Abstract
Renal cell carcinoma (RCC) is a malignant tumor, which severely threatens human’s life, moreover, the multi-drug resistance (MDR) under RCC undoubtedly strengthen the difficulties in the treatment. MiR-451 has been considered to play an important role in regulation of MDR in several cancers, but the role of it in MDR of RCC has not been explored. This study aims to explore the mechanism of miR-451 as a target to regulate chemotherapy resistance, which is crucial for further exploring novel therapy for RCC. Two human cell lines (ACHN and GRC-1) were performed in this study and adriamycin (ADM) was used to construct MDR cell lines. qRT-PCR was used to determine the mRNA expression of miR-451 and ATF-2. Weston blot was used to determine protein expression. MTT assay and flow cytometry were used for assessing cell viability and apoptosis, individually. Luciferase reporter assay was used to detect the targeting of miR-451 and ATF-2. Results presented that the expression of miR-451 was higher in low MDR cell line (ACHN) comparing with the high MDR cell line (GRC-1), while the expression of ATF-2 revealed an opposite results. MiR-451 targeted ATF-2 and regulated its expression. Overexpression of miR-451 strengthened drug resistance, decreased cell viability, and increased cell apoptosis of GRC-1 pretreated by ADM, while overexpressed ATF-2 reversed the effect induced by miR-451 overexpression. Then miR-451 knockdown improved drug susceptibility, decreased cell apoptosis, and increased cell viability of ACHN induced by ADM, however, ATF-2 suppression reversed the low rate of cell apoptosis and high rate of cell viability induced by miR-451 knockdown. Our results revealed that miR-451 regulates the drug resistance of RCC by targeting ATF-2 gene, which might be critical for overcoming MDR in RCC patients.
Impact statement
This is the first study to emphasize the expression of miR-451 on regulating multi-drug resistance (MDR) in renal cell carcinoma (RCC). Our study found that miR-451 regulates the drug resistance of RCC by targeting ATF-2, which might be critical for overcoming MDR in RCC patients. This study not only provides solid theory foundation for the clinical therapy, but also offers unique insights for the further RCC research. Furthermore, the study helps us to understand the mechanism of MDR, which was crucial for identifying the chemoresistance on several related tumors.
Keywords: Renal cell carcinoma, multi-drug resistance, MiR-451, ATF-2, chemoresistance, urological cancer
Introduction
Renal cell carcinoma (RCC) is a malignant tumor of urological cancer accounting for about 2–3% of all adult cancers.1 Importantly, only 10% of RCC patients with metastases survive longer than five years,2,3 which seriously threatens human’s health and life. Every year, about 338,000 new cases of RCC are diagnosed worldwide, among those patients, about 50% were diagnosed with intermediate and terminal cancer. Chemotherapy due to its convenient and quick effects is always selected as the perfect method. However, the difficulties were that the tumor might not be sensitive to chemotherapy or gradually formed multi-drug resistance, which is the great challenge for RCC treatment. So far, no chemotherapy is valuable with drug response rates higher than 15% that suit the RCC therapy.4 Therefore, identifying the mechanism that contributes to multi-drug resistance (MDR) is crucial for improving RCC therapy.
Chemoresistance was widely studied on several kinds of tumors, such as ovarian cancer, breast cancer and colorectal cancer,5–7 the mechanism including drug inactivation, decreased intracellular drug accumulation, increased DNA repair, apoptosis-related chemoresistance, and enhanced efflux.8–10 However, to regulate several related-gene expression to control the MDR seemed to be crucial in tumor MDR study.11,12
MicroRNAs (miRNAs) are a set of short single stranded RNAs with the length of about 22 nucleotides without protein coding function; studies have reported that miRNAs played a crucial role in regulating several transcriptional level and modulating various biological processes, such as gene expression, cell growth, differentiation, and apoptosis.13–16 Previous studies have reported that dysregulation of miRNAs could induce several cancers with inhibition or elevation of its expression.17,18 Additionally, much miRNAs have been identified to play an important role in RCC, for example, the down-regulated miR-141, miR-200c,19 the tumor suppressive miR-23b, miR-27b,20 miR-135a,21 and the up-regulated miR-22122 were found in the RCC cells or tissues. However, in the potential mechanism of miRNAs regulation the MDR of RCC is not fully understandable. MiR-451 was down-regulated in many cancers, and plays a crucial role in the oncogenic mechanisms of several cancers, such as liver cancer, liposarcoma, gastric carcinoma, and neuroglioma.23–26 More importantly, it has been considered that miR-451 also acts as an anti-oncogene in RCC.27 Furthermore, miR-451 played a vital role in the chemoresistance of breast cancer,28 lung adenocarcinoma,29 and non-small cell lung cancer,30 but the role of it in the development of chemoresistance in human RCC and the underlying mechanism remained unclear.
