The 56th Thomas L. Petty Aspen Lung Conference meeting held at the Gant Conference Center from June 5–8, 2013 brought together the community of microbiologists, basic respiratory scientists, and pulmonary clinicians to present and discuss the latest developments in the emerging area of the lung microbiome. The conference organizers, Richard Martin, Monica Kraft, and Sonia C. Flores, convened and led an outstanding meeting. As noted by many previous conference summarizers, it is almost impossible to capture all the content and details presented over the 3-day meeting. At best, one can attempt to capture some of the key themes, recognized gaps, and notable areas for future research, which we have attempted to do in this summary.
Unlike some past Aspen Lung Conferences, which focused on specific respiratory diseases, this meeting covered a topic that cuts across both lung health and most all respiratory conditions. In just the last 3 to 5 years, there has been remarkable progress in understanding the human and specifically the lung microbiome. This meeting highlighted early scientific progress in understanding the lung microbiota in healthy and diseased subjects, addressed current challenges and opportunities, and discussed trends and future directions for functional studies to unravel the mechanism of disease and a more defined role of microorganisms in health. The title of the conference is most appropriate, as not only is it the first time this topic has been covered at the Aspen Lung meeting but also it is indeed a new frontier for pulmonary medicine. Ironically, the area is not completely new to the pulmonary community, as the first awareness of lung microbes was in relation to tuberculosis, a pulmonary disease that scourged the 19th and 20th centuries (1, 2). However, part of the new frontier is that microbes were originally considered the “enemy,” and the approach was to eradicate a bug and cure the disease. We now recognize this paradigm has evolved from microbes being enemies to being partners, and a new challenge is to understand the delicate balance and symbiosis of those communities in defining the role of the microbiome(s) in health and disease.
The keynote lectures emphasized the contributions of high-throughput sequencing and microbiome-driven research to this continuously evolving area with a focus on the role of microbes as part of the human “supra organism.” The overarching messages from many of the presentations centered on the resident microbial communities of the lungs, the environment and its role in lung development, and their combined impact on health and disease states. It is now well established that a considerable number of microbial species still cannot be grown in most defined culture conditions. Therefore, a massive pool of biodiversity and novel genes remains to be discovered and their functions examined and linked to their roles in human development and health.
In the opening talk, Dr. James Beck presented a summary of the challenges ahead in this new path for lung research and some of the common definitions, methodologies, and tools that would be common language used in many presentations and posters. Dr. Beck highlighted seven areas that would need concerted efforts to advance the field (3). These challenges were also articulated during the National Heart, Lung, and Blood Institute Workshop on Lung Microbiome held in December 2011, at which current knowledge and the state of research on the lung and related areas of human microbiome investigation were reviewed. The resulting recommendations for advancing the field were recently published (4). These challenges and recommendations are summarized in Table 1.
Table 1.
Key challenges and recommendations from the NHLBI Microbiome Workshop
| Challenges | Recommendations |
|---|---|
| The need to examine large, well-characterized cohorts | Procedural details; sample types obtained; sample processing |
| Evaluate clinical and geographic diversity | Gut–lung axis and role in respiratory disease |
| Compare upper and lower respiratory samples | Functional properties of the lung microbiome (omics approaches) |
| Correlate longitudinal samples with clinical data | Characterization of fungal and viral microbiota; interkingdom microbiome and host interactions |
| Consider influence of other microbiota on lung, including the gut | Use/development of relevant animal or other models to understand mechanisms of human microbiome observations |
| Compare divergent methodology and the use of variable 16S regions | Noninvasive biomarkers of lung microbiota and microbial burden; noninvasive imaging of lung microbiota |
| Achieve consistent presentation of data and statistical analysis | Integration of environmental, clinical, and -omics data |
It has been almost 2 decades since the first microbial genome was sequenced in 1995 (5), and since then, a total of 4,409 genome projects (source: Genomes Online Database, http://genomesonline.org/) have been completed or are in progress; of these, 4,034 are finished bacterial genomes (∼63%) or are in permanent draft stage (e.g., thoroughly sequenced and essentially complete, but gaps remain). More importantly, the science has moved from sequencing a single isolate to sequencing the entire communities through metagenomic approaches, opening the door to discovery of a tremendously complex and dynamic microbial world and uncovering the delicate balance of the human–microbe ecosystem that governs our physiology.
