Abstract
Prion diseases (PrD) are unique neurodegenerative conditions that can present with sporadic, genetic, and infectious etiologies. The agent responsible for these pathologies in humans and mammals is a misfolded conformation of the prion protein (PrP), a membrane-anchored protein highly expressed in the brain. Although a process of autocatalytic “conversion” is known to mediate disease transmission, important gaps still remain regarding the physiological function of PrP and its relevance to pathogenesis, the molecular and cellular mechanisms mediating neurotoxicity and transmission, and the PrP conformations responsible for neurotoxicity. New Drosophila models expressing mammalian PrP have recently revealed physiological insight into PrP function and opened the door to significant progress in prion transmission and PrP neurotoxicity. Flies expressing human PrP with a critical robust eye phenotype will allow the identification of modifiers through genetic or small molecule screens to uncover novel mechanisms mediating neurotoxicity and pharmacological inhibitors of PrP-mediated neurodegeneration.
Keywords: Drosophila, prion protein, disease models, prionopathies, neurodegeneration
The unique world of prion diseases
Prion diseases (PrD) are rare neurodegenerative conditions that cause several distinct pathologies in humans: Kuru, Creutzfeldt-Jakob disease (CJD), Gerstmann-Straussler-Scheinker (GSS) disease, and fatal insomnia. These brain disorders receive special consideration because they can present with sporadic, genetic, and/or infectious etiologies [1]. A unique aspect of PrD is that true equivalent diseases exist in several mammals, including endemic forms in ungulates (sheep, goat, deer). One of the typical features of ALL PrD is vacuolar brain degeneration, which accounts for the all-encompassing term transmissible spongiform encephalopathies or TSE. The other common link to all PrD is brain deposition of misfolded conformations of the prion protein (PrP) in insoluble amyloid fibers, making PrD a typical member of the protein misfolding disorders or proteinopathies. Remarkably, insoluble PrP deposits are transmissible (in most cases), making PrD highly unique among other common proteinopathies like Alzheimer’s and Parkinson’s disease. The term prion refers to a proteinaceous infectious material containing PrP and other molecules responsible for TSE transmission. Remember that PrP ≠ prion, this is a concept that causes some confusion outside the TSE field. Another source of confusion is the terminology and techniques borrowed from microbiology: prion strain, prion titer, infectious agent, passage, etc. Despite these biological and “cultural” barriers (thought silos), recent studies have revealed extensive similarities among all proteinopathies, including structural and biophysical features, the identification of amyloid-β42 and tau strains [2,3], transmissibility [4–6], and prion-like cell-to-cell spread [7]. It is critical to understand how these shared properties can contribute to design more effective therapeutic approaches for a large class of proteinopathies.
One big advantage of studying PrP is that it is easy to manipulate in the lab. Most amyloids are hard to solubilize and perform protein biochemistry studies, but all PrP isoforms are easily dissolved in SDS and characterized by polyacrylamide gel electrophoresis (PAGE). Native, cellular PrP (PrPC) is monomeric and soluble, contains a signal peptide for secretion in the N-terminus and a signal for glycosylphosphatidylinositol (GPI) anchoring on the C-terminus (Fig. 1A). Thus, PrP exists mainly as a secreted, membrane anchored protein. PrP has an unstructured N-terminus with an octarepeat sequence and a globular C-terminus that contains three helices and two short β-strands (Fig. 1). Two facultative N-glycosylation sites and numerous antibodies enable the identification of distinct strains, conformations, and topologies by electrophoretic characteristics. Despite the vast information on PrP conformation and structural dynamics, there is still robust debate regarding the conformations responsible for neurotoxicity. The classic prion hypothesis suggests that the transmissible conformation of PrP [scrapie (PrPSc) or resistant (PrPres)] is also responsible for the brain pathology [1,8]. However, accumulating evidence suggests that disease transmission and neurotoxicity are caused by different PrP conformations [9–12**]. Although a distinct neurotoxic conformation(s) (PrPLethal) has not been identified so far, this is a hot topic and recent attention and technical advances may lead to its characterization. The work by Safar and cols. suggests that PrPL could be composed of oligomeric PrP conformations that act as intermediaries during PrP conversion [13]. Animal models of prionopathies with neurotoxicity in the absence of PrPSc like Drosophila may play a key role in identifying the molecular mechanisms implicated in PrD pathogenesis. Although the specific structural features of protease sensitive PrP oligomers may differ from those purified from patients, understanding their structural commonalities may contribute to identify the molecular mechanisms mediating PrP neurotoxicity.