Herein, this study was performed to investigate the expression profiles of miR-451 in regulating gene expression and understanding how miR-451 regulates MDR of renal carcinoma cells. Our results might contribute to explore the effectiveness of anticancer drugs for RCC.
Methods
Cell culture
Human renal carcinoma cell lines: GRC-1 and ACHN were purchased from a human clear cell carcinoma of kidney in the Cell Bank the Chinese Academy of Sciences (Shanghai) and cultured at the First Affiliated Hospital of Nanchang University. Briefly, two cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, as well as definite amounts of penicillin, streptomycin, and glutamine. Cells were incubated at 37℃ with 5% CO2.
Cell viability
Cell viability was detected according to the manufacture’s instruction of a MTT Cell Proliferation and Cytotoxicity Assay Kit. About 1 × 105 cells were planted into a 96-well plate. Chemical drug adriamycin (ADM) was added onto the plate and incubated for 24 h or 48 h. After incubation, 20 µL of 5 mmol/L of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) was added to each well and kept for 4 h at 37℃. Dimethyl sulfoxide (DMSO) was used to dissolve the formazan crystals. The results were read on an absorbance value of 490 nm. Median lethal concentration (LC50) was estimated by the relative survival curve.
Quantitative real-time PCR
Total RNA isolation was performed according to manufacturer’s guidance (Invitrogen, USA) by using of a RNA Extraction kit. The quality of RNA was detected by a Nanodrop apparatus.8 Then 1 µL of RNA was taken out and reverse transcribed into cDNA according to a manufacturer’s instrument based on a reverse transcription kit (Invitrogen). Real-time PCR was carried out using SYBR Premix DimerEraser (Takara, Dalian, China). All data obtained from qRT-PCR were repeated for three times. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the internal control.
Western blotting
Proteins were extracted from the cells by using bioelectrical impedance analysis (BIA) with different treatments, and the quality of proteins was quantified by bicinchoninic acid assay (BCA). Then the proteins were resolved and separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride membranes. After blocking, the membranes were washed and then incubated with primary polyclonal antibodies with the blot at room temperature for one night. After washing, the membranes were incubated with secondary antibodies at 37℃ for 1 h. Power Opti-ECL kit was used to observe the bands and ImageJ software was used to quantify the protein level; β-actin (Sigma) acted as the internal control.
Luciferase reporter assay
The sequence of ATF-2 was selected from Genebank, the ATF-2 3’ TUR was cloned into the selected vector (Genomeditech Co., Ltd., China). Lipofectamine 2000 transfection reagent (Invitrogen) was used for the co-transfection of miR-451 and Luc-ATF-2-3’UTR. Dual-Luciferase Reporter Assay (Promega) was used for the detection of firefly luciferase activity; Renilla served as the internal control.
Cell transfection
Cells were transfected by an instrument of Lipofectamine 2000 (Invitrogen, USA). Briefly, ADM was added to GRC-1 and ACHN cells for 24 h, then miR-451 mimic or miR-451 inhibitor was transfected into cells of GRC-1 or ACHN, separately. Later the cells were grown at 37℃ for 48 h. The cells were collected and qRT-PCR was used to determine the transfection efficiency.
Cell apoptosis
Annexin V-FITC Apoptosis Detection Kit (ThermoFisher) was used for detecting cells apoptosis. After transfection, cells were further incubated with ADM for 48 h. Then the cells were collected and washed twice with phosphate-buffered saline (PBS). Annexin V-FITC (5 µL) and propidium iodide (PI, 10 µL) were added and the cells were incubated at 25℃ for a quarter of an hour. The flow cytometry was used for counting the stained cells and FACScalibur8 was used for analyzing cell apoptosis. The experiments were repeated for three times.
Statistical analysis
Student’s t-test was used to test the differences between the variables of the groups. Statistical analyses were performed using the SPSS 15.0 program. Data were presented as mean ± SD. P < 0.05 was considered as significant difference.
