Abstract
O-GlcNAc-ylation is the post-translational addition of an O-linked β-N-acetylglucosamine to the serine and threonine residues of thousands of proteins in eukaryotic cells. Specifically, half of the thirty different types of protein components in the nuclear pore complex (NPC) are modified by O-GlcNAc, of which the majority are intrinsically disordered nucleoporins (Nups) containing multiple phenylalanine-glycine (FG) repeats. Moreover, these FG-Nups form a strict selectivity barrier with a high density of O-GlcNAc in the NPC to mediate bidirectional trafficking between the cytoplasm and nucleus. However, the roles that O-GlcNAc plays in the structure and function of the NPC remain obscure. In this review paper, we will discuss the current knowledge of O-GlcNAc-ylated Nups, highlight some new techniques used to probe O-GlcNAc's roles in the nuclear pore, and finally propose a new model for the effect of O-GlcNAc on the NPC's permeability.
Keywords: glycosylation, nucleoporins, nuclear pore complex
O-GlcNAc is a universal post-translation modification in all metazoans
O-GlcNAc-ylation is a post-translational modification (PTM) where an uncharged acetylated glucosamine is attached by glycosidic linkage to the hydroxyl group of serine and threonine residues1. Though not as well-known as other PTMs, such as phosphorylation or ubiquitination, O-GlcNAc-ylation diverts approximately 5% of all glucose from cellular metabolism to the nutrient dependent hexosamine pathway1,2. Being the nucleotide sugar substrate for O-GlcNAc-ylation, uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) is attached to and removed from the loops and intrinsically disordered regions of approximately 4000 target proteins1 by O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) in eukaryotic cells2,3. Splice variants of the OGT and OGA genes have been shown to sequester the enzymes to the cytoplasm, nucleus, and mitochondria4,5,6. OGT is a highly conserved glycosyltransferase, retaining up to 80% of its identity in a variety of eukaryotic organisms. However, there is little sequence or structural similarities between OGT and other glycosyltransferases7, suggesting an early evolutionary divergence and fundamental cellular function. Despite the dissimilarity with other glycosyltransferases, OGT does contain the common protein motif tetratricopeptide repeats (TPR), which is indicative of a regulatory complex7 and mediates protein-protein interactions8. In general, OGT has a strong preference for intrinsically disordered regions possibly due to the fact that there are many binding pockets along the enzyme's superhelical architecture composed of 11.5 TPR repeats. A subset of these pockets may be responsible for recognizing a single region on a substrate, which may not necessarily be structured in a distinct linear order. These intrinsically disordered regions are proposed to contort into a shape so that a sufficient number of its regions are recognized by the binding pockets on OGT9.
Extensive studies have been conducted to characterize a consensus sequence for O-GlcNAc-ylation. Approximately 1750 sites of O-GlcNAc-ylation have been experimentally determined in the murine synaptosome, in which a 19- and a 25-amino acid length sequence showed preference for O-GlcNAc-ylation with 439 and 130 occurrences respectively. Additionally, in a moderate number of O-GlcNAc-ylation sites, there is a proline two or three resides towards the amino-terminal or, less frequently, a valine one or three residues amino-terminal. Previously, PVXS/T had been suggested as a possible O-GlcNAc-ylation motif, however, this motif occurred at less than 20% of all identified sites10,11. The above results might suggest the absence of a universally consensus motif1,12. Another factor that makes it challenging to identify such a motif is the transience of the O-GlcNAc modification. Previous studies revealed that the turnover rate of the O-GlcNAc modification is much faster than that of the modified proteins, but still is not as fast as phosphorylation in signaling pathways7,12,13,14.
Many cellular processes are known to be targeted by both OGT and OGA, including higher order chromatin structure, histones, RNA polymerase II, transcription factors, lysosomal degradation, T cell activation, insulin signaling, glucose metabolism, and cell cycle progression1,12. One of the most widely studied processes affected by O-GlcNAc-ylation is the cellular stress response. In model organisms, such as Drosophila, C. elegans, and D. rerio, intracellular O-GlcNAc levels have been shown to increase in response to oxidative stress, osmotic stress, ethanol, UV light, and heat shock2,15,16,17,18 as well as improve cell survival during the normal life cycle15 and during development17. Furthermore, dysregulated O-GlcNAc-ylation has also been linked to diabetes, cardiovascular disease, neurodegenerative disorders, and cancer19,20,21,22,23,24. For instance, upregulated O-GlcNAc-ylation resulting from chronically high glucose has been identified as a key player in insulin resistance, a main indicator of diabetes25. It most likely interferes downstream of O-GlcNAcase26 in the insulin signaling cascade, resulting in increased O-GlcNAc-ylation of insulin receptor substrate 1 and beta-catenin27, as well as in glucose metabolism, at the level of glycogen synthase28.
