Abstract
Candida auris is an emerging fungus that presents a serious threat to global health. The organism is difficult to identify using conventional biochemical methods. C. auris has also attracted attention because of its reduced susceptibility to azoles, polyenes, and echinocandins, with a few strains even resistant to all three classes of antifungals. In this review paper we discuss the trends in emergence of C. auris in different parts of the world, associated risk factors, drug resistance, and diagnostic challenges. Strategies for prevention and therapeutic options for such infections is also addressed.
Keywords: Candidemia, Candida haemulonii, outbreak, drug resistance
Introduction
Fungi have recently emerged as a major cause of human diseases, especially among patients who are hospitalized for a long duration or are immunocompromised.1 Candida spp. belong to the normal microbiota of an individual’s mucosal oral cavity, gastrointestinal tract, and vagina.2 They are responsible for various clinical manifestations from mucocutaneous overgrowth to bloodstream infections.3 The pathogenicity of Candida spp. has been attributed to virulence factors such as ability to evade host defenses, biofilm formation, adherence, and the production of tissue-damaging hydrolytic enzymes such as proteases, phospholipases, and hemolysin.4 Non-albicans Candida spp. with potential to develop antifungal resistance have been reported from various institutions, cities, countries, and geographic regions5,6 (Table 1).
Table 1.
MICs of commonly isolated drug-resistant Candida spp.
| S No | Species | Antifungal agents | MIC ranges (μg/mL) |
|---|---|---|---|
| 1 | C. parapsilosis | FLU | 0.25–837 |
| VRC | 0.03–0.12537 | ||
| CAS | 0.06–137 | ||
| AMB | 0.06–0.538 | ||
| 2 | C. glabrata | FLU | 1- >6437 |
| VRC | 0.03–237 | ||
| CAS | 0.003–0.12537 | ||
| AMB | 0.06–1.038 | ||
| 3 | C. krusei | FLU | 16–1637 |
| VRC | 0.125–0.2537 | ||
| CAS | 0.06–0.2537 | ||
| AMB | 0.25–438 | ||
| 4 | C. guilliermondii | FLU | 1–837 |
| VRC | 0.03–0.12537 | ||
| CAS | 0.125–137 | ||
| AMB | 0.5–238 | ||
| 5 | C. dubliniensis | FLU | 0.12–0.2538 |
| VRC | 0.03–438 | ||
| CAS | 0.25–238 | ||
| AMB | 0.06–138 | ||
| 6 | C. haemulonii | FLU | >6439 |
| VRC | >839 | ||
| CAS | 0.25–>1639 | ||
| AMB | 0.25–239 | ||
| 7 | C. duobushaemulonii | FLU | 8–>6439 |
| VRC | 0.12–>839 | ||
| CAS | 0.5–>1639 |
Abbreviations: MIC, minimum inhibitory concentration; AMB, amphoterecin B; FLU, fluconazole; CAS, capsofungin; VRC, voriconazole.
Candida auris (C. auris) has been recognized as an emerging multidrug-resistant (MDR) yeast worldwide. In June 2016, government agencies in the USA and the UK issued alerts requesting clinicians, laboratory technicians, infection control practitioners, and public health authorities to report isolation of C. auris in their patients.7–9 In 2016, an interim guideline for the management of C. auris in health care facilities in South Africa was issued by the National Institute of Communicable diseases, requesting notification of new outbreaks.10
In this review, we discuss the trends in emergence of C. auris in different parts of the world, associated risk factors, drug resistance, and diagnostic challenges. We also address the strategies for prevention and therapeutic options for such infections.
Study selection
We systematically reviewed the published work on C. auris isolates in various specimens and their susceptibility pattern. We searched PubMed, NCBI, Researchgate, Sciencedirect, and World Health Organization, Centers for Disease Control and Prevention databases up to January 2017 and also included bibliographies of relevant studies.
Data extraction and synthesis
A total of 39 studies were referred to for this review. We extracted C. auris antifungal susceptibility data as reported in various studies or presented in tables or relevant graphs according to the criteria and methods used in each study. For studies in which more than one method was used for identification and antifungal susceptibility testing (AFST), we extracted the relevant data preferentially obtained by use of sequencing, Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF), VITEK 2, or chromagar for identification and, Clinical Laboratory Standard Institute – Broth Microdilution method (CLSI-BMD), Etest, VITEK 2 for susceptibility testing.
Epidemiology
C. auris was first reported in 2009 as an infectious agent from a patient’s ear (auris means “ear” in Latin) in Japan.9,11 Subsequently, it was recovered from 15 ear samples from 5 Korean hospitals and identified as a causative agent of otitis media.9 These yeast isolates had phenotypic similarity to C. haemulonii and were less susceptible to amphotericin B (AMB) and fluconazole (FLU) than most other Candida spp.9,12 However, a retrospective review of Candida strain collections in South Korea revealed that the earliest known strain of C. auris dated back to 1996 and was isolated from the blood of a pediatric patient.7,9
The potential of C. auris to cause invasive infection was recognized after it was isolated from the blood of 3 South Korean patients with septicemia in 2011.12 Around the same time, 15 isolates of C. auris were recovered from 15 patients in a tertiary care hospital in Northern India.13 These isolates were initially identified as C. haemulonii, but were later confirmed as C. auris upon sequencing. Further reports have been published regularly from health care centers from North India and some centers in Southern India.14–17
The first outbreak of C. auris in the region of the Americas was reported in Venezuela from March 2012 to July 2013. The outbreak occurred in the intensive care unit of a tertiary care hospital in Maracaibo. Eighteen patients were involved, and all the isolates were initially identified as C. haemulonii but later confirmed as C. auris following sequencing. The isolates were resistant to fluconazole and voriconazole (VRC), and half of the isolates showed elevated minimum inhibitory concentration (MIC) to AMB.18 In USA, a C. auris isolate was detected during an ongoing surveillance program in 2013.7,18
In Colombia, isolated cases of infection by C. auris have been reported from various cities since 2013. Between 2015 and 2016, the city of Barranquilla reported 27 isolates of C. auris. In August 2016, an outbreak was reported in a pediatric intensive care unit in Cartagena. Five cases of disseminated infection due to C. auris were identified. These isolates were initially identified as C. albicans, C. guillermondii, and Rhodotorula rubra (R. rubra), but later confirmed as C. auris.18,19
Sporadic cases of C. auris have been identified throughout England since August 2013. The largest outbreak of C. auris in Europe occurred in a cardiothoracic center in London between April 2015 and July 2016.20 Infections due to C. auris have also been reported from countries like South Africa, Kuwait, Pakistan, Kenya, and Israel.7,21,22 Hospital outbreaks have been confirmed in five countries from which it has been reported so far. The CDC speculates that C. auris may be prevalent in many other countries, but that these are under reported due to lack of specialized laboratory methods needed for correct identification and drug sensitivity.