Results
Effects of ADM on cells’ inhibition rates
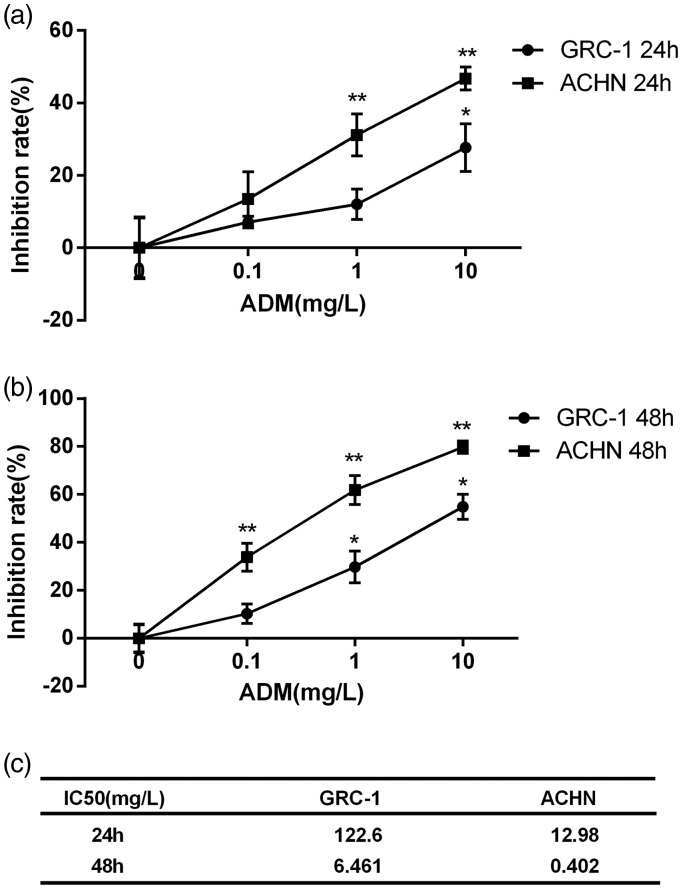
In order to investigate the drug resistance of human renal carcinoma cell lines GRC-1 and ACHN, a set of concentrations of ADM were tested on GRC-1 and ACHN. According to Figure 1(a and b), after ADM treatment, cell inhibition rates were measured. Briefly, the cell inhibition rates were concentration-dependent, and with the increased concentration of ADM the cell inhibition rates were significantly increased. At the same time, the inhibition rates of ADM on GRC-1 were significantly higher than that on ACHN. The 50% inhibitory concentration (lC50) was used to measure the drug resistance, and the results revealed that the values markedly varied between the cells of GRC-1 and ACNH with ADM for 24 h or 48 h, as shown in Figure 1(c); after the cells were treated with ADM for 24 h, lC50 values of GRC-1 and ACNH were 122.6 mg/L and 12.98 mg/L, respectively. And after the cells were treated with ADM for 48 h, lC50 values of GRC-1 and ACNH were 6.461 mg/L and 0.402 mg/L, respectively.
Figure 1.
Inhibition rates of ADM on cell lines of GRC-1 and ACHN. Cells inhibition rates were detected after the two cell lines were treated with a set of concentrations of ADM (0, 0.1 mg/L, 1 mg/L, 10 mg/L). (a) After cells were treated with ADM for 24 h, the inhibition rates were concentration-dependent, which presented a higher inhibition rates of ADM on GRC-1 cells rather than on ACHN cells. (b) After cells were treated with ADM for 48 h, the cells inhibition rates were increased comparing with the cells treated with ADM for 24 h, and the trends of cells inhibition rates were also increased along with the increased concentration of ADM. (c) lC50 of cells treated with ADM for 24 h and 48 h. Error bars represent SEM. **P < 0.01
The expression of miR-451 and ATF-2 in GRC-1 and ACHN cell lines
To profile miRNA expression, we performed the miR-451 expression in the GRC-1 and ACHN cell lines. The results revealed that miR-451 was significantly more highly expressed in ACHN than in GRC-1 (Figure 2(a)). Then we detected the relative expression of ATF-2, the results revealed that the expression of ATF-2 was significantly lower in ACHN than in GRC-1 (Figure 2(b) and (c)). The results revealed markedly negative correlation of miR-451 and ATF-2 expression in GRC-1 and ACHN.
