Abstract
Recent advances in MRI and increasing knowledge on the characterization and anatomical variability of medial temporal lobe (MTL) anatomy have paved the way for more specific subdivisions of the MTL in humans. In addition, recent studies suggest that early changes in many neurodegenerative and neuropsychiatric diseases are better detected in smaller subregions of the MTL rather than with whole structure analyses. Here, we developed a new protocol using 7 Tesla (T) MRI incorporating novel anatomical findings for the manual segmentation of entorhinal cortex (ErC), perirhinal cortex (PrC; divided into area 35 and 36), parahippocampal cortex (PhC), and hippocampus; which includes the subfields subiculum (Sub), CA1, CA2, as well as CA3 and dentate gyrus (DG) which are separated by the endfolial pathway covering most of the long axis of the hippocampus. We provide detailed instructions alongside slice-by-slice segmentations to ease learning for the untrained but also more experienced raters. Twenty-two subjects were scanned (19–32 yrs, mean age = 26 years, 12 females) with a turbo spin echo (TSE) T2-weighted MRI sequence with high-resolution oblique coronal slices oriented orthogonal to the long axis of the hippocampus (in-plane resolution 0.44 × 0.44 mm2) and 1.0 mm slice thickness. The scans were manually delineated by two experienced raters, to assess intra- and inter-rater reliability. The Dice Similarity Index (DSI) was above 0.78 for all regions and the Intraclass Correlation Coefficients (ICC) were between 0.76 to 0.99 both for intra- and inter-rater reliability. In conclusion, this study presents a fine-grained and comprehensive segmentation protocol for MTL structures at 7 T MRI that closely follows recent knowledge from anatomical studies. More specific subdivisions (e.g. area 35 and 36 in PrC, and the separation of DG and CA3) may pave the way for more precise delineations thereby enabling the detection of early volumetric changes in dementia and neuropsychiatric diseases.
Abbreviations: AG, Ambient Gyrus; CaS, Calcarine sulcus; CSF, Cerebrospinal Fluid; CS, Collateral Sulcus; CSa, anterior; CSp, posterior; CA1, Cornu Ammonis 1; CA2, Cornu Ammonis 2; CA3, Cornu Ammonis 3; DG, Dentate Gyrus; ErC, Entorhinal Cortex; FG, Fusiform Gyrus; HB, Hippocampal Body; HH, Hippocampal Head; HT, Hippocampal Tail; MTL, Medial Temporal Lobe; OTS, Occipito-temporal Sulcus; PhC, Parahippocampal Cortex; PhG, Parahippocampal Gyrus; PrC, Perirhinal Cortex; SaS, Semiannular Sulcus; SRLM, Stratum radiatum lacunosum-moleculare; Sub, Subiculum
Highlights
-
•
We present a new segmentation protocol for medial temporal lobe subregions at 7 T MRI.
-
•
Novel neuroanatomical knowledge from the work of Ding and Van Hoesen is incorporated.
-
•
Individual anatomical variability is covered by digitation- and depth-specific rules.
-
•
We provide detailed descriptions and slice-by-slice plots to guide the novice rater.
-
•
Reliability analyses provide compelling evidence that the rules can be replicated.
1. Introduction
The human hippocampus and the adjacent medial temporal lobe (MTL) regions have been implicated in a number of cognitive functions including episodic memory (Eichenbaum et al., 2007), spatial navigation (Ekstrom et al., 2003, Wolbers and Büchel, 2005, Epstein, 2008) and perception (Lee et al., 2005, Graham et al., 2010, Aly et al., 2013). At the same time, MTL regions are also affected by a number of pathological conditions such as depression (Huang et al., 2013), posttraumatic stress disorder (Wang et al., 2010), epilepsy (Bernasconi et al., 2003), and neurodegenerative diseases like Alzheimer's Disease (Dickerson et al., 2004, Rusinek et al., 2004, Du et al., 2007). Analyses of subregions in the MTL have thereby shown the potential to reveal subregion-specific aging and disease effects (e.g. de Flores et al., 2015) as well as distinct functional roles of specific subregions, for example, perirhinal cortex (PrC) in object processing (for a recent spotlight, see Inhoff and Ranganath, 2015), parahippocampal cortex (PhC) in contextual association (Aminoff et al., 2013), CA1 in memory integration (Schlichting et al., 2014), CA3 in memory recall (Chadwick et al., 2014), and dentate gyrus (DG) in pattern separation (Baker et al., 2016, Berron et al., 2016).
Recently, novel neuroanatomical findings have increased our knowledge of boundaries between MTL subregions and their variability within and across subjects (Ding and Van Hoesen, 2010, Ding and Van Hoesen, 2015; for a recent atlas, see Ding et al., 2016). This could improve in vivo segmentation and advance research on these MTL subregions because the sparsity of available anatomical reference information from histological studies, such as the presentation of only a few slices from a very small number of cases, has been a major barrier. Ding and Van Hoesen, however, addressed this by providing several cases to account for anatomical variation and showing multiple example slices along the longitudinal axis (Ding and Van Hoesen, 2010, Ding and Van Hoesen, 2015). More specifically, their work revealed novel insights on the boundaries between subdivisions in the PrC - area 35 and area 36 – which depend on sulcal patterns that differ between hemispheres in continuity and depth (Ding and Van Hoesen, 2010). Further on, they showed that the boundaries of Sub and CA1 subfields in the hippocampal head (HH) feature anatomical variations between individuals that depend on the number of hippocampal digitations and provide more anatomical detail for subfield boundaries in the hippocampal head (Ding and Van Hoesen, 2015). Additionally, in the latter study (Ding and Van Hoesen, 2015), data was sectioned perpendicular to the long axis of the hippocampus, thereby matching MR images commonly used for MTL subfield segmentation in vivo (Yushkevich et al., 2015a). Usually, most of the extant histological reference material is based on samples sectioned at orientations different from the slice orientation in these MR images, and it is unclear how much this difference in orientation affects the translation of anatomical boundaries to in vivo MR images, especially in the more complex head region of the hippocampus (these issues were also mentioned in Wisse et al., 2016b). To our knowledge, there is currently no segmentation protocol that incorporates these novel findings. Consequently, the aim of this paper was to develop an MRI segmentation protocol leveraging all the new information presented by Ding and Van Hoesen, 2010, Ding and Van Hoesen, 2015 using 7 Tesla (T) magnetic resonance imaging (MRI).
7 T MRI offers ultra-high resolution and increased signal-to-noise ratio thereby allowing for a more consistent slice-by-slice visualization of internal features, while maintaining a smaller slice thickness (of up to 1 mm), which is especially needed to resolve the complex anatomy in the hippocampal head. Specifically, T2-weighted images with high in-plane resolution were used to delineate MTL subregions because of the visualization of the stratum radiatum lacunosum moleculare (SRLM), which appears as a thin dark band on these scans and can be used to define borders between some of the subregions.
Several segmentation protocols have been published for 7 T (Wisse et al., 2012, Boutet et al., 2014, Goubran et al., 2014, Maass et al., 2014, Maass et al., 2015, Parekh et al., 2015, Suthana et al., 2015) utilizing the improved resolution for the distinction of small subfields such as the DG and CA3 in the hippocampus (Parekh et al., 2015), the SRLM (Kerchner et al., 2012), and allowing for specific analyses of subregions of the ErC (Maass et al., 2015) and even entorhinal layers (Maass et al., 2014). However, most 7 T protocols limit the segmentation to the hippocampal body and do not include the head and tail of the hippocampus, with the exception of Wisse et al. (2012) and Suthana et al. (2015). Critically, most published 7 T protocols have not reported inter- or intra-rater reliability, which is necessary to ensure that the described rules can be reliably applied.
At lower MRI field strengths, a greater number of segmentation protocols have been developed for hippocampal subfields (Zeineh et al., 2001, Mueller et al., 2007, Malykhin et al., 2010, La Joie et al., 2013, Winterburn et al., 2013, Daugherty et al., 2015) as well as extrahippocampal regions ErC, PrC and PhC (Insausti et al., 1998, Pruessner et al., 2002, Feczko et al., 2009, Kivisaari et al., 2013, Yushkevich et al., 2015b). However, these protocols often lump together hippocampal subfields, mostly do not include individual variability, and provide less detailed descriptions for manual segmentation. Crucially, none of these protocols at 7 T or lower field strengths have incorporated the anatomical variations dependent on hippocampal indentations (Ding and Van Hoesen, 2015) or individual sulcal patterns in the parahippocampal gyrus described above (Ding et al., 2009, Ding and Van Hoesen, 2010) to provide precise subvariant-specific and depth-dependent rules that account for the variability across and within subjects.
Reference should be made to a large multi-investigator effort currently underway to develop a harmonized protocol for hippocampal subfields and extrahippocampal subregions (Wisse et al., 2016a), following the harmonized protocol for the total hippocampus (Apostolova et al., 2015, Frisoni et al., 2015). This harmonization effort was launched to overcome significant differences reported between extant segmentation protocols (Yushkevich et al., 2015a). However, the harmonization effort is currently aimed at 3 T MRI (first limited to the hippocampal body, to be followed by expansion to the head and tail), and the protocol for 7 T and extrahippocampal regions is not anticipated for several more years.
