Abstract
Flap endonuclease 1 (FEN1) is a structure selective endonuclease required for proficient DNA replication and the repair of DNA damage. Cellularly active inhibitors of this enzyme have previously been shown to induce a DNA damage response and, ultimately, cell death. High-throughput screens of human cancer cell-lines identify colorectal and gastric cell-lines with microsatellite instability (MSI) as enriched for cellular sensitivity to N-hydroxyurea series inhibitors of FEN1, but not the PARP inhibitor olaparib or other inhibitors of the DNA damage response. This sensitivity is due to a synthetic lethal interaction between FEN1 and MRE11A, which is often mutated in MSI cancers through instabilities at a poly(T) microsatellite repeat. Disruption of ATM is similarly synthetic lethal with FEN1 inhibition, suggesting that disruption of FEN1 function leads to the accumulation of DNA double-strand breaks. These are likely a result of the accumulation of aberrant replication forks, that accumulate as a consequence of a failure in Okazaki fragment maturation, as inhibition of FEN1 is toxic in cells disrupted for the Fanconi anemia pathway and post-replication repair. Furthermore, RAD51 foci accumulate as a consequence of FEN1 inhibition and the toxicity of FEN1 inhibitors increases in cells disrupted for the homologous recombination pathway, suggesting a role for homologous recombination in the resolution of damage induced by FEN1 inhibition. Finally, FEN1 appears to be required for the repair of damage induced by olaparib and cisplatin within the Fanconi anemia pathway, and may play a role in the repair of damage associated with its own disruption.
Introduction
Flap endonuclease 1 (FEN1) is a structure-specific endonuclease and prototypical member of the RAD2-superfamily [1–3], required for the removal of 5’ flaps that arise as a consequence of Okazaki fragment displacement by replicative polymerases during lagging strand synthesis [4, 5]. This process is critical for proficient and processive replication, with many cancer cells showing over-expression of FEN1 [6–9]. Haploinsufficiency of FEN1 is associated with abnormal cell-cycle progression and cancer predisposition with decreased survival, driven by an accumulation of replication-associated alterations in DNA, such as microsatellite instabilities (MSI) and tri-nucleotide repeat expansion [10–12]. FEN1 also plays a role in the maintenance of telomeres in the absence of telomerase [13], the processing of stalled replication forks [14, 15], and in a number of DNA damage repair processes, including base excision repair (BER) [16], alternative end-joining (alt-EJ) [17] and homologous recombination (HR) [18]. As a result, cells defective for FEN1 activity are sensitive to many DNA lesions [15, 19–24] and, therefore, FEN1 is an attractive target for drug discovery.
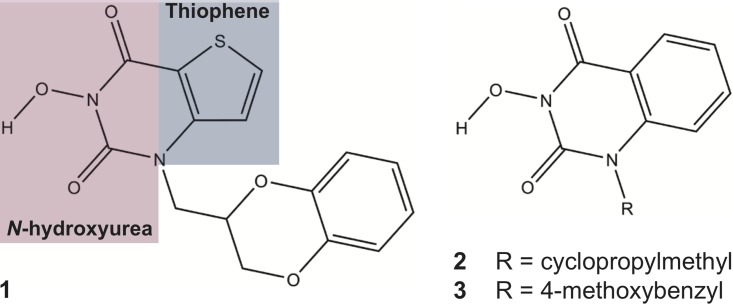
Previously it has been shown that the N-hydroxyurea series small molecule 1-(2,3-dihydro-1,4-benzodioxin-2-ylmethyl)-3-hydroxythieno[2,3-e]pyrimidine-2,4-dione (compound 1; Fig 1) can inhibit FEN1 activity in vitro [25, 26]. We have shown that compound 1 co-crystallizes within the active site of FEN1 in vitro, with inhibition achieved partly through the co-ordination of Mg2+ ions by the N-hydroxyurea moiety of 1 (shaded purple in Fig 1) [25]. These metal ions are critical for base un-pairing; the disruption of base pairing within the substrate, close to the junction of single-strand to double-strand DNA, which induces an essential conformational change within the substrate duplex to allow for enzymatic cleavage [2, 27]. Further, the thiophene ring of 1 (shaded blue in Fig 1) sits within a hydrophobic pocket of FEN1 containing conserved active site residues Y40, D181 and R100 essential for substrate positioning [2, 27, 28]. Binding of 1 to the active site occurs in both a competitive and non-competitive manner with DNA, and, potentially, induces a dominant negative effect, with a FEN1-inhibitor ‘dead-end’ complex associating into replication and repair complexes, such as the PCNA-Pol∂-FEN1 complex that stimulates Okazaki fragment maturation [29, 30]. Further members of this N-hydroxyurea series (compounds 2–3; Fig 1) have similarly been shown to inhibit FEN1 activity in biochemical assays [31]. This inhibitory activity is largely competitive with substrate, suggesting a different mode of action to 1 [25]. In the present study, we investigate further the therapeutic potential of N-hydroxyurea series inhibitors of FEN1.
Fig 1. Chemical structure of N-hydroxyurea series inhibitors of FEN1 used in this study.
Disruption or inhibition of FEN1 in both higher and lower eukaryotes induces a DNA damage response. Saccharomyces cerevisiae cells deficient for the FEN1 homologue RAD27 display temperature-dependent hyper-activation of post-replication repair (PRR) and DNA double-strand break (DSB) repair pathways following accumulation of unprocessed Okazaki fragments [19, 32, 33]. Previously [25] we demonstrated that N-hydroxyurea series molecules bind FEN1 protein ex vivo and that this binding translates to cellular activity, with mammalian cells treated with 1 initiating a DNA damage response in a dose-dependent manner. Similarly, we demonstrated that FEN1 inhibition activated the ATM checkpoint signalling pathway, the phosphorylation of histone H2AX and the ubiquitination of FANCD2 [25], suggesting the initiation of the Fanconi anemia (FA) pathway. The FA pathway is required for the stabilisation of stalled replication forks (Fig 2) and these data suggest that 1 induces replication-associated DNA damage. Moreover, it has been shown that inhibition of FEN1 is synthetic lethal with deficiencies in MRE11A [34], a member of the MRN (MRE11A-RAD50-NBS1) complex required for the sensing of DSBs and the activation of cell-cycle checkpoints (Fig 2) [35–38]. Following the sensing of DSBs by the MRN complex the kinases ATM and ATR induce DNA damage checkpoint signalling and repair is conducted via two main pathways: HR and non-homologous end-joining (NHEJ) (Fig 2). Previous data has suggested that mutation of FEN1 in Saccharomyces cerevisiae and humans is synthetically lethal with HR factors [32, 39–41]. In the current study, we further investigate the activity of N-hydroxyurea series inhibitors of FEN1 in human cancer cells by high-throughput and targeted means. We identify synthetic genetic interactions between these inhibitors and disruption of DNA damage repair genes, and our data suggests FEN1 as a potential target for drug discovery in DNA repair-deficient cancers.
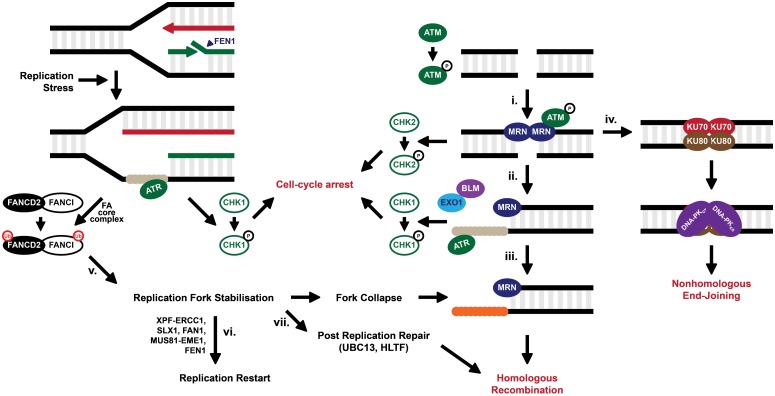
Fig 2. Model for the repair of DNA double-strand breaks.
DSBs are recognised by the MRN complex (i.) which binds to the blunt DNA ends, holding them in close proximity. The exonuclease activity of MRE11A (or EXO1 in concert with BLM) is able to resect blunt ends, creating a 3’ ssDNA overhang, which becomes coated by the single-strand binding protein RPA (ii.), signalling for cell-cycle checkpoint arrest via ATR. RPA is displaced by RAD51 (iii.) to allow for HR. Alternatively, the MRN complex can be replaced by KU78/KU80 complex (iv.), protecting DNA ends from resection and promoting NHEJ through the binding of the DNA-PK catalytic subunit (DNA-PKcs). DSBs formed as a consequence of replication fork collapse require HR for their repair. Fork stalling, following replication stress for example, activates the FA pathway in an attempt to stabilise and protect the fork (v.). The FA core complex recognises the stalled fork and ubiquitinates the FANCD2-FANCI heterodimer. Stalled forks can be further processed by structure selective endonucleases to restore the replication fork (vi.) or cleave the fork to produce a DSB (vii.). Alternatively, the post-replication machinery can bypass damaged bases at stalled forks. One such pathway leads to template switching in a HR-mediated pathway.
