Abstract
Purpose of review
Hematopoietic stem/progenitor cell fate decision during hematopoiesis is regulated by intracellular and extracellular signals such as transcription factors, growth factors, and cell-to-cell interactions. In this review we explore the function of DEK, a nuclear phosphoprotein, on gene regulation. We also examine how DEK is secreted and internalized by cells, and discuss how both endogenous and extracellular DEK regulates hematopoiesis. Finally, we explore what currently is known about the regulation of DEK during inflammation.
Recent findings
DEK negatively regulates the proliferation of early myeloid progenitor cells but has a positive effect on the differentiation of mature myeloid cells. Inflammation regulates intracellular DEK concentrations with inflammatory stimuli enhancing DEK expression. Inflammation-induced NF-κB activation is regulated by DEK, resulting in changes in the production of other inflammatory molecules such as IL-8. Inflammatory stimuli (i.e. DEK) in turn regulates DEK secretion by cells of hematopoietic origin. However, how inflammation-induced expression and secretion of DEK regulates hematopoiesis remains unknown.
Summary
Understanding how DEK regulates hematopoiesis under both homeostatic and inflammatory conditions may lead to a better understanding of the biology of HSCs and HPCs. Furthering our knowledge of the regulation of hematopoiesis will ultimately lead to new therapeutics that may increase the efficacy of hematopoietic stem cell transplantation.
Keywords: Hematopoiesis, DEK, Inflammation
Introduction
All cellular blood forming elements mature from the hematopoietic stem (HSC) and progenitor (HPC) cell populations. HSC and HPCs from the bone marrow, mobilized to the peripheral blood, or found in umbilical cord blood have been used in hematopoietic stem cell transplantation (HSCT) for the treatment of malignant and non-malignant diseases. Understanding the elements that regulate how these cell populations are maintained, differentiate, and mature ultimately will lead to improved methods of HSCT allowing for a more efficient and effective means to treat disease. One possible factor that influences HSC and HPC cell fate is the nuclear phosphoprotein DEK. DEK was originally discovered as part of the fusion protein DEK-NUP214 (i.e. DEK-CAN), a result of a t(6;9) chromosome translocation in a subset of patients with acute myelogenous leukemia [1–4*], and as an oncoprotein in various neoplasms including melanoma [5–7*], breast cancer [8], glioblastoma [9], hepatocellular carcinoma [10, 11*], retinoblastoma [12, 13], gastric adenocarcinoma [14*], non-small cell lung cancer [15*], pancreatic ductal adenocarcinoma [16*], and bladder cancer [17]. DEK was found to be a regulator of hematopoiesis controlling HSC and HPC numbers/fate decision [18, 19]. These findings were further supported by the fact that DEK plays an important role in mediating the activation of the granulocyte-colony stimulating factor receptor 3 (GCSFR3) promoter which regulates myeloid cell differentiation [20]. Even though many have explored the role of DEK under malignant conditions, little is known how DEK functions to regulate HSCs, HPCs, and hematopoiesis. In this review we will explore possible mechanisms how both intracellular and extracellular DEK may regulate hematopoiesis under homeostatic and stress/inflammatory conditions.
Intracellular and Extracellular DEK Functions
Intracellular/Endogenous DEK Function
The DEK gene encodes a conserved and structurally unique DNA-binding nuclear protein expressed in higher eukaryote cells and is the only known member of its protein class [21]. Although DEK bears little resemblance to other proteins, DEK does contain a conserved region called the Saf/Actinus/PARP-box (SAP-box) that has a helix-turn-helix motif that binds to DNA [22] further facilitated by a second DNA binding structure found in the C-terminal end of the protein [22, 23**]. There is some debate on how DEK recognizes where to bind to DNA. Either DEK functions by binding to specific DNA structures or in a sequence-dependent manner [23**] since DEK both accumulates onto specific chromatin structures (such as four-way DNA junctions [24]) and to sequence-specific areas (such as TG-rich peri-ets sites of HIV-2 enhancer regions [25]). However, there remains some controversy on whether DEK recognizes specific DNA sequences as DEK has been shown to non-discriminately bind to DNA of various sequences in the absence of other proteins [26, 27].
DEK is involved in various nuclear processes including transcription, DNA replication and DNA repair implicating a role for DEK in gene regulation. However, whether DEK promotes or represses gene expression remains controversial. DEK is associated with both activating (e.g. H3K4me2/3) and repressive (e.g. H3K9me3) histone modifications [28, 29]. Immunofluorescent imaging studies showed that DEK consistently localized to euchromatin [28, 30, 31]; however, knocking-down DEK in various cell types drastically reduces the distribution of heterochromatin [29, 32]. The later findings were further supported by the fact that DEK associates with heterochromatin binding protein (HP)1α allowing for binding to H3K9me3 and the chromatin remodeling complex B-WICH, both involved in the replication and stabilization of heterochromatin [29, 33]. Information on the role of DEK in gene regulation is reviewed in detail in Sanden and Gulberg, 2015 [23**].