Increasingly cheaper high-throughput sequencing technologies have facilitated the identification of uncultivable microbes in the human body, raising awareness of the incredible diversity of the human microbiota. Historically considered sterile, the lungs were initially excluded from the four body sites (gastrointestinal tract, mouth, vagina, and skin) in the original goals of the Human Microbiome Project (HMP, http://commonfund.nih.gov/hmp/). However, a growing body of research now shows that the lower respiratory tract may contain a diverse community of microbes. The biological and clinical significance of these findings remains understudied. The Aspen meeting encompassed a wide spectrum of lung diseases unified under the umbrella of the new microbiome paradigm. In the following paragraphs, we discuss the several themes that emerged from the collection of presentations and discussions.
The Microbiome, the Environment, and Lung Health
The importance of the environment to maintain a healthy state and in the development of lung disease was perhaps the main topic omnipresent throughout the conference. We are beginning to understand that we are not alone and that our concept of “self” goes beyond our own genetic makeup to also embrace our microbial partners. Past research highlighted the symbiotic nature of the relationship between “us” and our microbiome and the sometimes serious consequences of disrupting this homeostasis.
Dr. Fernando Martinez reminded us that the concept of the “human microbiome” has been with us for a long time but may be far from being something good. Metchnikoff wrote in his 1903 publication, The Nature of Man, that our bacterial flora “contribute[s] nothing to the well-being of man” (6). We now know that these bacteria, living not only on the surface of our skin, are inside us and part of our genetic makeup. Indeed, finding the link between microbial metabolism and disease (or its prevention) is an important future direction for the field. For example, the recent work by Tang and colleagues on the role of the microbiota on trimethylamine N oxide (TMAO) production and coronary artery disease illustrates the link between the host genome, diet, and the gut microbiota (7). However, Dr. Martinez challenged the group with the concept of “human microbiome domestication.” Cleaner living conditions, rich modern diets, and antibiotics have undoubtedly saved billions of lives, but with unsuspected side effects: a generalized commensal microbial imbalance, very different from that of our ancestors. How can we recover our original balance from our hunter-gatherer ancestors? Dr. Martinez suggests that we are a composite of our own genetic background and that of our microbiome, placing our health needs under a large polygenic control, rendering the idea of “controlling” our microbiome quite challenging.
Dr. von Mutius presented the effect of the environment on asthma outcomes in farm-raised versus non–farm-raised children, showing how children exposed to environmental factors (endotoxin, muramic acid, and ergosterol markers) are far less likely to suffer from asthma and allergies. She highlighted the concept that allergies may develop early in life and may even occur before birth. The microbial farm environment appears to have a profound impact on the airway microbiome by educating the immune system by stimulating innate immunity through increased expression of TLRs and CD4 as well as increased IFN-γ production at birth in newborns of exposed mothers. Dr. Huang’s abstract on the microbiome in severe asthma provided evidence that specific bacterial communities are associated with clinical and inflammatory features of severe conditions. Although the microbial diversity in the exposure cocktail matters (8), recent work from Calışkan and colleagues indicates that not all can be attributed to the sole exposure and diversity of the microbial environment, as the interaction between wheezing illness and a gene (or genes) in a region of chromosome 17 has been shown to determine childhood asthma risk (9).
Dr. LeVan’s abstract pointed out that not all farming environments are the same. In contrast to the Swiss or Amish farming environments, highly industrialized swine farms in the United States reveal a different problem and represent a major respiratory threat (rather than a protective environment) to the exposed individuals. Metagenomic analyses of these dusts (which present proinflammatory properties and high levels of endotoxin and muramic acid) indicate a higher prevalence of bacterial species from the Clostridiales order (a potential pathogen) when compared with grain elevators and house dust. A future direction is to determine if agricultural dusts modify the lung microbiome in healthy subjects and subjects with chronic obstructive pulmonary disease (COPD). A better understanding of dust composition may lead to prevention strategies.