Results and discussion
A high-throughput screen identifies potential cancer sub-populations sensitive to chemical inhibitors of FEN1
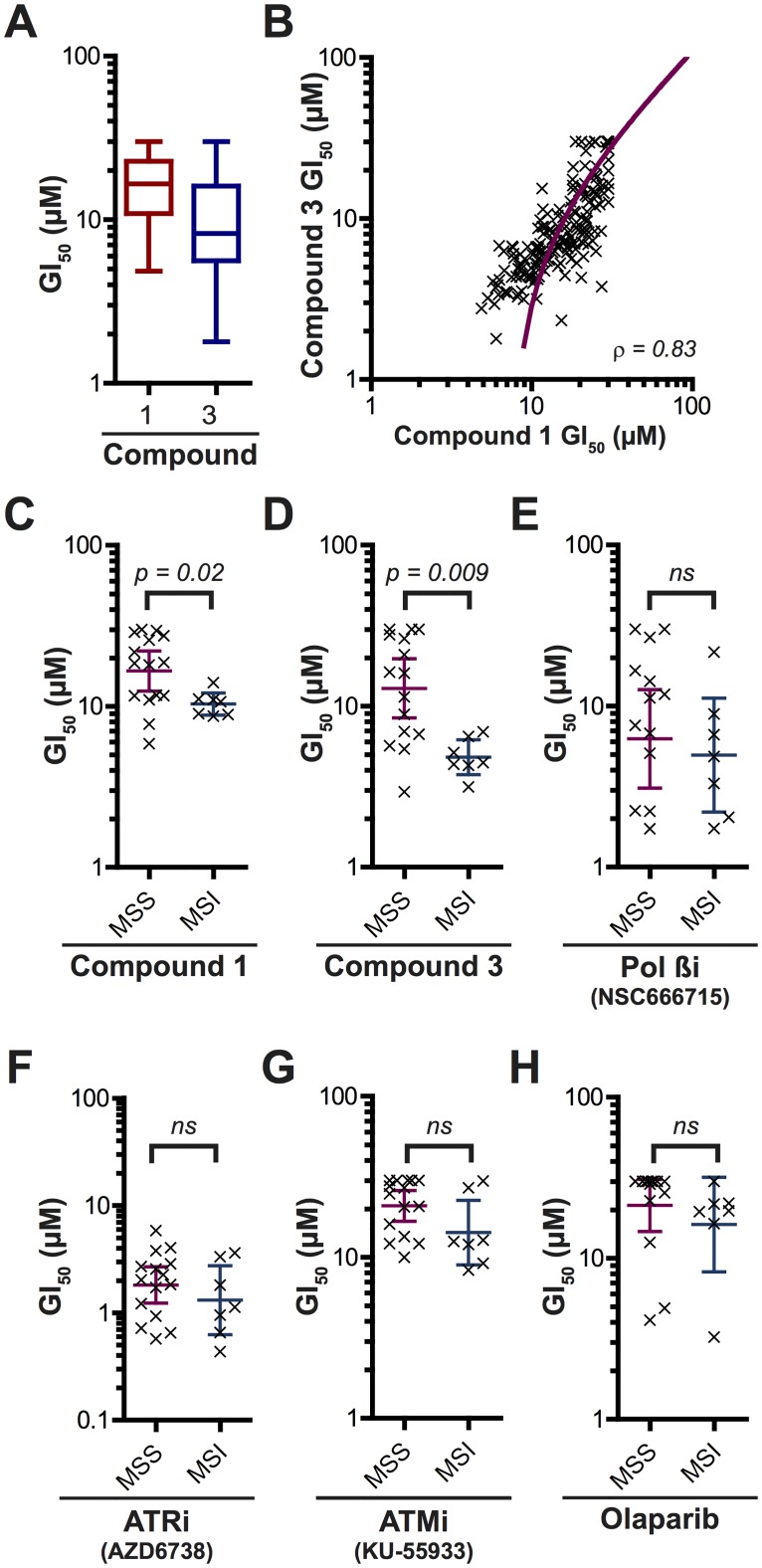
Previously we have shown that chemical inhibition of FEN1 leads to DNA damage accumulation and ultimately cell death. We wanted to further examine the cytotoxic efficacy of FEN1 inhibition, to determine whether there was any tissue or genetic influence on those cells sensitive to treatment with these compounds. Initially, we conducted high-throughput screens across a panel of 280 cancer cell-lines derived from solid tumours from a number of tissue types. We continuously dosed each cell-line with 0–30 μM of three previously identified FEN1 inhibitors (compounds 1–3) from the N-hydroxyurea series for 3 days and measured their growth inhibition effect (Table 1, S1 Table and Fig 3A). Dose-response results were obtained from 212 cell-lines treated with 1 with a mean GI50 of 15.5 μM. We were unable to calculate GI50 values for 26 of these cell-lines within the dose-range tested (i.e. the GI50 was greater than 30 μM), and these cell lines were deemed to be ‘resistant’ by this assay. N-hydroxyurea series compounds 2–3 similarly are inhibitors of FEN1 shown to have activity in biochemical assays [25, 26, 31]. FEN1 binding and inhibition by these molecules fit a competitive model while compound 1 fits a mixed non-competitive/competitive model, binding to FEN1 either in the presence or absence of DNA substrate [25]. We see little cellular activity for 2 in our high-throughput screen, with 79% cell-lines showing resistance to treatment with the dose-range specified. 3 is a structural relative of 2, replacing a cyclopropylmethyl side-chain with a longer 4-methoxyphenyl. This modification enhanced cellular activity with a mean GI50 of 9.0 μM in the 195 cell-lines for which data was obtained, and 30 cell-lines (15%) showing resistance (Fig 3A). There was good Pearson’s correlation (ρ = 0.86) between sensitivity to 1 and 3 (Fig 3B), suggesting that cellular activity is likely due to the common inhibition of FEN1 despite the differing modes of inhibition.
Table 1. Summary of a high-throughput study to identify cell-lines sensitive to FEN1 inhibitors.
| Compound | 1 | 2 | 3 |
|---|---|---|---|
| Cell-lines screened | 212 | 180 | 195 |
| Resistant cell-linesa | 26 (12%) | 143 (79%) | 30 (15%) |
| Geometric mean | 15.5 | 27.3 | 9.0 |
| 95% Confidence interval | 1.0 | 0.7 | 1.2 |
a Resistance was defined as GI50 greater than the maximum dose used in this study (30 μM).
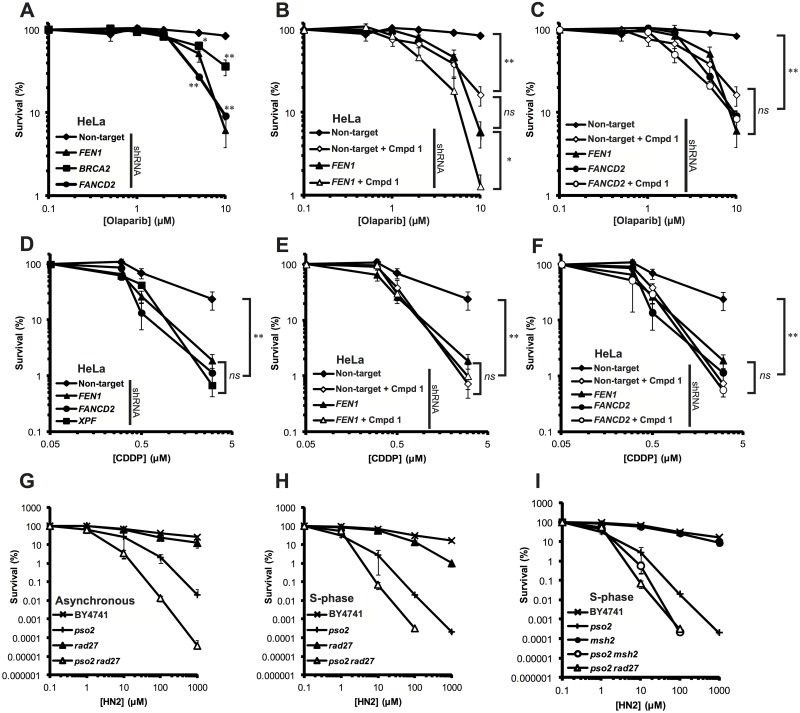
Fig 3. Cell-lines with MSI are specifically sensitive to chemical inhibition of FEN1 by compounds 1 and 3.
A. Box and whisker diagram to show the variation in GI50 values for cell-lines subject to treatment with 1 and 3. B. Correlation between the sensitivity of cell-lines to 1 and 3. C-H. The influence of MSI on sensitivity to DNA repair inhibitors. ns = not significant * p < 0.05. ** p < 0.005.
We wanted to determine whether there was any tissue-type specificity to FEN1 inhibitor sensitivity. We therefore split the cell-lines into broad panels by tissue of origin and compared GI50 (Tables 2–4 and S1–S3 Figs). While cell-lines derived from head and neck cancers appear to be more resistant to treatment with 3 than other panels, there was otherwise very little difference between the mean GI50 across the tissue-types with any of the compounds tested. Subsequently, we further separated the cell-lines according to known cancer sub-types, with no significant difference in the mean GI50 of these cancer sub-types for any of the compounds tested (see S1–S3 Figs). Next we examined the sensitivity in cell-lines with mutations in genes associated with cancer (KRAS, p53 and PTEN), again with no significant difference in sensitivity to FEN1 inhibition for all compounds tested (see S1–S3 Figs). Finally, we compared cell-lines with known MSI with cells shown to have stable microsatellites (MSS) from the same tissue-type (Table 5). MSI is a form of genetic hypermutability associated with variations in polynucleotide tracts. Approximately 10–15% of colorectal cancers (CRC) and ovarian cancers display MSI associated with a failure in genome surveillance by the mismatch repair (MMR) machinery, while MSI is also evident in head and neck, lung, oesophageal, pancreatic and prostate cancers [42–45]. Waterfall plots suggest that cell-lines with MSI were, largely, more sensitive to treatment with 1 and 3, falling below the mean GI50 for all cell-lines tested (green bars, panel H in S1 Fig and panel H in S3 Fig) and showed a statistically significant increase in sensitivity compared with MSS cell-lines (Fig 3C and 3D). These data suggest MSI as a potential cancer subset with selective sensitivity to FEN1 inhibition. We considered that this effect may be through the inhibition of BER, a DNA repair pathway in which FEN1 plays a role. We therefore screened MSI and MSS cell-lines against another BER inhibitor, the cellular active DNA Polymerase β inhibitor NSC666715 [46, 47] (Fig 3E). No significant difference between the two cell populations was seen. Similarly, we screened these cell panels for sensitivity to inhibitors of other DNA damage response elements, including the ATR inhibitor AZD6738 [48], ATM inhibitor KU-55933 [49] or PARP inhibitor olaparib [50] (Fig 3F–3H). No significant difference in sensitivity to these inhibitors was seen between the two populations. These data would suggest that toxicity of compounds 1–3 in MSI cells is specific and not due to an addiction to the DNA damage response more generally.