Therefore, we aimed to establish a segmentation protocol to manually delineate subregions in the parahippocampal gyrus as well as hippocampal subfields in 7 T MRI images leveraging the information on subfield boundaries and anatomical variability presented in Ding and Van Hoesen, 2010, Ding and Van Hoesen, 2015 and Ding et al. (2016), which can also inform high-resolution studies at 3 T. To ensure that the rules can be reliably replicated, we performed intra- and inter-rater reliability analyses in the left and right MTL of 22 younger adults. Critically, this manuscript provides very detailed and comprehensive descriptions alongside slice-by-slice plots to facilitate the application of the segmentation rules.
2. Materials and methods
2.1. Participants
Participants were included using baseline data from a study investigating the effects of physical exercise on the brain. Exclusion criteria were reports of regular sports activities that improve cardiovascular fitness as well as high physical activity levels. In addition, participants were screened for known metabolic disorders and neurological or psychiatric history, and excluded from further examination in case of incidents reported during history taking. Participants were recruited from the Otto-von-Guericke University campus in Magdeburg. Fifteen young and healthy individuals (16 hemispheres) were included from the baseline scan before any intervention. Seven additional subjects (8 hemispheres) were included after refining the rules for sulcus depth measurements (age range 19–32; mean age = 26, 12 female; see 2.6 and 3.1). In total, we used 24 hemispheres of 22 subjects. All subjects gave informed and written consent for their participation in accordance with ethic and data security guidelines of the Otto-von-Guericke University Magdeburg. The study was approved by the local ethics committee.
2.2. Workshop
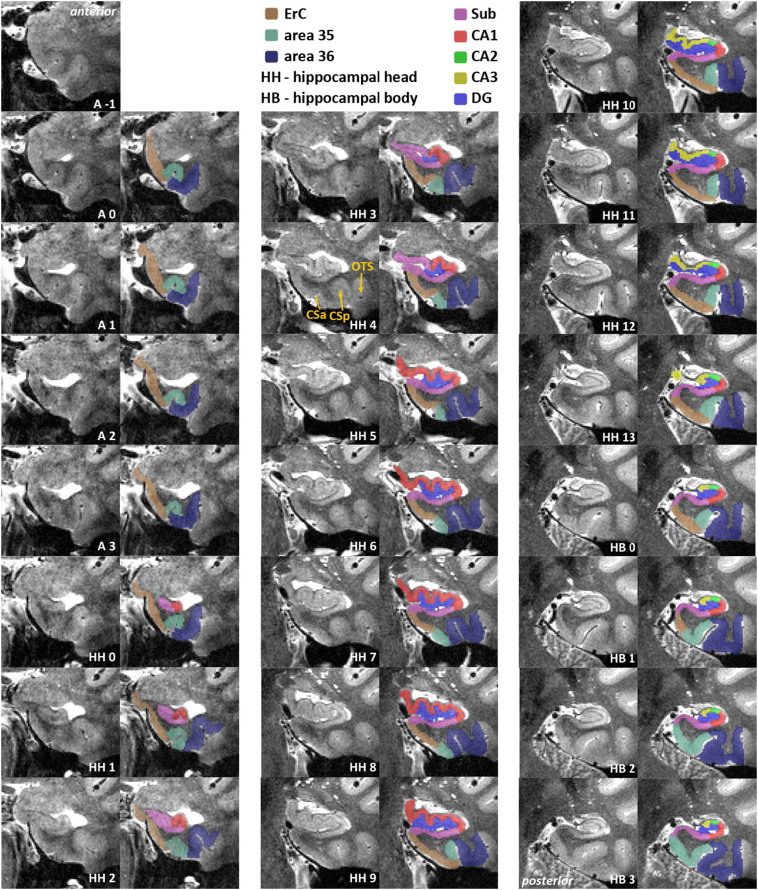
In order to test the usability of the manual segmentation protocol, we hosted a segmentation workshop for 35 participants who were mostly novices (29 out of 35). The protocol was sent out four weeks prior to the workshop in combination with example MR images in order to give participants the opportunity to familiarize themselves with the segmentation approach. On site, we presented the protocol followed by an intensive hands-on session. From that, we used the given feedback and most commonly occurring problems to refine the protocol and improve comprehensibility for novice raters. This includes figures that provide a quick overview of the rules (Fig. 9), more detailed annotations of the slice-by-slice plots (Fig. 3, Fig. 4, Fig. 6, Fig. 7) as well as supplemental material (see Appendix A) of cases with rare anatomic variants.
Fig. 9.
Exemplary segmentation profile. This anterior-to-posterior axis (i.e. along the longitudinal axis of the hippocampus) illustrates the key decision points of this protocol (numbers indicate mm distance from the first anterior slice in the protocol). Extrahippocampal regions and hippocampal head (HH), body (HB) and tail (HT) divisions are on the left; hippocampal subfields are depicted on the right. The start and end of each structure are depicted in the same color; often they depend on certain landmarks, e.g. the start and end of HH are used as a reference for the occurrence of the entorhinal cortex (ErC), area 35 and area 36, and the parahippocampal cortex (PhC). Most of these points are variable between brains but usually fall into a similar range as shown here. We recommend identifying these points prior to segmentation.
Fig. 3.
Slice-by-slice segmentation for a type 1 collateral sulcus (CS) – anterior part. Slices are 1.1 mm apart. Included are entorhinal cortex (ErC; brown), perirhinal cortex (area 35 in mint green, area 36 in dark blue), subiculum (pink), CA1 (red), CA2 (green), CA3 (yellow) and dentate gyrus (blue). Shown in HH2, ErC covers the ambient gyrus (AG) and superiorly ends at the semiannular sulcus (SaS). SaS constitutes the superior border of ErC and should be extrapolated to anterior slices when it cannot be identified there. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Continuation of Fig. 3 - Slice-by-slice segmentation for a type 1 collateral sulcus (CS) – posterior part. Slices are 1.1 mm apart. Included are parahippocampal cortex (PhC; dark pink), subiculum (pink), CA1 (red), CA2 (green), CA3 (yellow), dentate gyrus (blue), and the hippocampal tail which, is not divided into subfields. In HT7, the subsplenial gyrus starts medially blending into the hippocampus. As soon as it is detached from the hippocampus, it is excluded from segmentation (HT + 1). Delineation of PhC stops at the calcarine sulcus (CaS) in HT2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Slice-by-slice segmentation for a type II collateral sulcus (CS) – anterior part. Slices are 1.1 mm apart. Included are entorhinal cortex (ErC; brown), perirhinal cortex (area 35 in mint green, area 36 in dark blue), subiculum (pink), CA1 (red), CA2 (green), CA3 (yellow) and dentate gyrus (blue). HH4 is an example of a transition slice between anterior (CSa) and posterior CS (CSp) and the corresponding segmentation of area 35. The occipitotemporal sulcus (OTS) establishes the lateral border of area 36. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
Rules for hippocampal subfield segmentation shown on the relevant slices from anterior to posterior. Schematic descriptions of all rules are depicted in the first column. Specific rule changes or new borders are indicated in red. Dashed lines are used, when the rule in question is inferred from another slice; e.g. the inferior Sub/CA1 border is defined in B, but the superior Sub/CA1 border is defined in B' and extrapolated anteriorly. The relevant anatomical changes are indicated by white labels and arrows in the middle column, e.g. when the uncus separates from the hippocampal body (HB), or the colliculi (Col) disappear. The resulting segmentation is shown in the last column; subiculum (Sub) in pink, CA1 in red, CA2 in green, CA3 in yellow and dentate gyrus (DG) in blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Image acquisition
Imaging data were collected at the Leibniz Institute for Neurobiology in Magdeburg on a 7 T MR scanner (Siemens, Erlangen, Germany) with a 32-channel head coil (Nova Medical, Wilmington, MA). We acquired partial turbo spin echo (TSE) T2-weighted images oriented orthogonal to the long axis of the hippocampus (in-plane resolution = 0.44 × 0.44 mm, 55 slices, slice thickness = 1 mm, distance factor = 10%, TE = 76 ms, TR = 8000 ms, flip angle = 60°, FOV = 224 mm, bandwidth = 155 Hz/Px, echo spacing = 15.1 ms, TSE factor = 9, echo trains per slice = 57). The slice thickness of 1 mm together with the 10% distance factor results in a distance of 1.1 mm between slices. Scan-time was 7:46 min.
2.4. Segmentation software
Structures were manually traced by two experienced raters (A.H. and A.L., see 2.6 for details) on oblique coronal slices using ITK-SNAP (Version 3.4; www.itksnap.org; (Yushkevich et al., 2006)). The images were adjusted for equivalent contrast range prior to segmentation (by capping the contrast curve at a maximum of 500). ITK-SNAP provides very useful features for implementing this protocol, such as an annotation tool for drawing lines and measuring distances.
2.5. Manual segmentation protocol
The protocol describes rules for manual segmentation of structures in the MTL in coronal MR images. The segmentation guidelines for the parahippocampal cortex (PhC), perirhinal cortex (PrC; area 35 and 36), entorhinal cortex (ErC) as well as the outer contours of the hippocampus are described in the first part (2.5.2), and further subdivision of the hippocampus into subfields are described in the second part (2.5.3; for a segmentation hierarchy see Fig. 1). Boundary rules are based on recent data from neuroanatomical atlases (Ding and Van Hoesen, 2010, Ding and Van Hoesen, 2015, Mai et al., 2015, Ding et al., 2016). In this protocol we separately report neuroanatomical evidence and resulting rules, which can be applied to MR images. Boundary rules are provided in millimeters in order to make the protocol applicable to scans of different resolution and facilitate comparisons with the neuroanatomical literature. The protocol is particularly focused on T2-weighted images acquired at 7 T with 0.44 × 0.44 mm2 in-plane resolution and 1 mm slice-thickness with 0.1 mm spacing. Some inner boundaries described in the section about hippocampal subfields, especially the boundaries of CA3 and DG that rely on the visualization of the endfolial pathway (Lim et al., 1997), are likely only applicable to 7 T high-resolution T2 images. However, the described protocol could potentially also be applicable to other images that are acquired orthogonally to the long axis of the hippocampus with similar in-plane resolution and larger slice-thickness (e.g. 2 mm slice thickness).