Figure 2.
The difference in the expression of miR-451 and ATF-2. (a) The expression level of miR-451 was higher in ACHN cells than in GRC-1 cells. (b and c) The expression level of ATF-2 was higher in GRC-1 cells than in ACHN cells. Error bars represent SEM. *P < 0.05
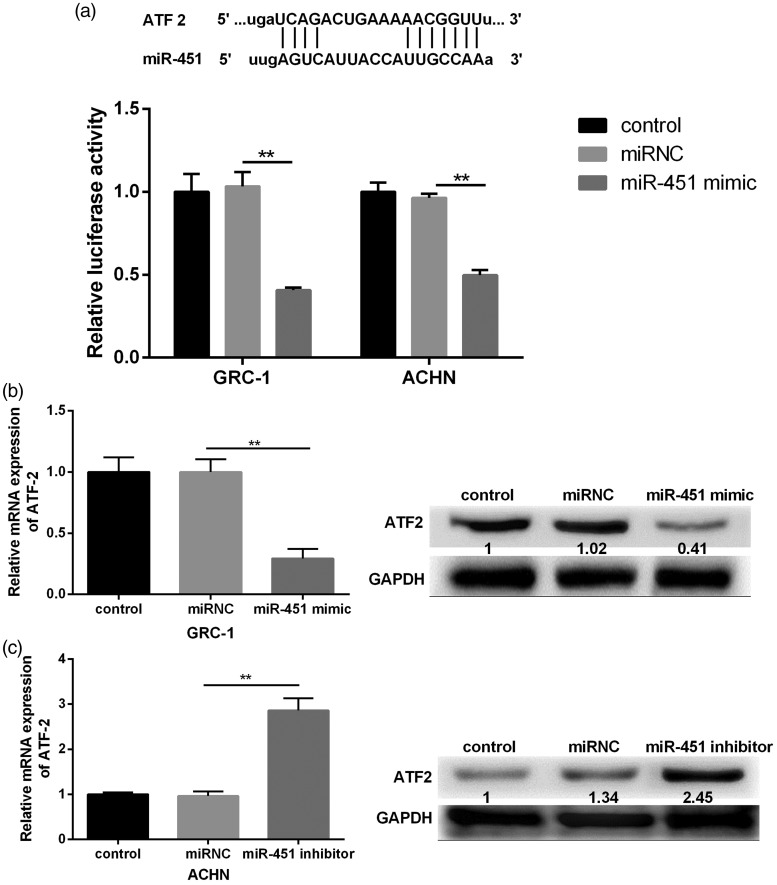
miR-451 targets ATF-2 to regulate its expression
To verify miR-451 targeted 3’ UTR of ATF-2, we used a targetScan database for the prediction. The results revealed that miR-451 had highly conserved target sequence with ATF-2 at 3’UTR. When the 3’ UTR of ATF-2 luciferase reporter vector and miR-451 mimic were co-transfected into GRC-1 and ACHN, we found that the luciferase activity was significantly decreased (Figure 3(a)). In order to verify the results, miR-451 mimic or miR-451 inhibitor was constructed and transfected into GRC-1 or ACHN. Results revealed that the overexpression of miR-451 dramatically decreased the expression of ATF-2 (Figure 3(b)). Reversely, miR-451 suppression significantly increased the expression of ATF-2 (Figure 3(c)).
Figure 3.
