Abstract
High-throughput sequencing technology, coupled with the use of conserved marker genes, has allowed for the understanding of communities of microbes (both culturable and unculturable) as well as their phylogenetic placement. The recent explosion of sequencing data prompted the development of software that could process the vast amount of data generated and phylogenetically differentiate groups of samples. Host-associated microbial studies have revealed that microbes are highly varied between individuals and fluctuate within an individual. Large-scale studies are being undertaken that include collection of extensive environmental data to help uncover the forces that shape microbial communities.
Keywords: microbiome, metagenomics, sequencing, microbial ecology, phylogenetics
When Antonie van Leeuwenhoek peered through a handcrafted microscope at a complex community of “animalcules” from a tooth scraping, the science of microbiology was born (1). The rich diversity and interactions between these microbes and their environment is still being explored over three centuries since Leeuwenhoek’s initial observation. A human can be considered to be a super organism (2): the human cells and genomic content are far outweighed by microbial cells (3) and microbial genes (4). Consequently, understanding the relationships between these complex communities and human physiology will be vital to progress in treating a range of human disease. In this review, we discuss some of the classical techniques for analyzing microbial communities, as well as current methods and future directions.
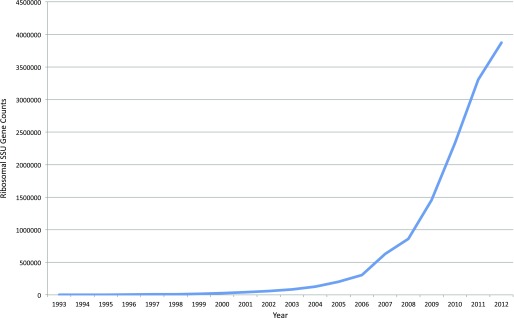
Classical microbiology primarily consisted of isolating microbes, growing these as pure cultures, and identifying biochemical properties of these organisms, such as cell wall structure by gram staining, oxygen tolerance, and carbon or nitrogen sources that supported their growth. The entirety of bacteria and archaea were joined together as a grouping under “prokaryotes” on the basis of these observations, and the phylogenetic relationships between these microbes and eukaryotes was unknown. Carl Woese and George Fox (5) changed this picture by using the ribosomal small subunit (SSU) gene as a phylogenetic marker. The variants of this gene, which makes up a critical component of the ribosome that is found in all cells, are named according to their size: 16S ribosomal RNA (rRNA) in bacteria and archaea, 18S rRNA in eukaryotes. By comparing shared nucleotide fragments in the SSU gene, Woese and Fox showed that archaea (then called archaebacteria) was a distinct domain of life. This study involved laborious purification of SSU RNA, followed by RNase digestion and two-dimensional gel electrophoresis, to establish shared SSU sequence identity by the resulting fragment locations. Acquiring SSU genes soon became much easier. The dideoxynucleotide termination sequencing method (i.e., “Sanger”) (6) became available in 1977, and, along with other advances, such as the PCR in 1985 (7), the number of known SSU sequences grew exponentially (Figure 1). Norman Pace and colleagues (8) observed that the SSU gene was not only found in all organisms, but also contained sites that were either universally conserved, or conserved in large groups of organisms. This led to the important discovery that organisms not amenable to cultivation can still be detected in the environment by amplifying and sequencing the SSU gene from environmental samples directly.
Figure 1.
Total ribosomal small subunit sequences in GenBank by year. The nucleotide database was queried with ribosomal rRNA (rRNA; Feature key), “X” (publication date), and 16S (title) for each year from 1993 to 2012.