Fig. 1.
Segmentation hierarchy. Segmentation of entorhinal cortex (ErC), area 35 and 36 of the perirhinal cortex (PrC), parahippocampal cortex (PhC) and the whole hippocampus separated into head (HH), body (HB) and tail (HT) are described in 2.5.2 (dark blue) and segmentation of hippocampal subfields is described in 2.5.3 (light blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.5.1. Anatomical labels used in the protocol
In this protocol, we segment ErC, PrC, PhC and the hippocampus. We differentiate between area 35 and 36, which are frequently considered together as constituting the PrC in manual segmentation protocols (Zeineh et al., 2001, Ekstrom et al., 2009, Preston et al., 2010, Olsen et al., 2013, Duncan et al., 2014), except for (Kivisaari et al., 2013, Yushkevich et al., 2015b). However, these regions constitute different neuroanatomical parts of PrC (Ding and Van Hoesen, 2010). Therefore, following the terminology of Ding and Van Hoesen, we refer to these regions as area 35 and 36. Note that these regions are slightly different from Brodmann areas 35 and 36 as the latter extend more posterior than area 35 and 36 in our study (for discussion see Ding and Van Hoesen, 2010). We note that area 35 roughly corresponds to the transentorhinal region (Braak and Braak, 1991) and also to the medial PrC (Kivisaari et al., 2013). Detailed guidelines for hippocampal subfields involve the boundaries between the subiculum (Sub), CA fields 1–3 and the dentate gyrus (DG). Note that our definition of Sub includes subiculum proper, prosubiculum, presubiculum and parasubiculum (Ding, 2013). Also, the DG here includes the hippocampal hilus or region CA4, as these cannot be separated at this field strength. The SRLM is equally divided between its surrounding structures and not segmented separately. Hippocampal subfield segmentation encompasses the whole hippocampal head (HH) and body (HB) and is not performed in the tail (HT) because of the limited information with regard to the subfield boundaries in this region.
2.5.2. Hippocampus and subregions in the parahippocampal gyrus
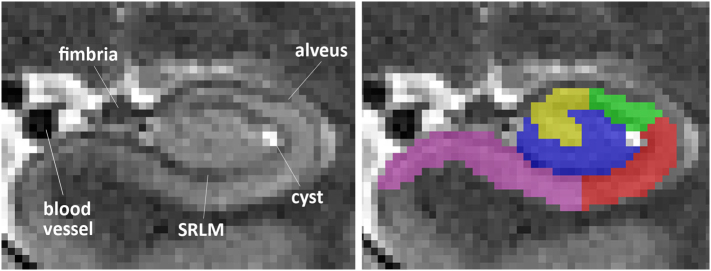
2.5.2.1. Exclusions: Alveus, fimbria, cerebrospinal fluid and blood vessels
Fimbria and alveus as well as blood vessels, all appearing hypointense in T2-weighted images (see Fig. 2), are excluded from anatomical masks as they do not belong to any particular subfield (Duvernoy et al., 2013). In general, the hippocampus is enclosed by white matter, visible as a hypointense line surrounding it. This line is spared from segmentation in this protocol. Additionally, there are several blood vessels within and close to the hippocampus. Both blood vessels and potential concomitant signal dropout should be excluded from the segmentation. Cerebrospinal fluid (CSF) and cysts appear hyperintense on T2-weighted MRI. Cysts, often located in the hippocampal sulcus (hippocampal fissure) at the ventrolateral flexion point of CA1 (van Veluw et al., 2013) are given a separate label. CSF - either surrounding the hippocampus or along a whole sulcus (e.g. hippocampal, uncal, collateral, occipito-temporal sulci) - are entirely excluded from the anatomical masks. CSF in sulci can be given a separate label as CSF (see explanation below).
Fig. 2.
Structures excluded from segmentation in a coronal view. Anterior hippocampal body slice from a T2 MRI scan including alveus, fimbria, SRLM, a blood vessel and a cyst in the ventrolateral flexion point of CA1 in the vestigial hippocampal sulcus.
2.5.2.2. Hippocampal formation
In the following paragraph we provide segmentation rules separately for the hippocampal head, body and tail. This is done to structure the following section rather than to construct independent masks of head, body and tail portions.
2.5.2.2.1. Hippocampal head
The anterior tip of the hippocampal head (HH) can be easily identified without additional landmarks (see Fig. 3, HH0). Once the uncal sulcus can be followed from its fundus to the medial surface, the ErC becomes the inferior boundary of the HH, which is segmented by connecting the most medial point of the white matter to the most medial point of the grey matter (see Fig. 3, HH4;(Wisse et al., 2012)). At the posterior end of the HH, the uncus separates from the hippocampus (see Fig. 3, HH14). While the uncus is still connected to the rest of the HH (via grey matter), the hippocampus is segmented as one structure (see Fig. 3, HH13). Once the uncus is separated (e.g. only connected via the fimbria), the HH and uncus are segmented as separate structures in the coronal plane (see Fig. 3, HH15).
2.5.2.2.2. Hippocampal body
The hippocampal body (HB) begins when the uncus has disappeared (1 slice posterior to the uncal apex; see Fig. 3, HB 0). White matter and CSF surround the HB superiorly, medially and laterally. The medial-inferior boundary of the HB is the connection of the most medial point of the white matter to the most medial part of the grey matter, where it successively borders ErC, area 35 and PhC (see Fig. 3 HB 0 - HB 3, e.g. (Ding and Van Hoesen, 2010). Sometimes, in more posterior slices, a small sulcus (the anterior tip of calcarine sulcus; CaS) appears medially between HB and PhG. In this case, the lateral and medial banks of the CaS are spared from segmentation (see Supplementary Fig. 1). However, often the CaS only appears in HT. The HB is segmented as long as the inferior and superior colliculi are visible (medial butterfly-shaped structures) (Wisse et al., 2016c). Segmentation does not stop before the colliculi have disappeared entirely. This rule is applied for each hemisphere separately (see Supplementary Fig. 2).
2.5.2.2.3. Hippocampal tail
The hippocampal tail (HT) is a structure that is surrounded by white matter laterally, superiorly and ventrally. Most of these white matter structures are represented by alveus, fimbria and fornix, and are therefore excluded from segmentation. The medial-inferior boundary is constructed in the same way as that for the HB (see Fig. 4, e.g. HT0–6). In more posterior slices, the HT (supero) laterally neighbors CSF in the trigone of the lateral ventricle (see Fig. 4, e.g. HT3). The last slice of the hippocampus is the last slice where the HT is clearly visible (see Fig. 4, HT11) which can also be checked on sagittal slices. It should be noted that at the very end of HT the hippocampus might medially blend with a gyrus, sometimes referred to as subsplenial gyrus (Ding et al., 2016). This gyrus is included in the hippocampal mask until it is no longer connected to the rest of the hippocampal grey matter.
Note that different definitions of the body/tail border exist. Here, we chose the colliculi as they are easily identifiable, and are intended to provide a reliable posterior border for subfield segmentation.
2.5.2.3. Entorhinal cortex
Segmentation of the ErC (as well as area 35 and area 36) begins 4.4 mm (= 4 slices here) anterior to the first slice of HH. That is, 4 slices have to be counted anterior to the hippocampus to define the starting slice. Although the ErC extends through most of the anterior temporal lobe (Ding and Van Hoesen, 2010, Kivisaari et al., 2013) we chose this border because it is easily identifiable, and high-resolution structural imaging protocols often do not cover the entire anterior MTL. The superior border in anterior slices is the semiannular sulcus (Mai et al., 2015, Ding et al., 2016). Sometimes, this sulcus is not visible from the most anterior end of ErC, in which case it should be extrapolated from more posterior slices where it can be clearly identified (see Fig. 3, HH2). The ErC covers the ambient gyrus (AG; see Fig. 3, HH0–3). Note that the ambient gyrus is made up of different subfields in an anterior-to-posterior direction. While the ambient gyrus is occupied by the ErC in more anterior slices (Insausti and Amaral, 2012), it consists of Sub and CA1 in more posterior sections (Ding and Van Hoesen, 2015). Moving posteriorly, at the point where the uncal sulcus can be followed from its fundus to the medial surface, Sub becomes the new superior border (see Fig. 3, HH4). It is constructed by drawing a line from the most medial part of the white matter to the most medial part of the grey matter (Mueller et al., 2007, Wisse et al., 2012, Yushkevich et al., 2015b). This rule applies until the posterior end of ErC. The lateral border of ErC mainly consists of white matter. With respect to the inferomedial border, in some subjects CSF can be discerned between the ErC and the laterally located meninges (Xie et al., 2016, Xie et al., 2017). Therefore, bright voxels medial to the ErC have to be spared from the segmentation (see Fig. 3, HH1–7). It should be noted that the intensity can depend on how much space there is between the meninges and the cortex. Sometimes these voxels appear slightly darker than CSF at other locations because of partial voluming with surrounding voxels. Inferolaterally, the ErC is bordered by area 35. This boundary is constructed at ¼ of the longest expansion of CS (from edge to top of the grey matter) as the shortest connection between CS and white matter (see Fig. 5). The only exception from this rule occurs when CS is < 4 mm deep (very shallow CS); in that case the boundary between ErC and area 35 moves more lateral to the extension of the fundus of the CS. ErC disappears approximately 2 mm after the HH (Insausti and Amaral, 2012). Segmentation of the ErC stops therefore after 2.2 mm (= 2 slices here) into the HB, i.e. after 2.2 mm posterior to the uncal apex (see Fig. 3, HB0). The last slice of ErC serves as an intermediate step between ErC and the increasing size of area 35. Therefore, the lateral border of the ErC shifts by dividing ErC in half (Insausti et al., 1998, Ding and Van Hoesen, 2010, Mai et al., 2015).