Candidemia surveillance
The ongoing international surveillance program SENTRY, which has around 15,271 Candida isolates were looked into to identify C. auris isolates that may have been overlooked or misidentified prior to 2009. These isolates were collected from 2004 to 2015. Only four isolates were identified as C. auris (from 2009, 2013, 2014, and 2015), one of which had been previously misidentified as C. haemulonii. This data rules out the possibility of C. auris emerging prior to 2009.23,24 Interestingly, in a multicenter study involving three continents, an isolate from Pakistan collected in 2008 (unpublished) was identified as C. auris.23
Epidemiological relatedness
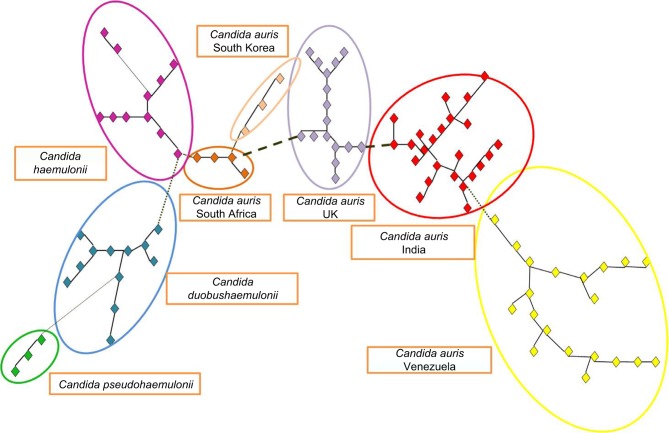
Molecular analyses of the strains collected from different geographic regions have revealed that C. auris have emerged independently in multiple regions of the world at roughly the same time. Strains from India, Japan, South Africa, South Korea, and Venezuela were compared with strains from the European outbreak. Isolates from closely related sibling species C. haemulonii, C. duobushaemulonii, and C. pseudohemulonii were also included as an outgroup. Cluster analysis showed that all species formed distinct clusters based on amplified fragment length polymorphism. C. auris isolates that came from the same geographic region were clustered together20 (Figure 1).
Figure 1.
AFLP-derived minimum spanning tree of Candida auris isolates.
Notes: AFLP-derived minimum spanning tree of C. auris isolates from the UK compared to those from India, Japan, South Africa, South Korea, and Venezuela isolate. C. hemulonii, C. duobushaemulonii, and C. pseudohaemulonii were included to serve as an outgroup. The branch lengths indicate the similarity between isolates with thick solid lines, thin solid line, thick dashed lines, thin dashed lines, and thin dotted lines. C. auris that came from the same geographic region are clustered together. The figure was based on data from Schelenz et al.20
Abbreviation: AFLP, amplified fragment length polymorphism.
M 13 polymerase chain reaction fingerprinting analysis done on strains from India showed distinct banding pattern in relation to isolates from Japan and South Korea strains. In a phylogenetic tree based on ITS sequences, South African isolates formed a cluster with Indian and Kuwait isolates. Moreover, C. auris isolates from India and South Africa assimilated N-acetylglucosamine in contrast to the isolates from Japan and South Korea.10,14
Molecular typing of many of the international strains (Eastern Asia, Southern Asia, Southern Africa, and South America) performed by the CDC suggests that isolates are highly related within countries and regions but distinct between continents.7
In a multicenter study involving three continents during 2012–2015, 47 isolates of C. auris were subjected to whole-genome sequencing analysis. Phylogenetic analysis identified a strong phylogeographic structure comprising 4 distinct C. auris clades. These clades were comprised exclusively of isolates from Pakistan, India, South Africa, Venezuela, or Japan. The clades were separated by tens of thousands of single-nucleotide polymorphisms and represented distinct geographic regions. Within each clade, isolates were clonal, and fewer single-nucleotide polymorphisms were identified within each cluster. This suggests simultaneous emergence of C. auris in more than 4 locations independently, and almost simultaneously, rather than recent spread worldwide of a dominant clone. However, these data also indicate that clonal isolates are distributed over large distances within countries and continents.23,24
Whole-genome sequencing has identified 3 different amino acid substitutions in the ERG11 gene of C. auris. These substitutions were strongly associated with geographic clades: F126T with South Africa, Y132F with Venezuela, and Y132F or K143F with India and Pakistan. Each mutation associated with isolates from a different continent, implying that resistance to fluconazole might be acquired rather than intrinsic.12,23,25
Who is at risk?
