Abstract
The Brazilian AIDS epidemic has been characterized by an increasing rate of BF1 recombinants and so far eight circulating recombinant forms/CRFs_BF1 have been described countrywide. In this study, pol sequences (protease/PR, reverse transcriptase/RT) of 87 BF1 mosaic isolates identified among 828 patients living in six Brazilian States from three geographic regions (Central West, North, Northeast) were analyzed. Phylogenetic and bootscan analyses were performed to investigate the evolutionary relationship and mosaic structure of BF1 isolates. Those analyses showed that 20.7% of mosaics (18 out of 87) were CRFs-like isolates, mostly represented by CRF28/CRF29_BF-like viruses (14 out of 18). We also identified five highly supported clusters that together comprise 42 out of 87 (48.3%) BF1 sequences, each cluster containing at least five sequences sharing a similar mosaic structure, suggesting possible new unidentified CRFs_BF1. The divergence time of these five potential new CRFs_BF1 clusters was estimated using a Bayesian approach and indicate that they probably originated between the middle 1980s and the middle 1990s. DNA was extracted from whole blood and four overlapping fragments were amplified by PCR providing full/near full length genomes (FLG/NFLG) and partial genomes. Eleven HIV-1 isolates from Cluster # 5 identified in epidemiologically unlinked individuals living in Central West and North regions provided FLG/NFLG/partial genome sequences with identical mosaic structure. These viruses differ from any known CRF_BF1 reported to date and were named CRF90_BF1 by the Los Alamos National Laboratory. This is the 9th CRF_BF1 described in Brazil and the first one identified in Central West and North regions. Our results highlight the importance of continued molecular screening and surveillance studies, especially of full genome sequences to understand the evolutionary dynamics of the HIV-1 epidemic in a country of continental dimensions as Brazil.
Introduction
Human Immunodeficiency Virus-1 (HIV-1) is a highly polymorphic and fast evolving pathogen [1]. Worldwide HIV-1 can be classified into groups (M, N, O and P), and the pandemic group M is classified in subtypes (A-D, F-H, J and K) and sub-subtypes (A1-A4, F1-F2) [2,3]. While mutation rates are similar to other RNA viruses, HIV-1 has a high recombinogenic capacity and intersubtype recombination events are frequent in coinfected or superinfected individuals from areas where two or multiple variants cocirculate [4]. Recombinant strains exhibiting identical mosaic patterns identified in at least three epidemiologically unlinked individuals have been classified as circulating recombinant forms (CRFs), while the ones displaying unique mosaic structures or only infecting individuals with epidemiological link are known as unique recombinant forms (URFs) [5,6]. Recombination has been recognized as a driving force in shaping the diversity of HIV-1 globally since the mid 90´s [7]. Currently, 88 CRFs have been assigned and 81 of them have been published with public data available at the Los Alamos HIV database [http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html]. CRFs together with URFs are estimated to account for at least 20% of HIV-1 infections worldwide [8].
The Brazilian AIDS epidemic is characterized by the cocirculation of multiple HIV-1 subtypes. Subtype B predominates in most regions followed by subtypes F1, C, and recombinants among these subtypes [9–15]. The first Brazilian BF1 mosaics were identified in the early 90´s in the Southeast region which is considered the epicenter of the epidemic [16,17]. Among the 14 CRFs_BF1 described so far, eight originated in Brazil (CRF28_BF, CRF29_BF, CRF39_BF, CRF40_BF, CRF46_BF, CRF70_BF, CRF71_BF and CRF72_BF) [18–22]. The importance of BF1 recombinants in Brazil is further corroborated by the description of countless URFs in all country regions [23–26]. Previous studies from our research group in different study populations in Central West, North and Northeast Brazilian States showed variable prevalence of BF1 recombinants in the pol subgenomic fragment: (Goiás: 3.7–18.1%, Mato Grosso: 11.9%, Mato Grosso do Sul: 8.2–25.9%, Tocantins: 7.7%, Maranhão: 7.5%, Piauí: 4.5% [11,27–37].
In this study, previously produced pol sequences of BF1 mosaic isolates circulating in Central West, North and Northeast Brazil were reclassified into possible CRFs or URFs. Full/near full-length genome (FLG/NFLG) and partial genome sequences were obtained for the most representative potential CRF detected. These analyzes allowed the identification of the novel CRF90_BF1 that is circulating in Central West and North Brazil, away from the epicenter of the epidemic. Other putative novel CRFs_BF1 are also described. The median time of origin of these mosaics was also estimated. The detailed molecular characterization of recombinant forms circulating countrywide contributes to the mapping of HIV-1 diversity in Brazil.
Material and methods
Study population
Previous studies from our group recruited from 2003 to 2013 a total of 828 individuals infected with HIV-1 residing in six Brazilian States located in three geographic regions (Central West: Goiás/GO, Mato Grosso/MT, Mato Grosso do Sul/MS; North: Tocantins/TO; Northeast: Maranhão/MA, Piauí/PI) (S1 Table) [11,27–37]. These studies have identified a total of 87 (10.5%) BF1 recombinant isolates based on sequencing of pol subgenomic fragment covering the protease (PR) and partial reverse-transcriptase (RT) (positions 2253–3251 relative to HXB2 genome). The related research protocols were approved by the institutional Ethics Committee review boards (Goiás: protocols #073/05, #003/2008, #163/2010 at CEPMHA/HC/UFG, Mato Grosso: protocol #435/07 at Universidade Federal do Mato Grosso/UFMT, Mato Grosso do Sul: protocol #1143 at Universidade Federal do Mato Grosso do Sul/UFMS, Piauí; protocol #022/2011 at Universidade Estadual do Piauí/UESPI, Maranhão: protocol #16/2011 at Hospital de Doenças Tropicais Dr Natan Portela). All patients signed an informed consent form before blood collection for HIV-1 molecular studies.
Amplification of HIV-1 PR/RT
RNA extraction, reverse transcription into complementary DNA (cDNA) and amplification by nested polymerase chain reaction (nested-PCR) of the PR/RT regions were previously described [11,27–37].
Amplification of HIV-1 full length genomes
Genomic DNA was extracted from whole blood samples (QIAamp® DNA Blood Mini Kit/QIAGEN, Qiagen, Hilden, Germany). The complete HIV-1 genome was amplified by nested-PCR employing Platinum Taq DNA polymerase enzyme (Invitrogen, Carlsbad, CA) into four overlapping fragments using HIV-1 specific primers, as following: fragment 1- SCAOSD/LR51 external primers and SCANSD/DP11 internal primers (408–2594), fragment 2- DP10/SCCNAS external primers and DP16/SCCOAS internal primers (2253–4830); fragment 3- MMINT8/ED14 external primers and MMINT3/ED12 internal primers (4653–7811); fragment 4- ED5/SCDOAD external primers and JH44/LTR2 internal primers (6954–9625) (S2 Table) [38–40], all positions were relative to HXB2 genome. Isolates with all four fragments completely sequenced were considered full length genomes (FLG); isolates with three complete fragments were considered near full length genomes (NFLG), and isolates with one or two fragment sequences were referred as partial genomes.
