Abstract
Ionizing radiation from natural sources or anthropogenic activity has the potential to cause oxidative stress or genetic damage in living organisms, through the ionization and excitation of molecules and the subsequent production of free radicals and reactive oxygen species (ROS). The present work focuses on radiation-induced biological effects using the zebrafish (Danio rerio) vertebrate model. Changes in developmental traits and gene expression in zebrafish were assessed after continuous external gamma irradiation (0.4, 3.9, 15 and 38 mGy/h) with corresponding controls, starting at 2.5 hours post fertilization (hpf) and lasting through embryogenesis and the early larval stage. The lowest dose rate corresponded to recommended benchmarks at which adverse effects are not expected to occur in aquatic ecosystems (2–10 mGy/day). The survival observed at 96 hours post fertilization (hpf) in the 38 mGy/h group was significantly lower, while other groups showed no significant difference compared to controls. The total hatching was significantly lower from controls in the 15 mGy/h group and a delay in hatching onset in the 0.4 mGy/h group was observed. The deformity frequency was significantly increased by prolonged exposure duration at dose rates ≥ 0.4 mGy/h. Molecular responses analyzed by RNA-seq at gastrulation (5.5 hpf transcriptome) indicate that the radiation induced adverse effects occurred during the earliest stages of development. A dose-response relationship was found in the numbers of differentially regulated genes in exposure groups compared to controls at a total dose as low as 1.62 mGy. Ingenuity Pathway Analysis identified retinoic acid receptor activation, apoptosis, and glutathione mediated detoxification signaling as the most affected pathways in the lower dose rate (0.54 mGy/h), while eif2 and mTOR, i.e., involved in the modulation of angiogenesis, were most affected in higher dose rates (5.4 and 10.9 mGy/h). By comparing gene expression data, myc was found to be the most significant upstream regulator, followed by tp53, TNF, hnf4a, TGFb1 and cebpa, while crabp2b and vegfab were identified as most frequent downstream target genes. These genes are associated with various developmental processes. The present findings show that continuous gamma irradiation (≥ 0.54 mGy/h) during early gastrula causes gene expression changes that are linked to developmental defects in zebrafish embryos.
Introduction
Living organisms are continuously exposed to ionizing radiation from naturally occurring radionuclides (e.g., radon daughters), cosmic radiation and from various anthropogenic activities (weapon testing, nuclear fuel reprocessing, nuclear accidents). Ionizing radiation interacts with matter by excitation and ionization of molecules, thereby producing free radicals and subsequently reactive oxygen species (ROS) and reactive nitrogen species (RNS) which can attack cell membranes or break chemical bonds in biological molecules, leading to oxidative stress or DNA damage [1]. Proliferating cells are specifically sensitive to radiation [2].
It is established that humans and animals are most vulnerable to ionizing radiation during early life stages such as gametogenesis, embryogenesis and organogenesis [3,4], due to the high rate of cell division, proliferation and differentiation. Ionizing radiation can affect all organs and biological systems, and can induce non-cancer effects as well as cancer [5]. Experimental studies have documented that exposure to ionizing radiation during critical periods of development may alter (reprogram) the differentiation signals leading to permanent toxic effects which can manifest later in life [5,6]. Permanent (irreversible) “developmental programming” is, among other mechanisms, attributed to epigenetic modification of gene transcription [7,8]. For aquatic organisms exposed to ionizing radiation, dose rates lower than 0.42 mGy/h (corresponding to 10 mGy/d) have been proposed as benchmark levels that are not likely to produce adverse effects at the population level [9]. Recently, a much lower predicted no effect dose rate (PNEDR) of 0.01 mGy/h (0.24 mGy/d) has been proposed as a risk assessment screening value below which one could be confident that exposures would not lead to adverse effects [10]. Protection criteria is based mostly on data from acute exposure experiments of adult organisms (IAEA), and the information on effects of ionizing radiation during sensitive life stages such as the embryonic and early larval development is scarce.
The zebrafish model offers many practical benefits, which may contribute to a better understanding of biological effects of radiation in both humans and non-human biota. Age-synchronized and optically transparent zebrafish embryos enable the visualization of major organ system development within all stages of the embryonic and early larval period. The available genomic resources in zebrafish, including a fully sequenced genome, have been proven valuable for providing various biological insights, including into human diseases [11]. Transcriptome analysis allows registration of changes in gene expression related to various biological processes and can be used to reveal potential mechanisms of radiotoxicity. The genome of the zebrafish is roughly half the size of the human genome and in comparison to it, shares about 70% of human gene orthologs and 82% of cancer gene orthologs [12,13].
This study aimed to assess biological effects such as survival, hatching and the occurrence of deformities in zebrafish exposed to gamma radiation (dose rates 0.4, 3.9, 15, and 38 mGy/h) and controls during embryogenesis and larval development (2.5 to 96 hpf). In order to elucidate the changes in gene expression with accompanying functional network analyses, RNA sequencing of total RNA extracted from irradiated (0.54, 5.4 and 10.9 mGy/h) pooled embryo samples and controls was performed. The embryos were exposed during 2.5–5.5 hpf, corresponding to the blastula period until the onset of the gastrula stage of development (256-cell stage until approximately 50% epiboly) [14]. This timeframe also includes the zygotic genome activation (ZGA) and onset of cell specification takes place [15–17]. The early molecular events initiated by a very low total dose of gamma radiation at 5.5 hpf and analyzed by transcriptomics were shown to be consistent with the observed developmental adversity in later stages.
Materials and methods
Ethics statement
The research was carried out according to the Norwegian Animal Protection Act (implemented EU Directive 2010/63/EU) and approved on December 12, 2013 by IACUC at Norwegian School of Veterinary Science (since 2014 Norwegian University of Life Sciences, Faculty of Veterinary Medicine and Biosciences, Oslo, Norway), under approval number FOTS ID 5793.
Zebrafish maintenance and embryo handling
Embryos from the AB wild type strain were obtained from the NMBU zebrafish facility (Norwegian Zebrafish Platform) and maintained according to standard operating procedures. Zebrafish were kept at 28 ± 1°C on a 14–10 hour light-dark cycle at a density of 5–10 fish/L. The system water (SW) was prepared from particle and active charcoal filtered reverse osmosis (RO) deionized tap water, kept sterile by UV irradiation. To generate a conductivity of 500 μS/cm, general hardness (GH) of 4–5 and pH 7.5 (adjusted with 1M HCl), 155 mg synthetic sea salt (Instant Ocean, Blacksburg, USA), 53 mg sodium carbonate and 15 mg calcium chloride (Sigma-Aldrich) was added per liter RO water. Adult fish were fed with Gemma Micro 300 (Skretting, Stavanger, Norway) dry feed twice a day and live artemia (Scanbur, Copenhagen, Denmark) once a day. Health was monitored by daily inspection, sentinel fish were sent to ZIRC for pathology every six month and water sent for microbiology analysis (NMBU Vetbio, Oslo). Adult fish were allowed to mate for 30 min in standard 1 L breeding tanks (Aquatic Habitats, Apopka, FL). For gamma radiation experiments, embryos were collected immediately after breeding and individually placed in 2 first rows of replicate 96 well plates (Nunc™, Thermo-Fisher Scientific, Waltham, Massachusetts, USA) with 200 μl of egg water (temperate autoclaved system water). A second group of embryos was placed in 2.5 ml tubes (Thermo-Fisher Scientific, Waltham, Massachusetts, USA) (50 embryos/ tube) with 2 ml egg water.
Embryo irradiation and dosimetry
After collection, embryos were transported to the FIGARO experimental irradiation facility at NMBU, Ås, Norway (60Co source, activity ~ 420 GBq). For both the toxicity endpoints and transcriptomic analyses, external gamma irradiation of zebrafish embryos commenced at 2.5 hpf with total doses to water ranging from 1.62 mGy– 3496 mGy during a 3 hour, 43.8 hour and 92 hour irradiation timespan (Table 1). Dose rates of 0.4, 3.9, 15 and 38 mGy/h were used for general toxicity analyses, and 0.54, 5.4 and 10.9 mGy/h for the transcriptomic analyses (Table 1). The experiments for both analyses were performed at separate time intervals. All exposures included corresponding controls. For the adverse effect observations and RNA-seq, 96-well plates and 2.5 ml tubes, respectively, were positioned at different distances from the gamma source corresponding to the dose rates to water (DWater) presented in Table 1.
Table 1. Exposure groups and dosimetry.
Total doses from measured dose rates during different time periods of exposure. (A): 43.8 hours; (B): 92 hours and (C): 3 hours.
| Developmental traits | Dose rate DWater (mGy/h)* | 0.4 | 3.9 | 15 | 38 | |
| Total dose DWater (mGy) | (A) | 17.5 | 171 | 657 | 1664 | |
| (B) | 36.8 | 359 | 1380 | 3496 | ||
| RNA-seq | Dose rate DWater (mGy/h)* | 0.54 | 5.4 | 10.9 | ||
| Total dose DWater (mGy) | (C) | 1.62 | 16.2 | 32.7 | ||
*Uncertainty (K = 2) for dose rate estimates is ~10% (Bjerke and Hetland, 2014).
Field dosimetry (air kerma rates measured with an ionization chamber) was traceable to the Norwegian Secondary Standard Dosimetry Laboratory (Norwegian Radiation Protection Authority, NRPA, Oslo, Norway) [18]. Average dose rates to water in the first and second rows of microplate wells were estimated according to established technical guidelines [19] and used as a proxy for dose rates to the fish embryos. Controls were placed in the same room, outside the beam cone and shielded by lead reducing the external (background) dose rate to ≤ 0.35 μGy/h (Thermo Eberline FHT6020). The irradiation room was thermostatically heated (28 ± 2°C), and had a 14–10 hours light-dark cycle (250–320 lx). To minimize variation in temperature, 2 control groups were used for the transcriptomic analyses.
Sampling procedure and experimental analysis of general toxicity endpoints
At approximately 48 hpf, half of the 96-well plates were removed from exposure (Table 1, Group “A”), while the remaining embryos were irradiated until 96 hpf (Table 1, Group “B”), n ≥ 145/ group. To determine the general toxicity in terms of adverse effects on survival and hatching, the embryos and larva were manually observed in a stereo microscope (3.5–45x) at 48 and 96 hpf in group “A”, and at 96 hpf in group in “B” (S1 Table). Additionally, the occurrence of deformities was observed at 96 hpf in both “A” and “B”. Analysis of endpoints was performed according to observations guidelines [20]. After observations, the larva used in this study was euthanized (prior to independent feeding at 120 hpf) using Tricaine (MS-222) (Sigma-Aldrich) overdose followed by rapid freezing (-70°C). For RNA extraction, embryos were sampled at 5.5 hpf (Table 1, Group “C”) in 2.5 ml tubes (n = 50/ sample).