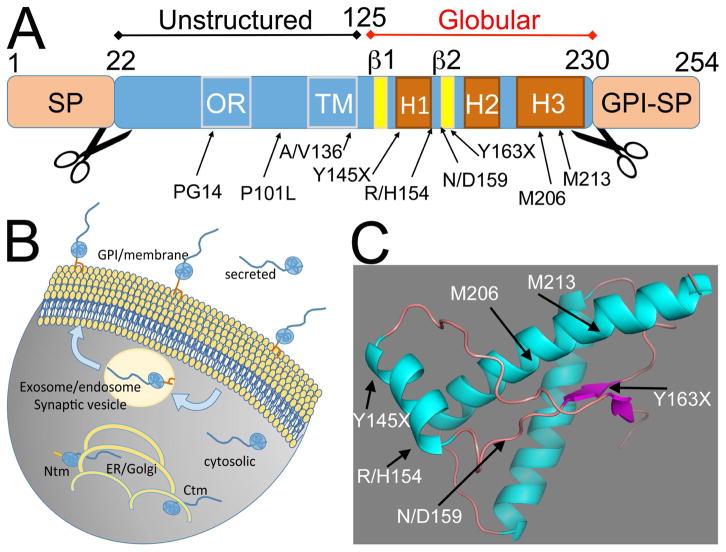
Figure 1. PrP domains, isoforms and topologies.
A, Mammalian PrP is small protein with a N-terminal signal peptide (SP) for secretion and an C-terminal signal for GPI anchoring. The mature protein has an unstructured N-terminal domain that contains an octarepeat region (OR) with five repeats and a cryptic transmembrane domain (TM). The C-terminal domain contains three helices and two short β-strands. The position of mutants described in the text are indicated in A and C. B, Subcellular distribution of PrP topologies and isoforms. Membrane PrP is attached to the external face of the membrane by a GPI anchor, but is internalized through endosomes. Secreted PrP lacks the GPI anchor and cytosolic PrP is retrotranslocated from the ER to the cytosol. The two TM topologies, Ctm and Ntm typically accumulate in the ER/Golgi. Note that Ctm is first processed in the ER and lacks the SP, but Ntm retains the SP. C, 3D model of the globular domain of hamster PrP indicating the position of the mutants described in the text.
The hard road to modeling PrD in flies
Two groups demonstrated in 1998 that the fruit fly Drosophila melanogaster could faithfully model key features of Huntington’s disease and spinocerebellar ataxia type 3 [14,15]. Since then, many other proteinopathies have been modeled in flies and used for genetic screens and testing candidate genes [16]. The Drosophila genome does not contain a PrP orthologue, making flies ideal for identifying the gain-of-function (GOF) mechanisms associated with PrP misfolding. Interestingly, a prior model expressing hamster PrP under the control of a heat shock promoter produced discouraging results [17]. The next attempt took advantage of the UAS/Gal4 system [18] to express mouse PrP either WT or carrying the pathogenic mutant PG14 (14 octarepeats), but these flies showed high turnover of mutant PrP and no neurodegenerative phenotypes [19]. A few years later, S. Supattapone created fruit flies expressing robust levels of WT and GSS-linked P101L in the mouse PrP scaffold, which showed neurodegenerative changes and misfolded PrP [20*]. We later demonstrated that robust expression of WT hamster PrP induced vacuolar brain degeneration and progressive accumulation of pathogenic conformations [21*]. The lesson from these initial studies is that WT and mutant PrP from mouse and hamster are easily cleared during biogenesis in Drosophila, but robust PrP levels enable the accumulation of PrP, and the study of conformational changes and neurotoxicity. Also, since these flies do not spontaneously produce prions based on the lack of PK-resistant PrP, work with PrP in flies should be considered relatively safe.
A more recent model introduced interesting upgrades [22]: the insertion of WT and P101L mouse PrP constructs in the same attP locus [23] to match their expression and the use of Gal80TS [24] for conditional expression in adult cholinergic neurons. This model confirmed the progressive locomotor dysfunction of both WT and P101L PrP, and reported a decrease in the frequency of action potentials in adult brains. Interestingly, halting PrP expression after two days reversed these phenotypes and PrP levels decreased over time, suggesting that Drosophila efficiently clears PrP. It will be even more interesting to replicate these experiments in flies expressing PrP for 10 or 20 days to examine the fate of insoluble, pathogenic conformations.
Is PrP a synaptic protein?
PrP is one of the most abundant proteins in the mammalian brain. Strikingly, the absence of PrP (Prnp−/−) results in viable mice with mild behavioral phenotypes [25–27]. What is the function of this abundant brain protein that mice can live without? Histological and ultrastructural studies have detected PrP in presynaptic terminals, but mammalian studies have made limited progress towards understanding the function of PrP at the synapse. Two Drosophila studies have made significant progress in this area. The first paper reported the accumulation of PrP in presynaptic terminals of motor neurons and several structural alterations due to the expression of mouse PrP-P101L, including increased sprouting and decreased expression of active zone markers [28]. A more detailed electrophysiological study suggested that PrP has profound physiological effects on synaptic activity [29**]. The authors found that expression of WT mouse PrP in the larval motor neurons had no effect on bouton morphology and active zone density. However, they described enlarged presynaptic vesicles, and increased synaptic activity and locomotor activity (Fig. 2). This activity is consistent with known interactions of PrP with several synaptic proteins, including scaffolding (laminin B), ion channels (voltage-gated Ca+ channel, VGCC), and synaptic trafficking proteins (synapsin), among others. It seems that PrP plays a role in vesicle dynamics (fusion, recycling, storage) (Fig. 2) and this physiological function would be relevant in pathogenesis because the progressive misfolding of PrP would lead to a loss-of-function (LOF) and a reduction in synaptic activity. This LOF component has been proposed for several amyloids with a known neuronal function, (e.g., tau, huntingtin, amyloid precursor protein, α-synuclein). However, the LOF hypothesis has not been proven yet in any proteinopathy. Proteinopathies are more consistent with gain-of-function mechanisms due to the accumulation of highly toxic oligomeric assemblies.
