Abstract
Recently, the World Health Organization confirmed 120 new human cases of avian H7N9 influenza in China resulting in 37 deaths, highlighting the concern for a potential pandemic and the need for an effective, safe, and high-speed vaccine production platform. Production speed and scale of mRNA-based vaccines make them ideally suited to impede potential pandemic threats. Here we show that lipid nanoparticle (LNP)-formulated, modified mRNA vaccines, encoding hemagglutinin (HA) proteins of H10N8 (A/Jiangxi-Donghu/346/2013) or H7N9 (A/Anhui/1/2013), generated rapid and robust immune responses in mice, ferrets, and nonhuman primates, as measured by hemagglutination inhibition (HAI) and microneutralization (MN) assays. A single dose of H7N9 mRNA protected mice from a lethal challenge and reduced lung viral titers in ferrets. Interim results from a first-in-human, escalating-dose, phase 1 H10N8 study show very high seroconversion rates, demonstrating robust prophylactic immunity in humans. Adverse events (AEs) were mild or moderate with only a few severe and no serious events. These data show that LNP-formulated, modified mRNA vaccines can induce protective immunogenicity with acceptable tolerability profiles.
Keywords: mRNA vaccines, influenza, immunogenicity, pandemic, vaccines, H10N8, H7N9, mRNA
Potential pandemic influenzas and the need for an effective, safe, and high-speed vaccine production platform have been widely discussed in the scientific community. Bahl et al. report the rapid and robust immune responses achieved against H10N8 and H7N9 viruses from modified mRNA vaccines with an acceptable safety profile.
Introduction
Several avian influenza A viruses (H5N1, H10N8, H7N9, and H1N1) have crossed the species barrier, causing severe and often fatal respiratory disease in humans. Fortunately, most of these strains are not able to sustain person-to-person transmission.1 However, lessons learned from these outbreaks demonstrated that new approaches are needed to address potential future pandemic influenza outbreaks.2
Two major glycoproteins, crucial for influenza infection, are hemagglutinin (HA) and neuraminidase (NA); both are expressed on the surface of the influenza A virion.3 HA mediates viral entry into host cells by binding to sialic acid-containing receptors on the cell mucosal surface and the fusion of viral and host endosomal membranes.4
The segmented influenza A genome permits re-assortment and exchange of HA (or NA) segments between different influenza strain subtypes during concomitant host-cell infection. Generation of novel antigenic proteins (antigenic shift) and sustainable person-to-person transmission are hallmarks of pandemic influenza strains.5 Such strains can spread quickly and cause widespread morbidity and mortality in humans due to high pathogenicity and little to no pre-existing immunity. Recent cases (2013) of avian-to-human transmission of avian influenza A virus subtypes included H7N9, H6N1, and H10N8.6, 7, 8 The case-fatality rate in over 600 cases of H7N9 infections was ∼30%.1, 9 Most recently, the World Health Organization reported another 120 cases since September 2016 resulting in 37 deaths.10 To date, H10N8 infection in man has been limited; yet, of the three reported cases, two were fatal.11
The limited efficacy of existing antiviral therapeutics (i.e., oseltamivir and zanamivir) makes vaccination the most effective means of protection against influenza.12 Conventional influenza vaccines induce protection by generating HA-specific neutralizing antibodies, the major correlate of protection, against the globular head domain.13, 14, 15 Such vaccines utilize the HA protein, administered as a subunit, split virion, inactivated whole virus, or live-attenuated virus. A majority of approved influenza vaccines are produced in embryonated chicken eggs or cell substrates. This process takes several months and relies on the availability of sufficient supplies of pathogen-free eggs and adaptation of the virus to grow within its substrate.16, 17 The 5–6 months required to produce enough vaccine to protect a substantial proportion of the population consumes much of the duration of the often-devastating first wave of a pandemic.18 This mismatch between the speeds of vaccine production and epidemic spread drives the search for vaccine platforms that can respond faster.19
Using mRNA complexed with protamine (RNActive, Curevac), Petsch et al.20 demonstrated that intradermal (ID) vaccination of mice with RNActive encoding full-length HA from influenza virus H1N1 (A/Puerto Rico/8/1934) induced effective seroconversion and virus-neutralizing antibodies in all vaccinated animals. Immunity was long lasting and protected both young and old animals from lethal challenge with the H1N1, H3N2, and H5N1 strains of the influenza A virus.20 Efficacy of these RNActive vaccines was also shown in ferrets and pigs.21
The use of a delivery system can dramatically reduce the doses needed to generate potent immune responses, without an additional conventional adjuvant. Lipid nanoparticles (LNPs) have been used extensively for the delivery of small interfering RNA (siRNA), and they are currently being evaluated in late-stage clinical trials via intravenous administration.22
Exogenous mRNA can stimulate innate immunity through Toll-like receptors (TLRs) 3, 7, and 8 and cytoplasmic signal-recognition proteins RIG-I and MDA5.23, 24 The adjuvant effect of stimulating innate immunity may be advantageous for purified protein vaccines, but indiscriminate immune activation can inhibit mRNA translation, reducing antigen expression and subsequent immunogenicity.25, 26 This can be overcome by replacing uridine nucleosides with naturally occurring base modifications, such as pseudouridine and 5-methylcytidine.27, 28, 29 Recently, we30 and others31 have shown how LNP-encapsulated modified mRNA vaccines can induce extraordinary levels of neutralizing immune responses against the Zika virus in mice and nonhuman primates, respectively.
In this study, we evaluated the immunogenicity of two LNP-formulated, modified mRNA-based influenza A vaccines encoding the HA of H10N8 (A/Jiangxi-Donghu/346/2013) and H7N9 (A/Anhui/1/2013) in animals and H10N8 HA mRNA in humans from an ongoing trial. In the animal studies, we show that both vaccines generated potent neutralizing antibody titers in mice, ferrets, and cynomolgus monkeys (cynos) after a single dose. Additionally, a single dose of H7N9 HA mRNA protected mice from an autologous lethal challenge and reduced lung viral titers in ferrets. Encouraged by these findings, a first-in-human, dose-escalating, phase 1 trial is ongoing, with interim results reported here that confirm the observed, preclinical immunogenicity data with a safety profile consistent with other non-live vaccines.
Results
H10N8 and H7N9 HA mRNA Immunogenicity in Mice
In vitro protein expression for both H10N8 HA (H10) and H7N9 HA (H7) mRNA vaccines were confirmed by transfection of HeLa cells. Western blot of resulting cell lysates demonstrated a 75-kDa band for both constructs using the corresponding HA-specific antibodies (Figure S1), consistent with previous reports for other HAs.22 Due to a lack of glycosylation, both H10 HA and H7 HA protein controls had a molecular weight of 62 kDa.
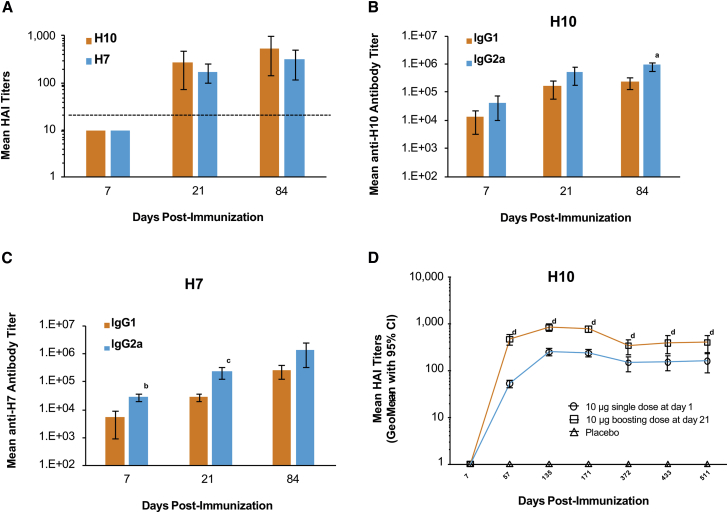
Hemagglutination inhibition (HAI), IgG1, and IgG2a titers were measured after a single 10-μg dose of either formulated H10 or H7 mRNA in BALB/c mice immunized ID. HAI titers were below the limit of detection (<10) at day 7 but increased well above baseline by day 21 (Figure 1A). Unlike HAI, both anti-H10 and anti-H7 IgG1 and IgG2a titers were detected on day 7 (Figures 1B and 1C). For H10, IgG1 and IgG2a titers continued to increase until day 21 and were maintained at day 84. For H7, both IgG1 and IgG2a antibody titers increased 10-fold between day 21 and day 84 (Figure 1C). IgG2a titers were greater than IgG1 titers at all time points following formulated H10 or H7 mRNA immunization, suggesting a TH1-skewed immune response. For H10, these differences were significant at day 84 (p = 0.0070) and for H7 at day 7 (p = 0.0017) and day 21 (p = 0.0185). A 10-μg H10 mRNA-boosting immunization (21 days post-prime) resulted in a 2- to 5-fold increase in HAI titers, compared to a single dose at all time points tested (p < 0.05) (Figure 1D). Titers remained stable for more than a year, regardless of the number of doses.