DNA sequencing
The amplified DNA fragments from the nested-PCR products were separated by gel electrophoresis, purified (kit QIAquick® PCR Purification Kit/QIAGEN, Qiagen, Hilden, Germany) and sequenced with the Big Dye Terminator Sequencing Kit v. 3.1 (Applied Biosystems, Foster City, CA) in an automated ABI Prism 3100 Genetic Analyzer (Applied Biosystems, USA). Chromatograms were analyzed and edited using the SeqMan software from the package DNASTAR Lasergene (MA, USA).
Phylogenetic and recombination analyses
Sequences were aligned using Clustal X 2.0 implemented in BioEdit 7.2.0 program [41]. Reference sequences of HIV-1 group M subtypes (A-D, F–H, J and K) and CRF-BF1 sequences were obtained from the Los Alamos HIV database (http://hiv.lanl.gov/). Phylogenetic trees were generated using the neighbor-joining (NJ) method [42] under the Kimura two-parameter model [43] using MEGA 6.0 software [44]. Bootstrap values (BP, 1.000 replicates) above 70% were considered significant. Recombination analyses were performed in all viral isolates using bootscan implemented in Simplot v3.5.1 software with the following parameters: 200nt or 300nt window, 20nt increments, NJ method under Kimura’s two-parameter correction with 100 bootstrap replicates [45]. In this study the parameters used for bootscan analyses of recombinant viruses differed for smaller and larger fragments: for the analyses of pol fragments (998nt) a smaller sliding window of 200nt was used whereas for larger fragments of near full-genomes (>6670nt) a larger sliding window of 300nt was adopted. To better characterize the recombination breakpoints suggested in the previous analyses, the putative recombinants were subjected to informative site analyses as described elsewhere [39]. For this purpose, consensus sequences from Brazilian HIV-1 subtypes B and F were generated in the DAMBE program [46]. Fragments of sequences assigned to specific HIV-1 subtypes were finally confirmed by separate NJ phylogenetic analysis as described above.
Representative samples from the HIV-1 BF1 Brazilian clusters herein identified were submitted to a Basic Local Alignment Search Tool (BLAST) analysis in order to recover other Brazilian sequences with high similarity (>95%) and probably similar recombination profile. The BLAST analysis was done sequences using sequences obtained from the Los Alamos HIV database (http://hiv.lanl.gov/).
Evolutionary analyses of BF1 recombinants
The time of the most recent common ancestor (TMRCA) of HIV-1 BF1 clades was estimated using a Bayesian Markov Chain Monte Carlo (MCMC) approach implemented in BEAST v1.8 [47,48] with BEAGLE to improve run-time [49]. Analyses were performed using the GTR+I+G nucleotide substitution model, a Bayesian Skyline coalescent tree prior [50] and a relaxed uncorrelated lognormal molecular clock model [51] with an informative uniform prior interval (1.0–3.0 x 10−3 nucleotide substitutions per site per year). One MCMC chain was run for 1x107 generations. Convergence and uncertainty of parameter estimates were assessed by calculating the effective sample size (ESS) and the 95% highest probability density (HPD) values, respectively using Tracer v1.6 [52]. The maximum clade credibility (MCC) tree was summarized with TreeAnnotator v1.8 and visualized with FigTree v1.4.0.
Data availability
All HIV-1 sequences generated in this study were deposited in the GenBank database (KY628215-KY628225).
Results
Phylogenetic and evolutionary analyses of BF1 pol recombinants
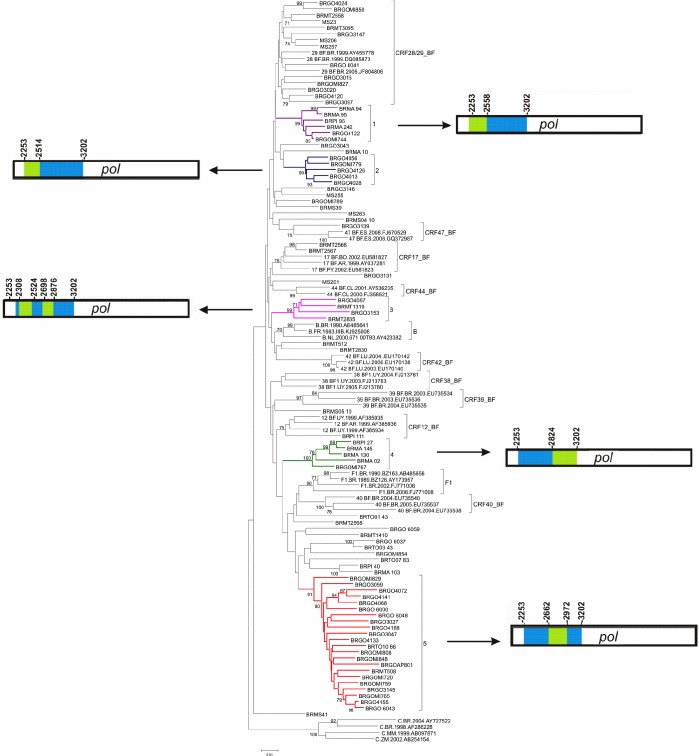
Initial phylogenetic analyses of 87 HIV-1 isolates previously characterized as BF1 recombinants in the PR/RT region (S1 Table) classified 18 (21%) sequences as CRF_BF-like (14 CRF28/CRF29_BF-like, two CRF17_BF-like, one CRF12_BF-like and one CRF47_BF-like) and 27 (31%) sequences as URFs_BF (Fig 1). The remaining 42 (48%) sequences were distributed in five clusters comprising between five and 22 sequences, sharing the same mosaic structure and were classified as potential news CRFs_BF1 (Fig 1). Clusters # 1, 3 and 4 displayed high supports (BP ≥ 99%) at initial analysis. For Clusters # 2 and 5, however, high supports were obtained only after exclusion of the URFs_BF MS251, BRGO3127 and BRGO4162 sequences (Fig 1). Cluster # 1 had six sequences, from three different States (two from Goiás, three from Maranhão and one from Piauí). Cluster # 2 had five sequences, all from Goiás State. Cluster # 3 comprised four sequences from two States (two from Mato Grosso and two from Goiás). Cluster # 4 had five sequences from three States (one from Goiás, three from Maranhão and one from Piauí). Cluster # 5 contained 22 sequences from three States (20 from Goiás, one from Mato Grosso and one from Tocantins).