Fig. 5.
Different depths of the collateral sulcus (CS) with respective segmentation rules applied. Sulcus depth is measured from edge to fundus of CS as indicated by the red arrows. Edge, fundus, crown and bank are indicated for easy anatomical descriptions of the gyral and sulcal patterns. Quartiles for segmentation rules are defined by measuring the full extent of grey matter from edge to top along the respective bank as indicated by the white two-sided arrows in the images on the right. Entorhinal cortex (brown) ends laterally at ¼ of the grey matter bank medial to CS, when CS is deeper than 4 mm. For very shallow CS (< 4 mm), entorhinal cortex covers the whole medial bank of CS and ends at the extension of the fundus of CS. Segmentation rules for area 35 (green) change depending on the depth of CS: very deep – area 35 covers the middle part from ¼ to ¾ of the grey matter bank medial to CS; deep – area 35 covers the whole superior ¾ of the grey matter bank medial to CS; shallow - area 35 extends up to half of the lateral bank of CS; very shallow - area 35 extends up to half of the crown of the fusiform gyrus (FG). Area 36 (blue) directly neighbors area 35 laterally, and extends towards the entire bank medial to occipitotemporal sulcus (OTS). The hippocampal head (HH) is depicted in turquoise. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.5.2.4. Perirhinal cortex
Segmentation of area 35 and area 36 of the PrC is dependent on the sulcal pattern within the MTL – especially the collateral sulcal patterns are highly variable between brains but can also differ between hemispheres of the same brain. There are two main types of MTL anatomy – one deep CS (Type 1; 45%), and a discontinuous CS (Type II; 52%), which can be divided into an anterior (CSa) and a posterior section (CSp) (Ding and Van Hoesen, 2010). CSp is usually longer and deeper than CSa. Studies have found a negative correlation between the depth of the CS and the depth of the occipito-temporal sulcus (OTS). In subjects with a shallow CS, the OTS is often deep and vice versa (see Fig. 5; (Ding and Van Hoesen, 2010)). In some cases, the CS is bifurcated, i.e. it appears to have two conjoined sulci; the more medial sulcus is used here in this case (i.e. for evaluating the depth of CS). When it is difficult to identify the sulcal pattern in one slice, we recommend to check in adjoining slices and interpolate to the difficult slices.
Given the differences in anatomy, different segmentation guidelines have to be applied for the different sulcal patterns as well as the depths of the CS. It is highly recommended to first define the sulcal pattern for each hemisphere before starting the manual tracing. The following descriptions are visualized in Fig. 5.
2.5.2.4.1. Area 35
Segmentation of area 35 starts at the same artificially chosen slice as ErC, i.e. 4.4 mm (= 4 slices) anterior to the first HH slice. Neuroanatomical atlases indicate that the posterior border of area 35 falls within 5 mm of the anterior portion of the HB. Segmentations therefore end 4.4 mm (= 4 slices) after the start of HB, which is also 2.2 mm posterior to ErC (Insausti et al., 1998, Ding and Van Hoesen, 2010). In the most posterior 2.2 mm, area 35 borders the Sub medially (see Fig. 3, HB1-HB3); in all anterior slices it borders ErC. The superolateral and inferomedial borders are in accordance with those of ErC (e.g. white matter and CSF or meninges). The lateral border of area 35 depends on the depths of the sulci, and is measured from edge to fundus of the respective sulcus on each individual slice. For that purpose, the edges adjacent to the sulcus are connected via a tangent line. The depth of the sulcus is now measured from the middle of this line to the fundus of the sulcus (see Supplementary Fig. 3A; also schematics in Fig. 5). If the sulcus bends, the depth is measured in separate legs along the middle of the sulcus (see Supplementary Fig. 3B).
Very deep CS (> 10 mm) Area 35 occupies the two middle fourths of the medial bank of the CS. Its lateral boundary with area 36 is constructed at ¾ of the medial bank of the CS (see Fig. 5).
Deep CS (7–10 mm) From the border to ErC, area 35 occupies the remaining ¾ of the medial bank of the CS (see Fig. 5); i.e. from ¼ of the medial bank up to the top of grey matter.
Shallow CS (4–7 mm) From the border to ErC, area 35 extends up to half of the lateral bank of the CS (see Fig. 5).
Very shallow CS (< 4 mm) From the border to ErC, area 35 extends up to half of the crown of the fusiform gyrus (FG; see Fig. 5).
When both CSa and CSp are visible on the same slice, the lateral boundary of area 35 is constructed at half of the crown between the two CS (see Fig. 6, HH4). As soon as the CSa has disappeared, the same depth rules apply to CSp as shown in Fig. 5. A decision tree can be used to facilitate the necessary decisions (see Supplementary Fig. 4). The relationship of area 35 to area 36 length in histological studies roughly resembles a 1:3 ratio. Our rules are designed in order to approximate this ratio.
2.5.2.4.2. Area 36
Segmentation of area 36 is done in the same slices as area 35, that is, starting 4.4 mm anterior to the first HH slice, and ending 2.2 mm posterior to ErC. Area 36 directly borders area 35, thus its medial boundary depends on the different sulcal patterns described for area 35. Its lateral border is defined by the next lateral sulcus – the OTS. This border extends previous protocols (Insausti et al., 1998, Pruessner et al., 2002, Feczko et al., 2009, Kivisaari et al., 2013) and is specifically based on Ding and Van Hoesen (2010). It is constructed by following the longest expansion of OTS, from medial edge to top of the grey matter, thereby including the whole medial bank of OTS (see Fig. 5). It should be noted here that OTS is very variable, i.e. it can be bifurcated, or there could be two OTS. In these cases, the more medial OTS should be used as the border (see Supplementary Figure 5). Generally, OTS is rather deep and shows a reciprocal relationship with CS (see Section 2.5.2.4.1 on area 35 for more detail), and thus can be differentiated from other small sulci that sometimes appear in-between CS and OTS, e.g. the mid-fusiform sulcus in posterior slices (for reference, see (Ding et al., 2016), and Supplementary Fig. 6).
Sometimes, another small sulcus, the rhinal sulcus, is visible in very anterior slices. Generally, it is medial to CS and more shallow; it often ‘travels’ up the CS (see Supplementary Fig. 7). In very rare cases, the rhinal sulcus can be separate from CS so far posterior that it affects segmentation. That is, when the rhinal sulcus is separate and visible on the medial cortical surface, the rules change in a way as if one were to substitute the CS with the rhinal sulcus and the OTS with the CS. The boundaries follow the same depth rules as above but are applied to the rhinal sulcus. The very lateral border of area 36 is now the CS and not the OTS. When the rhinal sulcus disappears, area 35 and area 36 change to the usual patterns.
2.5.2.5. Parahippocampal cortex
Anteriorly, the segmentation of the PhC directly adjoins the posterior end of area 35. Thus, it begins 5.5 mm (= 5 slices) posterior to the uncal apex (see Fig. 3, HB4; (Insausti et al., 1998, Ding and Van Hoesen, 2010)). As with area 35, the PhC has a medial-superior boundary with Sub (see Fig. 3, starting HB3). The superolateral and inferomedial borders are in accordance with those of ErC (e.g. white matter and CSF). The lateral boundary is the fundus of the CS extended to the top of grey matter. Posterior regions of the parahippocampal and fusiform gyrus include areas TH, TL and TF (Ding and Van Hoesen, 2010). While TH and TL cover regions in the parahippocampal gyrus, TF occupies parts of the fusiform gyrus. The PhC in this study only covers temporal areas TH and TL, but not TF (confer (Ding and Van Hoesen, 2010, Ding et al., 2016)). Segmentation stops when the anterior tip of the CaS appears medially - a small sulcus that mostly folds in a superior-to-inferior direction (see Fig. 4, HT2; Supplementary Fig. 1). Since little anatomical literature is available on the PhC, we based this decision on Song-Lin Ding's expertise annotating this region in histology samples, and because the CaS can be reliably identified in every subject. There is another small sulcus lateral to CaS, the newly discovered parahippocampal-ligual sulcus (PhligS; (Ding et al., 2016); see Supplementary Fig. 8), which would be a better indicator of PhC's borders. However, it was not possible to reliably distinguish this sulcus in every subject's MRI and we therefore chose to use the CaS as a landmark.