DEK undergoes several different post-transcriptional modifications that influence its activity. Phosphorylation of DEK is mediated by casein kinase 2 (CK2) in a cell cycle-dependent manner peaking in cells undergoing the G1 phase of the cell cycle [34]. Phosphorylation by CK2 reduces binding of DEK to histones and decreases the affinity of DEK binding to DNA [30, 34]. However, when DEK is unphosphorylated, it dimerizes and remains bound to chromatin [30]. Poly(ADP-ribosyl)ation of DEK by poly(ADP-ribose) polymerase (PARP)1 also leads to its removal from chromatin and is associated with its accumulation during apoptosis [35, 36]. Phosphorylation and poly(ADP-ribosyl)ation of DEK are two possible mechanisms regulating whether DEK activates or suppresses gene expression. Acetylation of DEK can also reduce the ability of DEK to bind to DNA. This disruption is associated with the relocalization of DEK to RNA processing machinery where DEK plays a role in intron removal [37–40]; however, due to issues with the specificity of the DEK antibodies used in these studies the role of DEK in RNA processing remains controversial [23**].
Extracellular DEK
Both phosphorylation by CK2 and poly(ADP-ribosyl)ation by PARP1 of DEK removes it from chromatin [30, 34, 36, 41]. These post-transcriptional modifications led to one of the most surprising observations about DEK, its ability to be secreted by cells [32, 41]. DEK is passively secreted by cells undergoing apoptosis (a process associated with poly(ADP-ribosyl)ation) and actively secreted by macrophages upon stimulation with interleukin (IL)-8 in a Golgi-apparatus-independent manner either in a free form or within exosomes [41]. This process can be blocked by the immunosuppressants dexamethasone and cyclosporine A suggesting that DEK plays a role in inflammation. Secreted DEK both in its free-form (by acting as a chemoattractant for neutrophils, cytotoxic T lymphocytes, and natural killer cells) and in exosomes is associated with autoimmunity. Circulating auto-antibodies against DEK have been found in juvenile idiopathic arthritis (JIA), sarcoidosis, and systemic lupus erythematosus [41, 42]. In JIA, both DEK and DEK auto-antibodies have been found in synovial fluids [41]. It is hypothesized that the secreted DEK in exosomes fuses with the membrane of antigen presenting cells where DEK is processed and presented to B cells allowing for DEK auto-antibody production. These findings suggest an important role for DEK in autoimmune disease pathogenesis.
The fact that DEK can act as a chemoattractant for immune cells suggests that DEK can be recognized by a yet unidentified extracellular receptor; however, DEK can also penetrate cells [32, 41]. DEK is a bulky, charged, and hydrophilic protein that is unable to cross the plasma membrane by diffusion. DEK can bind to the negatively charged, translocatory protein heparan sulfate-type proteoglycan (HSPG) allowing DEK to be endocytosed via invaginations in the plasma membrane [32]. Once internalized DEK can relocate to the nucleus and perform its normal intracellular/exogenous activity.
DEK and Hematopoiesis
Endogenous DEK and Hematopoiesis
DEK expression in human and mouse hematopoietic cells greatly changed with the level of maturity of the cell population. Upon examination of DEK mRNA levels in HSCs/HPCs and mature myeloid lineage cells, it was discovered that DEK expression was elevated in HSCs and HPCs when compared to mature granulocytic and monocytic populations in both human and mice [1, 43]. However, peak DEK expression levels varied between mice and man [1]. The greatest DEK expression in humans occurred in HSCs with a steady decline in expression levels as the cells differentiated into HPCs then matured into polymononuclear cells (PMNs) and monocytes. DEK expression levels were higher in monocytes then PMNs. In mice, DEK expression was greater in HPCs followed by HSCs, PMNs, and monocytes. Altered DEK expression levels during hematopoiesis indicate that DEK has specific functions in different cell types which may vary between humans and mice; however, this hypothesis requires further exploration.
To determine whether DEK regulates hematopoiesis, C57Bl/6 DEK−/− bone marrow was utilized to examine HSC/HPC numbers and function (see Figure 1A; [18, 19]). Increased numbers of colony forming units (CFU)-granulocyte monocyte (GM; granulocyte-monocyte progenitors), blast-forming unit-erythroid (BFU-E; erythroid progenitors), and CFU-granulocyte, erythrocyte, monocyte, megakaryocyte (CFU-GEMM; multipotent progenitors for myeloid cells) was seen using DEK−/− bone marrow when compared to wildtype bone marrow cells. Increases in DEK−/− bone marrow colony formation were associated with increased HPC cycling suggesting that DEK decreases HPC numbers by regulating proliferation. These findings were also seen in DBA DEK−/− bone marrow suggesting that the effect of DEK was not limited to the C57Bl/6 mouse strain [19]. C57Bl/6 DEK−/− bone marrow also demonstrated decreased long but not short-term repopulating capacity in a competitive bone marrow transplantation model when compared to transplantation using wildtype bone marrow [18]. This difference was further exasperated in secondary, non-competitive bone marrow transplantation suggesting that DEK is important in the self-renewal and long-term repopulating capacity of HSCs.
