Abstract
It has long been believed that most female mammalian species lose the ability to generate oocytes in postnatal ovaries. Recent evidence has demonstrated the isolation and culture of female germline stem cells (FGSCs) from adult mice and humans. However, the process and mechanisms of FGSC differentiation in vivo following transplantation have not yet been studied. Here, we isolated and characterized FGSCs from a single EGFP-transgenic mouse, and traced the development and behavior of transplanted FGSCs (F-TFs) in vivo. Comparisons of folliculogenesis between recipients with FGSC transplantation and wild-type (WT) mice were performed by single follicle RNA-sequencing (RNA-seq). Results showed that FGSCs exhibited a homing ability and began to differentiate into early-stage oocytes only when they reached the edge of the ovarian cortex. The F-TFs restored function of premature ovarian failure (gdf9iCre; PtenloxP/loxP genotype) and generated offspring. Furthermore, results demonstrated that the developmental mechanisms of follicles derived from F-TFs were similar to that of WT follicles. Weighted gene co-expression network analysis identified two potential sub-networks and core genes that played a critical role in follicular development. These findings provide a theoretical basis and lay a technology platform for specific or personalized medical treatment of ovarian failure or other ovarian diseases.
Keywords: stem cell tracing, stem cell therapy, RNA-seq of single follicle
Wu et al. isolated and characterized female germline stem cells (FGSCs) from a single EGFP-transgenic mouse and traced the development and behavior of transplanted FGSCs (F-TFs) in vivo. The F-TFs exhibited homing and began to differentiate into early-stage oocytes when they reached the edge of the ovarian cortex. The F-TFs restored function of mouse ovaries exhibiting premature ovarian failure and generated offspring, suggesting a possible strategy to treat ovarian failure or other ovarian diseases.
Introduction
Germ stem cells are the only stem cells that can produce gametes in vivo. These cells undergo meiosis, as well as mitosis. A century ago, spermatogonial stem cells (SSCs) were postulated to exist in male mammals,1 and the location and molecular characteristics of SSCs have been well documented.2, 3 However, in female mammals, it has long been believed that female germ cells arrested at the diplotene stage of meiotic prophase I prior to birth. Until recently, a number of studies have challenged this belief.4, 5, 6, 7, 8, 9, 10, 11, 12 Female germline stem cells (FGSCs) were isolated from ovaries of 5-day-old and adult mice using immunomagnetic sorting for the DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 (DDX4). The FGSCs were cultured for more than 15 months or for 6 months, respectively, and exhibited a proliferation capacity and a normal karyotype. Furthermore, the cell line produces fertile offspring after transplantation into ovaries.5 Subsequently, FGSCs were demonstrated in the rat and human.7, 13, 14, 15
Stem cells are thought to provide viable options in the field of cell therapy. Ovarian failure is induced by chemotherapy, radiation, genetic induction, or hormonal stress.16, 17, 18 Additionally, premature ovarian failure (POF) is one of the most common causes of ovarian function cessation prior to the age of 40 and affects 1% of young women.19 However, very little progress has been made to solve this problem. The findings that FGSCs play a role in fertility provide promising application for clinical therapies, as well as a strategy for studying germ cell development and its mechanism.8, 20, 21 Although a previous study showed that transplanted FGSCs (F-TFs) differentiate into oocytes in chemotherapy-treated sterile ovaries and are capable of generating offspring,10 very little is known about whether FGSCs can restore POF function (gdf9iCre; PtenloxP/loxP genotype). Furthermore, the mechanisms involved in the developmental patterns of F-TFs into oocytes in vivo remain poorly understood.
In mammalian germ cell development, meiosis is a crucial event, which is a highly regulated process.22, 23 Meiosis has been shown to start soon after the primordial germ cells (PGCs) arrive at the developing gonads in female mammals. Expression of Stra8, which is required for the retinoic acid pathway, leads to meiosis initiation.24, 25 Oogenesis is then followed by a series of progressive changes in gene expression profiles until meiosis arrests in dictyate.26, 27 During oogenesis, the follicle is formed and becomes a developmental unit, which consists of follicular somatic cells and a central oocyte. Bidirectional communications between oocytes and follicular somatic cells in follicles maintain the oocytes in meiotic arrest.28 The follicular antrum formation is an important event for oocyte development or folliculogenesis because preantral-early antral follicle transition in vivo is a necessary step for female fertility.29, 30 Moreover, oocytes resume meiosis in conjunction with the downregulation of many genes (Aurka, Plk1, Btrc, Arpp19, Prkaca, etc.).31, 32, 33, 34, 35 RNA-sequencing (RNA-seq) has been employed for transcriptional profiling of oocytes during folliculogenesis, as well as granulosa cells.33 However, granulosa cells from different regions play different roles in follicular development.36, 37 Furthermore, the follicle is a functional unit, and folliculogenesis is a complex process requiring interaction between follicular cells.38 Thus, exploring the single follicle expression profile is important to fully understand follicular developmental mechanisms. However, transcriptional profile analysis of single follicles at different stages, especially in preantral and antral formation stages, has not been performed.
In this study, we isolate and characterize FGSCs from a single EGFP-transgenic mouse and transplant them into infertile recipients: chemotherapy-treated and POF (gdf9iCre; PtenloxP/loxP genotype) female mice. In the recipients, ovarian function is restored and they are able to generate offspring. We then trace the migration of F-TFs and developmental patterns in vivo. Results show that the transplanted cells differentiate into oocytes after they migrate into the edge of the ovarian cortex. Folliculogenesis is also analyzed by RNA-seq in a single follicle at different stages. Results show that developmental mechanisms of follicles derived from F-TFs are similar to that of WT follicles. This study provides a theoretical basis and lays a technological platform for clinical translational research and application of FGSCs.
Results
Isolation and Characterization of FGSCs from a Single Mouse
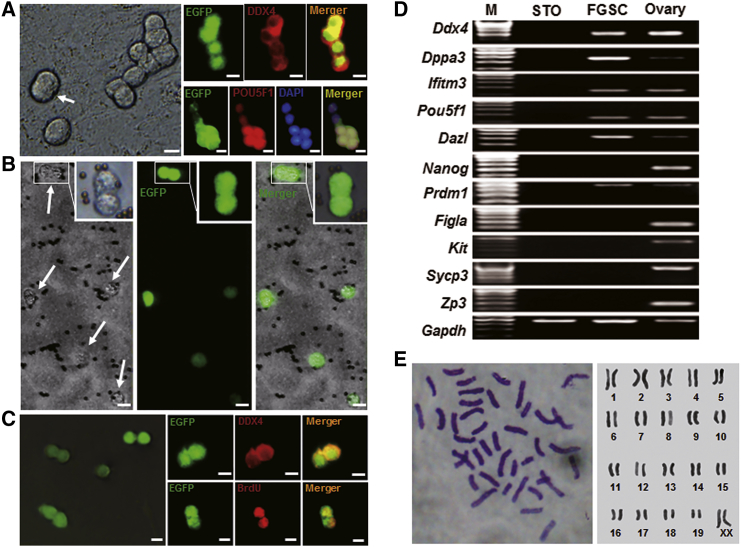
FGSCs were isolated from ovaries of a single neonatal CAG (cytomegalovirus [CMV] early-enhancer/chicken β-actin promoter)-EGFP transgenic mouse for each time using two-step enzymatic digestion following the described protocols.39 After 3 days in primary culture, the cells grew into grape-like clusters consisting of two to eight cells and exhibited typical division morphology. Furthermore, immunofluorescence analysis showed that these cells expressed DDX4 (exclusively expressed in germ cells) and Pou5f1 (a germ cell-specific transcriptional factor) (Figure 1A). After the initial passage, FGSCs were purified by DDX4-based immunomagnetic sorting (MACS) and were cultured. The sorted cells were round with a high nuclear-to-cytoplasm ratio (Figure 1B). The cells were then subcultured every 4–7 days at a 1:2 ratio for more than 2 months, and finally four independent long-term cultured FGSCs were established (n = 4). The proliferative potential and DDX4 expression of these cells were confirmed by immunofluorescence (Figure 1C). Subsequently, we assessed the gene expression profile of these cells using the following markers for germ cells or pluripotency stem cells: Ddx4, Dppa3 (a maternal effect gene maintenance of pluripotency), Ifitm3 (a marker of germ cells, especially early germ cells), Pou5f1, Dazl (a marker of germ cells), Nanog (a pluripotency sustaining factor), Prdm1 (a known transcriptional repressor and marker of early germ cells), Figla (a meiosis-specific marker), Kit (a stem cell factor receptor), Sycp3 (synaptonemal complex protein 3), and Zp3 (zona pellucida glycoprotein 3). RT-PCR results showed that the cells expressed Ddx4, Dppa3, Ifitm3, Pou5f1, Dazl, and Prdm1, but not Nanog, Figla, Kit, Sycp3, or Zp3 (Figure 1D). Moreover, karyotype analysis showed a normal karyotype (40, XX) in approximately 65% of cells (Figure 1E). These findings suggested that the cells possessed characteristics of germline stem cells with normal chromosomes, which was consistent with previously identified FGSCs.5, 40, 41 Furthermore, we also isolated long-term cultured FGSCs from a single 8-day-old (n = 3) or 10-day-old mouse (n = 3) (Figure S1).