Fig 1. Phylogenetic analysis of 87 pol sequences of B/F1 HIV-1 isolates presenting five highly supported clusters and the mosaic pattern of recombination in each cluster (neighbor-joining method, Kimura 2-parameters evolutive model/1000 replicate bootstrap values).
Bootscanning analyses of BF1 inter-subtype recombinant clusters (# 1–5) are represented. The five clusters identified in our study are indicated by different colors: Cluster # 1: purple, Cluster # 2: blue, Cluster # 3: pink, Cluster # 4: green and Cluster # 5: red. Bootscan analysis was performed in a 200nt sliding window advanced in 20nt step size increments (1.000 replicates). All CRF_BF depicting recombination breakpoints in pol region were included in the analysis. In the mosaic structure representations of BF1 isolates, the breakpoint positions according to HXB2 genome numeration are shown on the right and left sides of the clusters, blue stands for subtype B and green stands for subtype F.
A Blast search analysis was performed to identify sequences similar to the five potential new CRF_BF1 Brazilian clusters. The recovered sequences were included in the phylogenetic and recombinant analysis, bootstrap values higher than 87% and similar mosaic profiles compared to those previously classified in Clusters # 3, 4 and 5 was verified (Fig 2). Eighteen sequences branching within Custer # 3 were recovered from patients recruited in four States from the North region (seven from Amazonas, five from Rondônia, three from Roraima and one from Acre) along with two sequences from the South region (Paraná) (Fig 2). Two sequences from the North region (Amapá) classified in Cluster # 4 and three sequences classified in Cluster # 5 were recovered from patients from the North region (Rondônia) (Fig 2).
Fig 2. Phylogenetic tree of study BF1 isolates from Central West, North, Northeast and South Brazil and BF1 sequences from GenBank sharing over 95% similarity with study isolates.
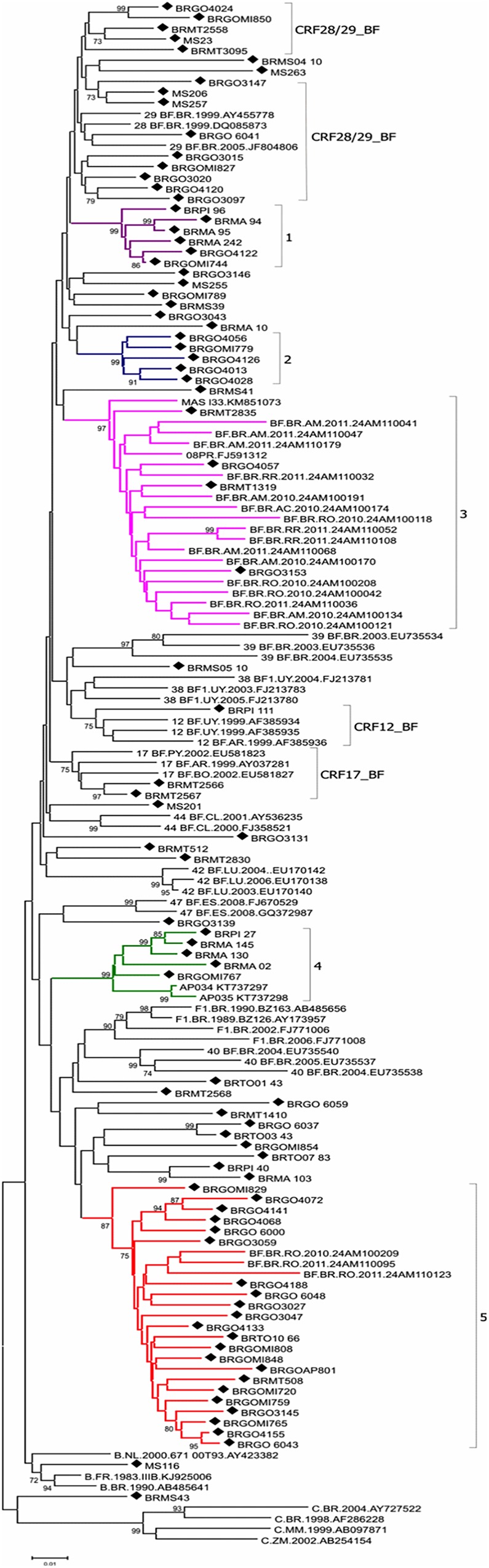
Trees were constructed using MEGA software, 6.0 version under neighbor-joining and Kimura 2 parameters methods (Bootstrap value over 70%). The sequences described in our study are distinguished from the sequences retrieved from the GenBank by a diamond signal. The five clusters identified in our study are indicated by different colors: Cluster # 1: purple, Cluster # 2: blue, Cluster # 3: pink, Cluster # 4: green and Cluster # 5: red.
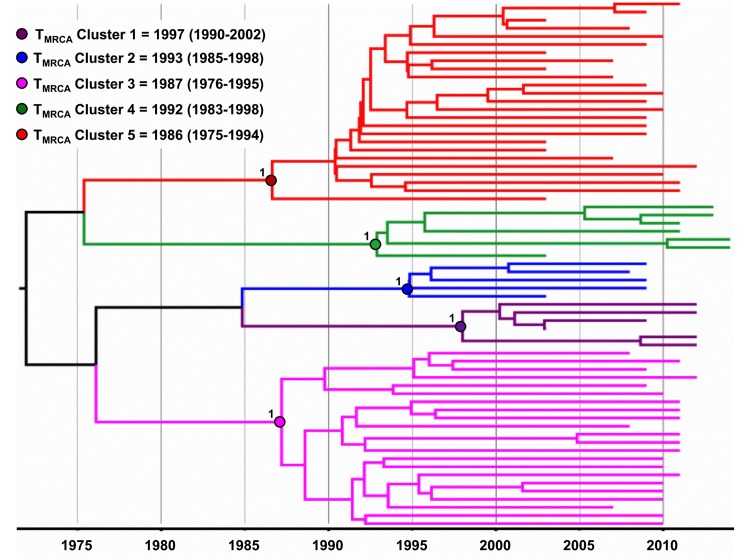
The Bayesian MCC tree displayed the same topology of the NJ tree, thus confirming the five BF1 phylogenetic clusters initially described (Fig 3). According to this analysis, the median TMRCA of the five potential new Brazilian CRFs_BF identified was estimated between the middle 1980s and the middle 1990s (Fig 3).
Fig 3. Time-scaled Bayesian MCMC tree of 65 pol sequences of BF1 HIV-1 isolates that grouped into five clusters from Central West, North, Norhteast and South Brazil.