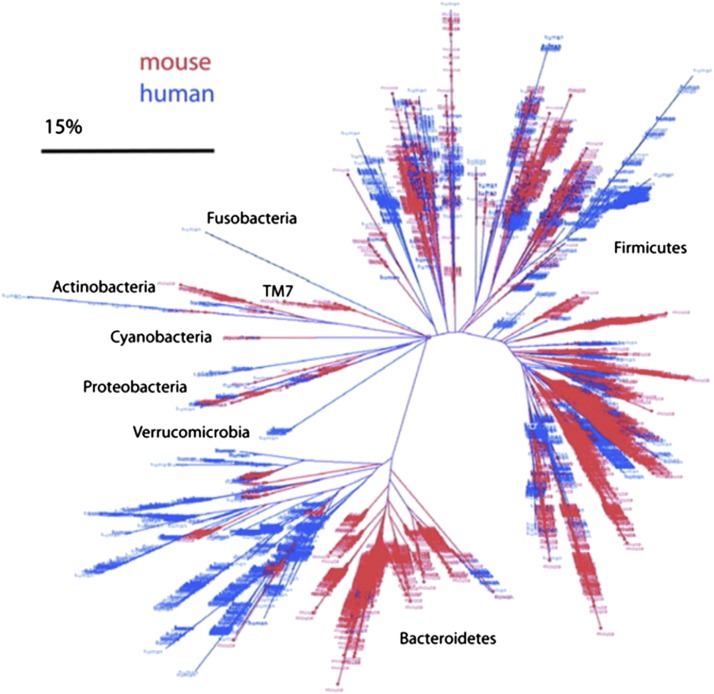
Throughout the latter part of the 20th century, and beginning of the 21st, Sanger sequencing was the dominant sequencing technology, and had been improved by parallelization and by automated fluorescent detection of nucleotide termination. Bacterial cloning of the SSU PCR amplicon product is used for Sanger sequencing, which has the practical effect of limiting these sorts of microbial studies to the range of tens to hundreds of sequences due to its expense and the need to clone each sequence into a separate bacterial culture. This limitation was escaped in the middle of the first decade of the 21st century, when newer high-throughput sequencing became commercially available, such as 454 (now Roche [F. Hoffman-La Roche AG, Basel, Switzerland]) company’s GS20 and the Illumina sequencing-by-synthesis Genome Analyzer (Illumina, San Diego, CA), which yielded orders of magnitude greater sequencing depth than was previously available (9, 10). A comparison of the capabilities of these next-generation technologies is shown in Table 1 (modified from Ref. 11). These new technologies caused an explosion in the number of SSU-based studies and, as a result, the sequenced SSU genes, as shown in Figure 1 (note that this figure only shows deposition into GenBank, not into the Short Read Archive where much next-generation sequencing data is deposited). Sanger-based studies would often group sequences using closest BLAST hits in GenBank, or using alignments and phylogenetic trees to illustrate the placement of detected novel sequences compared with known taxa. Barring specific knowledge of the taxa present, these trees can be difficult to glean meaning from (e.g., Figures 3–12 in the article be Hongoh and colleagues [12]). A study by Ley and colleagues (13) yielded a phylogenetic tree, which shows a clear qualitative pattern in phylogenetic differences between mice and human gut communities as shown in Figure 2. However, this tree only shows the differences between humans and mice as a whole, not the differences between the 40 individual samples from humans and mice included in this study, which are not accessible to visual inspection. Consequently, there was a clear need to understand the similarities and differences among communities, not just to detect that two communities were statistically significantly different. Large Sanger-based surveys, like that of Ley and colleagues’ and a plethora of next-generation SSU sequences, motivated the development of a metric to compare microbial communities while taking into account phylogeny. The UniFrac (14) metric addressed this issue, and uses unique- and shared-branch length to calculate distances between communities. Nonphylogenetic metrics have existed for quite some time, but a phylogenetic metric is arguably more accurate in that it does not assume that all taxa are equally different. This was illustrated by patterns observed in environmental samples clustering by salinity, which were obscured when using nonphylogenetic metrics, but were very clear with UniFrac (15).
Table 1.
Comparison of high-throughput sequencing technologies
| Read Length | Maximum Insert Size | Run Time | Reads Per Run | Relative Cost Factor (Per Mb) | Scale of Reads Per Sample | Scale of Samples Per Run | Raw Error Rate (%) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Insertions | Deletions | Mismatches | ||||||||
| ABI 3730 | 800 b | >1 Kb | 2 h | 96 | 100 | 102 | 101 | 0.001 | <<0.1 | <<0.1 | <<0.1 |
| 454 FLX Titanium | 300 to 400 b | 800 b | 9 h | 106 | 1 | 103 | 102 | 1 | <1 | <<0.1 | <<1 |
| 454 FLX+ | 700 to 1000 b | 1200 b | 23 h | 106 | 0.7 | 103 | 102 | — | — | — | — |
| Illumina GAIIx | 76 to 101 b | 500 b | 6–9 d | 4 × 108 | 0.1 | 105 to 106 | 103 to 104 | <1 | <<1 | <<1 | <1 |
| Illumina HiSeq 2000 | 101 to 151 b | 500 b | 9–15 d | 3 × 109 | 0.002 | 105 to 106 | 103 to 104 | — | — | — | — |
| Illumina MiSeq | up to 250 b | 500 b | 4–39 h | 1.0 × 107 | 0.06 | 104 | 102 | — | — | — | — |
| PacBio | 1100 b | >1 Kb | 1.5 h | 3.5 × 107 | 1.5 | 103 | 101 | 15 | 13 | 1 | 1 |
| IonTorrent | 200 b | 400 b | 2–3 h | 1.5 × 106 to 3 × 106 | 0.4 | 103 | 102 | 2 | 1 | 1 | <1 |
Updated from Table 1 of Reference 11 where newer data were available.
