Abstract
Animals partition their daily activity rhythms through their internal circadian clocks, which are synchronized by oscillating day-night cycles of light. The fruit fly, Drosophila melanogaster, senses day/night cycles in part through rhodopsin-dependent light reception in the compound eye, and photoreceptor cells in the Hofbauer-Buchner (H-B) eyelet1. However, a more significant light entrainment pathway is mediated in central pacemaker neurons in the brain. The Drosophila circadian clock is extremely light sensitive. However, the only known light sensor in pacemaker neurons, the flavoprotein, cryptochrome (Cry)2,3, responds only to high levels of light in vitro4. These observations indicate the existence of an additional light-sensing pathway in fly pacemaker neurons5. Here, we identified an uncharacterized rhodopsin, Rh7, which functions in circadian light entrainment through circadian pacemaker neurons in the brain. The pacemaker neurons respond to violet light, which was dependent on Rh7. While loss of either cry or rh7 caused minor affects on photoentrainment, the defects in the double mutant were profound. The circadian photoresponse to constant light was impaired in the rh7 mutant, especially under dim light. The demonstration that Rh7 functions in circadian pacemaker neurons represents the first role for an opsin in the central brain.
Cry is a light detector in central pacemaker neurons that contributes to phototrainment3,6. However, the cry mutant still entrains to light/dark (L/D) cycles3,6. Therefore, we screened for an additional light sensor that functions in circadian photoentrainment using Drosophila Activity Monitors. We entrained flies under 12-hour light/12-hour dark (light/dark; L/D) cycles for 4 days, and switched them to dark-dark (D/D) conditions. Control animals (w1118) display two daily activity peaks during the dawn and dusk, termed morning and evening peaks (Fig. 1a; note these are double plots). Activity increased before the light and dark transitions, indicating a capacity to anticipate changes in light, which is one hallmark of the circadian clock. Another is the ability to maintain activity patterns established under L/D cycles, after being transferred to constant darkness (dark/dark; D/D) (Fig. 1a and Extended Data Fig. 1a, h).
Figure 1. Rh7 is a light receptor.
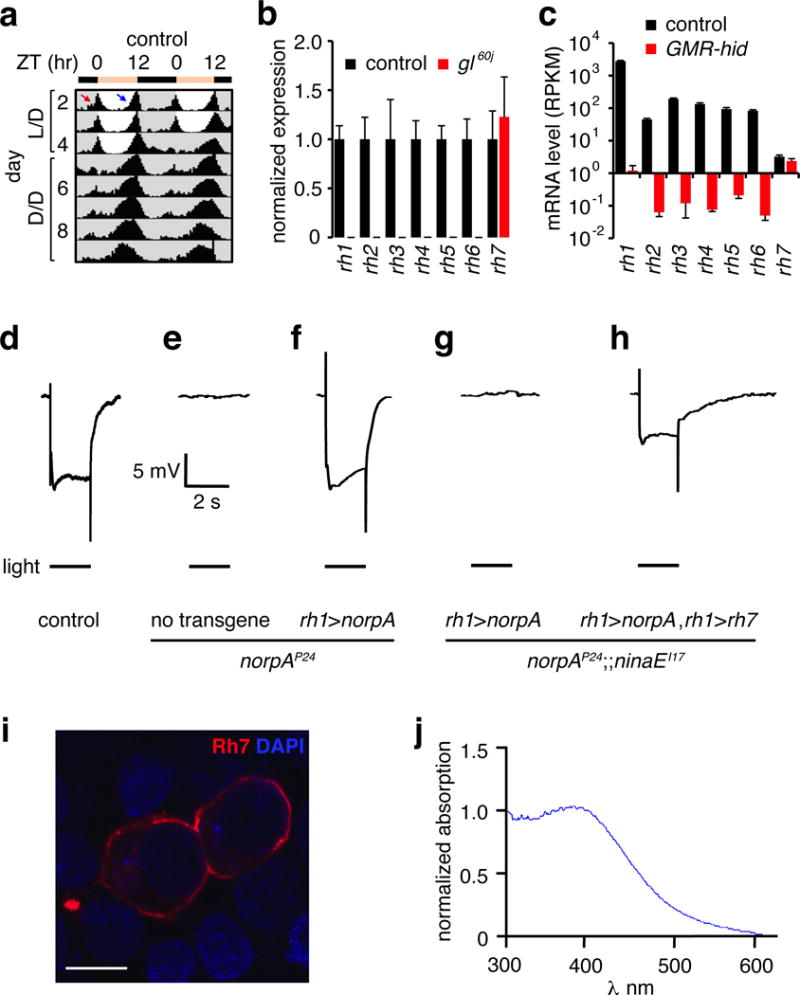
a, Actogram obtained with control flies entrained under L/D cycles and released to constant darkness (D/D). The red and blue arrows indicate morning and evening anticipation, respectively. n=62. b, Quantitative real-time PCR analysis of opsin genes using RNA from heads. Error bars indicate S.E.M.s. n=3/genotype. c, Opsin RNA-seq mRNA levels were quantified as Reads Per Kilobase of transcript per Million mapped reads (RPKM). Error bars indicate S.E.M.s. d–h, ERG responses using 2 sec light. d, Control. e, norpAP24. f, Expression of norpA using the rh1 promoter (rh1>norpA) in a norpAP24 background. g, Expression of rh1>norpA in a norpAP24;;ninaEI17 background. h, Expression of rh1>norpA and UAS-rh7 under the control of the rh1-Gal4 (rh1>norpA and rh1>rh7, respectively) in a norpAP24;;ninaEI17 background. i, HEK293T cells expressing Rh7 and stained with anti-Rh7. The DAPI stain indicates nuclei. Scale bar indicates 10 μm. j, Absorbance spectrum of Rh7 from HEK293T cells expressing Rh7.
Mutation of cry causes only subtle effects on circadian behavior (Extended Data Fig. 1a, b, h)3. Flies also show rhythmic behavior after photoentrainment if they are missing the phospholipase C (PLC) NORPA2,5 required for phototransduction in the compound eye, or if they are doubly mutant for norpA and cry (Extended Data Fig. 1c, d, h)2,5. Phototransduction in H-B eyelet photoreceptors couples to Rh6 and the TRPL channel7–9, but is NORPA independent5. Flies triply mutant for rh5, rh6 and cry5, for norpA, cry and trp, or for norpA, trpl and cry are entrained by L/D cycles (Extended Data Fig. 1e–h). Thus, as proposed5, there is likely to be an additional light input pathway that impacts on the circadian clock preceding exposure to D/D.
Drosophila encodes an uncharacterized opsin, Rh7 (Extended Data Fig. 2a), which shares 27–30% amino acid identities with other opsins in Drosophila melanogaster. Rh7 is conserved in other Drosophila species (79–99% identities) and in Aedes aegypti and Anopheles gambiae (49–52% identities)10. Photoreceptor cells in the compound eye and ocelli express six opsins (Rh1-Rh6; Extended Data Fig. 2b–c). However, a mutation (gl60j) eliminating ocular photoreceptor cells did not reduce rh7 RNA levels, in contrast to rh1-rh6 (Fig. 1b). We performed RNA-seq using RNA from flies expressing a cell death gene in ocular photoreceptor cells (GMR-hid). Transcript numbers of rh1-rh6 were reduced dramatically, while rh7 was unchanged (Fig. 1c). We did not detect Rh7 in the compound eye with Rh7 antibodies (see below; Extended Data Fig. 2b–e). We generated an rh7 null allele, rh71 (Extended Data Fig. 2f–g) and tested their light responses by performing electroretinogram (ERG) recordings. The control and rh71 ERGs were indistinguishable (Extended Data Fig. 2h–j). Thus, Rh7 was neither expressed nor functioned in known photoreceptor cells.
To address whether Rh7 is a light receptor, we tested whether it could substitute for Rh1 in R1-6 photoreceptor cells. Indeed, we rescued a wild-type-like ERG in the rh1 mutant (ninaEI17; Extended Data Fig. 2k–m). To assess the light response due to Rh7 only, we eliminated the light responses from the remaining two photoreceptor cells (R7/R8), which express other rhodopsins. Phototransduction is abolished in norpAP24 (Fig. 1d–e). We restored a photoresponse in R1-6 cells of norpAP24 by expressing a norpA+ transgene using the rh1 (ninaE) promoter (rh1>norpA; Fig. 1f). Upon eliminating rh1 (ninaEI17) from the norpAP24;rh1>norpA flies, the animals were unresponsive to light (Fig. 1g). We recovered a light response by expressing rh7 in the R1-6 cells (rh1>rh7; Fig. 1h). Thus, Rh7 is a light sensor and is capable of coupling to a Gq/PLC signaling pathway. We expressed Rh7 in HEK293T cells and found that it responded to light with a peak at 397 nm (Fig. 1i, j).
We raised Rh7 antibodies, which stained two groups of cells in the brain indicative of a subset of central pacemakers neurons (Fig. 2a and Extended Data Fig. 3a, c). The ~150 pacemaker neurons express Period (Per), a core component of the circadian clock11–13, and are classified as dorsal and lateral neurons (DNs and LNs, respectively; Extended Data Fig. 3a). The 15–16 LNs include 5–6 dorsal lateral neurons (LNds), 4–5 large ventrolateral neurons (l-LNvs), and 4–5 small ventrolateral neurons (s-LNvs)14. Rh7 and Per were co-expressed in the LNvs (Fig. 2a–c), which express the neuropeptide, Pigment Dispersing Factor (PDF)15 (Fig. 2d–f). We also detected Rh7-positive neurons in the vicinity of DN1s (Extended Data Fig. 3a, c), half of which express cry (Extended Data Fig. 3d, e)16. However, these neurons did not co-stain with the cry reporter (cry-Gal4.E13 2,17; Extended Data Fig. 3e). We did not detect anti-Rh7 in other central pacemaker neurons, or in rh71 flies (Fig. 2g–i and Extended Data Fig. 3b).
Figure 2. Rh7 contributes to light sensitivity of circadian pacemaker neurons.