Figure 1.
Mice Immunized with H10 or H7 mRNA Generate Robust and Stable Antibody Responses Consistent with a TH1 Profile
BALB/c mice were vaccinated ID with a single 10-μg dose of formulated H10 or H7 mRNA. (A) H10 and H7 indicate mean HAI titers (limit of detection is 1:10). Dotted line indicates the correlate of protection in humans (1:40). (B and C) IgG1 and IgG2a titers were measured for both H10 (B) and H7 (C) via ELISA (n = 5/group). ap = 0.0070, bp = 0.0017, and cp = 0.0185 versus IgG2a at the same time point. (D) BALB/c mice were immunized ID with a single 10-μg dose of formulated H10 mRNA. A subset of these mice received a 10-μg boost on day 21. Serum was collected at the indicated time points, and neutralizing antibody titers were determined by HAI (n = 15/group). Placebo controls were also included. dp < 0.05 single dose versus boosting dose at the same time point. Error bars indicate standard mean error.
While most vaccines are delivered via an intramuscular (IM) or subcutaneous administration,32 the ID route of administration has the potential to be dose sparing. Therefore, to examine the effect of administration route on immunogenicity, BALB/c mice were immunized ID or IM with formulated H10 or H7 mRNA at four different dose levels. All animals received a boosting immunization on day 21, and serum was collected 28 days post-boost (day 49). Immune responses were observed for both vaccines at all dose levels tested (Figures S2A and S2B). Titers were slightly higher following IM administration at 2 and 0.4 μg for H10, but this difference was only significant at the 2-μg dose (p = 0.0038) (Figure S2A). The differences in H10 HAI titers were significant between some of the dose levels following IM administration: 10 versus 0.4 μg, p = 0.0247; 10 versus 0.08 μg, p = 0.0002; 2 versus 0.08 μg, p = 0.0013; and 0.4 versus 0.08 μg, p = 0.0279. HAI titers following H7 immunization trended higher as the dose increased although no significance was detected. In addition, there was no significant difference between IM and ID immunization (Figure S2B). T cell responses, as measured by IFNɣ ELISpot, were observed for both H10 and H7 at all doses tested (Figures S2C and S2D). Similar to H7 HAI titers, T cell responses trended higher following IM administration, especially for H7. However, significance could not be established due to pooling of the samples by group. Overall, after two doses, immunization with either H10 or H7 mRNA elicited an immune response at all doses tested with both ID and IM administration.
Given this innovative vaccine platform, we examined the biodistribution of the mRNA vaccines for both routes of administration. Male CD-1 mice received 6 μg formulated H10 mRNA either IM or ID. Following IM administration, the maximum concentration (Cmax) of the injection site muscle was 5,680 ng/mL, and the level declined with an estimated t1/2 of 18.8 hr (Table 1). Proximal lymph nodes had the second highest concentration at 2,120 ng/mL (tmax of 8 hr with a relatively long t1/2 of 25.4 hr), suggesting that H10 mRNA distributes from the injection site to systemic circulation through the lymphatic system. The spleen and liver had a mean Cmax of 86.9 ng/mL (area under the curve [AUC]0–264 of 2,270 ng.hr/mL) and 47.2 ng/mL (AUC0–264 of 276 ng.hr/mL), respectively. In the remaining tissues and plasma, H10 mRNA was found at 100- to 1,000-fold lower levels.
Table 1.
Biodistribution of H10 mRNA in Plasma and Tissue after IM Administration in Mice
| Matrix | tmax (hr) | Cmax (ng/mL) |
AUC0–264 h (ng.hr/mL) |
t1/2 (h) | ||
|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | |||
| Bone marrow | 2.0 | 3.35 | 1.87 | NA | NC | |
| Brain | 8.0 | 0.429 | 0.0447 | 13.9 | 1.61 | NR |
| Cecum | 8.0 | 0.886 | 0.464 | 11.1 | 5.120 | NC |
| Colon | 8.0 | 1.11 | 0.501 | 13.5 | 5.51 | NC |
| Distal lymph nodes | 8.0 | 177.0 | 170.0 | 4,050 | 2,060 | 28.0 |
| Heart | 2.0 | 0.799 | 0.225 | 6.76 | 1.98 | 3.50 |
| Ileum | 2.0 | 3.54 | 2.60 | 22.6 | 10.8 | 5.42 |
| Jejunum | 2.0 | 0.330 | 0.120 | 5.24 | 0.931 | 8.24 |
| Kidney | 2.0 | 1.31 | 0.273 | 9.72 | 1.44 | 11.4 |
| Liver | 2.0 | 47.2 | 8.56 | 276 | 37.4 | NC |
| Lung | 2.0 | 1.82 | 0.555 | 12.7 | 2.92 | 16.0 |
| Muscle (injection site) | 2.0 | 5,680 | 2,870 | 95,100 | 20,000 | 18.8 |
| Plasma | 2.0 | 5.47 | 0.829 | 35.5 | 5.41 | 9.67 |
| Proximal lymph nodes | 8.0 | 2,120 | 1,970 | 38,600 | 22,000 | 25.4 |
| Rectum | 2.0 | 1.03 | 0.423 | 14.7 | 3.67 | NR |
| Spleen | 2.0 | 86.9 | 29.1 | 2,270 | 585 | 25.4 |
| Stomach | 2.0 | 0.626 | 0.121 | 11.6 | 1.32 | 12.7 |
| Testes | 8.0 | 2.37 | 1.03 | 36.6 | 11.8 | NR |
Male CD-1 mice received 300 μg/kg (6 μg) formulated H10 mRNA via IM immunization. Two replicates of bone marrow, lung, liver, heart, right kidney, inguinal- and popliteal-draining lymph nodes, axillary distal lymph nodes, spleen, brain, stomach, ileum, jejunum, cecum, colon, rectum, testes (bilateral), and injection site muscle were collected for bDNA analysis at 0, 2, 8, 24, 48, 72, 120, 168, and 264 hr after dosing (n = 3 mice/time point). NA, not applicable AUC with less than three quantifiable concentrations; NC, not calculated; NR, not reported because extrapolation exceeds 20% or R-squared is less than 0.80.
Following ID administration, Cmax within the skin at the injection site was 18.2 μg/mL. Levels declined by 24 hr with an estimated t1/2 of 23.4 hr, suggesting that the H10 mRNA likely dissipated to systemic circulation via the proximal draining lymph node, as seen for the IM dosing. Consistent with this, the spleen, with a Cmax of 1.66 ng/mL (1,663.52 pg/mL; AUC0–96 of 114.25 ng.hr/mL), had the highest levels among distal tissues. Only trace amounts of H10 mRNA were found in the heart, kidney, liver, and lung. Overall, whether administered ID or IM, the biodistribution of this vaccine was consistent with that observed for other vaccines,33 where a local deposition effect was observed followed by draining to the local lymph nodes and subsequent circulation in the lymphatic system (Table 1; Table S1).