Figure 1.
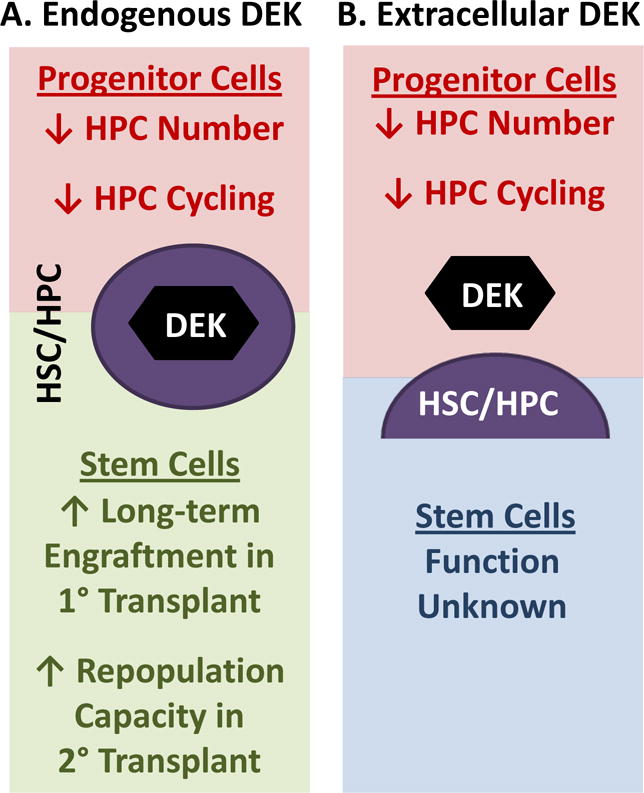
Effects of endogenous/intracellular and extracellular DEK on hematopoiesis. (A) Endogenous/intracellular DEK decreases hematopoietic progenitor cell (HPC) numbers and inhibits HPC cycling while increasing the long-term competitive repopulation capacity in lethally irradiated congenic mice and increasing the repopulation of lethally-irradiated secondary mouse recipients in a non-competitive transplantation model indicating that endogenous DEK enhances hematopoietic stem cell (HSC) repopulating and/or self-renewal capability. (B) Extracellular DEK, as determine through the co-culture of either mouse bone marrow cells and/or low-density human cord blood cells with recombinant DEK, decreases HPC numbers and inhibits HPC cycling. The effect of extracellular DEK on HSC function is currently unknown.
Further supporting the idea that DEK regulates hematopoiesis, especially during differentiation of myeloid cells, is the fact that DEK can form a complex with the transcription factor CCAAT enhancer binding protein (C/EBP)α [20]. Fetal liver cells from C/EBPα−/− mice demonstrate decreased granulocyte-monocyte progenitor (GMP) cell numbers with the inability of these cells to terminally differentiate into mature myeloid cells [44]. The decrease in GMPs seen in C/EBPα−/− mice is associated with an increase in the self-renewal capacity of HSCs and is partially restored with the reintroduction of C/EBPα [20, 45–48]. DEK is recruited specifically to chromatin with C/EBPα enhancing granulocyte colony-stimulating factor receptor 3 (GCSFR3) promotor activation [20]. Phosphorylation of serine 21 on C/EBPα inhibits DEK from forming a complex with C/EBPα ultimately disrupting C/EBPα driven, G-CSF-mediated granulocyte differentiation.
In addition to C/EBPα, DEK interacts with essential upstream enhancer elements of the erythroid Krüppel-like factor (EKLF) promoter enhancing EKLF expression [49]. EKLF is a zinc finger transcription factor that plays a role in the global regulation of erythroid gene expression by binding to DNA and recruiting chromatin-remodeling proteins and histone modifiers such as DEK [49–52]. Although murine embryonic stem cells deficient in EKLF can produce erythroid colonies in vitro, colonies were poorly hemoglobinized and enucleated erythrocytes in these colonies contained numerous Heinz bodies, inclusions within red blood cells composed of denatured hemoglobin [53]. EKLF−/− erythrocytes in mice appeared to be short-lived [49, 53]. Therefore, if DEK is essential in regulating EKLF expression, then DEK may also regulate mature, healthy erythrocyte numbers. These findings correlate well with the observations made using the DEK−/− mouse model that DEK regulates the function of myeloid progenitor cells.