Table 2. Tissue-specific sensitivity to compound 1.
a Resistance was defined as GI50 greater than the maximum dose used in this study (30 μM).
| Compound 1 | Bladder | Breast | Colorectal | Gastric | Liver | Lung | Pancreatic | Prostate | Gynae | Head & Neck |
|---|---|---|---|---|---|---|---|---|---|---|
| Cell-lines screened | 19 | 22 | 21 | 25 | 21 | 37 | 24 | 7 | 21 | 15 |
| Resistant cell-linesa | 0 (0%) | 2 (9%) | 4 (19%) | 5 (20%) | 3 (14%) | 2 (5%) | 3 (13%) | 0 (0%) | 7 (33%) | 0 (0%) |
| Geometric meanb | 13.6 | 13.1 | 16.7 | 15.0 | 20.1 | 12.1 | 16.5 | 17.1 | 23.1 | 20.0 |
| 95% Confidence interval | 2.8 | 3.0 | 3.8 | 3.5 | 2.7 | 2.1 | 2.8 | 3.6 | 2.5 | 2.6 |
Table 4. Tissue-specific sensitivity to compound 3.
a Resistance was defined as GI50 greater than the maximum dose used in this study (30 μM).
| Compound 3 | Bladder | Breast | Colorectal | Gastric | Liver | Lung | Pancreatic | Prostate | Gynae | Head & Neck |
|---|---|---|---|---|---|---|---|---|---|---|
| Cell-lines screened | 19 | 22 | 21 | 25 | 21 | 40 | 24 | 7 | 1 | 15 |
| Resistant cell-linesa | 1 (5%) | 1 (5%) | 5 (24%) | 6 (24%) | 4 (19%) | 2 (5%) | 2 (8%) | 0 (0%) | 0 (0%) | 9 (60%) |
| Geometric meanb | 8.2 | 6.4 | 11.0 | 11.5 | 11.0 | 7.4 | 8.1 | 9.2 | 5.2 | 19.8 |
| 95% Confidence interval | 2.9 | 2.6 | 4.6 | 4.2 | 3.8 | 2.0 | 3.2 | 3.0 | 4.9 |
Table 5. MRE11A poly(T)11 mutations associated with MSI.
| Cell-Line | MSI Status | MMR Defect | MRE11A poly(T)11 Intron 4 1 |
|---|---|---|---|
| HCT 116 | MSI-H | MLH1 | T10/T9 |
| HCT-15/DLD-1 | MSI-H | MSH6 | T11/T9 |
| LoVo | MSI-H | MSH2 | T10/T10 |
| RKO | MSI-H | MLH1 | T10/T9/T8 |
| SW48 | MSI-H | MLH1 | T10/T10 |
| SNU-1 | MSI-H | MLH1 | - |
| SNU-638 | MSI-H | MLH1 | - |
| COLO 205 | MSS | None | T11/T11 |
| COLO 320DM | MSS | None | - |
| HT-29/WiDr | MSS | None | T11/T11 |
| SK-CO-1 | MSS | None | - |
| SW403 | MSS | None | T11/T11 |
| SW480 | MSS | None | T11/T11 |
| SW620 | MSS | None | T11/T11 |
| SW948 | MSS | None | - |
| SNU-16 | MSS | None | - |
| SNU-216 | MSS | None | - |
| SNU-484 | MSS | None | - |
| SNU-5 | MSS | None | - |
| SNU-601 | MSS | None | - |
| SNU-620 | MSS | None | - |
| SNU-668 | MSS | None | - |
| HCT116 CHR3 | MSS | None2 | T10/T9 |
Table 3. Tissue-specific sensitivity to compound 2.
a Resistance was defined as GI50 greater than the maximum dose used in this study (30 μM).
| Compound 2 | Bladder | Breast | Colorectal | Gastric | Liver | Lung | Pancreatic | Prostate | Gynae |
|---|---|---|---|---|---|---|---|---|---|
| Cell-lines screened | 19 | 22 | 21 | 25 | 21 | 40 | 24 | 7 | 1 |
| Resistant cell-linesa | 18 (95%) | 14 (64%) | 15 (71%) | 17 (68%) | 19 (90%) | 32 (80%) | 21 (88%) | 6 (86%) | 1 (100%) |
| Geometric meanb | 29.0 | 27.6 | 28.9 | 26.9 | 29.6 | 28.9 | 28.8 | 29.3 | |
| 95% Confidence interval | 1.5 | 1.4 | 0.9 | 1.8 | 0.5 | 0.8 | 1.2 | 1.2 |
Colorectal cancer cell-lines with microsatellite instability-induced MRE11A deficiency are sensitised to down-regulation of FEN1
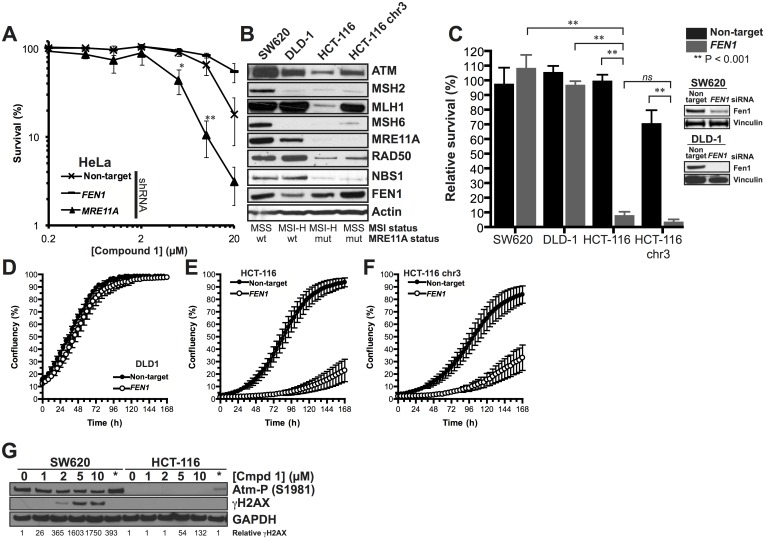
DNA repair genes are frequently co-mutated with MMR due to polymorphisms at microsatellites resulting from an accumulation of replication errors. MRE11A for example is commonly mutated at a microsatellite comprising a tract of 11 thymine residues (poly(T)11 microsatellite) in intron 4 [51–54]. Previously, MRE11A has been shown to be synthetic lethal with FEN1 [33, 35] and we therefore wanted to investigate the possibility that the selective sensitivity of FEN1 inhibitors is due to a loss of MRE11A. Of the seven confirmed MSI cell-lines examined in our high-throughput screen, five have been shown to have deleterious poly(T)11 mutations in one or more alleles of MRE11A (Table 5). We examined the role of MRE11A in the tolerance of 1 by initially treating cells stably expressing shRNA against MRE11A with the compound and compared sensitivity to an isogenic control cell-line expressing a non-targeting shRNA (Fig 4A). Cells disrupted for MRE11A were more sensitive to a dose escalation of 1, and at 10 μM 1 we saw 11% survival in the MRE11A deficient cell-line compared to 66% in the control cell-line.
Fig 4. Sensitivity of MSI cell-lines to FEN1 inhibition is through MRE11A deficiency.
A. Cells down-regulated for MRE11A by shRNA are sensitive to FEN1 inhibitor 1. B. Confirmation that the MSI cell-line HCT-116 is disrupted for the MRN complex. Re-introduction of chromosome 3 (HCT-116 chr3) restores MLH1 without affecting MRE11A protein level. C-F. Down-regulation of FEN1 by siRNA is toxic in MSI cells devoid of MRN complex by clonogenic survival (C) or by measuring proliferation (D-F). Each data point is the mean of at least 3 individual repeats and the error bars represent the standard error. Significance was determined by student t-test. ns = not significant * p < 0.05. ** p < 0.005 G. HCT-116 cells fail to signal for the DNA damage response upon treatment with 1.
Next, we wanted to examine further whether inhibition of FEN1 was responsible for lethality in MRE11A deficient MSI cells. We constructed a mini-panel of colorectal cancer cell-lines that differ by MSI-status and the mutation status of the MRE11A poly(T)11 microsatellite (Fig 4B), subject them to siRNA against FEN1 and scored for clonogenic survival (Fig 4C) or ability to proliferate (Fig 4D–4F). SW620, an MSS cell-line [55], MRE11A expressing cell-line showed no sensitivity to FEN1 disruption (Fig 4C & 4D). DLD-1 cells have MSI [55], yet these cells retain MRE11A protein as only one MRE11A poly(T)11 allele is mutated. Again, these cells are insensitive to FEN1 siRNA treatment (Fig 4C). HCT-116 cells are similarly MSI [56], however, mutation of both MRE11A poly(T)11 alleles means this cell-line is devoid of MRE11A protein (Fig 4B). These cells are sensitive to FEN1 siRNA treatment (Fig 4C & 4E), suggesting that disruption of FEN1 function by 1 is responsible for toxicity in this cell line. To examine the role played by MMR in sensitivity to FEN1 disruption, we also examined HCT-116 cells with chromosome 3 re-introduced, restoring MLH1. This has been shown to restore MMR function in these cells [57], without affecting MRE11A levels (Fig 4B). Both MMR-deficient and MMR-proficient cells were similarly sensitive to siRNA against FEN1 (Fig 4C & 4F), suggesting that the toxicity is specific MRE11A defects and not MMR.
Our previous data suggested that 1 inhibits both FEN1 and EXO1 function in vitro but we were unable to determine whether this corresponded with cellular activity [26]. In an attempt to determine whether disruption of EXO1 had an effect on the survival of MRE11A-deficient MSI cells, we also knocked-down EXO1 in SW620 and HCT-116 cells and examined clonogenic potential. In both cases, disruption of EXO1 had little effect on viability when compared to a non-target control (panel A in S4 Fig). Finally, we wanted to examine whether the disruption of EXO1 played a role in sensitivity to 1, explaining the severe and non-specific toxicity of 1 at high doses [26], we down-regulated EXO1 by siRNA and subject the cells to increasing concentrations of 1 (panel B in S4 Fig). We find that cells disrupted for EXO1 are no more sensitive to cells treated with a non-target control. This would suggest that the cytotoxicity induced by 1 is unlikely to be due to inhibition of EXO1 activity.
Finally, we wanted to examine the induction of DNA damage signalling pathways in these cells. Previously we have shown that treatment with 1 induces a DNA damage response [26]. In cells devoid of MRE11A, this signal is lost, with HCT-116 cells failing to accumulate γH2AX or activated ATM following treatment with 1 or olaparib (Fig 4G), suggesting that MRE11A is required for the activation of signalling pathways induced by damage associated with FEN1 or PARP inhibition.
Cells devoid of the DNA double-strand break sensing machinery are sensitive to chemical inhibition of FEN1
The MRN complex recognises DSBs and forms part of the signalling cascade that leads to damage induced cell-cycle arrest (Fig 2). This signalling cascade involves the activation of the kinase ATM by MRN [58]. ATM is activated when FEN1 is inhibited [26] and we hypothesised that cells deficient for ATM would likely be sensitive to 1. Moreover, while the majority of MSI cell-lines identified above as sensitive to 1 are mutated for MRE11A, two have mutations in exon 6 of ATM resulting in a 22 bp deletion in exon 6 (SNU-1 and SNU-638) [59]. When we examined cells expressing shRNA against ATM for sensitivity to 1, we found that the down-regulation of ATM lead to a potentiation of the toxic effects of FEN1 inhibition, phenocopying cells disrupted for MRE11A (Fig 5A). We further examined sensitivity in FaDu cells with all copies of ATM knocked out using the transcription activator-like effector nuclease (TALEN) system [60] with compound 1. These cells also displayed greater toxicity than their parental cell-line (Fig 5B). Similarly, knock-down of FEN1 by siRNA was toxic in these cells (Fig 5C), suggesting specificity in the activity of 1 in ATM deficient cells, further confirming synthetic lethality between FEN1 and ATM.
Fig 5. Cells disrupted for pathways required to maintain replication fork stability are sensitive to inhibition of FEN1.