2.5.2.6. Transitions and labeling of the sulci
In order to maintain smooth transitions between slices that resemble the anatomy more closely we introduce transitions. Whenever there are sudden changes from one rule to the other, or sudden appearances of anatomical structures, one intermediate slice serves as a transition. Thus, the last slice where the anatomy fulfills the criteria of one rule serves as a transition slice to the next rule by applying an intermediate step in the middle between both rules (e.g. see Fig. 3, HB1; Fig. 6, HH0 and HB1). This procedure should be used in the following cases: (1) when ErC ends posteriorly, (2) when only CSa changes to only CSp without both being visible on the same slice (see also Supplementary Fig. 9) and (3) when the OTS “jumps” (appears/disappears) from one slice to the next. An optional additional label for the CS and OTS can be added to facilitate thickness measurements using automated tools (e.g. ASHS (Yushkevich et al., 2015b). In case the CSF within the sulci is visible, these voxels can be labelled as sulcus. If the sulcus is not completely visible, there are usually some hints to it, such as an indentation on the inferior portion or a patch of CSF in the middle. If the sulcus cannot be identified, it can be estimated based on the thickness of the medial and lateral grey matter banks on surrounding slices. Inferring the sulcus in this way ensures that all voxels labelled as sulcus have adjacent edges (that is: not 1 voxel thick diagonal). In addition, if the gyri around CS and OTS touch, i.e. if no white matter in-between their grey matter banks is visible, the line of voxels in the middle between the two sulci should be artificially excluded from segmentation to allow meaningful thickness measurements. Alternatively, if the separation of the two banks can be inferred from the surrounding slices, a voxel line approximating that separation should be used instead.
2.5.3. Hippocampal subfields
These guidelines are mostly based on ex-vivo parcellations by Ding and Van Hoesen (2015), and on comparative, additional information derived from other publications, such as the Mai atlas (Mai et al., 2015) and the protocol from Wisse et al. (2012).
2.5.3.1. Sub and CA1 segmentation starts
Mostly, the first anterior slice of the HH appears as one structure. Sub is then assigned to all of it (Fig. 7A). Approximately 1–2 mm posterior to that, a hypointense line appears (i.e. uncal sulcus/SRLM; (Ding and Van Hoesen, 2015)) dividing the hippocampus into a superior and an inferior part and shaping the hippocampus similar to a lip (Fig. 7B). From here, the SRLM is equally divided between the regions it separates unless it is only 1 voxel wide, in which case it is segmented such that it always belongs to the superiorly located structure. Also at this point, the segmentation of CA1 starts. The guidelines can be more readily understood by looking at Fig. 7B and B’. The inferior boundary (i.e. on the “lower lip”) between Sub and CA1 is an orthogonal line to the longitudinal Sub. It is positioned by finding the most lateral voxel of the SRLM, moving to the next medial one, and is constructed there from inner to outer side of the structure. The superior boundary between Sub and CA1 is extrapolated from a posterior slice where the digitation of the HH can be clearly identified, i.e. when the “upper lip” has at least two dents (Fig. 7B’). At the second indentation counted from lateral to medial, a straight line is constructed orthogonal to the structure, and copied to anterior slices. Posteriorly, the border is positioned at this same indentation on each individual slice. This border closely approximates what is observed in the hippocampal subvariants with two and three indentations, as described by Ding et al. (see Fig. 6, Fig. 7 in Ding and Van Hoesen, 2015). Once the uncal sulcus opens, the separation of CA1 and Sub continues along the uncal sulcus (MR image in Fig. 7B’). This may coincide with the appearance of DG, although it may also occur slightly more posterior.
2.5.3.2. DG segmentation starts
When DG appears, and does not yet stretch to the most lateral extension of the uncal sulcus (confer Fig. 6, HH4), the inferior boundary between Sub and CA1 is constructed exactly like before. If, however, DG extends towards the most lateral point of the uncal sulcus (confer Fig. 6, HH5), the reference point changes from one voxel medial from the most lateral SRLM to the most lateral DG voxel (Fig. 7C). It is crucial to not confuse DG with cysts (which are brighter). However, if there is a cyst within DG that establishes the most lateral border, the cyst is used instead of DG to identify the CA1/Sub border (Fig. 7D). Based on Ding and Van Hoesen (2015), the superior part of the subiculum disappears 1.2–1.8 mm after the appearance of the DG. We therefore chose to end segmentation of the superior part of the subiculum 2.2 mm (= 2 slices) after the first appearance of DG and this portion is then occupied by CA1 (Fig. 7D). These borders are identified in the same way again on all following slices, although they often just stay the same as on the previous slices. From here, the SRLM is equally divided if thicker than 1 voxel, and otherwise segmented so it always belongs to the outer structure (i.e. CA1/Sub/etc., but not DG). It should be noted that contrary to the white matter surrounding the hippocampus, the hypointense line on the superior side of Sub is always included in the segmentation as it consists of the molecular layer of the Sub. Additionally, the inferior side of Sub is prone to signal drop-out due to the crossing perforant path; therefore voxels of intermediate intensity on the inferior side of Sub should be included because a very conservative visual segmentation of only the brightest voxels might result in an underestimation of Sub (see (Bronen and Cheung, 1991, Wisse et al., 2016c)).
2.5.3.3. CA2 and CA3 segmentation starts
Neuroanatomical data indicate that the anterior border of CA3 in the head falls within 3–5.4 mm relative to the start of the body (Ding and Van Hoesen, 2015). The segmentation of CA2 and CA3 therefore begins in the last 4.4 mm (= 4 slices) of the HH. Although CA2 generally appears before CA3 (Ding and Van Hoesen, 2015), there is limited information on the exact distance between the two and on potential differences between subjects. Therefore, we chose to start segmenting CA2 at the same slice as CA3. Additionally, CA2 and CA3 show an alternating pattern in the most anterior slices; we chose to simplify this and count all medial grey matter towards CA3. Although we realize that we may count some portions of CA2 towards CA1 or CA3, we chose for these simplifications to achieve high reliability. Again, it might help to consider Fig. 7E and E’ alongside this description. The border between CA1 and CA2 is constructed orthogonal to the CA structures at one voxel medial of the lateral boundary of DG; this is identical to the determination of the previous CA1/Sub border rule only on the superior instead of the inferior side. As in the previous section, if there is a cyst within DG that establishes the most lateral border, the cyst is used instead of DG to identify the CA1/CA2 border. The next step is to identify the point where the uncus separates from hippocampus. In some cases, only the fimbria is attached to both (Fig. 7E’). The border between CA2 and CA3 is extrapolated from that slice to more anterior slices (to include the last 4.4 mm of HH). It is constructed halfway between the most medial point of the CA fields and the most lateral point of DG (it can therefore only be determined after the medial border of CA3 is determined). For all posterior slices, this border is determined slice by slice as a line orthogonal to the structure. The detached uncus is defined as CA3 unless there is a hypointense line, which can be used to differentiate between CA3 superiorly and DG inferiorly (Duvernoy et al., 2013) (Fig. 7E’). The Sub/CA1 border also changes within the last 4.4 mm of HH (Ding and Van Hoesen, 2015). As soon as the uncus separates from hippocampus, the new border is marked at 1/4 from most lateral DG to most medial hippocampus proper. This line is extrapolated anteriorly to include the last 4 mm of HH (Fig. 7E’). In the HB, i.e. when the uncus has disappeared, this border shifts to 1/2 from most lateral DG to most medial hippocampus proper (Fig. 7F). This boundary is identified in the same way on all posterior slices.
2.5.3.4. CA3 and DG differentiation
Unique to our protocol is the delineation of CA3 and DG. Depending on image quality and resolution, we propose two different rules. Both rules apply to the whole HB and the most posterior HH slices where the uncus is only connected via the fimbria. Many protocols have defined everything on the inner side of the SRLM as DG. Based on ex-vivo segmentations (Ding and Van Hoesen, 2015, Mai et al., 2015) and the better contrast of T2 images and higher resolution of 7 T imaging, the visualization of the endfolial pathway is possible, which can be used to more clearly differentiate between CA3 and DG (see the full HB segmentations in Fig. 3; also in (Parekh et al., 2015, Wisse et al., 2016c). The endfolial pathway is followed from the medial edge of CA3 towards the point where it intersects the SRLM. All voxels that lie supero-medially to this line belong to CA3 (see Fig. 7F). However, if the endfolial pathway is not identifiable, or the aim of the research project is a comparison of groups where the endfolial pathway cannot be reliably distinguished in one group, an approximation can be achieved pursuing the following rules alongside Fig. 8. First, construct a line from the middle and most superior part of the hippocampus to the medial edge of DG touching Sub (Fig. 8, Fig. 1). From the latter point, draw a line laterally along the dark band until Sub starts curving (Fig. 8, Fig. 2). Then, compose a line parallel to this which centrally intersects the first line (Fig. 8, Fig. 3) between the outermost extent of hippocampus until it intersects the SRLM. On the halfway point construct an orthogonal line towards the superior SRLM and close CA3 infero-laterally (Fig. 8, Fig. 4). All voxels lying superiorly to those lines belong to CA3 (Fig. 8, Fig. 5).
Fig. 8.

Heuristic rules for separation of DG and CA3 if the endfolial pathway is not visible. (1) construct a line from middle most superior part of the hippocampus to medial DG touching Sub; (2) from that point, draw a line laterally along the dark band until Sub starts curving, (3) parallel to this intersect line 1 centrally between SRLM and outermost extent of the hippocampus proper,; (4) centrally intersect line 3 orthogonally; (5) CA3 assigned to voxels superior to lines 3 and 4. Applied rules are shown in the lower panel; unspecific hippocampal body (HB) in turquoise, subiculum (Sub) in pink, CA1 in red, CA2 in green, CA3 in yellow and dentate gyrus (DG) in blue. Compare the right panels for visual segmentation based on the endfolial pathway on the same slice. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.5.3.5. Subfield segmentation ends
As described for the end of HB above, subfield segmentation stops when the colliculi (see Fig. 7F’) have disappeared entirely. This rule applies hemisphere-specific (Fig. 7G; also Supplementary Fig. 2). Afterwards, manual subfield segmentation is no longer reliable.