Figure 1.
FGSC Characteristics
(A) Morphology (left image) and immunofluorescence analysis of DDX4 and Pou5f1 (right images) from isolated FGSCs cultured for 3 days. The cells exhibit a grape-like morphology. Arrow shows a dividing cell. Scale bars, 10 μm. (B) FGSCs cultured for 1 day after immunomagnetic sorting. Arrows show EGFP-positive cells in bright field. The inset shows a divided cell. Scale bars, 10 μm. (C) Morphology (left image) and immunofluorescence for DDX4 or BrdU (right images) of FGSCs cultured for 8 weeks. Scale bars, 10 μm. (D) Gene expression profile of FGSCs. Lane M: 100 bp DNA ladder marker. (E) Cytogenetic analysis shows FGSCs with a normal karyotype.
Tracing the Development and Behavior of F-TFs In Vivo
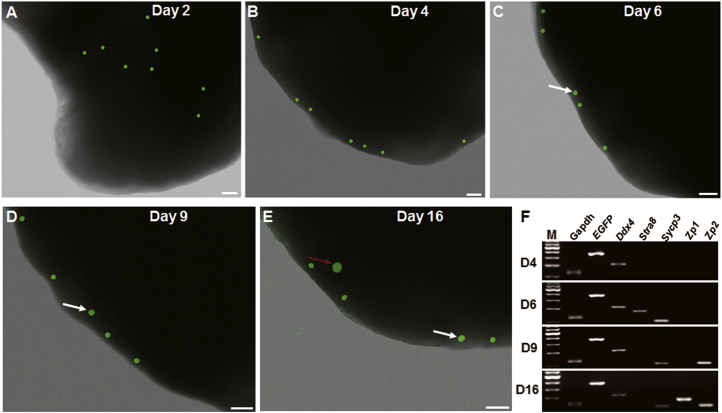
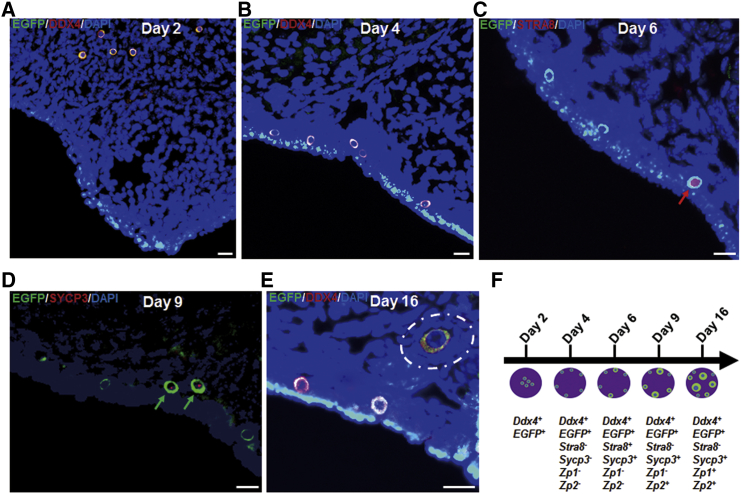
To trace differentiation of FGSCs in vivo, the EGFP-expressing FGSCs were transplanted into ovaries of mice sterilized by busulfan and cyclophosphamide, and monitored by a laser confocal microscope (Olympus). Results showed scattered FGSCs in the ovary on the second day after transplantation (Figure 2A). On day 4 post-transplantation, FGSCs migrated to the edge of the ovarian cortex (Figure 2B). Some transplanted cells were slightly larger than others on day 6 post-transplantation (Figure 2C), suggesting that the transplanted cells settled in the edge of the ovarian cortex and began to differentiate into early oocyte stages. On day 9 or 16 post-transplantation, some cells left the edge of the ovarian cortex and reached the depth of the ovary, becoming 1.5–3 times larger than other cells (Figures 2D and 2E), suggesting that the transplanted cells continued to differentiate into oocytes at different stages of development. To confirm this, we conducted RT-PCR of a single EGFP-positive cell to detect expression of EGFP, Ddx4, Stra8, Sycp3, Zp1, and Zp2. On day 6 post-transplantation, some EGFP-positive cells expressed Stra8 and Sycp3, suggesting meiosis. On day 9, Sycp3 and Zp2 were detected; on day 16, Zp1, Sycp3, and Zp2 were detected (Figure 2F). These results were confirmed by dual-immunofluorescence analysis of expression of DDX4/EGFP, STRA8/EGFP, and SYCP3/EGFP in transplanted cells (Figures 3A–3F). Interestingly, on day 16 post-transplantation, primary or secondary follicles were found in recipient ovaries (Figure 3E). Furthermore, RT-PCR and immunofluorescence analysis showed Ybx2, a crucial regulator in diplotene stage,42 was expressed on days 9 and 16, suggesting F-TFs developed gradually and reached meiosis arrest on day 9 (Figure S2).
Figure 2.
Tracing of Transplanted FGSCs In Vivo
(A) FGSCs scattered throughout the ovary at 2 days after FGSC transplantation. Scale bar, 50 μm. (B) FGSCs migrated to the edge of the ovary by day 4 post-transplantation. Scale bar, 50 μm. (C–E) Cells (white arrows) are slightly larger than others on days 6 (C) and 9 (D), and significantly larger on day 16 (E) (red arrow) post-transplantation. Scale bars, 50 μm. (F) Gene expression profile of a single EGFP-positive FGSC on days 4, 6, 9, and 16 post-transplantation. The image shows Stra8 and Sycp3 expression in cells on day 6, Sycp3 and Zp2 expression on days 9 and 16, and Zp1 expression on day 16 post-transplantation. Lane M: 100 bp DNA ladder marker.
Figure 3.
Development of Transplanted FGSCs In Vivo
(A and B) Immunofluorescence analysis of DDX4 and EGFP on days 2 (A) and 4 (B) after FGSC transplantation. Images show FGSCs scattered in the ovary by day 2, which migrated to the ovarian cortex surface by day 4 post-transplantation. Scale bars, 25 μm. (C and D) Immunofluorescence analysis shows STRA8 expression (red arrow) in germ cells on day 6 (C) and SYCP3 expression (green arrows) on day 9 (D) post-transplantation. Scale bars, 25 μm. (E) A representative view of immunofluorescence analysis with DDX4 and EGFP on day 16 post-transplantation. White dashed oval indicates that the follicle contains an EGFP-labeled oocyte and somatic cells (n = 5). Scale bar, 25 μm. (F) Development model of transplanted FGSCs in vivo. Arrow represents development process. Short line segments on arrow represent development time. Purple pans represent ovary. Blue and green represent the nucleus and cytoplasm of female germ cells (FGSCs), respectively. Gene expression profiles of FGSCs at different stages are listed below.
Restoration of Ovarian Function and the Generation of Offspring from FGSCs of a Single Mouse
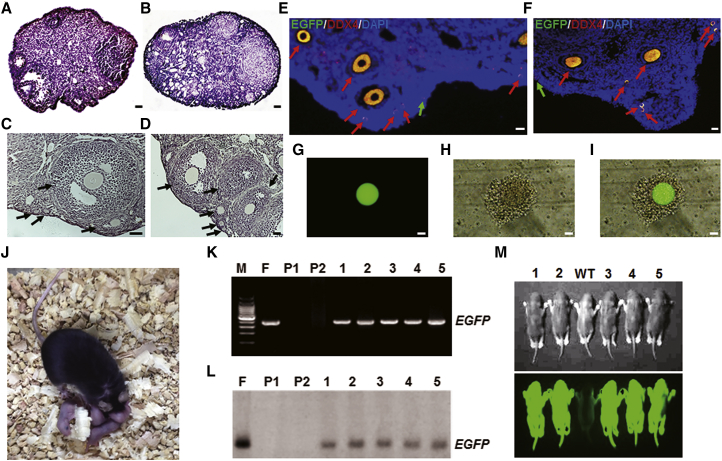
To determine whether FGSCs from a single mouse can recover ovary function and generate offspring, we transplanted FGSCs into infertile recipients: chemotherapy-treated and POF (gdf9iCre; PtenloxP/loxP genotype) female mice. The supernatant from the wash PBS was injected into the ovaries of infertile recipients as controls. Two months after transplantation, recipient ovaries were collected and evaluated for morphology and dual expression of EGFP and DDX4. Histological analysis showed that recipient ovaries injected with FGSCs contained numerous female germ cells at all stages of development, including FGSCs and oocytes expressing both EGFP and DDX4 (Figures 4C–4F) or EGFP (Figures 4G–4I). However, control ovaries from chemotherapeutically treated mice or from POF mice consisted of stromal and interstitial cells, as well as atretic follicles (Figures 4A and 4B). To determine whether FGSC oocytes from a single mouse could produce offspring, we mated sterilized recipient females with wild-type (WT) adult males 5–8 weeks after cell transplantation. Results showed that 75% of transplanted recipients produced offspring (Figure 4J; Table S1). PCR and Southern blot analysis showed EGFP expression in the offspring, but not in the recipient mice (Figures 4K and 4L). Furthermore, fluorescence detected by live imaging under a Lumazone imaging system (Mag Biosystems) also showed strong green fluorescence signals in the offspring (Figure 4M). These results indicate that FGSCs from a single mouse can restore ovary function and generate offspring.