A. Clonogenic survival of cells stably expressing shRNA against MRE11A, ATM or a non-targeting control when treated with 1. B. Clonogenic survival of FaDu cells or FaDu cells with all three ATM alleles knocked-out when treated with 1. C. Clonogenic survival of FaDu cells or FaDu cells with all three ATM alleles knocked-out when treated with siRNA against FEN1. D. DNA damage response induced in cells expressing shRNA against FEN1, ATM BRCA2, FANCD2 and a non-target control. E-F. Clonogenic survival of cells disrupted for FANCD2 by shRNA (E) or genes required for PRR (UBC13, HLTF) by siRNA (F) compared to a non-target control. (G) Accumulation of Rad51 foci in cells treated with 1. Data is collected from at least 500 cells per treatment. H. Clonogenic survival of cells disrupted for genes required for HR (BRCA2, BLM) and NHEJ (DNA-PKcs) by shRNA compared to a non-target control. In each clonogenic assay, data points represent the mean of at least 3 individual repeats and the error bars represent the standard error. Significance was determined by Student t-test. ns = not significant * p < 0.05. ** p < 0.005 D. DNA damage response induced in cells expressing shRNA against FEN1, ATM and a non-target control.
Above we have shown that disruption of the MRN complex leads to both a loss in the activation of ATM and the accumulation of γH2AX. We examined 1-induced γH2AX accumulation in cells disrupted for ATM by shRNA and found that, in contrast to MRE11A disruption, γH2AX accumulated more readily in ATM disrupted cells (Fig 5D). These data would suggest that the MRN complex activates ATM in response to FEN1 inhibition but ATM is not required for H2AX phosphorylation in this situation.
A role for replication-coupled repair pathways in the tolerance of FEN1 inhibition
The MRN-ATM pathway has a role in the repair of DSBs repaired by NHEJ and canonical HR, such as those described in Fig 2i.–iv., and it is possible that the activation of this pathway in cells inhibited for FEN1 is due to an accumulation of such DNA breaks. However, this pathway has similarly been shown to be activated following the accumulation of more complex DNA secondary structures, such as stalled replication forks [61] and has been shown to promote replication restart following the collapse of the stalled forks into DSBs [62, 63]. Stalled replication forks are recognised by members of the FA pathway, which bind and stabilise the fork while co-ordinating downstream repair processes through monoubiquitinated FANCD2 (FANCD2-Ub) (Fig 2) [64–70]. An accumulation of ubiquitinated FANCD2 is therefore used as a marker for replication fork instability. Inhibition of FEN1, by treating with compound 1 or shRNA against FEN1, leads to an increase in FANCD2 mono-ubiquitination (FANCD2-Ub; panel F in S4 Fig and [26]), suggesting that inhibition of FEN1 leads to the accumulation of unstable replication forks. We hypothesised therefore that disruption of the FA pathway would lead to increased sensitivity to 1. We obtained cells downregulated for FANCD2 by shRNA and treated them with increasing doses of 1. We find that, not only are these cells sensitive to 1, this sensitivity is far greater than in any other cell-line examined (Fig 5E). At 10 μM 1, 1% of FANCD2 deficient cells survive compared with 66% in non-target control cells and approximately 10% in cells disrupted for MRE11A or ATM. Moreover, these cells accumulate DNA damage markers far more readily than in cells devoid of ATM (Fig 5D), suggesting that failure to stabilise stalled replication forks leads to increased DNA damage associate with 1 treatment. These data suggest that chemical inhibition of FEN1 leads to replication fork instabilities that, if left unrepaired, can lead to cell death.
In lower eukaryotes, disruption of RAD27/FEN1 is synthetic lethal with disruption of the post-replication repair (PRR) damage tolerance pathway [34, 71]. PRR is required to overcome physical replication blocks or nicks in the DNA and is activated by regions of ssDNA (Fig 2) [72]. In the absence of RA27, PRR is required to prevent trinucleotide repeat expansion [73], which is often associated with FEN1 haploinsufficiency in vertebrates [13]. It has previously been demonstrated that ubiquitination of PCNA occurs following RAD27 inactivation, a phenotype supressed by overexpression of EXO1 [34]. Inactivation of RAD27/FEN1 leads to an increase in length of 5’ flaps formed by replicative polymerases dissociating Okazaki fragments [74]. These longer flaps become coated in RPA and it has been postulated that these longer RPA-bound flaps signal for PRR [34]. Furthermore, disruption of RAD27 leads to an accumulation of un-ligated single-strand nicks at Okazaki fragment [20, 33], which may also signal for PRR during subsequent rounds of replication. We wanted to examine this in our mammalian system so disrupted UBC13 and the Rad5 homologue HLTF [75], which are PRR factors required for HR-mediated template switching [76–78], by siRNA and exposed cells to 1. We found that disruption of either gene led to increased sensitivity to 1, phenocopying the knock-down of MRE11A by the same method (Fig 5F). It is possible, therefore, that inhibition of FEN1 by this method leads to an accumulation of aberrant DNA replication structures on the lagging strand that engages PRR pathway for repair. In the absence of PRR, persistent replication fork instability ultimately results in cell death.
A role for homologous recombination proteins in the repair of damage induced by FEN1 inhibition
Disruption of RAD27 in lower eukaryotes is synthetically lethal with disruption of the HR pathway [32,39–40,79–80]. Since we have shown that inhibition of FEN1 largely recapitulates the phenotype seen in a yeast rad27 cell, we wanted to examine the role played by HR factors in the tolerance of chemical inhibition of FEN1. HR is primed by the resection of DNA ends by MRE11A, resulting in a 3’ ssDNA overhang that rapidly becomes coated in RPA (Fig 2). RPA is displaced by RAD51 to form protein-DNA filaments that promote recombination. To determine whether HR is activated by 1, we examined the accumulation of RAD51 foci post treatment, using the CellInsight high-content screening platform to count all cells containing defined RAD51 foci within the nucleus. We find that there is a dose-dependent increase in RAD51 foci following treatment with compound 1 (Fig 5G). The resulting foci are large, forming as a consequence of dynamic nuclear reorganisation post DNA damage [81, 82]. These foci are entirely nuclear (examples are shown in panel C of S4 Fig), however it is possible that they are foci prepared for sequestration to micronuclei, suggesting irreparable DNA damage [83]. This dose-dependent increase coincided with an accumulation of γH2AX foci (panel D of S4 Fig). Furthermore, when cells disrupted for BRCA2 (which is required, among other DNA repair processes, for canonical HR) by shRNA were subject to increasing concentrations of 1, we found increased sensitivity that phenocopied MRE11A and ATM disruption (Fig 5H). Disruption of BRCA2, however, did not increase γH2AX accumulation (Fig 5D), suggesting a role for BRCA2 downstream of MRE11A. Cells disrupted for DNA-PKcs, a gene required for the non-homologous end-joining (NHEJ)–a pathway utilised frequently for the repair of simple DSBs that supresses HR (see also Fig 2)–were no more sensitive to 1 than a non-target control.
Above we have shown that disruption of DNA damage repair proteins contributes to cellular sensitivity to FEN1 inhibition. We therefore re-examined our high-throughput cell killing data set to determine whether gene expression and copy number influence sensitivity to our compounds. We found that cellular sensitivity to N-hydroxyurea series compounds did not correlate with expression of FANCD2, MRE11A, ATM or BRCA2 with any statistical significance (S6 Fig).
FEN1 is epistatic with FANCD2 for the repair of olaparib- and cisplatin-induced DNA damage
FEN1 has been implicated in the repair of damage induced by PARP inhibitors [19, 84]. Such inhibitors both prevent the repair of alkylated bases through inhibition of BER or the trapping of PARP1/2 to DNA breaks. Such lesions stall replication forks and, if left unrepair, are converted to DSBs following fork collapse. Disruption of the DSB recognition machinery, HR or the FA pathways sensitise cells to PARP inhibitors, such as olaparib or veliparib [53, 79, 80, 84–88]. Since the data above suggests a synthetic lethal interaction between FEN1 inhibition and the disruption of genes known to be required for the repair of damage induced by PARP inhibitors, we wanted to examine whether FEN1 inhibition would have an effect on olaparib sensitivity in cells disrupted for these pathways, particularly the FA pathway and BRCA2. Consistent with previous data, we found that cells down-regulated for FEN1, FANCD2 or BRCA2 with shRNA were profoundly more sensitive to olaparib than non-target control cells (Fig 6A). Next we examined whether treatment with 1 was able to recapitulate this sensitivity in cells otherwise wild-type for FEN1. We found that, treatment with 10 μM 1 sensitised cells to a similar extent to FEN1 knockdown by shRNA (panel A in S5 Fig and Fig 6B). Interestingly, cells disrupted for FEN1 by shRNA were sensitised to olaparib further by treatment with 1. This could be a result of the inactivation of remaining FEN1, the inactivation of EXO1 or through a dominant negative effect. To determine whether FEN1 inhibition would potentiate the toxic effects of olaparib in BRCA2 and FANCD2 deficient cells, we pre-treated cells expressing shRNA against either gene with 10 μM 1 before adding varying concentrations of olaparib. While BRCA2 disrupted cells showed intermediate sensitivity to olaparib, co-treatment with 1 led to an increase in sensitivity that phenocopied a FEN1 disrupted cell-line (panel B of S5 Fig). Similarly, FANCD2 disrupted cells treated with 1 were no more sensitive to olaparib than FANCD2 cells treated with DMSO alone (Fig 6C). These data would suggest that FEN1 is epistatic with BRCA2 and the FA pathway for the repair of olaparib-induced DNA damage, while it was shown above that disruption of FEN1 induces damage that requires the FA pathway for its repair. To examine whether this epistasis was specific for olaparib or for other replication-coupled repair, we repeated these experiments replacing olaparib with the cross-linking agent cisplatin (CDDP). The Saccharomyces cerevisiae homologue of FEN1, Rad27, plays a role in the repair of DNA interstrand cross-links (ICLs) in a pathway independent of the 5’-3’ exonuclease Pso2 [22, 25]. Similarly, FEN1 has been implicated in the repair of cisplatin (CDDP)-induced ICLs [8], however no formal role for FEN1 has been assigned in this pathway. We treated cells disrupted for FEN1 by shRNA with CDDP and compared sensitivity to cells disrupted for known ICL-repair genes (FANCD2 and XPF) (Fig 6D). We found that, not only were FEN1 disrupted cells sensitive to CDDP, this sensitivity phenocopied cells disrupted for FANCD2 and XPF. Pre-treatment with 1, again, sensitised FEN1 proficient cells to a level consistent with inhibition of the enzyme (Fig 6E) and was epistatic with down-regulation of FANCD2 (Fig 6F). These data, together with the data described above, suggest a role for FEN1 in the FA pathway for the repair of replication-associated DNA damage. Since S. cerevisiae has a prototypical FA pathway that functions in the absence of Pso2 [89–92], we wanted to examine whether this putative role for FEN1 was conserved. S. cerevisiae RAD27 was deleted in a pso2 background and the resulting strains treated with nitrogen mustard (HN2) in asynchronous culture or cultures held in S-phase. Consistent with previous data [22, 25], we found that rad27 cells were insensitive to HN2 in asynchronous culture, yet co-disruption of pso2 led to a synergistic increase in sensitivity (Fig 6G). This phenotype persisted in cells held in S-phase (Fig 6H) with pso2 rad27 phenocopying pso2 msh2 (Fig 6I), suggesting a conserved role for Rad27/FEN1 in the Saccharomyces cerevisiae FA pathway. These data together hint at a role for FEN1 in a FA-related pathway. This is somewhat consistent with previous data as FEN1 activity has been shown to be stimulated by the FA core-complex member FANCA [93], and has been shown to be involved in replication fork processing and restart [15, 94]. Collectively, our data suggest that FEN1 inhibition results in accumulation of DNA damage, which requires the FA pathway for repair. This damage is most likely associated with a failure to process 5’ flaps created during lagging-strand synthesis, leading to replication fork instability and, ultimately, collapse into a DSB. Given that FEN1 appears to have a role in the FA pathway for the repair of olaparib or CDDP-induced DNA damage, it is likely that FEN1 itself would be required for the repair of damage induced by chemical inhibition of FEN1.