To understand the expression profile of mRNA after IM and ID administration, BALB/c mice were injected on day 0 with formulated luciferase mRNA at four different dose levels (10, 2, 0.4, and 0.08 μg). Expression was found to be dose dependent. As the dose increased, expression was found in distal tissues, with peak expression observed 6 hr after dosing. There were no significant differences when comparing maximum expression and time of maximal expression across IM and ID routes (Figure S3A). The time course of expression was also similar with both routes (Figures S3B and S3C). However, the distribution of expression changed slightly when the two routes were compared. Expression outside of the site of administration was observed across all dose levels, but it was more pronounced following IM administration, which is consistent with the biodistribution data (Figures S4A–S4E; Table 1; Table S1).34
H7 mRNA Vaccine Provides Protection against Lethal Influenza H7N9, A/Anhui/1/2013, in Mice and Ferrets
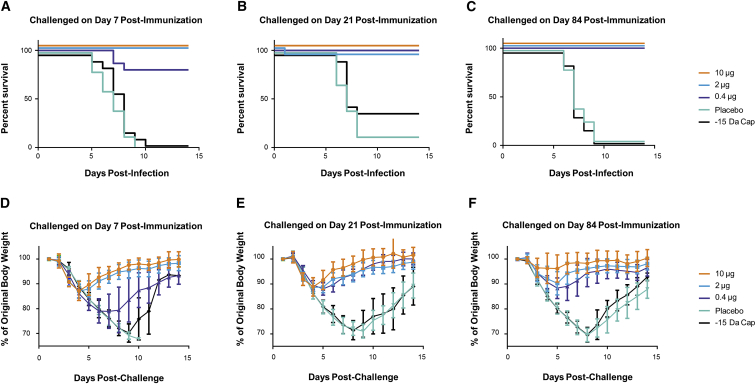
To determine the time to onset and duration of immunity to influenza H7N9 (A/Anhui/1/2013) lethal challenge, BALB/c mice were immunized ID with 10, 2, or 0.4 μg formulated H7 mRNA. For negative controls, placebo and 10 μg formulated H7 mRNA deficient in expression, due to the removal of a methyl group on the 2′-O position of the first nucleotide adjacent to the cap 1 structure at the 5′ end of the mRNA (−15 Da cap), were included. Serum was collected on days 6, 20, and 83, and mice were challenged via intranasal (IN) instillation with a target dose of 2.5 × 105 tissue culture infectious dose (TCID50) on days 7, 21, and 84. Changes in body weight and clinical signs of disease were monitored for 14 days post-challenge. A single vaccination was found to be protective against H7N9 challenge (2.5 × 105 TCID50; Figures 2A–2C). There was a significant increase in survival for animals in the three vaccine dose groups compared to the animals from the two control groups (p < 0.0001). Clinical observations in influenza-infected mice included rough coat, hunched posture, orbital tightening, and, in some cases, labored breathing. Weight loss (incidence and duration) was more prevalent for animals in the control groups and seen to a lesser extent in the low-dose vaccine group (Figures 2D–2F). HAI titers were below the limit of detection until day 20 for both the 10- and 2-μg dose groups (Figure S5). There was a 5- to 7-fold increase in HAI titers from day 20 to day 83 at all doses tested (p < 0.0001). Day-83 titers were dose dependent with mean titers of 224, 112, and 53 for the 10-μg dose, 2-μg dose, and 0.4-μg dose groups, respectively (p < 0.0001). Interestingly, despite complete protection to challenge at the 0.4-μg dose at day 21 (Figure 2B), a protective HAI titer (≥40) was not detected until day 83 at this dose, suggesting additional mechanism(s) of protection.
Figure 2.
A Single Injection of an H7 mRNA Vaccine Achieves Rapid and Sustained Protection in Mice
BALB/c mice were vaccinated ID with 10, 2, or 0.4 μg formulated H7 mRNA. Placebo and 10 μg formulated H7 mRNA with a reduced 5′ cap structure (−15 Da cap) were included as negative controls. On day 7, 21, or 84 post-immunization, mice were challenged via intranasal (IN) instillation with a target dose of 2.5 × 105 TCID50 of influenza A/Anhui/1/2013 (H7N9). Serum was collected prior to challenge (days 6, 20, and 83). (A–C) Survival curves of mice challenged on day 7 (A), day 21 (B), or day 84 (C) post-immunization at the indicated doses. p < 0.0001 10-, 2-, and 0.4-μg dose groups versus placebo or −15 Da cap at days 7, 21, and 84 post-immunization. (D–F) Weight curves of mice challenged on day 7 (D), day 21 (E), or day 84 (F) post-immunization at the indicated doses (n = 15/group). Error bars indicate standard mean error.
The negative mRNA control unexpectedly showed some delayed efficacy by day 21. However, this group of animals appeared to have received a dose lower than the day 7 and day 84 groups, based on back titer calculation (6.2 × 103 TCID50 versus 3.8 × 105 and 6.1 × 105, respectively.), which was only ∼3-fold higher than the LD50 of 1.88 × 103 (95% confidence interval [CI] = 8.02 × 102–5.51 × 103). Nonetheless, this group had comparable weight loss to the placebo group, and it was just above the threshold for euthanasia (30%) for some of the animals, thus confirming the significant protection observed in the positive vaccine groups. Additionally, it is not possible to rule out a low level of protein expression from the de-methylated cap of the negative mRNA control.35
Unlike mice, ferrets are naturally susceptible to human influenza virus isolates. Human and avian influenza viruses both replicate efficiently in the respiratory tract of ferrets, and numerous clinical signs found in humans following seasonal or avian influenza virus infection are also present in the ferrets.36, 37 Ferrets (n = 8/group) were vaccinated ID on day 0 with 200-, 50-, or 10-μg doses of formulated H7 mRNA. Formulated H7 mRNA with a −15 Da cap and placebo were included as negative controls. A subset of ferrets received a second ID vaccination on day 21. All groups were exposed to influenza H7N9 via IN challenge (1 × 106 TCID50). The primary endpoint for this study was viral burden determined by TCID50 in the lung at 3 days post-challenge, which is when the peak viral load is seen in control animals (data not shown). A reduction in lung viral titers was observed when ferrets were challenged 7 days post-immunization at all doses tested (Figures S6A–S6C). Ferrets immunized with 200 μg and challenged on day 49 had viral loads below the level of detection (Figure S6C). Antibody titers, as measured by HAI, increased significantly by day 21 for all dose groups (p < 0.05); as measured by microneutralization (MN), significant increases were observed by day 49 for all dose groups (p < 0.05) (Figures S7A and S7B). A second immunization increased titers but showed no statistical benefit compared to a single immunization, likely due to the two to four log reduction in viral lung titers seen in both the single- and double-immunization groups (Figures S7A–S7D). Two immunizations with 50-μg doses significantly increased HAI and MN titers compared to placebo (p < 0.05), and two immunizations with 200-μg doses generated significant HAI and MN titers versus placebo and all other doses (p < 0.0001) (Figures S7C and S7D).
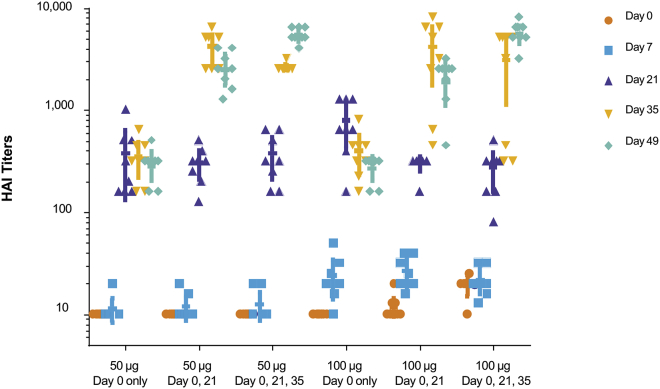
In the absence of an H10N8 (A/Jiangxi-Donghu/346/2013) challenge model, the onset and duration of immunity to formulated H10 mRNA in ferrets was tested by HAI. Groups of ferrets were immunized ID once, twice, or three times with 50 or 100 μg H10 mRNA. Immunization with a single dose of 50 or 100 μg resulted in significant and comparable increases in HAI titers at days 21, 35, and 49 (p < 0.0001; Figure 3). Immunization with a 100-μg dose resulted in only slightly elevated antibody responses on day 7 compared to day 0 (p < 0.0001), with minimal differences observed with the 50-μg dose on day 7 compared to day 0 (p < 0.3251). Subsequent boosts with either a 50- or 100-μg dose (delivered on day 21 or on both days 21 and 35) resulted in significant and comparable increases in HAI titers on days 35 and 49 (p < 0.0001). Overall, the H10 mRNA administered at a 50- or 100-μg dose yielded significant increases in HAI antibody titers as compared with prevaccination baseline values and controls (p < 0.0001). A single booster vaccination provided a significant increase in titers, but a second booster dose did not yield an additional increase (Figure 3).