Figure 4.
FGSC Transplantation into Infertile Recipient Mice
(A–D) Representative ovary morphology from chemical-treated mice without (A) or with (C) FGSC transplantation and POF mice without (B) or with (D) FGSC transplantation for 2 months. Arrows indicate different follicle stages. n = 3. Scale bars, 1 mm (A and B) and 50 μm (C and D). (E and F) Immunofluorescence analysis of DDX4 and EGFP for ovaries of chemical-treated (E) and POF (F) mice 2 months post-transplantation. Red arrows indicate different follicle stages. Green arrows indicate potential FGSCs. Scale bars, 50 μm. (G–I) Representative oocyte separated from recipient ovaries at 2 months post-FGSC transplantation. Images were captured in fluorescence (G), bright field (H), and merged (I) for (G) and (H), respectively. n = 5. Scale bars, 25 μm. (J–L) Representative offspring from FGSC-transplanted mice (J); n = 8. Offspring and recipients were detected by PCR (K) and Southern blotting (L). Lane M: 100 bp DNA ladder marker; lane F: mouse used for isolation FGSCs; lane P1: female recipient mouse; lane P2: male mouse used for mating; lanes 1–5: offspring. (M) Offspring were identified in bright field (top) and fluorescence (bottom). Lanes 1–5: offspring. WT, wild-type.
Characterizing Development of Follicles Derived from F-TFs
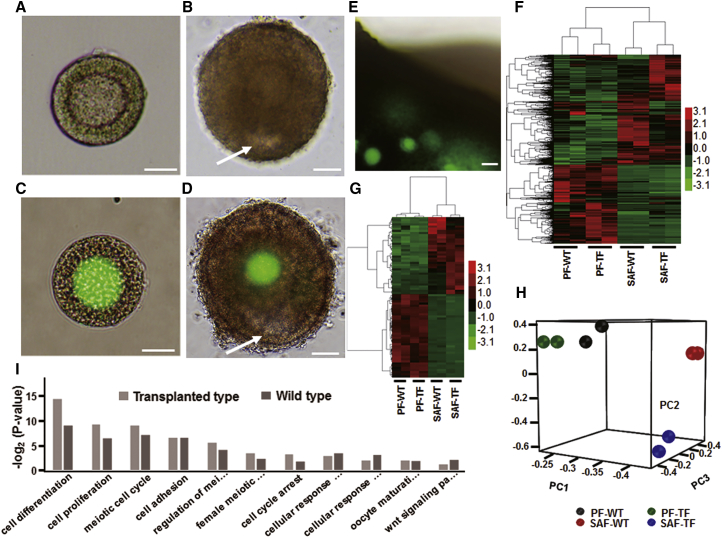
To determine the molecular characteristics during development of F-TFs in vivo, we used preantral follicle (PF) and small antral follicle (SAF) derived from F-TFs or from WT ovaries (Figures 5A–5E) to perform single-follicle RNA-seq. Using an Illumina HiSeq2500 sequencer, we generated 51.5 Gb of sequencing data from the eight samples. On average, we detected expression of 20,966 (47%) out of 44,788 RefSeq genes. Thus, half of the known mouse genes were expressed in the samples. To determine whether these gene expression profiles correlated with developmental stages of follicles, we used unsupervised and hierarchical clustering to analyze the RNA-seq data from follicles at different developmental stages. Results showed an obvious difference in transcriptional profiles between PF and SAF with 7,005 or 5,764 differential expression genes (DEGs), respectively, using the criteria fold change (FC) >2 or <0.5 (Figure S3). However, gene expression profiles of PF derived from F-TFs (PF-TF) were similar to those of PF from WT (PF-WT). Furthermore, SAFs from F-TFs (SAF-TF) and WT (SAF-WT) shared similar transcriptional profiles (Figures 5F and 5G; Table S2). Similar within-stage and different between-stage expression patterns were also confirmed by principal component analysis (PCA) (Figure 5H). To validate the RNA-seq analysis, we monitored expression of a subset of genes using qRT-PCR. Oocyte-specific genes (Bmp15 and Wee2), granule cell-specific genes (Amh and Nr5a1), and other genes (Asz1, Pdk4, and Actg2) were selected. Results confirmed a differential expression during the folliculogenesis process (Table S3). To investigate the biological functions associated with folliculogenesis, we conducted gene ontology (GO) analysis for DEGs on cell processes and signal pathways involved in meiosis. Results showed similar biological functions in follicles derived from F-TFs compared with WT (F-WT) (Figure 5I). Further DEG analysis revealed that F-TF and F-WT shared 3,896 DEGs during development (Figure S3). Some genes were associated with oocyte meiotic arrest (Aurka, Plk, Btrc, etc.) or resumption (Creb1 and Creb3).32, 33, 43 Additionally, meiotic arrest-related genes were downregulated and meiotic resumption-related genes were upregulated, suggesting that F-TF developmental mechanisms were similar to F-WT. Kyoto encyclopedia of genes and genomes (KEGG) analysis of transcriptional profiles in follicular development showed that steroid biosynthesis upregulated antrum formation of follicles. Furthermore, 118 or 105 genes in the phosphatidylinositol 3-kinase (PI3K)-Akt signaling pathway participated in protein synthesis, cell proliferation, DNA repair, and cell cycle, suggesting an important role for the PI3K-Akt signaling pathway in folliculogenesis (Figure S4). Multiple genes (syk, pten, etc.) were also upregulated in the PI3K pathway. Together, these findings demonstrated that F-TF developmental mechanisms were similar to F-WT.
Figure 5.
Global Expression Patterns of PF and SAF
(A–D) Representative PF and SAF used for RNA-seq: PF-WT (A), PF-TF (C), SAF-WT (B), and SAF-TF (D). n = 3. Arrows indicate a follicular antrum. Scale bars, 10 μm (A and C) and 50 μm (B and D). (E) Representative ovary for isolating follicles from FGSC-transplanted mice. n = 3. Scale bar, 25 μm. (F and G) Heatmap display of unsupervised hierarchical clustering of DEGs (F) and folliculogenesis-associated genes (G) between compartments. Red, black, and green represent high, mean, and low expression levels, respectively. (H) Principal component analysis (PCA) of all samples. (I) GO analysis. Bars represent p values at a logarithmic scale. Full information lists of GO terms are shown in Table S6.
Exploring the Potential Regulatory Network of Folliculogenesis from F-TFs
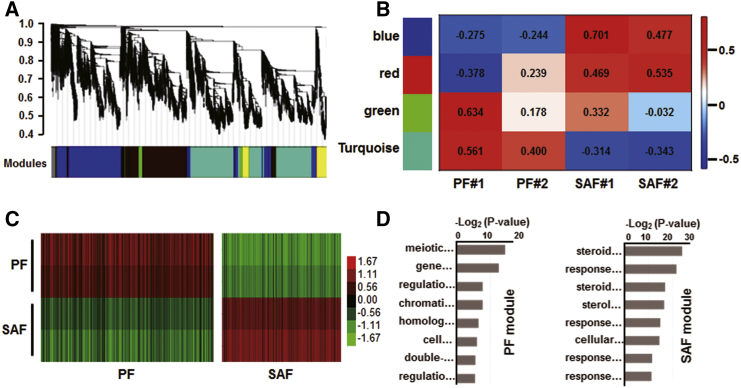
Weighted gene co-expression network analysis (WGCNA) revealed a crucial shift in folliculogenesis gene networks at the systems level in TF. With this unsupervised and unbiased analysis, we identified distinct co-expression modules corresponding to clusters of correlated transcripts (Figure 6A). Notably, 4 of 10 co-expression modules exhibited stage-specific expression values. These modules consisted of genes that tended to be overexpressed in a single developmental stage (Figure 6B).