2.5.4. General advice for manual segmentation
Segmentation of all regions is accomplished by tracing along white-to-grey matter boundaries, and several hypointense lines. These lines are not always continuous; we therefore recommend attempting smooth curvature even if the hypointense lines are discontinuous. Additionally, switching back and forth between coronal slices should ensure smooth transitions between slices, and avoid sudden jumps between regions. This is most important along the SRLM between Sub and CA1 in HH, at the endfolial pathway between CA3 and DG in HB, and for better identification of the sulcal pattern in PrC. Furthermore, special care is needed when measuring the depth of CS, because only slight variations can lead to different rule sets being required, i.e. at 7 mm rules for a shallow sulcus apply and at 7.1 mm rules for deep sulci apply (see Supplementary Fig. 3, see rules 2.5.2.4.1; for impact of incorrect measurements see also results 3.1).
As shown above, there are many cross-references between areas, therefore we recommend defining certain key decision points prior to segmentation (see Fig. 9). For example, the beginning and end of HH are needed as a reference for the start and end of ErC and areas 35 and 36. Additionally, we advise to check the full segmentation at the end (for a checklist, see Supplementary Fig. 10).
2.6. Statistical analyses
Two experienced raters (A.H. and A.L.) traced all subregions in the same 16 hemispheres independently (8 left and 8 right, 8 type I and 8 type II CS patterns). Both raters have each segmented around 40 subjects with the current rules prior to reliability testing. During that time, they met twice a week to discuss difficult cases, rule exceptions, to implement rule changes and confer with S.-L.D. (e.g. the very shallow CS category was only introduced after specific feedback from S.-L.D.). From 14 subjects only one hemisphere was included, but from one subject two hemispheres were included to reach an equal number of type I and II CS patterns. In addition all subregions were segmented for a second time by one rater (A.H.) after 4 weeks.
The intra-rater reliability was assessed in 16 hemispheres in terms of relative overlap between the two time-points using the Dice similarity index (DSI) (Dice, 1945). The DSI was calculated for each MTL subregion. The consistency of volume measurements within one rater was assessed using intraclass correlation coefficients (ICC) using SPSS 22 (IBM SPSS Statistics for Macintosh, Version 22.0. Armonk, NY: IBM Corp.). The ICC variant that measured absolute agreement under a 2-way mixed ANOVA model was used (ICC(3), (Shrout and Fleiss, 1979)).
The agreement of both raters was assessed in terms of relative overlap using the DSI and was calculated as before. The consistency of volume measurements between both raters was assessed using the ICC. This time, the ICC variant that measured absolute agreement under a 2-way random ANOVA model was used (ICC(2), (Shrout and Fleiss, 1979)). Due to the low ICC values for area 35 in type II CS patterns in the first inter-rater reliability analysis, 8 additional hemispheres with type II CS were segmented by both raters, after sulcus depth measurements had been made more concrete in the protocol following careful evaluation of the mismatches encountered during the first round.
Average subregion volumes (mean and standard deviation) were calculated for the final 16 hemispheres for both raters (i.e. the 8 type I hemispheres and the 8 type II hemispheres from the second iteration after refinement of depth measurement).
3. Results
3.1. Reliability
3.1.1. Intra-rater reliability
Table 1 shows the intra-rater reliability of a single rater (A.H.) for 16 hemispheres (all from the first iteration). Almost all DSI values were above 0.9. Regions that were smaller and more complicated, such as CA2, CA3 and area 35, showed slightly lower values but were still over 0.85. ICCs were all above 0.95 with the exception of CA3, which was at 0.78 and may be explained by the more difficult but anatomically valid separation from DG along the endfolial pathway.
Table 1.
Intra-rater reliability of a single rater: dice similarity coefficient (DSI) and intraclass-correlation coefficient (ICC).
| DSI (mean ± SD) | ICC | |
|---|---|---|
| ErC | 0.91 ± 0.01 | 0.98 |
| Area 35 | 0.88 ± 0.02 | 0.97 |
| Area 36 | 0.91 ± 0.02 | 0.96 |
| PhC | 0.93 ± 0.03 | 0.99 |
| CA1 | 0.91 ± 0.02 | 0.98 |
| CA2 | 0.87 ± 0.05 | 0.97 |
| CA3 | 0.85 ± 0.03 | 0.78 |
| DG | 0.90 ± 0.02 | 0.98 |
| Sub | 0.92 ± 0.02 | 0.95 |
| Hippocampus total | 0.96 ± 0.01 | 0.97 |
ErC = entorhinal cortex, PhC = parahippocampal cortex, CA = cornu ammonis, DG = dentate gyrus, Sub = subiculum.
3.1.2. Inter-rater reliability
In the first iteration, almost all DSI values were above 0.84. The DSI for smaller, more complicated regions CA2, CA3 and area 35 was slightly lower, though still over 0.77. Similarly, ICCs were above 0.87 for almost all subregions. The ICCs for DG and subiculum were slightly lower though still above 0.76. However, the ICC for area 35 was 0.68, and 0.47 for CA3. Since this number was discrepant from the remaining values, all segmentations were checked to find out whether the rules for CA3 were unclear and therefore could not be reliably implemented. An error was found in one subject by one rater. In this subject, the number of head slices in which CA3 was segmented was miscounted. As the last head slice was correctly identified and implemented for other labels depending on the most posterior head slice, we therefore concluded that this was a counting error rather than misinterpretation of the image or lack of clarity in the segmentation protocol with regard to CA3. This error was corrected and the ICC increased to 0.78.
As an additional exploratory experiment, we performed reliability analyses of area 35 and 36 separately for the type I and type II sulcal variants. For area 36 the DSI was similar for the two types, 0.87 ± 0.02 for type I and 0.86 ± 0.04 for type II (see Table 2). The ICC for area 36 for type II was 0.99, higher than 0.84 for type I. For area 35, the DSI for type I was slightly higher than type II; 0.84 ± 0.06 vs. 0.78 ± 0.07, but the difference was more notable for ICC of 0.87 for type I and − 0.12 for type II. Although the ICC for area 35 is higher for type I as compared to type II, the absolute difference, or ‘measurement error’, between the two raters is similar for both sulcal pattern types (mean absolute difference: type I: 0.06 mL, type II: 0.07 mL) while the range of volumes for type II is only a third of the range of type I (range type I: 0.49–0.81 mL, type II: 0.59–0.72 mL). Thus, the absolute difference between the raters relative to the normal variation in the population (the range) for type II is larger than for type I. This is further illustrated in Bland-Altman plots in Supplementary Fig. 11. Additionally, these plots show that neither rater had a bias as the differences lie around 0.
Table 2.
Inter-rater reliability for ErC, area 35 and 36 in type 1 and type 2 CS patterns separately: dice similarity coefficient (DSI) and intraclass-correlation coefficient (ICC) for 8 hemispheres in each category.
| DSI (mean ± SD) | ICC | |||||
|---|---|---|---|---|---|---|
| Type I | Type II | Type IIa | Type I | Type II | Type IIa | |
| ErC | 0.88 ± 0.02 | 0.87 ± 0.03 | 0.86 ± 0.02 | 0.94 | 0.80 | 0.87 |
| Area 35 | 0.84 ± 0.06 | 0.78 ± 0.07 | 0.83 ± 0.04 | 0.87 | − 0.12 | 0.90 |
| Area 36 | 0.87 ± 0.02 | 0.86 ± 0.05 | 0.88 ± 0.03 | 0.84 | 0.99 | 0.88 |
ErC = entorhinal cortex.
Results from a second inter-rater reliability analysis following refinement of segmentation rules.
Further inspection of the segmentation of area 35 type II cases revealed that a difference in segmentation between the two raters mainly resulted from measuring the sulcal depth. A small difference in sulcal depth, as can be seen in Fig. 5, can lead to a different segmentation rule. We therefore refined the segmentation protocol with regard to the sulcal depth measurements (see Supplementary Fig. 3). Following this refinement of the protocol, ERC, area 35 and 36 were segmented in 8 new type II hemispheres by both raters. The results of the reliability analyses for area 35 and 36 for these new type II hemispheres are presented in Table 2. ICC and DSI for ErC were 0.86 and 0.87, for area 35 they were 0.83 and 0.9, and for area 36 0.88 for both.
As a result, the ICC and DSI for the combined type I and II cases improved to over 0.84 for all three regions.
Table 3 shows the final results of all subregions for the comparison of both raters (i.e. average of the 8 type I hemispheres and the 8 type II hemispheres from the second iteration).
Table 3.
Inter-rater reliability between two raters: dice similarity coefficient (DSI) and intraclass-correlation coefficient (ICC).
| DSI (mean ± SD) | ICC | |||
|---|---|---|---|---|
| ErC◊ | 0.87 ± 0.02 | 0.94 | ||
| Area 35◊ | 0.84 ± 0.05 | 0.87 | ||
| Area 36◊ | 0.87 ± 0.02 | 0.88 | ||
| PhC | 0.86 ± 0.12 | 0.94 | ||
| CA1 | 0.84 ± 0.04 | 0.89 | ||
| CA2 | 0.81 ± 0.06 | 0.92 | ||
| CA3 | 0.78 ± 0.05 | *0.79 ± 0.05 | 0.76 | *0.78 |
| DG | 0.86 ± 0.03 | 0.76 | ||
| Sub | 0.85 ± 0.04 | 0.78 | ||
| Hippocampus total | 0.94 ± 0.01 | 0.98 | ||
ErC = entorhinal cortex, PhC = parahippocampal cortex, CA = cornu ammonis, DG = dentate gyrus, Sub = subiculum.