Figure 2.
Phylogenetic tree of 5,088 mouse (red) ceca-associated 16S rRNA sequences reported in this study and 11,831 human (blue) colon–associated 16S rRNA sequences. Scale bar indicates sequence convergence. Adapted by permission from Reference 13.
Another issue that arose from the deluge of next generation sequences was the handling of the sequences themselves. Processing a few hundred sequences by hand was possible, but not tenable with tens of thousands or millions of sequences. Replicating results across laboratories with in-house methods for processing data is also unviable. Automated pipelines for processing marker genes, such as mothur (16) and QIIME (17), as well as more metagenomic-oriented software (e.g., Megan [18] and MG-RAST [19]), answered this issue by providing pipelines that both democratized access to these analyses and allowed for reproducibility between groups. An outline of the quality filtering and processing pipeline that these software packages generally follow is described by Kuczynski and colleagues (11). Briefly, raw input sequences are assigned to samples according to barcodes, quality filtered, clustered according to sequence identity into operational taxonomic units (OTUs), compared against a reference database to establish taxonomic identity, and put into a table form of counts per OTU. This OTU table is then used for downstream analyses of diversity and of statistically significant differences in taxa among groups. Alternative reference databases may be used for clustering and taxonomic assignment, and one should keep this in mind when comparing results across multiple studies.
Using deep sequencing data of microbial communities, many important discoveries have been made. A “core” set of human gut- or skin-associated microbes shared by most humans does not appear to exist at the genus or OTU level (20, 21), and essentially any taxon that is common in or on one person is absent in others when enough people are examined. There is variability over time for adult individuals, and the extent of this variation depends upon the body site (22). The mode of delivery (Caesarian section versus vaginal) has been associated with the immune disorder asthma, and the initial colonization of newborn babies is largely dependent upon the mode of birth, as shown by Dominguez and colleagues (23), which could play a role in training the immune system. These studies suggest that a combination of stochastic, microbial exposure, and selective pressure (e.g., diet, immune system, gut environment) shapes microbial communities in human hosts: this combination is perhaps unsurprising, and a key challenge moving forward is to quantify the relative importance of these factors, both for the human microbiota overall and specifically for the components that affect health, which may differ in different people. Efforts to uncover the defining factors that shape the available microbial and overall community structure are being undertaken in large-scale projects like the Earth Microbiome Project (24) and the American Gut Project (25). These projects include extensive and standardized metadata collection, sampling, extraction, and sequencing protocols. Once there is a clear understanding of the defining forces that shape particular communities, we should have the ability to perturb these systems to our advantage.
Looking to the future, a common question is whether SSU rRNA gene sequence analysis is sufficient, or whether we are missing important information from this level of analysis. Shotgun metagenomics, in which total DNA is extracted and sequenced, has the advantage that all the genes can be observed directly (this is especially important in cases where gene content is highly plastic, because of pathogenicity islands or other mobile genetic elements), and is often thought to be the future of sequencing (but see Ref. 26 for a dissenting view). Interestingly, in the cases in which direct comparisons have been made, the SSU rRNA gene profiles identify very similar patterns of clustering to the shotgun metagenomics (e.g., Refs. 27–29). Shotgun metagenomics is especially difficult in heavily host-contaminated samples, such as those from lung biopsies, because most of the DNA is human, and many studies of environments with low bacterial biomass, including studies of indoor air, have mostly resequenced the human genome in a very expensive way. Metatranscriptomics, the study of RNA transcripts, perhaps holds more promise, and has identified functional patterns where SSU rRNA gene sequencing and shotgun metagenomics failed (e.g., Ref. 30). Metabolomics, the study of small-molecule metabolites produced by a community, is also very exciting, and has, for example, been used to link microbial communities to individual responses to drugs (31, 32). The appropriate level of analysis for a given study is, however, still a very active topic of investigation, and likely depends on the timescale and nature of the phenomena being investigated.