Figure 6.
WGCNA of Folliculogenesis
(A) Hierarchical cluster tree shows co-expression modules identified using WGCNA. Modules correspond to branches and are labeled by colors. (B) Heatmap reporting correlation between modules and stage indicators in FGSC-transplanted mice. Color legend indicates correlation level between gene co-expressions with stage-specific expression. (C) Heatmap display of relative expression of two representative stage-specific modules. The modules are assigned to highly correlated stages. (D) GO analysis of PF and SAF modules. Full information lists of GO terms are shown in Table S6.
The stage-specific modules (in particular, those with high r value variations) likely represented core gene networks operating at different stages of follicular development. The SAF module underwent a sharp activation over the course of development, whereas the PF module exhibited a sharp degradation (Figure 6C; Figure S5). In total, 5,468 genes belonged in the PF to SAF stage-specific modules, revealing a stepwise requirement of new transcripts involved in steroid- and cytokine-regulating genes, followed by chromatin- and centriole-regulating genes (Figure 6D).
To identify core genes, we chose potential follicle-associated mRNAs and significantly different lon non-coding RNA (lncRNAs) to perform gene co-expression network analysis (Table S4). Results revealed 42 core mRNAs and 6 core lncRNAs with a k-core = 39 in the SAF module, and 56 core mRNAs and 12 core lncRNAs with a k-core = 42 in the PF module (Figures S6 and S7).
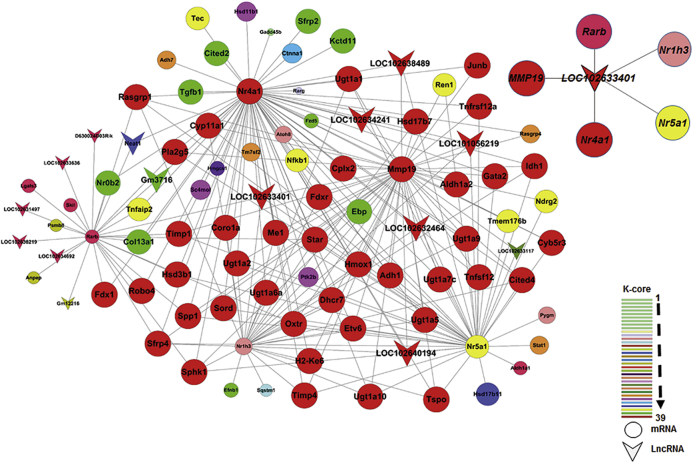
The potential networks of the two modules were explored, identifying hormone-associated genes (Nrlh3, Nr5a1, Nr4a1, Mmp19, and Rarb) in the SAF module and meiosis-associated genes (Mos, Pde3a, Fbxo43, Gpr3, Aurka, and Dicer1) in the PF module. We obtained two sub-networks that consisted of most core genes. One of the sub-networks showed that lncRNA LOC102633401 was the single core lncRNA that was co-expressed with Mmp19, Rarb, Nr1h3, Nr5a1, and Nr4a1 in the SAF module. Another one showed that lncRNA Gm10857 was the single core lncRNA that was co-expressed with aurka, Pde3a, Mos, Gpr3, Fbxo43, and Dicer1 in the PF module (Figures 7 and S8).
Figure 7.
Sub-network Analysis in SAF Module
Sub-network consists of Nr1h3, Nr5a1, Nr4a1, Mmp19, and Rarb in the SAF module. Top right corner image represents potential network extracted from sub-network. Color legend in bottom right corner indicates k-core values from 1 to 39. Circle with color represents mRNA, and arrowhead with color represents lncRNA.
Dynamic Patterns of Alternative Splicing in Folliculogenesis
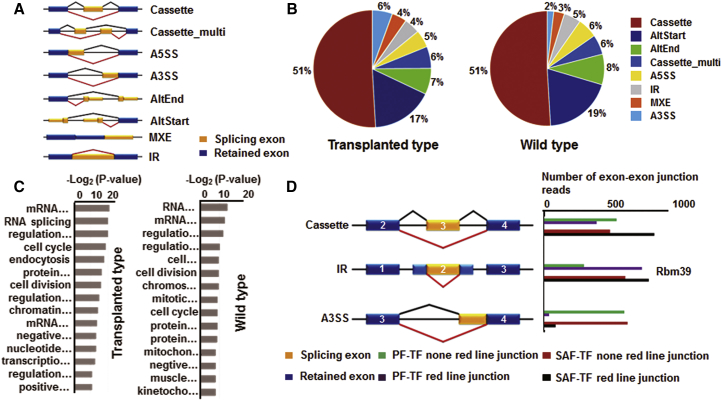
Alternative splicing (AS) is an important mechanism for controlling follicular development.44 However, the global AS pattern of folliculogenesis remains unclear. We evaluated known genes with two or more AS patterns. After comparing SAF with PF, 611 kinds of AS events were identified in F-TF and 267 kinds of AS events were identified in F-WT (FDR < 0.05). In total, eight kinds of ASs were identified in both F-TF and F-WT (Figure 8A).
Figure 8.
AS Analysis
(A) All AS patterns. Yellow box represents splicing exon. (B) Constitution of eight kinds of AS patterns in transplanted and wild-type ovaries. (C) GO analysis of AS genes. x bars represent p values at a logarithmic scale. Full information lists of GO terms are shown in Table S6. (D) Numbers of junction reads in three splicing events of Rbm39. Green and purple bars: AS of PF-TF reads. Red and black bars: AS of SAF-TF reads.
Notably, Cassette and altStart were the primary AS patterns in folliculogenesis, with 51% of genes representing the cassette AS pattern and >17% genes representing the altStart AS pattern (Figure 8B). These data suggest an important role for Cassette and altStart in follicular development. Biology function analysis showed that these genes control folliculogenesis by RNA splicing and mRNA processing (Figure 8C). Indeed, rbm39 is an RNA binding protein located in the nucleus. This protein co-localizes with core spliceosomal protein, suggesting that it is a possible splicing factor.45 Furthermore, this gene exhibited three kinds of AS patterns in F-TF-Cassette, IR, and A3SS. Among these AS patterns, there was a significant difference in A3SS expression between PF-TF and SAF-TF (Figure 8D). Interestingly, rbm39 functions as a transcriptional co-activator for JUN/AP-1 and estrogen receptors,46 suggesting an important role in follicular development.
Discussion
Because stem cells are undifferentiated biological cells that can differentiate into specialized cells and are capable of self-renewal, they play a key role in tissue maintenance and regeneration. They are also located in a specific microenvironment known as a niche that has both anatomical and functional dimensions.47 For example, mammalian SSCs are located in the seminiferous tube base-membrane and can home to the niche after transplantation into a recipient testis.48, 49 Stem cell characteristics show great advantages and promise in the field of regenerative medicine.50 Through the use of dual immunofluorescence of DDX4 or IFITM3 and bromodeoxyuridine (BrdU) in mammalian species, researchers showed that the FGSCs retain the basic properties of stem cells in the mouse and rat.4, 11, 13, 51 Results from the present study showed that isolated and cultured FGSCs migrated toward the ovarian cortex surface following transplantation into sterile ovaries. On day 4 post-transplantation, the FGSCs reached the ovarian cortex surface beneath the epithelium. We observed migrated cells that expressed Stra8 and Sycp3 on day 6 post-transplantation, suggesting that these cells began to differentiate into early oocyte stages. Three days later, these germ cells expressed Sycp3, Ybx2, and Zp2, suggesting the cells were preparing for recombination, meiosis arrest, and fertilization.52, 53, 54 On day 16 post-transplantation, the germ cells expressed Zp1, as well as Sycp3, Ybx2, and Zp2, suggesting a gradual differentiation into oocytes. Interestingly, when the FGSCs began to differentiate into the early oocyte stages, the cells gradually left the surface of ovarian cortex and moved toward the depths of the ovary. These results indicated that the ovarian cortex surface beneath the epithelium is the niche for FGSCs.