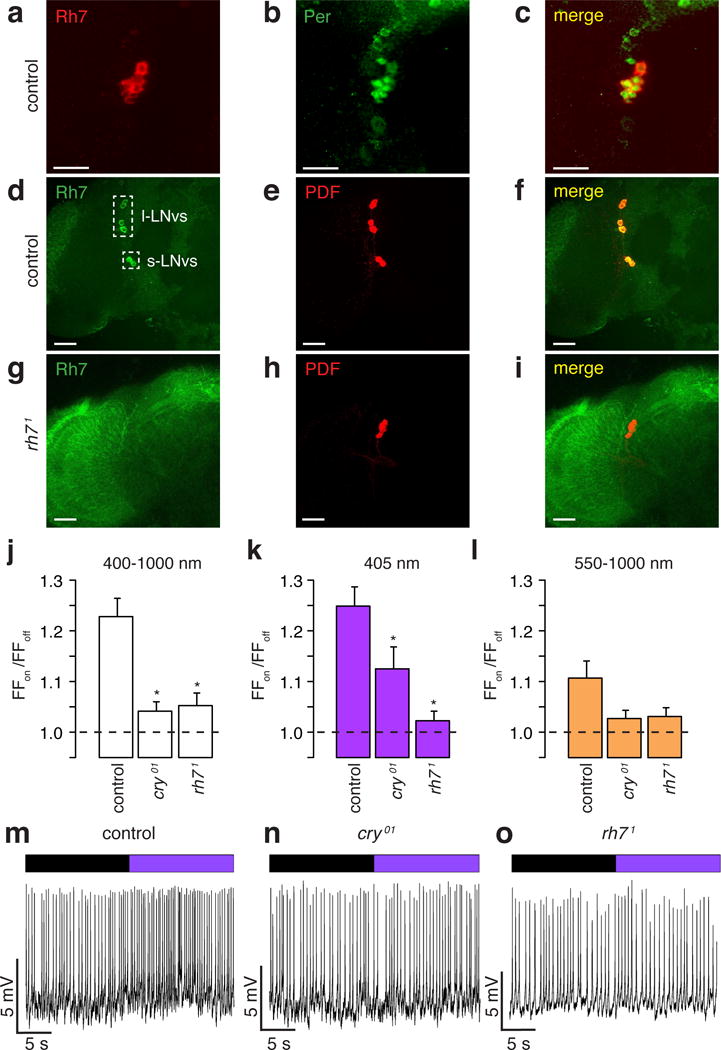
a–i, Control and rh71 brains stained with the indicated antibodies Merged images are to the right. Scale bars indicate 20 μm. j–l, Average firing frequencies of l-LNvs during “lights on” relative to the firing frequencies during “lights off” (FFon/FFoff). * Significant differences from the control (p<0.05). j, White light. Control (n =74), cry01 (n =80, p<0.05 vs. control) and rh71 (n =60, p<0.05 vs. control). k, 405 nm violet light. Control (n=76), cry01 (n=89, p<0.05 vs. control) and rh71 (n=66, p<0.05 vs. control). l, Orange light. Control (n=58), cry01 (n=65), and rh71 (n=46). m–o, Representative recordings showing responses of l-LNv neurons to 405 nm light. Purple bar=405 nm light; black bar= no light.
Cry mediates rapid increases in blue light (450 nm peak) evoked action potentials in l-LNvs18–20. We compared the electrophysiological responsiveness to white (400–1000 nm) and violet (405 nm) light in control and rh71 l-LNvs. The l-LNv responses to white and violet light were greatly diminished in cry01 and rh71 flies (Fig. 2j, k, m, n, o). Control, cry01 and rh71 flies had minimal or no response to orange light (550–1000nm; Fig. 2l).
To address the importance of Rh7 to entrainment, we first investigated its contribution to circadian phase changes in response to a nighttime light pulse, which shifts the phase of the clock21. The direction of the effect depends when the light is presented21–23. Lights are turned on and off at ZT0 and ZT12, respectively. An early night light pulse (ZT14–18) produces a phase delay in controls, while a late night light pulse (ZT20–22) causes a phase advance (Fig. 3a)21. As previously reported3,22, cry01 displays severely impaired phase shifting to early or late night pulses (Fig. 3a).
Figure 3. Rh7 is a circadian light receptor.
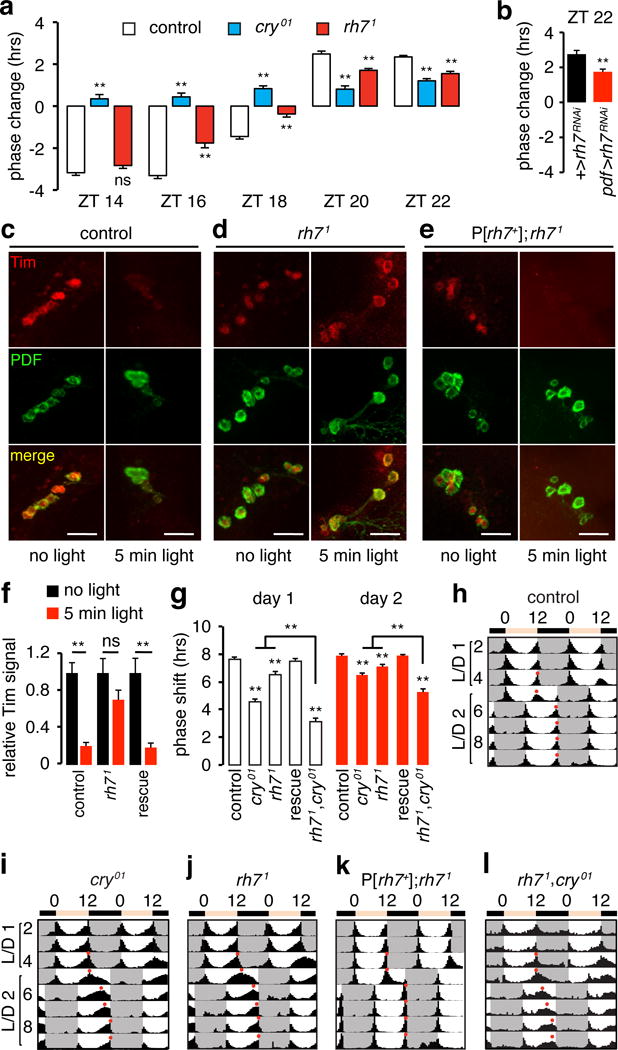
a, Phase response of flies to a 5-min white light at the indicated ZT. Negative and positive phase changes indicate phase delays and phase advances, respectively. Three independent assays/genotype (n=8–32/experiment). Total flies tested: ZT14, control, n=56; cry01, n=46; rh71, n=55. ZT16, control, n=61; cry01, n=52; rh71, n= 57. ZT18, control, n=61; cry01, n=51; rh71, n=77. ZT20, control, n=45; cry01, n=39; rh71, n=48. ZT22, control, n=54; cry01, n=60; rh71, n=48. b, Phase responses to a 5-min white light stimulation at ZT22. Three independent assays/genotype (n=8–24/experiment). +>rh7RNAi (UAS-rh7RNAi), n=40; pdf>rh7RNAi (UAS-rh7RNAi and pdf-Gal4), n=52. **p<0.01, unpaired Student’s t-test. c–e, Flies were exposed to a 5-min light stimuli at ZT22, and the LNvs were stained with anti-Tim and anti-PDF at ZT23. Scale bars indicate 40 μm. f, Quantification of light-mediated degradation of Tim protein in LNvs. The rescue flies are P[rh7+];rh71. Animals tested: control, dark n=6, light n=4; rh71, dark n=5, light n=4; rescue, dark n=5, light n=4. n.s. not significant. **p<0.01, unpaired Student’s t-test. g, Quantification of the phase shifts on days 1 and 2 after the 8-hr phase delay. 3–4 independent assays/genotype (n=10–30/experiment). Control, n=95; cry01, n=96; rh71, n=68; P[rh7 +];rh71, n=60; rh71,cry01, n=66. Error bars indicate S.E.M.s. One-away ANOVA test (Kruskal-Wallis test) followed by the Dunn’s test. **p<0.01. h–l, Actograms. Flies were entrained under L/D cycles. On day 5, the day cycle was extended by 8 hours. Red dots indicate the evening peaks.
We exposed rh71 flies to nighttime light pulses. The phase delay was normal if the stimulus was presented early (ZT14; Fig. 3a; Extended Data Fig. 4). However if the light occurred later, the degree of phase shift was significantly reduced (ZT16-ZT22; Fig. 3a; Extended Data Fig. 4). We knocked down of rh7 RNA using a pdf-Gal4 (Extended Data Fig. 3f–i), and then exposed the flies to light at ZT22. These flies exhibited the same phase advance defect as rh71 (Fig. 3b), indicating that Rh7 is required in PDF-positive neurons for normal photoentrainment.
Phase advances due to light pulses late at night (ZT22) correlate with light-induced degradation of the core clock protein Timeless (Tim)24. Tim degradation depends on binding to Cry24, or results from neuronal activation25. Since light-induced neuronal firing is reduced in rh71, light-induced degradation of Tim might be impaired. To test this hypothesis, we applied a light pulse at ZT22, and assayed anti-Tim signals in PDF neurons 55 min later (LNvs). In controls, light caused a 4.8-fold decline in Tim (Fig. 3c, f). However, in rh71 LNvs, light caused only a slight reduction in anti-Tim staining, which was not statistically significant (Fig. 3d, f). We rescued this defect with a genomic transgene (Fig. 3e, f).
Another test of photoentrainment is to assess the number of days required to adjust to a delay in the light-to-dark transition. If control flies are entrained under L/D cycles, and then the transition from light to darkness is delayed 8 hours, they re-entrain quickly to the new L/D cycle1 (Fig. 3g, h). The evening peak shifts 7.6 ±0.1 hours on day 1 and 7.9 ±0.1 hours on day 2 (Fig. 3g). Consistent with a previous study1, cry01 flies shifted only 4.6 ±0.2 hours on day 1 and 6.5 ±0.1 hours on day 2, and required 3 days to establish stable peak activity (Fig. 3g, i). We found that rh71 displayed significant delays in phase shift (day 1, 6.5 ±0.2 hours; day 2, 7.1 ±0.1 hours; Fig. 3g, j) consistent with a contribution of Rh7 to photoentrainment. We rescued the defect with an rh7+ genomic transgene (Fig. 3g, k). The impairment in the phase-shift exhibited by the rh71,cry01 double mutant (day 1, 3.1 ±0.2 hours; day 2, 5.3 ±0.2 hours) was more severe than the single mutants (Fig. 3g, l).