The effects of miR-451 alteration on the activity of ATF-2 3’UTR. (a) The predicted position of miR-451 base paring with the ATF-2 3’UTR. The relative luciferase activity was significantly decreased after GRC-1 or ACHN cells were transfected with miR-451 mimic. (b) GRC cells transfected with miR-451 mimic significantly decreased the expression of ATF-2. (c) ACHN cells transfected with miR-451 inhibitor significantly increased the expression of ATF-2
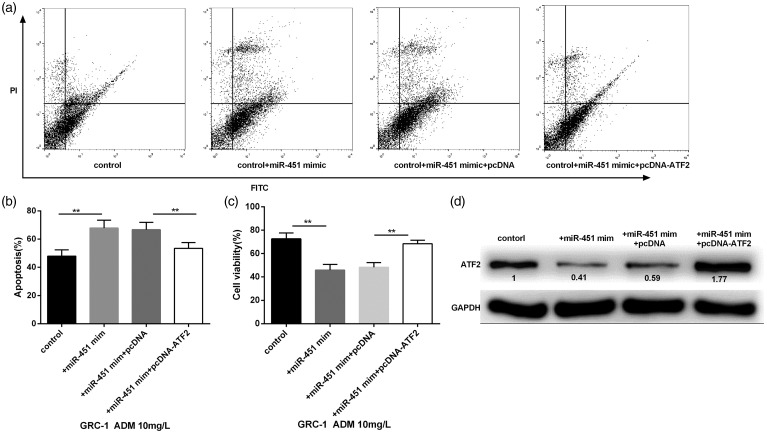
Interaction of miR-451 and ATF-2 overexpression on cell viability and death
To detect the overexpression of miR-451 and ATF-2 on cell viability and apoptosis, GRC-1 cells were randomly divided into four groups (control, control + miR-451 mimic, control + miR-451 mimic + pcDNA, control + miR-451 mimic + pcDNA-ATF-2). Then the cells of the four groups were stimulated by 10 mg/L ADM for 24 h. After stained with Annexin V and PI, cell apoptosis was detected by flow cytometry. On the dual parameter fluorescent dot, viable cells were presented in the lower-left quadrant and apoptotic cells were presented in the right quadrant (Figure 4(a)). Results revealed that overexpressed miR-451 significantly increased cell apoptosis, while, overexpressed ATF-2 significantly reversed the high ratio of cell apoptosis induced by miR-451 mimic (Figure 4(b)). MTT assay found that miR-451 overexpression significantly decreased the cell viability, then ATF-2 overexpression significantly reversed this effect (Figure 4(c)). Moreover, overexpression of miR-451 significantly decreased the expression of ATF-2 (Figure 4(d)).
Figure 4.
The effects of miR-451 and ATF-2 overexpression on ADM (10 mg/L)-induced cell viability and apoptosis of GRC. (a) The two-dimensional quadrant of cell apoptosis was detected by flow cytometry. (b and c) GRC cells transfected with miR-451 mimic significantly increased cell apoptosis and decreased cell viability, while the cells then transfected with pcDNA-ATF-2 significantly reversed the effects of miR-451 overexpression. (d) The cells transfected with miR-451 mimic significantly decreased the expression of ATF-2, however, ATF-2 overexpression significantly increased the expression of ATF-2. *P < 0.05
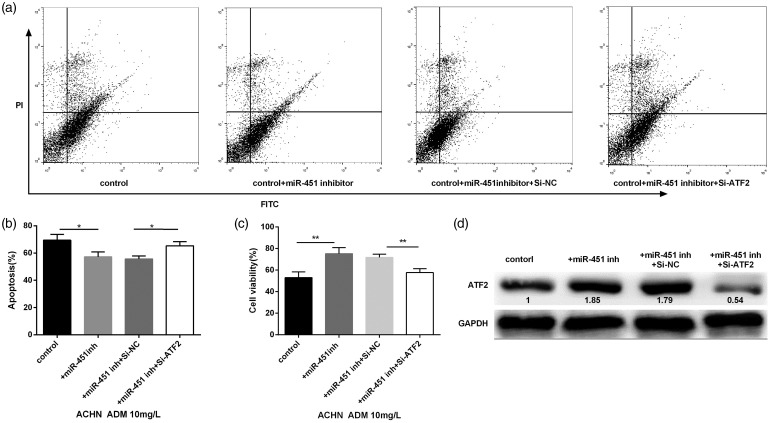
Interaction of miR-451 and ATF-2 suppression on cell viability and death
In order to detect the role of miR-451 and ATF-2 on cell viability and apoptosis, ACHN cells were randomly divided into four groups: control, control + miR-451 inhibitor, control + miR-451 inhibitor + si-NC, and control + miR-451 inhibitor + si-ATF-2. The cells were then stimulated by 10 mg/L ADM for 24 h. After staining with Annexin V and PI, cell apoptosis was detected by flow cytometry. The two-dimensional quadrant of cell apoptosis was shown in Figure 5(a). The results showed that miR-451 knockdown significantly decreased cell apoptosis, while ATF-2 suppression reversed the effect induced by miR-451 knockdown (Figure 5(b)). At the same time, cell viability of ACHN was significantly increased with the transfection of miR-451 inhibitor, then ATF-2 suppression reversed the effect induced by miR-451 knockdown (Figure 5(c)). Western blot revealed that miR-451 knockdown significantly increased the expression of ATF-2, and cells transfected with si-ATF-2 significantly decreased the expression of ATF-2 (Figure 5(d)).