To determine the mechanisms of F-TF development in vivo, we used single-follicle RNA-seq analysis to identify the transcriptome landscape during the preantral-to-antral follicle transition. There was an obvious difference in transcriptional profiles between PF and SAF derived from F-TFs, which was similar to WT. The DEGs primarily regulated antrum formation of follicles, cell cycle, or cell differentiation, and only some were related to meiosis arrest. For instance, transforming growth factor β (TGF-β) promotes antrum formation by promoting proliferation of granulosa cells, PF growth, progesterone production, and follicle-stimulating hormone (FSH)-induced estrogen synthesis.29, 55 Furthermore, pathway analysis showed that the steroid biosynthesis pathway might play an important role in antrum formation of follicles in transplanted and WT ovaries. Recent studies indicated that aquaporins 1, 7, and 8 regulate water permeability during antrum formation.56 Aurka, which is a centrosome-localized serine/threonine kinase, plays a vital role in cell cycle regulation. In meiosis I, Aurka is a critical microtubule organizing center component involved in meiosis resumption, and Aurka overexpression leads to an abnormal spindle.32 Cpeb1 controls meiosis progress by activating maternal mRNAs. The upregulation of Cpeb1 increases the rate of meiotic resumption in porcine.57 However, Cpeb1 is degraded during the first meiotic in Xenopus oocytes.58 In our study, Cpeb1 expression was low, suggesting a similar function between mouse and Xenopus. The findings demonstrated that once the oocyte reaches the antral compartment, it is fully capable of restarting the meiotic process,59 confirming that antrum formation is an important event for female germ cell development.60, 61
Moreover, AS showed that Cassette and altStart were the primary splicing patterns in folliculogenesis. Furthermore, GO analysis confirmed that RNA splicing and mRNA processing were the primary AS mechanisms for regulating follicle development in transplanted and WT ovaries. These results suggested that folliculogenesis of F-TFs was similar to WT in vivo.
Folliculogenesis is a complex process that involves proliferation and differentiation of both germ cells and somatic cells. Single-follicle RNA-seq and WGCNA are powerful tools for fully understanding the intricate processes in follicle development, revealing potential core genes. Some core genes have been shown to be important in follicular development. For example, Junb is a transcription factor binding protein involved in follicle-stimulating hormone activity in the SAF module,62 and Fgf8 is a growth factor that plays a potentially important role in regulating follicular activation in the PF module.63 Furthermore, most core genes associated with folliculogenesis have been analyzed, such as Mos, Wee2, Dazl, Nanos2, Gpr3, Fbxo43, Fgf8, Asz1, Tet3, Zfp57, Bmp6, and Aurka.25, 32, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72 Interestingly, the PF module showed that the entire gene expression was downregulated, suggesting that downregulation was the main regulation manner for follicular development after the PF stage. Using WGCNA, we, for first time, identified regulation networks between potential core genes and lncRNAs; in particular, the sub-network consisted of Mmp19, lncRNA LOC102633401, Rarb, Nr1h3, Nr5a1, and Nr4a1, or Aurka, lncRNA Gm10857, Pde3a, Mos, Gpr3, Fbxo43, and Dicer1. Furthermore, lncRNA Gm10857, which is the single core lncRNA in the sub-network of the PF module, was downregulated in follicle development. Additionally, Gm10857 was co-expressed with meiosis-associated genes, such as Mos and Pde3a. This suggested that Gm10857 might be crucial in regulating meiosis resumption. This provides a basis for studying the role of lncRNAs in follicular development.
In the present study, we isolated, purified, and cultured FGSCs long term from a single EGFP-transgenic mouse. Furthermore, we found that these FGSCs possessed characteristics of germline stem cells with normal chromosomes, demonstrating that these cells were consistent with previously identified FGSCs. More importantly, the FGSCs from a single mouse restored ovarian function and generated offspring when transplanted into POF mouse ovaries. After tracing FGSC migrations and developmental patterns in vivo, we found that FGSCs gradually differentiated into oocytes, but not somatic cells, suggesting the unipotency characteristic. These findings provide the theoretical basis and lay a technology platform for specific or personalized medical treatment of ovarian failure or other ovarian diseases.
Materials and Methods
Animals
CAG-EGFP female mice were obtained from the Model Animal Research Center of Nanjing University. The CAG-EGFP female mice, also named C57BL/6-Tg(CAG-EGFP)C14-Y01-FM131-Osb, and the mice were generated by Okabe et al.73 in 1997. Gdf9-iCre mice were obtained from Austin J. Cooney’s Lab (Baylor College of Medicine). Ptenloxp/loxp mice, with BALB/c; 129S4 genomic background, were purchased from Jackson Laboratory. To obtain (Gdf9-iCre; PtenloxP/loxP) genotype POF female mice, we first generated Gdf9-iCre/Pten+/loxP male mice using PtenloxP/loxP genotype female mice crossed with Gdf9-iCre/Pten+/+ male mice. Then, the Gdf9-iCre/Pten+/loxP male mice were crossed with PtenloxP/loxP genotype female mice. After multiple rounds of crossing, the (Gdf9-iCre; PtenloxP/loxP) genotype POF mice were identified by genotyping as described by Reddy et al.74 Six-week-old females were used as recipients and were sterilized by intraperitoneal injection of busulfan (30 mg/kg) and cyclophosphamide (120 mg/kg). All animal experiments were approved by the Institutional Animal Care and Use Committee of Shanghai and were performed according to the National Research Council Guide for Care and Use of Laboratory Animals.
Isolation and Culture of FGSCs
Ovaries from one neonatal female mouse (6 days old) were collected in ice-cold Hank’s balanced salt solution (HBSS) without calcium or magnesium, and cells were isolated using the two-step enzymatic method.39 After initial culture, FGSCs were purified using DDX4-based immunomagnetic beads. The sorted FGSCs were then cultured on mitotically inactivated STO (SIM mouse embryo-derived thioguanine- and ouabain-resistant) cell feeders in minimum essential medium alpha (MEMα; Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Life Technologies), 1 mM non-essential amino acids (NEAA) (Life Technologies), 2 mM L-glutamine (Sigma), 30 μg/mL pyruvate (Amresco), 50 μM β-mercaptoethanol (Biotech), 20 ng/mL mouse epidermal growth factor (EGF) (PeproTech), 10 ng/mL basic fibroblast growth factor (bFGF) (PeproTech), 10 ng/mL mouse glial cell line-derived neurotrophic factor (GDNF) (PeproTech), and 10 ng/mL mouse leukemia inhibitory factor (LIF) (Santa Cruz Biotechnology). The FGSCs were subcultured every 4–7 days at a 1:2 ratio.
BrdU Labeling
BrdU (50 μg/mL) was added to cultured medium and incubated for 5 hr. Immunofluorescence of BrdU was performed as described below.
Immunofluorescence
Samples were washed with PBST (PBS containing 0.1% Triton X-100), blocked in 5% BSA, and then incubated with primary antibodies at 4°C overnight.75 After washing twice with PBST, secondary fluorescence antibodies were used, including tetraethyl rhodamineisothiocyanate (TRITC)-conjugated goat anti-rabbit IgG (1:100; Proteintec), fluorescein isothiocyanate (FITC)-conjugated donkey anti-goat IgG (1:100; Proteintec), and TRITC-conjugated goat anti-mouse IgG (1:100; Proteintec). Images were acquired using a Leica digital camera under a fluorescent microscope (DM2500, DMI3000B; Leica). The following primary antibodies were used: rabbit polyclonal anti-DDX4 (1:200; Abcam), anti-Pou5f1 (1:200; Chemicon), anti-SYCP3 (1:100; Santa Cruz), anti-STRA8 (1:100; Abcam), anti-YBX2 (1:100; Abcam), goat polyclonal anti-EGFP (1:1000; Abcam), or mouse anti-BrdU (1:100; Lab Vision Corporation).
Karyotype Analysis
Cultured cells were treated with colchicine (100 ng/mL) for 2 hr, hypotonically treated with 75 mM KCl for 20 min at 37°C, and fixed twice in methanol-acetic acid (3:1) for 30 min at room temperature. The fixed cells were then dropped onto a glass slide, air-dried, and stained with Giemsa.
RNA Isolation and PCR
Total RNA from FGSCs was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions.76, 77 After treating with DNase I, total RNA was reverse-transcribed to cDNA with Reverse Transcriptase moloneymurine leukemia virus (M-MLV; Takara). Single cell or follicle RT-PCR was performed according to a previously described method.78 Single sample was collected into RNase-free PCR tubes using a mouth pipette with 5 μL of ice-cold lysis buffer (50 mM Tris-HCl, 75 mM KCl, and 3 mM MgCl2). HiScript II Q RT SuperMix for qPCR kit (Vazyme Biotech) was used to obtain cDNA. Briefly, samples were incubated for 10 min at 65°C, then 10 min at 42°C with 2 μL of DNA-wiper, followed by the addition of 2 μL of 5X qRT SuperMix II for 30 min at 42°C, followed by 85°C for 2 min to inactivate the enzyme.
For single-cell PCR, 45 PCR cycles were performed using Taq polymerase with instruction. For real-time PCR, cDNA products were diluted at 1:5 and then performed with 7500 real-time PCR amplification system using SYBR Green PCR mix (Roche). All primers are listed in Table S5.