Constant light (L/L) leads to arrhythmic circadian behavior in wild-type flies (Fig. 4a, d)6. However, cry mutant flies remain rhythmic in constant light6 (93.1%; Fig. 4d and Extended Data Fig. 5a). Under L/L conditions, 19.0% of rh71 flies are also rhythmic, and this phenotype is rescued by a wild-type rh7+ transgene (Fig. 4b–d and Extended Data Fig. 5b).
Figure 4. Effects of rh7 mutation on circadian behavior in response to constant light, constant darkness, and on arousal in response to a light pulse.
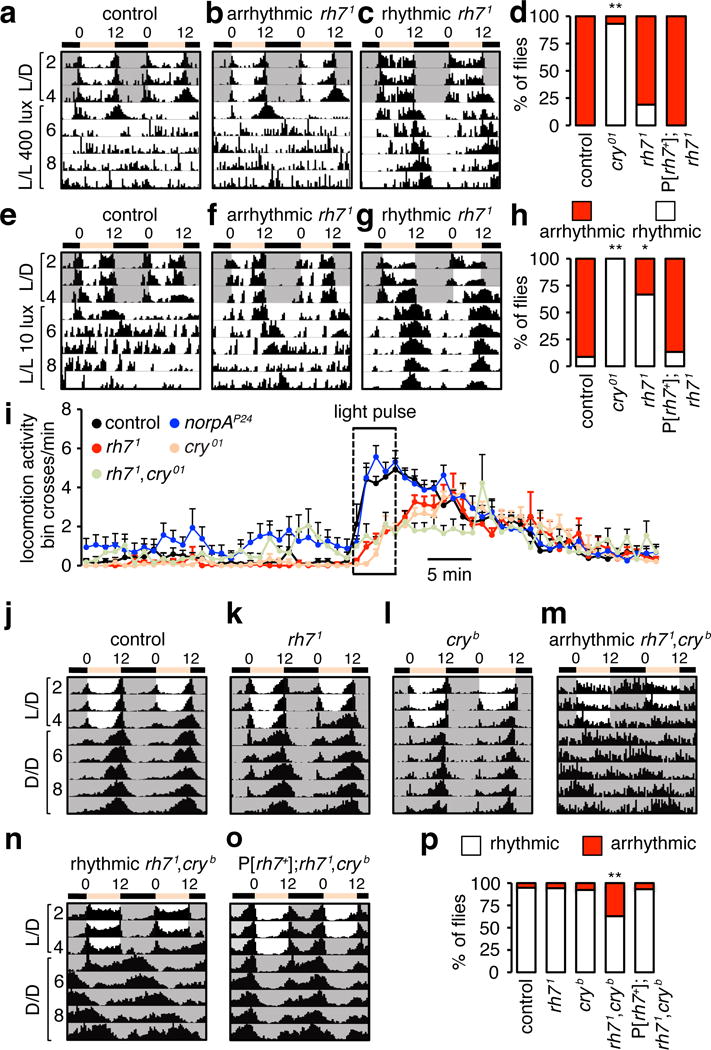
a–c, Actograms showing circadian responses of flies entrained to L/D cycles and released to constant light (L/L) of ~400 lux. d, Percentages of rhythmic and arrhythmic flies. Fisher’s exact test. **p<0.01. Control, n=48; cry01, n=29; rh71, n=58; P[rh7+];rh71, n=16. e–g, Actograms showing circadian responses of flies entrained under L/D cycles and released to dim L/L of ~10 lux. h, Percentages of rhythmic and arrhythmic flies. Fisher’s exact test. *p<0.05. **p<0.01. Control, n=23; cry01, n=30; rh71, n=27; and P[rh7 +];rh71, n=15. i, Light-dependent arousal impaired by the rh71 mutation. Average locomotion activities (bin crosses per min) at ZT22. The box indicates the 5-min white light pulse. j–o, Average actograms showing synergistic effect of the cryb and rh71 mutations on circadian behavior. The flies were entrained to L/D cycles and released to constant darkness (D/D). rh7+ genomic transgene, P[rh7+]. p, Percentages of rhythmic and arrhythmic flies. **p<0.01. Fisher’s exact test. control, n=76; rh71 n=29; cryb n=39; rh71,cryb, n=78 and P[rh7 +]; rh71,cryb,n=29.
Because phototransduction promotes signal amplification and sensitivity to low light, we tested the effects on rhythmicity, after photoentraining rh71, and then exposing the flies to constant dim light (10 lux). Few control flies maintained rhythmicity even under dim light, while all cry01 flies were rhythmic (Fig. 4e, h and Extended Data Fig. 5c). Of significance, the majority of rh71 flies maintained rhythmicity under constant dim light (66.7%; Fig. 4f–h). We restored wild-type responses with an rh7+ genomic transgene (Extended Data Fig. 5d). These data suggest that Rh7 is required for sensitizing the Cry-dependent circadian photoresponse under dim light conditions.
PDF-expressing LNvs promote light-dependent arousal26,27. To test if Rh7 functions in arousal, we stimulated the flies at night (ZT22) with a 5-minute white light pulse. The rh71 flies displayed decreased light-coincident arousal (Fig. 4i and Extended Data Fig. 5e) and a longer arousal delay than control flies (Extended Data Fig. 5f), which were at least as great as those exhibited by cry01 flies18,20 (Fig. 4i and Extended Data Fig. 5e, f). The responses to red light were not impaired significantly in the mutant flies (Extended Data Fig. 5g). The rh71,cry01 double mutant but not the single mutants exhibited a violet light arousal deficit (405 nm; Extended Data Fig. 5h), indicating that the two light sensors compensate for each other during the arousal response.
We tested for a potential role of Rh7 for maintaining rhythmic behavior during constant darkness (D/D) following L/D entrainment. The mutant flies showed rhythmic diurnal and circadian behavior similar to wild-type (Fig. 4j, k, p). As previously shown1, cryb flies also photoentrained and exhibited rhythmic diurnal and circadian behavior (Fig. 4l, p). However, rh71,cryb double mutants had profound deficits. 37.2% of the animals were arrhythmic (Fig. 4m, p), possibly due to insufficient synchronization of the molecular clock between different groups of central pacemaker neurons, in the absence of both light sensors intrinsic to these cells. The remaining rhythmic flies displayed much longer periodicity during D/D than controls (control, 23.8 ±0.08 hours; rh71,cryb, 27.3 ±0.07 hours, p<0.01; Fig. 4j, n). rh71,cry01 double mutants displayed similar impairments (Extended Data Fig. 6a–c). Thus, at least one of the two light receptors is required in pacemaker neurons for normal rhythmic behavior. We rescued these defects with a rh7+ genomic transgene (Fig. 4o, p).
While gl60j, rh71 and cry single mutants display photoentrainment impairments1, they are capable of circadian rhythmicity. Thus, any two of these three photoreceptors (Cry, Rh7 and gl-dependent) are sufficient for rhythmic circadian behavior. However, Rh7 is insufficient on its own since gl60j,cryb flies are circadian blind1. To test whether cry alone is sufficient for rhythmic behavior, we subjected the rh71,gl60j double mutant to a L/D regime followed by D/D, and found that 25% were arrhythmic (Extended Data Fig. 6d–h). Thus, while Cry alone cannot preserve fully wild-type circadian behavior, it enables flies to maintain greater rhythmicity than just Rh7 or gl photoreceptor cells. We exposed rh71,gl60j flies to a 5-minute light pulse at ZT22 to test their circadian phase-response. While rh71 and gl60j single mutants showed a decreased phase-advance, the rh71,gl60j double mutant displayed a greater impairment (Extended Data Fig. 6i). Since cry01 flies also showed defects in the phase-response to a 5-minute pulse (Fig. 3a), multiple light-input pathways contribute to circadian light sensation.
To test whether rh7 functioned in LNvs neurons, we performed rescue experiments. Expression of rh7 (UAS-rh7) using either the tim-GAL4 or an LNv-specific driver, pdf-GAL4, rescued the increase in arrhythmicity exhibited by rh71,cryb flies (Extended Data Fig. 7). Introduction of other rhodopsins in pdf-positive neurons also restored normal rhythmicity (Extended Data Fig. 8), further supporting a rhodopsin-based mechanism for circadian function in LNvs.
In s-LNvs and l-LNvs, Per protein peaks during dawn (ZT22-ZT2) and enters a trough (Extended Data Fig. 9a–c)28. In rh71 flies, Per also displayed oscillations in LNvs,(Extended Data Fig. 9d–f), consistent with their rhythmic behavior under L/D cycles. Per oscillations in LNvs are altered in cryb 1,3. There is a delayed trough and peak in s-LNVs, and a large reduction in oscillation amplitude in l-LNVs (Extended Data Fig. 9g–i). In rh71,cryb, Per levels remained high from ZT2-ZT14 in s-LNvs and peaked shortly after dusk (ZT14) in l-LNvs (Extended Data Fig. 9j–l). PDF does not oscillate in control LNvs 29, or rh71 (Extended Data Fig. 9a, b).
In the eye, rhodopsins signal through the PLC encoded by norpA. However, norpA and cry double mutant flies exhibit normal rhythmicity during constant darkness following a L/D regime2,5 (Extended Data Fig. 1d, h), indicating that Rh7 signaling in LNvs is NORPA-independent. To address if the other PLCβ (PLC21C) functions in PDF neurons, we combined two UAS-plc21C RNAi lines with the pdf-Gal4 (Extended Data Fig. 10a), and then exposed the flies to a white-light pulse for 5 minutes at ZT22. Knockdown of plc21C caused reductions in phase-advance at ZT22 (Extended Data Fig. 10b–g), similar to the rh71 mutation (Fig. 3a).
In summary, our work establishes a role for a rhodopsin in the central brain. Rh7 is strategically expressed in the PDF-positive cells, which appear to be master light input clock neurons that also receive input from the optical lobes26. The fact that PDF-positive neurons express two distinct light sensors (Rh7 and Cry) highlights their key role in circadian photoentrainment. In the mammalian retina, ~1% of the retinal ganglion cells are intrinsically photosensitive (ipRGCs), and function in circadian photoentrainment. ipRGCs are directly light sensitive due to expression of melanopsin and also receives light information from rods and cones. The striking similarities between Drosophila PDF-positive neurons and ipRGCs indicate a common strategy for circadian photoentrainment. Opsins are also expressed in the mammalian brain30, although their functions are unknown. Because light penetrates the mammalian skull, our findings raise questions as whether neurons in the mammalian brain also sense light and contribute to photoentrainment of circadian rhythms.