Due to the close relationship with glucose metabolism, it is likely that cellular nutrition status influences the level of O-GlcNAc-ylation of target proteins, such as histones and non-histone proteins like Polycomb and Trithorax which modify chromatin structure29,30,31. In fact, OGT is a Polycomb group, originally characterized as sxc in D. melanogaster, whose chromatin targets coincide with Polycomb binding sites32. Counterintuitively, glucose deprivation has also been shown to cause an increase in O-GlcNAc modification in HepG2 human liver cells33, indicating not a linear relationship between O-GlcNAc and glucose but rather a homeostatic relationship where O-GlcNAc-ylation compensates for disruptions in glucose abundance. Further complicating this relationship is the fact that there is a class of proteins that form the majority of the nuclear pore complex (NPC) that appear to be O-GlcNAc-ylated to some degree regardless of glucose abundance34,35. They would therefore not be good nutrition sensors without extremely sensitive tools and most likely serve some other function in addition to the metabolism of glucose.
The NPC is a gatekeeper between the nuclear and cytoplasmic compartments
The NPC is a 120 – 125 MDa36,37 protein complex structure composed of approximately 30 unique nucleoporins (Nups) in multiple copies embedded in the nuclear envelope of eukaryotic cells. The NPC allows for efficient trafficking, up to 1000 transport events per second38, of soluble proteins and RNAs between the cytoplasmic and nuclear compartments36. With eight-fold radial symmetry36, the NPC possesses an hourglass-shaped central channel with ∼50 nm in diameter at its narrowest width36 and ∼200 nm in total length37. The central channel of NPC is filled with flexible intrinsically disordered Nups containing many phenylalanine-glycine (FG) repeats39. Critically, these FG Nups form a selectivity barrier allowing for the passive diffusion of signal-independent cargo (< 40 kDa)36 and the facilitated transport of signal-dependent cargo by the assistance of transport receptors. Both transport modes could account for the translocation of various proteins, mRNPs, snRNPs, pre-ribosomal subunits, and tRNAs between the nucleus and cytoplasm in eukaryotic cells36. Unfortunately, it is still unclear specifically how the FG Nups establish the NPC's selectivity barrier. It appears that hydrophobic interactions between the FG Nups are required to make it energetically unfavorable for large passively diffusing molecules to pass through the NPC40. While, if large cargos contain a nuclear localization sequence or nuclear export signal, they will be recognized by transport receptors and these transport receptors can directly interact with FG Nups to complete the nucleocytoplasmic transport for these cargos36.
Glycosylation in the NPC
Based on early immunostaining and electron microscopy studies, the FG Nups in the central scaffold of the NPC were revealed to be heavily O-GlcNAc-ylated34,41,42. Soon after, protein purification and mass spectrometry studies further showed that all FG Nups34, and also some structural Nups were O-GlcNAc modified in the NPC34,43. It is believed that fundamentally O-GlcNAc-ylation is necessary in the NPC to prevent degradation of modified Nups35. Evidently, inhibition or knock out of OGT caused the loss of commonly O-GlcNAc-ylated Nup93 and several FG Nups, including Nup62, Nup153, Nup214, and Nup358, from the NPC by increasing their ubiquitination and degradation35. The fundamental mechanism suggested is that O-GlcNAc could prevent degradation by disrupting inappropriate aggregation of the Nups35. Lending credibility to this conclusion is the fact that in vitro experiments showed that a hydrogel composed of O-GlcNAc-ylated Nup98 was devoid of amyloid-like beta structures, which were characteristic of Nup-hydrogels without O-GlcNAc44. Consequently, loss or inhibition of OGT and the subsequent loss of Nups led to an inability of NPCs to exclude 70-kDa dextran molecules, and a decrease in the initial rate of facilitated transport as well35. Similar results were obtained in studies of X. laevis nuclei depleted of O-GlcNAc-ylated FG Nups45. Another early study by Finlay et al focused on facilitated transport through NPCs that were devoid of O-GlcNAc-ylated Nups. They showed that cargos facilitated by transport receptors could traverse these modified NPCs, but with a much longer time compared to untreated NPCs46. It is clear that O-GlcNAc is necessary for proper NPC permeability, however it is still difficult to discern whether these results are from the loss of O-GlcNAc or the loss of Nups caused by inhibiting or knocking out OGT. The former would lead to a more condensed and rigid FG-Nup barrier while the latter would result in a less crowded and leaky NPC channel.