Available data suggest that the risk factors for C. auris infections are no different from risk factors associated with infections due to other species of Candida. These include diabetes mellitus, presence of central venous catheter, immunosuppressive state, neutropenia, exposure to broad-spectrum antibiotics, parenteral nutrition, blood transfusion, hemodialysis, surgery within 30 days, intensive care, previous antifungal agents within 30 days, concomitant bacteremia, concomitant candidemia, indwelling urinary catheter, candiduria, chronic kidney disease, and chemotherapy. Infections have been reported in patients of all ages, from preterm infants to the elderly. Further study may be needed to learn more about risk factors associated with C. auris infection.13,26–29
Clinical conditions and mortality
C. auris has been reported to be isolated from clinical conditions including bloodstream infections (fungemia), urinary tract infection, otitis, surgical wound infections, skin abscesses related to insertion of the catheter, infection of the heart muscle, meningitis, bone infections, and wound infections (colonization and infection in burns). C. auris has also been reported to be isolated from urine and the respiratory tract samples, but it is difficult to differentiate whether these are due to true infections or just colonization.10,13,26–30
Invasive infections with any Candida spp. can be fatal. There is no data to prove whether patients with invasive C. auris infection are more likely to die than patients with other Candida spp. infections. There are reports that have documented mortality up to 72% in patients with C. auris infections.7,23 However, many of these patients had other serious underlying medical conditions that may have contributed to the high mortality.
Concerns
The emergence of C. auris raises several serious concerns for public health.7,31
The isolates are often MDR, with some strains having elevated MICs to drugs in all the 3 major classes of antifungal medications
The isolates are difficult to identify with standard laboratory methods. Identification requires specialized methods such as Matrix-Assisted Laser Desorption Ionization Time of Flight or molecular identification based on sequencing the D1–D2 region of the 28s ribosomal DNA. Misidentification may lead to inappropriate treatment. Many of these isolates have been misidentified as C. haemulonii, Rhodotorula glutinis, or Saccharomyces cerevisiae
C. auris has the propensity to cause outbreaks in the health care settings, as has already been reported from several countries worldwide.
C. auris virulence factors
C. auris shares numerous virulence attributes with C. albicans such as nutrient acquisition, histidine kinase-2 component system, iron acquisition, tissue invasion, enzyme secretion, multidrug efflux, and genes/pathways involved in cell wall modeling and nutrition acquisition. C. auris is closely related phylogenetically to C. krusei C. haemulonii, and C. lusitaniae, which are known to have intrinsic and inducible resistance to fluconazole, AMB, or both.23 Survival of C. auris in the hospital environments may be promoted by its capacity for salt tolerance and cell aggregation into large- and difficult-to-disperse aggregates. Moreover isolates exhibit thermotolerence up to 42°C and do not form biofilms on intravascular catheters, which is an added advantage for survival and pathogenesis. A draft genome of C. auris also revealed a large percentage of genes devoted to central metabolism, a property that is crucial for adaptation to highly divergent environments.12,14,24,32
Diagnosis of a C. auris infection
Like other Candida infections, C. auris infections are usually diagnosed by fungal culture of blood, body fluids, and pus from the affected site. However, C. auris is more difficult to identify from cultures compared with other Candida spp. There are many challenges in the identification of this isolate. Most laboratories worldwide use commercially available biochemical-based tests like analytical profile index strips and VITEK 2 for identification of yeasts. These cannot differentiate C. auris from related species as it has not been included in their database for identification. Moreover, diagnostic laboratories do not undertake molecular identification routinely, which has led to underestimation of the actual prevalence of this yeast.14,33
Phenotypically, C. auris is difficult to distinguish from other pathogenic species of Candida. On microscopy, the isolates are oval without pseudohyphae and are germ tube-negative. In contrast, C. haemulonii and C. duobushaemulonii isolates form pseudohyphae with blastoconidia.33 On culture, C. auris appears pale purple to pink on CHROM agar (Difco, Becton Dickinson, Baltimore, MD, USA) and grows at 37°C–42°C.32,33 This characteristic can help differentiate C. auris isolates from C. haemulonii, which does not grow at 42°C.33 C. auris appears as butyrous to viscous, white to gray, smooth, and glistening, with an entire margin after a month of incubation on malt extract agar at 25°C. Pseudohyphae are not produced on slide culture (cornmeal agar) at 25°C even after incubation for 59 days.11
C. auris ferments glucose, sucrose (weak), and trehalose (weak), but does not ferment galactose, maltose, lactose, or raffinose. Carbon assimilation is seen for glucose, sucrose, maltose, d-trehalose, d-raffinose, d-melezitose, inulin (weak), soluble starch, ribitol (weak), galactitol, d-mannitol, sorbitol, and citrate. Ammonium sulfate, cadaverine, and l-lysine are utilized as sole sources of nitrogen; sodium nitrite, potassium nitrate, and ethylamine are not utilized by C. auris. It can grow in vitamin-free media and 50% glucose and 10% NaCl/5% glucose media. Starch formation, urease activity, and diazonium blue B reaction are not shown by this species of Candida. It does not grow in the presence of 0.1% and 0.01% cycloheximide. The major ubiquinone is Q-9, and Mol% of G+C is 45.3%.11
The VITEK 2 YST (bioMérieux, Marcy-l’Étoile, France) currently misidentifies C. auris as C. haemulonii or C. famata, the API20C (bioMérieux) as R. glutinis or C. sake, and the Auxacolor (Bio-Rad, Hercules, CA, USA) misidentifies it as S. cerevisiae.14 Because of these challenges, clinical laboratories have misidentified the organism as C. haemulonii, R. glutinis, and S. cerevisiae. Some clinical laboratories do not fully identify all Candida to the species level, and C. auris isolates have been reported as “other Candida species”.