Figure 3.
A Single Dose of H10 mRNA in Ferrets Generates Robust HAI Titers, Which Are Significant and Comparable at All Time Points
Ferrets were vaccinated ID with 50 or 100 μg formulated H10 mRNA. p < 0.0001, days 21, 35, and 49 versus day 0 with single doses of 50 or 100 μg; p < 0.0001 100-μg single dose, day 7 versus day 0. A subset of immunized ferrets received a boost on day 21 and an additional subset received a second boost on day 35. HAI titers were measured on days 0, 7, 21, 35, and 49 (n = 8/group). p < 0.0001 50 or 100 μg boosting dose(s), days 35 and 49 versus day 0.
H10 HA and H7 HA mRNA Immunogenicity in Nonhuman Primates
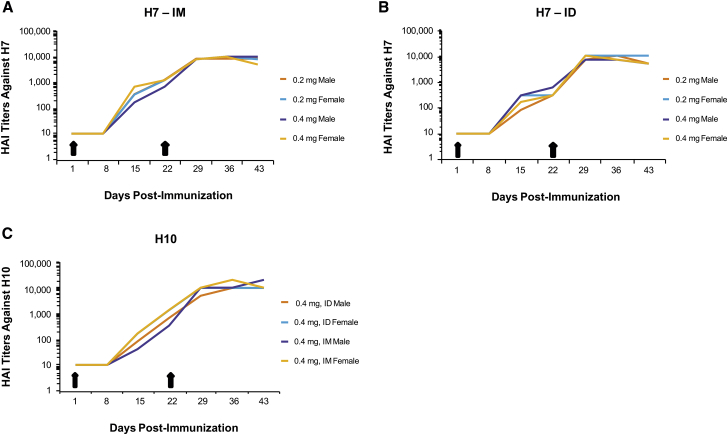
One of the major limitations with other nucleic acid-based technologies, such as plasmid DNA, has been translation to higher-order species, such as nonhuman primates. To evaluate the immune responses elicited in nonhuman primates, HAI titers were measured in cynos after two immunizations (days 1 and 22) at two dose levels (0.2 and 0.4 mg) of formulated H7 mRNA administered IM and ID (Figures 4A and 4B). Formulated H10 mRNA was tested with only the 0.4-mg dose delivered ID and IM with the same immunization schedule (days 1 and 22) (Figure 4C). Both H10 and H7 mRNA vaccines generated HAI titers between 100 and 1,000 after a single immunization (day 15). HAI titers of 10,000 were generated for both H10 and H7 at 3 weeks following the second immunization (day 43), regardless of dose or route of administration. At 0.4 mg, the cynos experienced some systemic symptoms, such as warm to touch pain at the injection site, minor injection site irritation, and, in some cases, decreased food consumption following either H10 or H7 immunization. All symptoms resolved within 48–72 hr. Overall, both ID and IM administration elicited similar HAI titers regardless of dose, suggesting that lower doses may generate a similar HAI titer.
Figure 4.
Vaccination with Either H10 or H7 mRNA Generates Strong HAI Titers in Nonhuman Primates following ID and IM Immunizations
(A and B) Male or female cynomolgus monkeys (cynos) were immunized on day 1 with 0.2 or 0.4 mg formulated H7 mRNA, both IM and ID, and received a boosting immunization on day 22. Serum was collected on days 1, 8, 15, 22, 29, 36, and 43 to determine HAI titers. (C) Male and female cynos were immunized with 0.4 mg formulated H10 mRNA via an IM or ID route and received a boosting immunization on day 22. Serum was collected on days 1, 8, 15, 22, 29, 36, and 43 to determine HAI titers (n = 1/group).
H10 mRNA Immunogenicity and Safety in Humans
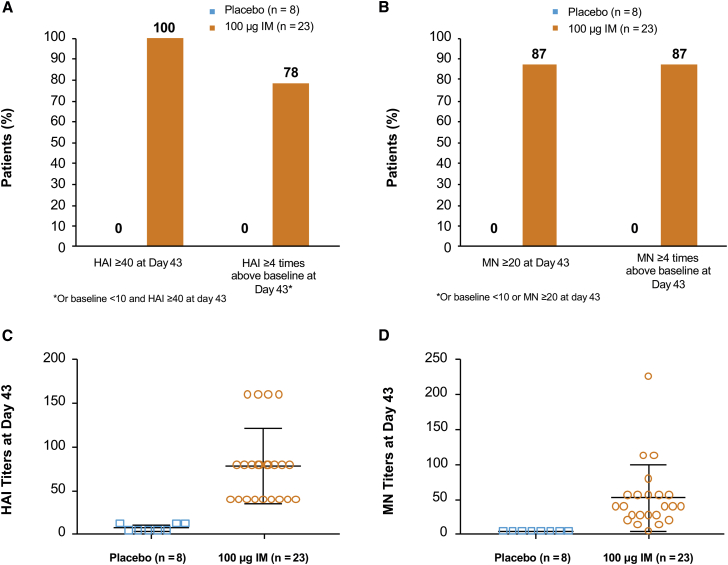
To evaluate the safety and immunogenicity of H10 mRNA in humans, a randomized, double-blind, placebo-controlled, dose-escalating phase 1 trial is ongoing (Clinical Trials Identifier NCT03076385). We report here interim results, obtained 43 days post-vaccination of 31 subjects (23 of whom received active H10 at 100 μg IM and eight of whom received placebo). Immunogenicity data show that 100% (n = 23) and 87% (n = 20) of subjects who received the H10 vaccine had an HAI ≥ 40 and MN ≥ 20 at day 43, respectively, compared to 0% of placebo subjects (Figures 5A and 5B). A total of 78% (n = 18) and 87% (n = 20) who received the H10 vaccine had an HAI baseline <10 and post-vaccination HAI ≥ 40 or HAI four or more times baseline, respectively, compared to 0% for placebo (Figures 5A and 5B). HAI geometric mean antibody titers of subjects given the H10 vaccine were 68.8 compared to 6.5 for placebo, and the MN geometric mean titers were 38.3 versus 5.0, respectively (Figures 5C and 5D).
Figure 5.
H10 mRNA Immunogenicity in Humans
(A and B) A greater percentage of subjects who received active vaccine had an HAI ≥ 40 (A) and MN ≥ 20 (B) compared to placebo. (C and D) HAI (C) and MN (D) titers of individual subjects were substantially more pronounced in those who received active vaccine compared to placebo. Error bars indicate SEM (100 μg IM, n = 23; placebo, n = 8).
The majority of adverse events (AEs) were mild (107/163 events; 66%) or moderate (52/163 events; 32%), using the Center for Biologics Evaluation and Research (CBER) severity scale.38 AEs were comparable in frequency, nature, and severity to unadjuvanted and adjuvanted H1N1 influenza vaccines.39 Twenty-three subjects who received 100 μg H10 IM reported 163 reactogenicity events with no idiosyncratic or persistent AEs observed. The majority of events were injection site pain, myalgia, headache, fatigue, and chills/common-cold-like symptoms (Table S2). Only four events (2.5%), reported by three subjects (13% of exposed subjects), were categorized as severe and included injection site erythema (1.2%), injection site induration (0.6%), and chills/common cold (0.6%) (Table 2; Table S2). No serious AE occurred and all events were expected and reversible. Overall, this reactogenicity profile is similar to that of a monovalent AS03-adjuvanted H1N1 vaccine, and it is comparable to that of meningococcal conjugate vaccine in healthy adults (19–55 years).40, 41
Table 2.