Other studies have been conducted to probe the relationship between O-GlcNAc-ylation and NPC permeability38,40,41,47,48. Labokha et al confirmed that O-GlcNAc-ylated Nup98 hydrogels efficiently admitted freely diffusing molecules smaller than 40-kDa, while excluding those larger than 40-kDa. In contrast, non-O-GlcNAc-ylated Nup98-hydrogels excluded all passively diffusing molecules regardless of size44. Also, O-GlcNAc-depleted Nup98-hydrogels almost completely inhibited the penetration of a major transport receptor, Importin β144.
A “balance” model for the roles of O-GlcNAc in the NPC
A critical question raised by the above in vitro facilitated transport studies is: is O-GlcNAc directly involved in the specific hydrophobic interactions between FG Nups and transport receptors? Interestingly, other studies showed that modification of FG Nups with a disaccharide, O-linked N-acetyllactosamine (O-LacNAc), instead of O-GlcNAc, had no effect on the permeability or selectivity capabilities of NPCs49,50. So it seems to suggest that the function of O-GlcNAc in the NPC could be less of a specific interaction but more to confer some qualitative characteristic to the FG Nups, namely, increasing the FG Nups' fluidity which could prevent them from congealing through hydrophobic forces. Very recently, single-molecule tracking and super-resolution three-dimensional mapping of passive diffusion and facilitated transport through the NPC have revealed that distinct spatial transport routes adopted by these two transport modes and the heterogeneous spatial distribution of FG domains inside the NPC51,52. Therefore, we propose a “balance” model for the roles of O-GlcNAc in the native NPC's permeability and selectivity as follows (Figure 1). Since the FG Nups are anchored in the wall of the NPC with a rotational symmetry and their FG domains expand inward, passive diffusion of small molecules occurs down the central axial channel where the FG domains are most flexible and sparsely populated. On the other hand, facilitated transport mainly happens in the peripheral regions around the central axial channel where the FG Nups are more dense and structured due to hydrophobic forces. Both of these transport routes are realized by the FG-Nup barrier in a configuration that is not too dense or too sparse, which could be a result of cooperation between FG-FG hydrophobic interactions and O-GlcNAc-ylation of FG Nups (Figure 1a). In a situation where there is too little O-GlcNAc-ylation of FG Nups in the NPC, the FG-Nup selectivity barrier might be altered or even collapsed to form a more rigid beta-amyloid structure, which will reduce their steric hindrance of the passive diffusion of signal-independent small molecules and shield hydrophobic interactions for facilitated transport of signal-dependent cargos through the NPC (Figure 1b).
Figure 1.
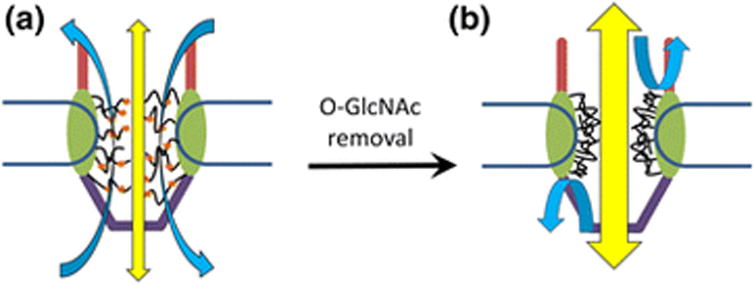
Model of O-GlcNAc's role in the NPC. a) O-GlcNAc-ylation (orange dots) of the NPC prevents excessive hydrophobic interactions between the FG Nups (black curves). This weakens the hydrophobic interactions enough for the FG domains to expand, which also allows the passive diffusion (yellow arrow) of signal-independent small molecules (< 40 kDa) and exposes FG repeats for interactions with karyopherins during facilitated transport (light blue arrow). b) If FG Nups in the NPC are too little O-GlcNAc-ylated, they bind to each other tightly through hydrophobic interactions and collapse into a tight secondary structure. The passive diffusion channel is widened admitting larger molecules (> 40 kDa) while facilitated transport is prevented due to a lack of exposed FG repeats.