Extracellular DEK and Hematopoiesis
To examine the role of extracellular DEK in regulating hematopoiesis, recombinant human (rh) DEK was utilized in experiments in vitro [18, 19]. Bone marrow and low-density human cord blood cells when in culture with rhDEK demonstrated inhibited formation of CFU-GM, BFU-E and CFU-GEMM in a dose-response manner and decreased the number of cycling HPCs (see Figure 1B). The negative regulatory role of rhDEK on colony numbers and cycling status in both mouse and humans in vitro further supports the findings from the DEK−/− mouse model that DEK is required for the maintenance of HPC numbers whether intracellular or extracellular; however, the role of extracellular DEK in regulating HSCs remains unknown.
How extracellular/recombinant DEK regulates HPC numbers and function in mouse bone marrow cells or low-density human cord blood cells is not yet well understood. DEK may function through several different pathways. First, extracellular DEK may become internalized, as it is known to do in other cell types, by binding to HSPGs, translocating to the nucleus, and then binding to chromatin thus having regulatory effects on cell proliferation and transcription. Second, extracellular DEK may bind to a yet unknown receptor and mediate a signal transduction pathway. Third, DEK internalization may be receptor-mediated in hematopoietic stem/progenitor cells. Finally, DEK may be working indirectly through another cell in the bone marrow or cord blood low-density population altering the cytokine milieu. This last possibility seems unlikely, however, as the suppressive effects of rhDEK on colony formation were apparent even when assayed using single human CD34+ cord blood cells [18]. Thus far, DEK is only known to be actively secreted under inflammatory/stress-mediated conditions [32, 36, 41]; therefore, any effect that DEK has on the inflammatory or stress response may also regulate hematopoiesis.
DEK and Inflammation
DEK is associated with inflammation as demonstrated by its function in autoimmune diseases (as discussed above), persistent viral infections (e.g. human immunodeficiency virus and Kaposi’s sarcoma-associated herpesvirus [25, 54–57]), and tumorigenesis (for a comprehensive review on these topics please read Pease et al., 2015 [58**]). DEK secretion is induced in response to inflammatory stimuli and can be inhibited by the immunosuppressive agents [41]. DEK expression levels are also increased following inflammatory stimuli. In vivo models where rodents ate diets containing organic pollutants demonstrated a significant increase in molecules persistent with chronic low-grade inflammation and in DEK expression [59, 60]. DEK expression levels were also increased in human bronchial epithelial cells exposed to fine particles found in industrial workplaces in an in vitro model of airway inflammation [61]. Although the mechanism is not clear why DEK expression increased following inflammation or whether using immunosuppressants can block this phenomenon, upon examination of the DEK promotor region several inflammation-associated transcription factor binding sites were found. These transcription factors include C/EBPβ, AP-1, Ets-1, NF-κB, STAT4, and NF-AT [58**]. Although not much is known about the relationship of most of these transcription factors and DEK, the interaction between DEK and NF-κB has been explored.
The NF-κB signaling pathway is a crucial pathway involved in inflammation and the immune response. Loss of DEK was associated with an increase in NF-κB activity [62–64]. Tumor necrosis factor (TNF)α-treated DEK−/− mouse embryonic fibroblast cells (MEFs) had increased NF-κB activity as indicated in a luciferase expression assay and had increased transcription of NF-κB target genes and increased NF-κB p65 (a subunit of NF-κB) translocation to the nucleus, an indicator of NF-κB activation [64]. Further studies in HeLa cells where DEK was silenced using shRNA demonstrated increased IκBα, an important NF-κB inhibitor, phosphorylation-induced degradation in association with increased NF-κB activity [62, 63]. These findings suggest that DEK is a negative regulator of NF-κB activation. However, the effect of DEK on NF-κB signaling is more complicated. In TNFα-treated, non-manipulated HeLa cells, DEK co-localized with NF-κB p65 to the promotor regions of the NF-κB target genes, IL-8 and c-IAP2, increasing their mRNA expression levels [64]. Although not examined, this could potentially be a critical pathway in regulating DEK secretion from cells since, as previously mentioned, IL-8 induced DEK secretion by macrophages thus possible forming a feedback loop [41]. Under physiological conditions (normal to slightly elevated levels of DEK) DEK is associated with NF-κB activation via the degradation of the NF-κB inhibitor, IκBα [58**, 62, 63]. Further complicating matters, overexpression of DEK in HeLa cells inhibits NF-κB activity in a dose-dependent manner [62, 64]. These data put together suggests that the concentration of DEK within the cell plays a large part on whether or not DEK inhibits or activates NF-κB activity.