Number and Percentage of Subjects Who Experienced a Solicited Reactogenicity Event after Receiving 100 μg H10N8 mRNA IM or Placebo
| Parameter | 100 μg IM H10N8 mRNA n (%) | Placebo n (%) |
|---|---|---|
| Total number of subjects | 23 (100) | 8 (100) |
| Any reactogenicity event | 23 (100) | 5 (62.5) |
| Mild | 23 (100) | 3 (37.5) |
| Moderate | 12 (52.2) | 1 (12.5) |
| Severe | 3 (13.0) | 1 (12.5) |
| Any local reactogenicity event | 12 (91.3) | 2 (25.0) |
| Mild | 20 (87.0) | 2 (25.0) |
| Moderate | 9 (39.1) | 0 |
| Severe | 2 (8.7) | 0 |
| Any systemic reactogenicity event | 21 (91.3) | 5 (62.5) |
| Mild | 21 (91.3) | 3 (37.5) |
| Moderate | 11 (47.8) | 1 (12.5) |
| Severe | 1 (4.3) | 1 (12.5) |
Reactogenicity was defined as selected AE signs and symptoms occurring after dose administration that were reported by the subject using diary cards during the day of and 6 days after each dose administration. Events were categorized according to the toxicity grading scale for heathy adult and adolescent volunteers enrolled in preventative vaccine clinical trials (CBER 2007). AEs were defined as any unfavorable and unintended medical occurrence. Mild AEs were defined as those having no limitations in normal daily activities, moderate AEs as causing some limitations, and severe AEs were defined as events causing inability to perform normal daily activities. The total number of patients are those who received at least one dose of treatment. Percentages are based on the number of patients who reported at least one solicited reactogenicity event after treatment.
Discussion
Nucleic acid vaccines (NAVs) offer the potential to accurately express any protein antigen, whether intracellular, membrane bound, or secreted. Although first identified in the early 1990s, mRNA vaccines were not advanced into the clinic until recently due to concerns around stability and production.42, 43 The mRNA vaccines are produced by a well-controlled, enzymatic, and well-characterized scalable process that is agnostic to the antigen being produced. Additionally, host cell production and presentation of the antigen more closely resemble viral antigen expression and presentation than compared to an exogenously produced, purified, and formulated protein antigen. They offer advantages in speed, precision, adaptability of antigen design and production control that cannot be replicated with conventional platforms. This may be especially valuable for emerging infections, such as potential pandemic influenza.44 The mRNA vaccine platform described here allows for rapid mRNA production and formulation, within a few weeks, at sufficient quantities to support typical-sized clinical trials. Moreover, this mRNA-based vaccine technology overcomes the challenges other nucleotide approaches pose, such as pre-existing antivector immunity for viral vectors, and concern for genome integration, or the high doses and devices needed (e.g., electroporation), for DNA-based vaccines.
Other mRNA vaccine approaches have previously been reported for influenza.20, 45, 46, 47 Unmodified, sequence-optimized mRNA was used to generate H1-specific responses in mice, ferrets, and pigs at dose levels ∼4- to 8-fold higher than tested by us.20 Brazzoli et al.45 evaluated a self-amplifying mRNA that expressed H1 HA from the 2009 pandemic formulated with a cationic nanoemulsion in ferrets. HAI titers were low but measurable for the 15-μg dose (two of six responders) and at the 45-μg dose (three of six responders) after a single immunization. Following a boost, titers were measurable in all animals and provided protection to a homologous challenge strain.45 In another study, mice singly immunized against H1N1 (A/WSN/33), receiving a self-amplifying mRNA, showed no IgG responses after 7 days. After a second immunization, responses were boosted and animals were protected against a homologous challenge.46 Immunization in mice against either H1 or H7, with a self-amplifying mRNA, induced HAI and IgG titers that were comparable to those achieved in our study at similar doses (Figure 1).47 Our platform, therefore, is surprisingly efficacious when compared to existing self-replicating RNA approaches. It also offers potential additional advantages in terms of rapid onset of immunity, as shown by the protection from challenge achieved after one immunization at low doses (Figure 2), and manufacturability, since it obviates the need to produce very large-sized mRNAs to accommodate the self-replicating portions of the vectors (typically 7–9 kb).
Modified mRNA has been shown to express more efficiently than unmodified mRNA, likely due to its reduced indiscriminate activation of innate immunity.29 When included in a vaccine formulation, our modified-mRNA technology balances immune stimulation and antigen expression, leading to very potent immune responses that are superior to unmodified mRNA approaches. The very high, transient levels of protein, expressed shortly after administration, are similar to what is seen during a viral infection. Indeed, the biodistribution we observed (Table 1; Table S1) is similar to an influenza virus, where virus could be measured outside the primary site of inoculation after 5 days.48 Importantly, there was no way for our vaccine to revert to a virulent form because key parts of the virus were missing, including any nonstructural elements or capsid structures.
We selected LNPs for delivery of the mRNA as they have been validated in the clinic for siRNA and are well tolerated compared to other nonviral delivery systems.22, 49 Other groups have relied on either exogenous RNA as an adjuvant or on the adjuvant properties generated during self-amplification of the mRNA. Using an LNP, we generate very high levels of transient expression without the need for additional immunostimulatory compounds.
In the studies summarized here, we demonstrated that the LNP-based, modified-mRNA vaccine technology is able to generate robust and protective immune responses in mice, ferrets, and cynomolgus monkeys. In animals, we showed that a range of doses of formulated mRNA encoding the HA protein of either H7N9 or H10N8 is able to stimulate rapid, robust, and long-lasting, immune responses, as measured by HAI, MN assay, and protection from viral challenge. A single vaccination on day 0 with as little as 0.4 μg was shown to protect mice against challenge with H7N9 on days 7, 21, and 84 (Figure 2), despite the fact that H7 HA has demonstrated relatively poor immunogenicity.50, 51 Increased survival of mice vaccinated with H7 HA and challenged with H7N9 (A/Anhui/1/2013) at early time points (Figure 2) suggests additional mechanism(s) of protection, since HAI titers were below the level of detection (Figure S5). T cells have been shown to elicit protection against pandemic influenza strains.52, 53 We detected T cell responses to both H10 and H7 vaccines at multiple doses (Figures S2C and S2D). Additional follow-up studies are ongoing, to determine whether T cell responses alone offer protective benefits, to lend insight into the specific mechanism of vaccine protection.
These interim results of H10 mRNA vaccination in humans are the first published example of a nucleic acid vaccine against an infectious disease working in man without the use of electroporation. Although strategies, such as electroporation, have been developed to increase the efficacy of DNA-based vaccines, they continue to have relatively poor immunogenicity compared to protein vaccines.54 Initial data from the first-in-human trial appear to confirm a robust immune response with a safe and well-tolerated profile. However, the full dataset from the trial will need to be evaluated in order to confirm this interim analysis. Nonetheless, these results are encouraging in that microgram-dose levels provided immunogenicity with a safety profile comparable to traditional vaccines.40, 41
The completion of these and additional clinical trials is needed to confirm whether mRNA vaccines will become an effective vaccine platform that can overcome many of the shortcomings of conventional vaccines. Our initial findings are nonetheless encouraging and provide support for further clinical exploration.
Materials and Methods
mRNA Synthesis and Formulation
Our mRNA was synthesized in vitro by T7 polymerase-mediated transcription from a linearized DNA template, which incorporates 5′ and 3′ UTRs, including a poly-A tail.55 The mRNA is purified and resuspended in a citrate buffer at the desired concentration. A donor methyl group S-adenosylmethionine (SAM) is added to methylated capped RNA (cap-0), resulting in a cap-1 to increase mRNA translation efficiency.56
LNP formulations were prepared using a modified procedure of a method previously described for siRNA.57 Briefly, lipids were dissolved in ethanol at molar ratios of 50:10:38.5:1.5 (ionizable lipid: 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC): cholesterol: PEG-lipid). The lipid mixture was combined with a 50 mM citrate buffer (pH 4.0) containing mRNA at a ratio of 3:1 (aqueous:ethanol) using a microfluidic mixer (Precision Nanosystems). Formulations were dialyzed against PBS (pH 7.4) in dialysis cassettes for at least 18 hr. Formulations were concentrated using Amicon ultra centrifugal filters (EMD Millipore), passed through a 0.22-μm filter, and stored at 4°C until use. All formulations were tested for particle size, RNA encapsulation, and endotoxin, and they were found to be between 80 and 100 nm in size, with >90% encapsulation and <1 EU/mL endotoxin.
