor 647 goat anti-rabbit IgG secondary antibody for 1 hour at room temperature. After washing, cells were stained with Alexa Fluor 568 anti-desmin antibody and DAPI was applied for nuclear counterstaining. All images were captured using an Olympus FV1000 or a Nikon 80i fluorescence microscope and processed with ImageJ software.
Whole-mount Immunofluorescence Staining
AV nodes from Cx3cr1GFP/+ mice were harvested as described above and SA nodes were microdissected as part of the right atrium at the junction of the crista terminalis with the superior and inferior vena cava, and the intercaval region between the two great veins. Both nodal tissues were fixed using PLP for 1 hour at room temperature, washed in PBS, and processed as whole or embedded in 4% agarose and cut in 300 μm sections using a Pelco 101 vibratome. Tissues were then washed in 1% Triton X-100 diluted in PBS, and blocked and permeabilized in blocking solution (PBS containing 20% goat or donkey serum, 1% Triton X-100 and 0.2% sodium azide) for 1 hour at room temperature. AV and SA nodes were stained with chicken anti-GFP, rabbit anti-mouse Cx43 (AV node) or goat anti-mouse Cx40 (SA node) and rat anti-mouse HCN4 (Abcam) antibodies in blocking solution for 7 days at 4°C. After washing, samples were incubated with Alexa Fluor 488 goat anti-chicken IgY, Alexa Fluor 568 goat anti-rabbit IgG (AV node) or Alexa Fluor 568 donkey anti-goat IgG (SA node) and Alexa Fluor 647 goat anti-rat IgG secondary antibodies for 7 days at 4°C. For fibrobl ast quantification, sections were incubated with PDGFRα-APC antibody overnight at 4°C and DAPI was applied for nuclear counterstaining. Human AV node tissues were fixed using 4% PFA for 24 hours at 4°C. Tissues were washed in PBS, embedded in 4% agarose and 500 μm sections were cut using a Pelco 101 vibratome. The sections were then washed in PBS containing 2% Triton X-100 and 20% DMSO, followed by blocking and permeabilization in blocking solution (PBS containing 20% goat serum, 2% Triton X-100, 20% DMSO and 0.2% sodium azide) for 1 hour at room temperature. Tissue sections were stained with mouse anti-human CD68 (clone EBM11) or mouse anti-human CD163 and rabbit anti-human Cx43 antibodies in blocking solution for 7 days at 4°C. After washing, samples were incubated with Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 568 goat anti-rabbit IgG secondary antibodies for 7 days at 4°C. To visualize nuclei and delineate cells in mouse and human AV nodes, tissues were incubated with DAPI and the membrane stain Alexa Fluor 647 WGA overnight at 4°C . Stained tissues were then optically cleared and imaged.
Optical Clearing
Mouse and human tissues were cleared using RapiClear 1.49 by immersion in the clearing solution for 1 hour at room temperature. The cleared tissues were then mounted on a custom-made sample holder and imaged using an Olympus FV1000 microscope. Collagen deposition in human tissues was investigated with second-harmonic generation (SHG) in multiphoton microscopy. Acquired images were processed with Amira 3D or ImageJ software.
Immunohistochemistry
Human and mouse AV node samples were stained with Masson’s trichrome to identify the cardiac conduction tissue and to determine collagen content after macrophage depletion in Cd11bDTR mice. Briefly, sections were fixed with 10% formalin for 30 minutes at room temperature and incubated in Bouin’s fixative solution overnight at room temperature. The nuclei were stained with the mixture of Weigert’s iron hematoxylin solution A and B, and Masson’s trichrome staining was carried out according to the manufacturer’s instructions. To identify human cardiac macrophages, the paraffin-embedded tissue was first deparaffinized and antigen retrieval was performed using sodium citrate, pH 6.0 (BD Biosciences). In order to block endogenous peroxidase activity, the tissue sections were incubated in 1% H2O2 diluted in dH2O for 10 minutes and rinsed in dH2O and PBS. The sections were then blocked with 4% horse serum in PBS for 30 minutes at room temperature and incubated with a mouse anti-human CD68 antibody (clone: KP1) overnight at 4°C. A biot inylated horse anti-mouse IgG antibody was applied for 30 minutes at room temperature. For color development, the VectaStain ABC kit and AEC substrate were used. All the slides were counterstained with Harris hematoxylin and scanned with NanoZoomer 2.0-RS (Hamamatsu). Sections were analyzed at 20x magnification using ImageJ or iVision software.
Electron Microscopy
Hearts from Cx3cr1GFP/+ mice were fixed using PLP solution and frozen 50 μm sections were incubated in 0.3% H2O2 diluted in PBS for 10 minutes, followed by incubation with PBS containing 1% BSA and 0.05% saponin for 1 hour at room temperature. A rabbit anti-GFP antibody was applied to the sections and incubated overnight at 4°C. The tissue sections were washed and incubated with a biotinylated goat anti-rabbit IgG antibody for 2 hours at room temperature. After washing, sections were incubated with VectaStain ABC reagent for 30 minutes at room temperature, washed and then fixed with PBS containing 1% glutaraldehyde and 5% sucrose for 30 minutes at room temperature. For color development, diaminobenzidine solution was applied followed by 1% H2O2 in dH2O. The sections were washed and incubated with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer on ice for 30 minutes. Prior to embedding, sections were dehydrated and allowed to pre-infiltrate in a 1:1 mix of Eponate resin and propylene oxide overnight at room temperature with gentle agitation. Sections were then infiltrated with fresh 100% Eponate resin and polymerized for 1–2 days at 60°C. Polymerized sections were trimmed and oriented such that the targeted AV node region would lie at the sectioning face. Thin sections were cut using a Leica EM UC7 ultramicrotome, collected onto formvar-coated grids, stained with uranyl acetate and Reynold’s lead citrate and examined in a JEOL JEM 1011 transmission electron microscope at 80 kV. Images were collected using an AMT digital imaging system (Advanced Microscopy Techniques).