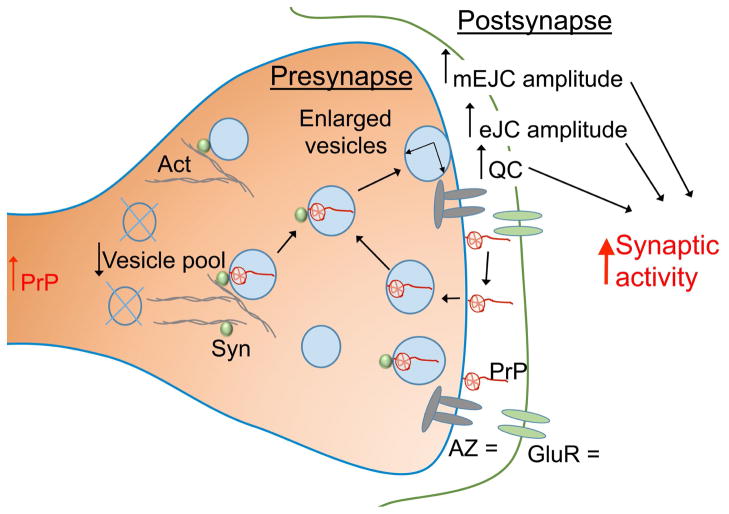
Figure 2. Potential function of PrP at the synapse.
Expression of PrP in Drosophila motor neurons alters synaptic physiology and result in synaptic activity and motor activity. PrP is abundant at synapses, particularly in the membrane and in synaptic vesicles, where it is known to interact with synapsin (Syn). The interaction of Syn with the actin cytoskeleton controls the vesicle pool available for release. Synaptic vesicles are fused with the synaptic membrane to release neurotransmitters and are recycled later to reuse the same neurotransmitters. PrP is recycled in a similar way and may play a role in the process of synaptic vesicle recycling (arrows). The amount and distribution of active zones (AZ), the site of release of neurotransmitters, and Glutamate receptors (GluR) are not affected by PrP. ↑ indicates enhanced activity, ↓ indicates decreased activity, = indicates no effect.
The PrP zoo
PrP is a highly conserved protein in mammals and small changes in its sequence may alter PrP pathogenic potential. In fact, a wide spectrum of TSE susceptibility in wild animals may reveal key insight into PrP conformational dynamics and pathogenesis. Sheep, goats, and cervids develop endemic forms of PrD (scrapie and chronic wasting disease [CWD]). Bovine spongiform encephalopathy (BSE) seems to originate from scrapie-contaminated bone meal, although anecdotal cases of endemic BSE have been reported. Several other mammals, including rodents, felines, and mustelids (e.g., ferrets), are also susceptible to PrD transmitted in the lab or in zoos during the “mad-cow” epidemics of the 1980’s. On the other end of the spectrum, rabbits, dogs, and horses seem to be resistant to PrD. Rabbits were inoculated with prions in classic studies [30], and dogs and horses were exposed to the same contaminated feed as other domestic and farm animals, but no single case of TSE has been reported for these well-cared animals. We expressed WT rabbit PrP in Drosophila to demonstrate that a few changes in the sequence of PrP conferred structural stability and prevented neurotoxicity [31, 32]. In a more recent study, we demonstrated that an amino acid only present in PrP from dog and other canids (D158) prevented the accumulation of pathogenic conformations and suppressed neurotoxicity in mouse PrP-N158D [33*]. The side chain of D159 (dog PrP scaffold) faces outward and changes the surface charge, which is proposed to alter PrP interactions with other factors [34]. These studies indicate that natural variants in mammalian PrP can reveal key clues to understand PrP misfolding and susceptibility to TSE.
The broad spectrum of natural PrP variants enabled the Bujdoso group to study ovine PrP misfolding [35]. Sheep can spontaneously develop scrapie, but natural PrP variants influence the disease risk. This can be exploited to select for variants that protect flocks and to understand the impact on PrP conversion dynamics. In one study, Bujdoso compared the R/H polymorphism at 154 (Fig. 1A) and found that flies expressing ovine PrP-H154 showed more pronounced neurotoxicity than flies expressing R154, suggesting that H154 promotes the accumulation of neurotoxic conformations [36].
Topological variants of PrP
Among the many unique features of PrP is the existence of several topological conformations, all of them apparently produced by normal physiological processes. Although most PrP accumulates as a membrane-bound secreted protein, totally secreted (unbound) and cytosolic isoforms are also detected. The secreted isoform can originate from a failure to form the GPI anchor, whereas the highly toxic cytosolic isoform [37] can be a consequence of retro-translocation during ER synthesis. The Bujdoso group compared the toxicity of membrane-bound, secreted, and cytosolic ovine PrP expressed from the same genomic locus. They found that the secreted form was the most toxic and the cytosolic accumulated at lower levels, likely due to higher turnover [38*]. This heightened toxicity is consistent with the identification of pathogenic PrP mutations lacking the C-terminus that are highly amyloidogenic and show similarities to the amyloid-β peptide (e.g., Y145X, Y163X) (Fig. 1).