The circles indicate the positions of the MRCA of each BF1 cluster. Branch lengths are depicted in units of time (years). The tree was automatically rooted under the assumption of a relaxed molecular clock. The five clusters identified in our study are indicated by different colors: Cluster # 1: purple, Cluster # 2: blue, Cluster # 3: pink, Cluster # 4: green and Cluster # 5: red.
Analysis of FLG, NFLG and partial genomes
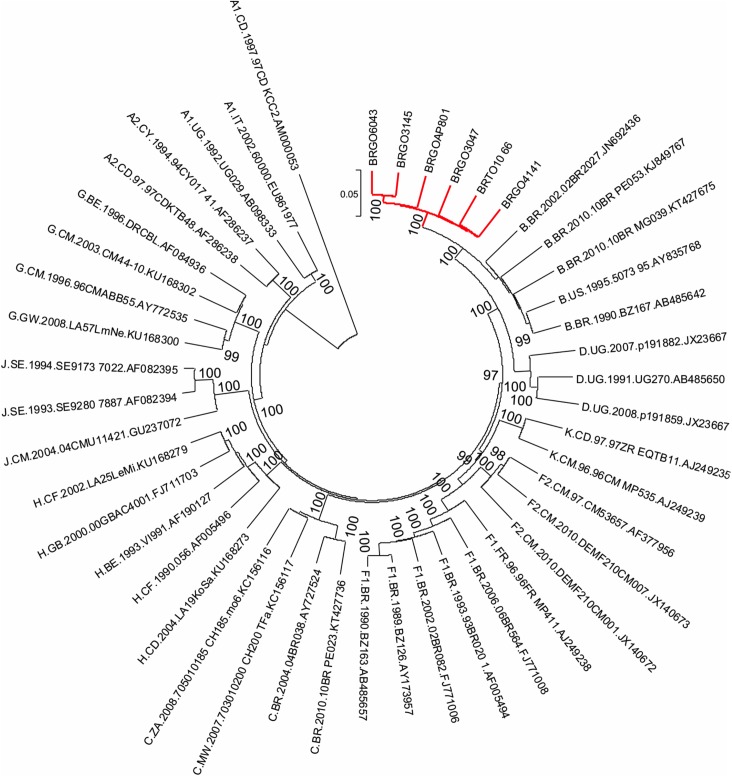
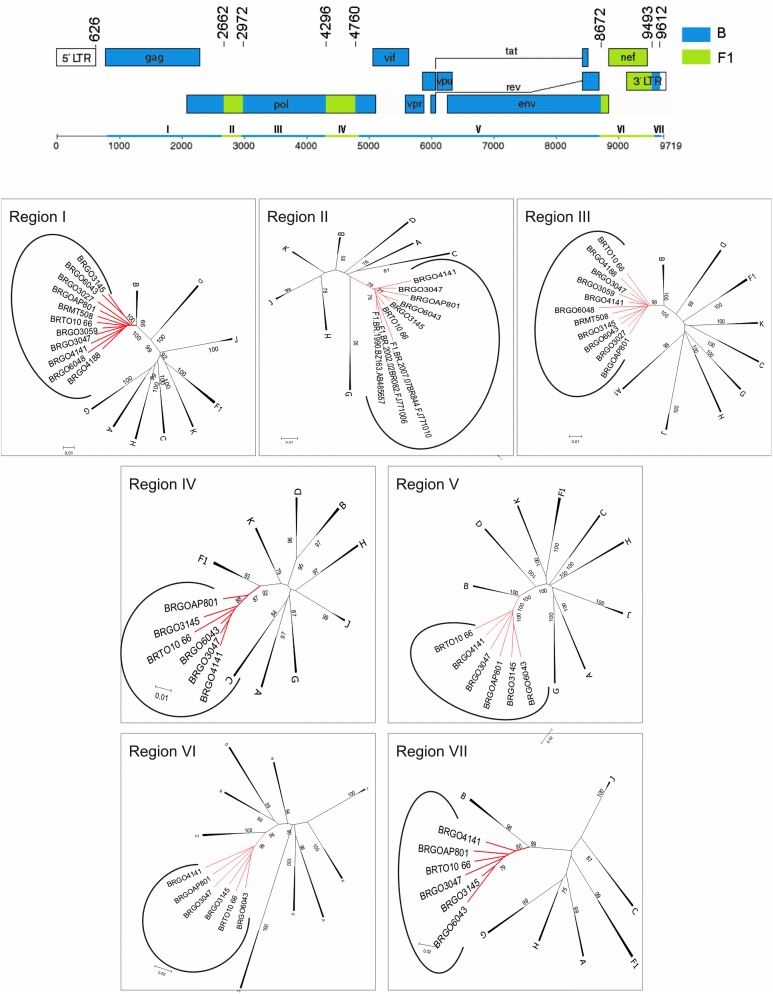
Phylogenetic (Fig 4) and bootscan analyses of six full length genomes (BRGOAP801, BRGO6043, BRTO10_66, BRGO4141, BRGO3145 and BRGO3047) obtained from isolates classified in Cluster # 5 allowed the description of a new recombinant lineage designated CRF90_BF1 by the Los Alamos HIV Sequence Database (Los Alamos National Laboratory) according to the standardized nomenclature [2]. We also obtained one NFLG and four partial genomes for isolates from this Cluster that share the same mosaic structure (Fig 5). The mosaic structures inferred from the analyses of these FLG, NFLG and partial genomes showed a genome predominantly of subtype B, which can be divided into seven subregions alternating subtypes B and F1. These seven subregions were named I (626–2.661), II (2.662–2.971), III (2.972–4.295), IV (4.296–4.759), V (4.760–8.671), VI (8.672–9.492) and VII (9.493–9.612) all positions relative to HXB2 genome. Subregion NJ analyses also confirmed the putative parental HIV-1 subtype (Fig 5). Fully coincident intersubtype breakpoint locations at I-III sub regions were also observed in the NFLG of BRGO4188 isolate and in the partial genome sequences of BRMT508, BRGO3027, BRGO3059 and BRGO6048 isolates (Fig 5 and Table 1).
Fig 4. Phylogenetic analyses on the full length/near-full-length genome sequences of HIV-1 BF1 isolates from cluster # 5 belonging to a new CRF90_BF1 identified in patients from Goiás and Tocantins States in the Central West and North Brazilian regions.
The HIV group M reference sequences of subtypes were obtained from the Los Alamos database. The scale bar represents 0.02 nucleotide substitutions per site. Phylogenic analyses were constructed with Mega software version 6.0.
Fig 5. Mosaic structure of the new CRF composed by subtypes B and F1.