YFP Target-to-Background Ratio (TBR) Measurement
Cx3cr1wt/wt and Cx3cr1wt/CreER mice were intravenously injected with 4 μg of CX3CR1-PE and Sca1-APC antibodies to label tissue-resident macrophages and endothelial cells, respectively. After 30 minutes of in vivo labeling, mice were perfused through the LV with 10 mL of ice-cold PBS. Hearts were then mounted between two long coverslips and imaged using an Olympus IV100 microscope. Z-stack images acquired at 1 μm steps were analyzed in Matlab with custom developed functions. Semi-automatic thresholding-based algorithms were used for TBR measurements. A BM3D filter method was implemented for noise reduction to increase the overall signal-to-noise ratio.
Western Blot
Total protein was extracted from heart tissue in RIPA lysis buffer supplemented with a protease/phosphatase inhibitor cocktail. Protein concentration was measured using a BCA assay. Lysates of 3 μg were then subjected to electrophoresis using the NuPAGE Novex Gel system (Thermo Fisher Scientific) and were blotted to nitrocellulose membrane using the iBlot Gel Transfer system (Thermo Fisher Scientific) according to the manufacturer’s instructions. Anti-mouse Cx43 antibody, anti-mouse GAPDH antibody and HRP-coupled secondary antibodies were used. Signals were visualized with chemiluminescent substrate and densitometric analysis was performed with ImageJ.
PCR Confirmation of the Deletion of the Cx43 Allele
Genomic DNA from FACS-purified cardiac macrophages was isolated with the DNeasy Blood & Tissue kit and used in PCR with two pairs of Cx43-specific primers for detecting Cx43fl or Cx43wt alleles, and for detecting the Cx43 allele lacking the floxed fragment. To normalize the amount of input DNA, specific primers to the Cx3cr1wt gene were used.
qPCR
Total RNA from whole AV node tissue was extracted using the RNeasy Micro kit or from FACS-purified cells using the PicoPure RNA isolation kit according to the manufacturer’s protocol. First-strand cDNA was synthesized using the High-Capacity RNA-to-cDNA kit and pre-amplified using the TaqMan PreAmp Master Mix kit according to the manufacturer’s instructions. TaqMan gene expression assays were used to quantify target genes. The relative changes were normalized to Gapdh mRNA using the 2−ΔΔCT method.
Bulk RNA-Seq
Total RNA from whole AV node tissue was extracted using the RNeasy Micro kit according to the manufacturer’s protocol. The RNA quality was assessed with the RNA 6000 Pico assay kit using the Agilent Bioanalyzer. Sequencing-ready cDNA libraries were prepared using the NEBNext Ultra RNA Directional Library Prep kit for Illumina following the manufacturer’s protocol. Bioanalyzer traces were used to confirm library size distribution. The libraries were quantified by qPCR using the KAPA Library Quantification kit and then sequenced as single-end 50 base reads on a Illumina HiSeq 2000 in high-output mode.
Single-Cell RNA-Seq
AV node macrophages were FACS-purified from whole AV node tissue as described above, which included two singlet gates to avoid collection of doublets. Single macrophages were then captured using the Fluidigm C1 microfluidic chip designed for 5 to 10 μm cells according to the manufacturer’s protocol. A concentration of 1.8x105 cells per mL was used for chip loading. After cell capture, chips were examined visually to identify empty chambers and chambers containing more than one cell, which were excluded from later analysis. Cell lysis and cDNA synthesis were performed on-chip with the SMARTer Ultra Low RNA kit for the Fluidigm C1 system. Amplified cDNA was validated and quantified on an Agilent Bioanalyzer with the High Sensitivity DNA chip. Illumina libraries were then constructed in 96-well plates using the Nextera XT DNA Sample Preparation kit according to a modified protocol supplied by Fluidigm. Constructed libraries were validated and quantified with the High Sensitivity DNA chip, and subsequently normalized and pooled to equal concentrations. The pooled libraries were quantified by qPCR and sequenced as single-end 50 base reads on a Illumina HiSeq 2000 in high-output mode.
RNA-Seq and Microarray Data Analysis
Bulk RNA-Seq
Transcriptome mapping was performed with STAR v2.3.0 (Dobin et al., 2013) using the Ensembl 67 release exon/splice-junction annotations. Approximately 65–78% of reads mapped uniquely. Read counts for individual genes were calculated using the unstranded count feature in HTSeq v0.6.0 (Anders et al., 2015). Differential expression analysis was performed using the exactTest routine of the edgeR R package (Robinson et al., 2010) after normalizing read counts and including only those genes with counts per million (cpm) > 1 for two or more replicates. Differentially expressed genes were then defined as those genes with > 2-fold change in expression and false discovery rate (FDR) < 0.05. Hierarchical clustering of differentially expressed genes was performed with the heatmap.2 function in the R gplots library. Gene set enrichment analysis (GSEA) was performed as described previously (Mootha et al., 2003). Input rankings were based on the sign of the fold change multiplied by the inverse of the p value. Genes involved in cardiac conduction (gene ontology term GO:0061337, 38 unique members) were downloaded from the QuickGO Browser (http://www.ebi.ac.uk/QuickGO/).
Single-Cell RNA-Seq
Transcriptome mapping (73–87% reads were uniquely mapped) and counts per gene calculations were performed in the same manner as with the bulk RNA-seq data. The 76 cells with the most reads (260K – 6.3M, median 2.1M) were selected for further analysis. Expression thresholding for detected genes and calculation of overdispersion (i.e., higher than expected variance) was performed with SCDE (Kharchenko et al., 2014) using the clean.counts and pagoda.varnorm routines, respectively, which resulted in 9,235 genes retained for further analysis. Hierarchical clustering of the 200 most overdispersed genes was performed using the heatmap.2 function in the R gplots library. To group cells into three co-expression categories based on H2 and Ccr2 expression levels, we performed spectral clustering on their joint distribution based on log2(cpm) values (specc command in the factoextra R library). Then, the two clusters with lowest average H2 expression were joined to form a larger cluster shown in orange in Figure S2B. Raw single-cell RNA-seq data of ventricular cardiomyocytes were downloaded from Gene Expression Omnibus database, accession number GSE56638 (Dueck et al., 2015), and normalized using the cpm() routine in edgeR (Robinson et al., 2010).