Methods
No statistical methods were used to predetermine sample sizes. All behavior data were collected in a random manner. No blinding method was used in assessing experimental outcomes.
Fly stocks
The following flies were obtained from Bloomington Stock Center: isogenized w1118 (BL5905), norpAP24 (BL9048), ninaE-norpA (rh1>norpA; this is a direct fusion of the ninaE promoter to the norpA coding region; BL52276), ninaE-Gal4 (rh1-Gal4; BL8691), trpMB (BL23636), trplMB (BL29314), UAS-mcherry-NLS (BL38425), gl60j (BL 509), pdf-Gal4 (BL6900), and two UAS-plc21C RNAi lines (01210, BL 31269 and 01211, BL31270). GMR-hid 31 was obtained from the Drosophila Genetic Resource Center, Kyoto (108419). We used the w1118 as the control strain. The UAS-rh7 RNAi line (v1478) was from VDRC Stock Center. The tim-Gal4 32 was provided by A. Sehgal (University of Pennsylvania). The cry-Gal4.E132 was from M. Rosbash (Brandeis University). The cryb 2 and cry01 33 flies were provided by Mark Wu (The Johns Hopkins University School of Medicine) and the rh502 34, rh601 35, UAS-rh3 35, UAS-rh4 35 and UAS-rh5 35 lines were provided by C. Desplan (New York University). We also used ninaEI17 flies 36.
Cloning of the rh7 cDNA and generation of transgenic flies
To clone the rh7 coding region, we prepared mRNA from w1118 heads, performed reverse transcription (RT)-PCR using the following primers, and cloned the cDNA into TOPO vector (pCR2.1-TOPO, Invitrogen): rh7 forward, GCGGCCGCCACCATGGAGGCCATCATCATGACG; rh7 reverse, GCGGCCGCTCAGAACTTACTCTGTTCCATGAC. To generate the UAS-rh7 transgene, we subcloned the rh7 open reading frame into the NotI site of the pUAST vector. To construct the plasmid for expression of Rh7 in HEK293T cells, we subcloned the rh7 open reading frame between the BamH1 and Xba1 sites of the pCS2+MT vector using the following primers: rh7 forward, ATCAGATCTCACCATGGAGGCCATCATCATGACG; rh7 reverse ATCTCTAGATCAGAACTTACTCTGTTCCATGAC.
To generate transgenic flies expressing a Rh7::FLAG fusion protein, we first constructed the pUAST-FLAG vector using the following two oligonucleotides, which we annealed and cloned into the XhoI and XbaI site of the pUAST vector: FLAG5’-XbaI, TCGAGGGGATTACAAGGATGACGACGATAAGTAAT and FLAG3’-XhoI, CTAGATTACTTATCGTCGTCATCCTTGTAATCCCC. We amplified the rh7 coding region using the same forward primer as above, in conjunction with the following reverse primer to eliminate the stop codon: rh7 reverseno-stop, GCGGCCGCGAACTTACTCTGTTCCATGAC. Both the UAS-rh7 and UAS-rh7-FLAG transgenic flies were obtained by germline transformation using w1118 embryos (Bestgene Inc.).
To generate flies expressing a rh7+ genomic transgene (P[rh7+]), a BAC genomic DNA clone (CH322180G19) was obtained from the P[acman] collection 37. The germline transformation took advantage of site-specific integration using the Φ31-attB/attP system (Bestgene Inc).
Generation of rh71 mutant flies by homologous recombination
We produced the plasmid for knocking out rh7 by ends-out homologous recombination 38 as follows. We PCR amplified two homologous arms (left, 3.2 kb and right, 3.3 kb) using the following primers: left arm forward, AATTGCTGGGATCCCTCAATTGGCCTAATCGGTTTCTG; left arm reverse, AATTGCTGGGTACCGACTGACTTGGCCAAATATTTACG; right arm forward, AATGCTGGCGGCCGCTTAAAATGCTGCCCGAGACT; right arm reverse, AATTGCTGGCGGCCGCTGGCTTATGAAGTTGCAAAAAG. We cloned the two arms into the targeting vector, pw35loxp-Gal4. This construct was designed to delete 540 base pairs (bp) 3′ to the rh7 translational start site, and was replaced with a cassette containing the mini-white marker and Gal4 flanked by two loxP sites. The upstream loxP sequence contained a translational start site that rendered the Gal4 coding region out of frame. Consequently, the Gal4 is not functional. To obtain the donor lines for generating the rh7 knockout (rh71 allele), the targeting vector was injected into w1118 embryos (Bestgene Inc.). We mobilized the donor insertion by crossing the donor line to y,w;P[70FLP]11 P[70I-SceI]2B nocSco/CyO flies (from the Bloomington Stock Center, BL6934). The progeny were screened for gene targeting in the rh7 locus by PCR using two pairs of primers. The first pair (P1 and P2) were the following two primers that annealed to the first and second coding exons, and produced a DNA product (885 bp) only in wild-type (Extended Data Fig. 2g): P1, CTCTCGCTCTCCGAGATGTT and P2, ACCACCGAAATCAGGCAATA. The following second pair of primers (P3 and P4) annealed to the mini-white gene and to a sequence 3’ to rh7, and therefore only generated a product in the rh71 mutant (4.4 kb; Extended Data Fig. 2g): P3, TGTACATAAAAGCGAACCGAACCT and P4, ACTGTGCGACAGAGTGAGAGAGCAATAGTA. After generating rh71, we outcrossed the flies to the control stain (w1118) for five generations.
Genotyping timeless, jetlag and period alleles
To determine whether or not the key fly lines used in this study harbored the perSLIH, timls, or jetc mutations in the genetic background, we performed DNA sequencing. We extracted genomic DNA from adult flies, and amplified the relevant regions in the per, tim and jet genes by PCR (Phusion High-Fidelity DNA Polymerase, NEB) using the following primers: 1) per: forward, GTCCACACACAACACCAAGG; reverse, TTGATGATCATGTCGCTGCT. 2) tim: forward, TGGCTGGGGATTGAAAATAA; reverse, TTACAGATACCGCGCAAATG, and 3) jet: forward, AGCCGATCATAGTGGAGTGC; reverse, AAGGCACGCACAGGTTTACT.
We purified the PCR products and subjected them to DNA sequencing (DNA Sequencing Facility at the University of California, Berkeley). The perSLIH allele has a C to A transversion at nucleotide 2688438. The control (per+) sequence encompassing this region (2688436–2688448, Drosophila genome release r6.14) is CTCCGGCAGCAGT. The perSLIH sequence is CTACGGCAGCAGT. All of the fly lines checked had sequences that matched per+. These include: 1) rh71, 2) rh71,cryb, 3) rh71,cry01, 4) pdf-Gal4, and 5) rh7-RNAi. The timls allele has a single nucleotide insertion (C) after nucleotide 3504474 relative to tims. The sequence spanning this region in the control (timls) is ATCAAAGTTCTGAT (3504473–3504486, Drosophila genome release r6.14) and in tims is ATAAAGTTCTGAT. We sequenced the following lines, all which had sequences that matched the control (tims): 1) cry01, 2) rh71, and 3) P[rh7+]; rh71. The jetc allele has a T to A transversion at nucleotide 4949048. The control (jet+) sequence spanning this region (4949059–4949047, Drosophila genome release r6.14) is CTTGATTATCTTC, while the jetc sequence is CTTGATTATCTAC. We sequenced the following lines, all of which had sequences that matched the control (jet+): 1) cry01, 2) rh71, and 3) P[rh7+]; rh71.
Quantitative RT-PCR and RNA-seq data for opsins
To quantify expression of opsin genes (Fig. 1b), we isolated total RNA from ~50 fly heads from each of the indicated fly stocks, and used 1 μg of total RNA from each sample as a temperate for reverse transcription using SuperScript™ III Reverse Transcriptase (ThermoFisher, cat. 18080093). Oligo dT primers were used for cDNA synthesis. cDNA preparation was subjected to real-time quantitative PCR (Roche, LightCycler 480 system) according to the LightCycler 480 SYBR Green 1 Master Mix (cat. 04707516001) protocol. The primers used for real-time quantitative PCR were:
rh1: forward CGCTACCAAGTGATCGTCAA, reverse GTATGAGCGTGGGTTCCAGT.
rh2: forward TCCGTGCTGGACAATGTG, reverse AATCATGCACATGGACCAGA.
rh3: forward CGAGCAAAAGAACAGGAAGC, reverse TCGATACGCGACTCTTTGTG.
rh4: forward GTAGCCCTCTGGCACGAAT, reverse TCTTCAGCACATCCAAGTCG.
rh5: forward TCCTGACCACCTGCTCCTTC, reverse GCTCCAGCTCCAGACGATAC.
rh6: forward CAAGGACTGGTGGAACAGGT, reverse GTACTTCGGGTGGCTCAATC.
rh7: forward GTTTCCACGGGTCTGACAAT, reverse GCTGTAGCACCAGATCAGCA.
rp49: forward GACGCTTCAAGGGACAGTATCTG, reverse AAACGCGGTTCTGCATGAG.
We also analyzed opsin gene expression using an RNA-seq data set (Fig. 1c). For each genotype, three independent RNA libraries were prepared from ~50 heads using the TruSeq Stranded mRNA Library Prep Kit. Pair-end sequencing was performed using the TruSeq platform (Illumina). Details of the RNA-seq experiments and data analysis will be presented elsewhere (Ni, J. D., Tekin, I. and Montell, C., in preparation). Opsin RNA-seq mRNA levels were quantified as Reads Per Kilobase of transcript per Million mapped reads (RPKM). RPKMs for each opsin were calculated independently and the average RPKMs are plotted.