Fig 6. FEN1 is epistatic with FANCD2 for the repair of olaparib- and cisplatin-induced DNA damage.
A. Olaparib sensitivity of cells disrupted for FEN1, FANCD2 or BRCA2 by shRNA compared to a non-target control. B. Sensitisation of cells to olaparib following treatment with 10 μM 1. C. Epistasis analysis of FEN1 inhibition and FANCD2 depletion following exposure to olaparib. D. Cisplatin sensitivity of cells disrupted for FEN1, XPF and FANCD2 by shRNA compared to a non-target control. E. Sensitisation of cells to cisplatin following treatment with 5 μM 1. F. Epistasis analysis of FEN1 inhibition and FANCD2 depletion following exposure to cisplatin. G-I. Sensitivity of Saccharomyces cerevisiae strains deleted for rad27, pso2, msh2 singularly and in combination either in asynchronous culture (G) or synchronised in S-phase (H-I). In all cases, each data point is the mean of at least 3 individual repeats and the error bars represent the standard error. Significance was determined by student t-test. ns = not significant * p < 0.05. ** p < 0.005.
A model for the induction and repair of DNA damage associated with chemical inhibition of FEN1
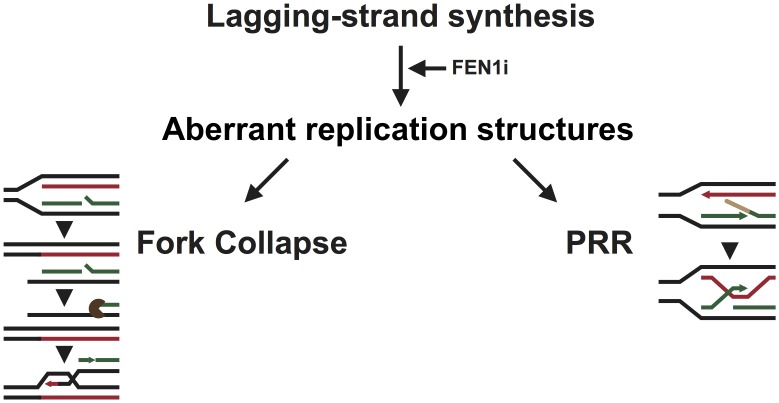
The data presented above shows that inhibition of FEN1 by N-hydroxyurea series inhibitors leads to specific cell killing in cells disrupted for a number of DNA damage pathways. These inhibitors activate the FA pathway (S4 Fig and [26]), suggesting an accumulation of aberrant replication structures following FEN1 inhibition (Fig 7). Failure to stabilise and resolve these structures appears to be extremely toxic as disruption of FANCD2 leads to extreme sensitivity to FEN1 inhibition (Fig 5E). It is currently unclear what the precise nature of these aberrant structures, however we have genetic evidence to suggest that they are generated behind the replication fork, since disruption of error-free PRR leads to increased sensitivity to FEN1 inhibitors (Fig 5F). In yeast cells inhibited for Okazaki fragment maturation, PRR is thought to be activated in response to the accumulation of long 5’ flaps [34], nicks [71] and abnormal secondary structures in the DNA such as hairpins [13, 73], and it is possible that these structures are generated by FEN1 inhibitors.
Fig 7. Current model for the formation and repair of DNA damage following FEN1 inhibition.
The inhibition of FEN1 leads to the accumulation of immature Okazaki fragments bound by RPA, accumulating aberrant replication structures that destabilise the replication fork. The PRR machinery is thought to allow for the tolerance of such structures by switching template strand, however failure to do so in a timely manner could lead to the stalling and, ultimately, collapse of the fork. The broken fork would require processing by endonucleases to create a 3’ overhang able to invade into back into the dsDNA and re-start replication. Persistent inhibition of FEN1 would ultimately lead to an overwhelming level of DNA damage and, ultimately, cell death.
BRCA2 and RAD51 are proteins with canonical roles in HR but they also play a role in the protection of forks against degradation [63, 95]. Both proteins are required in the absence of functional FEN1 (Fig 5G and 5H). It is possible that the role these proteins play in the tolerance of FEN1 inhibitors is solely in the protection of stalled replication forks, but it is equally possible that canonical HR is activated in the absence of FEN1 [33, 40–42]. Failure to repair replication forks leads to collapse and the generation of DSBs. These DSBs are repaired by non-reciprocal HR, in which the broken arm of the fork is processed by MRN complex to create a 3’overhang, which ultimately invades into the complete dsDNA to re-establish the replication fork. We and others [35] have shown that MRE11A is required in the absence of FEN1 activity (Fig 4) and here we have shown that cells with MSI that display disruption in MRN complex show selective sensitivity to FEN1 inhibitors (Fig 3). MRE11A and ATM act in the same pathway for the repair of DSBs (Fig 2) [96] and are both required for the repair of damage induced by FEN1 inhibition (Fig 5A–5D). ATM is activated by FEN1 inhibitors [26] in an MRE11A dependent manner (Fig 4G) suggesting the presence of DSBs. There is, however, no requirement for NHEJ in the repair of damage induced by FEN1 inhibitors (Fig 5H), suggesting that the damage induced is not a simple two-ended DSB. MRE11A, however, also plays a role in the processing and restart of stalled replication forks [63, 97] and it may be that the function of MRE11A is in the processing of aberrant replication structures.
Finally, we have shown that FEN1 operates in the repair of lesions induced by cisplatin, nitrogen mustard and olaparib in a pathway that also requires FANCD2 (Fig 6). This suggests a role for FEN1 in replication-coupled DNA repair, which could include damage induced by FEN1 inhibition. The role for FEN1 in the repair of these lesions is unclear, however there is evidence to suggest that FEN1 plays a role in replication restart and the processing of stalled replication forks [15, 16, 94, 98]. It would be interesting to determine the precise function of FEN1 in this pathway.
The work presented here suggests that inhibitors of FEN1 would be effective in targeting cells deleted, mutated or disrupted for a number of DNA damage response genes, many of which are associated with cancer [99]. Similarly, FEN1 inhibitors sensitise cells to chemotherapeutics such as cisplatin, and DNA repair inhibitors such as olaparib. The work presented here supports FEN1 as a possible target for oncology drug discovery.
Materials and methods
High-throughput cell-killing assay
Toxicity was calculated by using a modified, high-throughput version of the MTS assay. Cells were plated at an appropriate density in 96-well plates and allowed to adhere for 24 h. Compound, diluted in DMSO, was added to a final concentration of between 0 and 30 μM and plates were incubated at 37°C for 0 or 2 days. Subsequently, CellTiter96 AQueous MTS (Sigma) was added as per the manufacturers recommendations and plates were incubated at 37°C for 2 h. Absorbance was measured using a Tecan Ultra microplate reader (Tecan Life Sciences) and data validated and analysed as per internal data handling methodology.
Cell-lines and media
HeLa, FaDu, SW-620, DLD1, HCT-116 and HCT-116 chr3 were all obtained from internal stocks. The FaDu ATM triple knock-out cell-line was created using transcription activator-like effector nucleases (TALEN) by AstraZeneca (Discovery Sciences, Sweden). SilenciX cell-lines were all obtained from Tebu Bioscience and knockdown of the stated gene was confirmed externally by qPCR. All cell-lines were grown in DMEM (Sigma Aldrich) media. SilenciX cell-lines were maintained in DMEM media supplemented with 125 μg/mL hygromycin B to prevent the loss of shRNA-expression plasmids. All cell-lines were verified in-house by short tandem repeat (STR) fingerprinting and routinely tested for mycoplasma.
siRNA knockdown
siRNA-lipofectamine complexes were created using Lipofectamine RNAiMAX (Thermo Fisher Scientific) as per the manufacturer’s instructions. All siRNAs used in this study were Dharmacon ON-TARGETplus SMARTpools (GE Healthcare). siRNA was added at a final concentration of 20 nM and allowed to grow for 24 hours. Cells were subsequently washed and fresh media applied to avoid lipofectamine toxicity. For knock-down confirmation, cells were trypsinised after 3 days in fresh media, collected and lysed in Cell Panel Lysis Buffer (5 mM Tris-HCl, 3 mM EDTA, 3 mM EGTA, 50 mM NaF, 2 mM sodium orthovanadate, 0.27 M sucrose, 10 mM ß-glycerophosphate, 5 mM sodium pyrophosphate, and 0.5% Triton X-100) supplemented with complete protease and phosSTOP phosphotase inhibitors (both Roche). Examples of knock-down efficiency are shown with the figures. These cells were subsequently used for down-stream experiments discussed below.
Mammalian colony formation assay
Cells were plated at an appropriate density and allowed to settle for >24 hours in appropriate media. Media was supplemented with compounds (1, olaparib, cisplatin) at the stated doses and allowed to grow until colonies of >50 cells were achieved. For combination experiments, growth media was changed after 24 hours for media supplemented with 5 or 10 μM 1 (stated in the Fig legend) and cells were allowed to grow for 24 hours. Subsequently, media was further supplemented with drug at the stated dose and allowed to grow until colonies of >50 cells were achieved. For ionising radiation experiments, cells were seeded at the stated dose and allowed to settle for 24 hours. Media was supplemented with 5 μM 1 and allowed to grow for 24 hours. Cells were subsequently exposed to ionising radiation at the stated dose. For siRNA knockdown toxicity experiments, cells were treated with siRNA as described above and allowed to grow in fresh media until colonies of >50 cells were achieved.