Microarray
Raw microarray data from (Pinto et al., 2012) were downloaded from ArrayExpress (www.ebi.ac.uk/arrayexpress), accession number E-MEXP-3347, and normalized using the robust multi-array average (Irizarry et al., 2003). GSEA was performed using standard parameters (gene set permutation, signal-to-noise ratio as a ranking metric).
Computational Modeling
Macrophages were modeled as unexcitable cells based on a previously published model (Sachse et al., 2008), which was adjusted using the experimental whole-cell patch clamp data recorded for cardiac macrophages in this study (Figures 4G, 4H, S4F and S4G). The resulting macrophage model comprises an inwardly rectifying potassium current and an unspecific background current. Table S4 shows the constants of the model. Potassium concentrations were set to match experimental conditions. The remaining parameters Cm, Gb, and GKir were fitted to the experimental whole-cell patch clamp data. The membrane capacitance of the model, Cm, was set to the mean of the measured macrophage membrane capacitances (n = 18). The conductance of the unspecific background current, Gb, was set to the inverse of the mean of measured membrane resistances (n = 9). Finally, the maximal conductance of the potassium channel, GKir, was adapted such that the resulting resting membrane potential matched the measurements (n = 20). The resulting resting membrane potential also served as initial value for the membrane potential Vm of the model. A mathematical model of a rabbit AV bundle cardiomyocyte (Inada et al., 2009) was adapted to mouse cells to be able to estimate the effects of macrophage coupling to an AV bundle cardiomyocyte. The rabbit model was modified such that the action potential duration (APD90) was reduced from 48 ms to 30 ms, a physiological value for mouse atrial cardiomyocytes (Xu et al., 1999). For this purpose, we introduced two scaling factors for the time constants of gating variables that correspond to the currents ICa,L, and Ito. Namely, in the altered model it is for ∈ {d, r, pi} where is the corresponding original value from the unaltered model. The resulting scaling factors of the modified model were sd = 0.5182 and sr = 7.0239.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were conducted with GraphPad Prism software. Data are presented as mean ± SEM. The sample size for each experiment and the replicate number of experiments are reported in the text, Figures and Figure Legends. The data were tested for normality using the D’Agostino-Pearson normality test. Statistical significance was assessed by the two-sided Student’s t test for normally distributed data. If normal distribution assumption was not valid, statistical significance was evaluated using the two-sided Mann-Whitney test or the two-sided Wilcoxon rank-sum test. For multiple comparisons, nonparametric Kruskal-Wallis tests followed by Dunn’s posttest were performed. The Mantel-Cox test was used to compare onset of AV block in DT-treated mice. P values of 0.05 or less were considered to denote significance. Animal group sizes were as low as possible and empirically chosen. No statistical methods were used to predetermine sample size and animals were randomly assigned to treatment groups.
DATA AND SOFTWARE AVAILABILITY
Data Resources
The transcriptome sequencing data for whole AV node tissues and all single cells have been deposited in the Gene Expression Omnibus database under ID codes GSE86306 and GSE86310, respectively.
Supplementary Material
(A) Immunofluorescence images of GFP+ macrophages (green) in the AV bundle of a Cx3cr1GFP/+ mouse stained with WGA (red) and DAPI (blue). Arrowheads indicate GFP+ macrophages closely interacting with a conducting cardiomyocyte (dashed cell).
(B) Volumetric reconstruction of confocal microscopy after optical clearing of the sinoatrial (SA) node in a Cx3cr1GFP/+ mouse stained with Cx40 (red) and HCN4 (white). (Top) The dashed area delineates the SA node, which is orientated along the intercaval region between the superior and inferior vena cava; (bottom) higher magnification images of the inset in the top image.
(C) Electron microscopy images of a DAB+ macrophage in the proximity of the AV node of a Cx3cr1GFP/+ mouse stained with a primary antibody for GFP.
(D) Expression of CD64, CX3CR1, CD11c and CD103 on AV node and LV free wall macrophages. Representative histograms of 4 C57BL/6 mice are shown. Gray, isotype control antibody.
(E) Representative flow cytometry plots illustrating GFP expression in cardiac macrophages and fibroblasts in C57BL/6 (black) and Cx3cr1GFP/+ (green) mice. RMCs, resident mesenchymal cells.
(F and G) Macrophage-(F) and fibroblast-specific (G) gene expression by qPCR in FACS-purified cardiac CX3CR1+ cells, macrophages and fibroblasts (n = 6 mice per group). CX3CR1+ cardiac cells (green bars) were sorted from Cx3cr1GFP/+ mice; cardiac macrophages (grey bars) and fibroblasts (black bars) were sorted from C57BL/6 mice according to the gating strategy shown in (E).
(H and I) Representative immunofluorescence images of the AV node of a Cx3cr1GFP/+ mouse stained for macrophage- (H) and fibroblast-specific (I) markers and nuclei (blue).
n = 3 per group. cmp, counts per million; FC, fold change; FDR, f alse discovery rate.
(A) Unsupervised clustering of macrophage- and cardiomyocyte-specific gene expression levels (cpm, counts per million) from RNA-seq data of 19 cardiomyocytes (Dueck et al., 2015) and 76 AV node macrophages.
(B) Grouping of AV node macrophages according to their expression levels of H2 and Ccr2.
(C) Principal component (PC) analysis of 76 single-cell samples based on expression levels of overdispersed genes, color-coded according to the three subsets in (B).
(D) Variables factor map of the top 200 overdispersed genes highlighting H2 (red) and Ccr2 (blue). The arrow tip denotes the correlation coefficients of the respective gene with the first two principal components.
(E) Venn diagram illustrating the shared expression profile of conduction-related genes for the three AV node macrophage subsets by single-cell RNA-seq.
(F) Ion channel expression by qPCR in FACS-purified macrophages and whole AV node (n = 4 to 9 from 2 independent experiments).
(G) Gene set enrichment analysis shows that expression of genes involved in cardiac conduction (GO:0061337) is higher in cardiac macrophages than in brain- and spleen-derived macrophages (q value < 0.05).
(A) Immunofluorescence image of a CD163+ macrophage (green) in the central fibrous body surrounding the human AV bundle. Second-harmonic generation (SHG, magenta) in multiphoton microscopy was used for collagen visualization.