Transcriptome analysis at 5.5 hours post fertilization
RNA sequencing was conducted to compare gene expression profiles between the controls and the 0.54, 5.4 and 10.9 mGy/h exposed embryos. Total RNA was isolated from embryos exposed between 2.5 hours and 5.5 hpf with TRIzol Reagent (Invitrogen) and purified with RNeasy Mini Kit (Qiagen) according to manufactures’ instructions. Briefly, 1 ml TRIzol was added to each sample consisting of 50 embryos and homogenized using Magnalyser Beads (Roche Diagnostics). Isolated RNA was DNase I (Qiagen) treated for 20 min at 25°C before further purification. Each sample was eluted in 50 μl RNase-free water and stored at − 80°C until required. RNA purity and yield (A260/A280 > 1.8, A260/A230 > 2, yield > 200 ng/μl) was determined using NanoDrop-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE) and quality (RIN > 8.5) was assessed with Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) using RNA Nano LabChip Kit (Agilent Technologies). None of the samples showed any signs of degradation or impurities. Photometric parameters and RNA integrity number (Bioanalyzer; Agilent technologies, USA) determined the quality of RNA sequenced samples. The RNA was sequenced (Illumina HiSeq 2000) at BGI Tech Solutions Co., Ltd., Hong Kong. Three single-end libraries (biological replicates), in the 5.4 and 10.9 mGy/h groups and a duplicate per 0.54 exposure group were sequenced. The bioinformatics analysis pipeline of the RNA sequencing data is presented in S1 Fig. Quality assessment of raw reads (49 nt long) and adapter trimming was performed using Trim Galore! v0.3.7, a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files [21,22]. Only reads with Phred score > 20 were kept. Afterwards, using TopHat v2.0.9 [23] with bowtie1, reads were mapped to the ZF genome (version Zv9, release 76) downloaded from Ensembl (http://www.ensembl.org/Danio_rerio/Info/Index). Options -g (maximum multihits number) was modified from its default value to 1, --no-coverage-search was allowed, --library type was set to “fr-unstranded” and -p (number of threads) was restricted to 4. As for bowtie1 options, -q (fastq files), -v (report end to end), -k 20 (report up to 20 good alignments), -m 20 (suppress all alignments if > 20), -S (to use SAM format) were used. BAM files were uploaded into Seqmonk [24] for visualization of aligned and mapped reads and read counting. Reads were counted as reads exactly overlapping with exons and the resulting count table was analyzed for gene expression under edgeR v3.4.2 Bioconductor [25]. The RNA-seq experiment was deposited in SRA database (https://www.ncbi.nlm.nih.gov/) and is publically available under accession SRP096352.
Quantitative real-time PCR (qPCR) analysis
To verify the RNA-sequencing results, eight differently expressed genes were selected for qPCR analysis, based on their common differential expression in the exposure groups. The DNA Sequence information for each gene was retrieved from genebank (http://www.ncbi.nlm.nih.gov/Genbank). The Primer3Plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/) was used to design primers. These primers were analyzed for oligo duplex and primer dimers. Amplicons which are shorter than 130 bp and spanned over different exons were selected (S1 Table). The cDNA was prepared from 1 μg of same total RNA used for RNA sequencing analyses (n = 3). For cDNA synthesis, Superscript III reverse transcriptase (Invitrogen, USA) and random hexamer primers were used according to product specifications. The qPCR was performed on a LightCycler® 96 Real-Time PCR system (Roche, Mannheim, Germany) using LightCycler® 480 SYBR Green I Master (Roche). Each cDNA was analyzed in a duplicate and composed of 5 μL mastermix, 2 μL primer mix (5 μM of each forward and reverse), and 3 μL of each 10× diluted cDNA sample in a total volume of 10 μL. The cycling parameters were 10 min pre-incubation at 95°C, followed by 45 cycles of amplification at 95°C for 10 sec, 60°C for 10 sec and 72°C for 8 sec, followed by a melting curve from 60°C to 95°C. The qPCR assay was performed in three biological replicates. RefFinder analysis tool (http://fulxie.0fees.us/) [26] was used to find the best candidate reference genes. Analyzed reference genes were hmbs (hydroxymethylbilane synthase), b-actin (beta-actin) and rps18 (ribosomal protein S18). For all exposure groups, hmbs was found to be the most stable housekeeping gene. The expression of each target gene transcript was normalized to hmbs and the fold change was calculated using the ΔΔCT method.
Ingenuity pathway analysis
For predicted networks/pathways and biological function analyses of differently transcribed genes, IPA software (http://www.ingenuity.com, Ingenuity Systems Inc., Redwood City, CA) was used. The Core analysis and comparison sub analysis blocks were used to determine the interaction networks of up- and down-regulated genes, upstream regulators and biological states (diseases and bio functions) in each and across the three exposure doses. A right-tailed Fisher’s exact test was used to determine the probability that each biological function is due to chance alone and the association identified as statistically significant and non-random (p < 0.05). The results in gene regulation are given as negative logarithms of the p-value computed by numbers of genes participating in the process, number of genes from the reference dataset mapped to the network and the size of the entire network in the Ingenuity knowledge database. The upstream regulator analysis examines how many known targets of each transcription regulator are present in the dataset, comparing their expression to what is known from the literature. In the present study, ranking by overlap p-value (cutoff p-value ≤ 0.001) and filtering for genes, RNAs and proteins in order to predict the most relevant transcriptional regulators was used. For the predicted activation state of the transcription regulators, a z-score describing the quantity of activated (z-score > 0) or inhibited predictions (z-score < 0) was calculated. However, this prediction is not available for upstream regulators with less target genes in the datasets (i.e. in lower dose-rates), and could not be considered to determine the most likely relevant regulators where the value of the correction for the z-score was too high (bias > 0.25).
Statistical analyses
After establishing the database for the general toxicity observations, tabulating and checking for errors in Excel®, data were transferred to Stata (MP/14 for Windows, StatCorp, College Station, TX). Confidence intervals were calculated using the proportion command for each of the outcomes survival, hatching and deformities at dose levels and the two exposure durations. Further logistic regression reported as odds ratios (OR) was used to estimate the effect of the treatments on hatching, survival and deformities and standard methods were used to check model fit. If significant, multiple comparisons were conducted using Tukey’s or Dunnett’s tests (Graphpad Prism 6, La Jolla, USA). Statistical significance was set to p < 0.05.
For analysis of gene expression, the dataset was TMM normalized first (trimmed mean of M-values, edgeR v3.4.2 Bioconductor, Robinson, McCarthy, and Smyth 2010), followed by data exploration using the statistical package R v3.0.2 [27]. Data was explored for descriptive statistics such as: minimum, maximum, 1st quantile, 3rd quantile, median, mean, standard deviation, also the similarity among samples was determined by correlation analysis and hclust (ward method) analysis to determine the distance between samples. The statistical analysis of differentially expressed genes (DEGs) was based on pairwise comparison between treatment and control RNA-seq samples (biological replicates) with a cut off set to ± 0.40 log2 fold change (1.3 FC). The FDR (false discovery rate) was set up to a significance of p ≤ 0.05. Venn diagram (Venny v2.1, Oliveros, (2007–2015)) was used to explore overlapping differential expressed genes among radiation treatments. For qPCR, obtained mean relative gene expression values (exposed vs. control) were compared to mean relative gene expression values for the same genes from RNA-seq and a Pearson’s correlation coefficient was calculated (p < 0.05) for all three exposure groups (Graphpad Prism 6, La Jolla, USA).
Results
General toxicity
To determine the effects of gamma radiation on the embryonic and larval development, the survival, hatching rate and deformities were assessed at 48 and 96 hpf. Compared to controls, a decrease in survival was observed in all exposed groups, albeit only the 38 mGy/h group was statistically significant, both after a 43.8-hour and 92-hour exposure (Table 2, S2 Table). The timing of hatching was significantly affected by irradiation, as a premature onset of hatching in the 0.4 mGy/h group (p < 0.0001) and a delayed onset of hatching in the 38 mGy/h group (p = 0.0072), respectively, were observed (S2 Table). The total hatching was above 95% in all exposure groups, however, with significantly lower total hatching in fish exposed to 15 mGy/h compared to controls (Table 2, S2 Table). The deformity frequency at 96 hpf increased linearly in response to dose for both the 43.8- and 92-hour exposure (linear regression, R2 = 0.93 and R2 = 0.99, respectively) and was significantly higher than in controls (p < 0.05) in all exposure groups, except from the 43.8-hour exposure to 0.4 mGy/h and 3.9 mGy/h (Fig 1, Table 2). The lowest dose rate (0.4 mGy/h) caused significant increase in deformities (p = 0.049) only in the group exposed for 92 hours (Fig 1, Table 2). The most frequently observed deformities were retardation in development manifested as failed hatching and absence of pigmentation, irregularities in formation of the head and eyes, as well as a short tail or lack of a tail (S15 Fig). In summary, a significant dose dependent response was observed for deformities and mortality, whereas hatching showed a non-monotonic dose-response.
Table 2. Adverse effects.
Adverse effects on total hatching, survival and deformities at 96 hpf, reported as Odds Ratios with 95% confidence intervals and related p-values compared to the base level (OR = 1). The OR describes the risk for occurrence of an adverse effect, given the two variables: dose rate and duration of exposure to the specified dose-rates. Significance denoted with (*).
| Odds ratio (95% CI); p-values compared to base level | ||||
|---|---|---|---|---|
| Variables | Hatching | Survival | Deformities | |
| Dose rate (mGy/h) | Control | 1.00 (-) | 1.00 (-) | 1.00 (-) |
| 0.4 | 0.40 (0.08–2.10); 0.28 | 0.65 (0.41–1.04); 0.07 | 5.00 (1.09–23.0); 0.04* | |
| 3.9 | 0.39 (0.08–2.03); 0.26 | 0.66 (0.42–1.06); 0.09 | 8.44 (1.93–37.0); 0.005* | |
| 15 | 0.13 (0.03–0.59); 0.008* | 0.75 (0.47–1.20); 0.23 | 13.43 (3.16–57.0); <0.001* | |
| 38 | 0.26 (0.05–1.24); 0.09 | 0.46 (0.29–0.73); 0.001* | 18.4 (4.37–77.6); <0.001* | |
| Duration of exposure (hours) | 43.8 | 1.00 (-) | 1.00 (-) | 1.00 (-) |
| 92 | 0.77 (0.45–1.33); 0.35 | 0.99 (0.78–1.27); 0.98 | 1.61 (1.09–2.37); 0.015* | |
Fig 1. Deformities.