MRN complex and MMR status determination
SW620, DLD1, HCT-116 and HCT-116 chr3 cells were trypsinised, collected and lysed in Cell Panel Lysis Buffer with complete protease and phosphotase inhibitors. Proteins were separated by gel electrophoresis and transferred to nitrocellulose membrane by Western blot. Membranes were probed, at a concentration of 1:1000 unless stated otherwise, for ATM (sc-23921, Santa Cruz Biotechnology), MSH2 (556349, BD Pharmigen), MLH1 (WH0004292M2, Sigma Aldrich), MSH6 (610918, BD Transduction), MRE11A (ab214, Abcam), RAD50 (611010, BD Transduction), NBS1 (NB100-143, Novus Biologicals), FEN1 (ab109132, Abcam) and β-Actin (A5441, Sigma Aldrich).
Damage induction assay
DNA damage induction was determined as described previously [26]. Briefly, cells were plated at an appropriate density and allowed to settle overnight. Media was supplemented with 1 at the dose stated and cells were allowed to grow for 4 days with constant exposure. Cells were trypsinised, collected and lysed in Cell Panel Lysis Buffer with complete protease and phosphotase inhibitors. Proteins were separated by gel electrophoresis and transferred to nitrocellulose membrane by Western blot. Membranes were probed, at a concentration of 1:1000 unless stated otherwise, for γH2AX (#2577, Cell Signaling Technology; 1:500), GAPDH (#3683, Cell Signaling Technology; 1:5000), phospho-ATM (Ser1981) (ab81292, Abcam) and FANCD2 (sc-20022, Santa Cruz Biotechnology).
Immunofluorescence
Cells were seeded in 96-well plates at an appropriate density and allowed to settle for 24 hours. Media was supplemented with compound at the stated dose and allowed to grow with constant dosing for 4 days. Cells were fixed in a final concentration of 4% paraformaldehyde for 20 min and washed 3 times in PBSA. Cells were permeablized for 4 min in 0.5% triton X-100 in PBSA, washed and probed with anti-γH2AX (JBW301, Millipore) or anti-RAD51 (ABE257, Millipore) at 1:500 for 48 hours at 4°C. Cells were washed in 0.05% Tween in PBSA 3 times for 5 min before probing for 1 hour at room temperature with AlexaFluor 488 or AlexaFlour 546-conjugated secondary antibodies (both Life Technologies) at a concentration of 1:500, and Hoechst DNA stain at 1:2000. Immunoflourescence was examined using CellInsight CX5 (Thermo Fisher) and foci were quantified using the software and algorithms provided to specifically count foci formed within the nucleus. Example images of foci are shown in panels C and E of S4 Fig.
Yeast strains
Strains used in this study (S2 Table) were created as previously described [89]. Gene deletion was carried out by micro-homology targeted gene disruption using the pFA6a vector series and its derivatives [100–102] or generated by synthetic genetic analysis methodology [103]. Deletions were confirmed by PCR analysis and restriction enzyme digestion. Cells were grown in YPD media and plated onto YPD agar.
Yeast nitrogen mustard sensitivity
Nitrogen mustard sensitivity was determined as previously described [25, 89]. Exponentially growing cells were treated with increasing doses of nitrogen mustard in PBSA for 1 h at 30°C. Cells were washed and plated at an appropriate density and colonies were scored after 3 days incubation at 30°C. Percent survival is calculated as a fraction of an untreated control.
Replication-dependent nitrogen mustard sensitivity
Replication-dependent sensitivity was determined as previously described [25]. Cells were synchronised in G1-phase of the cell-cycle with alpha mating factor for 2 h before being washed in PBSA and re-suspended in fresh YPD. Cells allowed to enter S-phase for 10 min before being treated with nitrogen mustard as described above. In pilot experiments, cell-cycle synchronisation was conducted by FACS.
Supporting information
Scatter plots show cell-line sensitivities to treatment with 1, broadly grouped by tissue of origin (A), tissue sub-group (B-D) or oncogene mutation status (E-G). Waterfall plot (H) shows all cell-line sensitivities to 1, with those confirmed to be MSI highlighted in green.
(TIF)
Scatter plots show cell-line sensitivities to treatment with 2, broadly grouped by tissue of origin (A), tissue sub-group (B-D) or oncogene mutation status (E-G). Waterfall plot (H) shows all cell-line sensitivities to 2, with those confirmed to be MSI highlighted in green.
(TIF)
Scatter plots show cell-line sensitivities to treatment with 3, broadly grouped by tissue of origin (A), tissue sub-group (B-D) or oncogene mutation status (E-G). Waterfall plot (H) shows all cell-line sensitivities to 3, with those confirmed to be MSI highlighted in green.
(TIF)
A. Clonogenic survival of SW620 and HCT-116 cells treated with siRNA against EXO1. B. Clonogeneic survival of HeLa cells disrupted for EXO1 and MRE11 following treatment with 1. C. Example of γH2AX foci induced by 1 or olaparib. D. Dose dependent increase in cells with at least 5 γH2AX foci. Data is a quantification from at least 500 cells. E. Example of RAD51 foci induced by 1 or 3. G. Activation of the Fanconi anemia pathway in cells disrupted for FEN1 compared to a non-target control.
(TIF)
A. Effect treatment with 5 μM and 10 μM 1 has on sensitivity to olaparib in cells with wild-type levels of FEN1. B. Epistasis analysis of FEN1 inhibition and BRCA2 depletion following exposure to olaparib.
(TIF)
ρ denotes the Pearson’s correlation coefficient.
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOCX)
* Strain kindly donated by the Boone Lab. # Strains purchased from Open Biosystems.
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Work performed in the laboratory of PJM was supported by Cancer Research UK Programme Grant CA9047/A10111 (http://science.cancerresearchuk.org/). Work performed by TAW at AstraZeneca Pharmaceuticals was funded by the AstraZeneca Postdoctoral Research Program. AstraZeneca provided financial support in the form of salaries and research materials for TAW and STD, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Harrington JJ, Lieber MR. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev. 1994;8(11):1344–55. Epub 1994/06/01. . [DOI] [PubMed] [Google Scholar]
- 2.Grasby JA, Finger LD, Tsutakawa SE, Atack JM, Tainer JA. Unpairing and gating: sequence-independent substrate recognition by FEN superfamily nucleases. Trends BiochemSci. 2012;37(2):74–84. doi: 10.1016/j.tibs.2011.10.003. WOS:000300923200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, et al. Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily. Cell. 2011;145(2):198–211. doi: 10.1016/j.cell.2011.03.004. WOS:000289549900010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 SNA replication with purified replication factors. J Biol Chem. 1994;269(14):10923–34. WOS:A1994NF01700107. [PubMed] [Google Scholar]
- 5.Bambara RA, Murante RS, Henricksen LA. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272(8):4647–50. WOS:A1997WJ85500001. [DOI] [PubMed] [Google Scholar]
- 6.Lam JS, Seligson DB, Yu H, Li A, Eeva M, Pantuck AJ, et al. Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score. BJU Int. 2006;98(2):445–51. doi: 10.1111/j.1464-410X.2006.06224.x. WOS:000238790900039. [DOI] [PubMed] [Google Scholar]
- 7.Nikolova T, Christmann M, Kaina B. FEN1 is Overexpressed in Testis, Lung and Brain Tumors. Anticancer Res. 2009;29(7):2453–9. WOS:000268104100005. [PubMed] [Google Scholar]
- 8.Singh P, Yang M, Dai HF, Yu DK, Huang Q, Tan W, et al. Overexpression and Hypomethylation of Flap Endonuclease 1 Gene in Breast and Other Cancers. Mol Cancer Res. 2008;6(11):1710–7. WOS:000261134500006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdel-Fatah TM, Russell R, Albarakati N, Maloney DJ, Dorjsuren D, Rueda OM, et al. Genomic and protein expression analysis reveals flap endonuclease 1 (FEN1) as a key biomarker in breast and ovarian cancer. Mol Oncol. 2014;8(7):1326–38. Epub 2014/06/02. doi: 10.1016/j.molonc.2014.04.009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kucherlapati M, Yang K, Kuraguchi M, Zhao J, Lia M, Heyer J, et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc Natl Acad Sci U S A. 2002;99(15):9924–9. Epub 2002/07/18. doi: 10.1073/pnas.152321699 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kokoska RJ, Stefanovic L, Tran HT, Resnick MA, Gordenin DA, Petes TD. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t). Mol Cell Biol. 1998;18(5):2779–88. Epub 1998/05/05. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, Freudenreich CH. Haploinsufficiency of yeast FEN1 causes instability of expanded CAG/CTG tracts in a length-dependent manner. Gene. 2007;393(1–2):110–5. Epub 2007/03/27. doi: 10.1016/j.gene.2007.01.025 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durant ST. Telomerase-independent paths to immortality in predictable cancer subtypes. J Cancer. 2012;3:67–82. Epub 2012/02/09. doi: 10.7150/jca.3965 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng L, Zhou M, Chai Q, Parrish J, Xue D, Patrick SM, et al. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 2005;6(1):83–9. Epub 2004/12/14. doi: 10.1038/sj.embor.7400313 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahata C, Masuda Y, Takedachi A, Tanaka K, Iwai S, Kuraoka I. Repair synthesis step involving ERCC1-XPF participates in DNA repair of the Top1-DNA damage complex. Carcinogenesis. 2015;36(8):841–51. Epub 2015/05/31. doi: 10.1093/carcin/bgv078 . [DOI] [PubMed] [Google Scholar]
- 16.Balakrishnan L, Bambara RA. Flap endonuclease 1. Annu Rev Biochem. 2013;82:119–38. Epub 2013/03/05. doi: 10.1146/annurev-biochem-072511-122603 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tadi SK, Sebastian R, Dahal S, Babu RK, Choudhary B, Raghavan SC. Microhomology Mediated End Joining is the Principal Mediator of Double-strand Break Repair During Mitochondrial DNA Lesions. Mol Biol Cell. 2015. Epub 2015/11/27. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fehrmann RS, Karjalainen JM, Krajewska M, Westra HJ, Maloney D, Simeonov A, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47(2):115–25. Epub 2015/01/13. doi: 10.1038/ng.3173 . [DOI] [PubMed] [Google Scholar]