(B) Whole-mount immunofluorescence microscopy of the human AV bundle stained with CD68 (green), WGA (red) and DAPI (blue). Arrowheads indicate CD68+ macrophages closely interacting with a single conducting cardiomyocyte (star) at different z axis positions.
(A and B) Macrophage- (A) and cardiomyocyte-specific (B) gene expression by qPCR in FACS-purified macrophages and whole AV node (n = 5 to 9 from 2 independent experiments).
(C) Number of Cx43+ contact points between adjacent HCN4+ cardiomyocytes and between CX3CR1+ macrophages and HCN4+ cardiomyocytes in the AV bundle. Data are mean ± SEM, n = 27–31 in 5 mice.
(D and E) Whole-mount immunofluorescence microscopy of the human AV bundle stained with CD163 (green) and Cx43 (red). Arrowheads indicate Cx43 colocalization with macrophages; (D) autofluorescence signal (AF, white) or (E) WGA (white) and DAPI (blue) were used for visualization of cell morphology.
(F and G) Representative spontaneous recordings (F) and corresponding resting membrane potentials (G) of cardiac macrophages attached to co-cultured neonatal mouse cardiomyocytes show no activity (n = 23), irregular depolarization (n = 10) and regular depolarization (n = 10). Data are mean ± SEM from 13 independent experiments, *p < 0.05, Kruskal-Wallis followed by Dunn’s posttest.
(H) Immunofluorescence images of a GFP+ cardiac macrophage (green) and cardiomyocyte both loaded with ANNINE-6plus voltage-sensitive dye (red). Arrow and arrowhead demark the positions of simultaneous line scan data acquisition in macrophage and cardiomyocyte, respectively.
(I) Spontaneous, simultaneous recordings of action potential-related fluorescence changes (ΔF/F0) in the cardiac macrophage (green) and cardiomyocyte (red) depicted in (H).
(J) Resting membrane potential of solitary cardiomyocytes (n = 3) before and after adding the Cx43 inhibitor Gap26. Data are mean ± SEM from 3 independent experiments, Wilcoxon rank-sum test.
(K) Spontaneous, simultaneous recordings of action potential-related fluorescence changes (ΔF/F0) in a cardiac macrophage (green) and cardiomyocyte (red) before and after adding the Cx43 inhibitor Gap26.
(L) Simulated membrane potential of an AV bundle cardiomyocyte uncoupled or coupled to one cardiac macrophage at increasing junctional conductance (Ggap).
(A) (Top) Representative immunofluorescence images of Cx3cr1wt/wt and Cx3cr1wt/CreER heart tissue illustrating the endogenous YFP signal (green), which is co-expressed with CreER, and additional staining for CX3CR1+ macrophages (red) and endothelial cells (ECs, white); (bottom) YFP target-to-background ratio (TBR) of cardiomyocytes and CX3CR1+ macrophages (targets) in comparison with ECs (background). Data are mean ± SEM, n = 5 to 20 z-stack images per group, **p < 0.01, nonparametric Mann-Whitney test.
(B) Representative immunofluorescence images of a Cx3cr1 ChR2 AV node showing the endogenous YFP signal (green), which is fused with ChR2, and additional staining for HCN4+ cardiomyocytes (red) and nuclei (blue).
(C) Bar graphs showing the number of conducted atrial stimuli between two non-conducted impulses of a Wenckebach period during light off and on cycles when the fiber optic cannula was focused on the right atrium or LV free wall of a Cx3cr1 ChR2 heart. Data are mean ± SEM, Kruskal-Wallis test followed by Dunn’s posttest.
(D) Diagram illustrating the mathematical model of macrophage-mediated passive action potential conduction (‘double-sided coupling’). Two strands of 10 cardiomyocytes with intercellular conductance of 167 nS are connected via one macrophage. The outer half of the proximal (left) cardiomyocyte strand is stimulated with 2 nA per cell at 3 Hz and the minimum heterocellular junctional conductance (Ggap) that can support macrophage-mediated passive conduction of sufficient amplitude to stimulate an action potential at the distal strand is determined by modeling.
(E) Minimum junctional conductance between macrophage and cardiomyocyte strands sufficient to bridge action potential propagation between cardiomyocyte strands that are connected via a single macrophage only. The required junctional conductance decreases with macrophage depolarization, i.e. the likelihood of conduction increases with a more positive macrophage resting membrane potential.
(A) PCR analysis of FACS-purified Cx3cr1wt/wt and Cx3cr1wt/CreER cardiac macrophages seven days post-tamoxifen for the presence of wild-type (Cx43wt) and conditional undeleted (Cx43fl) or deleted (Cx43Δ) Cx43 alleles.
(B) Cx43 mRNA levels in FACS-purified control and Cx3cr1 Cx43−/− cardiac macrophages by qPCR (mean ± SEM, n = 3 mice per group).
(C) Western blot showing Cx43 expression in heart tissue of control and Cx3cr1 Cx43−/− mice. Data are mean ± SEM, n = 4 mice per group, nonparametric Mann-Whitney test.
(A) Representative flow cytometry plots illustrating GFP expression in CD11b− and CD11b+ cardiac cells in C57BL/6 (black) and Cx3cr1GFP/+ (green) mice. Lin, lineage; PMNs, polymorphonuclear leukocytes.
(B) (Left) Flow cytometric quantification of AV node macrophages four days after intraperitoneal injection of different doses of DT in Cd11bDTR mice. Data are mean ± SEM, n = 5 to 16 mice per group. (Right) Degree of AV block evaluated by surface ECG in Cd11bDTR mice injected with different doses of DT.
(C) Representative immunofluorescence image of the AV node in a macrophage-depleted Cd11bDTR mouse (DT dose: 25 ng/g) stained with HCN4 and TUNEL.
(D) (Left) Representative Masson’s trichrome stain of the AV bundle in control and macrophage-depleted Cd11bDTR mice (DT dose: 25 ng/g). Dashed area indicates the AV bundle. (Right) Percentage of positive collagen staining per region of interest (ROI). Data are mean ± SEM, n = 5 to 8 mice per group, nonparametric Mann-Whitney test.