FGSC Transplantation
FGSCs were collected, washed, and then resuspended in PBS at a concentration of 1 × 103/μL. Recipient mice were anaesthetized with a sodium pentobarbitone injection (45 mg/kg body weight, intraperitoneal). Approximately 10 μL of cell suspension containing about 10,000 cells was injected into each ovary. Approximately 10 μL of supernatant from the wash PBS was injected into each ovary for the control.
Southern Blotting
Genomic DNA was extracted from mouse tails and digested with EcoRI for 22 hr. Samples were separated by 0.8% agarose gels. Following denaturation and neutralization, DNA was transferred to a nylon membrane via capillary blotting. After pre-hybridization, hybridization was performed with an EGFP probe overnight in a hybridization oven at 55°C. Signals were then detected using a CDP-Start kit (GE Healthcare).
Library Construction and Transcriptional Profile of Single Follicles
A single follicle was separated by a hypodermic needle; then a PF or SAF was collected according to size and morphology using a finely drawn glass pipette and was stored at −80°C. The cDNA library was prepared using SMARTer UltraLow Input RNA for Sequencing (Clontech Laboratories). In brief, first-strand cDNA was synthesized using SMARTScribe Reverse Transcriptase and then purified using SPRI Ampure Beads. After amplification using the Polymerase Mix, double-stranded complementary DNA (ds cDNA) was purified using SPRI Ampure Beads and stored at −20°C.
After sequencing using HiSeq 2500, high-quality clean reads were obtained from raw reads by removing the adaptor sequences, reads with >5% ambiguous bases, and low-quality reads. The clean reads were then aligned to the mouse genome (version: mm10_GRCm38) using TopHat program. Gene expression was calculated using the FPKM method (fragments per kilobase per million mapped reads) and adjusted using the upper-quartile algorithm. DEG analysis was performed using DESeq. Raw sequence data and processed data were submitted to the GEO and are accessible through the accession number GEO: GSE85412.
GO
GO analysis was applied to analyze the main function of DEGs according to GO, which is the Wey functional classification of NCBI. Generally, the Fisher’s exact test and the χ2 test were used to classify the GO category, and false discovery rate (FDR) was used to adjust the p value.
WGCNA
A signed weighted correlation network was conducted by first establishing the correlation matrix that contained pairwise correlations of all DEG pairs. The adjacency matrix was calculated by raising the co-expression to the power of 19, which was the soft threshold of the correlation matrix. Based on the resulting adjacency matrix, a topological overlap measurement was used to cluster genes with a highly similar co-expression relationship into a network module.
RNA AS
After mapping, information regarding chromosome position for exonic reads and exon-exon junction reads was generated. All junction reads were selected using the following criteria: a potential junction site must match perfectly with at least 6 nt of a read to each of the two flanking regions, and filter the site with more than three non-redundant reads in both samples. ASs were then identified in the software AS detector (ASD) using the Fisher’s exact test.79 RNA AS events were adjusted using JSD (Jensen-Shannon divergence) (FDR < 0.05).
Statistics
Generally, one-sided Student’s t test was used in real-time PCR analysis; the Fisher’s exact test and the χ2 test were used in GO and pathway analysis. Pearson correlation was used for the gene co-expression network analysis, and FDR was used to adjust the p value. Fisher’s exact test was used for ASs analysis. A p value less than 0.05 was considered significant, and a p value less than 0.005 was considered extremely significant.
Author Contributions
C.W. conducted all the major experiments, data analysis, and wrote the manuscript. B.X., X.L., W.M., P.Z., and X.C. characterized FGSCs and traced the development of transplanted FGSCs. J.W. initiated and supervised the entire project, carried out FGSC transplantation, analyzed data, and wrote the manuscript.
Acknowledgments
This work was supported by the National Basic Research Program of China (grant 2013CB967401), the National Nature Science Foundation of China (grants 81370675 and 81421061), and the Shanghai Jiao Tong University Medicine-Engineering Fund (grant YG2014ZD08).
Footnotes
Supplemental Information includes eight figures and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.04.019.
Supplemental Information
References
- 1.Regaud C. Etudes sur la structure des tubes seminiferes et sur la spermatogenese chez les mammiferes [Studies on the structure of seminiferous tubules and on spermatogenesis in mammals]. Part 1. Archives d’Anatomie Microscopiques et de Morphologie Experimentale. 1901;4:101–156. [Google Scholar]
- 2.He Z., Kokkinaki M., Dym M. Signaling molecules and pathways regulating the fate of spermatogonial stem cells. Microsc. Res. Tech. 2009;72:586–595. doi: 10.1002/jemt.20698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boitani C., Di Persio S., Esposito V., Vicini E. Spermatogonial cells: mouse, monkey and man comparison. Semin. Cell Dev. Biol. 2016;59:79–88. doi: 10.1016/j.semcdb.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Johnson J., Canning J., Kaneko T., Pru J.K., Tilly J.L. Germline stem cells and follicular renewal in the postnatal mammalian ovary. Nature. 2004;428:145–150. doi: 10.1038/nature02316. [DOI] [PubMed] [Google Scholar]
- 5.Zou K., Yuan Z., Yang Z., Luo H., Sun K., Zhou L., Xiang J., Shi L., Yu Q., Zhang Y. Production of offspring from a germline stem cell line derived from neonatal ovaries. Nat. Cell Biol. 2009;11:631–636. doi: 10.1038/ncb1869. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y., Yang Z., Yang Y., Wang S., Shi L., Xie W., Sun K., Zou K., Wang L., Xiong J. Production of transgenic mice by random recombination of targeted genes in female germline stem cells. J. Mol. Cell Biol. 2011;3:132–141. doi: 10.1093/jmcb/mjq043. [DOI] [PubMed] [Google Scholar]
- 7.White Y.A., Woods D.C., Takai Y., Ishihara O., Seki H., Tilly J.L. Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nat. Med. 2012;18:413–421. doi: 10.1038/nm.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woods D.C., Tilly J.L. The next (re)generation of ovarian biology and fertility in women: is current science tomorrow’s practice? Fertil. Steril. 2012;98:3–10. doi: 10.1016/j.fertnstert.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H., Jiang M., Bi H., Chen X., He L., Li X., Wu J. Conversion of female germline stem cells from neonatal and prepubertal mice into pluripotent stem cells. J. Mol. Cell Biol. 2014;6:164–171. doi: 10.1093/jmcb/mju004. [DOI] [PubMed] [Google Scholar]
- 10.Xiong J., Lu Z., Wu M., Zhang J., Cheng J., Luo A., Shen W., Fang L., Zhou S., Wang S. Intraovarian transplantation of female germline stem cells rescue ovarian function in chemotherapy-injured ovaries. PLoS ONE. 2015;10:e0139824. doi: 10.1371/journal.pone.0139824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo K., Li C.H., Wang X.Y., He D.J., Zheng P. Germ stem cells are active in postnatal mouse ovary under physiological conditions. Mol. Hum. Reprod. 2016;22:316–328. doi: 10.1093/molehr/gaw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C., Wu J. Production of offspring from a germline stem cell line derived from prepubertal ovaries of germline reporter mice. Mol. Hum. Reprod. 2016;22:457–464. doi: 10.1093/molehr/gaw030. [DOI] [PubMed] [Google Scholar]
- 13.Zhou L., Wang L., Kang J.X., Xie W., Li X., Wu C., Xu B., Wu J. Production of fat-1 transgenic rats using a post-natal female germline stem cell line. Mol. Hum. Reprod. 2014;20:271–281. doi: 10.1093/molehr/gat081. [DOI] [PubMed] [Google Scholar]
- 14.Ding X., Liu G., Xu B., Wu C., Hui N., Ni X., Wang J., Du M., Teng X., Wu J. Human GV oocytes generated by mitotically active germ cells obtained from follicular aspirates. Sci. Rep. 2016;6:28218. doi: 10.1038/srep28218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hummitzsch K., Anderson R.A., Wilhelm D., Wu J., Telfer E.E., Russell D.L., Robertson S.A., Rodgers R.J. Stem cells, progenitor cells, and lineage decisions in the ovary. Endocr. Rev. 2015;36:65–91. doi: 10.1210/er.2014-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X.F., Zhang L., Wu Q.H., Min J.X., Ma N., Luo L.C. Biological mechanisms of premature ovarian failure caused by psychological stress based on support vector regression. Int. J. Clin. Exp. Med. 2015;8:21393–21399. [PMC free article] [PubMed] [Google Scholar]