Conclusion
DEK regulates hematopoiesis by increasing the repopulating and self-renewal capacity of HSCs while decreasing HPC cycling and numbers; however, the molecular mechanisms of how DEK differentially regulates HSCs and HPCs remains unknown. Understanding how DEK regulates hematopoiesis under homeostatic conditions may lead to new methods of enhancing HSC numbers allowing for more efficient HSCT. This would also help us to understand how DEK may function under inflammatory and/or stress conditions. DEK, released extracellularly by immune cells following inflammatory stimuli and cell death, may act as a danger signal influencing inflammation/disease pathogenesis and ‘emergency’ hematopoiesis (see figure 2). Recently it has been shown that DEK-targeting DNA aptamers can reduce joint inflammation in an animal model of arthritis [65**]. Modulating DEK expression, secretion and/or function during hematopoiesis may be a potential target to modulate chronic inflammatory conditions thus augmenting disease pathogenesis.
Figure 2.
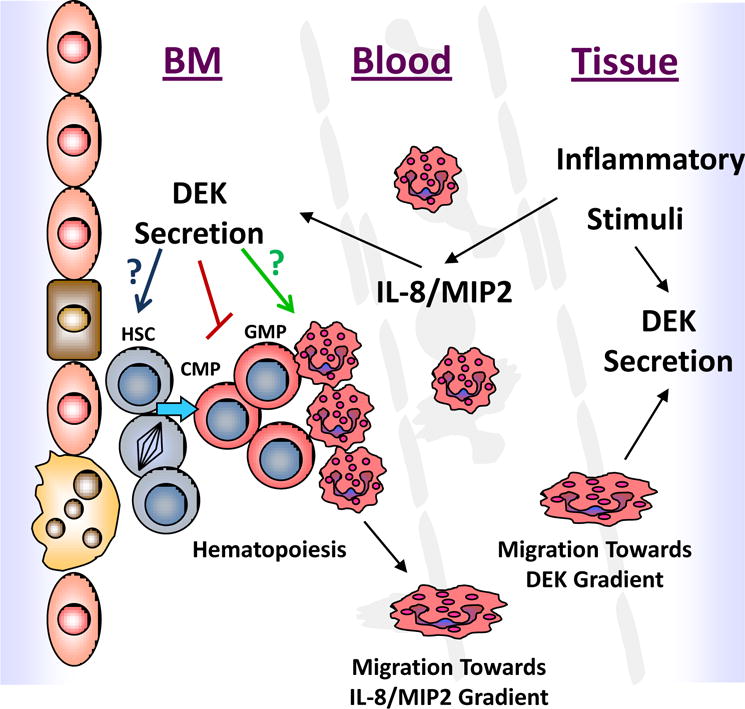
The possible role DEK may play during inflammation and how changes in DEK expression/secretion regulate hematopoiesis. Inflammatory stimuli, such as IL-8, can lead to the secretion of DEK. Extracellular DEK is known to inhibit early myeloid progenitor cells; however, whether it can increase G-CSF-mediated granulocytic differentiation or alter hematopoietic stem cell function like endogenous DEK remains unknown. Hematopoietic cells (e.g. granulocytes) also migrate towards secreted DEK. The migration of mature hematopoietic cells into secondary tissues/blood will ultimately also influence hematopoiesis.
Key Points.
DEK, a protein abundantly found in the nucleus, plays an important role in both enhancing and repressing gene expression by altering chromatin structure.
Endogenous DEK can regulate hematopoiesis by increasing the self-renewal and repopulating capacity of hematopoietic stem cells and by decreasing the cycling and number of hematopoietic progenitor cells.
Inflammatory stimuli enhance DEK expression levels and its ability to be secreted extracellularly.
Extracellular DEK functions as a chemoattractant for immune cells but also regulates hematopoietic progenitor cell cycling and numbers.
Acknowledgments
The authors wish to extend their apologies to our colleagues whose work we were unable to cite due to space limitations. The authors are supported by National Institutes of Health under awards R01 HL056416, R01 HL67384, R01 HL112669, R01 DK109188, P01 DK090948, and T32 DK07519.
Disclosure of Funding: Research done by the Broxmeyer laboratory reported in this review article was supported by the National Institutes of Health under awards # R01 HL056416, R01 HL67384, R01 HL112669, R01 DK109188, P01 DK090948, and T32 DK07519.