Identification of C. auris should currently be confirmed using accepted methods such as MALDI-TOF or molecular identification techniques like polymerase chain reaction, sequencing, and amplified fragment length polymorphism fingerprinting.20,33
However, the disadvantage of MALDI-TOF is that the biotyper library has 3 isolates from Japan and South Korea in its database. In the absence of sequences in the database (eg, US Food and Drug Administration database), isolates will be identified as yeast without score.24
Drug resistance
For guiding therapy and determining the prognosis of the patient, both species-level identification and AFST of the isolates should be done using reference methods. Since the routine laboratories mostly rely on commercial systems for identification and antifungal susceptibility testing for yeasts, a cautionary approach is recommended for isolates showing elevated MICs to antifungals with these systems.
C. auris is usually resistant to fluconazole, but recent reports have also documented high MICs to AMB, voricon-azole, and caspofungin (CAS). Some strains of C. auris have been reported to be resistant to all 3 major classes of anti-fungal drugs12,15,32 (Table 2). This type of MDR has not been seen before in other species of Candida, thus limiting the treatment options.30
Table 2.
Antifungal susceptibility pattern of Candida auris isolates published till December 2016
| Reference | No of isolates tested | Method of susceptibility | MIC Range (μg/mL)
|
||||
|---|---|---|---|---|---|---|---|
| FLU | VRC | AMB | CAS | 5-FC | |||
| Satoh et al11 (2009) | 1 | Not mentioned | 2 | 0.03 | – | – | 0.5 |
| Kim et al9 (2009) | 15 | Etest method | 2–128 | 0.03–2 | 0.38–1.5 | 0.125–0.25 | – |
| Lee et al27 (2011) | 6 | CLSI (2008) | 2–128 | 0.03–1 | 0.5–1 | 0.06 | – |
| Sarma et al13 (2012) | 15 | Vitek 2 compact YST (MIC50/90) | 64/64 | 1/2 | 8/16 | – | 1/1 |
| Chowdhary et al14 (2013) | 12 | CLSI (2008) | 16–64 | 0.125–0.25 | 0.25–1 | 0.125–0.5 | 0.06–0.125 |
| Chowdhary t al14 (2013) | 15 | CLSI (2008) | 64 | 0.5–4 | 0.25–1 | 0.25–1 | 0.25–64 |
| Khillan et al15 (2014) | 4 | CLSI (2008) | >64 | 0.06–0.125 | 0.125–0.5 | 1 | 0.125–4 |
| Shallu Kathuria et al33 (2016) | 90 | CLSI (2008) | 4–>64 | <0.03–16 | 0.125–8 | 0.125–8 | <0.125–>64 |
| Schelenz et al20 (2016) | 50 | Sensititre YeastOne | >256 | – | 0.5–2 | 0.06–0.25 | 0.06–0.12 |
| Sharma et al34 (2016) | 5 | CLSI (2008) | ≥64 | 0.125–16 | 0.25–4 | 0.25–8 | 0.125–64 |
The commonly used methods worldwide for testing antifungal sensitivity are the CLSI-BMD, the VITEK 2 antifungal susceptibility test, and the Etest method. There are no established MIC breakpoints at present for C. auris drug susceptibility interpretation. The CDC has applied conservative breakpoints developed for other Candida spp. to C. auris for epidemiological purposes. The breakpoint for fluconazole was arbitrarily set at ≥32 μg/mL, while it was ≥2 μg/mL for voriconazole, ≥ 8 μg/mL for the echinocandins, ≥128 μg/mL for flucytocine, and ≥2 μg/mL for AMB.23
Using these breakpoints, CDC has demonstrated that, of the global outbreaks that they have been investigating, nearly all isolates are highly resistant to fluconazole. More than half of C. auris isolates were resistant to voriconazole, one-third were resistant to AMB (MIC ≥2 mg/L), and a few were resistant to echinocandins. However, these breakpoints may not necessarily be clinically relevant at an individual patient level.33
In a study from India, MICs using all the 3 abovementioned methods were compared to identify the most consistent method that can be used (Table 3). Uniformly elevated MICs for fluconazole were seen by all the methods, whereas elevated caspofungin MIC was observed in 37% of the isolates by the CLSI-BMD method only. Elevated MICs for AMB, fluconazole, caspofungin, and voriconazole were observed in 10% of the isolates only by the CLSI-BMD method. 34% of the isolates had coexisting elevated MICs for fluconazole and voriconazole (MICs of >2 ug/mL), and 10% of the isolates had elevated coexisting MICs (≥1 μg/mL) to posaconazole and isavuconazole. A major concern of the authors in this study was the high MICs of amphotericin B with VITEK 2 automated readings, which was in contrast to the observations of low MIC by the CLSI-BMD method. This observation was statistically significant, but the essential agreement between the 2 methods was low (10%). Interestingly, 37% of the isolates showing elevated MICs for caspofungin by CLSI-BMD method reduced to 12% using the Etest. The performance of the caspofungin Etest based on the revised CLSI breakpoints for Candida isolates showed that 13.1% were either misclassified as intermediate or resistant.32 Moreover, there have been several reports of marked interlaboratory variation with both CLSI-BMD and the European Committee on Antimicrobial Susceptibility Testing method for capsofungin susceptibility.32,33
Table 3.