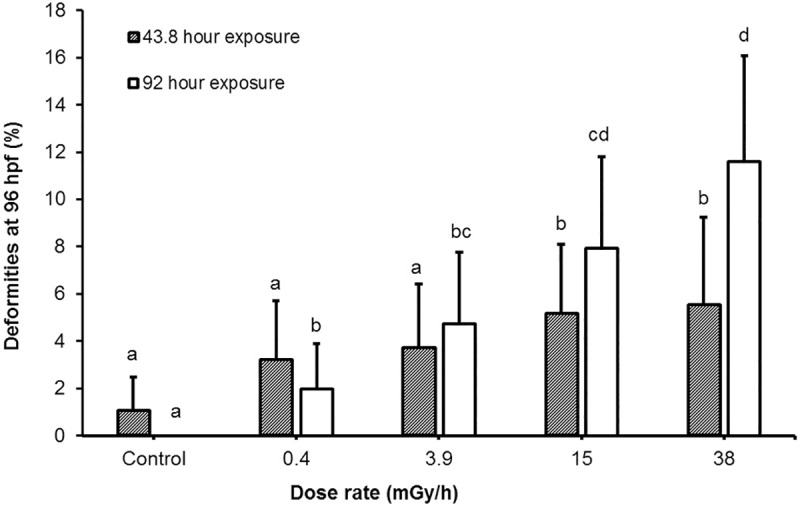
Deformities observed at 96 hpf which occurred after a 43.8- and 92-hour exposure to the specified dose rates. The exposures had separate controls. Values presented as mean percentage ± 95% confidence interval (p < 0.05).
Gene expression analysis
Transcriptional analysis was performed at the gastrula stage 5.5 hpf in order to identify potential changes to the transcriptional program induced by the gamma exposures. An average of 27 million reads (49 nt long) were obtained in both the treated and control groups. The mapping statistics showed a high grade of similarity among all samples (S2 Fig, S3 Table). The expression dataset analysis for replicability and distribution by means of multidimensional scaling plot (MDS) showed a clear difference between exposed and their respective controls (S3 Fig). A total number of ~10000 genes was expressed in all samples, while the number of differentially expressed genes (DEGs) showed a clear dose rate dependency (Fig 2 and S4 Fig and the full list of DEGs is available in S4 Table).
Fig 2. Expressed and differentially expressed genes in each exposure.
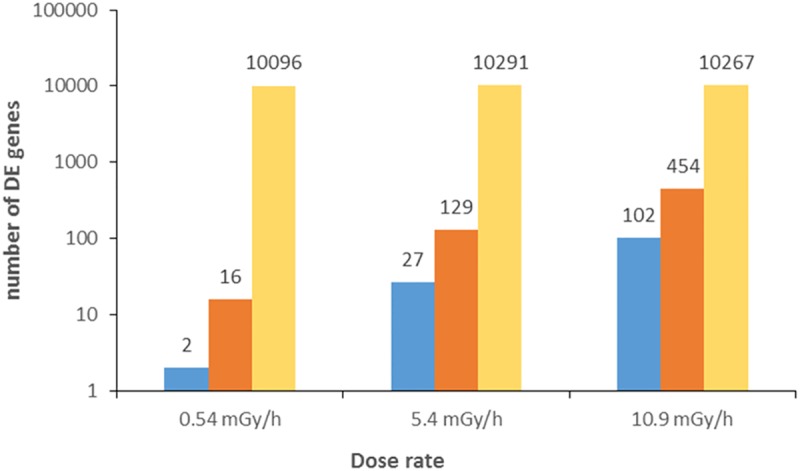
Threshold set to FC ±1.3 FDR < 0.05; down-regulated genes (blue), up-regulated genes (red) and total number of expressed genes (yellow).
In the 0.54 mGy/h exposure group, 16 genes were up-regulated (FC from 1.3 to 2.2) and two genes down-regulated with FC from 1.3 to 1.7 (Fig 2, S4 Table). In the 5.4 mGy/h exposure group, 129 genes were up-regulated with FCs from 1.3 to 674, while 27 were down-regulated with FCs from 1.3 to 2 (Fig 2, S4 Table). In the 10.9 mGy/h exposure group 556 DEGs were split between 454 up-regulated with FCs from 1.3 to 3.2 and 102 down-regulated genes with FCs of 1.3 to 2.4 (Fig 2, S4 Table). Among the DEGs, two were found to be differentially expressed in all three exposure groups: pfkfb3 (6-phosphofructo-2-kinase-fructose-2,6-biphosphatase 3) up-regulated in 0.54, but down-regulated in the 5.4 and 10.9 mGy/h; crabp2b (cellular retinoic acid binding protein 2b) which is similarly up-regulated in all exposure groups (Fig 3A, S4 Table).
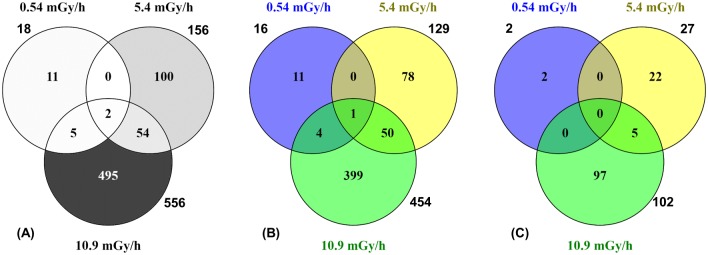
Fig 3. Venn diagram showing common and unique sets of differentially expressed genes between exposure treatments.
Total number of (A) common genes between 0.54, 5.4 and 10.9 mGy/h after pairwise comparison to controls (FC ± 1.3, FDR < 0.05); (B) Up-regulated genes; (C) Down-regulated genes.
In addition, five and 54 DEGs were overlapping between the 0.54/10.9 and 5.4/10.9 mGy/h groups, respectively (Fig 3A). As for the up-regulated DEGs, four genes were overlapping between the 0.54 and 10.9 mGy/h, while 50 genes were overlapping between the 5.4 and 10.9 mGy/h group (Fig 3B). Furthermore, down-regulated overlapping genes were found (five genes) only between the 5.4 and 10.9 mGy/h exposure (Fig 3C). The most up-regulated common gene in the 10.9 mGy/h group, tfa (transferrin-a), was also highly up-regulated in the 5.4 mGy/h group (S4 Table), although the FC values differed between the groups (S4 Fig). In addition, lipoprotein genes: apoBb (apolipoprotein Bb), apoA1a and apoA1b (apolipoprotein A-Ia/Ib), and common with the 10.9 mGy/h group, apoA-IV (apolipoprotein A-IV) were amid the top up-regulated in the 5.4 mGy/h group. The most down-regulated common gene between 5.4 and 10.9 mGy/h was vegfab (vascular endothelial factor Ab (S4 Table). The expression levels for up and down-regulated genes overlapping between the three dose rates are presented in Fig 4.
Fig 4. Expression levels for up and down-regulated overlapping genes between exposures.
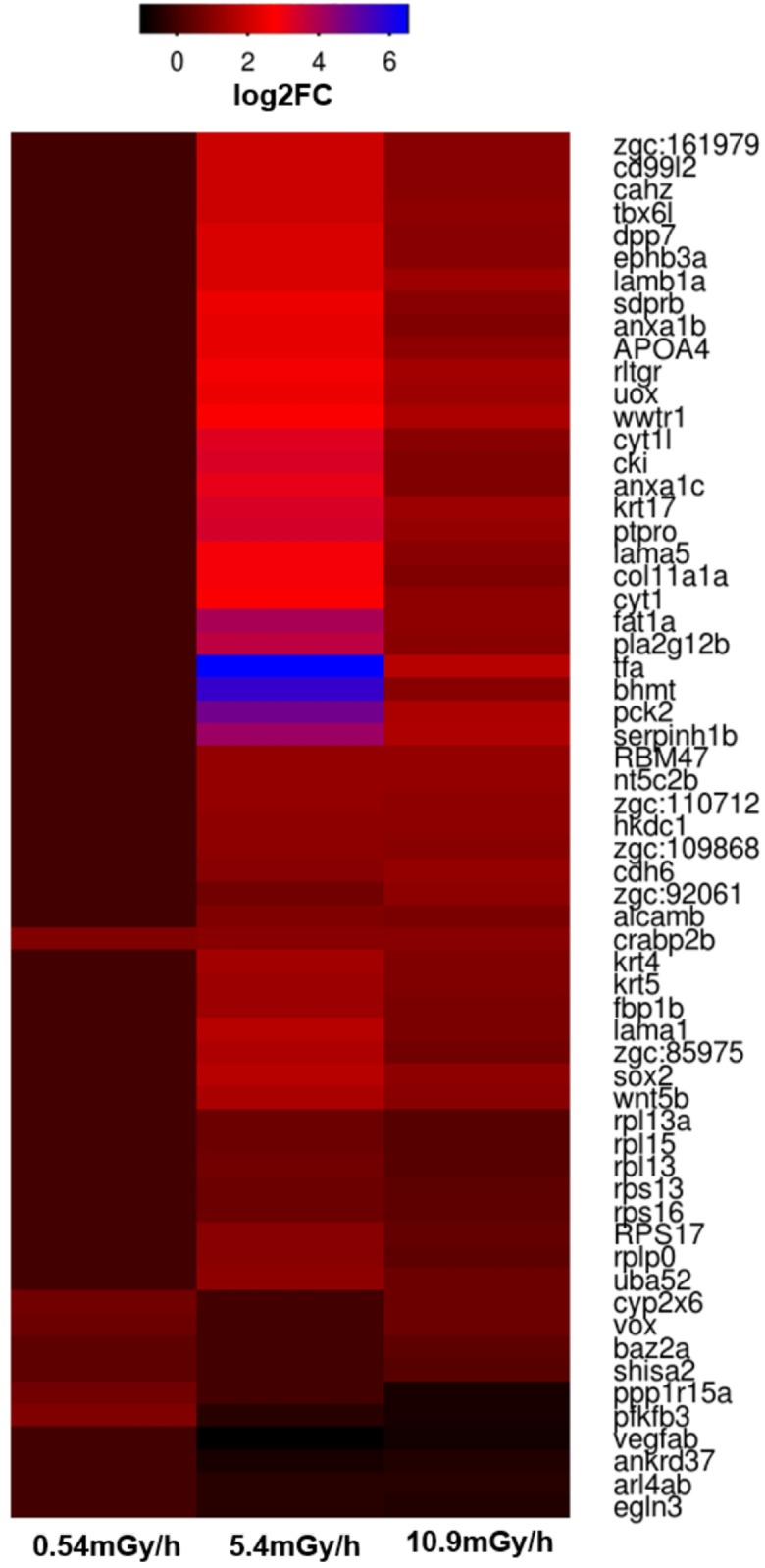
Expression levels in the 0.54, 5.4 and 10.9 mGy/h groups are given in log2 of the fold changes (FC).