(E) Blood electrolytes of Cd11bDTR and C57BL/6 animals three days after DT injection (DT dose: 25 ng/g, mean ± SEM, n = 5 mice per group, nonparametric Mann-Whitney test).
(F) Surface ECG of Cd11bDTR mice with second and third degree AV block (DT dose: 25 ng/g) after intravenous isoproterenol, atropine or epinephrine administration. Arrows indicate non-conducted P waves in second degree AV block.
(G) Heat map of expression values (cpm) among the top 50 dysregulated genes in microdissected AV nodes of control (n = 3), Cx3cr1 Cx43−/− (n = 3) and macrophage-depleted Cd11bDTR (DT dose: 25 ng/g, n = 3) mice by RNA-seq.
(H) Gene set enrichment analysis shows that expression of genes involved in cardiac conduction (GO:0061337) is lower in macrophage-depleted AV nodes than in control AV nodes (q value < 0.0001).
Cells were grown on a glass-bottom dish coated with fibronectin in a line pattern, and real-time acquisition was performed with an Olympus VivaView FL incubator microscope at 40x magnification.
Acknowledgments
We thank M. Weglarz, M. Handley and A. Galvin for assistance with cell sorting; I. Klier for help with telemetric ECG analysis and K. Joyes for editing the manuscript. This work was funded in part by federal funds from the National Institutes of Health NS084863, HL128264, HL114477, HL117829, HL092577, HL105780 and HL096576. The General Hospital Corporation has filed a patent application (62/294,765) with the U.S. Patent and Trademark Office. The application is entitled “Targeting Macrophages to Modulate Electrical Conduction in the Heart” and names M.H., M.N. and R.W. as inventors. RNA sequencing and data analysis were performed at the Massachusetts General Hospital NextGen Core. Electron microscopy was performed in the Microscopy Core of the Center for Systems Biology/Program in Membrane Biology, which is partially supported by an Inflammatory Bowel Disease Grant DK43351 and a Boston Area Diabetes and Endocrinology Research Center (BADERC) Award DK57521. M.H. was supported by the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen. S.C. was supported by a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Program (PIOF-GA-2012-328352) and by the German Center for Cardiovascular Research (DZHK). A.D.A. was supported by the American Heart Association (14FTF20380185). H.B.S. was funded by Deutsche Forschungsgemeinschaft (SA1668/2-1). P.K. was supported by the European Research Council Advanced Grant CardioNect and the Baden Wurttemberg ‘Sonderlinie Medizin’. P.T.E was supported by an Established Investigator Award from the American Heart Association (13EIA14220013) and by the Fondation Leducq (14CVD01). M.N. was supported by the MGH Research Scholar Program.
Footnotes
AUTHOR CONTRIBUTIONS
M.H., S.C., L.X., A.D.A and K.R.K. performed experiments, collected, analyzed and discussed data. A.H., W.J.H., E.M.W., G.S., G.C., Y.I., Y.S., A.J.S., H.B.S., K.J.L., D.E.C., N.D.S., L.M., K.N. and C.V. performed experiments and collected data. G.A.F. and R.N.M. provided human tissues. H.K., C.E.S., J.G.S., R.I.S., D.B., P.L., R.W., F.K.S., P.K., C.V., D.J.M. and P.T.E. conceived experiments and discussed results and strategy. M.N. conceived, designed and directed the study. M.H. and M.N. wrote the manuscript, which was revised and approved by all authors.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel M, Wahl-Schott C, Michalakis S, Zong X. Hyperpolarization-activated cation channels: from genes to function. Physiol Rev. 2009;89:847–885. doi: 10.1152/physrev.00029.2008. [DOI] [PubMed] [Google Scholar]
- Billette J. Atrioventricular nodal activation during periodic premature stimulation of the atrium. Am J Physiol. 1987;252:H163–77. doi: 10.1152/ajpheart.1987.252.1.H163. [DOI] [PubMed] [Google Scholar]
- Bu G, Adams H, Berbari EJ, Rubart M. Uniform action potential repolarization within the sarcolemma of in situ ventricular cardiomyocytes. Biophys J. 2009;96:2532–2546. doi: 10.1016/j.bpj.2008.12.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch TJ, Hohnloser SH, Gersh BJ. Mechanisms of sudden cardiac death in myocardial infarction survivors: insights from the randomized trials of implantable cardioverter-defibrillators. Circulation. 2007;115:2451–2457. doi: 10.1161/CIRCULATIONAHA.106.683235. [DOI] [PubMed] [Google Scholar]
- Catterall WA. Voltage-gated calcium channels. Cold Spring Harb Perspect Biol. 2011;3:a003947. doi: 10.1101/cshperspect.a003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini MG, Dominguez MG, Mocci S, Wetterwald A, Felix R, Fleisch H, Chisholm O, Hofstetter W, Pollard JW, Stanley ER. Role of colony stimulating factor-1 in the establishment and regulation of tissue macrophages during postnatal development of the mouse. Development. 1994;120:1357–1372. doi: 10.1242/dev.120.6.1357. [DOI] [PubMed] [Google Scholar]
- Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol. 2013;14:986–995. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueck H, Khaladkar M, Kim TK, Spaethling JM, Francis C, Suresh S, Fisher SA, Seale P, Beck SG, Bartfai T, et al. Deep sequencing reveals cell-type-specific patterns of single-cell transcriptome variation. Genome Biol. 2015;16:122. doi: 10.1186/s13059-015-0683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, Brija T, Gautier EL, Ivanov S, Satpathy AT, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40:91–104. doi: 10.1016/j.immuni.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallin EK, Gallin JI. Interaction of chemotactic factors with human macrophages. Induction of transmembrane potential changes. J Cell Biol. 1977;75:277–289. doi: 10.1083/jcb.75.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallin EK, Livengood DR. Nonlinear current-voltage relationships in cultured macrophages. J Cell Biol. 1980;85:160–165. doi: 10.1083/jcb.85.