In Vitro Expression
The day before transfection, 400,000 HeLa cells (ATCC) were seeded in a six-well cell culture plate, and 2.5 μg of either H10 or H7 HA mRNA was transfected using the Transit mRNA transfection kit (Mirus Bio). Recovered protein lysate, 30 μg, was resolved on a NuPage Novex 4%–12% Bis-Tris Protein Gel and transferred onto nitrocellulose using an iBlot 2 (7-min transfer). Blots were incubated with either anti-H10 HA polyclonal antibody (rabbit, 11693; Sino Biological) or anti-H7 HA monoclonal antibody (mouse, 11082-MM04; Sino Biological) overnight at 4°C. Included as positive controls were 0.5 μg recombinant H10 HA protein (1505-001; IBT) and recombinant H7 HA protein (1502-001; IBT). A polyclonal antibody against actin was also included as a loading control (rabbit, A2066; Sigma-Aldrich). Blots were scanned and analyzed on an Odyssey CLx (LI-COR Biosciences).
Animal Studies
Female BALB/c mice 5–8 weeks old were purchased from Charles River Laboratories and housed at the study site (Noble Life Sciences or Moderna Therapeutics,). For mouse H7N9 challenge studies, female BALB/c mice 7–8 weeks old were purchased from Harlan Laboratories and housed at MRIGlobal’s ABSL-3 facility.
Male ferrets 13–15 weeks old (Triple F Farms) with a baseline HAI titer of ≤20 to influenza virus, A/California/07/2009 (H1N1), A/Wisconsin/15/2009 (H3N2), and B/Massachusetts/2/2012, were used for studies at MRIGlobal’s ABSL-3 facility.
Nonhuman primate studies were conducted at Charles River Laboratories using naive cynomolgus monkeys (cynos), 2–4 years old, weighing 2–6 kg. Animals were housed in stainless steel, perforated-floor cages, in a temperature- and humidity-controlled environment (21°–26°C and 30%–70%, respectively), with an automatic 12-hr dark/light cycle. Animals were fed PMI Nutrition Certified Primate Chow No. 5048 twice daily. Tuberculin tests were carried out on arrival at the test facility. The study plan and procedures were approved by PCS-SHB Institutional Animal Care and Use Committee (IACUC). Animal experiments and husbandry followed the NIH (NIH Publications No. 8023, eighth edition) and the USA National Research Council and the Canadian Council on Animal Care (CCAC) guidelines. No treatment randomization or blinding methods were used for any of the animal studies. Sample sizes were determined by the resource equation method.
First-in-Human Phase 1 Study
A single-center, randomized, double-blind, placebo-controlled, dose-ranging study is ongoing to evaluate the safety and immunogenicity of H10N8 antigen mRNA in humans between the ages of 18 and 64 (Clinical Trials Identifier NCT03076385). Subjects are being followed for up to 1 year post-vaccination for safety and immunogenicity. Only interim analysis (day 43) of one dose-group cohort (100 μg IM) in healthy adults is reported (all other analyses are ongoing).
Briefly, males and females were eligible for this study if they had a body mass index between 18.0 and 30.0 kg/m2, were considered in general good health with no ongoing acute or chronic illness, did not have any asymptomatic (e.g., mild hypertension) or any suspected immunosuppressive condition, or and did not have a history of serious reactions to influenza vaccinations or Guillain-Barre Syndrome. Eligible adults were randomized at a ratio of 3:1 to receive either H10N8 mRNA 100 μg IM or placebo. All study personnel who conducted assessments were blinded to treatment. Immunogenicity was determined by HAI and MN assays.
Safety was assessed from solicited (local and systemic reactogenicity events) and unsolicited AEs via scheduled clinic visit (vital signs, laboratory assessments, and physical examinations), subject diaries, and follow-up telephone calls at specific intervals. AEs were defined as any problematic medical occurrence even if seemingly unrelated to treatment and graded by the Toxicity Grading Scale and defined as mild (transient with no normal daily activity limitations), moderate (some normal daily limitations), and severe (unable to perform normal daily activities).38 Serious AEs were defined as any occurrence of death, a life-threatening situation, hospitalization, persistent or significant disability/incapacity, congenital anomaly/birth defect, or any medical event that jeopardizes the subject or requires medical intervention. A safety review committee reviewed safety data at key intervals throughout the study before allowing dose expansion or dose escalation. Prior to study enrollment, all subjects completed a written informed consent in accordance with all applicable local- and country-specific regulations. This study was conducted by PAREXEL International and was reviewed and approved by an Independent Ethics Committee. This study was conducted in compliance with the International Conference on Harmonization Good Clinical Practice guidelines and the ethical principles of the Declaration of Helsinki.
Immunizations
For mouse IM immunizations, 50 μL was injected in either the left or right quadriceps. For ferret and mouse ID immunizations, the needle was inserted bevel up with the point visualized through the skin. The vaccine (50 μL) was administered slowly, creating a blister-like formation.
For ID delivery to cynos, the material was injected into the lumbar region in a 100 μL vol for the 0.2-mg dose and delivered at two sites for the 0.4-mg dose (0.2 mg in 100 μL per site). For IM delivery to cynos, the material was injected into the left thigh in a 100 μL vol for the 0.2-mg dose or a 200 μL vol for the 0.4-mg dose.
In the human study, each subject in the 100-μg IM cohort received two treatment doses on day 1 and day 22. Each subject received their vaccine via IM administration according to standard procedures in their deltoid muscle, with the second dose administered in the same arm.
Viral Challenges
Influenza strain A/Anhui/1/2013 (H7N9) was grown and characterized at MRIGlobal to a concentration of 3.3 × 108 TCID50/mL. BALB/c mice were challenged via IN instillation (2.5 × 105 TCID50 in 50 μl Dulbecco’s phosphate-buffered saline [DPBS]). Anesthetized ferrets were inoculated IN with 1 × 106 TCID50 with 2 × 250 μL per nostril.
Serum Collection
Approximately 200 μL blood was collected from mice via tail vein or retro-orbital bleed (1 mL for terminal bleeds) and centrifuged at 1,200 × g for serum isolation (10 min at 4°C). Collected blood (1–3 mL) from the ferrets’ cranial vena cava was processed to serum using a serum separator tube (SST). Blood collected from the peripheral vein of cynos (0.5 mL) was centrifuged at 1,200 × g (10 min at 4°C). All serum was frozen immediately and stored at −80°C.
In the human study, blood samples for immunogenicity analysis were collected via intravenous cannula or by direct venipuncture of the forearm. Serum samples were stored and transported under controlled conditions to Synexa Life Sciences for HAI analysis and to Southern Research Institute for MN testing.
Lung Homogenate
Ferrets were then euthanized by intraperitoneal injection of Euthasol, and lungs (1 cm3 of the lower part of each of the three right lung lobes), nasal turbinates, and a portion of the trachea were collected. The lung portions were weighed and immediately homogenized and tested in the TCID50 assay.
TCID50 Assay
Influenza virus levels in nasal washes and lung homogenates were determined by TCID50 assay. Madin-Darby canine kidney (MDCK) cells were seeded in 96-well plates in serum-free media and incubated at 37°C with 5% CO2. Nasal washes and lung homogenates (four to eight replicates) were serially diluted in serum-free media and added to plates that were ≥95% confluent after a single wash. Cytopathic effects (CPEs) were determined after 3–5 days at 37°C with 5% CO2. The TCID50/mL was calculated using the lowest dilution at which CPE was observed. Lung homogenate results were reported as TCID50/g lung tissue.
Biodistribution Studies
Male CD-1 mice received 300 μg/kg (6 μg) H10 HA mRNA (50 uL vol) via ID or IM (left side) administration. Blood, heart, lung, spleen, kidney, liver, and skin injection sites were collected pre-dose and 2, 4, 8, 24, 48, 72, and 96 hr post-ID dosing (n = 4 mice/time point). Two replicates each of bone marrow (left and right femur), lung, liver, heart, right kidney, inguinal- and popliteal-draining lymph nodes, axillary distal lymph nodes, spleen, brain, stomach, ileum, jejunum, cecum, colon, rectum, testes (bilateral), and injection site muscle were collected pre-dose and 2, 8, 24, 48, 72, 120, 168, and 264 hr post-IM dosing (n = 3 mice/time point). Blood samples were collected from jugular venipuncture at study termination.