Quantitative RT-PCR following plc21C RNAi knockdown
To knock-down plc21C expression, we combined each UAS-plc21C RNAi transgene (01210 and 01211) with UAS-Dicer2;;actin-Gal4. To quantify the efficacy of the RNAi, we extracted total RNA from 10 adult flies (5 males and 5 females), and used 1 μg of total RNA from each sample as a temperate for reverse transcription using SuperScript™ III Reverse Transcriptase (ThermoFisher, cat. 18080093). Oligo dT primers were used for cDNA synthesis. cDNA preparation was subjected to quantitative PCR (Roche, LightCycler 480 system) according to the LightCycler 480 SYBR Green 1 Master Mix (cat. 04707516001) protocol. The plc21C primers used were: forward, GGATCTCTCCAAGTCGTTCG; reverse, TAGCCGCTTCACCAGCTTAT. The rp49 primers were: forward, GACGCTTCAAGGGACAGTATCTG; reverse, AAACGCGGTTCTGCATGAG. In each reaction, we normalized expression of plc21C transcripts to rp49.
Generation or Rh7 antibodies
To obtain Rh7 antibodies, we generated a GST-Rh7 fusion protein by subcloning the region encoding the N-terminal 80 amino acids into the pGEX6P-1 vector (GE Healthcare Life Science). We expressed the fusion protein in E. coli (BL21), purified the recombinant protein using glutathione Sepharose beads (GE Healthcare Life Science) and generated antiserum in a rabbit (Covance). We affinity purified the antibodies by conjugating the antigen to Affi-Gel 10 (Bio-Rad).
Immunohistochemistry on whole-mounts of Drosophila brains
We performed immunohistochemistry using whole-mount fly brains as described previously 39. Briefly, we fixed dissected brains for 15–20 min at 4°C in 4% paraformaldehyde in phosphate buffer (0.1 M Na3PO4, pH 7.4) with 0.3% Triton-X100 (Sigma), hereafter referred to as PBT. The brains were blocked with 5% normal goat serum (Sigma) in phosphate buffer for 1 hr at 4°C. We then incubated the tissue with primary antibodies at 4°C for ≥24 hrs. After three washes in PBT, the brains were incubated overnight at 4°C with the following secondary antibodies from Life Technologies: anti-mouse Alexa Fluor 488 or 568 Dyes, anti-rabbit Alexa Fluor 488 or 568 Dyes or Alexa dyes. The brains were washed three times with PBT and mounted in VECTASHIELD mounting medium (Vector Labs) for imaging. For Rh7 and PDF co-staining (Fig. 2d–i), four brains were examined.
Immunohistochemistry for circadian clock proteins (Tim and Per)
To analyze light-mediated degradation of Tim (Fig. 3c–f), we entrained the flies for 3 days under 12-hr light/12-hr dark cycles (~600 lux LED white light). The flies were then exposed to a 5-min LED light stimulation (~600 lux) at ZT22, kept in the dark for 55 minutes, fixed at ZT23 under a red photographic safety light (for 45 min), and dissected for whole-mount immunostaining. Flies that were not exposed to the nocturnal light treatment were fixed and stained at the same time.
To examine Per staining at different ZT points (Extended Fig. 9), flies were entrained for 4 days under 12-hr light (~400 lux)/12-hr dark cycles, and were collected at the indicated ZTs. For nighttime samples, we handled the flies under a red photographic safety light. We prefixed whole flies at 4°C with 4% paraformaldehyde in PBT for 45 min before dissecting out the brains. After the dissections, he brains were fixed again for 15–20 min at 4°C in 4% paraformaldehyde in PBT.
We used the following primary antibodies: anti-Rh7 (1:250, rabbit), anti-Per (1:1000, guinea pig), anti-Tim (1:1000, rat) 40, anti-PDF (1:1000, c7 mouse monoclonal antibody from the Developmental Studies Hybridoma Bank), anti-dsRed (1: 500, mouse, Clontech Catalog # 632392). The Per and Tim antibodies were contributed by A. Sehgal. The secondary antibodies (Thermo Fisher Scientific) were anti-rat Alexa Fluor 568 Dye and anti-guinea pig Alexa Fluor 555 Dye. We acquired the images using a Zeiss LSM 700 confocal microscope.
Immunostaining of whole-mounts of the Drosophila retina
To perform whole-mount staining of the retina, we dissected the retina (within the eye cup) and fixed the tissue at 4°C in 4% paraformaldehyde in PBT for 20 minutes. After washing briefly in PBT, we blocked the retina for 1 hr in PBT plus with 5% normal goat serum. We used the following primary antibodies: anti-Rh7 (1:250, rabbit), anti-Rh3 (1:200, mouse, gift from S. Britt, University of Colorado, Denver) and anti-Rh5 (1:200, mouse, gift from S. Britt, University of Colorado, Denver). The secondary antibodies were: anti-rabbit Alexa Fluor 568 Dye (1:1000) and anti-mouse Alexa Fluor 488 Dye (1:1000).
Circadian behavior to access rhythmicity and periodicity
Circadian experiments were performed at 25°C using the Drosophila Activity Monitoring (DAM) System (Trikinetics). Individual 3–7-day-old male flies were loaded into monitoring tubes, which contained 1% agarose (Invitrogen) and 5% sucrose (Sigma) as the food source. The flies were entrained to 12-hr light/12-hr dark cycles for four days and released to constant darkness or constant light (10 lux for dim light conditions and 400 lux for bright light conditions, unless indicated otherwise) for at least six days to measure the periodicities.
Data collection and analyses were performed using Clocklab (Actimetrics). Activity data for each fly was binned every 30 min for the circadian analyses. To obtain the periodicities, data from constant darkness were subjected to χ2 periodograms and Fast Fourier Transfer analysis using Clocklab software. The rhythm strength of a fly was measured as the power minus the significance (p-s). Flies were considered arrhythmic based on either p-s value < 10 or an FFT value (< 0.03). Actograms of weakly rhythmic flies were visually inspected to confirm rhythmicity.
Circadian behavior to access light mediated phase-shift and phase delay
To assay the effects on activity due to 5-min light pulses at night (Fig. 3a, b; Extended Data Fig. 4, 10), we first entrained the flies for 3 days under 12-hr light/12-hr dark cycles (~600 lux LED white light). During the night of the fourth L/D cycle (at ZT14, ZT16, ZT18, ZT20 or ZT22 respectively), we exposed the flies to a single 5-min light pulse (LED white light, ~600 lux), and then maintained the flies under constant darkness. The phase shift was calculated as the phase difference of the evening peaks before and after the light pulse. Negative and positive phase changes indicate phase delays and phase advances, respectively.
To conduct the phase delay experiments (Fig. 3g–l), we first entrained the flies for 4 days under 12-hr light/12-hr dark cycles (~400 lux LED white light). To obtain a phase delay of 8 hrs, on day 5 we extended the light phase to 20 hrs, and then returned the flies to normal 12-hr dark/12-hr light cycles. The phase shift magnitude was calculated as the phase difference between the evening peak of the day before the phase shift and the indicated day after the phase shift.
Circadian behavior to access light-mediated arousal
To assay light-dependent arousal, we entrained the flies for 4 days under 12-hr light/12-hr dark cycles and then exposed the flies to a 5-min white light pulse (~600 lux LED lights) at ZT22. We binned the activity data for each fly every min. “Light-coincident arousal” is the increase in locomotion activity (bin-crosses) during the 5-minute stimulation compared to the previous 5 minutes. “Arousal delay” is the time between lights on, and when maximum activity occurs.
Cell transfection, membrane extraction and spectral photometry
HEK293T (ATCC) cells were cultured to 70% confluency and transfected with 2 μg of the pCS2+MT-rh7 plasmid per 10 cm dish. We used the FuGENE® HD Transfection Reagent (Cat.E2311) for performing the transfections. Cells were harvested 24–36 hrs after transfection and stored at −80°C. For reconstitution of Rh7 with the chromophore, the HEK293T cells were resuspended in cold PBS (phosphate buffered saline pH 7.4,Quality Biological Inc.) supplemented with a protease inhibitor cocktail (Sigma P8340) and incubated with 40 μM 11-cis-retinal in the dark for 4 hrs. We prepared membrane protein extracts by resuspending membrane pellets in 0.1% CHAPs in PBS, rotating for 2 hrs at 4°C, then centrifuged (14,000 × g) for 20 min at 4°C. The supernatants were removed and analyzed with a UV3600 UV-Nir-NIR Spectrometer (Shimadzu). To obtain the spectral absorption for Rh7, we used membrane extracts from untransfected cells as the blank.
ERG recordings
ERG recordings were performed by filling two glass electrodes with Drosophila Ringer’s solution (3 mM CaCl2, 182 mM KCl, 46 mM NaCl, 10 mM Tris pH 7.2) and placing small droplets of electrode cream on the surface of the compound eye and the thorax to increase conductance. We inserted the recording electrode into the cream on the surface of the compound eye and the reference electrode into the cream on the thorax. Flies were dark adapted for 1 min before stimulating with a 2-sec pulse using a halogen light (~30 mV/cm2 unless indicated otherwise). The ERG signals were amplified with a Warner electrometer and recorded with a Powerlab 4/30 analog-to-digital converter (AD Instruments). Data were collected and analyzed with the Lab Chart version 6.1 program (AD Instruments).
Patch-clamp recordings and pharmacology
Patch-clamp measurements were performed on acutely dissected adult fly brains as described previously18,19. Briefly, all patch-clamp recordings were performed during the daytime to avoid clock-dependent variance in firing rate. All l-LNvs were recorded within a relatively narrow daytime window, which for each genotype tested were normally distributed for the time of day recorded and did not significantly vary between all three genotypes. l-LNv recordings were made in the whole-cell current clamp mode. After allowing the membrane properties to stabilize after whole cell break-in, we recorded for 30–60 sec in the current clamp configuration (unless otherwise stated) under nearly dark conditions (~0.05 mW/cm2) before the lights were turned on. Lights-on data were collected for 5–20 sec and followed by 60–120 sec of darkness.
Multiple light sources were used for these studies. We used a standard halogen light source on an Olympus BX51 WI microscope (Olympus USA, Center Valley, PA) for all experiments with white light (400–1000 nm, 4 mW/cm2). Orange light (550–1000 nm; 4 mW/cm2) for electrophysiological recordings was achieved by placing appropriate combinations of 25 mm long- and short-pass filters (Edmund Industrial Optics, Barrington, NJ) over the halogen light source directly beneath the recording chamber. We changed the filters during the recordings to internally match the neuronal responses to different wavelengths of light. Recordings using 405 nm violet light (0.8 mW/cm2) were obtained using the LEDs obtained from Prizmatix 405 LED (UHP-Mic-LED-405), which provides >2 W collimated purple light (405nm peak, 15 nm spectrum half width). Light was measured for all sources using a Newport 818-UV Sensor and the Optical Power/Energy Meter (842-PE, Newport Corporation, Irvine, CA) and expressed as mW/cm2.