This model could be used to further understand how the cell upregulates mass flow between the nucleus and cytoplasm, for example, in the transcription factor re-localization during the oxidative stress response53. Recently, it has been shown that several critical FG Nups involved in facilitated transport, including Nup62, Nup214 and Nup153, were heavily O-GlcNAc-ylated in response to oxidative stress54,55. Crampton et al also concluded that there was net inhibition of Crm1-mediated nuclear export following oxidative stress, suggesting some key interactions in the NPC were altered. Specifically, the Crm1/Nup62 and Crm1/Nup153 interactions were both strengthened while the interactions between FG Nups were weakened due to oxidative stress followed by O-GlcNAc-ylation55. As shown in our model, a balance between O-GlcNAc-ylation of FG Nups and FG-FG hydrophobic interactions is critical for cells to handle oxidative stress.
Approaches for the detection of O-GlcNAc in the NPC
As is the case with many fields of biological research, technological advancement in the tools used to study O-GlcNAc is one of the main impediments to progress25. Early contributions to the field of O-GlcNAc biology especially as it pertains to the NPC were made primarily using affinity labeling techniques coupled with mass spectrometry for identification or electron microscopy for spatial localization56,57,58,59. Wheat germ agglutinin (WGA) was often used as the affinity reagent57,58 due to its higher affinity for O-GlcNAc residues versus sialic acid residues57. However, an array of monoclonal antibodies (mAbs), with more specificity against single O-GlcNAc-ylated proteins, have since replaced WGA to avoid WGA's non-selective affinity and binding for multiple O-GlcNAc residues17,18,59,60,61. Electron microscopy studies have shown that these mAb-reactive Nups primarily reside on the nuclear and cytoplasmic faces of the NPC59.
Another popular method of O-GlcNAc identification is a combination of metabolic and biochemical approaches. Since there is flexibility in the type of substrates that OGT can use, O-linked sugars bound to a chemically reactive azide, alkyne, or diazirine group can be incorporated by OGT post-translationally into proteins. The reactive groups can then be chemically modified in several ways such as attaching a fluorophore for visualization62,63,64, an affinity agent such as biotin for purification64, or photocrosslinking the diazirine with nearby molecules to determine novel interactions by mass spectrometry34. Several of these “click” chemistry approaches, termed as such because of their ease of use and speed, have offered femtomolar levels of sensitivity63 and high throughput volumes64.
Also by utilizing click chemistry labeling, super-resolution fluorescence microscopy, direct Stochastic Optical Reconstruction Microscopy (dSTORM)65 in particular, has been adapted to study the O-GlcNAc distribution on the plasma membrane with high localization precision. It was found that O-GlcNAc distributed homogenously across the plasma membrane in contrast to the view that glycosylated nanodomains exist on the membrane66. Based on the localization precision, it is promising that super-resolution microscopy could be adapted to study O-GlcNAc labeling at the level of the NPC and determine its in vivo localization.
There are several bioinformatics tools available to predict O-GlcNAc-ylation sites67. An early approach called YinOYang extracted amino acid sequence features from 40 experimentally determined O-GlcNAc sites and found that the serine and threonine residues modified with O-GlcNAc are often in close proximity to proline and valine, distant from leucine and glutamine, and upstream of other serines68. As more experimentally determined O-GlcNAc sites were found, O-GlcNAcScan was developed which used a training data set of about 400 positive and 30,000 negative sites of O-GlcNAc-ylation to determine potential amino acid sequences whose serine/threonine residues are O-GlcNAc-ylated69. While these two approaches have accurately identified O-GlcNAc-ylation sites, they are limited because no consensus site has been determined and the negative training data set is comprised of every other serine and threonine residue that have not been experimentally shown to be O-GlcNAc-ylated. There will inevitably be sites in this data set that have yet to be identified as positive70. Recently, a newer approach called O-GlcNAcPRED, which uses a more efficient algorithm for amino acid sequence feature extraction and another parameter that looks at the physiochemical properties of the that amino acid sequence, has been shown to outperform both previous predictors70. It appears that there might actually be no consensus sequence for O-GlcNAc-ylation, but rather a combination of amino acid sequence and biochemical context which marks a site for O-GlcNAc-ylation.