References
- 1.Logan GE, Mor-Vaknin N, Braunschweig T, et al. DEK oncogene expression during normal hematopoiesis and in Acute Myeloid Leukemia (AML) Blood Cells Mol Dis. 2015;54(1):123–31. doi: 10.1016/j.bcmd.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 2.von Lindern M, Breems D, van Baal S, et al. Characterization of the translocation breakpoint sequences of two DEK-CAN fusion genes present in t(6;9) acute myeloid leukemia and a SET-CAN fusion gene found in a case of acute undifferentiated leukemia. Genes Chromosomes Cancer. 1992;5(3):227–34. doi: 10.1002/gcc.2870050309. [DOI] [PubMed] [Google Scholar]
- 3.von Lindern M, Poustka A, Lerach H, Grosveld G. The (6;9) chromosome translocation, associated with a specific subtype of acute nonlymphocytic leukemia, leads to aberrant transcription of a target gene on 9q34. Mol Cell Biol. 1990;10(8):4016–26. doi: 10.1128/mcb.10.8.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *4.Mohamed AM, Balsat M, Thenoz M, et al. Oncogene- and drug resistance-associated alternative exon usage in acute myeloid leukemia (AML) Oncotarget. 2016;7(3):2889–909. doi: 10.18632/oncotarget.3898. This paper shows a possible new role for DEK in mediating drug-resistance in AML. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kappes F, Khodadoust MS, Yu L, et al. DEK expression in melanocytic lesions. Hum Pathol. 2011;42(7):932–8. doi: 10.1016/j.humpath.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khodadoust MS, Verhaegen M, Kappes F, et al. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69(16):6405–13. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *7.Riveiro-Falkenbach E, Ruano Y, Garcia-Martin RM, et al. DEK oncogene is overexpressed during melanoma progression. Pigment Cell Melanoma Res. 2016 doi: 10.1111/pcmr.12563. This paper demonstrates that DEK may be a potential biomarker for melanoma progression. [DOI] [PubMed] [Google Scholar]
- 8.Privette Vinnedge LM, McClaine R, Wagh PK, et al. The human DEK oncogene stimulates beta-catenin signaling, invasion and mammosphere formation in breast cancer. Oncogene. 2011;30(24):2741–52. doi: 10.1038/onc.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroes RA, Jastrow A, McLone MG, et al. The identification of novel therapeutic targets for the treatment of malignant brain tumors. Cancer Lett. 2000;156(2):191–8. doi: 10.1016/s0304-3835(00)00462-6. [DOI] [PubMed] [Google Scholar]
- 10.Kondoh N, Wakatsuki T, Ryo A, et al. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999;59(19):4990–6. [PubMed] [Google Scholar]
- *11.Yu L, Huang X, Zhang W, et al. Critical role of DEK and its regulation in tumorigenesis and metastasis of hepatocellular carcinoma. Oncotarget. 2016;7(18):26844–55. doi: 10.18632/oncotarget.8565. This paper explores the role DEK has in the pathogenesis and metastisis of hepatocellular carcinoma cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grasemann C, Gratias S, Stephan H, et al. Gains and overexpression identify DEK and E2F3 as targets of chromosome 6p gains in retinoblastoma. Oncogene. 2005;24(42):6441–9. doi: 10.1038/sj.onc.1208792. [DOI] [PubMed] [Google Scholar]
- 13.Orlic M, Spencer CE, Wang L, Gallie BL. Expression analysis of 6p22 genomic gain in retinoblastoma. Genes Chromosomes Cancer. 2006;45(1):72–82. doi: 10.1002/gcc.20263. [DOI] [PubMed] [Google Scholar]
- *14.Ou Y, Xia R, Kong F, et al. Overexpression of DEK is an indicator of poor prognosis in patients with gastric adenocarcinoma. Oncol Lett. 2016;11(3):1823–8. doi: 10.3892/ol.2016.4147. This paper explores whether DEK may be a good biomarker for gastric adenocarcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *15.Liu X, Qi D, Qi J, et al. Significance of DEK overexpression for the prognostic evaluation of non-small cell lung carcinoma. Oncol Rep. 2016;35(1):155–62. doi: 10.3892/or.2015.4365. One of the pioneering papers exploring whether DEK is a prognostic factor for small cell lung cancer. [DOI] [PubMed] [Google Scholar]
- *16.Sun J, Bi F, Yang Y, Zhang Y, et al. DEK protein overexpression predicts poor prognosis in pancreatic ductal adenocarcinoma. Oncol Rep. 2017;37(2):857–64. doi: 10.3892/or.2016.5302. The paper examines whether DEK expression can predict the prognosis of patients with pancreatic ductal adenocarcinoma. [DOI] [PubMed] [Google Scholar]