The control genotype for the electrophysiological recordings was w;pdf-Gal4-dORK-NC1-GFP. The cry01 and rh71 recordings were performed using w;pdf-Gal4-dORK-NC1-GFP;cry01 and w;pdf-Gal4-dORK-NC1-GFP;rh71.
Statistical analyses
To analyze two sets of data, we used the unpaired Student’s t-test. To compare multiple sets of behavioral data, we used a one-away ANOVA test (Kruskal-Wallis test) followed by the Dunn’s test. Data are presented as mean ±S.E.M.s. We used the Fisher’s exact test for analyzing the percentages of rhythmic flies. For the patch-clamp recordings, the data are presented as mean ±S.E.M.s. Values of “n” refer to number of measured lights on/off cycles. In all cases the “n” values were obtained from ≥5 separate recordings (see details in the figure legends). Statistical tests of ANOVA were performed with SigmaPlot 11 (Systat Software Inc) or Prism 6 (Graphpad Software). The data were first tested for normal distributions. If the data were not normally distributed, we performed Kruskal-Wallis One Way Analysis of Variance on Ranks, followed by the Dunn’s test. ANOVA on normally-distributed data was followed by Tukey’s test to determine significant differences between genotypes.
Data availability
All data are available from the corresponding author upon reasonable request.
Extended Data
Extended Data Figure 1. Circadian photoentrainment in flies lacking cryptochrome and proteins required for phototransduction in the compound eye.
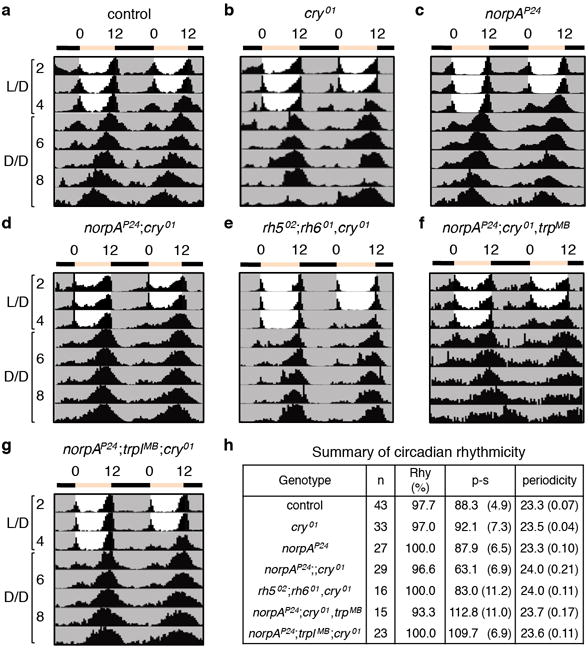
a–g, Average actograms exhibited by flies of the indicated genotypes maintained under 12-hr light/12-hr dark (L/D) for four days and then released to constant darkness (D/D). h, Summary of circadian rhythmicity of flies in a–g. Rhythm strength of a fly was measured as the power minus the significance (p-s).
Extended Data Figure 2. Rh7 is an extra-retinal opsin.
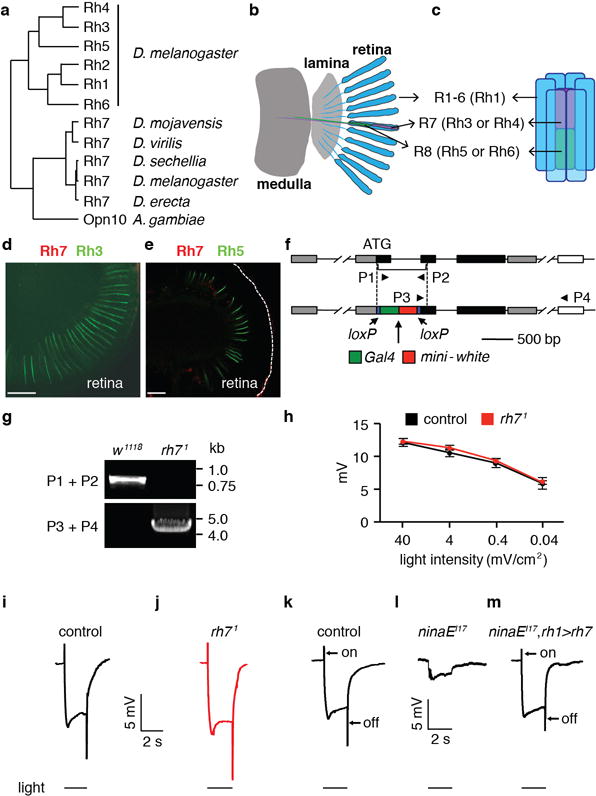
a, Phylogenetic tree constructed with protein sequences corresponding to the indicated opsins. The full name for Ae. aegypti OP10 is GPROP10 (VectorBase). b, Cartoon showing a longitudinal view of the main structures in the visual system, including the retina, lamina and medulla. The blue units represent ommatidia, which are comprised of eight photoreceptor cells (R1-8) and support cells. c, Cartoon showing the photoreceptor cells in a single ommatidium. The six outer photoreceptor cells (R1-R6) are represented in blue and express Rh136,41. The central R7 photoreceptor cell (purple) expresses either Rh3 or Rh442,43, while the R8 photoreceptor cell (green) expresses Rh5 or Rh644–46. d, A wild-type retina stained with anti-Rh7 (red) and anti-Rh3 (green). e, A wild-type retina stained with anti-Rh7 (red) and anti-Rh5 (green). The scale bars in d and e indicate 30 μm. No Rh7 positive staining was detected in the retina. f, Generation of rh71 by homologous recombination. Shown are cartoons of the wild-type rh7 locus (top) and the genomic organization of the rh71 allele (bottom). The triangles (P1-P4) indicate the primers used to verify the rh71 mutation. g, Confirmation of the rh71 mutation by PCR. We prepared genomic DNA from control (w1118) and rh71 flies and performed PCR using the P1/P2 and P3/P4 primer pairs. The positions of DNA markers (kb) are indicated to the right. See Supplementary Information for the raw images of the PCR gels. h, ERG amplitudes of control and rh71 flies using 2-sec white light pulses of the indicated intensities. The error bars indicate S.E.M.s. n=4. i–m, ERGs from flies of the indicated genotypes. The event markers below the ERGs indicate the initiation and cessation of the light stimuli. The scale bars indicate 5 mV and 2 sec. i, Control flies. j, rh71. k–m, Testing for rescue of the reduced ERG amplitude and loss of on- and off-transients in ninaEI17 flies with an rh7+ transgene. k, Control flies. l, ninaEI17 (rh1 mutant). m, ninaEI17 fly expressing UAS-rh7 in R1-R6 cells under control of the rh1-Gal4 (ninaEI17,rh1>rh7).
Extended Data Figure 3. Expression of Rh7 in non-clock neurons located in the dorsal part of the brain.
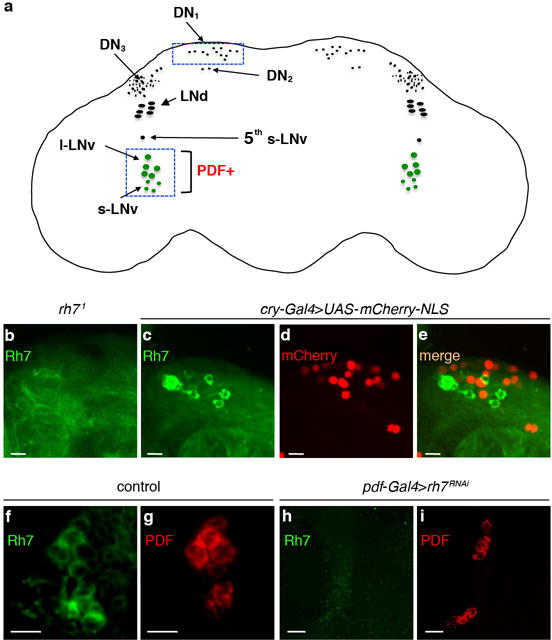
a, Cartoon of a fly brain showing different groups of clock neurons. The boxed areas indicate locations of two groups of Rh7-positive cells. b, rh71 brain stained with anti-Rh7. c–e, Double labeling of the dorsal region of the brain with a clock neuron reporter (cry-Gal4E13>UAS-mCherry-NLS) and anti-Rh7. The scale bars in b–e indicate 20 μm. c, Anti-Rh7. d, Anti-mCherry. e, Merge of c and d. f, Control brain stained with anti-Rh7. g, Control brain stained with anti-PDF. h, pdf-Gal4>rh7RNAi brain stained with anti-Rh7. i, pdf-Gal4>rh7RNAi brain stained with anti-PDF. The scale bars in f–i indicate 10 μm.
Extended Data Figure 4. Actograms showing representative behaviors of control and mutant flies before and after the 5-minute light pulse at the indicated ZT.
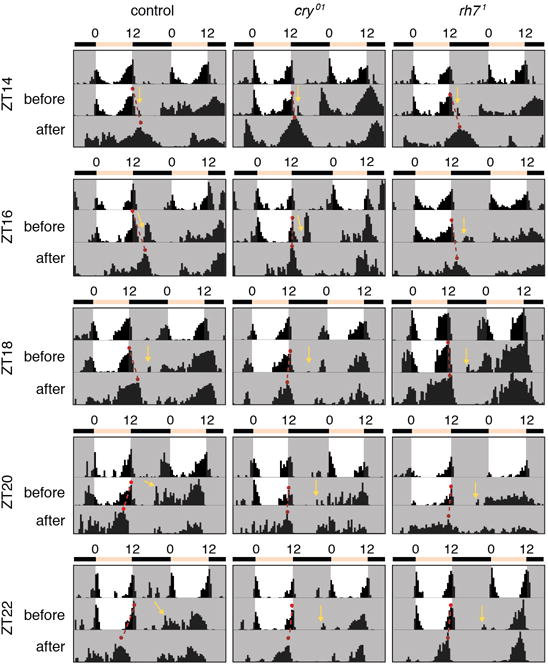
The red dots connected by the dashed red lines indicate the evening peaks before and after the light pulse. Each yellow arrow indicates the times that the animals were exposed to the 5-minute ~600 lux LED light pulse.