Pathway analysis
General pathways analysis
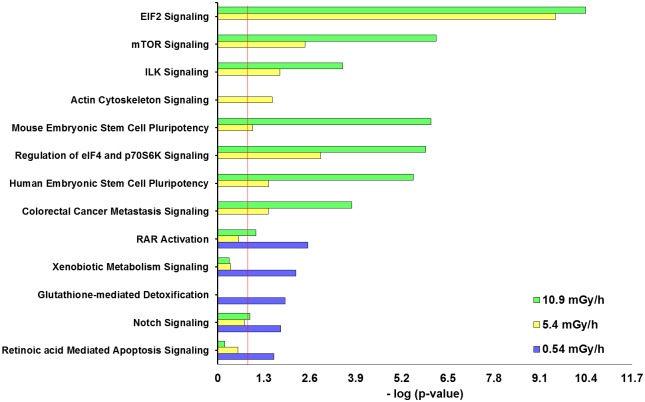
The core analysis IPA software tool was used to find the most significantly affected biological signaling (canonical) pathways by the DEGs in the three exposure groups. A statistically significant difference between the signaling pathways in the 0.54 mGy/h exposure group compared to the 5.4 and 10.9 mGy/h was found. In the lowest dose rate, the most affected signaling pathway was retinoic acid receptor activation (RARa), followed by RA mediated apoptosis and glutathione mediated detoxification signaling (Fig 5).
Fig 5. DEG functional analysis (IPA).
Top signaling pathways in canonical pathway comparison between all exposure groups: 0.54 mGy/h, 5.4 mGy/h and 10.9 mGy/h. The rankings were based on Fisher's exact test and high-ranking categories are displayed along in a decreasing order of significance from top. The threshold line (red) denotes the cut-off for significance (p-value 0.05).
Interestingly, compared to the signaling pathways in 0.54 mGy/h, the higher doses demonstrated some RA pathway activity, but this was below the significance threshold (Fig 5, S11 Fig). In the two higher dose rates, the most significantly affected signaling pathways were eif2 (eukaryotic initiation factor 2) and mTOR (mechanistic target of rapamycin), which were not affected (p-value > 0.05) in the lowest dose rate group (Fig 5, S12 and S13 Figs).
Toxicological pathways
To identify the top diseases and biological functions of altered genes in each exposure group, the gene expression data sets were compared between all exposure groups in IPA. The DEGs in the datasets were shown to be involved in gene networks associated with various embryonic developmental processes and cell functions (Fig 6).
Fig 6. The most affected biological function and diseases networks.

IPA predictions in comparison analysis between 0.54 (1) 5.4 (2) and 10.9 mGy/h (3) group. The heat map is based on the activation z-score, consistent with the particular activation state: “activated” (orange) or “inhibited” (blue). Z-score cut off set to ± 2.5 (arbitrary).
In the 0.54 mGy/h exposure group, gene networks associated with apoptosis and other cell death mechanisms were active, while gene networks associated with organismal death and proliferation of tumor cell lines (Fig 6) were inhibited. In contrast, in the 5.4 and 10.9 mGy/h groups, gene networks associated with apoptosis were inhibited and gene networks related to proliferation of tumor cell lines were active. Similarly to the lower dose rate exposure, the gene networks related to organismal death in these groups were inhibited. Comparison of expression of apoptosis genes showed that of total 129 DEGs found in the network, 5 were found in the 0.54 mGy/h group (all up-regulated), 40 in the 5.4 mGy/h group (34 up- and 6 down-regulated) and 101 in the 10.9 mGy/h (83 up- and 18 down-regulated) (S14 Fig). The one common and similarly expressed gene between all exposures in the apoptosis network was crabp2b, while expression levels of 16 common genes between 5.4 and 10.9 mG/h groups differed (S14 Fig). Additionally, networks associated with cell movement, growth, cardiovascular developmental processes and cancer development were significantly activated in the two higher dose rate exposure groups; albeit more significantly in the highest dose (Fig 6).
Key regulators
A transcription factor enrichment analysis was conducted to identify upstream regulators of transcriptional networks modulated by ionizing radiation. A total of 159, 632 and 939 transcription regulators in the 0.54, 5.4 and 10.9 mGy/h exposures were identified, respectively (S5 Table). Myc (v-myc avian myelocytomatosis viral oncogene derived homolog), TNF (tumor necrosis factor), tp53 (tumor protein p53) and hnf4a (hepatic nuclear factor 4, alpha) were identified as upstream regulator genes in all exposure groups (S5 Table, S5–S8 Figs). In the two higher dose rates, TGFb1 (transforming growth factor, beta 1) and cebpa (CCAAT/enhancer binding protein C/EBP, subunit alpha) were found to be significant upstream regulators (S5 Table, S9 and S10 Figs).
Validation by quantitative real-time PCR (qPCR)
In order to validate the RNA sequencing results, eight differently expressed genes were selected for quantitative real-time polymerase chain reaction (qPCR) analyses in the groups exposed to 0.54, 5.4, 10.9 mGy/h and controls. The selected genes and their respective fold changes are presented in Table 3. The data from real-time qPCR and the RNA-sequencing showed a good correlation (Pearson’s linear correlation coefficient r = 0.89, p < 0.0001).
Table 3. Real time qPCR verification of RNA sequencing.
Results presented as fold change (FC) for eight genes. The (n.a.*) refers to not differentially expressed, while the fold change was not available for this gene in this group.
| Gene ID | FC RNA-seq | FC RT qPCR | |||||
|---|---|---|---|---|---|---|---|
| Dose rate (mGy/h) | 0.54 | 5.4 | 10.9 | 0.54 | 5.4 | 10.9 | |
| pfkfb3 | 1.8 | 1.3 | 1.5 | 1.8 | 1.4 | 0.9 | |
| crabp2b | 1.9 | 2.1 | 2.0 | 2.9 | 2.3 | 2.0 | |
| vox | 1.5 | 1 | 1.5 | 1.6 | 2.2 | 0.7 | |
| ppp1r15a | 1.6 | 0.8 | 1.5 | 1.8 | 1.3 | 0.6 | |
| shisa2 | 1.3 | 1 | 1.3 | 1.5 | 1.9 | 0.7 | |
| sox2 | n.a.* | 3.3 | 2.2 | 3.0 | 1.6 | 0.8 | |
| tfa | n.a.* | 93.2 | 3.2 | 1.2 | 40.7 | 20.7 | |
| eef2b | 1.1 | 1.3 | 1.2 | 0.6 | 1.6 | 1.4 | |
Two of the selected genes are common between all three exposure groups (pfkfb3 and crabp2b). Three are common between 0.54 and 10.9 mGy/h groups (vox, ppp1r15a and shisa2) and between 5.4 and 10.9 mGy/h (sox2, tfa and eef2b). Only two genes were found to have an opposite regulation at one of the dose rates; pfkfb3 in the 5.4 mGy/h group was up-regulated, while shisa2 in the 10.9 mGy/h was down-regulated (Table 3).
Discussion
Previous studies in zebrafish reported underlying molecular mechanisms responsible for adverse biological effects such as DNA damage [28,29], ROS, oxidative stress, apoptosis, bystander effects [30–32] and also genetic [32–34] and epigenetic changes [8] following exposure to ionizing radiation. However, most of the genetic responses were studied following acute exposures.
In this study, we focused on potential adverse effects on the embryonic development caused by low dose and dose rate ionizing radiation. To this end, we investigated the developmental and toxicological effects of continuous gamma irradiation (doses between 17.5–3496 mGy) during early blastula (2.5 hpf; 256-cell stage), through to the hatching period (48–72 hpf) and early larval development, i.e., life stages associated with numerous delicate morphological changes [35].
To investigate molecular initial events associated to effects of ionizing radiation later in development, analysis of the gastrula stage 5.5 hpf embryo transcriptome was carried out using RNA sequencing combined with a functional gene network analysis software.
Adverse effects of radiation in developing embryos
The results from the observations of survival, deformities and total hatching at 96 hpf showed that radiation caused a significant dose-dependent reduced survival, affected the total hatching and increased the number of deformities. (Table 2, Fig 1). The exposure dose rates for evaluating the phenotypic effects used in the present work (0.4, 3.9, 15, and 38 mGy/h) were higher than the ERICA screening value of 10 μGy/h (0.24 mGy/d) [10]. However, the dose-rates span the proposed level of 0.42 mGy/h (10 mGy/day), which is considered to be a level below which there is not likely to be any detrimental effect on aquatic populations (UNSCEAR Report, 1996) and the derived consideration reference levels (DCRL) for fish (~0.42 mGy/h– 40 mGy/h), at which there are “likely to be some observable adverse effects occurring to individuals” [36].
The lowest dose rate in the present work at which deformities were observed was 0.4 mGy/h (total dose 36.8 mGy). The onset of hatching was premature in the 0.4 mGy/h exposure group (17.5 mGy total dose, Table 1), and significantly delayed in the 38 mGy/h group (1664 mGy total) (S2 Table). The total hatching in these groups was unaffected (Table 2). Interestingly, in a previous study of hatching intervals following X-ray exposure during the blastula stage, earlier hatching was associated with low dose (25 mGy at 0.43 Gy/min), while higher doses (250–500 mGy) delayed the onset of hatching [37]. In addition, other studies report that both low and high doses had an accelerating effect on the hatching interval [28,32]. In the 15 mGy/h group, the total hatching was significantly decreased (Table 2). A similar result was reported after X-rays exposure to 500 mGy [37], which is close to the total dose (657 mGy) in the present 15 mGy/h exposure group (Table 1). The survival, although exceeding 82% in all groups (S2 Table), was significantly lower than control in the 38 mGy/h group (Table 2) after both 43.8 and 92 hours of exposure. Previously, mortality in zebrafish embryos was reported only for acute exposures from 1 to 24 hpf (1–10 Gy, X-rays) [38]. Although embryo mortality from the 43.8 h exposure was observed at 48 hpf, no further increase was observed at 96 hpf (S2 Table). Collectively, these observations might indicate that the early developmental stages, prior to the hatching interval, are more sensitive to the effects of ionizing radiation, resulting in mortality. Other studies have reported no significant differences in embryo viability after receiving acute ionizing radiation doses ranging from 1–10 Gy [34,39], nor following continuous exposures up to 24 mGy/h (2280 mGy) [32], although the latter induced multiple deformities. Generally, the adverse effects on embryo development from the continuous exposure in the present study showed considerable variability in response to lower and higher doses, and in order to elucidate potential molecular mechanisms behind the observed effects, this variability was further studied by transcriptomics.