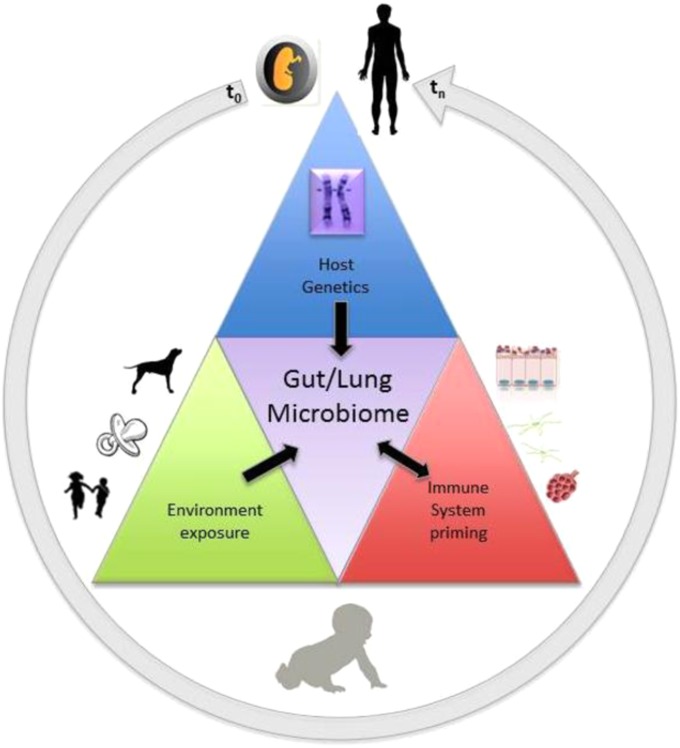
Dr. Homer Boushey brought together many common elements about the microbiome and asthma, emphasizing the importance of the environment in promoting beneficial and harmful effects during the early stages of development. He discussed the importance of bacteria colonizing the gut during early life. He revisited the hygiene hypothesis and reminded the group that diversity of the microbiome per se may not be as important as having the “right” microbes, ideally provided through vaginal delivery, breastfeeding, pet ownership, and siblings. Dr. Boushey introduced the role of the microbial flora in antiviral activity (10), emphasizing the concept of “gut preparedness” to combat viral infections. He showed that when infants were provided probiotics (Lactobacillus and Bifidobacterium) before virus season they were 72% more “resistant” to the virus symptoms. Figure 1 (11) summarizes the key concepts of the interplay between host genetics, environment, the microbiome, and the immune system, where time is a key element because environmental exposure is important very early as well as later in life to determine health status.
Figure 1.
Selective pressures shaping the microbiome. Graphic representation of the interplay among environment, the microbiota of humans, the host genetics, and the immune system. Exposure to animals, foods, and siblings are the vehicle environmental bacteria use to access the human gut/lung axis. Disturbance in the balanced state by lack of exposure (extreme hygiene), inappropriate exposure (smoke), or viral infections may lead to disease. Adapted by permission from Reference 13.
Probiotics were discussed in depth by Dr. Forsyth. He summarized the potential therapeutic effect of microbes in allergic airway inflammation and respiratory infection and described the current understanding of the underlying mechanisms of immunomodulation by probiotics. Like other speakers, his talk emphasized the role of the gut microbiota on T regulatory responses in the airway and argued that functional changes in dendritic cells after interaction with the microbiota are critical in coordinating these responses. In addition, he presented evidence that probiotic usage can modulate immune responses in the lung. The concept of the effect of microbiota immunity and homeostasis in a systemic fashion and taxa-specific effects was also presented by Dr. Neish, who proposed that the microbiome may be executing a beneficial “cross-talk” with the host. Although animal models show promising results in the use of certain lactobacillus species as probiotics, the outcome of clinical trials has been variable and consequently in need of more exploration, particularly bidirectional communication along the gut–lung axis.
How does the microbiome protect us? Dr. Susan Lynch proposed that protection can occur in two ways: (1) through competitive colonization (e.g., biofilm formation or antimicrobial production), and (2) through immune modulation via local down-regulation of inflammatory responses. When the microbiome is disturbed, for example by a respiratory viral infection, this perturbation of the mucosal microbiome can lead to colonization by organisms with traits that facilitate translocation, rapid response to environmental changes, competitive colonization, and metabolic plasticity, often associated with pathogenicity. Our microbiome represents, in this sense, our personal probiotic load, which when present in optimal amounts and quality confers a health benefit.