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidt T, Courties G, Dutta P, Sager HB, Sebas M, Iwamoto Y, Sun Y, Da Silva N, Panizzi P, van der Laan AM, et al. Differential contribution of monocytes to heart macrophages in steady-state and after myocardial infarction. Circ Res. 2014;115:284–295. doi: 10.1161/CIRCRESAHA.115.303567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucker WJ, McCain ML, Laughner JI, Iaizzo PA, Efimov IR. Connexin 43 expression delineates two discrete pathways in the human atrioventricular junction. Anat Rec (Hoboken) 2008;291:204–215. doi: 10.1002/ar.20631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada S, Hancox JC, Zhang H, Boyett MR. One-dimensional mathematical model of the atrioventricular node including atrio-nodal, nodal, and nodal-his cells. Biophys J. 2009;97:2117–2127. doi: 10.1016/j.bpj.2009.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl P, Gourdie RG. Fibroblast-myocyte electrotonic coupling: does it occur in native cardiac tissue. J Mol Cell Cardiol. 2014;70:37–46. doi: 10.1016/j.yjmcc.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1+ macrophages to CD103+ dendritic cells. Immunity. 2014;40:248–261. doi: 10.1016/j.immuni.2013.12.012. [DOI] [PubMed] [Google Scholar]
- Metchnikoff É. Leçons Sur La Pathologie Comparée De L’inflammation. Paris: Masson; 1892. [Google Scholar]
- Molawi K, Wolf Y, Kandalla PK, Favret J, Hagemeyer N, Frenzel K, Pinto AR, Klapproth K, Henri S, Malissen B, et al. Progressive replacement of embryo-derived cardiac macrophages with age. J Exp Med. 2014;211:2151–2158. doi: 10.1084/jem.20140639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnerat G, Alarcón ML, Vasconcellos LR, Hochman-Mendez C, Brasil G, Bassani RA, Casis O, Malan D, Travassos LH, Sepúlveda M, et al. Macrophage-dependent IL-1β production induces cardiac arrhythmias in diabetic mice. Nat Commun. 2016;7:13344. doi: 10.1038/ncomms13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, et al. A molecular pathway including Id2, Tbx5, and Nkx2–5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neijssen J, Pang B, Neefjes J. Gap junction-mediated intercellular communication in the immune system. Prog Biophys Mol Biol. 2007;94:207–218. doi: 10.1016/j.pbiomolbio.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Nerbonne JM, Nichols CG, Schwarz TL, Escande D. Genetic manipulation of cardiac K(+) channel function in mice: what have we learned, and where do we go from here. Circ Res. 2001;89:944–956. doi: 10.1161/hh2301.100349. [DOI] [PubMed] [Google Scholar]
- Nguyen KD, Qiu Y, Cui X, Goh YP, Mwangi J, David T, Mukundan L, Brombacher F, Locksley RM, Chawla A. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480:104–108. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolski VP, Jones SA, Lancaster MK, Boyett MR, Efimov IR. Cx43 and dual-pathway electrophysiology of the atrioventricular node and atrioventricular nodal reentry. Circ Res. 2003;92:469–475. doi: 10.1161/01.RES.0000059304.97120.2F. [DOI] [PubMed] [Google Scholar]
- Nisbet AM, Camelliti P, Walker NL, Burton FL, Cobbe SM, Kohl P, Smith GL. Prolongation of atrio-ventricular node conduction in a rabbit model of ischaemic cardiomyopathy: Role of fibrosis and connexin remodelling. J Mol Cell Cardiol. 2016;94:54–64. doi: 10.1016/j.yjmcc.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongstad E, Kohl P. Fibroblast-myocyte coupling in the heart: Potential relevance for therapeutic interventions. J Mol Cell Cardiol. 2016;91:238–246. doi: 10.1016/j.yjmcc.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oviedo-Orta E, Howard Evans W. Gap junctions and connexin-mediated communication in the immune system. Biochim Biophys Acta. 2004;1662:102–112. doi: 10.1016/j.bbamem.2003.10.021. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Park DS, Fishman GI. The cardiac conduction system. Circulation. 2011;123:904–915. doi: 10.1161/CIRCULATIONAHA.110.942284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AR, Paolicelli R, Salimova E, Gospocic J, Slonimsky E, Bilbao-Cortes D, Godwin JW, Rosenthal NA. An abundant tissue macrophage population in the adult murine heart with a distinct alternatively-activated macrophage profile. PLoS One. 2012;7:e36814. doi: 10.1371/journal.pone.0036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook MB, van Ginneken AC, de Jonge B, el Aoumari A, Gros D, Jongsma HJ. Differences in gap junction channels between cardiac myocytes, fibroblasts, and heterologous pairs. Am J Physiol. 1992;263:C959–77. doi: 10.1152/ajpcell.1992.263.5.C959. [DOI] [PubMed] [Google Scholar]
- Rosas M, Davies LC, Giles PJ, Liao CT, Kharfan B, Stone TC, O’Donnell VB, Fraser DJ, Jones SA, Taylor PR. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 2014;344:645–648. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubart M, Zipes DP. Arrhythmias, sudden death and syncope. In: Libby P, Bonow RO, Mann DL, Zipes D, editors. Braunwald’s Heart Disease. Philadelphia: Suanders Elsevier; 2008. pp. 909–921. [Google Scholar]
- Sachse FB, Moreno AP, Abildskov JA. Electrophysiological modeling of fibroblasts and their interaction with myocytes. Ann Biomed Eng. 2008;36:41–56. doi: 10.1007/s10439-007-9405-8. [DOI] [PubMed] [Google Scholar]
- Sager HB, Hulsmans M, Lavine KJ, Beltrami Moreira MB, Heidt T, Courties G, Sun Y, Iwamoto Y, Tricot B, Khan OF, et al. Proliferation and recruitment contribute to myocardial macrophage expansion in chronic heart failure. Circ Res. 2016;119:853–864. doi: 10.1161/CIRCRESAHA.116.309001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Yamamoto T. Gap junctions in the cardiac muscle cells of the lamprey. Cell Tissue Res. 1977;178:477–482. doi: 10.1007/BF00219569. [DOI] [PubMed] [Google Scholar]