Distribution of MICs of amphotericin B, caspofungin, and voriconazole obtained by 3 different methods for Candida auris (n=90)
| Data tested | Test methods | No. of isolates at MIC (μg/mL)
|
MIC (μg/mL)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <0.03 | 0.03 | 0.06 | 0.13 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | >16 | MIC50 | MIC90 | ||
| AMB | CLSI-BMD | 2 | 16 | 23 | 35 | 4 | 6 | 4 | 1 | 4 | |||||
| Vitek 2 | 1 | 48 | 41 | 8 | 16 | ||||||||||
| Etest | 5 | 1 | 4 | 25 | 54 | 1 | 0.5 | 1 | |||||||
| CAS | CLSI-BMD | 1 | 29 | 27 | 25 | 1 | 4 | 3 | 0.5 | 1 | |||||
| Vitek 2 | 21 | 34 | 28 | 7 | 0.5 | 4 | |||||||||
| Etest | 9 | 1 | 9 | 22 | 33 | 5 | 4 | 7 | 0.25 | 2 | |||||
| VRC | CLSI-BMD | 1 | 4 | 7 | 8 | 18 | 17 | 18 | 6 | 3 | 3 | 5 | 1 | 8 | |
| Vitek 2 | 3 | 5 | 12 | 28 | 16 | 14 | 10 | 2 | 1 | 4 | |||||
| Etest | 1 | 3 | 2 | 8 | 15 | 36 | 12 | 3 | 7 | 3 | 1 | 16 | |||
Note: Reproduced from Genome Announc. 2015;3(4):e00722–15. Doi: 10.1128/JCM.00367-15. Amended with permission from American Society for Microbiology.32
Abbreviations: AMB, amphotericin B; CAS, caspofungin; CLSI-BMD, Clinical Laboratory Standard Institute–Broth Microdilution Method; MIC, minimum inhibitory concentration; VRC, voriconazole.
Similar reports of low MICs (0.25–1 μg/mL) for amphotericin B by the CLSI-BMD method was reported from South Korea, but with an essential agreement of 100% between the 2 methods. Authors from Columbia not only observed elevated MICs for amphotericin B by VITEK card but also observed discrepancies with E-strip. Therefore they suggested the use of more than one method for drug sensitivity testing to prevent inappropriate use of antifungal therapy.34
The isolates from the London outbreak showed high level resistance to fluconazole (MIC ≥256 mg/L), but the majority of the isolates were susceptible to echinocandins (MIC 0.06–0.25 mg/L), 5-flucytosine (MIC <0.06–0.12 mg/L), and had variable susceptibility to amphotericin B (0.5–2 mg/L).20 Till June 2016, no MDR strains have been reported from the UK. All the isolates were resistant to fluconazole, and some of them showed cross-resistance to other azoles.35
It is noteworthy that C. haemulonii and C. auris differ in susceptibility to amphotericin B, with C. auris mostly susceptible and C. haemulonii being remarkably resistant to amphotericin B and azoles.9,16 The recommended methods for drug susceptibility testing as suggested by several authors are the CLSI-BMD and the Etest method. Studies on complete genomic analysis are warranted to detect true antifungal resistance to this pathogen.
Therapeutic options
First-line therapy remains an echinocandin, pending specific susceptibility testing, which should be undertaken as soon as possible.36 However, the drug of choice will depend on the drug susceptibility report of the isolate. This requires appropriate identification of the infecting strain and laboratory determination of the strain’s drug susceptibility. Some clinicians prefer to use >1 antifungal drug to treat these MDR invasive organisms. There is currently no evidence or experience to support combination therapy in invasive infections with this organism, and clinicians are advised to make decisions on a case-by-case basis.
Duration of antifungal treatment is similar to that used for infections caused by other Candida spp. Treatment for candidemia should be continued for 14 days after documented clearance of Candida from the bloodstream and resolution of symptoms attributable for candidemia. The time to clearance of infection after starting treatment is not clear; however, persistent fungemia for up to 3 weeks has been observed by some authors.14,23 Wherever feasible, an effort should be made to remove devices such as central venous catheters and urinary catheters.
If an isolate is found to be resistant to azoles, echinocandins, or amphotericin B, the laboratory should also test for susceptibility to flucytosine, nystatin, and terbinafine. Currently, UK strains remain susceptible to topical agents nystatin and terbinafine. For the treatment of any further MDR strains, a regimen incorporating oral terbinafine could be considered.8
Risk for acquiring infections on travel to endemic countries
It is unlikely that routine travel to countries with documented C. auris infections would increase the chance of someone getting sick from C. auris. Infections have occurred primarily in patients who were already in the hospital for other reasons. People who travel to endemic countries to seek medical care or who are hospitalized there for a long time may have an increased risk for C. auris infection.7
Infection prevention and control (IPC)
Candida was previously thought to be a colonizer within the gastrointestinal tract, and later acquired into the hospital environment. Early evidence suggests that the organism might spread through contact with contaminated environmental surfaces or equipments, or from person to person. The majority of infected patients have had a recent exposure to an indwelling device or have undergone some invasive procedures.23 Infections have been observed several days to weeks (median time 19 days) after hospitalization in susceptible patients, suggesting an exogenous source associated with breach in infection control practices.30 Reports from India, Pakistan, and Venezuela have described health care outbreaks of C. auris infection and colonization.7,8,13,14 The precise mode of transmission of the organism within the health care environment is not known. Apart from direct transmission from fomites (such as blood pressure cuffs, stethoscopes, and other equipment in contact with the patient), infections can be transmitted indirectly via the contaminated hands of health care workers. The evidence for hospital outbreaks and clonal spread suggests that C. auris infections may differ from invasive candidiasis due to other Candida spp. Infection due to C. auris is usually sporadic and is caused by genetically distinct, endogenous isolates that are normal colonizers in the patient’s skin and mucosal surfaces.8,24
Route cause analysis following the London outbreak revealed that the minimum contact period for transmission of infection with a positive case or a contaminated environment was ≥4 h. No single point source of transmission could be identified. The persistence and propagation of the fungus in spite of all the infection prevention measures indicate an innate resilience of C. auris for survival and persistence in the environment, high transmissibility, and the ability to rapidly colonzie the patient’s skin and environment.20
The CDC is conducting studies to understand the contagiousness of this organism as it has been found on the skin of several patients and on other surfaces in the patient’s rooms. Where possible, equipment used for the infected/colonized patient should not be shared with other patients on the ward unless between-patient cleaning can be assured. It is essential that all health care providers work in a multidisciplinary team with their clinical microbiologists and under the direction of their specialist IPC team.