We introduced artificial mutations that perturb the stability of helix 3 on hamster PrP by replacing two highly conserved Met at 206 and 212 with polar residues (Ser) (Fig. 1). These Met are deeply buried in a hydrophobic core that maintains helices 2 and 3 in close contact. Introducing the two Ser at 206 and 212 in hamster PrP had a dramatic effect on PrP biogenesis [39]. The polar substitutions prevented the formation of the cysteine bridge between helices 2 and 3, exposing a weak transmembrane domain (residues 111–135), and causing its insertion into the membrane with the C-terminus in the ER lumen and the N-terminus in the cytosol (Ctm topology). Ctm PrP and the inverse Ntm PrP can be normally detected in small amounts as a consequence of errors during PrP translation and folding, suggesting the challenging nature of PrP biogenesis [40]. Ctm PrP remains stuck in the ER, but it is not toxic in Drosophila [39]. However, Ctm PrP accelerated the conformational change and toxicity of WT PrP, suggesting a potential role for age-related aberrant PrP biogenesis in sporadic PrP pathogenesis.
Transmission studies: are we there yet?
The main purpose for generating flies expressing ovine PrP was to determine whether flies could replicate mammalian prions. If so, Drosophila could be used to uncover the molecular mechanisms mediating this unique process and as a diagnostic bioassay for flock surveillance. Flies expressing ovine PrP and exposed to scrapie brain extracts from sheep in the food exhibited accelerated locomotor dysfunction and reduced survival (Fig. 3) [36,38*]. Although these flies showed no vacuolar pathology, they accumulated pathogenic PrP conformations recognized by the conformational antibody 2G11 that were sensitive to protease digestion [36]. These results are suggestive of prion-mediated conversion of transgenic ovine PrP. Recent studies showed that the toxicity of membrane-bound and cytosolic, but not secreted, ovine PrP increased in the presence of sheep prions [38*,41], suggesting their ability to promote PrP conversion in Drosophila. Flies expressing cytosolic PrP showed the most dramatic increase in toxicity in the presence of prions in the absence of PK-resistant PrP. In their most recent work, the authors used the revolutionary and sensitive PMCA (protein misfolding cyclic amplification) assay [42**] to detect PK-resistant PrP in flies expressing membrane-bound PrP, but not in cytosolic and secreted PrP. PMCA uses the principles of PCR to amplify PrPSc in vitro through cycles of incubation and sonication that break aggregates to create new seeds that exponentially grow over time to make small amounts of PrPSc visible by western blot. These PMCA results suggest that the robust effect of scrapie extract on cytosolic PrP is apparently not due to prion transmission. More importantly, since PMCA is extremely sensitive and prone to contamination, careful experiments should be designed to unambiguously determine whether flies produce and accumulate small amounts of PrPSc.
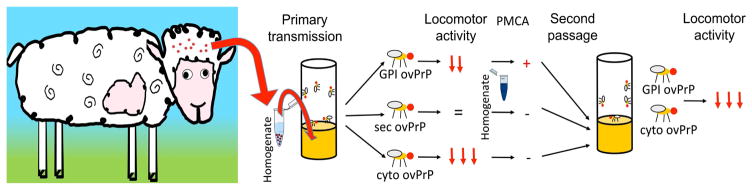
Figure 3. Prion transmission experiments in Drosophila.
Brain homogenate from scrapie sheep was added to the media of flies expressing three forms of ovine PrP: membrane-bound (GPI), secreted (sec), and cytosolic (cyto) [38*,41]. The locomotor activity of these flies was tested to determine the ability of scrapie prions to accelerate the toxicity of Drosophila-expressed ovine PrP. The arrows indicate the degree of locomotor dysfunction induced by the scrapie prions (=, no effect). These flies were then homogenated and used to detect protease-resistant PrP by PMCA. Only GPI ovine PrP was positive in PMCA. The same fly homogenates were used to feed transgenic flies in second passage to determine the transmissibility of Drosophila-produced material. Flies expressing GPI and secreted ovine PrP exhibited increased locomotor dysfunction suggesting the “transmission of prions” from flies to flies.
To further test prion transmission in flies, the Bujdoso lab also performed fly-to-fly secondary transmission (second passage) studies [38*]. Since cytosolic PrP was apparently the most efficient substrate in the scrapie-transmission studies, extracts from 30 day-old flies expressing membrane-bound, cytosolic, and secreted ovine PrP exposed to scrapie prions were homogenized and fed to flies expressing cytosolic ovine PrP (Fig. 3). Interestingly, all extracts accelerated locomotor dysfunction in host flies expressing cytosolic and membrane-bound PrP. Moreover, extracts from secreted PrP flies not exposed to prions also accelerated locomotor dysfunction in host flies expressing cytosolic PrP, suggesting a potential case of sporadic prion generation. Overall, these studies set up the basis for future transmission studies, but fall short of demonstrating conclusively the replication of mammalian prions and the spontaneous generation of prions in Drosophila. Such demonstration would require the detection of PrPSc in Drosophila extracts without amplifying techniques (PMCA) and transmission back to mammals (in mice expressing ovine PrP) to demonstrate strain preservation and species barrier features of the original prion.