Conclusion
The O-GlcNAc modification is an essential and evolutionarily conserved post-translational modification that has housekeeping duties and functions as a component of the cellular stress response. At the level of the NPC, it may maintain the fluidity of FG Nups which promotes steric interference of passively diffusing molecules while exposing FG Nups for interactions with transport receptors in facilitated transport. With further development of new tools to study O-GlcNAc-ylation, it will be possible to distinguish the full complement of responsibilities O-GlcNAc has in the NPCs and beyond.
Acknowledgments
The project was supported by grants from the National Institutes of Health (NIH GM094041, GM097037 and GM116204).
Footnotes
Conflicts of Interest: Andrew Ruba and Weidong Yang declare that they have no conflicts of interest.
Ethical Standards: No human studies were carried out by the authors of this article. No animal studies were carried out by the authors of this article.
References
- 1.Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. J Cell Biol. 2015;2087:869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butkinaree C, Park K, et al. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 2010;18002:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slawson C, Lakshmanan T, et al. A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Mol Biol Cell. 2008;1910:4130–4140. doi: 10.1091/mbc.E07-11-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harwood KR, Hanover JA. Nutrient-driven O-GlcNAc cycling - think globally but act locally. J Cell Sci. 2014;127Pt 9:1857–1867. doi: 10.1242/jcs.113233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Love DC, Kochan J, et al. Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J Cell Sci. 2003;116Pt 4:647–654. doi: 10.1242/jcs.00246. [DOI] [PubMed] [Google Scholar]
- 6.Hanover JA, Yu S, et al. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch Biochem Biophys. 2003;4092:287–297. doi: 10.1016/s0003-9861(02)00578-7. [DOI] [PubMed] [Google Scholar]
- 7.Kreppel LK, Blomberg MA, et al. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;27214:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 8.Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;2111:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 9.Jinek M, Rehwinkel J, et al. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 2004;1110:1001–1007. doi: 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- 10.Trinidad JC, Barkan DT, et al. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics. 2012;118:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dieterich DC, Kreutz MR. Proteomics of the synapse - A quantitative approach to neuronal plasticity. Mol Cell Proteomics. 2015 doi: 10.1074/mcp.R115.051482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wells L, Vosseller K, et al. Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science. 2001;2915512:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 13.Roquemore EP, Chevrier MR, et al. Dynamic O-GlcNAcylation of the small heat shock protein alpha B-crystallin. Biochemistry. 1996;3511:3578–3586. doi: 10.1021/bi951918j. [DOI] [PubMed] [Google Scholar]
- 14.Chou CF, Smith AJ, et al. Characterization and dynamics of O-linked glycosylation of human cytokeratin 8 and 18. J Biol Chem. 1992;2676:3901–3906. [PubMed] [Google Scholar]
- 15.Zachara NE, O'Donnell N, et al. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J Biol Chem. 2004;27929:30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- 16.Kodiha M, Crampton N, et al. Traffic control at the nuclear pore. Nucleus. 2010;13:237–244. doi: 10.4161/nucl.1.3.11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radermacher PT, Myachina F, et al. O-GlcNAc reports ambient temperature and confers heat resistance on ectotherm development. Proc Natl Acad Sci U S A. 2014;11115:5592–5597. doi: 10.1073/pnas.1322396111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeves RA, Lee A, et al. Characterization of the specificity of O-GlcNAc reactive antibodies under conditions of starvation and stress. Anal Biochem. 2014;457:8–18. doi: 10.1016/j.ab.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozcan S, Andrali SS, et al. Modulation of transcription factor function by O-GlcNAc modification. Biochim Biophys Acta. 2010;17995-6:353–364. doi: 10.1016/j.bbagrm.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laczy B, Hill BG, et al. Protein O-GlcNAcylation: a new signaling paradigm for the cardiovascular system. Am J Physiol Heart Circ Physiol. 2009;2961:H13–28. doi: 10.1152/ajpheart.01056.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazarus BD, Love DC, et al. O-GlcNAc cycling: implications for neurodegenerative disorders. Int J Biochem Cell Biol. 2009;4111:2134–2146. doi: 10.1016/j.biocel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou TY, Hart GW. O-linked N-acetylglucosamine and cancer: messages from the glycosylation of c-Myc. Adv Exp Med Biol. 2001;491:413–418. doi: 10.1007/978-1-4615-1267-7_26. [DOI] [PubMed] [Google Scholar]