- 17.Evans AJ, Gallie BL, Jewett MA, et al. Defining a 0.5-mb region of genomic gain on chromosome 6p22 in bladder cancer by quantitative-multiplex polymerase chain reaction. Am J Pathol. 2004;164(1):285–93. doi: 10.1016/S0002-9440(10)63118-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broxmeyer HE, Kappes F, Mor-Vaknin N, et al. DEK regulates hematopoietic stem engraftment and progenitor cell proliferation. Stem Cells Dev. 2012;21(9):1449–54. doi: 10.1089/scd.2011.0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broxmeyer HE, Mor-Vaknin N, Kappes F, et al. Concise review: role of DEK in stem/progenitor cell biology. Stem Cells. 2013;31(8):1447–53. doi: 10.1002/stem.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koleva RI, Ficarro SB, Radomska HS, et al. C/EBPalpha and DEK coordinately regulate myeloid differentiation. Blood. 2012;119(21):4878–88. doi: 10.1182/blood-2011-10-383083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waldmann T, Scholten I, Kappes F, et al. The DEK protein–an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343(1):1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 22.Kappes F, Scholten I, Richter N, et al. Functional domains of the ubiquitous chromatin protein DEK. Mol Cell Biol. 2004;24(13):6000–10. doi: 10.1128/MCB.24.13.6000-6010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **23.Sanden C, Gullberg U. The DEK oncoprotein and its emerging roles in gene regulation. Leukemia. 2015;29(8):1632–6. doi: 10.1038/leu.2015.72. An excellent and detailed review on the controversial and often opposing functions of DEK. [DOI] [PubMed] [Google Scholar]
- 24.Waldmann T, Baack M, Richter N, Gruss C. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003;31(23):7003–10. doi: 10.1093/nar/gkg864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu GK, Grosveld G, Markovitz DM. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc Natl Acad Sci U S A. 1997;94(5):1811–5. doi: 10.1073/pnas.94.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexiadis V, Waldmann T, Andersen J, et al. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 2000;14(11):1308–12. [PMC free article] [PubMed] [Google Scholar]
- 27.Waldmann T, Eckerich C, Baack M, Gruss C. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J Biol Chem. 2002;277(28):24988–94. doi: 10.1074/jbc.M204045200. [DOI] [PubMed] [Google Scholar]
- 28.Hu HG, Scholten I, Gruss C, Knippers R. The distribution of the DEK protein in mammalian chromatin. Biochem Biophys Res Commun. 2007;358(4):1008–14. doi: 10.1016/j.bbrc.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 29.Kappes F, Waldmann T, Mathew V, et al. The DEK oncoprotein is a Su(var) that is essential to heterochromatin integrity. Genes Dev. 2011;25(7):673–8. doi: 10.1101/gad.2036411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawatsubashi S, Murata T, Lim J, et al. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010;24(2):159–70. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takata H, Nishijima H, Ogura S, et al. Proteome analysis of human nuclear insoluble fractions. Genes Cells. 2009;14(8):975–90. doi: 10.1111/j.1365-2443.2009.01324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saha AK, Kappes F, Mundade A, et al. Intercellular trafficking of the nuclear oncoprotein DEK. Proc Natl Acad Sci U S A. 2013;110(17):6847–52. doi: 10.1073/pnas.1220751110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavellan E, Asp P, Percipalle P, Farrants AK. The WSTF-SNF2h chromatin remodeling complex interacts with several nuclear proteins in transcription. J Biol Chem. 2006;281(24):16264–71. doi: 10.1074/jbc.M600233200. [DOI] [PubMed] [Google Scholar]
- 34.Kappes F, Damoc C, Knippers R, et al. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol Cell Biol. 2004;24(13):6011–20. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat Struct Mol Biol. 2007;14(6):548–55. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- 36.Kappes F, Fahrer J, Khodadoust MS, et al. DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol. 2008;28(10):3245–57. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cleary J, Sitwala KV, Khodadoust MS, et al. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J Biol Chem. 2005;280(36):31760–7. doi: 10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]
- 38.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19(24):6860–9. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGarvey T, Rosonina E, McCracken S, et al. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J Cell Biol. 2000;150(2):309–20. doi: 10.1083/jcb.150.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soares LM, Zanier K, Mackereth C, et al. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312(5782):1961–5. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 41.Mor-Vaknin N, Punturieri A, Sitwala K, et al. The DEK nuclear autoantigen is a secreted chemotactic factor. Mol Cell Biol. 2006;26(24):9484–96. doi: 10.1128/MCB.01030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong X, Michelis MA, Wang J, et al. Autoantibodies to DEK oncoprotein in a patient with systemic lupus erythematosus and sarcoidosis. Arthritis Rheum. 1998;41(8):1505–10. doi: 10.1002/1529-0131(199808)41:8<1505::AID-ART23>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 43.Ageberg M, Gullberg U, Lindmark A. The involvement of cellular proliferation status in the expression of the human proto-oncogene DEK. Haematologica. 2006;91(2):268–9. [PubMed] [Google Scholar]
- 44.Heath V, Suh HC, Holman M, et al. C/EBPalpha deficiency results in hyperproliferation of hematopoietic progenitor cells and disrupts macrophage development in vitro and in vivo. Blood. 2004;104(6):1639–47. doi: 10.1182/blood-2003-11-3963. [DOI] [PubMed] [Google Scholar]