Genetic discoveries in prion disease models: a dry well
One of the main advantages of generating Drosophila models of human diseases is the ability of conducting fast and efficient tests for candidate genes as well as large, unbiased genetic screens. In our first study expressing hamster PrP, we identified the protective activity of the potent and multifunctional molecular chaperone Hsp70 in adult fly locomotion and PrP conformation [21*]. Moreover, activation of Hsp70 with a two-drug cocktail (17-DMAG and dexamethasone) prevented PrP misfolding and neurotoxicity [43]. No other candidate genes have been identified so far due to the limitations of studying PrP toxicity in time-consuming assays.
The main reason for these limited discoveries is the lack of an easy-to-score phenotype, like the eye. Almost all the Drosophila models of proteinopathies display robust and unique eye phenotypes [15] that support their specific toxicity and facilitate efficient genetic tests in adult eyes. But none of the models expressing WT or mutant PrP form mouse, hamster, or sheep show eye perturbations. To overcome this limitation, we examined the ability of PrP from other animals to induce an eye phenotype. Our rationale was that within the spectrum of TSE susceptibility, some mammals should express highly toxic PrP. First, we created flies expressing PrP from bank vole, a rodent highly susceptible to most prions, but these flies exhibited similar features to those expressing hamster PrP (PFF, JSG, DERL, unpublished observations). Next, we considered bovine and human PrP, but since they had similar biosafety concerns, we decided to work with human PrP because its involvement in several sporadic and genetic PrD suggests increased neurotoxic potential. After obtaining institutional permission to work in a BSL3 facility, we created transgenic flies carrying human PrP-V129 and expressed it in the eye. V/M129 are polymorphisms in human PrP relevant for the transmission of BSE to humans as variant CJD since all cases have been reported in M/M homozygous individuals. This polymorphism is not known to affect other relevant aspects of PrP conformation and pathogenesis. As we predicted, human PrP induced a robust eye phenotype: small, highly disorganized, and glassy (Fig. 2A), a phenotype distinct from many other amyloids [16], suggesting that human PrP perturbs specific pathways. Hence, we can use this human PrP model to uncover the molecular mechanisms perturbed by aberrant conformations of PrP.
To demonstrate the sensitivity of this robust eye phenotype to known genetic modifiers of PrP, we simultaneously modulated the unfolded protein response (UPR). Human patients and mouse models of prion diseases show ER stress and activation of the unfolded protein response [44**,45]. Activation of the ER stress sensor PERK results in the phosphorylation of eIF2α, a translational repressor that restores proteostasis. But sustained translational repression can also be deleterious by limiting the availability of critical proteins. A recent paper demonstrated that overexpression of the eIF2α phosphatase GADD34 rescues prion-induced pathogenesis in mice, supporting the key role of the PERK-eIF2α pathway in PrD [41**]. In flies, PERK overexpression enhanced human PrP toxicity (Fig. 3A–C), whereas PERK LOF completely suppressed the glassy eye of human PrP (Fig. 3D). Additionally, LOF of ATF4, another PERK effector, also suppressed human PrP toxicity (Fig. 3E). These results support (1) the role of the two PERK pathways, eIF2α and ATF4, in human PrP toxicity, (2) the conserved mechanisms of PrP pathogenesis from mice to flies, and (3) the feasibility of a genetic screen, since known mediators of PrP toxicity rescue human PrP toxicity in flies.
Conclusions and future studies
As we have discussed, Drosophila models of PrD present some limitations and many opportunities for investigating molecular and cellular mechanisms relevant to TSE and, potentially, to other proteinopathies. Overall, Drosophila has proved an excellent model for studying PrP conformational dynamics and progressive phenotypes in the CNS. The impact of exogenous PrP on motor neuron electrophysiology supports previous studies suggesting a role for PrP in synaptic activity, but more work is needed to determine the molecular effects of PrP at the synapse. The ovine PrP models are very encouraging towards probing the capacity of Drosophila to support prion transmission, which could open the door to several relevant applications, including bioassays and dissecting the molecular mechanisms regulating prion conversion. Our new model expressing human PrP will overcome the limitations for conducting large genetic screens thanks to the robust eye phenotype. Overall, the Drosophila models of PrD are poised to make big impacts in several relevant areas of PrP pathobiology taking advantage of the easy manipulation and extraordinary resources for genetic exploration.
Brief methods
The cDNA for human PrP carrying the V129 polymorphism was codon-optimized for Drosophila, synthesized by GeneScript, and cloned into the pUAST vector to generate transgenic flies [18]. The construct was injected into yw embryos at Rainbow Transgenics following standard procedures [43] to generate multiple independent transgenic lines. The driver line GMR-Gal4 (all differentiating eye cells) and the line overexpressing PERK were obtained from the Bloomington Drosophila Stock Center and the RNAi lines for PERK and ATF4 were obtained from the Vienna Drosophila RNAi Collection (VDRC). PrP constructs and combinations were expressed in the eye at 27.5 C.