Figure 5.
Effects of miR-451 knockdown and ATF-2 suppression on ADM (10 mg/L)-induced cell viability and apoptosis of ACHN. (a) The two-dimensional quadrant of cell apoptosis was detected by flow cytometry. (b and c) The cells transfected with miR-451 inhibitor significantly decreased the cell apoptosis but increased the cell viability, ATF-2 suppression significantly reversed the effects induced by miR-451 knockdown. (d) miR-451 knockdown dramatically increased the expression of ATF-2, however, ATF-2 suppression reversed the increased expression induced by miR-451 knockdown. *P < 0.05
Discussion
MDR is an important problem on RCC therapy, which could resist drugs’ effects and lead to failure chemotherapy. In this study, two cell lines GRC-1 and ACHN were used as high-MDR and low-MDR cells, and the ATF-2 expression was investigated to evaluate the chemoresistance mechanism in RCC.
Previous studies have indicated that miR-451 was related to several cancers, such as liver cancer and gastric carcinoma.31,25 Researchers also found the expression of miR-451 was remarkably changed in RCC tissues as well as RCC serum.32,23 However, several studies had assessed the relationship of miR-451 expression and MDR, and the results indicated that the expression of miR-451 was higher in drug resistance tissue than normal tissue in breast cancer as well as in lung cancer.33,29,30 As our results presented that high-MDR cell line resulted in lower miR-451 expression and low-MDR cell line caused higher miR-451 expression. Based on the reports and our observation, our results might indicate that the expression of miR-451 has a close relationship with drug resistance of RCC.
To further demonstrate how miR-451 modulates MDR in RCC, we analyzed drug-triggered cells death in GRC-1 transfected miR-451 mimic and in ACHN cells transfected miR-451 inhibitor. Among the results, miR-451 mimic significantly repressed cell viability and accelerated cell apoptosis of GRC induced by ADM, but miR-451 inhibitor dramatically increased cell viability and reduced cell apoptosis of ACHN induced by ADM. The results indicated that ATF-2 overexpression could significantly reverse the effects induced by overexpressed miR-451, moreover, ATF-2 suppression significantly reversed the effect induced by miR-451 knockdown. However, the results might strengthen the results that the expression of miR-451 regulated MDR, with overexpression of miR-451 strengthened drug resistance, while miR-451 knockdown promoted drug susceptibility in MDR.
ATF2 is a kind of transcription factor and functions as carcinogenesis and tumor suppressive, which mainly depends on its location and cell context.34 Studies have revealed that ATF2 was related to cancer ontogenesis through activating target genes, and then promoting tumor cells proliferation, and finally forming chemoresistance.35–37 Previous studies have indicated that ATF2 contributes to chemoresistance in breast cancer, and melatonine in non-small cell lung cancer.38,39 This study obtained the results that the expression of ATF2 was significantly higher in high-MDR cells rather than in low-MDR cells. Our results also proved that ATF-2 was target regulated by miR-451, then we inferred that miR-451 regulated cell MDR in RCC probably through targeted regulating the expression of ATF2.
In conclusion, this study was first to illustrate miR-451 regulates the drug resistance of RCC by regulating ATF-2 expression. Our findings suggested that both miR-451 and ATF2 acted as biological markers and regulated the chemoresistance of RCC. The study helps us to understand the mechanism of MDR and also provides future therapeutic strategy for RCC treatment.
Author contributions
LW conceived of the study, and participated in its design. LL and KZ performed the experiments and analyzed the data. XS interpreted the data and wrote the article.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The work was supported by the Research Project of Science and Technology in Jiangxi Provincial Education Department (GJJ160136).