Breakpoint positions according to HXB2 genome numbering system are indicated. The phylogenetic trees for each of the seven mosaic segments (I-VII) were constructed with Mega software v6.0 and the trees were midpoint rooted. The stability of each node was confirmed by bootstrapping with 1.000 replicates and only significant bootstrap values >70% are shown at the corresponding nodes. The genetic distance corresponding to the length of the branches is shown by the line at the bottom. The red color represents the CRF90_BF1 identified in this study.
Table 1. Demographic characteristics of study subjects infected by HIV-1 CRF90_BF1.
| Sample ID | ARV status | Gender | Age | Risk Group | HIV Diagnosis (Year) | Sample Collection (Year) | HXB2 location (nt) | Accession Number |
|---|---|---|---|---|---|---|---|---|
| BRGOAP801 | HAART | M | 59 | Heterosexual | 2007 | 2010 | 407–9626 | KY628223 |
| BRGO6043 | Naïve | F | 30 | Heterosexual | 2011 | 2011 | 407–9626 | KY628221 |
| BRTO10_66 | Naïve | F | 27 | Heterosexual | 2009 | 2009 | 407–9616 | KY628225 |
| BRGO4141 | Prophylaxis | F | 24 | Heterosexual | 2010 | 2010 | 408–9615 | KY628219 |
| BRGO3145 | HAART | F | 35 | Heterosexual | 2004 | 2007 | 407–9612 | KY628218 |
| BRGO3047 | HAART | M | 47 | NA | 2002 | 2007 | 474–9589 | KY628216 |
| BRGO4188 | Prophylaxis | F | 27 | Heterosexual | 2006 | 2010 | 407–7080* | KY628220 |
| BRGO3027 | Naïve | M | 27 | IDU | 2002 | 2007 | 453–5924* | KY628215 |
| BRMT508 | Naïve | M | 41 | Heterosexual | 2009 | 2009 | 414–5919* | KY628224 |
| BRGO3059 | Naïve | M | 51 | Heterosexual | 2007 | 2007 | 407–4778* | KY628217 |
| BRGO6048 | Naïve | F | 30 | Heterosexual | 2009 | 2012 | 454–4122* | KY628222 |
M: Male; F: Female; Naïve: antiretroviral naïve patients; HAART: Patients under highly active antiretroviral therapy; Prophylaxis: mother-to-child-transmission antiretroviral prophylaxis (MTCT ARV prophylaxis); IDU: intravenous drug user; N.A.: not available
* isolates with partial genome sequences; Isolates are listed according to the size of sequenced fragments. nt: nucleotide position
The epidemiological features of the 11 patients presenting the newly described CRF90_BF1 lineage included six females (four of them pregnant) and five males (two of them prisoners) (Table 1). The prevailing risk category was heterosexual sex reported by nine patients while one prisoner patient reported intravenous drug use. Six patients were ARV naïve and five had been exposed to ARV drugs either as highly active antiretroviral therapy (HAART) or temporary mother-to-child-transmission (MTCT) prophylaxis. Most patients were from the Central West region (Goiás State: isolates BRGO3027, BRGO3047, BRGO3145, BRGO3059, BRGO4188, BRGO4141, BRGO6048, BRGOAP801 and BRGO6043; Mato Grosso State: isolate BRMT508) and one patient lived in the North region (Tocantins State: isolate BRTO10_66).
Discussion
In this study, we report the characterization of a novel HIV-1 CRF_BF1, named CRF90_BF1 based on six FLG, one NFLG and four partial genome sequences. These isolates shared identical mosaic structures and were identified in individuals without any epidemiological link that live in two distinct geographic regions in Brazil (Central West and North) located around 800–900 km apart. These criteria fulfill the requirements to define a new CRF, which is circulating in distant interior urban areas in Brazil. This novel CRF is the 9th CRF involving subtypes B and F1 described in Brazil and the 14th reported in South America. The estimated frequency of the CRF90_BF1 in our sample set was 1.3% (11/828), with predominant detection in the Central West region. However, the actual prevalence of this new CRF in these geographic regions cannot be accurately estimated since there is limited molecular data on HIV-1 isolates especially from the States of Mato Grosso, Mato Grosso do Sul and Tocantins.
The CRF12_BF, the first CRF identified in the Americas was described in 2001 in patients from Argentina and Uruguay and its origin was estimated around the early 80s [53,54], while BF1 recombinants were first reported in Brazil in the early 90’s [16,17]. Patients harboring the CRF90_BF1 were diagnosed between 2002 and 2011. The median estimated TMRCA of the CRF90_BF1 and of other putative CRF_BF1 clusters identified in our study is not recent and ranges from middle 80’s to middle 90’s, similar to that previously estimated for Brazilian CRF28_BF and CRF29_BF [55]. These estimates indicate that CRFs_BF1 have been probably circulating in Brazil for three to four decades.
Besides its early generation, we have evidences, as shown by blast search analyses, that the CRF90_BF1 and also the other putative CRFs_BF1 clades identified here have a wide geographic circulation (Fig 6). The CRF90_BF1 that we identified in Central West (Goiás and Mato Grosso) and North Brazil (Tocantins) is probably also circulating in Rondônia, another State in the North region which borders Bolivia in the South/East. HIV-1 BF1 isolates with the same recombination pattern of isolates from Cluster # 3 detected in Central West were also identified in several North Brazilian States (Amazonas, Rondônia, Roraima and Acre), and in the South State of Paraná. Isolates with similar recombination profile of isolates from Cluster # 4 were also identified in the North region (Amapá State) besides the Central West (Goiás) and Northeast Brazil (Maranhão and Piauí). These results suggest the existence of novel CRFs_BF1 circulating in Brazil.
Fig 6. South America map highlighting Brazil.
Color marks indicate the possible geographical area of circulation of BF1 isolates identified in Central West, North, Northeast and South Brazil and BF1 sequences from GenBank sharing over 95% similarity with study isolates from Clusters # 1–5. In the Brazilian map, each colored mark represents the geographic area of potential circulation of: Cluster # 1-purple, Cluster # 2-blue, Cluster # 3-pink, Cluster # 4-green, Cluster # 5-red.
CRF28_BF and CRF29_BF described in the Southeast in 2006 (Santos, São Paulo State) represent the first Brazilian CRFs, and their origin date to 1988–1989 [18,55]. Studies have shown a low prevalence of CRF28_BF and CRF29_BF [14,56], outside São Paulo except in Salvador, Bahia State, Northeast where prevalence ranged from 10%-21% [57,58]. Among all BF1 isolates identified in our study we have found a moderate rate of CRF28/CRF29_BF-like isolates (16.1%, 14 out of 87) and an overall rate of 1.7% (14 out of 828) which represent one of the highest frequencies of these CRFs identified outside São Paulo State.