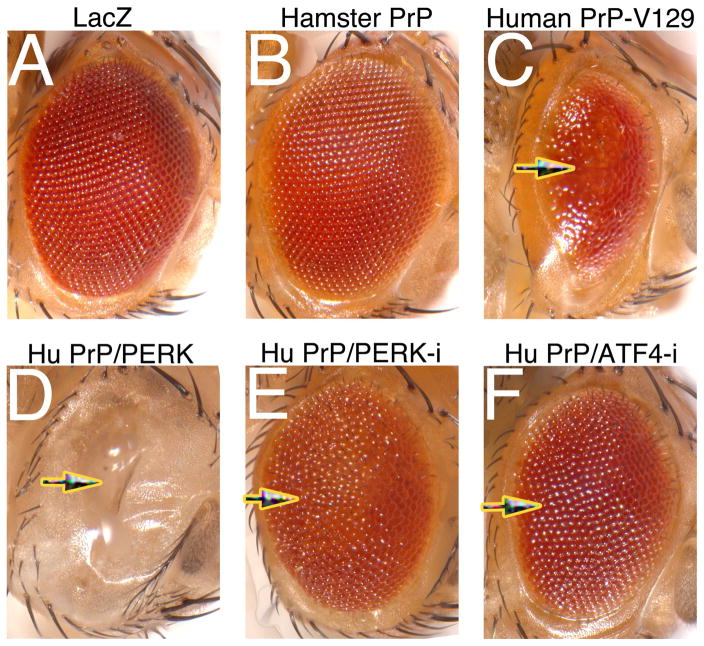
Figure 4. Human PrP induces a robust eye phenotype that is rescued by PERK inhibition.
A, Control expressing LacZ in the eye. B, Fly expressing hamster PrP in the eye showing a normal eye. C, Fly expressing human PrP (HuPrP) in the eye at comparable levels to those of hamster PrP. The eye is smaller, disorganized, and glassy. D, Co-expression of HuPrP and PERK completely prevents the development of the eye. E and F. Co-expression of PERK or ATF4 RNAi suppresses the toxicity of HuPrP, resulting in large and organized eyes.
Highlights.
Expression of mammalian prion protein (PrP) in Drosophila induces progressive neurodegeneration and accumulation of pathogenic PrP conformations
Early expression of PrP in developing motor neurons induces developmental and physiological perturbations associated with PrP endogenous function
Natural and pathogenic variants in PrP sequence show different conformational dynamics and toxicity profiles
New Drosophila models expressing ovine or human PrP may be critical for dissecting transmission and neurotoxicity mechanisms
Acknowledgments
We want to thank the Bloomington Drosophila Stock Center (NIH P40OD018537) for Drosophila strains and Javier A. Fernandez-Ambite for assistance with the figures. This work was completed in part with the support of the NIH grant R21 NS096627-01A1 to PFF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95(23):13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stohr J, Watts JC, Mensinger ZL, Oehler A, Grillo SK, DeArmond SJ, Prusiner SB, Giles K. Purified and synthetic alzheimer’s amyloid beta (abeta) prions. Proc Natl Acad Sci U S A. 2012;109(27):11025–11030. doi: 10.1073/pnas.1206555109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirbaha H, Li A, Barker SJ, Foley AC, Thorpe JR, Serpell LC, Miller TM, et al. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron. 2014;82(6):1271–1288. doi: 10.1016/j.neuron.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisele YS, Obermuller U, Heilbronner G, Baumann F, Kaeser SA, Wolburg H, Walker LC, Staufenbiel M, Heikenwalder M, Jucker M. Peripherally applied abeta-containing inoculates induce cerebral beta-amyloidosis. Science. 2010;330(6006):980–982. doi: 10.1126/science.1194516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jucker M, Walker LC. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. 2013;501(7465):45–51. doi: 10.1038/nature12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morales R, Duran-Aniotz C, Castilla J, Estrada LD, Soto C. De novo induction of amyloid-beta deposition in vivo. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.120. [DOI] [PubMed] [Google Scholar]
- 7.Desplats P, Lee HJ, Bae EJ, Patrick C, Rockenstein E, Crews L, Spencer B, Masliah E, Lee SJ. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci U S A. 2009;106(31):13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffith JS. Self-replication and scrapie. Nature. 1967;215(5105):1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 9.Sandberg MK, Al-Doujaily H, Sharps B, Clarke AR, Collinge J. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature. 2011;470(7335):540–542. doi: 10.1038/nature09768. [DOI] [PubMed] [Google Scholar]
- 10**.Sandberg MK, Al-Doujaily H, Sharps B, De Oliveira MW, Schmidt C, Richard-Londt A, Lyall S, Linehan JM, Brandner S, Wadsworth JD, Clarke AR, et al. Prion neuropathology follows the accumulation of alternate prion protein isoforms after infective titre has peaked. Nat Commun. 2014;5(4347) doi: 10.1038/ncomms5347. Describes critical observations that uncouple the accumulation of prions and neurotoxicity, suggesting that a different PrP entites are responsible for neurodegeneration and transmission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solomon IH, Schepker JA, Harris DA. Prion neurotoxicity: Insights from prion protein mutants. Current issues in molecular biology. 2010;12(2):51–61. [PMC free article] [PubMed] [Google Scholar]