- Swirski FK, Nahrendorf M. Leukocyte behavior in atherosclerosis, myocardial infarction, and heart failure. Science. 2013;339:161–166. doi: 10.1126/science.1230719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawara S. Das Reizleitungssystem Des Säugetierherzens: Eine Anatomisch-Histologische Studie Über Das Atrioventrikularbündel Und Der Purkinjeschen Fäden. Jena: Verlag von Gustav Fischer; 1906. [Google Scholar]
- Temple IP, Inada S, Dobrzynski H, Boyett MR. Connexins and the atrioventricular node. Heart Rhythm. 2013;10:297–304. doi: 10.1016/j.hrthm.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurl I, Hilgendorf I, Nairz M, Tymoszuk P, Haschka D, Asshoff M, He S, Gerhardt LM, Holderried TA, Seifert M, et al. On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat Med. 2016;22:945–951. doi: 10.1038/nm.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger VM, Kumar NM, Gilula NB, Yeager M. Three-dimensional structure of a recombinant gap junction membrane channel. Science. 1999;283:1176–1180. doi: 10.1126/science.283.5405.1176. [DOI] [PubMed] [Google Scholar]
- Xu H, Li H, Nerbonne JM. Elimination of the transient outward current and action potential prolongation in mouse atrial myocytes expressing a dominant negative Kv4 alpha subunit. J Physiol. 1999;519(Pt 1):11–21. doi: 10.1111/j.1469-7793.1999.0011o.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Dobrzynski H, Fedorov VV, Xu SZ, Yamanushi TT, Jones SA, Yamamoto M, Nikolski VP, Efimov IR, Boyett MR. Localization of Na+ channel isoforms at the atrioventricular junction and atrioventricular node in the rat. Circulation. 2006;114:1360–1371. doi: 10.1161/CIRCULATIONAHA.106.613182. [DOI] [PubMed] [Google Scholar]
- Zoob M, Smith KS. The Aetiology of complete heart-block. Br Med J. 1963;2:1149–1153. doi: 10.1136/bmj.2.5366.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Immunofluorescence images of GFP+ macrophages (green) in the AV bundle of a Cx3cr1GFP/+ mouse stained with WGA (red) and DAPI (blue). Arrowheads indicate GFP+ macrophages closely interacting with a conducting cardiomyocyte (dashed cell).
(B) Volumetric reconstruction of confocal microscopy after optical clearing of the sinoatrial (SA) node in a Cx3cr1GFP/+ mouse stained with Cx40 (red) and HCN4 (white). (Top) The dashed area delineates the SA node, which is orientated along the intercaval region between the superior and inferior vena cava; (bottom) higher magnification images of the inset in the top image.
(C) Electron microscopy images of a DAB+ macrophage in the proximity of the AV node of a Cx3cr1GFP/+ mouse stained with a primary antibody for GFP.
(D) Expression of CD64, CX3CR1, CD11c and CD103 on AV node and LV free wall macrophages. Representative histograms of 4 C57BL/6 mice are shown. Gray, isotype control antibody.
(E) Representative flow cytometry plots illustrating GFP expression in cardiac macrophages and fibroblasts in C57BL/6 (black) and Cx3cr1GFP/+ (green) mice. RMCs, resident mesenchymal cells.
(F and G) Macrophage-(F) and fibroblast-specific (G) gene expression by qPCR in FACS-purified cardiac CX3CR1+ cells, macrophages and fibroblasts (n = 6 mice per group). CX3CR1+ cardiac cells (green bars) were sorted from Cx3cr1GFP/+ mice; cardiac macrophages (grey bars) and fibroblasts (black bars) were sorted from C57BL/6 mice according to the gating strategy shown in (E).
(H and I) Representative immunofluorescence images of the AV node of a Cx3cr1GFP/+ mouse stained for macrophage- (H) and fibroblast-specific (I) markers and nuclei (blue).
n = 3 per group. cmp, counts per million; FC, fold change; FDR, f alse discovery rate.
(A) Unsupervised clustering of macrophage- and cardiomyocyte-specific gene expression levels (cpm, counts per million) from RNA-seq data of 19 cardiomyocytes (Dueck et al., 2015) and 76 AV node macrophages.
(B) Grouping of AV node macrophages according to their expression levels of H2 and Ccr2.
(C) Principal component (PC) analysis of 76 single-cell samples based on expression levels of overdispersed genes, color-coded according to the three subsets in (B).
(D) Variables factor map of the top 200 overdispersed genes highlighting H2 (red) and Ccr2 (blue). The arrow tip denotes the correlation coefficients of the respective gene with the first two principal components.
(E) Venn diagram illustrating the shared expression profile of conduction-related genes for the three AV node macrophage subsets by single-cell RNA-seq.
(F) Ion channel expression by qPCR in FACS-purified macrophages and whole AV node (n = 4 to 9 from 2 independent experiments).
(G) Gene set enrichment analysis shows that expression of genes involved in cardiac conduction (GO:0061337) is higher in cardiac macrophages than in brain- and spleen-derived macrophages (q value < 0.05).
(A) Immunofluorescence image of a CD163+ macrophage (green) in the central fibrous body surrounding the human AV bundle. Second-harmonic generation (SHG, magenta) in multiphoton microscopy was used for collagen visualization.
(B) Whole-mount immunofluorescence microscopy of the human AV bundle stained with CD68 (green), WGA (red) and DAPI (blue). Arrowheads indicate CD68+ macrophages closely interacting with a single conducting cardiomyocyte (star) at different z axis positions.
(A and B) Macrophage- (A) and cardiomyocyte-specific (B) gene expression by qPCR in FACS-purified macrophages and whole AV node (n = 5 to 9 from 2 independent experiments).
(C) Number of Cx43+ contact points between adjacent HCN4+ cardiomyocytes and between CX3CR1+ macrophages and HCN4+ cardiomyocytes in the AV bundle. Data are mean ± SEM, n = 27–31 in 5 mice.
(D and E) Whole-mount immunofluorescence microscopy of the human AV bundle stained with CD163 (green) and Cx43 (red). Arrowheads indicate Cx43 colocalization with macrophages; (D) autofluorescence signal (AF, white) or (E) WGA (white) and DAPI (blue) were used for visualization of cell morphology.