The 5.5 hours post fertilization embryo transcriptome
The gene expression analysis was performed at the late blastula / early gastrula stage (~ 5.5 hpf), a critical stage of embryogenesis, characterized by intensive cell proliferation and specification [17,35]. At this stage the zygotic genome is activated, while the inherited maternal transcript (synthesized during oogenesis and stored in the egg) is degraded [15]. Thus, changes in transcriptome profile can be attributed to radiation effects on the transcriptional program of the embryo’s own genome. The choice of this stage served two major aims: early toxic effects and accompanying stress or defense mechanisms would be reflected at the transcriptional level, and deviation of the transcriptional program at this stage could be indicative or predictive to adverse outcome observed later during embryogenesis. The applied dose rates were selected to both encompass a toxic effects dose response and to be environmentally relevant. The RNAseq analysis was thus conducted on low total doses, which consequently should produce only low level of DNA damage. This strategy enables investigation of more subtle and less well-described molecular effects of ionizing radiation in addition to genotoxicity. The fact that significant transcriptional changes could be observed from a 3 hour exposure to total doses from 1.6 to 33 mGy corroborates the validity of the approach. Moreover, the observed responses were intelligibly connected to the adverse outcomes observed at the phenotype level. This correlation is important with respect to the level of dose rates and total doses that would be required to elicit changes at the molecular level.
The number of similarly and differently expressed genes, as well as overlapping DEGs, showed a clear dose-response effect in the gamma exposed embryos with the lowest number of modulated genes in the 0.54 mGy/h group and with an increasing number in the two highest exposure groups (5.4 and 10.9 mGy/h) (Fig 2). A considerable variation in FC between the 5.4 and 10.9 mGy/h groups was observed (S4 Fig), but a total of 56 DEGs were common in these exposure groups.
Two genes, pfkfb3 and crabp2b, were found to be differentially expressed in all exposures. The pfkfb3 gene is involved in regulating the expression of cyclin-dependent kinase 1, which promotes proliferation and survival in tumor cells [40] by protecting cancer cells against oxidative stress through S-glutathionylation and glucose metabolism switch to the pentose phosphate pathway [41,42], and thereby counteracting ionizing radiation generated ROS. The crabp2b gene is a one of the two zebrafish crabp2 genes orthologous known to encode retinoic acid (RA) protein family and lipocalin/cytosolic fatty acid binding protein family. Interestingly, the crabp2b was found to be similarly up-regulated (FC ~ 2) in all three irradiation treatments in both the RNA-seq and the qPCR (Table 3, Fig 4). Retinoic acid is the biological active metabolite of Vitamin A and crabp2 regulates the access of retinoic acid by binding with the nuclear retinoic acid receptor alpha (RARa) [43] and helps modulating the RA gradient, which is important for the development of vertebrates, including humans [43,44]. Deficient or excess levels of vitamin A have induced malformations in experimental animals and humans, indicating that the concentration must be kept within a narrow range [45,46]. Furthermore, crabp2b is associated with regulation of the hindbrain anterior-posterior axis development [47], and is expressed in structures requiring the retinoic acid during embryonic development, such as the CNS, dorsal retina, branchial arches, epidermis, otic vesicle and pectoral fins [43]. Considering the increased number of malformations observed in irradiated fish, it could be hypothesized that this is in part induced by modification of the crabp2b gene.
Among the common genes modulated by 5.4 and 10.9 mGy/h exposures, the most significantly up-regulated gene is tfa (Fig 4). This gene is critical for iron transport and iron regulated hormone expression [48], and is involved in the immune response to bacterial infection [49]. A decrease in concentration of the transferrin protein was found in blood plasma of radiological accident victims compared to blood plasma from non-irradiated individuals, and reported as a possible mutagenic factor [50]. However, a protective role of the transferrin pathway for antioxidant repair and sequestering metals was also suggested [51]. Additionally, increased chromosomal damage combined with increased transferrin was demonstrated in lymphocyte cultures following exposure to 1 Gy of ionizing radiation, suggesting that transferrin is affected by radiation [52].
The highly up-regulated apolipoprotein genes in the two higher dose rate exposure groups, and notably the 5.4 mGy/h group (apoBb, apoA1a, apoA1b and apoA-IV), could point to radiation affecting mechanisms behind the lipid metabolism and transport from yolk cells to the embryo (S4 Table) [53]. Apolipoprotein genes play a role in reducing fat intake during embryonic development, as previously shown in zebrafish [53] and humans [54], causing malnutrition of the embryo, which may have disrupted normal development. In addition, apolipoprotein genes were reported to negatively regulate (apoB) [55], or even inhibit the angiogenesis (apoA1), in a vegf down-regulation dependent pathway [56]. Relatedly, among the common genes modulated by 5.4 and the 10.9 mGy/h treatments, the most significantly down-regulated gene in both data sets is vegfab (FC 1.6–2), an isoform of the human ortholog VEGF-A (Fig 4) [57]. At early life stages, this gene mediates differentiation of endothelial cells and early vascular development and angiogenesis (formation of new blood vessels) [58], including retinal angiogenesis [59]. In developed individuals, vegf stimulates the angiogenesis [60], either in a physiological (such as tissue repair processes) or pathological states (such as tumor growth), and vegf activity has been shown to be stimulated through an intracellular increase in ROS generated as a result of exposure to ionizing radiation [61]. In an experimental study of radiation effects in mice, vegf together with eif2 was modulated in bladder tissue [62].
Molecular pathways—Potential mechanisms of radiotoxicity
A transcription factor enrichment analysis was performed to investigate whether gamma induced pathways or gene networks could be ascribed to master regulators. IPA analyzes of the datasets identified upstream regulator genes, which were not necessarily significantly affected, but may play key roles in the regulation of DEGs. The transcription factors myc, TNF, tp53 and hnf4a were found to be in central positions of functional networks of modulated genes in comparison between the three exposure groups (S5 Table). Additionally, TGFb1 and cebpa were identified as key regulators at the two higher dose-rates (5.4 and 10.9 mGy/h) (S5 Table).
Myc was found to be one of the top upstream regulators, in all three exposures (S5 Table, S5 Fig) and is implicated in the regulation of various processes in the cell, such as growth and proliferation, migration, differentiation and cell death. Up-regulation of the oncogenes myc and mycn is associated with poor outcomes of several cancers, such as aggressive neuroblastoma [63], large B cell lymphoma [64], acute myeloid leukemia (AML) [65] and nephroblastoma (Wilms tumor) [66]. Combined Myc up-regulation with an altered retinoic acid (RA) pathway activity worsens the prognosis of such cancers [67]. Furthermore, TNF was found to regulate a high number of molecules in the datasets (S5 Table, S6 Fig). This cytokine was previously shown to be strongly protective at lower ionizing radiation doses for the hematopoietic stem cell system [68] and via selective destruction of blood vessels in T-cell tumors [69]. Interestingly, the activity of tumor necrosis factor-alpha (TNF-α) in cell lines was found to be antagonistic to the activity of TGFb [70]. Another identified upstream regulator, tp53 (S5 Table, S7 Fig), is known to regulate apoptosis in response to DNA damage [71], but was also demonstrated to be a critical factor for normal development and survival in zebrafish embryos after exposure to ionizing radiation [72,73]. Tp53 was found to decrease, but also to concomitantly regulate tumor suppressive TGFb responses through Smad2/3 DNA complexes [74]. Although not differentially expressed in the 0.54 and 5.4 mGy/h datasets, hnf4a is found to be a transcription regulator for a large number of DEGs in all exposure groups (S3 Table, S8 Fig). This transcription regulator was found to be up-regulated in the blood of patients exposed to ionizing radiation [75], and in a human tissue model exposed to low dose gamma radiation [76]. Hnf4a regulates the gastrulation [77], the developmental period during which the morphogenetic cell movements, along with production of the three primary germ layers (ectoderm, mesoderm and endoderm) and the embryonic axis (> 5.25 hpf) occur [78]. It is mainly expressed in the digestive system and in the brain. This data propose hnf4a as a factor involved in the induction of biological effects of radiation in humans as well as in other vertebrate species.
An activated predicted upstream regulator in both the 5.4 and 10.9 mGy/h, but not in the 0.54 mGy/h exposure was TGFb1 (S3 Table, S9 Fig). The TGFb1 cytokine regulates a variety of functions, and is known to be a mediator of the apoptosis, redox homeostasis and bystander effects in tissues and cells in response to radiation [69,79–81]. In addition, TGFb was found to co-regulate angiogenesis in tumors with vegf [82]. IPA also identified cebpa as a regulator gene among the common DEG in the two higher dose rates (5.4 and 10.9 mGy/h) (S10 Fig). In the study of B-cell chronic lymphocytic leukemia (B-CLL) patients in the post-Chernobyl period, similar key regulator genes, gene networks and signaling pathways were altered [83]. Cebpa is associated with regulation of hematopoiesis, hematopoietic stem cell migration, liver development and regulation of transcription [84]. It is predominantly found in mature myeloid cells and is required for the differentiation of myeloid cells in order to prevent the occurrence of myeloproliferative diseases [85]. Diseases associated with a down-regulation of cebpa include acute myeloid leukemia with cebpa somatic mutations [86]. Moreover, other studies have reported that ionizing radiation caused increased expression of cebpa, which was associated with a reduction of hematopoietic stem cells and the self-renewal of multipotent hematopoietic progenitor cells [87]. The similar regulation of these genes in mammals and zebrafish may suggest that similar mechanisms might be behind the molecular changes following exposure to radiation.
The signaling pathways affected most significantly by the 0.54 mGy/h exposure (RAR activation, RA mediated apoptosis and glutathione mediated detoxification seem to be consistent with the described repair mechanisms occurring at low doses. This adaptive response to low doses of ionizing radiation in biological systems is mainly characterized by antioxidant mediated detoxification of ROS, more rapid DNA repair, apoptosis signaling and stimulated immune response [88,89].