Despite the predominance of subtype B in most geographic Brazilian regions, except in the South where subtype C prevails, studies have shown that the prevalence of non-B subtypes, particularly URFs_BF1 and URFs_BC has increased in the last decade [15,25,40,59,60]. Our studies have shown a significant percentage of recombinant BF1 forms (3.7–25.9%) in the Central West, North and Northeast Brazilian regions [11,27–37]. The most recently described Brazilian CRFs_BF1 (CRF70_BF1 and CRF71_BF1) were identified among blood donors from Pernambuco State, Northeast region [22]. The CRF72_BF1 was identified among blood donors from five public blood banks in Minas Gerais State, Southeast region [21]. These recent data point out the increasing generation and spread of CRFs, especially involving subtypes B and F1 which play an important role in the Brazilian AIDS epidemic. However, the number of complete genome sequences available is still limited, especially sequences from areas away from the epicenter, as our study areas (Central West, North and Northeast) suggesting that the actual proportion of HIV-1 recombinant forms in the Brazilian pandemic is probably underestimated.
Conclusions
In summary, we identified the novel CRF90_BF1 among heterosexual patients living in two geographic regions in Brazil, away from the epicenter of the epidemic. This is the 9th CRF_BF1 described in Brazil indicating that continued molecular screening and surveillance are necessary to fully understand the evolutionary dynamics of the HIV-1 epidemic in such a country of continental dimensions. Our results also underscore the importance of full-length genome sequencing of HIV-1 isolates obtained from patients infected by different transmission routes and in different country regions to fully understand the diversity and complexity of the HIV-1 epidemic in Brazil.
Supporting information
Pregnant: women infected with HIV-1 attending a regional antenatal care; Naïve: antiretroviral naïve patients; HAART: Patients under highly active antiretroviral therapy. * Ref 29: the study group (n = 27) comprises prisoner patients recruited in Goiania/GO (n = 7) and in Campo Grande (n = 20).
(DOCX)
*Some primers had their original sequence modified based on the alignment of subtypes B, C and F1 HIV sequence compendium (2005) from HIV Los Alamos Database.
(DOCX)
Acknowledgments
We are thankful to all participants of this study and to the institutions where they were recruited. We are also grateful to Dr. Vera Saddi (PUC/GO) for sharing sequencing equipment with us.
Data Availability
All full, near full and partial genome sequences of HIV-1 are available from the GenBank database accession numbers KY628215-KY628225.
Funding Statement
This work was supported by the "Programa de Apoio a Núcleos de Excelência/ PRONEX; Fundação de Amparo à Pesquisa do Estado de Goiás/FAPEG; Conselho Nacional de Desenvolvimento Científico e Tecnológico/ CNPq (grant number:201210267000801 to MMAS) and by the "Conselho Nacional de DesenvolvimentoCientífico e Tecnológico/CNPq Universal grant number: 481208/2012-7 to MMAS). MNGR was supported by a scholarship from FAPEG (grant number:201410267000598). MMAS is a recipient of a fellowship from CNPq (grant number 308381/2015-7). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Robertson DL, Hahn BH, Sharp PM. Recombination in AIDS viruses. J Mol Evol. 1995; 40(3):249–59. [DOI] [PubMed] [Google Scholar]
- 2.Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, et al. HIV-1 nomenclature proposal. Science. 2000; 288(5463):55–56 [DOI] [PubMed] [Google Scholar]
- 3.Tebit DM and Arts EJ. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis. 2011; 11(1):45–56. doi: 10.1016/S1473-3099(10)70186-9 [DOI] [PubMed] [Google Scholar]
- 4.Thomson MM, Nájera R. Molecular epidemiology of HIV-1 variants in the global AIDS pandemic: an update. AIDS Rev. 2005;7(4):210–24. [PubMed] [Google Scholar]
- 5.Thomson MM, Pérez-Alvarez L, Nájera R. Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy. Lancet Infect Dis. 2002; 2(8):461–471. Review. [DOI] [PubMed] [Google Scholar]
- 6.Nájera R, Delgado E, Pérez-Alvarez L, Thomson MM. Genetic recombination and its role in the development of the HIV-1 pandemic. AIDS. 2002; Suppl 4:S3–16. [DOI] [PubMed] [Google Scholar]
- 7.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, et al. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996; 70(10):7013–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemelaar J, Gouws E, Ghys PD, Osmanov S. WHO-UNAIDS Network for HIV Isolation and Characterization 2011. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS. 2011; 25(5):679–689. doi: 10.1097/QAD.0b013e328342ff93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brindeiro RM, Diaz RS, Sabino EC, Morgado MG, Pires IL, Brigido L, et al. Brazilian Network for HIV Drug Resistance Surveillance (HIV-BResNet): a survey of chronically infected individuals. AIDS. 2003; 17(7):1063–9. doi: 10.1097/01.aids.0000060345.12269.d7 [DOI] [PubMed] [Google Scholar]
- 10.Inocencio LA, Pereira AA, Sucupira MC, Fernandez JC, Jorge CP, Souza DF, et al. Brazilian Network for HIV Drug Resistance Surveillance: a survey of individuals recently diagnosed with HIV. J Int AIDS Soc. 2009; 18;12:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardoso LP, Queiroz BB, Stefani MM. HIV-1 pol phylogenetic diversity and antiretroviral resistance mutations in treatment naïve patients from Central West Brazil. J Clin Virol. 2009; 46(2):134–139. doi: 10.1016/j.jcv.2009.07.009 [DOI] [PubMed] [Google Scholar]
- 12.Alencar CS, Sabino EC, Carvalho SM, Leao SC, Carneiro-Proietti AB, Capuani L, et al. HIV genotypes and primary drug resistance among HIV-seropositive blood donors in Brazil: role of infected blood donors as sentinel populations for molecular surveillance of HIV. J Acquir Immune Defic Syndr. 2013;1;63(3):387–92. doi: 10.1097/QAI.0b013e31828ff979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silveira J, Santos AF, Martínez AM, Góes LR, Mendoza-Sassi R, Muniz CP, et al. Heterosexual transmission of human immunodeficiency virus type 1 subtype C in southern Brazil. J Clin Virol. 2012; 54(1):36–41. doi: 10.1016/j.jcv.2012.01.017 [DOI] [PubMed] [Google Scholar]