Extended Data Figure 5. Circadian responses to constant light and light-dependent arousal in rh71 flies.
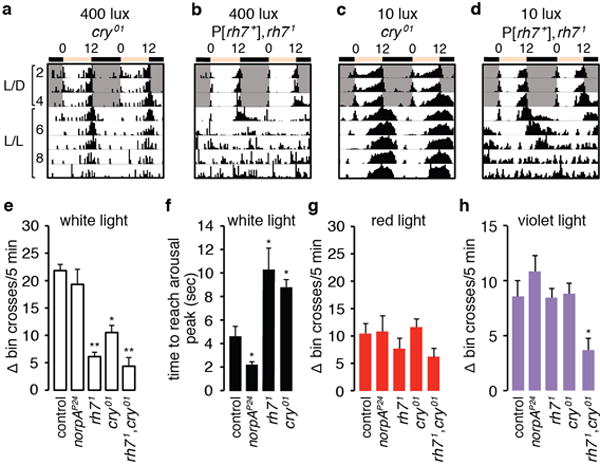
a, b, Flies of the indicated genotypes were entrained under 12-hr light/12-hr dark (L/D) cycles and subsequently released to constant ~400 lux light (L/L). c, d, Flies of the indicated genotypes were entrained under 12-hr light/12-hr dark (L/D) cycles and subsequently released to constant ~10 lux light (L/L). e, Quantification of the effect of a 5-min white light pulse on arousal. Arousal was quantified as increases in total bin crosses during the 5-min light stimulation compared to the total bin crosses during the 5 min before the light stimulation. f, Quantification of the time required to reach the maximum activity after white light stimulation. g, h, Quantification of the effects on arousal due to a 5-min exposure to red (625 nm) LED lights (g) or violet (405 nm) LED lights (h). The error bars indicate S.E.M.s. One-away ANOVA test (Kruskal-Wallis test) followed by the Dunn’s test. *p<0.05, **p<0.01. Number of flies tested: norpAP24, n=16, other genotypes, n=24.
Extended Data Figure 6. Effects of multiple light input pathways on circadian behavior.
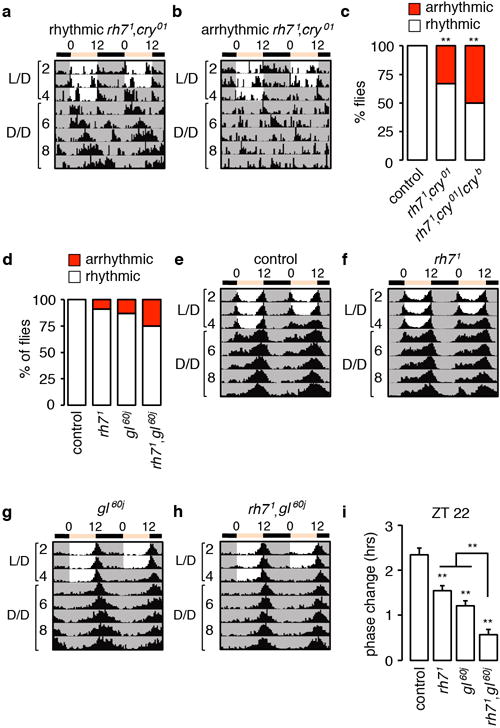
a, b, Actograms showing rhythmic and arrhythmic rh71,cry01 double mutants. The flies were entrained under 12-hr light/12-hr dark (L/D) cycles and subsequently released to constant darkness (D/D). c, Percentages of rhythmic and arrhythmic flies. Fisher’s exact test. **p<0.01. Number of flies tested: control, n=16; other genotypes, n=30. d–h Circadian behavior of gl60j and rh71,gl60j double mutant flies. The flies were entrained to 12-hr light/12 hr dark cycles (L/D) for 4 days and subsequently released to constant darkness (D/D). d, Percentages of rhythmic and arrhythmic flies. e–h, Average actograms showing the activities of flies of the indicated genotypes. Number of flies tested: control, n=46; rh71 n=34; gl60j n=38; rh71,gl60j, n=40. i, Phase response of the indicated genotypes to a 5-min white light stimulation at ZT22. The error bars indicate S.E.M.s. One-way ANOVA test (Kruskal-Wallis test) followed by the Dunn’s test. **p<0.01. Flies tested: control, n=54; rh71 n=49; gl60j n=53; rh71,gl60j, n=57.
Extended Data Figure 7. Rescue of the rh71,cryb photoentrainment defect by expression of rh7 in pacemaker neurons.
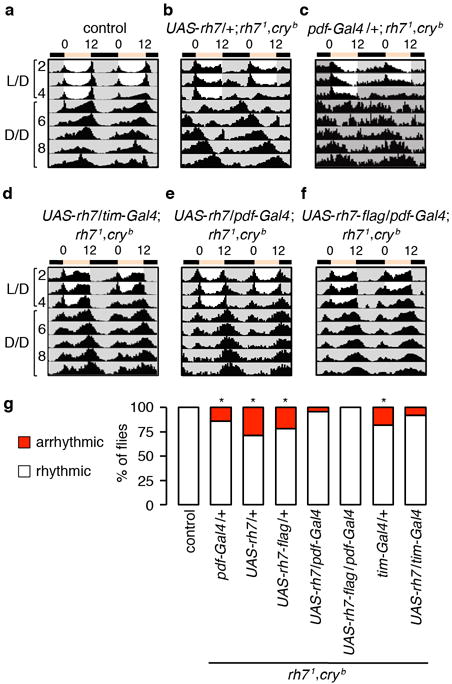
a–c, Actograms of control flies and rh71,cryb flies harboring only the UAS-rh7 or pdf-Gal4 transgenes. Number of flies tested: control, n=16; UAS-rh7/+; rh71,cryb, n=52; pdf-Gal4/+; rh71,cryb, n=21. d–f, Actograms of rh71,cryb flies expressing UAS-rh7 in pacemaker neurons under the control of the tim-Gal4 or pdf-Gal4 as indicated. Number of flies tested: UAS-rh7/tim-Gal4; rh71,cryb, n=37; UAS-rh7/pdf-Gal4; rh71,cryb, n=23; UAS-rh7-flag/pdf-Gal4; rh71,cryb, n=21. g, Percentages of rhythmic and arrhythmic flies of the indicated genotypes. Fisher’s exact test. *p<0.05.
Extended Data Figure 8. Rescue of rh71,cryb photoentrainment defect by expression of other fly rhodopsins.
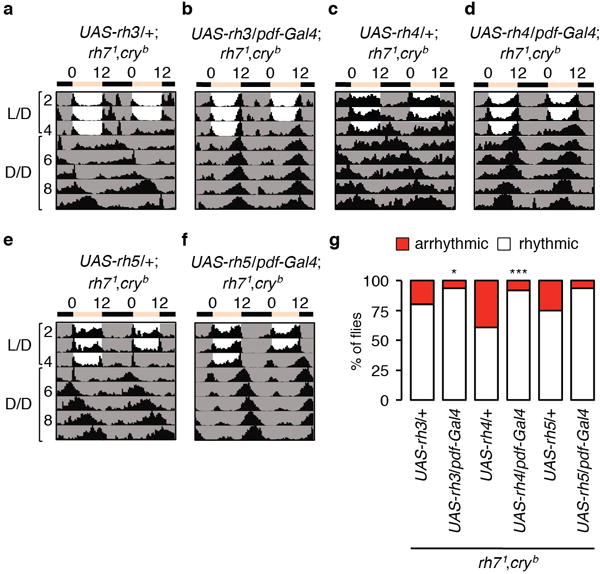
a–f, Controls showing actograms of rh71,cryb flies harboring UAS-rhodopsin transgenes only, and of rh71,cryb flies expressing the indicated rhodopsin genes in pacemaker neurons under control of the pdf-Gal4. Number of flies tested: UAS-rh3/+; rh71,cryb, n=56; UAS-rh4/+; rh71,cryb, n=46; UAS-rh5/+; rh71,cryb, n=24; UAS-rh3/pdf-Gal4; rh71,cryb, n=64; UAS-rh4/pdf-Gal4; rh71,cryb, n=25; UAS-rh5/pdf-Gal4; rh71,cryb, n=16. g, Percentages of rhythmic and arrhythmic flies of the indicated genotypes. Fisher’s exact test. *p<0.05. ***p<0.001.
Extended Data Figure 9. Per oscillates in control, rh71, cryb and rh71,cryb flies.
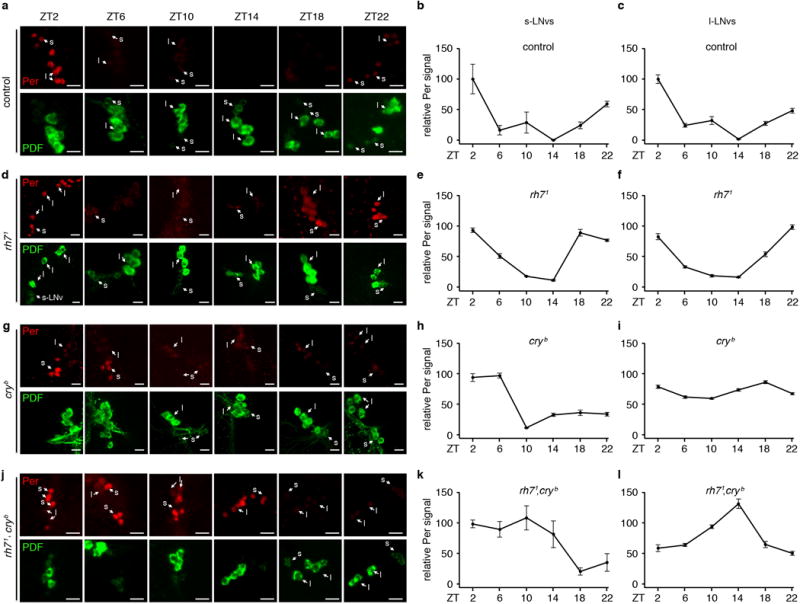
a, d, g, j, Flies of the indicated genotypes were entrained under L/D cycles for 4 days and the brains were dissected on the 5th day. The ZT times indicate when the brains were fixed and dissected for staining with anti-Per (Per, upper rows, red) and anti-PDF (PDF, lower rows, green) as indicated. At least one s-LNV (s) and one l-LNV (l) are labeled in the images obtained at each ZT to facilitate identification of LNVs. The scale bars indicate 10 μm. b, c, e, f, h, i, k, l, Quantification of relative Per levels in s-LNvs and L-LNVs of flies of the indicated genotypes. The image quantification was performed using ImageJ. The Y-axes indicate relative Per intensities. The Per intensities in ZT2 of the control flies were designated as 100. For control flies, ZT10, n=6; ZT22, n=8, and n=5 for all other time points. For rh71, ZT2, n=9; ZT6, n=8; ZT10, n=6; ZT14, n=8; ZT18, n=8; ZT22, n=7. For cryb, ZT2, n=8; ZT6, n=9; ZT10, n=8; ZT14, n=7; ZT18, n=10; ZT22, n=8. For rh71,cryb, n=5 for all time points. Error bars indicate S.E.M.s.