- 23.Copeland RJ, Bullen JW, et al. Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am J Physiol Endocrinol Metab. 2008;2951:E17–28. doi: 10.1152/ajpendo.90281.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buse MG. Hexosamines, insulin resistance, and the complications of diabetes: current status. Am J Physiol Endocrinol Metab. 2006;2901:E1–E8. doi: 10.1152/ajpendo.00329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hart GW, Housley MP, et al. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;4467139:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 26.Macauley MS, He Y, et al. Inhibition of O-GlcNAcase using a potent and cell-permeable inhibitor does not induce insulin resistance in 3T3-L1 adipocytes. Chem Biol. 2010;179:937–948. doi: 10.1016/j.chembiol.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vosseller K, Wells L, et al. Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc Natl Acad Sci U S A. 2002;998:5313–5318. doi: 10.1073/pnas.072072399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker G, Taylor R, et al. Hyperglycemia and inhibition of glycogen synthase in streptozotocin-treated mice: role of O-linked N-acetylglucosamine. J Biol Chem. 2004;27920:20636–20642. doi: 10.1074/jbc.M312139200. [DOI] [PubMed] [Google Scholar]
- 29.Gut P, Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature. 2013;5027472:489–498. doi: 10.1038/nature12752. [DOI] [PubMed] [Google Scholar]
- 30.Gambetta MC, Oktaba K, et al. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;3255936:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 31.Chikanishi T, Fujiki R, et al. Glucose-induced expression of MIP-1 genes requires O-GlcNAc transferase in monocytes. Biochem Biophys Res Commun. 2010;3944:865–870. doi: 10.1016/j.bbrc.2010.02.167. [DOI] [PubMed] [Google Scholar]
- 32.Sinclair DA, Syrzycka M, et al. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci U S A. 2009;10632:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor RP, Parker GJ, et al. Glucose deprivation stimulates O-GlcNAc modification of proteins through up-regulation of O-linked N-acetylglucosaminyltransferase. J Biol Chem. 2008;28310:6050–6057. doi: 10.1074/jbc.M707328200. [DOI] [PubMed] [Google Scholar]
- 34.Li B, Kohler JJ. Glycosylation of the nuclear pore. Traffic. 2014;154:347–361. doi: 10.1111/tra.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Y, Liu TW, et al. Post-translational O-GlcNAcylation is essential for nuclear pore integrity and maintenance of the pore selectivity filter. J Mol Cell Biol. 2015 doi: 10.1093/jmcb/mjv033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akey CW, Radermacher M. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol. 1993;1221:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beck M, Forster F, et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;3065700:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- 38.Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;206:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;1810:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ribbeck K, Gorlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 2002;2111:2664–2671. doi: 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinshaw JE, Carragher BO, et al. Architecture and design of the nuclear pore complex. Cell. 1992;697:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- 42.Favreau C, Worman HJ, et al. Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein Gp210. Biochemistry. 1996;3524:8035–8044. doi: 10.1021/bi9600660. [DOI] [PubMed] [Google Scholar]
- 43.Wang S, Huang X, et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates Akt signaling. PLoS One. 2012;75:e37427. doi: 10.1371/journal.pone.0037427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Labokha AA, Gradmann S, et al. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 2013;322:204–218. doi: 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hulsmann BB, Labokha AA, et al. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell. 2012;1504:738–751. doi: 10.1016/j.cell.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 46.Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;601:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- 47.Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;1303:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 48.Sterne-Marr R, Blevitt JM, et al. O-linked glycoproteins of the nuclear pore complex interact with a cytosolic factor required for nuclear protein import. J Cell Biol. 1992;1162:271–280. doi: 10.1083/jcb.116.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hicks GR, Raikhel NV. Protein import into the nucleus: an integrated view. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 50.Miller MW, Hanover JA. Functional nuclear pores reconstituted with beta 1-4 galactose-modified O-linked N-acetylglucosamine glycoproteins. J Biol Chem. 1994;26912:9289–9297. [PubMed] [Google Scholar]