◊These values contain 8 type 1 hemispheres from the first and 8 type 2 hemispheres from the second iteration following a refinement of the segmentation rules.
*These values correspond to the alternative heuristic rule (see Fig. 8).
In addition, we tested the inter-rater reliability of the heuristic rule for CA3 (see Fig. 8). ICC was 0.78, and average DSI was 0.79 indicating that the heuristic rule could be applied as reliably as using the endfolial pathway. We also compared the overlap between the two rules, therefore calculating an inter-rule DSI for all 32 hemispheres segmented by the two raters, which was 0.63 ± 0.09. While this value is relatively low, the upper limit for the “between-rule” DSI is given by the DSI values of the inter-rater reliability for the two different sets of rules, i.e. even with perfect agreement between the heuristic and anatomy-based rules, the DSI would still only be 0.79. Volume comparisons revealed that using the heuristic rule slightly underestimates CA3 (0.24 mL compared to 0.31 mL) and overestimates DG (1.0 mL compared to 0.93 mL) as compared to a separation at the endfolial pathway. This confirms that the heuristic rule is a good alternative to the anatomical landmark. That is, if divergent from the endfolial pathway, in the majority of the slices CA3 will be underestimated, similar to all current segmentation protocols for CA3.
3.2. Volumes in comparison to anatomy
Mean volumes across both raters are shown in Table 4. Due to very different segmentation schemes for PrC as well as PhC, we did not compare volumes of those regions to earlier studies (Insausti et al., 1998, Pruessner et al., 2002). Volumes of hippocampal subfields are compared to studies that used manual segmentation procedures at 7 T (Wisse et al., 2016c), automated approaches at 3 T (Iglesias et al., 2015, Yushkevich et al., 2015b) and histological techniques (Simić et al., 1997). The comparison of the mean volumes found in the current study with the other manually derived volumes from Wisse et al. (2016c) highlights the changes in the recent protocol and reflects the novel anatomical findings from Ding et al. While CA1 has less volume compared to earlier estimations, subiculum shows an increase in volume. This probably relates to the new rule of subiculum segmentation in the hippocampal head as well as the new boundary between CA1 and subiculum in the hippocampal body. On the other hand, CA3 has increased while DG has reduced volume compared to Wisse et al. (2016c). Again, this most probably highlights our new rule which follows the endfolial pathway to separate DG and CA3 more accurately. Using this approach the portion of CA3 that folds into the DG is also segmented as CA3, which should result in an increase in CA3 volume. Additionally, hippocampal subfield volumes obtained with the current protocol approximate those obtained from post mortem studies (Iglesias et al., 2015, Simić et al., 1997), especially for the subfields in the hippocampus proper.
Table 4.
Volumes of ErC, area 35, area 36, PhC and hippocampal subfields in mL.
| Current study | Simić et al. (1997)a | Iglesias et al. (2015)b | Yushkevich et al. (2015b)c | Wisse et al. (2016c)d | |
|---|---|---|---|---|---|
| ErC | 0.99 ± 0.2 | – | – | – | 0.53 |
| Area 35 | 0.64 ± 0.11 | – | – | – | – |
| Area 36 | 2.22 ± 0.39 | – | – | – | – |
| PhC | 0.58 ± 0.24 | – | – | – | – |
| Sub | 1.07 ± 0.16 | –e | 0.64f | 0.34 | 0.63 |
| CA1 | 0.82 ± 0.15 | 0.64 | 0.52 | 1.25 | 1.48 |
| CA2 | 0.07 ± 0.02 | – | – | 0.018 | 0.07 |
| CA3 | 0.17 ± 0.02 | – | – | 0.067 | 0.12 |
| CA2 & 3 | 0.24 | 0.14 | 0.18 | 0.085 | 0.19 |
| DG(&CA4) | 0.50 ± 0.09 | 0.31 | 0.46 | 0.76 | 0.80 |
| Hippocampus total | 3.16 ± 0.40 | 1.54 | 2.26 | 2.44 | 3.1 |
ErC = entorhinal cortex, PhC = parahippocampal cortex, CA = cornu ammonis, DG = dentate gyrus, Sub = subiculum. We provide standard deviations for the data from the current study.
Data derived from Table 2 in Simić et al. (1997), ‘Normal’.
Data derived from Table 3 in Iglesias et al. (2015), ‘Ex vivo atlas’.
Data derived from Table 6 in Yushkevich et al. (2015b), ‘ASHS’, mean of right and left side.
Data derived from Table 1 in Wisse et al. (2016c), ‘Manual segmentation’, mean of right and left side.
Not shown because only entails subiculum and prosubiculum.
Values for parasubiculum, presubiculum and subiculum were summed up from Table 3 in Iglesias et al. (2015).
4. Discussion
We have developed and tested a new protocol for manual segmentation of the entorhinal cortex, perirhinal cortex (distinguishing area 35 and 36), parahippocampal cortex, and hippocampus as well as its subfields including subiculum, CA1, CA2, CA3, and dentate gyrus, in-vivo at 0.44 × 0.44 mm in-plane resolution using 7 T MRI. We showed that our protocol had an intra-rater reliability ICC higher than 0.95, except CA3 (0.78) and DSI higher than 0.85 and an inter-rater reliability ICC higher than 0.76 and DSI higher than 0.81, except CA3 (> 0.78) for all regions in young adults. The strengths of the protocol are outlined as follows.
First, we leveraged recent developments in neuroanatomy. This has enabled us to incorporate more distinct rules than previously known. Chiefly, subdivisions in HH and HB (Ding and Van Hoesen, 2015), PrC (Ding and Van Hoesen, 2010) and PhC (Ding et al., 2016) have substantially extended earlier work as they provide more details on the order of appearance and location of the subregions and additionally provide information on between-subject variability in some of the regions. These findings were incorporated in the current segmentation protocol, e.g. the location of subregions in relation to different numbers of digitations in the hippocampal head or folding patterns of the collateral sulcus. Also note, that sectioning in one article (Ding and Van Hoesen, 2015) was done perpendicular to the long axis of the hippocampus making it more comparable to the commonly used T2-weighted images. The other two atlases (Ding and Van Hoesen, 2010, Ding et al., 2016) were based on histology data sectioned in a coronal plane. However, no histological data on extrahippocampal regions is currently available with slices perpendicular to the long axis of the hippocampus. Additionally, earlier protocols mostly collapsed across subregions of the PrC instead of differentiating between medial and lateral parts (Insausti et al., 1998, Pruessner et al., 2002) but see (Kivisaari et al., 2013, Yushkevich et al., 2015b). We have extended that framework by differentiating more specifically between area 35 and 36 using available data from neuroanatomy (Ding and Van Hoesen, 2010). Another example is the PhC, where studies have mostly included the posterior PhG up to CS across the whole length of the hippocampal tail because of lack of a well-established boundary (Pruessner et al., 2002, Yushkevich et al., 2015a). However, a recent histological atlas (Ding et al., 2016) disentangles the subdivisions of the posterior PhC. That is, the posterior PhC not only consists of areas TH and TL of the PhG, but also area TF of the fusiform gyrus. In addition, areas TL and TF extend further posteriorly than area TH, which in most cases disappears (replaced with ventral visual area V2) after the shallow parahippocampal-ligual sulcus (PhligS) appears. Although the newly identified PhligS would be anatomically the most valid landmark, it could not be reliably identified in all subjects. We observed that the anterior part of the CaS in close proximity to the PhligS can serve as a landmark, which can be distinguished reliably. The CaS is a rather conservative border and leads to an exclusion of a portion of posterior PhC. However, as this posterior portion also consists of visual area V2, its exclusion may benefit the study of parahippocampal function. This fine-tuning of the segmentation protocol with more detailed information on the borders and anatomical variability may further facilitate research on memory such as different memory pathways in the MTL (Ranganath and Ritchey, 2012, Reagh and Yassa, 2014, Das et al., 2015). Additionally, if the protocol is validated in older populations, it may facilitate research on aging and neurodegenerative diseases. For example, early stages of tau pathology in Alzheimer's Disease constitute especially in the transentorhinal region and the entorhinal cortex (Braak and Braak, 1991, Ding et al., 2009). The transentorhinal region as described by Braak and Braak corresponds roughly to area 35 in this protocol. Therefore, detailed measurements of these regions that closely follow the anatomy are critical to detect early disease effects in volume and regional thickness measures (Wolk et al., 2017, Xie et al., 2017).