Extended Data Figure 10. Knockdown of plc21C in PDF-positive neurons impaired circadian phase response.
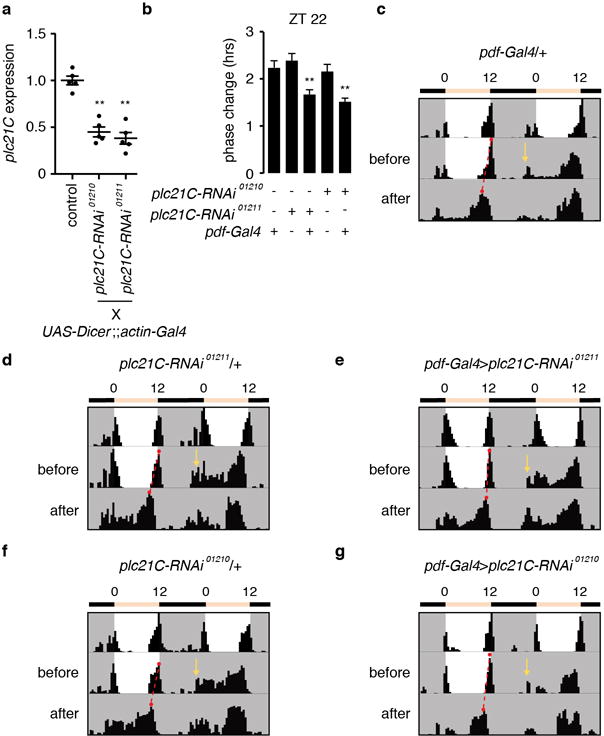
a, Quantitative real-time PCR analysis of plc21C mRNA using RNA prepared from whole adults. The plc21C expression levels in each sample were normalized using rp49 expression. The control was w1118. b, Phase response of the indicated genotypes to a 5-min white light stimulation at ZT22. One-away ANOVA test (Kruskal-Wallis test) followed by the Dunn’s test. **p<0.01. pdf-Gal4/+, n=32; plc21C-RNAi01211/+, n=31; pdf-Gal4>plc21C-RNAi01211, n=37; plc21C-RNAi01210/+, n= 15; pdf-Gal4>plc21C-RNAi01210, n=32. The error bars indicate S.E.M.s. c–g, Examples of behavior before and after the 5-minute light pulse. The yellow arrows indicate the times of the 5-minute white light pulses (~600 lux). The red dots connected by the dashed red lines indicate the evening peaks before and after the light pulse.
Supplementary Material
Acknowledgments
We thank A. Sehgal, M. Rosbash, M. Wu, and P. Emery for fly stocks, and A. Sehgal and S. Britt for antibodies. We thank E. Guzman, H. Zhou and the Next Generation Sequencing Core at the UCSB Biological Nanostructures Lab for help with the RNA-seq. This work was supported by grants to C.M. from the National Eye Institute (EY008117) and the National Institute on Deafness and other Communication Disorders (DC007864), and to T.C.H. from National Institute of General Medical Sciences (GM102965 and GM107405).
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions
J.N. T.H. and C.M. designed the study. J.N. and L.B. performed experiments and all authors analyzed the data. J.N. and C.M. wrote the manuscript with input from T.H. and L.B.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Helfrich-Förster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron. 2001;30:249–261. doi: 10.1016/s0896-6273(01)00277-x. [DOI] [PubMed] [Google Scholar]
- 2.Emery P, et al. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 3.Stanewsky R, et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 4.Ozturk N, Selby CP, Annayev Y, Zhong D, Sancar A. Reaction mechanism of Drosophila cryptochrome. Proc Natl Acad Sci USA. 2011;108:516–521. doi: 10.1073/pnas.1017093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szular J, et al. Rhodopsin 5- and Rhodopsin 6-mediated clock synchronization in Drosophila melanogaster is independent of retinal phospholipase C-ß signaling. J Biol Rhythms. 2012;27:25–36. doi: 10.1177/0748730411431673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emery P, Stanewsky R, Hall JC, Rosbash M. A unique circadian-rhythm photoreceptor. Nature. 2000;404:456–457. doi: 10.1038/35006558. [DOI] [PubMed] [Google Scholar]
- 7.Helfrich-Förster C, et al. The extraretinal eyelet of Drosophila: development, ultrastructure, and putative circadian function. J Neurosci. 2002;22:9255–9266. doi: 10.1523/JNEUROSCI.22-21-09255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malpel S, Klarsfeld A, Rouyer F. Larval optic nerve and adult extra-retinal photoreceptors sequentially associate with clock neurons during Drosophila brain development. Development. 2002;129:1443–1453. doi: 10.1242/dev.129.6.1443. [DOI] [PubMed] [Google Scholar]
- 9.Sprecher SG, Desplan C. Switch of rhodopsin expression in terminally differentiated Drosophila sensory neurons. Nature. 2008;454:533–537. doi: 10.1038/nature07062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senthilan PR, Helfrich-Forster C. Rhodopsin 7-The unusual Rhodopsin in Drosophila. PeerJ. 2016;4:e2427. doi: 10.7717/peerj.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 13.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helfrich-Förster C. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J Comp Physiol A. 1998;182:435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- 15.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 16.Yoshii T, Todo T, Wulbeck C, Stanewsky R, Helfrich-Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol. 2008;508:952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
- 17.Helfrich-Förster C, et al. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J Comp Neurol. 2007;500:47–70. doi: 10.1002/cne.21146. [DOI] [PubMed] [Google Scholar]
- 18.Fogle KJ, Parson KG, Dahm NA, Holmes TC. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science. 2011;331:1409–1413. doi: 10.1126/science.1199702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheeba V, Gu H, Sharma VK, O’Dowd DK, Holmes TC. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol. 2008;99:976–988. doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fogle KJ, et al. CRYPTOCHROME-mediated phototransduction by modulation of the potassium ion channel ß-subunit redox sensor. Proc Natl Acad Sci USA. 2015;112:2245–2250. doi: 10.1073/pnas.1416586112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dushay MS, et al. Phenotypic and genetic analysis of Clock, a new circadian rhythm mutant in Drosophila melanogaster. Genetics. 1990;125:557–578. doi: 10.1093/genetics/125.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kistenpfennig C, Hirsh J, Yoshii T, Helfrich-Förster C. Phase-shifting the fruit fly clock without cryptochrome. J Biol Rhythms. 2012;27:117–125. doi: 10.1177/0748730411434390. [DOI] [PubMed] [Google Scholar]
- 23.Vinayak P, et al. Exquisite light sensitivity of Drosophila melanogaster cryptochrome. PLoS Genet. 2013;9:e1003615. doi: 10.1371/journal.pgen.1003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo F, Cerullo I, Chen X, Rosbash M. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. Elife. 2014;3 doi: 10.7554/eLife.02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheeba V, et al. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol. 2008;18:1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci USA. 2008;105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- 29.Nitabach MN, et al. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blackshaw S, Snyder SH. Encephalopsin: a novel mammalian extraretinal opsin discretely localized in the brain. J Neurosci. 1999;19:3681–3690. doi: 10.1523/JNEUROSCI.19-10-03681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 32.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 33.Dolezelova E, Dolezel D, Hall JC. Rhythm defects caused by newly engineered null mutations in Drosophila's cryptochrome gene. Genetics. 2007;177:329–345. doi: 10.1534/genetics.107.076513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamaguchi S, Wolf R, Desplan C, Heisenberg M. Motion vision is independent of color in Drosophila. Proc Natl Acad Sci USA. 2008;105:4910–4915. doi: 10.1073/pnas.0711484105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasiliauskas D, et al. Feedback from rhodopsin controls rhodopsin exclusion in Drosophila photoreceptors. Nature. 2011;479:108–112. doi: 10.1038/nature10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Tousa JE, et al. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40:839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- 37.Venken KJ, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci USA. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y, Montell C. Drosophila TRPA1 functions in temperature control of circadian rhythm in pacemaker neurons. J Neurosci. 2013;33:6716–6725. doi: 10.1523/JNEUROSCI.4237-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jang AR, Moravcevic K, Saez L, Young MW, Sehgal A. Drosophila TIM binds importin α1, and acts as an adapter to transport PER to the nucleus. PLoS Genet. 2015;11:e1004974. doi: 10.1371/journal.pgen.1004974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuker CS, Cowman AF, Rubin GM. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985;40:851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]
- 42.Montell C, Jones K, Zuker C, Rubin G. A second opsin gene expressed in the ultraviolet-sensitive R7 photoreceptor cells of Drosophila melanogaster. J Neurosci. 1987;7:1558–1566. doi: 10.1523/JNEUROSCI.07-05-01558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker CS, Montell C, Jones K, Laverty T, Rubin GM. A rhodopsin gene expressed in photoreceptor cell R7 of the Drosophila eye: homologies with other signal-transducing molecules. J Neurosci. 1987;7:1550–1557. doi: 10.1523/JNEUROSCI.07-05-01550.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chou WH, et al. Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron. 1996;17:1101–1115. doi: 10.1016/s0896-6273(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 45.Papatsenko D, Sheng G, Desplan C. A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development. 1997;124:1665–1673. doi: 10.1242/dev.124.9.1665. [DOI] [PubMed] [Google Scholar]
- 46.Chou WH, et al. Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development. 1999;126:607–616. doi: 10.1242/dev.126.4.607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding author upon reasonable request.