(F and G) Representative spontaneous recordings (F) and corresponding resting membrane potentials (G) of cardiac macrophages attached to co-cultured neonatal mouse cardiomyocytes show no activity (n = 23), irregular depolarization (n = 10) and regular depolarization (n = 10). Data are mean ± SEM from 13 independent experiments, *p < 0.05, Kruskal-Wallis followed by Dunn’s posttest.
(H) Immunofluorescence images of a GFP+ cardiac macrophage (green) and cardiomyocyte both loaded with ANNINE-6plus voltage-sensitive dye (red). Arrow and arrowhead demark the positions of simultaneous line scan data acquisition in macrophage and cardiomyocyte, respectively.
(I) Spontaneous, simultaneous recordings of action potential-related fluorescence changes (ΔF/F0) in the cardiac macrophage (green) and cardiomyocyte (red) depicted in (H).
(J) Resting membrane potential of solitary cardiomyocytes (n = 3) before and after adding the Cx43 inhibitor Gap26. Data are mean ± SEM from 3 independent experiments, Wilcoxon rank-sum test.
(K) Spontaneous, simultaneous recordings of action potential-related fluorescence changes (ΔF/F0) in a cardiac macrophage (green) and cardiomyocyte (red) before and after adding the Cx43 inhibitor Gap26.
(L) Simulated membrane potential of an AV bundle cardiomyocyte uncoupled or coupled to one cardiac macrophage at increasing junctional conductance (Ggap).
(A) (Top) Representative immunofluorescence images of Cx3cr1wt/wt and Cx3cr1wt/CreER heart tissue illustrating the endogenous YFP signal (green), which is co-expressed with CreER, and additional staining for CX3CR1+ macrophages (red) and endothelial cells (ECs, white); (bottom) YFP target-to-background ratio (TBR) of cardiomyocytes and CX3CR1+ macrophages (targets) in comparison with ECs (background). Data are mean ± SEM, n = 5 to 20 z-stack images per group, **p < 0.01, nonparametric Mann-Whitney test.
(B) Representative immunofluorescence images of a Cx3cr1 ChR2 AV node showing the endogenous YFP signal (green), which is fused with ChR2, and additional staining for HCN4+ cardiomyocytes (red) and nuclei (blue).
(C) Bar graphs showing the number of conducted atrial stimuli between two non-conducted impulses of a Wenckebach period during light off and on cycles when the fiber optic cannula was focused on the right atrium or LV free wall of a Cx3cr1 ChR2 heart. Data are mean ± SEM, Kruskal-Wallis test followed by Dunn’s posttest.
(D) Diagram illustrating the mathematical model of macrophage-mediated passive action potential conduction (‘double-sided coupling’). Two strands of 10 cardiomyocytes with intercellular conductance of 167 nS are connected via one macrophage. The outer half of the proximal (left) cardiomyocyte strand is stimulated with 2 nA per cell at 3 Hz and the minimum heterocellular junctional conductance (Ggap) that can support macrophage-mediated passive conduction of sufficient amplitude to stimulate an action potential at the distal strand is determined by modeling.
(E) Minimum junctional conductance between macrophage and cardiomyocyte strands sufficient to bridge action potential propagation between cardiomyocyte strands that are connected via a single macrophage only. The required junctional conductance decreases with macrophage depolarization, i.e. the likelihood of conduction increases with a more positive macrophage resting membrane potential.
(A) PCR analysis of FACS-purified Cx3cr1wt/wt and Cx3cr1wt/CreER cardiac macrophages seven days post-tamoxifen for the presence of wild-type (Cx43wt) and conditional undeleted (Cx43fl) or deleted (Cx43Δ) Cx43 alleles.
(B) Cx43 mRNA levels in FACS-purified control and Cx3cr1 Cx43−/− cardiac macrophages by qPCR (mean ± SEM, n = 3 mice per group).
(C) Western blot showing Cx43 expression in heart tissue of control and Cx3cr1 Cx43−/− mice. Data are mean ± SEM, n = 4 mice per group, nonparametric Mann-Whitney test.
(A) Representative flow cytometry plots illustrating GFP expression in CD11b− and CD11b+ cardiac cells in C57BL/6 (black) and Cx3cr1GFP/+ (green) mice. Lin, lineage; PMNs, polymorphonuclear leukocytes.
(B) (Left) Flow cytometric quantification of AV node macrophages four days after intraperitoneal injection of different doses of DT in Cd11bDTR mice. Data are mean ± SEM, n = 5 to 16 mice per group. (Right) Degree of AV block evaluated by surface ECG in Cd11bDTR mice injected with different doses of DT.
(C) Representative immunofluorescence image of the AV node in a macrophage-depleted Cd11bDTR mouse (DT dose: 25 ng/g) stained with HCN4 and TUNEL.
(D) (Left) Representative Masson’s trichrome stain of the AV bundle in control and macrophage-depleted Cd11bDTR mice (DT dose: 25 ng/g). Dashed area indicates the AV bundle. (Right) Percentage of positive collagen staining per region of interest (ROI). Data are mean ± SEM, n = 5 to 8 mice per group, nonparametric Mann-Whitney test.
(E) Blood electrolytes of Cd11bDTR and C57BL/6 animals three days after DT injection (DT dose: 25 ng/g, mean ± SEM, n = 5 mice per group, nonparametric Mann-Whitney test).
(F) Surface ECG of Cd11bDTR mice with second and third degree AV block (DT dose: 25 ng/g) after intravenous isoproterenol, atropine or epinephrine administration. Arrows indicate non-conducted P waves in second degree AV block.
(G) Heat map of expression values (cpm) among the top 50 dysregulated genes in microdissected AV nodes of control (n = 3), Cx3cr1 Cx43−/− (n = 3) and macrophage-depleted Cd11bDTR (DT dose: 25 ng/g, n = 3) mice by RNA-seq.
(H) Gene set enrichment analysis shows that expression of genes involved in cardiac conduction (GO:0061337) is lower in macrophage-depleted AV nodes than in control AV nodes (q value < 0.0001).
Cells were grown on a glass-bottom dish coated with fibronectin in a line pattern, and real-time acquisition was performed with an Olympus VivaView FL incubator microscope at 40x magnification.