- 19.Reagan MS, Pittenger C, Siede W, Friedberg EC. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177(2):364–71. Epub 1995/01/01. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim K, Biade S, Matsumoto Y. Involvement of flap endonuclease 1 in base excision DNA repair. J Biol Chem. 1998;273(15):8842–8. doi: 10.1074/jbc.273.15.8842. WOS:000072990800041. [DOI] [PubMed] [Google Scholar]
- 21.McHugh PJ, Sones WR, Hartley JA. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20(10):3425–33. Epub 2000/04/25. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuzaki Y, Adachi N, Koyama H. Vertebrate cells lacking FEN-1 endonuclease are viable but hypersensitive to methylating agents and H2O2. Nucleic Acids Res. 2002;30(14):3273–7. Epub 2002/07/24. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata Y, Nakamura T. Defective flap endonuclease 1 activity in mammalian cells is associated with impaired DNA repair and prolonged S phase delay. J Biol Chem. 2002;277(1):746–54. Epub 2001/11/01. doi: 10.1074/jbc.M109461200 . [DOI] [PubMed] [Google Scholar]
- 24.Barber LJ, Ward TA, Hartley JA, McHugh PJ. DNA interstrand cross-link repair in the Saccharomyces cerevisiae cell cycle: overlapping roles for PSO2 (SNM1) with MutS factors and EXO1 during S phase. Mol Cell Biol. 2005;25(6):2297–309. Epub 2005/03/04. doi: 10.1128/MCB.25.6.2297-2309.2005 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Exell JC, Thompson MJ, Finger LD, Shaw SJ, Debreczeni J, Ward TA, et al. Cellularly active N-hydroxyurea FEN1 inhibitors block substrate entry to the active site. Nat Chem Biol. 2016. doi: 10.1038/nchembio.2148 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tumey LN, Bom D, Huck B, Gleason E, Wang J, Silver D, et al. The identification and optimization of a N-hydroxy urea series of flap endonuclease 1 inhibitors. Bioorg Med Chem Lett. 2005;15(2):277–81. Epub 2004/12/18. doi: 10.1016/j.bmcl.2004.10.086 . [DOI] [PubMed] [Google Scholar]
- 27.Finger LD, Patel N, Beddows A, Ma L, Exell JC, Jardine E, et al. Observation of unpaired substrate DNA in the flap endonuclease-1 active site. Nucleic Acids Res. 2013;41(21):9839–47. Epub 2013/08/27. doi: 10.1093/nar/gkt737 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Algasaier SI, Exell JC, Bennet IA, Thompson MJ, Gotham VJ, Shaw SJ, et al. DNA and Protein Requirements for Substrate Conformational Changes Necessary for Human Flap Endonuclease-1 Catalyzed Reaction. J Biol Chem. 2016. Epub 2016/02/18. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dovrat D, Stodola JL, Burgers PM, Aharoni A. Sequential switching of binding partners on PCNA during in vitro Okazaki fragment maturation. Proc Natl Acad Sci U S A. 2014;111(39):14118–23. Epub 2014/09/18. doi: 10.1073/pnas.1321349111 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stodola JL, Burgers PM. Resolving individual steps of Okazaki-fragment maturation at a millisecond timescale. Nat Struct Mol Biol. 2016. Epub 2016/04/12. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McWhirter C, Tonge M, Plant H, Hardern I, Nissink W, Durant ST. Development of a high-throughput fluorescence polarization DNA cleavage assay for the identification of FEN1 inhibitors. J Biomol Screen. 2013;18(5):567–75. Epub 2013/02/22. doi: 10.1177/1087057113476551 . [DOI] [PubMed] [Google Scholar]
- 32.Symington LS. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 1998;26(24):5589–95. Epub 1998/12/05. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becker JR, Pons C, Nguyen HD, Costanzo M, Boone C, Myers CL, et al. Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing. PLoS Genet. 2015;11(11):e1005659 Epub 2015/11/07. doi: 10.1371/journal.pgen.1005659 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Pel DM, Barrett IJ, Shimizu Y, Sajesh BV, Guppy BJ, Pfeifer T, et al. An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLoS Genet. 2013;9(1):e1003254 Epub 2013/02/06. doi: 10.1371/journal.pgen.1003254 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173(2):195–206. Epub 2006/04/19. doi: 10.1083/jcb.200510130 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hopfner KP. ATP puts the brake on DNA double-strand break repair: a new study shows that ATP switches the Mre11-Rad50-NbS1 repair factor between signaling and processing of DNA ends. Bioessays. 2014;36(12):1170–8. Epub 2014/09/13. doi: 10.1002/bies.201400102 . [DOI] [PubMed] [Google Scholar]
- 37.Grenon M, Gilbert C, Lowndes NF. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat Cell Biol. 2001;3(9):844–7. Epub 2001/09/05. doi: 10.1038/ncb0901-844 . [DOI] [PubMed] [Google Scholar]
- 38.Porter-Goff ME, Rhind N. The role of MRN in the S-phase DNA damage checkpoint is independent of its Ctp1-dependent roles in double-strand break repair and checkpoint signaling. Mol Biol Cell. 2009;20(7):2096–107. Epub 2009/02/13. doi: 10.1091/mbc.E08-09-0986 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Debrauwere H, Loeillet S, Lin W, Lopes J, Nicolas A. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc Natl Acad Sci U S A. 2001;98(15):8263–9. Epub 2001/07/19. doi: 10.1073/pnas.121075598 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159(4):1423–33. Epub 2002/01/10. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McManus KJ, Barrett IJ, Nouhi Y, Hieter P. Specific synthetic lethal killing of RAD54B-deficient human colorectal cancer cells by FEN1 silencing. Proc Natl Acad Sci U S A. 2009;106(9):3276–81. doi: 10.1073/pnas.0813414106. WOS:000263844100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338(21):1481–7. Epub 1998/05/21. doi: 10.1056/NEJM199805213382101 . [DOI] [PubMed] [Google Scholar]
- 43.Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342(2):69–77. Epub 2000/01/13. doi: 10.1056/NEJM200001133420201 . [DOI] [PubMed] [Google Scholar]
- 44.Lawes DA, SenGupta S, Boulos PB. The clinical importance and prognostic implications of microsatellite instability in sporadic cancer. Eur J Surg Oncol. 2003;29(3):201–12. Epub 2003/03/27. . [DOI] [PubMed] [Google Scholar]
- 45.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med. 2005;352(18):1851–60. Epub 2005/05/06. doi: 10.1056/NEJMoa043146 . [DOI] [PubMed] [Google Scholar]
- 46.Jaiswal AS, Panda H, Law BK, Sharma J, Jani J, Hromas R, et al. NSC666715 and Its Analogs Inhibit Strand-Displacement Activity of DNA Polymerase beta and Potentiate Temozolomide-Induced DNA Damage, Senescence and Apoptosis in Colorectal Cancer Cells. PLoS One. 2015;10(5):e0123808 Epub 2015/05/02. doi: 10.1371/journal.pone.0123808 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaiswal AS, Banerjee S, Panda H, Bulkin CD, Izumi T, Sarkar FH, et al. A novel inhibitor of DNA polymerase beta enhances the ability of temozolomide to impair the growth of colon cancer cells. Mol Cancer Res. 2009;7(12):1973–83. Epub 2009/12/10. doi: 10.1158/1541-7786.MCR-09-0309 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vendetti FP, Lau A, Schamus S, Conrads TP, O'Connor MJ, Bakkenist CJ. The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget. 2015;6(42):44289–305. Epub 2015/10/31. doi: 10.18632/oncotarget.6247 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64(24):9152–9. Epub 2004/12/18. doi: 10.1158/0008-5472.CAN-04-2727 . [DOI] [PubMed] [Google Scholar]
- 50.Menear KA, Adcock C, Boulter R, Cockcroft XL, Copsey L, Cranston A, et al. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin- 1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem. 2008;51(20):6581–91. Epub 2008/09/20. doi: 10.1021/jm8001263 . [DOI] [PubMed] [Google Scholar]
- 51.Giannini G, Ristori E, Cerignoli F, Rinaldi C, Zani M, Viel A, et al. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep. 2002;3(3):248–54. Epub 2002/02/19. doi: 10.1093/embo-reports/kvf044 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li HR, Shagisultanova EI, Yamashita K, Piao Z, Perucho M, Malkhosyan SR. Hypersensitivity of tumor cell lines with microsatellite instability to DNA double strand break producing chemotherapeutic agent bleomycin. Cancer Res. 2004;64(14):4760–7. Epub 2004/07/17. doi: 10.1158/0008-5472.CAN-04-0975 . [DOI] [PubMed] [Google Scholar]
- 53.Vilar E, Bartnik CM, Stenzel SL, Raskin L, Ahn J, Moreno V, et al. MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Res. 2011;71(7):2632–42. Epub 2011/02/09. doi: 10.1158/0008-5472.CAN-10-1120 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giannini G, Rinaldi C, Ristori E, Ambrosini MI, Cerignoli F, Viel A, et al. Mutations of an intronic repeat induce impaired MRE11 expression in primary human cancer with microsatellite instability. Oncogene. 2004;23(15):2640–7. Epub 2004/03/30. doi: 10.1038/sj.onc.1207409 . [DOI] [PubMed] [Google Scholar]
- 55.Shibata D, Peinado MA, Ionov Y, Malkhosyan S, Perucho M. Genomic instability in repeated sequences is an early somatic event in colorectal tumorigenesis that persists after transformation. Nat Genet. 1994;6(3):273–81. Epub 1994/03/01. doi: 10.1038/ng0394-273 . [DOI] [PubMed] [Google Scholar]
- 56.Wheeler JM, Beck NE, Kim HC, Tomlinson IP, Mortensen NJ, Bodmer WF. Mechanisms of inactivation of mismatch repair genes in human colorectal cancer cell lines: the predominant role of hMLH1. Proc Natl Acad Sci U S A. 1999;96(18):10296–301. Epub 1999/09/01. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koi M, Umar A, Chauhan DP, Cherian SP, Carethers JM, Kunkel TA, et al. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N'-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54(16):4308–12. Epub 1994/08/15. . [PubMed] [Google Scholar]
- 58.Paull TT. Mechanisms of ATM Activation. Annu Rev Biochem. 2015;84:711–38. Epub 2015/01/13. doi: 10.1146/annurev-biochem-060614-034335 . [DOI] [PubMed] [Google Scholar]
- 59.Kim HS, Choi SI, Min HL, Kim MA, Kim WH. Mutation at intronic repeats of the ataxia-telangiectasia mutated (ATM) gene and ATM protein loss in primary gastric cancer with microsatellite instability. PLoS One. 2013;8(12):e82769 Epub 2013/12/11. doi: 10.1371/journal.pone.0082769 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boch J. TALEs of genome targeting. Nat Biotechnol. 2011;29(2):135–6. 10.1038/nbt.1767. doi: 10.1038/nbt.1767 [DOI] [PubMed] [Google Scholar]
- 61.Robison JG, Elliott J, Dixon K, Oakley GG. Replication protein A and the Mre11.Rad50.NbS1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem. 2004;279(33):34802–10. Epub 2004/06/08. doi: 10.1074/jbc.M404750200 . [DOI] [PubMed] [Google Scholar]