- 14.Guimarães ML, Marques BC, Bertoni N, Teixeira SL, Morgado MG, Bastos FI, et al. Assessing the HIV-1 Epidemic in Brazilian Drug Users: Molecular Epidemiology Approach. PLoS One. 2015;10(11):e0141372 doi: 10.1371/journal.pone.0141372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.da Costa CM, Costa de Oliveira CM, Chehuan de Melo YF, Delatorre E, Bello G, Couto-Fernandez JC. High HIV-1 Genetic Diversity in Patients from Northern Brazil. AIDS Res Hum Retroviruses. 2016; 32(9):918–22. doi: 10.1089/AID.2016.0044 [DOI] [PubMed] [Google Scholar]
- 16.Sabino EC, Shpaer EG, Morgado MG, Korber BT, Diaz RS, Bongertz V, et al. Identification of human immunodeficiency virus type 1 envelope genes recombinant between subtypes B and F in two epidemiologically linked individuals from Brazil.J Virol. 1994; 68(10):6340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos A, Tanuri A, Schechter M, Rayfield MA, Hu DJ, Cabral MC, et al. Dualand recombinant infections: an integral part of the HIV-1 epidemic in Brazil. Emerg Infect Dis. 1999; 5(1):65–74. doi: 10.3201/eid0501.990108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Sá Filho DJ, Sucupira MC, Caseiro MM, Sabino EC, Diaz RS, Janini LM. Identification of two HIV type 1 circulating recombinant forms in Brazil. AIDS Res Hum Retroviruses. 2006; 22(1):1–13. doi: 10.1089/aid.2006.22.1 [DOI] [PubMed] [Google Scholar]
- 19.Guimarães ML, Eyer-Silva WA, Couto-Fernandez JC, Morgado MG. Identification of two new CRF_BF in Rio de Janeiro State, Brazil. AIDS. 2008; 22(3):433–435. doi: 10.1097/QAD.0b013e3282f47ad0 [DOI] [PubMed] [Google Scholar]
- 20.Sanabani SS, Pastena ER, Neto WK, Martinez VP, Sabino EC. Characterization and frequency of a newly identified HIV-1 BF1 intersubtype circulating recombinant form in São Paulo, Brazil. Virol J. 2010; 16;7:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pessôa R, Carneiro Proietti AB, Busch MP, Sanabani SS. Identification of a Novel HIV-1 Circulating Recombinant Form (CRF72_BF1) in Deep Sequencing Data from Blood Donors in Southeastern Brazil. Genome Announc. 2014a; 12;2(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pessôa R, Watanabe JT, Calabria P, Felix AC, Loureiro P, Sabino EC, et al. Deep sequencing of HIV-1 near full-length proviral genomes identifies high rates of BF1 recombinants including two novel circulating recombinant forms (CRF) 70_BF1 and a disseminating 71_BF1 among blood donors in Pernambuco, Brazil. PLoS One. 2014b; 17;9(11):e112674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guimarães ML, Couto-Fernandez JC, Eyer-Silva Wde A, Teixeira SL, Chequer-Fernandez SL, Morgado MG. Analysis of HIV-1 BF pr/rt recombinant strains from Rio de Janeiro/Brazil reveals multiple unrelated mosaic structures. Infect Genet Evol. 2010; 10(7):1094–100. doi: 10.1016/j.meegid.2010.07.001 [DOI] [PubMed] [Google Scholar]
- 24.Sanabani SS, Pastena ÉR, da Costa AC, Martinez VP, Kleine-Neto W, de Oliveira AC, et al. Characterization of partial and near full length genomes of HIV-1 strains sampled from recently infected individuals in São Paulo, Brazil. PLoS One. 2011; 6(10):e25869 doi: 10.1371/journal.pone.0025869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanabani SS, Pessôa R, Soares de Oliveira AC, Martinez VP, Giret MT, de Menezes Succi RC, et al. Variability of HIV-1 genomes among children and adolescents from São Paulo, Brazil. PLoS One. 2013; 8(5):e62552 doi: 10.1371/journal.pone.0062552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pessôa R, Loureiro P, Esther Lopes M, Carneiro-Proietti AB, Sabino EC, Busch MP, et al. Ultra-Deep Sequencing of HIV-1 near Full-Length and Partial Proviral Genomes Reveals High Genetic Diversity among Brazilian Blood Donors. PLoS One. 2016; 11(3):e0152499 doi: 10.1371/journal.pone.0152499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardoso LPV, Stefani MMA. High level of multidrug resistance mutations in HIV type 1 pol gene and resistance-associated mutations to enfuvirtide (T-20) among antiretroviral-experienced patients from central Brazil. AIDS Res Hum Retroviruses. 2009; 25:943–950. doi: 10.1089/aid.2009.0060 [DOI] [PubMed] [Google Scholar]
- 28.Cardoso LPV, Pereira GAS, Viegas AA, Schmaltz LEPR, Stefani MMA. HIV-1 primary and secondary antiretroviral drug resistance and genetic diversity among pregnant women from Central Brazil. Journal of Medical Virology. 2010; 82:351–357. doi: 10.1002/jmv.21722 [DOI] [PubMed] [Google Scholar]
- 29.Cardoso LP, da Silveira AA, Francisco RB, da Guarda Reis MN, Stefani MM. Molecular characteristics of HIV type 1 infection among prisoners from Central Western Brazil. AIDS Res Hum Retroviruses. 2011; 27(12):1349–1353. doi: 10.1089/aid.2011.0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferreira AS, Cardoso LP, Stefani MM. Moderate prevalence of transmitted drug resistance and high HIV-1 genetic diversity in patients from Mato Grosso State, Central Western Brazil. J Med Virol. 2011; 83(8):1301–1307. doi: 10.1002/jmv.22128 [DOI] [PubMed] [Google Scholar]
- 31.Carvalho BC, Cardoso LP, Damasceno S, Stefani MM. Moderate prevalence of transmitted drug resistance and interiorization of HIV type 1 subtype C in the inland North State of Tocantins, Brazil. AIDS Res Hum Retroviruses. 2011; 27(10):1081–1087. doi: 10.1089/AID.2010.0334 [DOI] [PubMed] [Google Scholar]
- 32.Silveira AA, Cardoso LP, Francisco RB, de Araújo Stefani MM. HIV type 1 molecular epidemiology in pol and gp41 genes among naïve patients from Mato Grosso do Sul State, central western Brazil. AIDS Res Hum Retroviruses. 2012; 28(3):304–307. doi: 10.1089/aid.2011.0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alcântara KC, Lins JB, Albuquerque M, Aires LM, Cardoso LP, Minuzzi AL, et al. HIV-1 mother-to-child transmission and drug resistance among Brazilian pregnant with high Access to diagnosis and prophylactic measures. Journal of Clinical Virology. 2012; 54:15–20. doi: 10.1016/j.jcv.2012.01.011 [DOI] [PubMed] [Google Scholar]