- 51.Ma J, Yang W. Three-dimensional distribution of transient interactions in the nuclear pore complex obtained from single-molecule snapshots. Proc Natl Acad Sci U S A. 2010;10716:7305–7310. doi: 10.1073/pnas.0908269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma J, Goryaynov A, et al. Self-regulated viscous channel in the nuclear pore complex. Proc Natl Acad Sci U S A. 2012;10919:7326–7331. doi: 10.1073/pnas.1201724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kodiha M, Chu A, et al. Multiple mechanisms promote the inhibition of classical nuclear import upon exposure to severe oxidative stress. Cell Death Differ. 2004;118:862–874. doi: 10.1038/sj.cdd.4401432. [DOI] [PubMed] [Google Scholar]
- 54.Kodiha M, Tran D, et al. Dissecting the signaling events that impact classical nuclear import and target nuclear transport factors. PLoS One. 2009;412:e8420. doi: 10.1371/journal.pone.0008420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crampton N, Kodiha M, et al. Oxidative stress inhibits nuclear protein export by multiple mechanisms that target FG nucleoporins and Crm1. Mol Biol Cell. 2009;2024:5106–5116. doi: 10.1091/mbc.E09-05-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldberg MW, Allen TD. High resolution scanning electron microscopy of the nuclear envelope: demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J Cell Biol. 1992;1196:1429–1440. doi: 10.1083/jcb.119.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanover JA, Cohen CK, et al. O-linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J Biol Chem. 1987;26220:9887–9894. [PubMed] [Google Scholar]
- 58.Powers MA, Macaulay C, et al. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol. 1995;1285:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snow CM, Senior A, et al. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;1045:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holt GD, Snow CM, et al. Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J Cell Biol. 1987;1045:1157–1164. doi: 10.1083/jcb.104.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park MK, D'Onofrio M, et al. A monoclonal antibody against a family of nuclear pore proteins (nucleoporins): O-linked N-acetylglucosamine is part of the immunodeterminant. Proc Natl Acad Sci U S A. 1987;8418:6462–6466. doi: 10.1073/pnas.84.18.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vocadlo DJ, Hang HC, et al. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci U S A. 2003;10016:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clark PM, Dweck JF, et al. Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J Am Chem Soc. 2008;13035:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim EJ, Abramowitz LK, et al. Versatile O-GlcNAc transferase assay for high-throughput identification of enzyme variants, substrates, and inhibitors. Bioconjug Chem. 2014;256:1025–1030. doi: 10.1021/bc5001774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van de Linde S, Loschberger A, et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc. 2011;67:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 66.Letschert S, Gohler A, et al. Super-resolution imaging of plasma membrane glycans. Angew Chem Int Ed Engl. 2014;5341:10921–10924. doi: 10.1002/anie.201406045. [DOI] [PubMed] [Google Scholar]
- 67.Ma J, Hart GW. O-GlcNAc profiling: from proteins to proteomes. Clin Proteomics. 2014;111:8. doi: 10.1186/1559-0275-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gupta R, Brunak S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 2002:310–322. [PubMed] [Google Scholar]
- 69.Wang J, Torii M, et al. dbOGAP - an integrated bioinformatics resource for protein O-GlcNAcylation. BMC Bioinformatics. 2011;12:91. doi: 10.1186/1471-2105-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jia CZ, Liu T, et al. O-GlcNAcPRED: a sensitive predictor to capture protein O-GlcNAcylation sites. Mol Biosyst. 2013;911:2909–2913. doi: 10.1039/c3mb70326f. [DOI] [PubMed] [Google Scholar]