H10 HA mRNA quantification for both serum and tissues was performed by AxoLabs using the Quantigene 2.0 branched DNA (bDNA) Assay (Panomics/Affymetrix).57 A standard curve on each plate of known amounts of mRNA (added to untreated tissue samples) was used to quantitate the mRNA in treated tissues. The calculated amount in picograms (pg) was normalized to the amount of weighed tissue in the lysate applied to the plate.
Luciferase Studies
Female BALB/c mice 6–8 weeks old were dosed with formulated luciferase mRNA via IM or ID administration at four dose levels as follows: 10, 2, 0.4, and 0.08 μg (n = 6 per group). At 6, 24, 48, 72, and 96 hr post-dosing, animals were injected with 3 mg luciferin and imaged on an in vivo imaging system (IVIS Spectrum, PerkinElmer). At 6 hr post-dosing, three animals were sacrificed and dissected, and the muscle, skin, draining lymph nodes, liver, and spleen were imaged ex vivo.
MN Assay
Heat-inactivated serum was serially diluted on 96-well plates, and ∼2 × 103 TCID50/mL H7N9 (A/Anhui/1/2013) was added to each dilution. Following a 1-hr incubation at room temperature, the serum/virus mixtures from each well were transferred to plates containing MDCK cells and incubated at 37°C (5% CO2). After 3–5 days, the CPE titer was determined based on the most dilute sample at which no CPE was observed. Each sample was tested three times, and the geometric mean of the three replicates was reported as the overall titer.
HAI Assay
The HAI titers of serum samples in both the animal and human studies were determined using a protocol adapted from the World Health Organization protocol.18 Sera were first treated with receptor-destroying enzyme (RDE) to inactivate nonspecific inhibitors. RDE was inactivated by incubation at 56°C for 30 min. Treated sera were serially diluted in 96-well plates, mixed with a standardized amount of recombinant HA (eight HA units of H10N8 or H7N9 rHA; Medigen), and incubated for 30 min at room temperature. Turkey red blood cells (RBCs) (Lampire Biological Laboratories) were then added to the wells of the 96-well plates, mixed, and incubated at room temperature for 45 min. The most dilute serum sample that completely inhibited hemagglutination was the reported titer for that replicate. Each serum sample was analyzed in triplicate and the results are reported as the geometric mean of the three results.
IFNγ ELISpot
Mouse IFNγ ELISpot assays were performed using the IFNγ pre-coated ELISpot kit catalog 3321-4APW (MabTech), according to the manufacturer’s protocol. Briefly, the plates were blocked using complete RPMI (R10) and incubated for 30 min prior to plating cells. Peptide libraries for H7 or H10 were diluted to a final concentration of 10 μg/mL. Mouse splenocytes were pooled by group and plated at 600,000 cells/well, with peptide, phorbol myristate acetate (PMA) + Ionomycin or R10 media alone. Cells were stimulated in a total volume of 125 μL/well. Plates were then incubated at 37°C, 5% CO2 for 18–24 hr. Assay plates were developed and counted using the automated ELISpot reader CTL ImmunoSpot/FluoroSpot. Overlapping peptide libraries (15mers with ten amino acid overlaps) for H10 HA (A/Jiangxi-Donghu/346/2013) and H7 HA (A/Anhui/1/2013) were ordered from Genscript.
Statistical Analysis and Data Collection
In general, two datasets were compared by two sample t test and more than two groups were compared by ANOVA proc mixed model. Two-way ANOVA was used to analyze titers in lung tissue. Survival curves were compared via log-rank (Mantel-Cox) test. Statistical analyses for the animal studies were performed with GraphPad Prism 6. The phase 1 human clinical trial is being conducted by PAREXEL International, and data were collected utilizing their electronic records ClinBase system. All statistical analyses for the human trial are performed using SAS (SAS Institute, version 9.1 or higher).
Author Contributions
G.C. led and planned the studies. K.B. and G.C. contributed to the experimental design and analysis of all in vivo studies. O.Y. contributed to the HAI analysis. J.J.S. and A.B. contributed to the toxicology and biodistribution studies, respectively. L.A.B., K.J.H., and O.A. contributed to formulation design and expression optimization. M.E.L. and M.S. contributed to mRNA synthesis and process optimization. A.R., T.Z., and M.W. supervised the conduct of the human study. All authors drafted and revised the manuscript for critical intellectual content and have reviewed and approved the final paper.
Conflicts of Interest
This study was funded by Valera Therapeutics, a Moderna Therapeutics venture. Authors K.B., O.Y., K.J.H., A.R., M.W., and G.C. are employees of Valera Therapeutics. Authors J.J.S., A.B., L.A.B., M.E.L, M.S., O.A., J.T., and T.Z. are employees of Moderna Therapeutics.
Acknowledgments
Statistical analyses were conducted by Georges Carlettis of Strategiestat and Katherine Kacena of BioBridges. Editorial support was provided by Stephanie Eide of BioBridges.
Footnotes
Supplemental Information includes seven figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.03.035.
Supplemental Information
References
- 1.Pascua P.N., Choi Y.K. Zoonotic infections with avian influenza A viruses and vaccine preparedness: a game of “mix and match”. Clin. Exp. Vaccine Res. 2014;3:140–148. doi: 10.7774/cevr.2014.3.2.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fineberg H.V. Pandemic preparedness and response--lessons from the H1N1 influenza of 2009. N. Engl. J. Med. 2014;370:1335–1342. doi: 10.1056/NEJMra1208802. [DOI] [PubMed] [Google Scholar]
- 3.Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 5.Bouvier N.M., Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan J., Zhang L., Kan X., Jiang L., Yang J., Guo Z., Ren Q. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin. Infect. Dis. 2013;57:1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- 7.Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W., Chen J., Jie Z., Qiu H., Xu K. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 8.Chen H., Yuan H., Gao R., Zhang J., Wang D., Xiong Y., Fan G., Yang F., Li X., Zhou J. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 9.Su W., Wang C., Luo J., Zhao Y., Wu Y., Chen L., Zhao N., Li M., Xing C., Liu H. Testing the effect of internal genes derived from a wild-bird-origin H9N2 influenza A virus on the pathogenicity of an A/H7N9 Virus. Cell Rep. 2015;12:1831–1841. doi: 10.1016/j.celrep.2015.08.029. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization (2017). Influenza at the human-animal interface. http://www.who.int/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_01_16_2017_FINAL.pdf?ua=1.
- 11.Zhang W., Wan J., Qian K., Liu X., Xiao Z., Sun J., Zeng Z., Wang Q., Zhang J., Jiang G. Clinical characteristics of human infection with a novel avian-origin influenza A(H10N8) virus. Chin. Med. J. (Engl.) 2014;127:3238–3242. [PubMed] [Google Scholar]
- 12.Moscona A. Neuraminidase inhibitors for influenza. N. Engl. J. Med. 2005;353:1363–1373. doi: 10.1056/NEJMra050740. [DOI] [PubMed] [Google Scholar]
- 13.Rimmelzwaan G.F., McElhaney J.E. Correlates of protection: novel generations of influenza vaccines. Vaccine. 2008;26(Suppl 4):D41–D44. doi: 10.1016/j.vaccine.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 14.Li C.K., Rappuoli R., Xu X.N. Correlates of protection against influenza infection in humans--on the path to a universal vaccine? Curr. Opin. Immunol. 2013;25:470–476. doi: 10.1016/j.coi.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Reperant L.A., Rimmelzwaan G.F., Osterhaus A.D. Advances in influenza vaccination. F1000Prime Rep. 2014;6:47. doi: 10.12703/P6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soema P.C., Kompier R., Amorij J.P., Kersten G.F. Current and next generation influenza vaccines: Formulation and production strategies. Eur. J. Pharm. Biopharm. 2015;94:251–263. doi: 10.1016/j.ejpb.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 17.Houser K., Subbarao K. Influenza vaccines: challenges and solutions. Cell Host Microbe. 2015;17:295–300. doi: 10.1016/j.chom.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization (2009). Pandemic influenza vaccine manufacturing process and timeline. http://www.who.int/csr/disease/swineflu/notes/h1n1_vaccine_20090806/en/.