The Microbiome and Lung Disease
A number of presentations reviewed the role of the pulmonary microbial communities in diverse lung diseases such as rheumatoid arthritis (RA), COPD, and cystic fibrosis (CF). Dr. Demourelle presented evidence suggesting that microbes and lung mucosal inflammation are involved in the development of RA. She identified differences in the lung microbiota of arthritis-free but at-risk patients (positive for anti–cyclic citrullinated peptide antibody [anti-CCP]), raising the possibility of a mechanistic link between the lung microbiome and RA autoimmunity.
Several presented abstracts addressed the role of the microbiome in COPD, often in combination with the effect of smoke from cigarettes and other sources. Dr. Bezemer reminded the group that cigarette smoke acts as a carrier of microbes, and these may be responsible for inflammatory responses and disease progression. In particular, Dr. Rutebembewa identified the presence of specific genera, Novosphingobium spp, in samples of smoking patients with COPD with advanced disease. Mice challenged with this organism developed an inflammatory response in the lung, supporting the idea that the bacteria present in the lung of smokers interacts with the environmental stress triggered by smoke, leading to severe lung injury.
In CF, culture-independent approaches (from cross-sectional studies) mostly agree on the existence of a general microbiome composition in the disease. Diversity remains stable in clinically stable patients and decreases with disease progression, but more longitudinal studies are needed. Dr. Surette stressed the need for better understanding of the microbiota to inform treatment, specifically, what antibiotics are doing to these communities, because “normal” microbiota may include organisms that should be considered pathogens in the lower airways, acting as synergens. He argued that typical microbiome profiling does not have the resolution required to guide clinical management of airway disease (not only CF) and proposes that accurate quantitative microbiology is needed to guide therapy (whether molecular, culture, or a combination of methods).
Other Bug-omes
As a field of research still in its early stages, lung microbiome research has focused on the identification of bacterial phyla, whereas eukaryotes and viruses have been largely neglected, mainly due to technical difficulties, such as their significantly lower biomass and the lag in the development of databases such as the Ribosomal Database Project. However, the importance of the “mycobiome” in human health is significant, particularly in immune-compromised individuals. Dr. Elodie Ghedin’s group presented pilot work on the oral fungal microbiome, showing that HIV-positive patients as well as patients with COPD with complications appear to have several fungal genera specifically associated with the progression of disease. In addition to the mycobiome, Dr. Sonia C. Flores’s group presented a poster titled “There is More to Microbiome than Bacteria,” addressing the bacteria-centric view of the microbiome. They hypothesize that the complement of viruses or “virome” of patients with HIV with pulmonary hypertension may be different than that of the uninfected population. Using RNA from plasma they performed RNA-Seq and have been able to capture potential viral sequences. Although preliminary, their effort complements the broader effort to address the presence of important “bugs” other than bacteria.
Technical Aspects of Microbiome Research
Defining standard practices for sample collection to support the analyses of the lung microbiome is still a challenging area with technical issues. Studies of the lung microbiome are facing major hurdles that are unique to the lungs and not encountered in any of the four other body sites that have initially been the focus of the metagenomic studies funded under the HMP. Among these, the low microbial biomass, the difficult access and invasiveness of sampling procedures, distinguishing what is endogenous and what might be sample contamination, and how to determine whether bacteria are dead or alive remain unresolved and require additional research. Dr. Twigg presented results using acellular bronchoalveolar lavage (BAL) as a better indicator of the lower respiratory tract BAL than whole BAL. He argued that acellular BAL may be better suited to detect differences in the lung microbiome between populations as well as provide a better indicator of the lower respiratory tract immunologic/inflammatory milieu.
However, sampling is just one issue. Assuming optimal sampling techniques are achieved (or agreed on using standard operating procedures), data analysis presents a sizable challenge, because data manipulation, integration, and interpretation are not easy tasks. Dr. Rob Knight discussed old and new technologies and techniques used to analyze data, emphasizing the methods used to make substantial differences between samples, including sequencing platforms, choice of 16S regions, and metrics to reconstruct evolutionary history.
Future Directions: Can a Better Understanding of Lung Microbiome Help Us Predict and Prevent Lung Disease?