Guidance for C. auris reporting, detection, infection control, and environmental cleaning is available through various international health care regulatory bodies, some of which are discussed below.7,10,31,35
Role of screening
Although there is limited evidence supporting routine screening for C. auris at the time of hospital admission, screening policies can be designed based on local risk assessment and prevalence of infections. Screening may not be feasible in low-to-middle income developing countries, but it has been recommended in units having patients with ongoing infections or patients coming from other affected hospitals/units or in countries where cases have not been detected yet.
C. auris screening could also be considered for patients at risk for candidiasis, mainly patients in the intensive care units, pediatric patients, HIV/AIDS patients, and patients with malignancies, including those undergoing hematopoietic stem cell transplantation in endemic countries.35
Suggested screening sites based on the predilection of Candida spp. to colonzie the skin and mucosal surfaces are:
Nose, throat, and groin
Urine/urethral swab
Perineal or low vaginal swab (if appropriate)
Sputum/endotracheal secretions
Drain fluid (abdominal/pelvic/mediastinal)
Cannula entry sites (if clinically indicated)
Wounds
Routine wound swabs may be used to collect the screening sample. All screen-positive patients should be isolated or cohorted. A series of three negative screens taken 24 hours apart is suggested before deisolating the patient.20 As there is clinical experience of recurrence of colonization, the need for ongoing vigilance in the form of weekly screens in certain clinical environments should be considered following local risk assessments.
Decolonization
Colonization of inpatients has been reported from affected hospitals around the world. Clinical studies till date have shown that colonization is difficult to eradicate and that it tends to persist making infection prevention and control strategies even more important. Colonized patients could present opportunities for contamination of the health care environment both during admission as well as postdischarge follow-ups.
For decolonization of skin, patients can be prescribed twice-daily 2% chlorhexidine gluconate-containing wipes or aqueous 4% chlorhexidine formulations.20 Oral decolonization can be done by using 0.2% chlorhexidine mouthwash or 1% chlorhexidine dental gel in patients on ventilator support. Oral nystatin has been used in oropharyngeal colonization. Chlorhexidine-impregnated protective disks for central vascular catheter exit sites have been used to reduce line-associated C. auris bloodstream infections.10,20,24
In a recent report on C. auris outbreak, it was observed that in spite of daily chlorhexidine washes for decolonization, patients continued to be colonized. This could probably be due to reinfection from hospital environment like beddings and clothing. Reduced susceptibility to chlorhexidine could also have been a possibility, but there is not enough evidence that can establish whether C. auris is susceptible or resistant to chlorhexidine, and so more work needs to be done in this field.20,35
Patient care guidelines
Improved adherence to central line-associated bloodstream infection, catheter-associated urinary tract infection care bundles, as well as tracheostomy site care
Contact isolation of all patients colonized or infected with the organism, preferably in a single room
Strict adherence to standard precautions including hand hygiene using soap and water followed by alcohol hand rub
Personal protective equipment in the form of gloves and aprons (or gowns if there is a high risk of soiling with blood or body fluids). These should be donned after hand washing and before entering the room and should be removed and discarded in the room followed by a thorough handwash and application of alcohol hand rub
Visors and masks are not routinely required and should be worn only if there is a procedural risk of spillage or splashes
Visitors of infected or colonized patients need to be briefed about the infection and infection prevention and control precautions
Contact isolation of all patients transferred from other affected hospitals or a hospital abroad until screening results are available
In case the infected/colonized patients are transferred to other health care facilities, the receiving facilities should be notified of the presence of C. auris to ensure appropriate precautions are continued
If a patient needs to be taken out of the side room or bay to theater or for imaging, they should be scheduled last on the list for the day and the environment cleaned adequately
Clinicians and ancillary health professionals should be trained regarding IPC recommendations.
Environment and fomites
Despite a comprehensive review of modern technologies for environmental decontamination, there is currently no published data evaluating the effectiveness of cleaning agents or decontamination of the environment for C. auris specifically. Facilities should ensure daily and terminal cleaning of patient rooms with a US Environmental Protection Agency-registered disinfectant with a fungal claim. Chlorine-based agents, ultraviolet light, and hydrogen peroxide vaporization are reported to be effective for environmental cleaning.20,31 A high-strength chlorine-releasing agent is currently recommended for cleaning of the environment, and it should contain 1,000 ppm of available chlorine. However, medical centers should adopt a local cleaning policy and regimen depending on the level of contamination and case load. Domestic staff should be trained and supervised until declared competent.
Terminal clean
Once the patient has left the environment, a terminal cleaning should be undertaken either by using 1,000 ppm chlorine-based product or by using hydrogen peroxide vapors. All equipment should be cleaned in accordance with manufacturer’s instructions, and where relevant returned to the company for cleaning. Particular attention should be paid to cleaning of multiple-use equipment (eg, blood pressure cuffs, thermometers, computers on wheels, ultrasound machines) from the bed space of an infected/colonized patient.