- 12**.Mays CE, van der Merwe J, Kim C, Haldiman T, McKenzie D, Safar JG, Westaway D. Prion infectivity plateaus and conversion to symptomatic disease originate from falling precursor levels and increased levels of oligomeric prpsc species. J Virol. 2015;89(24):12418–12426. doi: 10.1128/JVI.02142-15. Recent discovery of a new PrP isoform that correlates with neurotoxicity and competes with infectious conformations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C, Haldiman T, Surewicz K, Cohen Y, Chen W, Blevins J, Sy MS, Cohen M, Kong Q, Telling GC, Surewicz WK, et al. Small protease sensitive oligomers of prpsc in distinct human prions determine conversion rate of prp(c) PLoS Pathog. 2012;8(8):e1002835. doi: 10.1371/journal.ppat.1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warrick JM, Paulson HL, Gray-Board GL, Bui QT, Fischbeck KH, Pittman RN, Bonini NM. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in drosophila. Cell. 1998;93(6):939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 15.Jackson GR, Salecker I, Dong X, Yao X, Arnheim N, Faber PW, MacDonald ME, Zipursky SL. Polyglutamine-expanded human huntingtin transgenes induce degeneration of drosophila photoreceptor neurons. Neuron. 1998;21(3):633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 16.Rincon-Limas DE, Jensen K, Fernandez-Funez P. Drosophila models of proteinopathies: The little fly that could. Curr Pharm Des. 2012;18(8):1108–1122. doi: 10.2174/138161212799315894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raeber AJ, Muramoto T, Kornberg TB, Prusiner SB. Expression and targeting of syrian hamster prion protein induced by heat shock in transgenic drosophila melanogaster. Mech Dev. 1995;51(2–3):317–327. doi: 10.1016/0925-4773(95)00379-7. [DOI] [PubMed] [Google Scholar]
- 18.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 19.Deleault NR, Dolph PJ, Feany MB, Cook ME, Nishina K, Harris DA, Supattapone S. Post-transcriptional suppression of pathogenic prion protein expression in drosophila neurons. J Neurochem. 2003;85(6):1614–1623. doi: 10.1046/j.1471-4159.2003.01819.x. [DOI] [PubMed] [Google Scholar]
- 20*.Gavin BA, Dolph MJ, Deleault NR, Geoghegan JC, Khurana V, Feany MB, Dolph PJ, Supattapone S. Accelerated accumulation of misfolded prion protein and spongiform degeneration in a drosophila model of gerstmann-straussler-scheinker syndrome. J Neurosci. 2006;26(48):12408–12414. doi: 10.1523/JNEUROSCI.3372-06.2006. First sucessful model with progressive degenerative phenotypes and conformational changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Fernandez-Funez P, Casas-Tinto S, Zhang Y, Gomez-Velazquez M, Morales-Garza MA, Cepeda-Nieto AC, Castilla J, Soto C, Rincon-Limas DE. In vivo generation of neurotoxic prion protein: Role for hsp70 in accumulation of misfolded isoforms. PLoS Genet. 2009;5(6):e1000507. doi: 10.1371/journal.pgen.1000507. First model with robust phenotypes induced by WT PrP, PrPSc-like conformations, and suppression of phenotypes by Hsp70 co-expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murali A, Maue RA, Dolph PJ. Reversible symptoms and clearance of mutant prion protein in an inducible model of a genetic prion disease in drosophila melanogaster. Neurobiol Dis. 2014;67:71–78. doi: 10.1016/j.nbd.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic drosophila by using the site-specific integrase from phage phic31. Genetics. 2004;166(4):1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 25.Bueler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, DeArmond SJ, Prusiner SB, Aguet M, Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface prp protein. Nature. 1992;356(6370):577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 26.Collinge J, Whittington MA, Sidle KC, Smith CJ, Palmer MS, Clarke AR, Jefferys JG. Prion protein is necessary for normal synaptic function. Nature. 1994;370(6487):295–297. doi: 10.1038/370295a0. [DOI] [PubMed] [Google Scholar]
- 27.Tobler I, Gaus SE, Deboer T, Achermann P, Fischer M, Rulicke T, Moser M, Oesch B, McBride PA, Manson JC. Altered circadian activity rhythms and sleep in mice devoid of prion protein. Nature. 1996;380(6575):639–642. doi: 10.1038/380639a0. [DOI] [PubMed] [Google Scholar]
- 28.Choi JK, Jeon YC, Lee DW, Oh JM, Lee HP, Jeong BH, Carp RI, Koh YH, Kim YS. A drosophila model of gss syndrome suggests defects in active zones are responsible for pathogenesis of gss syndrome. Hum Mol Genet. 2010;19(22):4474–4489. doi: 10.1093/hmg/ddq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29**.Robinson SW, Nugent ML, Dinsdale D, Steinert JR. Prion protein facilitates synaptic vesicle release by enhancing release probability. Hum Mol Genet. 2014;23(17):4581–4596. doi: 10.1093/hmg/ddu171. Detailed electrophysiological characterization of Drosophila motor neurons expressing WT and P101L mouse PrP. Describes several alterations in synaptic activity, suggesting that PrP is a protein with a critical regulatory function at the synapse. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibbs CJ, Jr, Gajdusek DC. Experimental subacute spongiform virus encephalopathies in primates and other laboratory animals. Science. 1973;182:67–68. doi: 10.1126/science.182.4107.67. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez-Funez P, Zhang Y, Casas-Tinto S, Xiao X, Zou WQ, Rincon-Limas DE. Sequence-dependent prion protein misfolding and neurotoxicity. J Biol Chem. 2010;285(47):36897–36908. doi: 10.1074/jbc.M110.174391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez-Funez P, Zhang Y, Sanchez-Garcia J, Jensen K, Zou WQ, Rincon-Limas DE. Pulling rabbits to reveal the secrets of the prion protein. Commun Integr Biol. 2011;4(3):262–266. doi: 10.4161/cib.4.3.15054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Sanchez-Garcia J, Jensen K, Zhang Y, Rincon-Limas DE, Fernandez-Funez P. A single amino acid (asp159) from the dog prion protein suppresses the toxicity of the mouse prion protein in drosophila. Neurobiol Dis. 2016;95:204–209. doi: 10.1016/j.nbd.2016.07.025. Demonstration that a single amino acid change from dog PrP is sufficient to stabilize mouse PrP and prevent its toxicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lysek DA, Schorn C, Nivon LG, Esteve-Moya V, Christen B, Calzolai L, von Schroetter C, Fiorito F, Herrmann T, Guntert P, Wuthrich K. Prion protein nmr structures of cats, dogs, pigs, and sheep. Proc Natl Acad Sci U S A. 2005;102(3):640–645. doi: 10.1073/pnas.0408937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thackray AM, Muhammad F, Zhang C, Denyer M, Spiropoulos J, Crowther DC, Bujdoso R. Prion-induced toxicity in prp transgenic drosophila. Exp Mol Pathol. 2012;92(2):194–201. doi: 10.1016/j.yexmp.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Thackray AM, Muhammad F, Zhang C, Di Y, Jahn TR, Landgraf M, Crowther DC, Evers JF, Bujdoso R. Ovine prp transgenic drosophila show reduced locomotor activity and decreased survival. Biochem J. 2012;444(3):487–495. doi: 10.1042/BJ20112141. [DOI] [PubMed] [Google Scholar]
- 37.Ma J, Wollmann R, Lindquist S. Neurotoxicity and neurodegeneration when prp accumulates in the cytosol. Science. 2002;298(5599):1781–1785. doi: 10.1126/science.1073725. [DOI] [PubMed] [Google Scholar]
- 38*.Thackray AM, Di Y, Zhang C, Wolf H, Pradl L, Vorberg I, Andreoletti O, Bujdoso R. Prion-induced and spontaneous formation of transmissible toxicity in prp transgenic drosophila. Biochem J. 2014;463(1):31–40. doi: 10.1042/BJ20140129. Experiments suggesting trasnmission of scrapie prions to Drosophila. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez-Garcia J, Arbelaez D, Jensen K, Rincon-Limas DE, Fernandez-Funez P. Polar substitutions in helix 3 of the prion protein produce transmembrane isoforms that disturb vesicle trafficking. Hum Mol Genet. 2013;22(21):4253–4266. doi: 10.1093/hmg/ddt276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279(5352):827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 41.Thackray AM, Zhang C, Arndt T, Bujdoso R. Cytosolic prp can participate in prion-mediated toxicity. J Virol. 2014;88(14):8129–8138. doi: 10.1128/JVI.00732-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42**.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411(6839):810–813. doi: 10.1038/35081095. Original description of the PMCA procedure. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Casas-Tinto S, Rincon-Limas DE, Fernandez-Funez P. Combined pharmacological induction of hsp70 suppresses prion protein neurotoxicity in drosophila. PLoS One. 2014;9(2):e88522. doi: 10.1371/journal.pone.0088522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44**.Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, Halliday M, Morgan J, Dinsdale D, Ortori CA, Barrett DA, et al. Sustained translational repression by eif2alpha-p mediates prion neurodegeneration. Nature. 2012;485(7399):507–511. doi: 10.1038/nature11058. Rescue of prion pathology in mice by reduced eif2alpha activity, demonstrating the role of ER stress and the UPR in prion-mediated neurotoxicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hetz C, Russelakis-Carneiro M, Maundrell K, Castilla J, Soto C. Caspase-12 and endoplasmic reticulum stress mediate neurotoxicity of pathological prion protein. Embo J. 2003;22(20):5435–5445. doi: 10.1093/emboj/cdg537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubin GM, Spradling AC. Genetic transformation of drosophila with transposable element vectors. Science. 1982;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]