In the 5.4 and 10.9 mGy/h treatments, eif2 and mTOR were the most significantly up-regulated signaling pathways. A significant role of the eif2 signaling pathway is the adaptive response to stress by regulating the formation of translation initiation complexes, which leads to reduced recognition of AUG start codons and therefore total translational inhibition and the induction of apoptosis [90]. The mTOR (mammalian target of rapamycin) signaling pathway is centrally involved in cell metabolism, growth, proliferation and survival via regulation of protein synthesis and mRNA stabilization [91]. Furthermore, it is activated during tumor formation and modulation of angiogenesis, development of diabetic retinopathy [92] and in radiation induced apoptosis [93]. A dysregulation of mTOR was reported to affect the premature aging of cells and destabilize the cytoskeletal structure after exposure to chronic ionizing radiation, in addition to changes in the eif2 signaling pathway [94]. The eif2 signaling pathway was in comparison to the present results found to be down-regulated in the blood of post Chernobyl leukemia patients [83]. The predicted top diseases and biological functions (IPA), suggest that the changes in signaling pathways and gene expression in the lower dose-rate (0.54 mGy/h) are activating gene networks associated with apoptosis and other cell death mechanisms in the embryos, while inhibiting proliferation of tumor cell lines (Fig 6). In the higher dose-rate exposure groups (5.4 and 10.9 mGy/h), gene networks involved in cell death and apoptosis were shown to be inhibited, while cell movement, cardiovascular development and tumor development were activated (Fig 6). The predictions from the gene expression suggest an early response of the developing embryos to continuous ionizing radiation and would be interesting to address in follow up studies using genetic, epigenetic and mutagenesis methods.
Conclusion
Continuous exposure to external gamma radiation at environmentally relevant dose-rates (from 0.4 mGy/h, total dose 17.5 mGy) resulted in severe consequences for the development and gene expression of zebrafish embryos and larva. Significant mortality compared to controls was observed in the groups exposed to the highest dose rate (38 mGy/h), while increased number of deformities and differences in the hatching was observed in groups exposed to lower doses ≥ 0.4 mGy/h (Tables 1 and 2 and S2 Table). Consistent with the observed adverse effects, the changes in gene transcription could be attributed to cell differentiation and morphological development. The results suggest that active repair mechanisms mediated by antioxidants could be the reason for the lack of phenotypic observable effects in the lower dose. The higher radiation dose rates instigate, among others, genes and networks involved in cell cycle control (tp53), translation and cell survival (eif2, mTOR), and disrupted development and cancer (myc, TGFb1, hnf4a, cebpa), which in sum increase the risk for an adverse effect. Thus, RNA sequencing enabled identification of molecular initiating events from a 3 hour gamma radiation exposure to 0.54, 5.4 and 10.9 mGy/h (total dose 1.6 to 33 mGy), which are consistent with the phenotype level adverse outcomes observed in 96 hpf stage larvae.
Supporting information
(XLSX)
Survival and hatching after 43.8 and 92 hours exposure to specified dose rates. Survival at the 43.8 hours exposure did not differ at 48 and 96 hpf. All values presented as mean percentage ± 95% confidence interval (CI).
(XLSX)
Mapping statistics presented separately for each replicate (A) and each exposure (B) with their respective controls. Approximately 60% of the reads were mapped to the reference genome. Of the mapped reads, ~ 40% were mapped when allowing no mismatch, while ~ 15% of the reads were mapped when ≤ 2 bp mismatches were allowed. On the other hand, ~ 56% out of the mapped reads were found to represent unique genome positions, with ~ 1.5% of reads mapping to multiple positions.
(XLSX)
(XLSX)
(XLSX)
(TIF)
Differential expression threshold is FC ± 1.3. A, B, C and D show the distribution of mapped reads at 0.54, 5.4 and 10.9 mGy/h. E and F represent the distribution of mapped reads in control groups for the lowest (0.54 mGy/h) and for higher dose rates 5.4 and 10.9 mGy/h, respectively. All libraries were mapped to the ZF genome (Zv9).
(TIF)
A) Group exposed at 0.54 mGy/h and the control group for the lowest dose. Two and three biological replicates of the exposed group and controls, respectively, were included in the analysis. B) and C) Groups exposed to 5.4 and 10.9 mGy/h and controls. Three replicates were included.
(TIF)
Analysis was conducted by pairwise comparison of exposed and their respective controls. A) 0.54 mGy/h, B) 5.4 mGy/h and C) 10.9 mGy/h. Expression values were log2 transformed. Black and red dots represent non-differential and differentially expressed genes respectively (FDR < 0.05) (edgeR v3.4.2 Bioconductor).
(TIF)
Myc target gene networks and interactions, presented in a subcellular layout as part of the 10.9 mGy/h group.
(TIF)
TNF target gene networks and interactions, presented in a subcellular layout as part of the 10.9 mGy/h group.
(TIF)
Tp53 target gene networks and interactions, presented in a subcellular layout as part of the 10.9 mGy/h group.
(TIF)
Hnf4a target gene networks and interactions, presented in a subcellular layout as part of the 10.9 mGy/h group.
(TIF)
TGFB1 target gene networks and interactions, presented in a subcellular layout as part of the 10.9 mGy/h group.
(TIF)
Cebpa target gene networks and interactions, presented in a subcellular layout as part of the 10.9 mGy/h group.
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
The observations were done at 96 hours post fertilization (hpf). A. Control larva showing normal development; B-C. Larvae exposed to 38 mGy/h for 92 hours (Group “B”), demonstrating general developmental defects and short-tails.
(TIF)
Acknowledgments
We would like to thank Prof. Dr Anton Gartner for valuable comments.
Data Availability
All relevant data are within the paper and its Supporting Information files. The RNA-seq experiment was deposited in SRA database (https://www.ncbi.nlm.nih.gov/) and is publically available under accession SRP096352.
Funding Statement
This work was supported by the Norwegian Research Council funded through the Centre of Excellence CERAD – Centre for Environmental Radioactivity (project 223268/F50).
References
- 1.Jarvis R., Knowles J. DNA damage in zebrafish larvae induced by exposure to low-dose rate γ-radiation: detection by the alkaline comet assay. Mutat Res Toxicol Environ Mutagen. 2003;541: 63–69. [DOI] [PubMed] [Google Scholar]
- 2.Bergonié J, Tribondeau L. Interpretation of some results from radiotherapy and an attempt to determine a rational treatment technique. 1906. Yale J Biol Med. Yale Journal of Biology and Medicine; 2003;76: 181–2. [PMC free article] [PubMed] [Google Scholar]
- 3.Jacquet P. Sensitivity of germ cells and embryos to ionizing radiation. J Biol Regul Homeost Agents. 2004;18: 106–14. [PubMed] [Google Scholar]
- 4.Streffer C. Bystander effects, adaptive response and genomic instability induced by prenatal irradiation. Mutat Res. 2004;568: 79–87. doi: 10.1016/j.mrfmmm.2004.07.014 [DOI] [PubMed] [Google Scholar]
- 5.Li CI, Nishi N, McDougall JA, Semmens EO, Sugiyama H, Soda M, et al. Relationship between radiation exposure and risk of second primary cancers among atomic bomb survivors. Cancer Res. 2010;70: 7187–98. doi: 10.1158/0008-5472.CAN-10-0276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hudson D, Kovalchuk I, Koturbash I, Kolb B, Martin OA, Kovalchuk O. Induction and persistence of radiation-induced DNA damage is more pronounced in young animals than in old animals. Aging (Albany NY). 2011;3: 609–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skinner MK. Role of epigenetics in developmental biology and transgenerational inheritance. Birth Defects Res C Embryo Today. 2011;93: 51–5. doi: 10.1002/bdrc.20199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilnytskyy Y, Kovalchuk O. Non-targeted radiation effects-an epigenetic connection. Mutat Res. 2011;714: 113–25. doi: 10.1016/j.mrfmmm.2011.06.014 [DOI] [PubMed] [Google Scholar]
- 9.UNSCEAR. United Nations Scientific Committee on the Effects of Atomic Radiation (UNSCEAR) Report. New York; 1996.
- 10.Garnier-Laplace J, Della-Vedova C, Andersson P, Copplestone D, Cailes C, Beresford NA, et al. A multi-criteria weight of evidence approach for deriving ecological benchmarks for radioactive substances. J Radiol Prot. IOP Publishing; 2010;30: 215–33. doi: 10.1088/0952-4746/30/2/S02 [DOI] [PubMed] [Google Scholar]
- 11.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8: 353–67. doi: 10.1038/nrg2091 [DOI] [PubMed] [Google Scholar]
- 12.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496: 498–503. doi: 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White R, Rose K, Zon L. Zebrafish cancer: the state of the art and the path forward. Nat Rev Cancer. 2013;13: 624–36. doi: 10.1038/nrc3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kane D, Kimmel C. The zebrafish midblastula transition. Development. 1993;119: 447–456. [DOI] [PubMed] [Google Scholar]
- 15.Aanes H, Østrup O, Andersen IS, Moen LF, Mathavan S, Collas P, et al. Differential transcript isoform usage pre- and post-zygotic genome activation in zebrafish. BMC Genomics. 2013;14: 331 doi: 10.1186/1471-2164-14-331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Østrup O, et al. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell. 2011;21: 993–1004. doi: 10.1016/j.devcel.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 17.Haberle V, Li N, Hadzhiev Y, Plessy C, Previti C, Nepal C, et al. Two independent transcription initiation codes overlap on vertebrate core promoters. Nature. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved.; 2014;507: 381–5. doi: 10.1038/nature12974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlsson Tedgren Å, Bjerke H, Grindborg J-E, Hetland P-O, Kosunen A, Hellebust TP, et al. Comparison of high-dose-rate 192Ir source strength measurements using equipment with traceability to different standards. Brachytherapy. 2014;13: 420–423. doi: 10.1016/j.brachy.2014.01.002 [DOI] [PubMed] [Google Scholar]
- 19.Bjerke H, Hetland PO. Dosimetry at Figaro—gamma irradiation facility at NMBU, Aas; Dosimetri ved Figaro—gammaanlaget ved NMBU, Aas. Østerås; 2014.