Waste and linen disposal
Current waste and soiled linen policies as for any other MDR health care-associated organism should be followed
Appropriate bagging and isolation of soiled linen and waste should be done to prevent contamination of the environment
Specific attention should be paid in the pediatric and neonatal units during disposal of soiled nappies
Soiled material should never be discarded or washed in the clinical handwashing sink.
Conclusion
C. auris is an emerging fungus that presents a serious threat to global health. Till date, it has been reported from more than 12 countries, involving 4 continents. The organism is difficult to identify using conventional biochemical methods. Accurate identification of this species is important for estimating the actual prevalence of this underreported pathogen in different geographical areas. C. auris has also attracted attention because of its reduced susceptibility to azoles, polyenes, and echinocandins, with a few strains even resistant to all these three classes of antifungals. Increased availability of antifungal agents may have played an important role in the emergence of resistance. However, antifungal selection is unlikely to be a sole determinant as amphotericin B has been available since 1954, fluconazole since 1991, and echinocandins since the early 2000s, but access to these drugs occurred much later in resource-limited settings.23,32 Although accurate data on use of antifungals are difficult to obtain, anecdotal evidence suggests increased use in recent years of triazoles and other antifungals for empiric treatment of surgical and medical conditions. Moreover most patients infected with C. auris were already on some form of antifungals, supporting the antifungal selection pressure hypothesis.23 Interestingly, 2 of the South African echinocandin-resistant C. auris isolates came from patients who did not have any history of echinocandin exposure.23
To date, infections with C. auris have been largely acquired in hospital settings, and horizontal spread of the pathogen has been demonstrated through clonality of isolates within a hospital. Changes in the ecological niches of C. auris have probably brought this fungus into contact with susceptible humans. C. auris lineages with MDR may continue to emerge independently and spread clonally in countries currently affected as well as those not affected. Major global outbreaks can occur if the intrinsic ability of C. auris to adapt in highly divergent environment is aligned with epidemiological factors supporting dissemination.24 Therefore, implementation of stringent infection prevention and control measures for all positive C. auris cases along with regular audits for compliance should be undertaken.20,23
Finally, the existence of other drug-resistant Candida spp. like C. haemulonii, C. glabrata, C. krusei, C. lusitaniae, and C. duobushaemulonii should not be overlooked. We need to stay alert and be vigilant in monitoring the epidemiology of these isolates globally. Implementation of IPC protocols following isolation of these strains can also be considered to prevent dissemination of these pathogens thereby preventing outbreaks.
More research needs to be undertaken to understand the factors promoting environmental resilience, transmission of infection, development of resistance mechanisms to antifungal drugs/disinfectants, and risk factors contributing to host colonization and infection.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Arendrup MC, Fuursted K, Gahrn-Hansen B, et al. Seminational surveillance of fungemia in Denmark: notably high rates of fungemia and numbers of isolates with reduced azole susceptibility. J Clin Microbiol. 2005;43(9):4434–4440. doi: 10.1128/JCM.43.9.4434-4440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shao LC, Sheng CQ, Zhang WN. Recent advances in the study of antifungal lead compounds with new chemical scaffolds. Yao Xue Xue Bao. 2007;42:1129–1136. Chinese. [PubMed] [Google Scholar]
- 3.Eggimann P, Garbino J, Pittet D. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect Dis. 2003;3:685–702. doi: 10.1016/s1473-3099(03)00801-6. [DOI] [PubMed] [Google Scholar]
- 4.Silva S, Henriques M, Martins A, Oliveira R, Williams D, Azeredo J. Biofilms of non-Candida albicans Candida species: quantification, structure and matrix composition. Med Mycol. 2009;47(7):681–689. doi: 10.3109/13693780802549594. [DOI] [PubMed] [Google Scholar]
- 5.Pfaller MA, Andes DR, Diekema DJ, et al. Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: data from the Prospective Antifungal Therapy (PATH) Registry 2004–2008. PLoS One. 2014;9(7):e101510. doi: 10.1371/journal.pone.0101510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfaller MA, Messer SA, Woosley LN, Jones RN, Castanheira M. Echinocandin and Triazole antifungal susceptibility profiles for clinical opportunistic yeast and mold isolates collected from 2010 to 2011: application of new CLSI clinical breakpoints and epidemiological cutoff values for characterization of geographic and temporal trends of antifungal resistance. J Clin Microbiol. 2013;51(8):2571–2581. doi: 10.1128/JCM.00308-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention . Global emergence of invasive infections caused by the multidrug-resistant yeast Candida auris. CDC; 2016. [updated June 24, 2016]. Available from: http://www.cdc.gov/fungal/diseases/candidiasis/candida-auris-alert.html. [Google Scholar]
- 8.Public Health England . Candida auris identified in England. Public Health England; 2016. [updated July 1, 2016]. Available from: https://www.gov.uk/government/organisations/public-health-england. [Google Scholar]
- 9.Kim MN, Shin JH, Sung H, et al. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis. 2009;48:e57–e61. doi: 10.1086/597108. [DOI] [PubMed] [Google Scholar]
- 10.Interim guidance for management of Candida auris infections in South African hospitals. Centre for Opportunistic, Tropical and Hospital Infections; Compiled December 2016. Available from: http://www.nicd.ac.za/assets/files/2016-12-22%20InterimNICDRecommdtnsCAuris.pdf. [Google Scholar]
- 11.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 12.Oh BJ, Shin JH, Kim MN, et al. Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Med Mycol. 2011;49:98–102. doi: 10.3109/13693786.2010.493563. [DOI] [PubMed] [Google Scholar]
- 13.Sarma S, Kumar N, Sharma S, et al. Candidemia caused by amphotericin B and Fluconazole resistant Candida auris. Indian J Med Microbiol. 2013;31(1):90–101. doi: 10.4103/0255-0857.108746. [DOI] [PubMed] [Google Scholar]
- 14.Chowdhary A, Sharma C, Duggal S, et al. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis. 2013;19(10):1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khillan V, Rathore N, Kathuria S, Chowdhary A. A rare case of breakthrough fungal pericarditis due to fluconazole-resistant Candida auris in a patient with chronic liver disease. JMM Case Reports. 2014;1(3) [Google Scholar]
- 16.Chowdhary A, Kumar AV, Sharma C, et al. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis. 2014;33(6):919–926. doi: 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 17.Girard V, Mailler S, Chetry M, et al. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses. 2016;59(8):535–538. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 18.Pan American Health Organization/World Health Organization . Epidemiological Alert: Candida auris outbreaks in health care services. Washington, DC: PAHO/WHO; Oct 3, 2016. [Google Scholar]
- 19.Information provided by the Colombia International Health Regulations National Focal Point, August 26, 2016.