References
- 1.Chow WH, Devesa SS, Warren JL, Fraumeni JF., Jr Rising incidence of renal cell cancer in the United States. JAMA 1999; 281: 1628–31. [DOI] [PubMed] [Google Scholar]
- 2.Campbell SC, Flanigan RC, Clark JI. Nephrectomy in metastatic renal cell carcinoma. Curr Treat Options Oncol 2003; 4: 363–72. [DOI] [PubMed] [Google Scholar]
- 3.Doberstein K, Steinmeyer N, Hartmetz AK, Eberhardt W, Mittelbronn M, Harter PN, Juengel E, Blaheta R, Pfeilschifter J, Gutwein P. MicroRNA-145 targets the metalloprotease ADAM17 and is suppressed in renal cell carcinoma patients. Neoplasia 2013; 15: 218–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zellweger T, Miyake H, July LV, Akbari M, Kiyama S, Gleave ME. Chemosensitization of human renal cell cancer using antisense oligonucleotides targeting the antiapoptotic gene clusterin. Neoplasia 2001; 3: 360–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon RI. Commentary: medical errors, sentinel events, and malpractice. J Am Acad Psych Law 2006; 34: 99–100. [PubMed] [Google Scholar]
- 6.Devarajan E, Sahin AA, Chen JS, Krishnamurthy RR, Aggarwal N, Brun AM, Sapino A, Zhang F, Sharma D, Yang XH. Down-regulation of caspase 3 in breast cancer: a possible mechanism for chemoresistance. Oncogene 2002; 21: 8843–51. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Kolar C, Lawson TA, Gmeiner WH. Targeted drug delivery to chemoresistant cells: folic acid derivatization of FdUMP[10] enhances cytotoxicity toward 5-FU-resistant human colorectal tumor cells. J Org Chem 2001; 66: 5655–63. [DOI] [PubMed] [Google Scholar]
- 8.Wang CY, Cusack JC, Jr, Liu R, Baldwin AS., Jr Control of inducible chemoresistance: enhanced anti-tumor therapy through increased apoptosis by inhibition of NF-kappaB. Nat Med 1999; 5: 412–7. [DOI] [PubMed] [Google Scholar]
- 9.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov 2006; 5: 219–34. [DOI] [PubMed] [Google Scholar]
- 10.Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev 2007; 33: 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu E, Copur SM, Ju J, Chen TM, Khleif S, Voeller DM, Mizunuma N, Patel M, Maley G, Maley F, Allegra C. Thymidylate synthase protein and p53 mRNA form an in vivo ribonucleoprotein complex. Mol Cell Biol 1999; 19: 1582–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu L, Minden MD, Benchimol S. Translational regulation of human p53 gene expression. EMBO J 1996; 15: 4392–401. [PMC free article] [PubMed] [Google Scholar]
- 13.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005; 65: 6029–33. [DOI] [PubMed] [Google Scholar]
- 14.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65: 7065–70. [DOI] [PubMed] [Google Scholar]
- 15.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 2006; 94: 776–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem 2013; 372: 35–45. [DOI] [PubMed] [Google Scholar]
- 17.Wang F, Li T, Zhang B, Li H, Wu Q, Yang L, Nie Y, Wu K, Shi Y, Fan D. MicroRNA-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochem Biophys Res Commun 2013; 434: 688–94. [DOI] [PubMed] [Google Scholar]
- 18.Pu Y, Yi Q, Zhao F, Wang H, Cai W, Cai S. MiR-20a-5p represses multi-drug resistance in osteosarcoma by targeting the KIF26B gene. Cancer Cell Int 2016; 16: 64–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakada C, Matsuura K, Tsukamoto Y, Tanigawa M, Yoshimoto T, Narimatsu T, Nguyen LT, Hijiya N, Uchida T, Sato F. Genome-wide microRNA expression profiling in renal cell carcinoma: significant down-regulation of miR-141 and miR-200c. J Pathol 2008; 216: 418–27. [DOI] [PubMed] [Google Scholar]
- 20.Ishihara T, Seki N, Inoguchi S, Yoshino H, Tatarano S, Yamada Y, Itesako T, Goto Y, Nishikawa R, Nakagawa M. Expression of the tumor suppressive miRNA-23b/27b cluster is a good prognostic marker in clear cell renal cell carcinoma. J Urol 2014; 192: 1822–30. [DOI] [PubMed] [Google Scholar]