- 20.OECD/OCDE. OECD GUIDELINES FOR THE TESTING OF CHEMICALS Fish Embryo Acute Toxicity (FET) Test. 2013. doi: 10.1787/20745761 [Google Scholar]
- 21.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17: 10 doi: 10.14806/ej.17.1.200 [Google Scholar]
- 22.Andrews S. FastQC A Quality Control tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 23.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25: 1105–11. doi: 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews SS mapped sequence analysis tool. (2012). Babraham Bioinformatics—SeqMonk Mapped Sequence Analysis Tool [Internet]. http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/
- 25.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26: 139–40. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 2012; doi: 10.1007/s11103-012-9885-2 [DOI] [PubMed] [Google Scholar]
- 27.R Foundation for Statistical Computing, Vienna A (2013). R: The R Project for Statistical Computing [Internet]. 2014. https://www.r-project.org/
- 28.Pereira S, Bourrachot S, Cavalie I, Plaire D, Dutilleul M, Gilbin R, et al. Genotoxicity of acute and chronic gamma-irradiation on zebrafish cells and consequences for embryo development. Environ Toxicol Chem. 2011;30: 2831–7. doi: 10.1002/etc.695 [DOI] [PubMed] [Google Scholar]
- 29.Simon O, Massarin S, Coppin F, Hinton TG, Gilbin R. Investigating the embryo/larval toxic and genotoxic effects of γ irradiation on zebrafish eggs. J Environ Radioact. Elsevier Ltd; 2011;102: 1039–44. doi: 10.1016/j.jenvrad.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 30.Bladen CL, Navarre S, Dynan WS, Kozlowski DJ. Expression of the Ku70 subunit (XRCC6) and protection from low dose ionizing radiation during zebrafish embryogenesis. Neurosci Lett. 2007;422: 97–102. doi: 10.1016/j.neulet.2007.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pereira S, Malard V, Ravanat J-L, Davin A-H, Armengaud J, Foray N, et al. Low doses of gamma-irradiation induce an early bystander effect in zebrafish cells which is sufficient to radioprotect cells. PLoS One. 2014;9: e92974 doi: 10.1371/journal.pone.0092974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gagnaire B, Cavalié I, Pereira S, Floriani M, Dubourg N, Camilleri V, et al. External gamma irradiation-induced effects in early-life stages of zebrafish, Danio rerio. Aquat Toxicol. 2015;169: 69–78. doi: 10.1016/j.aquatox.2015.10.005 [DOI] [PubMed] [Google Scholar]
- 33.Jaafar L, Podolsky RH, Dynan WS. Long-term effects of ionizing radiation on gene expression in a zebrafish model. PLoS One. 2013;8: e69445 doi: 10.1371/journal.pone.0069445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freeman JL, Weber GJ, Peterson SM, Nie LH. Embryonic ionizing radiation exposure results in expression alterations of genes associated with cardiovascular and neurological development, function, and disease and modified cardiovascular function in zebrafish. Front Genet. Frontiers; 2014;5: 268 doi: 10.3389/fgene.2014.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203: 253–310. doi: 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- 36.ICRP. ICRP 2012. Compend Dose Coefficients based ICRP Publ 60. 2012;0: 130 doi: 10.1016/j.icrp.2006.06.001 [Google Scholar]
- 37.Miyachi Y, Kanao T, Okamoto T. Marked depression of time interval between fertilization period and hatching period following exposure to low-dose X-rays in zebrafish. Environmental Research. 2003. [DOI] [PubMed] [Google Scholar]
- 38.McAleer MF, Davidson C, Davidson WR, Yentzer B, Farber SA, Rodeck U, et al. Novel use of zebrafish as a vertebrate model to screen radiation protectors and sensitizers. Int J Radiat Oncol Biol Phys. 2005;61: 10–3. doi: 10.1016/j.ijrobp.2004.09.046 [DOI] [PubMed] [Google Scholar]
- 39.Bladen CL, Kozlowski DJ, Dynan WS. Effects of low-dose ionizing radiation and menadione, an inducer of oxidative stress, alone and in combination in a vertebrate embryo model. Radiat Res. 2012;178: 499–503. doi: 10.1667/RR3042.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yalcin A, Clem BF, Simmons A, Lane A, Nelson K, Clem AL, et al. Nuclear Targeting of 6-Phosphofructo-2-kinase (PFKFB3) Increases Proliferation via Cyclin-dependent Kinases. J Biol Chem. 2009;284: 24223–24232. doi: 10.1074/jbc.M109.016816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seo M, Lee Y-H. PFKFB3 Regulates Oxidative Stress Homeostasis via Its S-Glutathionylation in Cancer. J Mol Biol. 2014;426: 830–842. doi: 10.1016/j.jmb.2013.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto T, Takano N, Ishiwata K, Ohmura M, Nagahata Y, Matsuura T, et al. Reduced methylation of PFKFB3 in cancer cells shunts glucose towards the pentose phosphate pathway. Nat Commun. 2014;5 doi: 10.1038/ncomms4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma MK, Saxena V, Liu R-Z, Thisse C, Thisse B, Denovan-Wright EM, et al. Differential expression of the duplicated cellular retinoic acid-binding protein 2 genes (crabp2a and crabp2b) during zebrafish embryonic development. Gene Expr Patterns. 2005;5: 371–9. doi: 10.1016/j.modgep.2004.09.010 [DOI] [PubMed] [Google Scholar]
- 44.Diez del Corral R, Storey KG. Opposing FGF and retinoid pathways: a signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. Bioessays. 2004;26: 857–69. doi: 10.1002/bies.20080 [DOI] [PubMed] [Google Scholar]
- 45.Blomhoff R, Beckman-Sundh, Brot C, Solvoll K, Steingrimsdóttir L C MH. Health risks related to high intake of preformed retinol (vitamin A) in the Nordic countries Tema Nord 2003:502 Nordic Council of Ministers, Copenhagen; 2003. [Google Scholar]
- 46.Gutierrez-Mazariegos J, Theodosiou M, Campo-Paysaa F, Schubert M. Vitamin A: a multifunctional tool for development. Semin Cell Dev Biol. 2011;22: 603–10. doi: 10.1016/j.semcdb.2011.06.001 [DOI] [PubMed] [Google Scholar]
- 47.Cai AQ, Radtke K, Linville A, Lander AD, Nie Q, Schilling TF. Cellular retinoic acid-binding proteins are essential for hindbrain patterning and signal robustness in zebrafish. Development. 2012;139: 2150–5. doi: 10.1242/dev.077065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fraenkel PG, Gibert Y, Holzheimer JL, Lattanzi VJ, Burnett SF, Dooley KA, et al. Transferrin-a modulates hepcidin expression in zebrafish embryos. Blood. 2009;113: 2843–50. doi: 10.1182/blood-2008-06-165340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rojo I, de Ilárduya OM, Estonba A, Pardo MA. Innate immune gene expression in individual zebrafish after Listonella anguillarum inoculation. Fish Shellfish Immunol. 2007;23: 1285–93. doi: 10.1016/j.fsi.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 50.Nylund R, Lemola E, Hartwig S, Lehr S, Acheva A, Jahns J, et al. Profiling of low molecular weight proteins in plasma from locally irradiated individuals. J Radiat Res. 2014;55: 674–82. doi: 10.1093/jrr/rru007 24570173 [Google Scholar]
- 51.Crichton RR, Wilmet S, Legssyer R, Ward RJ. Molecular and cellular mechanisms of iron homeostasis and toxicity in mammalian cells. J Inorg Biochem. 2002;91: 9–18. doi: 10.1016/S0162-0134(02)00461-0 [DOI] [PubMed] [Google Scholar]
- 52.Beckman L, Nordenson I. Transferrin C2 and radiation-induced chromosomal damage. Hum Hered. 1988;38: 56–8. [DOI] [PubMed] [Google Scholar]
- 53.Otis JP, Zeituni EM, Thierer JH, Anderson JL, Brown AC, Boehm ED, et al. Zebrafish as a model for apolipoprotein biology: comprehensive expression analysis and a role for ApoA-IV in regulating food intake. Dis Model Mech. Company of Biologists; 2015;8: 295–309. doi: 10.1242/dmm.018754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shoji H, Murano Y, Mori M, Matsunaga N, Ohkawa N, Suganuma H, et al. Lipid profile and atherogenic indices soon after birth in Japanese preterm infants. Acta Paediatr. 2014;103: 22–6. doi: 10.1111/apa.12436 [DOI] [PubMed] [Google Scholar]
- 55.Avraham-Davidi I, Ely Y, Pham VN, Castranova D, Grunspan M, Malkinson G, et al. ApoB-containing lipoproteins regulate angiogenesis by modulating expression of VEGF receptor 1. Nat Med. NIH Public Access; 2012;18: 967–73. doi: 10.1038/nm.2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang L, Choi S-H, Baek JS, Liu C, Almazan F, Ulrich F, et al. Control of angiogenesis by AIBP-mediated cholesterol efflux. Nature. Nature Research; 2013;498: 118–122. doi: 10.1038/nature12166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bahary N, Goishi K, Stuckenholz C, Weber G, Leblanc J, Schafer CA, et al. Duplicate VegfA genes and orthologues of the KDR receptor tyrosine kinase family mediate vascular development in the zebrafish. Blood. American Society of Hematology; 2007;110: 3627–36. doi: 10.1182/blood-2006-04-016378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goishi K, Klagsbrun M. Vascular endothelial growth factor and its receptors in embryonic zebrafish blood vessel development. Curr Top Dev Biol. 2004;62: 127–52. doi: 10.1016/S0070-2153(04)62005-9 [DOI] [PubMed] [Google Scholar]
- 59.Stalmans I. Role of the vascular endothelial growth factor isoforms in retinal angiogenesis and DiGeorge syndrome. Verh—K Acad voor Geneeskd van België. 2005;67: 229–76. [PubMed] [Google Scholar]
- 60.Ruhrberg C. Growing and shaping the vascular tree: multiple roles for VEGF. Bioessays. 2003;25: 1052–60. doi: 10.1002/bies.10351 [DOI] [PubMed] [Google Scholar]