- 19.Treanor J.J. CLINICAL PRACTICE. Influenza Vaccination. N. Engl. J. Med. 2016;375:1261–1268. doi: 10.1056/NEJMcp1512870. [DOI] [PubMed] [Google Scholar]
- 20.Petsch B., Schnee M., Vogel A.B., Lange E., Hoffmann B., Voss D., Schlake T., Thess A., Kallen K.J., Stitz L., Kramps T. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012;30:1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 21.Kallen K.J., Heidenreich R., Schnee M., Petsch B., Schlake T., Thess A., Baumhof P., Scheel B., Koch S.D., Fotin-Mleczek M. A novel, disruptive vaccination technology: self-adjuvanted RNActive(®) vaccines. Hum. Vaccin. Immunother. 2013;9:2263–2276. doi: 10.4161/hv.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coelho T., Adams D., Silva A., Lozeron P., Hawkins P.N., Mant T., Perez J., Chiesa J., Warrington S., Tranter E. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 23.Karikó K., Ni H., Capodici J., Lamphier M., Weissman D. mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 24.Desmet C.J., Ishii K.J. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat. Rev. Immunol. 2012;12:479–491. doi: 10.1038/nri3247. [DOI] [PubMed] [Google Scholar]
- 25.Coffman R.L., Sher A., Seder R.A. Vaccine adjuvants: putting innate immunity to work. Immunity. 2010;33:492–503. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cláudio N., Dalet A., Gatti E., Pierre P. Mapping the crossroads of immune activation and cellular stress response pathways. EMBO J. 2013;32:1214–1224. doi: 10.1038/emboj.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Karikó K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozenski J., Crain P.F., McCloskey J.A. The RNA modification database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell. 2017;168:1114–1125.e10. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hickling J.K., Jones K.R., Friede M., Zehrung D., Chen D., Kristensen D. Intradermal delivery of vaccines: potential benefits and current challenges. Bull. World Health Organ. 2011;89:221–226. doi: 10.2471/BLT.10.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riede O., Seifert K., Oswald D., Endmann A., Hock C., Winkler A., Salguero F.J., Schroff M., Croft S.L., Juhls C. Preclinical safety and tolerability of a repeatedly administered human leishmaniasis DNA vaccine. Gene Ther. 2015;22:628–635. doi: 10.1038/gt.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheets R.L., Stein J., Bailer R.T., Koup R.A., Andrews C., Nason M., He B., Koo E., Trotter H., Duffy C. Biodistribution and toxicological safety of adenovirus type 5 and type 35 vectored vaccines against human immunodeficiency virus-1 (HIV-1), Ebola, or Marburg are similar despite differing adenovirus serotype vector, manufacturer’s construct, or gene inserts. J. Immunotoxicol. 2008;5:315–335. doi: 10.1080/15376510802312464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terenin I.M., Andreev D.E., Dmitriev S.E., Shatsky I.N. A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent. Nucleic Acids Res. 2013;41:1807–1816. doi: 10.1093/nar/gks1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouvier N.M., Lowen A.C. Animal models for influenza virus pathogenesis and transmission. Viruses. 2010;2:1530–1563. doi: 10.3390/v20801530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belser J.A., Katz J.M., Tumpey T.M. The ferret as a model organism to study influenza A virus infection. Dis. Model. Mech. 2011;4:575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.US Department of Health and Human Services (2007). Guidance for industry toxicity grading scale for healthy adult and adolescent volunteers enrolled in preventive vaccine clinical trials. https://www.fda.gov/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/Vaccines/ucm074775.htm. [DOI] [PubMed]
- 39.Greenberg M.E., Lai M.H., Hartel G.F., Wichems C.H., Gittleson C., Bennet J., Dawson G., Hu W., Leggio C., Washington D., Basser R.L. Response to a monovalent 2009 influenza A (H1N1) vaccine. N. Engl. J. Med. 2009;361:2405–2413. doi: 10.1056/NEJMoa0907413. [DOI] [PubMed] [Google Scholar]
- 40.Reisinger K.S., Baxter R., Block S.L., Shah J., Bedell L., Dull P.M. Quadrivalent meningococcal vaccination of adults: phase III comparison of an investigational conjugate vaccine, MenACWY-CRM, with the licensed vaccine, Menactra. Clin. Vaccine Immunol. 2009;16:1810–1815. doi: 10.1128/CVI.00207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roman F., Vaman T., Kafeja F., Hanon E., Van Damme P. AS03(A)-Adjuvanted influenza A (H1N1) 2009 vaccine for adults up to 85 years of age. Clin. Infect. Dis. 2010;51:668–677. doi: 10.1086/655830. [DOI] [PubMed] [Google Scholar]
- 42.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 43.Weide B., Pascolo S., Scheel B., Derhovanessian E., Pflugfelder A., Eigentler T.K., Pawelec G., Hoerr I., Rammensee H.G., Garbe C. Direct injection of protamine-protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J. Immunother. 2009;32:498–507. doi: 10.1097/CJI.0b013e3181a00068. [DOI] [PubMed] [Google Scholar]
- 44.Partridge J., Kieny M.P., World Health Organization H1N1 influenza vaccine Task Force Global production of seasonal and pandemic (H1N1) influenza vaccines in 2009-2010 and comparison with previous estimates and global action plan targets. Vaccine. 2010;28:4709–4712. doi: 10.1016/j.vaccine.2010.04.083. [DOI] [PubMed] [Google Scholar]
- 45.Brazzoli M., Magini D., Bonci A., Buccato S., Giovani C., Kratzer R., Zurli V., Mangiavacchi S., Casini D., Brito L.M. Induction of broad-based immunity and protective efficacy by self-amplifying mRNA vaccines encoding influenza virus hemagglutinin. J. Virol. 2015;90:332–344. doi: 10.1128/JVI.01786-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chahal J.S., Khan O.F., Cooper C.L., McPartlan J.S., Tsosie J.K., Tilley L.D., Sidik S.M., Lourido S., Langer R., Bavari S. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl. Acad. Sci. USA. 2016;113:E4133–E4142. doi: 10.1073/pnas.1600299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hekele A., Bertholet S., Archer J., Gibson D.G., Palladino G., Brito L.A., Otten G.R., Brazzoli M., Buccato S., Bonci A. Rapidly produced SAM(®) vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2013;2:e52. doi: 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mori I., Komatsu T., Takeuchi K., Nakakuki K., Sudo M., Kimura Y. Viremia induced by influenza virus. Microb. Pathog. 1995;19:237–244. doi: 10.1016/s0882-4010(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 49.Kanasty R., Dorkin J.R., Vegas A., Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 50.Bart S.A., Hohenboken M., Della Cioppa G., Narasimhan V., Dormitzer P.R., Kanesa-Thasan N. A cell culture-derived MF59-adjuvanted pandemic A/H7N9 vaccine is immunogenic in adults. Sci. Transl. Med. 2014;6:234ra55. doi: 10.1126/scitranslmed.3008761. [DOI] [PubMed] [Google Scholar]
- 51.Fries L.F., Smith G.E., Glenn G.M. A recombinant viruslike particle influenza A (H7N9) vaccine. N Engl J Med. 2013;369:2564–2566. doi: 10.1056/NEJMc1313186. [DOI] [PubMed] [Google Scholar]
- 52.Lu I.N., Farinelle S., Sausy A., Muller C.P. Identification of a CD4 T-cell epitope in the hemagglutinin stalk domain of pandemic H1N1 influenza virus and its antigen-driven TCR usage signature in BALB/c mice. Cell. Mol. Immunol. 2016 doi: 10.1038/cmi.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weinfurter J.T., Brunner K., Capuano S.V., 3rd, Li C., Broman K.W., Kawaoka Y., Friedrich T.C. Cross-reactive T cells are involved in rapid clearance of 2009 pandemic H1N1 influenza virus in nonhuman primates. PLoS Pathog. 2011;7:e1002381. doi: 10.1371/journal.ppat.1002381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L., Saade F., Petrovsky N. The future of human DNA vaccines. J. Biotechnol. 2012;162:171–182. doi: 10.1016/j.jbiotec.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuge H., Brownlee G.G., Gershon P.D., Richter J.D. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 1998;26:3208–3214. doi: 10.1093/nar/26.13.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collins M.L., Irvine B., Tyner D., Fine E., Zayati C., Chang C., Horn T., Ahle D., Detmer J., Shen L.P. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.