- 62.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11(10):683–7. Epub 2010/09/16. doi: 10.1038/nrm2974 . [DOI] [PubMed] [Google Scholar]
- 63.Ying S, Hamdy FC, Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 2012;72(11):2814–21. Epub 2012/03/27. doi: 10.1158/0008-5472.CAN-11-3417 . [DOI] [PubMed] [Google Scholar]
- 64.Sobeck A, Stone S, Costanzo V, de Graaf B, Reuter T, de Winter J, et al. Fanconi Anemia Proteins Are Required To Prevent Accumulation of Replication-Associated DNA Double-Strand Breaks. Molecular and Cellular Biology. 2006;26(2):425–37. doi: 10.1128/MCB.26.2.425-437.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14(5):693–701. Epub 2005/01/22. doi: 10.1093/hmg/ddi065 . [DOI] [PubMed] [Google Scholar]
- 66.Matsushita N, Kitao H, Ishiai M, Nagashima N, Hirano S, Okawa K, et al. A FancD2-monoubiquitin fusion reveals hidden functions of Fanconi anemia core complex in DNA repair. Mol Cell. 2005;19(6):841–7. Epub 2005/09/20. doi: 10.1016/j.molcel.2005.08.018 . [DOI] [PubMed] [Google Scholar]
- 67.Lachaud C, Moreno A, Marchesi F, Toth R, Blow JJ, Rouse J. Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science. 2016;351(6275):846–9. Epub 2016/01/23. doi: 10.1126/science.aad5634 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, et al. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci U S A. 2011;108(16):6492–6. Epub 2011/04/06. doi: 10.1073/pnas.1018487108 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35(1):116–27. Epub 2009/07/15. doi: 10.1016/j.molcel.2009.06.020 . [DOI] [PubMed] [Google Scholar]
- 70.Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18(16):1958–63. Epub 2004/08/18. doi: 10.1101/gad.1196104 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen HD, Becker J, Thu YM, Costanzo M, Koch EN, Smith S, et al. Unligated Okazaki Fragments Induce PCNA Ubiquitination and a Requirement for Rad59-Dependent Replication Fork Progression. PLoS One. 2013;8(6):e66379 Epub 2013/07/05. doi: 10.1371/journal.pone.0066379 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 2008;29(5):625–36. Epub 2008/03/18. doi: 10.1016/j.molcel.2007.12.016 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daee DL, Mertz T, Lahue RS. Postreplication repair inhibits CAG.CTG repeat expansions in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27(1):102–10. Epub 2006/10/25. doi: 10.1128/MCB.01167-06 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng L, Shen B. Okazaki fragment maturation: nucleases take centre stage. J Mol Cell Biol. 2011;3(1):23–30. Epub 2011/02/01. doi: 10.1093/jmcb/mjq048 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Achar YJ, Balogh D, Neculai D, Juhasz S, Morocz M, Gali H, et al. Human HLTF mediates postreplication repair by its HIRAN domain-dependent replication fork remodelling. Nucleic Acids Res. 2015;43(21):10277–91. Epub 2015/09/10. doi: 10.1093/nar/gkv896 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419(6903):135–41. Epub 2002/09/13. doi: 10.1038/nature00991 . [DOI] [PubMed] [Google Scholar]
- 77.Ball LG, Zhang K, Cobb JA, Boone C, Xiao W. The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol Microbiol. 2009;73(1):89–102. Epub 2009/06/06. doi: 10.1111/j.1365-2958.2009.06748.x . [DOI] [PubMed] [Google Scholar]
- 78.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96(5):645–53. Epub 1999/03/25. . [DOI] [PubMed] [Google Scholar]
- 79.Koppensteiner R, Samartzis EP, Noske A, von Teichman A, Dedes I, Gwerder M, et al. Effect of MRE11 loss on PARP-inhibitor sensitivity in endometrial cancer in vitro. PLoS One. 2014;9(6):e100041 Epub 2014/06/14. doi: 10.1371/journal.pone.0100041 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weston VJ, Oldreive CE, Skowronska A, Oscier DG, Pratt G, Dyer MJ, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116(22):4578–87. Epub 2010/08/27. doi: 10.1182/blood-2010-01-265769 . [DOI] [PubMed] [Google Scholar]
- 81.Raderschall E, Golub EI, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci U S A. 1999;96(5):1921–6. Epub 1999/03/03. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci U S A. 1995;92(6):2298–302. Epub 1995/03/14. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haaf T, Raderschall E, Reddy G, Ward DC, Radding CM, Golub EI. Sequestration of mammalian Rad51-recombination protein into micronuclei. J Cell Biol. 1999;144(1):11–20. Epub 1999/01/13. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72(21):5588–99. Epub 2012/11/03. doi: 10.1158/0008-5472.CAN-12-2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–7. Epub 2005/04/15. doi: 10.1038/nature03443 . 10.1158/0008-5472.CAN-12-2753. 23118055; [DOI] [PubMed] [Google Scholar]
- 86.Duan W, Gao L, Aguila B, Kalvala A, Otterson GA, Villalona-Calero MA. Fanconi anemia repair pathway dysfunction, a potential therapeutic target in lung cancer. Front Oncol. 2014;4:368 Epub 2015/01/08. doi: 10.3389/fonc.2014.00368 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66(16):8109–15. Epub 2006/08/17. doi: 10.1158/0008-5472.CAN-06-0140 . [DOI] [PubMed] [Google Scholar]
- 88.McCabe N, Lord CJ, Tutt AN, Martin NM, Smith GC, Ashworth A. BRCA2-deficient CAPAN-1 cells are extremely sensitive to the inhibition of Poly (ADP-Ribose) polymerase: an issue of potency. Cancer Biol Ther. 2005;4(9):934–6. Epub 2005/10/28. . [DOI] [PubMed] [Google Scholar]
- 89.Ward TA, Dudasova Z, Sarkar S, Bhide MR, Vlasakova D, Chovanec M, et al. Components of a Fanconi-like pathway control Pso2-independent DNA interstrand crosslink repair in yeast. PLoS Genet. 2012;8(8):e1002884 Epub 2012/08/23. doi: 10.1371/journal.pgen.1002884 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McHugh PJ, Ward TA, Chovanec M. A prototypical Fanconi anemia pathway in lower eukaryotes? Cell Cycle. 2012;11(20):3739–44. Epub 2012/08/17. doi: 10.4161/cc.21727 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Daee DL, Myung K. Fanconi-like crosslink repair in yeast. Genome Integr. 2012;3(1):7 Epub 2012/10/16. doi: 10.1186/2041-9414-3-7 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Daee DL, Ferrari E, Longerich S, Zheng XF, Xue X, Branzei D, et al. Rad5-dependent DNA repair functions of the Saccharomyces cerevisiae FANCM protein homolog Mph1. J Biol Chem. 2012;287(32):26563–75. Epub 2012/06/15. doi: 10.1074/jbc.M112.369918 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qian L, Yuan F, Rodriguez-Tello P, Padgaonkar S, Zhang Y. Human Fanconi anemia complementation group a protein stimulates the 5' flap endonuclease activity of FEN1. PLoS One. 2013;8(12):e82666 Epub 2013/12/19. doi: 10.1371/journal.pone.0082666 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shen B, Singh P, Liu R, Qiu J, Zheng L, Finger LD, et al. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays. 2005;27(7):717–29. Epub 2005/06/15. doi: 10.1002/bies.20255 . [DOI] [PubMed] [Google Scholar]
- 95.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145(4):529–42. Epub 2011/05/14. doi: 10.1016/j.cell.2011.03.041 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26(56):7749–58. Epub 2007/12/11. doi: 10.1038/sj.onc.1210880 . [DOI] [PubMed] [Google Scholar]
- 97.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28(17):2601–15. Epub 2009/07/25. doi: 10.1038/emboj.2009.206 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pizzolato J, Mukherjee S, Scharer OD, Jiricny J. FANCD2-associated Nuclease 1, but Not Exonuclease 1 or Flap Endonuclease 1, Is Able to Unhook DNA Interstrand Cross-links in Vitro. J Biol Chem. 2015;290(37):22602–11. Epub 2015/07/30. doi: 10.1074/jbc.M115.663666 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cerbinskaite A, Mukhopadhyay A, Plummer ER, Curtin NJ, Edmondson RJ. Defective homologous recombination in human cancers. Cancer Treat Rev. 2012;38(2):89–100. Epub 2011/07/01. doi: 10.1016/j.ctrv.2011.04.015 . [DOI] [PubMed] [Google Scholar]
- 100.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10(13):1793–808. Epub 1994/12/01. . [DOI] [PubMed] [Google Scholar]
- 101.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–61. Epub 1998/08/26. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U . [DOI] [PubMed] [Google Scholar]
- 102.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15(14):1541–53. Epub 1999/10/09. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K . [DOI] [PubMed] [Google Scholar]
- 103.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294(5550):2364–8. Epub 2001/12/18. doi: 10.1126/science.1065810 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scatter plots show cell-line sensitivities to treatment with 1, broadly grouped by tissue of origin (A), tissue sub-group (B-D) or oncogene mutation status (E-G). Waterfall plot (H) shows all cell-line sensitivities to 1, with those confirmed to be MSI highlighted in green.
(TIF)
Scatter plots show cell-line sensitivities to treatment with 2, broadly grouped by tissue of origin (A), tissue sub-group (B-D) or oncogene mutation status (E-G). Waterfall plot (H) shows all cell-line sensitivities to 2, with those confirmed to be MSI highlighted in green.
(TIF)
Scatter plots show cell-line sensitivities to treatment with 3, broadly grouped by tissue of origin (A), tissue sub-group (B-D) or oncogene mutation status (E-G). Waterfall plot (H) shows all cell-line sensitivities to 3, with those confirmed to be MSI highlighted in green.
(TIF)
A. Clonogenic survival of SW620 and HCT-116 cells treated with siRNA against EXO1. B. Clonogeneic survival of HeLa cells disrupted for EXO1 and MRE11 following treatment with 1. C. Example of γH2AX foci induced by 1 or olaparib. D. Dose dependent increase in cells with at least 5 γH2AX foci. Data is a quantification from at least 500 cells. E. Example of RAD51 foci induced by 1 or 3. G. Activation of the Fanconi anemia pathway in cells disrupted for FEN1 compared to a non-target control.
(TIF)
A. Effect treatment with 5 μM and 10 μM 1 has on sensitivity to olaparib in cells with wild-type levels of FEN1. B. Epistasis analysis of FEN1 inhibition and BRCA2 depletion following exposure to olaparib.
(TIF)
ρ denotes the Pearson’s correlation coefficient.
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOCX)
* Strain kindly donated by the Boone Lab. # Strains purchased from Open Biosystems.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.