- 61.Kim Y-W, Byzova T V. Oxidative stress in angiogenesis and vascular disease. Blood. American Society of Hematology; 2014;123: 625–31. doi: 10.1182/blood-2013-09-512749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aktaş C, Kurtman C, Ozbilgin MK, Tek I, Toprak SK. An Experimental Study of Radiation Effect on Normal Tissue: Analysis of HIF-1α, VEGF, eIF2, TIA-1, and TSP-1 Expression. Turkish J Haematol Off J Turkish Soc Haematol. 2013;30: 371–8. doi: 10.4274/Tjh.2012.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang R, Cheung N-K V, Vider J, Cheung IY, Gerald WL, Tickoo SK, et al. MYCN and MYC regulate tumor proliferation and tumorigenesis directly through BMI1 in human neuroblastomas. FASEB J. The Federation of American Societies for Experimental Biology; 2011;25: 4138–49. doi: 10.1096/fj.11-185033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schuhmacher B, Rengstl B, Döring C, Bein J, Newrzela S, Brunnberg U, et al. A strong host response and lack of MYC expression are characteristic for diffuse large B cell lymphoma transformed from nodular lymphocyte predominant Hodgkin lymphoma. Oncotarget. 2016; doi: 10.18632/oncotarget.12363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen L-J, Chen F-Y, Zhang Y, Cao L-F, Kuang Y, Zhong M, et al. MYCN transgenic zebrafish model with the characterization of acute myeloid leukemia and altered hematopoiesis. PLoS One. 2013;8: e59070 doi: 10.1371/journal.pone.0059070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams RD, Chagtai T, Alcaide-German M, Apps J, Wegert J, Popov S, et al. Multiple mechanisms of MYCN dysregulation in Wilms tumour. Oncotarget. 2015;6: 7232–43. doi: 10.18632/oncotarget.3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wegert J, Bausenwein S, Kneitz S, Roth S, Graf N, Geissinger E, et al. Retinoic acid pathway activity in Wilms tumors and characterization of biological responses in vitro. Mol Cancer. BioMed Central; 2011;10: 136 doi: 10.1186/1476-4598-10-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karkanitsa L V., Komarovskaya ME, Krivenko SI. Abrogation of radiation injury to human hematopoietic stem cells with tumor necrosis factor-α. Stem Cells. 2009;15: 95–102. doi: 10.1002/stem.5530150714 [DOI] [PubMed] [Google Scholar]
- 69.Miao L, Holley AK, Zhao Y, St Clair WH, St Clair DK. Redox-mediated and ionizing-radiation-induced inflammatory mediators in prostate cancer development and treatment. Antioxid Redox Signal. 2014;20: 1481–500. doi: 10.1089/ars.2013.5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamane K, Ihn H, Asano Y, Jinnin M, Tamaki K. Antagonistic effects of TNF-alpha on TGF-beta signaling through down-regulation of TGF-beta receptor type II in human dermal fibroblasts. J Immunol. 2003;171: 3855–62. [DOI] [PubMed] [Google Scholar]
- 71.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer. Nature Publishing Group; 2009;9: 714 doi: 10.1038/nrc2716 [DOI] [PubMed] [Google Scholar]
- 72.Duffy KT, Wickstrom E. Zebrafish tp53 knockdown extends the survival of irradiated zebrafish embryos more effectively than the p53 inhibitor pifithrin-alpha. Cancer Biol Ther. 2007;6: 675–8. [DOI] [PubMed] [Google Scholar]
- 73.Guo L, Liew HP, Camus S, Goh AM, Chee LL, Lunny DP, et al. Ionizing radiation induces a dramatic persistence of p53 protein accumulation and DNA damage signaling in mutant p53 zebrafish. Oncogene. 2013;32: 4009–16. doi: 10.1038/onc.2012.409 [DOI] [PubMed] [Google Scholar]
- 74.Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S. Links between Tumor Suppressors: p53 Is Required for TGF-β Gene Responses by Cooperating with Smads. Cell. 2003;113: 301–314. doi: 10.1016/S0092-8674(03)00308-8 [DOI] [PubMed] [Google Scholar]
- 75.Savli H, Sunnetci D, Cine N, Gluzman DF, Zavelevich MP, Sklyarenko LM, et al. Gene expression profiling of B-CLL in Ukrainian patients in post-Chernobyl period. Exp Oncol. 2012;34: 57–63. [PubMed] [Google Scholar]
- 76.Mezentsev A, Amundson SA. Global gene expression responses to low- or high-dose radiation in a human three-dimensional tissue model. Radiat Res. 2011;175: 677–88. doi: 10.1667/RR2483.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, et al. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8: 2466–77. [DOI] [PubMed] [Google Scholar]
- 78.Warga RM, Kimmel CB. Cell movements during epiboly and gastrulation in zebrafish. Development. 1990;108: 569–80. [DOI] [PubMed] [Google Scholar]
- 79.Pardali K, Moustakas A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta. 2007;1775: 21–62. doi: 10.1016/j.bbcan.2006.06.004 [DOI] [PubMed] [Google Scholar]
- 80.Dancea HC, Shareef MM, Ahmed MM. Role of Radiation-induced TGF-beta Signaling in Cancer Therapy. Mol Cell Pharmacol. 2009;1: 44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu W, Xu S, Yao B, Hong M, Wu X, Pei H, et al. MiR-663 inhibits radiation-induced bystander effects by targeting TGFB1 in a feedback mode. RNA Biol. 2014;11: 1189–1198. doi: 10.4161/rna.34345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ferrari G, Cook BD, Terushkin V, Pintucci G, Mignatti P. Transforming growth factor-beta 1 (TGF-beta1) induces angiogenesis through vascular endothelial growth factor (VEGF)-mediated apoptosis. J Cell Physiol. NIH Public Access; 2009;219: 449–58. doi: 10.1002/jcp.21706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Savlı H, Akkoyunlu RU, Çine N, Gluzman DF, Zavelevich MP, Sklyarenko LM, et al. Deregulated Levels of the NF-κB1, NF-κB2, and Rel Genes in Ukrainian Patients with Leukemia and Lymphoma in the Post-Chernobyl Period. Turkish J Haematol Off J Turkish Soc Haematol. 2016;33: 8–14. doi: 10.4274/tjh.2014.0190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yuan H, Wen B, Liu X, Gao C, Yang R, Wang L, et al. CCAAT/enhancer-binding protein α is required for hepatic outgrowth via the p53 pathway in zebrafish. Sci Rep. 2015;5: 15838 doi: 10.1038/srep15838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu J, Patrick BA, Jaiswal AK. NRH:Quinone Oxidoreductase 2 (NQO2) Protein Competes with the 20 S Proteasome to Stabilize Transcription Factor CCAAT Enhancer-binding Protein (C/EBP), Leading to Protection against Radiation-induced Myeloproliferative Disease. J Biol Chem. 2013;288: 34799–34808. doi: 10.1074/jbc.M113.495580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tawana K, Fitzgibbon J. CEBPA-Associated Familial Acute Myeloid Leukemia (AML). University of Washington, Seattle; 2016. [PubMed] [Google Scholar]
- 87.Fleenor CJ, Rozhok AI, Zaberezhnyy V, Mathew D, Kim J, Tan A-C, et al. Contrasting roles for C/EBPα and Notch in irradiation-induced multipotent hematopoietic progenitor cell defects. Stem Cells. 2015;33: 1345–58. doi: 10.1002/stem.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Feinendegen LE, Pollycove M, Sondhaus CA. Responses to low doses of ionizing radiation in biological systems. Nonlinearity Biol Toxicol Med. 2004;2: 143–71. doi: 10.1080/15401420490507431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feinendegen LE, Pollycove M, Neumann RD. Whole-body responses to low-level radiation exposure: new concepts in mammalian radiobiology. Exp Hematol. 2007;35: 37–46. doi: 10.1016/j.exphem.2007.01.011 [DOI] [PubMed] [Google Scholar]
- 90.Rajesh K, Krishnamoorthy J, Kazimierczak U, Tenkerian C, Papadakis AI, Wang S, et al. Phosphorylation of the translation initiation factor eIF2α at serine 51 determines the cell fate decisions of Akt in response to oxidative stress. Cell Death Dis. Macmillan Publishers Limited; 2015;6: e1591 doi: 10.1038/cddis.2014.554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18: 1926–45. doi: 10.1101/gad.1212704 [DOI] [PubMed] [Google Scholar]
- 92.Jacot JL, Sherris D. Potential Therapeutic Roles for Inhibition of the PI3K/Akt/mTOR Pathway in the Pathophysiology of Diabetic Retinopathy. J Ophthalmol. 2011;2011: 589813 doi: 10.1155/2011/589813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sharlow ER, Leimgruber S, Lira A, McConnell MJ, Norambuena A, Bloom GS, et al. A Small Molecule Screen Exposes mTOR Signaling Pathway Involvement in Radiation-Induced Apoptosis. ACS Chem Biol. American Chemical Society; 2016; doi: 10.1021/acschembio.5b00909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yentrapalli R, Azimzadeh O, Sriharshan A, Malinowsky K, Merl J, Wojcik A, et al. The PI3K/Akt/mTOR pathway is implicated in the premature senescence of primary human endothelial cells exposed to chronic radiation. PLoS One. 2013;8: e70024 doi: 10.1371/journal.pone.0070024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Survival and hatching after 43.8 and 92 hours exposure to specified dose rates. Survival at the 43.8 hours exposure did not differ at 48 and 96 hpf. All values presented as mean percentage ± 95% confidence interval (CI).
(XLSX)
Mapping statistics presented separately for each replicate (A) and each exposure (B) with their respective controls. Approximately 60% of the reads were mapped to the reference genome. Of the mapped reads, ~ 40% were mapped when allowing no mismatch, while ~ 15% of the reads were mapped when ≤ 2 bp mismatches were allowed. On the other hand, ~ 56% out of the mapped reads were found to represent unique genome positions, with ~ 1.5% of reads mapping to multiple positions.
(XLSX)
(XLSX)
(XLSX)
(TIF)
Differential expression threshold is FC ± 1.3. A, B, C and D show the distribution of mapped reads at 0.54, 5.4 and 10.9 mGy/h. E and F represent the distribution of mapped reads in control groups for the lowest (0.54 mGy/h) and for higher dose rates 5.4 and 10.9 mGy/h, respectively. All libraries were mapped to the ZF genome (Zv9).
(TIF)
A) Group exposed at 0.54 mGy/h and the control group for the lowest dose. Two and three biological replicates of the exposed group and controls, respectively, were included in the analysis. B) and C) Groups exposed to 5.4 and 10.9 mGy/h and controls. Three replicates were included.
(TIF)
Analysis was conducted by pairwise comparison of exposed and their respective controls. A) 0.54 mGy/h, B) 5.4 mGy/h and C) 10.9 mGy/h. Expression values were log2 transformed. Black and red dots represent non-differential and differentially expressed genes respectively (FDR < 0.05) (edgeR v3.4.2 Bioconductor).
(TIF)
Myc target gene networks and interactions, presented in a subcellular layout as part of the 10.9 mGy/h group.
(TIF)
TNF target gene networks and interactions, presented in a subcellular layout as part of the 10.9 mGy/h group.
(TIF)
Tp53 target gene networks and interactions, presented in a subcellular layout as part of the 10.9 mGy/h group.
(TIF)
Hnf4a target gene networks and interactions, presented in a subcellular layout as part of the 10.9 mGy/h group.
(TIF)
TGFB1 target gene networks and interactions, presented in a subcellular layout as part of the 10.9 mGy/h group.
(TIF)
Cebpa target gene networks and interactions, presented in a subcellular layout as part of the 10.9 mGy/h group.
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
The observations were done at 96 hours post fertilization (hpf). A. Control larva showing normal development; B-C. Larvae exposed to 38 mGy/h for 92 hours (Group “B”), demonstrating general developmental defects and short-tails.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. The RNA-seq experiment was deposited in SRA database (https://www.ncbi.nlm.nih.gov/) and is publically available under accession SRP096352.