- 20.Schelenz S, Hagen F, Rhodes JL, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control. 2016;5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magobo RE, Corcoran C, Seetharam S, Govender NP. Candida auris-associated candidemia, South Africa. Emerg Infect Dis. 2014;20:1250–1251. doi: 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emara M, Ahmad S, Khan Z, et al. Candida auris candidemia in Kuwait, 2014. Emerg Infect Dis. 2015;21(6):1091–1092. doi: 10.3201/eid2106.150270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lockhart SR, Etienne KA, Vallbhaneni S, et al. Simultaneous emergence of multidrug-resistant Candida auris on three continents confirmed by whole genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clancy CJ, Nguyen MH. Emergence of Candida auris: an international call to arms. Clin Infect Dis. 2017;64(2):141–143. doi: 10.1093/cid/ciw696. [DOI] [PubMed] [Google Scholar]
- 25.Britz E, Govender NP. Global emergence of a multi-drug resistant fungal pathogen, Candida auris. S Afr J Infect Dis. 2016;31(3):69–70. [Google Scholar]
- 26.Sardi JCO, Pitangui NDS, Gullo FP, Almeida AMS, Mendes-Giannini MJS. A mini review of Candida species in hospital infection: epidemiology, virulence factor and drug resistance and prophylaxis. Trop Med Surg. 2013;1:141. [Google Scholar]
- 27.Lee WG, Shin JH, Uh Y, et al. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol. 2011;49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Ami R, Olshtain-Pops K, Krieger M, et al. Antibiotic exposure as a risk factor for fluconazole-resistant Candida bloodstream infection. Antimicrob Agents Chemother. 2012;56(5):2518–2523. doi: 10.1128/AAC.05947-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wey SB, Mori M, Pfaller MA, Woolson RF, Wenzel RP. Risk factors for hospital-acquired Candidemia, a matched case-control study. Arch Intern Med. 1989;149(10):2349–2353. [PubMed] [Google Scholar]
- 30.CIDRAP . CDC issues warning on multidrug-resistant yeast infection. CIDRAP; 2016. Available from: http://www.cidrap.umn.edu/news-perspective/2016/06/cdc-issues-warning-multidrug-resistant-yeast-infection. [Google Scholar]
- 31.Vallabhanemi S, Kallen A, Tsay S, et al. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus—United States, May 2013–August 2016. MMWR Morb Mortal Wkly Rep. 2016;65:1234–1237. doi: 10.15585/mmwr.mm6544e1. [DOI] [PubMed] [Google Scholar]
- 32.Sharma C, Kumar N, Meis JF, Pandey R, Chowdhary A. Draft genome sequence of a fluconazole-resistant Candida auris strain from a candidemia patient in India. Genome Announc. 2015;3(4):e00722–15. doi: 10.1128/genomeA.00722-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kathuria S, Singh PK, Sharma C, et al. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol. 2015;53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morales-López SE, Parra-Giraldo CM, Ceballos-Garzón A, et al. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg Infect Dis. 2017;23(1):162–164. doi: 10.3201/eid2301.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Public Health England . Guidance for the Laboratory Investigation, Management and Infection Prevention and Control for Cases of Candida auris. Public Health England; Jun, 2016. Available from: https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/532117/Guidance-candida-auris.pdf. [Google Scholar]
- 36.Pappas PG, Kauffman CA, Andes DR, et al. Clinical practice guideline for the management of candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin Infect Dis. 2016;62(4):e1–e50. doi: 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Won EJ, Shin JH, Choi MJ, et al. Antifungal susceptibilities of bloodstream isolates of Candida species from nine hospitals in Korea: application of new antifungal breakpoints and relationship to antifungal usage. PLoS One. 2015;10(2):e0118770. doi: 10.1371/journal.pone.0118770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimopoulos G, Velegraki A, Falagas ME. A 10-year survey of antifungal susceptibility of Candidemia isolates from intensive care unit patients in Greece. Antimicrob Agents Chemother. 2009;53(3):1242–1244. doi: 10.1128/AAC.01368-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, et al. Reclassification of the Candida haemulonii Complex as Candida haemulonii (C. haemulonii Group I), C. duobushaemulonii sp. nov. (C. haemulonii Group II), and C. haemulonii var. vulnera var. nov.: Three multiresistant human pathogenic yeasts. J Clin Microbiol. 2012;50(11):3641–3651. doi: 10.1128/JCM.02248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]