- 17.Jang H., Lee O.H., Lee Y., Yoon H., Chang E.M., Park M., Lee J.W., Hong K., Kim J.O., Kim N.K. Melatonin prevents cisplatin-induced primordial follicle loss via suppression of PTEN/AKT/FOXO3a pathway activation in the mouse ovary. J. Pineal Res. 2016;60:336–347. doi: 10.1111/jpi.12316. [DOI] [PubMed] [Google Scholar]
- 18.Meattini I., Saieva C., Meacci F., Scotti V., De Luca Cardillo C., Desideri I., Baldazzi V., Mangoni M., Scoccianti S., Detti B. Impact of age on cytotoxic-induced ovarian failure in breast cancer treated with adjuvant chemotherapy and triptorelin. Future Oncol. 2016;12:625–635. doi: 10.2217/fon.15.357. [DOI] [PubMed] [Google Scholar]
- 19.Hernández-Angeles C., Castelo-Branco C. Early menopause: a hazard to a woman’s health. Indian J. Med. Res. 2016;143:420–427. doi: 10.4103/0971-5916.184283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woods D.C., Tilly J.L. Autologous germline mitochondrial energy transfer (AUGMENT) in human assisted reproduction. Semin. Reprod. Med. 2015;33:410–421. doi: 10.1055/s-0035-1567826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oktay K., Baltaci V., Sonmezer M., Turan V., Unsal E., Baltaci A., Aktuna S., Moy F. Oogonial precursor cell-derived autologous mitochondria injection to improve outcomes in women with multiple IVF failures due to low oocyte quality: a clinical translation. Reprod. Sci. 2015;22:1612–1617. doi: 10.1177/1933719115612137. [DOI] [PubMed] [Google Scholar]
- 22.Koubova J., Hu Y.C., Bhattacharyya T., Soh Y.Q., Gill M.E., Goodheart M.L., Hogarth C.A., Griswold M.D., Page D.C. Retinoic acid activates two pathways required for meiosis in mice. PLoS Genet. 2014;10:e1004541. doi: 10.1371/journal.pgen.1004541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monniaux D. Driving folliculogenesis by the oocyte-somatic cell dialog: lessons from genetic models. Theriogenology. 2016;86:41–53. doi: 10.1016/j.theriogenology.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 24.Menke D.B., Koubova J., Page D.C. Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev. Biol. 2003;262:303–312. doi: 10.1016/s0012-1606(03)00391-9. [DOI] [PubMed] [Google Scholar]
- 25.Feng C.W., Bowles J., Koopman P. Control of mammalian germ cell entry into meiosis. Mol. Cell. Endocrinol. 2014;382:488–497. doi: 10.1016/j.mce.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Grive K.J., Freiman R.N. The developmental origins of the mammalian ovarian reserve. Development. 2015;142:2554–2563. doi: 10.1242/dev.125211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacLennan M., Crichton J.H., Playfoot C.J., Adams I.R. Oocyte development, meiosis and aneuploidy. Semin. Cell Dev. Biol. 2015;45:68–76. doi: 10.1016/j.semcdb.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wigglesworth K., Lee K.B., O’Brien M.J., Peng J., Matzuk M.M., Eppig J.J. Bidirectional communication between oocytes and ovarian follicular somatic cells is required for meiotic arrest of mammalian oocytes. Proc. Natl. Acad. Sci. USA. 2013;110:E3723–E3729. doi: 10.1073/pnas.1314829110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orisaka M., Tajima K., Tsang B.K., Kotsuji F. Oocyte-granulosa-theca cell interactions during preantral follicular development. J. Ovarian Res. 2009;2:9. doi: 10.1186/1757-2215-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orisaka M., Hattori K., Fukuda S., Mizutani T., Miyamoto K., Sato T., Tsang B.K., Kotsuji F., Yoshida Y. Dysregulation of ovarian follicular development in female rat: LH decreases FSH sensitivity during preantral-early antral transition. Endocrinology. 2013;154:2870–2880. doi: 10.1210/en.2012-2173. [DOI] [PubMed] [Google Scholar]
- 31.Nishimura Y., Endo T., Kano K., Naito K. Porcine Aurora A accelerates Cyclin B and Mos synthesis and promotes meiotic resumption of porcine oocytes. Anim. Reprod. Sci. 2009;113:114–124. doi: 10.1016/j.anireprosci.2008.05.074. [DOI] [PubMed] [Google Scholar]
- 32.Saskova A., Solc P., Baran V., Kubelka M., Schultz R.M., Motlik J. Aurora kinase A controls meiosis I progression in mouse oocytes. Cell Cycle. 2008;7:2368–2376. doi: 10.4161/cc.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonnet A., Cabau C., Bouchez O., Sarry J., Marsaud N., Foissac S., Woloszyn F., Mulsant P., Mandon-Pepin B. An overview of gene expression dynamics during early ovarian folliculogenesis: specificity of follicular compartments and bi-directional dialog. BMC Genomics. 2013;14:904. doi: 10.1186/1471-2164-14-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dupré A., Daldello E.M., Nairn A.C., Jessus C., Haccard O. Phosphorylation of ARPP19 by protein kinase A prevents meiosis resumption in Xenopus oocytes. Nat. Commun. 2014;5:3318. doi: 10.1038/ncomms4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui C., Zhao H., Zhang Z., Zong Z., Feng C., Zhang Y., Deng X., Xu X., Yu B. CDC25B acts as a potential target of PRKACA in fertilized mouse eggs. Biol. Reprod. 2008;79:991–998. doi: 10.1095/biolreprod.108.068205. [DOI] [PubMed] [Google Scholar]
- 36.Wigglesworth K., Lee K.B., Emori C., Sugiura K., Eppig J.J. Transcriptomic diversification of developing cumulus and mural granulosa cells in mouse ovarian follicles. Biol. Reprod. 2015;92:23. doi: 10.1095/biolreprod.114.121756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sumitomo J., Emori C., Matsuno Y., Ueno M., Kawasaki K., Endo T.A., Shiroguchi K., Fujii W., Naito K., Sugiura K. Mouse oocytes suppress miR-322-5p expression in ovarian granulosa cells. J. Reprod. Dev. 2016;62:393–399. doi: 10.1262/jrd.2015-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell D.L., Gilchrist R.B., Brown H.M., Thompson J.G. Bidirectional communication between cumulus cells and the oocyte: old hands and new players? Theriogenology. 2016;86:62–68. doi: 10.1016/j.theriogenology.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 39.Wang H., Shi L., Xiang J., Ding X., Luo H., Wang S., Wu J. Isolation, culture and transplantation of female germline stem cells from neonatal and prepubertal mice. Protoc. Exch. 2013 Published online January 2, 2013. [Google Scholar]
- 40.Xie W., Wang H., Wu J. Similar morphological and molecular signatures shared by female and male germline stem cells. Sci. Rep. 2014;4:5580. doi: 10.1038/srep05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X.L., Wu J., Wang J., Shen T., Li H., Lu J., Gu Y., Kang Y., Wong C.H., Ngan C.Y. Integrative epigenomic analysis reveals unique epigenetic signatures involved in unipotency of mouse female germline stem cells. Genome Biol. 2016;17:162. doi: 10.1186/s13059-016-1023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grive K.J., Gustafson E.A., Seymour K.A., Baddoo M., Schorl C., Golnoski K., Rajkovic A., Brodsky A.S., Freiman R.N. TAF4b regulates oocyte-specific genes essential for meiosis. PLoS Genet. 2016;12:e1006128. doi: 10.1371/journal.pgen.1006128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Q.Y., Miao Y.L., Schatten H. Towards a new understanding on the regulation of mammalian oocyte meiosis resumption. Cell Cycle. 2009;8:2741–2747. doi: 10.4161/cc.8.17.9471. [DOI] [PubMed] [Google Scholar]