- 34.da Costa ZB, de Lima YA, Martelli CM, Stefani MM. Transmitted HIV resistance among pregnant young women infected with HIV-1 in Brazil. AIDS Patient Care STDS. 2013; 27(8):439–41. doi: 10.1089/apc.2012.0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moura ME, Reis MN, Lima YA, Eulálio KD, Cardoso LP, Stefani MM. Low rate of transmitted drug resistance may indicate low access to antiretroviral treatment in Maranhão State, northeast Brazil. AIDS Res Hum Retroviruses. 2015a; 31(2):250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moura ME, da Guarda Reis MN, Lima YA, Eulálio KD, Cardoso LP, Stefani MM. HIV-1 transmitted drug resistance and genetic diversity among patients from Piauí State, Northeast Brazil. J Med Virol. 2015b; 87(5):798–806. [DOI] [PubMed] [Google Scholar]
- 37.Lima YA, Cardoso LP, Reis MN, Stefani MM. Incident and long term HIV-1 infection among pregnant women in Brazil: Transmitted drug resistance and mother-to-child transmission. J Med Virol. 2016; 88(11):1936–43. doi: 10.1002/jmv.24540 [DOI] [PubMed] [Google Scholar]
- 38.Delwart EL, Shpaer EG, Louwagie J, McCutchan FE, Grez M, Rubsamen-Waigmann H, et al. Genetic relationship determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993; 19;262(5137):1257–61. [DOI] [PubMed] [Google Scholar]
- 39.Sierra M, Thomson MM, Rios M, Casado G, Castro RO, Delgado E, et al. The analysis of near full-length genome sequences of human immunodeficiency virus type 1 BF intersubtype recombinant viruses from Chile, Venezuela and Spain reveals their relationship to diverse lineages of recombinant viruses related to CRF12_BF. Infect. Genet. Evol. 2005; 5,209–217. doi: 10.1016/j.meegid.2004.07.010 [DOI] [PubMed] [Google Scholar]
- 40.Passaes CP, Bello G, Lorete RS, Matos Almeida SE, Junqueira DM, Veloso VG, et al. Genetic characterization of HIV-1 BC recombinants and evolutionary history of the CRF31_BC in Southern Brazil. Infect Genet Evol. 2009; 9(4):474–82. doi: 10.1016/j.meegid.2009.01.008 [DOI] [PubMed] [Google Scholar]
- 41.Thompson JD, Higgins DS, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nei M & Kumar S. Molecular Phylogenetics and Evolution. Oxford University. 2002; 3:567–568 [Google Scholar]
- 43.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980; 16:111–120. [DOI] [PubMed] [Google Scholar]
- 44.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013; 30(12): 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999; 73(1):152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia X., Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 2001; 92,371–373. [DOI] [PubMed] [Google Scholar]
- 47.Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002; 161:1307–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007; 7:214 doi: 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suchard MA, Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics. 2009; 25:1370–1376. doi: 10.1093/bioinformatics/btp244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005; 22:1185–1192. doi: 10.1093/molbev/msi103 [DOI] [PubMed] [Google Scholar]
- 51.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006; 4:e88 doi: 10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rambaut A, Drummond A. 2007 Tracer v1.6. Available from http://treebioedacuk/software/tracer/
- 53.Carr JK, Avila M, Gomez Carrillo M, Salomon H, Hierholzer J, Watanaveeradej V, et al. Diverse BF recombinants have spread widely since the introduction of HIV-1 into South America. AIDS. 2001; 19;15(15):F41–7. [DOI] [PubMed] [Google Scholar]
- 54.Bello G, Aulicino PC, Ruchansky D, Guimarães ML, Lopez-Galindez C, Casado C, et al. Phylodynamics of HIV-1 Circulating Recombinant Forms 12_BF and 38_BF in Argentina and Uruguay. Retrovirology. 2010; 7:22 doi: 10.1186/1742-4690-7-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ristic N, Zukurov J, Alkimim W, Diaz RS, Janini LM, Chin MP. Analysis of the origin and evolutionary history of HIV-1 CRF28_BF and CRF29_BF reveals a decreasing prevalence in the AIDS epidemic of Brazil. PLoS One. 2011; 6(3):e17485 doi: 10.1371/journal.pone.0017485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Sa-Filho DJ, Ambar RF, Duarte NB, Matias RB, Candido V, Gagliani LH, et al. HIV type 1 diversity from newly diagnosed patients in Santos metropolitan area/Brazil. AIDS Res Hum Retroviruses. 2009; 25(9):925–9. doi: 10.1089/aid.2009.0073 [DOI] [PubMed] [Google Scholar]
- 57.Monteiro-Cunha JP, Araujo AF, Santos E, Galvao-Castro B, Alcantara LC. Lack of high-level resistance mutations in HIV type 1 BF recombinant strains circulating in northeast Brazil. AIDS Res Hum Retroviruses. 2011; 27(6):623–31. doi: 10.1089/AID.2010.0126 [DOI] [PubMed] [Google Scholar]
- 58.Santos LA, Monteiro-Cunha JP, Araujo AF, Brites C, Galvao-Castro B, Alcantara LC. Detection of distinct human immunodeficiency virus type 1 circulating recombinant forms in northeast Brazil. J Med Virol.2011; 83(12):2066–72. doi: 10.1002/jmv.22170 [DOI] [PubMed] [Google Scholar]
- 59.Barreto CC, Nishyia A, Araújo LV, Ferreira JE, Busch MP, Sabino EC. Trends in antiretroviral drug resistance and clade distributions among HIV-1 infected blood donors in Sao Paulo, Brazil. J Acquir Immune Defic Syndr. 2006; 41(3):338–41. doi: 10.1097/01.qai.0000199097.88344.50 [DOI] [PubMed] [Google Scholar]
- 60.Prellwitz IM, Alves BM, Ikeda ML, Kuhleis D, Picon PD, Jarczewski CA, et al. HIV behind bars: human immunodeficiency virus cluster analysis and drug resistance in a reference correctional unit from southern Brazil. PLoS One.2013; 9;8(7):e69033 doi: 10.1371/journal.pone.0069033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pregnant: women infected with HIV-1 attending a regional antenatal care; Naïve: antiretroviral naïve patients; HAART: Patients under highly active antiretroviral therapy. * Ref 29: the study group (n = 27) comprises prisoner patients recruited in Goiania/GO (n = 7) and in Campo Grande (n = 20).
(DOCX)
*Some primers had their original sequence modified based on the alignment of subtypes B, C and F1 HIV sequence compendium (2005) from HIV Los Alamos Database.
(DOCX)
Data Availability Statement
All full, near full and partial genome sequences of HIV-1 are available from the GenBank database accession numbers KY628215-KY628225.
All HIV-1 sequences generated in this study were deposited in the GenBank database (KY628215-KY628225).