- 21.Yamada Y, Hidaka H, Seki N, Yoshino H, Yamasaki T, Itesako T, Nakagawa M, Enokida H. Tumor-suppressive microRNA-135a inhibits cancer cell proliferation by targeting the c-MYC oncogene in renal cell carcinoma. Cancer Sci 2013; 104: 304–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teixeira AL, Ferreira M, Silva J, Gomes M, Dias F, Santos JI, Maurício J, Lobo F, Rui M. Higher circulating expression levels of miR-221 associated with poor overall survival in renal cell carcinoma patients. Tumour Biol 2014; 35: 4057–66. [DOI] [PubMed] [Google Scholar]
- 23.Yi Z, Fu Y, Zhao S, Zhang X, Ma C. Differential expression of miRNA patterns in renal cell carcinoma and nontumorous tissues. J Cancer Res Clin Oncol 2010; 136: 855–62. [DOI] [PubMed] [Google Scholar]
- 24.Li HP, Zeng XC, Zhang B, Long JT, Zhou B, Tan GS, Zeng WX, Chen W, Yang JY. miR-451 inhibits cell proliferation in human hepatocellular carcinoma through direct suppression of IKK-beta. Carcinogenesis 2013; 34: 2443–51. [DOI] [PubMed] [Google Scholar]
- 25.Gits CM, van Kuijk PF, Jonkers MB, Boersma AW, Smid M, van Ijcken WF, Coindre JM, Chibon F, Verhoef C, Mathijssen RHJ. MicroRNA expression profiles distinguish liposarcoma subtypes and implicate miR-145 and miR-451 as tumor suppressors. Int J Cancer 2014; 135: 348–61. [DOI] [PubMed] [Google Scholar]
- 26.Su Z, Zhao J, Rong Z, Geng W, Wang Z. MiR-451, a potential prognostic biomarker and tumor suppressor for gastric cancer. Int J Clin Exp Pathol 2015; 8: 9154–60. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu S, Huang Y, Su X. Mir-451 correlates with prognosis of renal cell carcinoma patients and inhibits cellular proliferation of renal cell carcinoma. Med Sci Monit 2016; 22: 183–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu X, Li JY, Guo J, Li PS, Zhang WH. Influence of MiR-451 on drug resistances of paclitaxel-resistant breast cancer cell line. Med Sci Monit 2015; 21: 3291–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen D, Huang J, Zhang K, Pan B, Chen J, De W, Wang R, Chen L. MicroRNA-451 induces epithelial-mesenchymal transition in docetaxel-resistant lung adenocarcinoma cells by targeting proto-oncogene c-Myc. Eur J Cancer 2014; 50: 3050–67. [DOI] [PubMed] [Google Scholar]
- 30.Bian HB, Pan X, Yang JS, Wang ZX, De W. Upregulation of microRNA-451 increases cisplatin sensitivity of non-small cell lung cancer cell line (A549). J Exp Clin Cancer Res 2011; 30: 20–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, Ma X, Han S, Zhang Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci 2012; 57: 897–904. [DOI] [PubMed] [Google Scholar]
- 32.Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, Lakomy R, Svoboda M, Vyzula R, Slaby O. Circulating miR-378 and miR-451 in serum are potential biomarkers for renal cell carcinoma. J Transl Med 2012; 10: 55–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu X, Xue JQ, Han SJ, Qian SY, Zhang WH. Circulating microRNA-451 as a predictor of resistance to neoadjuvant chemotherapy in breast cancer. Cancer Biomarkers 2016; 16: 395–403. [DOI] [PubMed] [Google Scholar]
- 34.Bhoumik A, Ronai Z. ATF2: a transcription factor that elicits oncogenic or tumor suppressor activities. Cell Cycle 2008; 7: 2341–5. [DOI] [PubMed] [Google Scholar]
- 35.Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell 2004; 119: 431–43. [DOI] [PubMed] [Google Scholar]
- 36.Passegue E, Jochum W, Schorpp-Kistner M, Mohle-Steinlein U, Wagner EF. Chronic myeloid leukemia with increased granulocyte progenitors in mice lacking junB expression in the myeloid lineage. Cell 2001; 104: 21–32.. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Bergami P, Lau E, Ronai Z. Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat Rev Cancer 2010; 10: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lo Iacono M, Monica V, Vavala T, Gisabella M, Saviozzi S, Bracco E, Novello S, Papotti M, Scagliotti GV. ATF2 contributes to cisplatin resistance in non-small cell lung cancer and celastrol induces cisplatin resensitization through inhibition of JNK/ATF2 pathway. Int J Cancer 2015; 136: 2598–609. [DOI] [PubMed] [Google Scholar]
- 39.Lau E, Sedy J, Sander C, Shaw MA, Feng Y, Scortegagna M, Claps G, Robinson S, Cheng P, Srivas R. Transcriptional repression of IFNbeta1 by ATF2 confers melanoma resistance to therapy. Oncogene 2015; 34: 5739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]