- 44.Wang F., Pan J., Liu Y., Meng Q., Lv P., Qu F., Ding G.L., Klausen C., Leung P.C., Chan H.C. Alternative splicing of the androgen receptor in polycystic ovary syndrome. Proc. Natl. Acad. Sci. USA. 2015;112:4743–4748. doi: 10.1073/pnas.1418216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imai H., Chan E.K., Kiyosawa K., Fu X.D., Tan E.M. Novel nuclear autoantigen with splicing factor motifs identified with antibody from hepatocellular carcinoma. J. Clin. Invest. 1993;92:2419–2426. doi: 10.1172/JCI116848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stepanyuk G.A., Serrano P., Peralta E., Farr C.L., Axelrod H.L., Geralt M., Das D., Chiu H.J., Jaroszewski L., Deacon A.M. UHM-ULM interactions in the RBM39-U2AF65 splicing-factor complex. Acta Crystallogr. D Struct. Biol. 2016;72:497–511. doi: 10.1107/S2059798316001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scadden D.T. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 48.Lee K.H., Lee W.Y., Kim D.H., Lee S.H., Do J.T., Park C., Kim J.H., Choi Y.S., Song H. Vitrified canine testicular cells allow the formation of spermatogonial stem cells and seminiferous tubules following their xenotransplantation into nude mice. Sci. Rep. 2016;6:21919. doi: 10.1038/srep21919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanatsu-Shinohara M., Takehashi M., Takashima S., Lee J., Morimoto H., Chuma S., Raducanu A., Nakatsuji N., Fässler R., Shinohara T. Homing of mouse spermatogonial stem cells to germline niche depends on beta1-integrin. Cell Stem Cell. 2008;3:533–542. doi: 10.1016/j.stem.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 50.Hagenhoff A., Bruns C.J., Zhao Y., von Lüttichau I., Niess H., Spitzweg C., Nelson P.J. Harnessing mesenchymal stem cell homing as an anticancer therapy. Expert Opin. Biol. Ther. 2016;16:1079–1092. doi: 10.1080/14712598.2016.1196179. [DOI] [PubMed] [Google Scholar]
- 51.Zou K., Hou L., Sun K., Xie W., Wu J. Improved efficiency of female germline stem cell purification using fragilis-based magnetic bead sorting. Stem Cells Dev. 2011;20:2197–2204. doi: 10.1089/scd.2011.0091. [DOI] [PubMed] [Google Scholar]
- 52.Schalk J.A., Dietrich A.J., Vink A.C., Offenberg H.H., van Aalderen M., Heyting C. Localization of SCP2 and SCP3 protein molecules within synaptonemal complexes of the rat. Chromosoma. 1998;107:540–548. doi: 10.1007/s004120050340. [DOI] [PubMed] [Google Scholar]
- 53.Avella M.A., Baibakov B., Dean J. A single domain of the ZP2 zona pellucida protein mediates gamete recognition in mice and humans. J. Cell Biol. 2014;205:801–809. doi: 10.1083/jcb.201404025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang M.Y., Fortune J.E. The capacity of primordial follicles in fetal bovine ovaries to initiate growth in vitro develops during mid-gestation and is associated with meiotic arrest of oocytes. Biol. Reprod. 2008;78:1153–1161. doi: 10.1095/biolreprod.107.066688. [DOI] [PubMed] [Google Scholar]
- 55.Sun J., Li X. Growth and antrum formation of bovine primary follicles in long-term culture in vitro. Reprod. Biol. 2013;13:221–228. doi: 10.1016/j.repbio.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 56.Rodgers R.J., Irving-Rodgers H.F. Formation of the ovarian follicular antrum and follicular fluid. Biol. Reprod. 2010;82:1021–1029. doi: 10.1095/biolreprod.109.082941. [DOI] [PubMed] [Google Scholar]
- 57.Nishimura Y., Kano K., Naito K. Porcine CPEB1 is involved in Cyclin B translation and meiotic resumption in porcine oocytes. Anim. Sci. J. 2010;81:444–452. doi: 10.1111/j.1740-0929.2010.00755.x. [DOI] [PubMed] [Google Scholar]
- 58.Igea A., Méndez R. Meiosis requires a translational positive loop where CPEB1 ensues its replacement by CPEB4. EMBO J. 2010;29:2182–2193. doi: 10.1038/emboj.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dokshin G.A., Baltus A.E., Eppig J.J., Page D.C. Oocyte differentiation is genetically dissociable from meiosis in mice. Nat. Genet. 2013;45:877–883. doi: 10.1038/ng.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elvin J.A., Yan C., Matzuk M.M. Oocyte-expressed TGF-beta superfamily members in female fertility. Mol. Cell. Endocrinol. 2000;159:1–5. doi: 10.1016/s0303-7207(99)00185-9. [DOI] [PubMed] [Google Scholar]
- 61.Elvin J.A., Matzuk M.M. Mouse models of ovarian failure. Rev. Reprod. 1998;3:183–195. doi: 10.1530/ror.0.0030183. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y., Fortin J., Lamba P., Bonomi M., Persani L., Roberson M.S., Bernard D.J. Activator protein-1 and smad proteins synergistically regulate human follicle-stimulating hormone beta-promoter activity. Endocrinology. 2008;149:5577–5591. doi: 10.1210/en.2008-0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Price C.A. Mechanisms of fibroblast growth factor signaling in the ovarian follicle. J. Endocrinol. 2016;228:R31–R43. doi: 10.1530/JOE-15-0414. [DOI] [PubMed] [Google Scholar]
- 64.Tang W., Wu J.Q., Guo Y., Hansen D.V., Perry J.A., Freel C.D., Nutt L., Jackson P.K., Kornbluth S. Cdc2 and Mos regulate Emi2 stability to promote the meiosis I-meiosis II transition. Mol. Biol. Cell. 2008;19:3536–3543. doi: 10.1091/mbc.E08-04-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lim A.K., Lorthongpanich C., Chew T.G., Tan C.W., Shue Y.T., Balu S., Gounko N., Kuramochi-Miyagawa S., Matzuk M.M., Chuma S. The nuage mediates retrotransposon silencing in mouse primordial ovarian follicles. Development. 2013;140:3819–3825. doi: 10.1242/dev.099184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu C., Zhang Y.L., Pan W.W., Li X.M., Wang Z.W., Ge Z.J., Zhou J.J., Cang Y., Tong C., Sun Q.Y., Fan H.Y. CRL4 complex regulates mammalian oocyte survival and reprogramming by activation of TET proteins. Science. 2013;342:1518–1521. doi: 10.1126/science.1244587. [DOI] [PubMed] [Google Scholar]
- 67.Agca C., Yakan A., Agca Y. Estrus synchronization and ovarian hyper-stimulation treatments have negligible effects on cumulus oocyte complex gene expression whereas induction of ovulation causes major expression changes. Mol. Reprod. Dev. 2013;80:102–117. doi: 10.1002/mrd.22141. [DOI] [PubMed] [Google Scholar]
- 68.Emori C., Sugiura K. Role of oocyte-derived paracrine factors in follicular development. Anim. Sci. J. 2014;85:627–633. doi: 10.1111/asj.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oh J.S., Han S.J., Conti M. Wee1B, Myt1, and Cdc25 function in distinct compartments of the mouse oocyte to control meiotic resumption. J. Cell Biol. 2010;188:199–207. doi: 10.1083/jcb.200907161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sousa Martins J.P., Liu X., Oke A., Arora R., Franciosi F., Viville S., Laird D.J., Fung J.C., Conti M. DAZL and CPEB1 regulate mRNA translation synergistically during oocyte maturation. J. Cell Sci. 2016;129:1271–1282. doi: 10.1242/jcs.179218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J., Mao G., Xia G. FSH modulates PKAI and GPR3 activities in mouse oocyte of COC in a gap junctional communication (GJC)-dependent manner to initiate meiotic resumption. PLoS ONE. 2012;7:e37835. doi: 10.1371/journal.pone.0037835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miyagaki Y., Kanemori Y., Tanaka F., Baba T. Possible role of p38 MAPK-MNK1-EMI2 cascade in metaphase-II arrest of mouse oocytes. Biol. Reprod. 2014;91:45. doi: 10.1095/biolreprod.113.116962. [DOI] [PubMed] [Google Scholar]
- 73.Okabe M., Ikawa M., Kominami K., Nakanishi T., Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 74.Reddy P., Liu L., Adhikari D., Jagarlamudi K., Rajareddy S., Shen Y., Du C., Tang W., Hämäläinen T., Peng S.L. Oocyte-specific deletion of Pten causes premature activation of the primordial follicle pool. Science. 2008;319:611–613. doi: 10.1126/science.1152257. [DOI] [PubMed] [Google Scholar]
- 75.Decembrini S., Martin C., Sennlaub F., Chemtob S., Biel M., Samardzija M., Moulin A., Behar-Cohen F., Arsenijevic Y. Cone genesis tracing by the Chrnb4-EGFP mouse line: evidences of cellular material fusion after cone precursor transplantation. Mol. Ther. 2017;25:634–653. doi: 10.1016/j.ymthe.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun M., Nie F.Q., Zang C., Wang Y., Hou J., Wei C., Li W., He X., Lu K.H. The pseudogene DUXAP8 promotes non-small-cell lung cancer cell proliferation and invasion by epigenetically silencing EGR1 and RHOB. Mol. Ther. 2017;25:739–751. doi: 10.1016/j.ymthe.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Liu Y., Zhao D., Qiu F., Zhang L.L., Liu S.K., Li Y.Y., Liu M.T., Wu D., Wang J.X. Manipulating PML SUMOylation via silencing UBC9 and RNF4 regulates cardiac fibrosis. Mol. Ther. 2017;25:666–678. doi: 10.1016/j.ymthe.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dulac C., Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 79.Zhou X., Wu W., Li H., Cheng Y., Wei N., Zong J., Feng X., Xie Z., Chen D., Manley J.L. Transcriptome analysis of alternative splicing events regulated by SRSF10 reveals position-dependent splicing modulation. Nucleic Acids Res. 2014;42:4019–4030. doi: 10.1093/nar/gkt1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.