As the cost of generating sequence data continues to decline, one very exciting shift in the field has been the transition from attempting to detect differences between sets of samples (e.g., healthy versus diseased people) toward predictive modeling and detailed spatial and temporal characterization of sites on the body (33, 34). In particular, showing that two groups of people differ in their microbial communities cannot reveal whether those microbial differences are causal, simply reflect a microbial response to pre-existing damage, or whether a feedback relationship exists in which a changed environment changes the microbes, which then changes the environment further. Identifying causality thus poses a major challenge for the field; however, the recent discovery that features of individual human metabolic phenotypes can be recaptured in germ-free mice that are then colonized with stool samples from those individual people (29) provides a powerful paradigm for mechanistic studies. Especially for studies of the lung, the combination of the ability to colonize animals with defined microbial communities, assay the transcription of those communities and the metabolites they produce, and deduce causal mechanistic pathways for how specific microbes may influence the lung itself or the host immune system provides very high potential for insight into a range of medical conditions.
Footnotes
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Madigan M, Martinko J, editors. Brock biology of microorganisms. 11th ed. New Jersey: Prentice Hall; 2006. [Google Scholar]
- 2.Sleator RD. The human superorganism—of microbes and men. Med Hypotheses. 2010;74:214–215. doi: 10.1016/j.mehy.2009.08.047. [DOI] [PubMed] [Google Scholar]
- 3.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 4.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 8.Pace NR, Stahl DA, Lane DJ, Olsen GJ. The analysis of natural microbial populations by ribosomal RNA sequences. Am Soc Microbiol News. 1985;51:4–12. [Google Scholar]
- 9.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors Nature 2005437376–380.Published erratum appears in Nature 2006;441:120. Ho, Chun He (corrected to Ho, Chun Heen) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2010;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet. 2012;13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hongoh Y, Ohkuma M, Kudo T. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae) FEMS Microbiol Ecol. 2003;44:231–242. doi: 10.1016/S0168-6496(03)00026-6. [DOI] [PubMed] [Google Scholar]
- 13.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight R, Gordon J. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lozupone C, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso J, Kuczynski J, Stombaugh J, Bittinger K, Bushman F, Costello EK, Fierer N, Peña A, Goodrich JK, Gordon J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huson H, Auch A, Qi J, Schuster SC. MEGAN Analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass E, Wilkening J, Wilke A, Antonopoulos D, Meyer F. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc. 2010;2010(1):pdb.prot5368. doi: 10.1101/pdb.prot5368. [DOI] [PubMed] [Google Scholar]
- 20.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominguez-Bello MG, Costella EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert JA. Constructing the microbial biomap for earth. [Internet] Argonne National Laboratory [updated 2013 Dec 12; accessed 2013 Dec 28]. Available from: http://www.earthmicrobiome.org/
- 25.Knight RD. World’s largest open-source science project to understand the microbial diversity of the human gut. [Internet] Biofrontiers Institute at the University of Colorado-Boulder [updated 2013 Oct 2, accessed 2013 Dec 28]. Available from http://humanfoodproject.com/americangut/
- 26.Tringe SG, Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol. 2008;11:442–446. doi: 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host–microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci USA. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want EJ, de Waziers I, Cloarec O, et al. Colonization-induced host–gut microbial metabolic interaction. MBio. 2011;2:e00271–10. doi: 10.1128/mBio.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez A, Knight R. Advancing analytical algorithms and pipelines for billions of microbial sequences. Curr Opin Biotechnol. 2012;23:64–71. doi: 10.1016/j.copbio.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez A, Clemente JC, Shade A, Metcalf JL, Song S, Prithiviraj B, Palmer BE, Knight R. Our microbial selves: what ecology can teach us. EMBO Rep. 2011;12:775–784. doi: 10.1038/embor.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]