It is now clear that the environment plays a fundamental role in lung development and the onset of immunity and health of the respiratory tract. However, there are still many unknowns: How do we define health in the context of the microbiome? Which are the microbiome markers associated with health? Can microbiome knowledge be exploited to promote health and to potentially diagnose, prevent, and treat lung disease? We are beginning to understand the role of our colonizing microbes, and the following statement, taken from the HMP, holds true: “Few examples yet exist where the overall composition, relative abundances, or microbial proportions of a microbiome are conclusively causal in human disease” (12). Composition does not say much about the metabolic processes; therefore, it becomes evident that the metabolic readout of the microbial system is what is important. How to achieve this knowledge is not a simple task: the microbiome is a dynamic, complex system that is highly individual and easily perturbed, and the paths that will lead functionality and the mechanisms behind the onset of disease are intricate. Future research will require an integrated/systems approach to study the many complex levels. We need to consider host-omics, bug-omics, pathophysiology, and clinical phenotypes to define models of lung disease. Given the complexity of the interactions, it is highly unlikely that one model will fit all. To achieve this multidimensional integration, it will be important to consider creative ways to implement existing tools, leverage existing animal models, and rethink the way we conduct future clinical studies, in particular how to leverage existing cohorts and foster collaboration across different fields. A multidisciplinary approach is necessary to embrace knowledge from the fields that intersect classic microbiome research with approaches from basic lung scientists, physician scientists, microbiologists, genomics experts focused on mucosal sites other than the lung, informaticians, and patients (4).
Conclusions
The use of state-of-the art culture-independent techniques has enabled tremendous growth in understanding the microbial communities that reside in the lungs and in the respiratory tract and the relationship between them. However, challenges with this growth and technology include the fact that we can now identify what may be ancestral bacterial sequences with no live organism to fulfill Koch’s postulates. Nonetheless, this meeting provided an overview of the tremendous progress made in the past 5 years by basic scientists, clinicians, and microbiologists moving away from the “sterile lung” paradigm and toward delineating the nature of the lung microbiome both in health and disease states and an appreciation of the impact of the environment on the establishment of the human microbiome. This understanding can only be expected to increase in this nascent but burgeoning area of lung research as technology continues to improve and become more accessible.
Footnotes
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Daniel TM. The impact of tuberculosis on civilization. Infect Dis Clin North Am. 2004;18:157–165. doi: 10.1016/S0891-5520(03)00096-5. [DOI] [PubMed] [Google Scholar]
- 2.Daniel TM. The origins and precolonial epidemiology of tuberculosis in the Americas: can we figure them out? Int J Tuberc Lung Dis. 2000;4:395–400. [PubMed] [Google Scholar]
- 3.Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Transl Res. 2012;160:258–266. doi: 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. The role of the lung microbiome in health and disease. A National Heart, Lung, and Blood Institute workshop report. Am J Respir Crit Care Med. 2013;187:1382–1387. doi: 10.1164/rccm.201303-0488WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 6.Metchnikoff E. The nature of man: studies in optimistic philosophy. In: Mitchell PC, editor. New York and London: G. P. Putnam’s Sons; 1903. [Google Scholar]
- 7.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrländer C, Heederik D, Piarroux R, von Mutius E GABRIELA Transregio 22 Study Group. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 9.Calışkan M, Bochkov YA, Kreiner-Møller E, Bønnelykke K, Stein MM, Du G, Bisgaard H, Jackson DJ, Gern JE, Lemanske RF, Jr, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leyer GJ, Li S, Mubasher ME, Reifer C, Ouwehand AC. Probiotic effects on cold and influenza-like symptom incidence and duration in children. Pediatrics. 2009;124:e172–e179. doi: 10.1542/peds.2008-2666. [DOI] [PubMed] [Google Scholar]
- 11.de Wouters T, Doré J, Lepage P. Does our food (environment) change our gut microbiome (‘in-vironment’): a potential role for inflammatory bowel disease? Dig Dis. 2012;30:33–39. doi: 10.1159/000342595. [DOI] [PubMed] [Google Scholar]
- 12.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martins dos Santos V, Muller M, de Vos WM. Systems biology of the gut: the interplay of food microbiota, and host at the mucosal interface. Curr Opin Biotechnol. 2010;21:539–555. doi: 10.1016/j.copbio.2010.08.003. [DOI] [PubMed] [Google Scholar]