- 45.Cammenga J, Mulloy JC, Berguido FJ, et al. Induction of C/EBPalpha activity alters gene expression and differentiation of human CD34+ cells. Blood. 2003;101(6):2206–14. doi: 10.1182/blood-2002-05-1546. [DOI] [PubMed] [Google Scholar]
- 46.Radomska HS, Huettner CS, Zhang P, et al. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18(7):4301–14. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schepers H, Wierenga AT, van Gosliga D, et al. Reintroduction of C/EBPalpha in leukemic CD34+ stem/progenitor cells impairs self-renewal and partially restores myelopoiesis. Blood. 2007;110(4):1317–25. doi: 10.1182/blood-2006-10-052175. [DOI] [PubMed] [Google Scholar]
- 48.Zhang DE, Zhang P, Wang ND, et al. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A. 1997;94(2):569–74. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lohmann F, Dangeti M, Soni S, et al. The DEK Oncoprotein Is a Critical Component of the EKLF/KLF1 Enhancer in Erythroid Cells. Mol Cell Biol. 2015;35(21):3726–38. doi: 10.1128/MCB.00382-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siatecka M, Bieker JJ. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood. 2011;118(8):2044–54. doi: 10.1182/blood-2011-03-331371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tallack MR, Perkins AC. KLF1 directly coordinates almost all aspects of terminal erythroid differentiation. IUBMB Life. 2010;62(12):886–90. doi: 10.1002/iub.404. [DOI] [PubMed] [Google Scholar]
- 52.Yien YY, Bieker JJ. EKLF/KLF1, a tissue-restricted integrator of transcriptional control, chromatin remodeling, and lineage determination. Mol Cell Biol. 2013;33(1):4–13. doi: 10.1128/MCB.01058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim SK, Bieker JJ, Lin CS, Costantini F. A shortened life span of EKLF−/− adult erythrocytes, due to a deficiency of beta-globin chains, is ameliorated by human gamma-globin chains. Blood. 1997;90(3):1291–9. [PubMed] [Google Scholar]
- 54.Faulkner NE, Hilfinger JM, Markovitz DM. Protein phosphatase 2A activates the HIV-2 promoter through enhancer elements that include the pets site. J Biol Chem. 2001;276(28):25804–12. doi: 10.1074/jbc.M006454200. [DOI] [PubMed] [Google Scholar]
- 55.Hilfinger JM, Clark N, Smith M, et al. Differential regulation of the human immunodeficiency virus type 2 enhancer in monocytes at various stages of differentiation. J Virol. 1993;67(7):4448–53. doi: 10.1128/jvi.67.7.4448-4453.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krithivas A, Fujimuro M, Weidner M, et al. Protein interactions targeting the latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus to cell chromosomes. J Virol. 2002;76(22):11596–604. doi: 10.1128/JVI.76.22.11596-11604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verma SC, Cai Q, Kreider E, et al. Comprehensive analysis of LANA interacting proteins essential for viral genome tethering and persistence. PLoS One. 2013;8(9):e74662. doi: 10.1371/journal.pone.0074662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **58.Pease NA, Wise-Draper T, Privette Vinnedge L. Dissecting the Potential Interplay of DEK Functions in Inflammation and Cancer. J Oncol. 2015;2015:106517. doi: 10.1155/2015/106517. A comprehensive review on DEK function in inflammation, presistant viral infection, autoimmunity and tumorogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ibrahim MM, Fjaere E, Lock EJ, et al. Chronic consumption of farmed salmon containing persistent organic pollutants causes insulin resistance and obesity in mice. PLoS One. 2011;6(9):e25170. doi: 10.1371/journal.pone.0025170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruzzin J, Petersen R, Meugnier E, et al. Persistent organic pollutant exposure leads to insulin resistance syndrome. Environ Health Perspect. 2010;118(4):465–71. doi: 10.1289/ehp.0901321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim TH, Shin SW, Park JS, Park CS. Genome wide identification and expression profile in epithelial cells exposed to TiO(2) particles. Environ Toxicol. 2015;30(3):293–300. doi: 10.1002/tox.21906. [DOI] [PubMed] [Google Scholar]
- 62.Kim DW, Kim JY, Choi S, et al. Transcriptional regulation of 1-cys peroxiredoxin by the proto-oncogene protein DEK. Mol Med Rep. 2010;3(5):877–81. doi: 10.3892/mmr.2010.346. [DOI] [PubMed] [Google Scholar]
- 63.Liu K, Feng T, Liu J, et al. Silencing of the DEK gene induces apoptosis and senescence in CaSki cervical carcinoma cells via the up-regulation of NF-kappaB p65. Biosci Rep. 2012;32(3):323–32. doi: 10.1042/BSR20100141. [DOI] [PubMed] [Google Scholar]
- 64.Sammons M, Wan SS, Vogel NL, et al. Negative regulation of the RelA/p65 transactivation function by the product of the DEK proto-oncogene. J Biol Chem. 2006;281(37):26802–12. doi: 10.1074/jbc.M600915200. [DOI] [PubMed] [Google Scholar]
- **65.Mor-Vaknin N, Saha A, Legendre M, et al. DEK-targeting DNA aptamers as therapeutics for inflammatory arthritis. Nat Commun. 2017;8:14252. doi: 10.1038/ncomms14252. This paper describes an exciting new therapeutic tool to augment the function of DEK in inflammation. [DOI] [PMC free article] [PubMed] [Google Scholar]