A second strength of this protocol is that we acquired more fine-grained images using ultra-high resolution 7 T MRI, which enables a more detailed delineation of smaller structures. In particular, the delineation of hippocampal subfields in the head as well as the visual distinction between CA3 and DG in the body benefit from the higher resolution. As can be seen from Fig. 3 and Fig. 6, the appearance of hippocampal head and presence of subfields can change drastically from slice to slice. The thinner slices obtained at 7 T allow us to establish more precise segmentation rules for the hippocampal head – that is, determining the distance between the appearance of subfields in the order of 1 mm rather than the more frequently reported thickness of 2 mm. In addition, it likely also allows for a more reliable segmentation. This may add value when investigating diseases or cognitive functions for which the anterior portion of the hippocampus is proposed to be specifically important (Sahay and Hen, 2007, Poppenk et al., 2013). Additionally, we propose the use of the endfolial pathway, a white matter band aligned with the actual border of CA3, to separate CA3 from the DG in the hippocampal body in populations where it is visible. This accurate distinction of DG and CA3 may enable functional MRI studies to dissociate the contributions of DG and CA3, because they are assumed to be involved in different cognitive processes (Neunuebel and Knierim, 2014). Although some of our rules are still geometrical in nature, the rules follow neuroanatomy more closely and take between-subject variability into account where possible. Additionally, most rules are independent of the in-plane orientation of the MTL; that is, most boundaries are drawn perpendicular to the structure rather than being based on the image orientation.
Thirdly, the protocol provides more detailed instructions for replication than previous protocols in the literature. We have included comprehensive slice-by-slice plots of high-resolution images that show the application of the rules along the full longitudinal axis for the most prevalent sulcal patterns – a continuous type 1 as well as a discontinuous type II CS (Ding and Van Hoesen, 2010). In addition, we provide practical segmentation tips, a checklist for segmentation (Supplementary Fig. 10), schematic descriptions of the rules throughout the protocol, a decision tree for the segmentation of area 35 and examples of some difficult cases in the supplemental material. To further facilitate the understanding of our rules, we incorporated specific feedback from a workshop on our protocol in Magdeburg. One of our main aims was to understand the difficulties that novice raters encounter while learning to apply the protocol rules. During the workshop we identified the most common difficulties and adjusted the protocol accordingly. For example, it became clear that the frequent cross-referencing to certain anatomical structures (e.g. uncal apex) was difficult to follow. Therefore, we included recommendations in what order to approach segmentation (exemplified in Fig. 9).
The high intra-rater reliability showed that the protocol could be reliably applied, with DSI values higher than 0.85 and ICC values higher than 0.95 with the exception of 0.78 for CA3. The latter is probably due to the fact that we are using a more complex separation along the endfolial pathway, or using the heuristic rule. Analyses of the reliability between two raters showed that we were able to apply this protocol in a consistent manner, with almost all DSI values above 0.84 and almost all ICC values over 0.89. Even for smaller regions and for regions, such as area 35, for which the segmentation protocol is more difficult to accommodate anatomical variants, the ICC and DSI were reasonable (ICC over 0.68 for area 35 and over 0.76 for the other regions and DSI over 0.77), showing that these smaller and more complicated regions can be segmented with reasonable reliability. It should be noted that the ICC for the DG and CA3 were also slightly lower, which can be explained by the more complex separation as already discussed for the intra-rater reliability. The overall high DSI values are encouraging for the application of this protocol to functional MRI studies since spatial overlap is critical in this context.
In comparison to other studies, the ICC values and DSI values reported here are well within the range of previously reported reliability values (Prasad et al., 2004, Mueller et al., 2007, Yushkevich et al., 2010, Bonnici et al., 2012, Palombo et al., 2013, Winterburn et al., 2013, Goubran et al., 2014, Lee et al., 2014, de Flores et al., 2015). Although some other studies reported slightly higher values for CA1 (Yushkevich et al., 2010, Shing et al., 2011, Lee et al., 2014) or subiculum (Travis et al., 2014, de Flores et al., 2015), our protocol includes more specific rules and may be more complicated. Additionally, the reliability for small regions such as CA2 and CA3 are among the highest reported in the literature. The ICCs for ErC, area 36 and PhC are also well in the range of previously reported reliability values (Pruessner et al., 2002, Feczko et al., 2009). The ICC value for area 35 was below the reliability estimates of earlier protocols (combining area 35 and 36). When splitting up the group in the two types of sulcal patterns, it became clear that this lower ICC value for area 35 was mainly driven by the type II variant. Importantly, our aim to incorporate the findings from histological studies (Ding and Van Hoesen, 2010) and match anatomy as closely as possible resulted in a slightly more detailed protocol with segmentation rules dependent on sulcal depth measurements. Small differences in sulcal depth measurement could result in different segmentation rules especially in the type II variant. After initial evaluation of the results, we therefore refined the guidelines for sulcal depth measurements (see Supplementary Fig. 3). A second reliability test in eight new type II hemispheres revealed an improved DSI of 0.83 and ICC of 0.90 which is similar compared to type I hemispheres. Although a learning effect could have affected the reliability measures, it seems unlikely that this played a large role as both raters had already segmented 40 subjects before the initial reliability test and this second reliability test was performed in 8 new hemispheres. These results indicate that with the refined segmentation protocol also a challenging region such as area 35 in the type II variant can be segmented reliably.
There are also limitations to the current study. First, by focusing specifically on anatomical validity and accounting for anatomical variability as much as possible, the resulting protocol is more elaborate and time-consuming compared to earlier approaches. However, we made considerable efforts to explain the protocol and make it understandable to novice raters. Additionally, we are planning to incorporate this segmentation protocol into the ASHS automatic MTL segmentation framework (Yushkevich et al., 2015b). A second limitation is that although we tried to match anatomy as closely as possible, for some of the borders we still use heuristic rules to improve reliability of the protocol. This means that portions of subregions may be included in the labels of adjacent subregions. Third, our protocol was mainly based on work from Ding et al. and might not be in agreement with work from other neuroanatomists. However, our protocol is largely consistent with other neuroanatomical references (Insausti et al., 1998, Duvernoy et al., 2013, Mai et al., 2015), and the volumes as obtained by the current protocol approximate the volumes from post mortem studies, as shown in Table 4 (Simić et al., 1997, Iglesias et al., 2015). The volume of the subiculum is slightly larger as compared to Iglesias et al., which may be due to the difference in age between the current study and the post mortem studies as subiculum volume is suggested to be affected by age (La Joie et al., 2010). It should be noted though that differences exist between these and other references in terms of nomenclature, for example the existence of the ‘prosubiculum’ and perhaps also in the placement of certain boundaries. Fourth, although we embrace the possibilities provided by higher resolution, we are fully aware that not all researchers have access to 7 T, which may limit the applicability of the current protocol. In order to facilitate application to 3 T protocols, we provided all segmentation rules in millimeters and have included heuristic rules to guide segmentation of CA3 and DG when the anatomical landmark, the endfolial pathway, is not visible. Indeed we are currently trying out this segmentation protocol in a set of older adults and patients with MCI for whom a high resolution 0.4 × 0.4 × 1.2 mm3 T2-weighted 3 T MRI was obtained. Of note, although most studies on MTL subregions are using 3 T imaging protocols, 7 T might play a more prominent role in the future with an increasing number of sites with access to a 7 T scanner. For example, the European Ultrahigh-Field Imaging Network for Neurodegenerative Diseases (EUFIND) was founded recently with the aim to summarize and investigate the potential of ultrahigh-field imaging in neurodegenerative research (http://www.neurodegenerationresearch.eu/initiatives/annual-calls-for-proposals/closed-calls/brain-imaging-working-groups-2016/brain-imaging-working-groups/). Finally, the distance between appearance of the different subfields is given in millimeters, although lengths of the MTL and hippocampus differ between individuals and might be affected by disease. It is unclear how this affects subfield measurements in the current protocol; a limitation true for all current segmentation protocols. Although a potential solution could be to provide relative distances rather than absolute distances between subregions, based on the total length of the MTL; this would further complicate the protocol. Additionally, the relative distance between subfields is not necessarily similar between subjects nor are they similarly affected by disease. Using this measure would therefore inherently also induce a measurement error.
The current protocol is not meant to replace the protocol of the harmonization effort for hippocampal subfields (www.hippocampalsubfields.com) or hamper the progress of this collaborative effort of many groups in various disciplines aiming to harmonize all the different protocols for hippocampal and parahippocampal subregions (Wisse et al., 2016a). Due to the iterative and thorough nature of the harmonization effort, the timeline for development of protocols for parahippocampal subregions and 7 T images are further down the road and the current protocol is therefore meant to facilitate the segmentation of MTL regions, and especially parahippocampal subregions, for centers with a 7 T scanner in the meantime.
In summary, we present a protocol to delineate medial temporal lobe structures as well as hippocampal subfields and provide evidence that it can be reliably applied. The inclusion of the most recent anatomical literature guiding the detailed subdivision of MTL regions and hippocampal subfields will make this an especially useful protocol for the investigation of the functional role of MTL subregions using fMRI, as well as research on the effect of exercise on MTL subregions and their differential relation with depression, autism, aging and neurodegenerative diseases.
Disclosures
D.A.W. has received consulting fees from Merck, Janssen, and Piramal and grant funding from Avid/Lilly, Merck, and Biogen. L.X. was a summer intern at Siemens Healthcare, Princeton from May 2016 to August 2016. None of the other authors has anything to disclose.
Acknowledgments
We would like to thank the participants of the segmentation workshop on MTL subregions for valuable feedback and insights on the applicability of our segmentation rules.
This work was supported by the National Institute on Aging grants: P30 AG010124 and R01 AG037376; National Institute of Biomedical Imaging and Bioengineering grants: R01 EB017255 and R03 EB016923; Grant # 10037893, Institute for Translational Medicine and Therapeutics Translational Bioimaging Center, University of Pennsylvania; and the donors of Alzheimer's Disease Research, a program of the BrightFocus Foundation (L.E.M.W.). P.V. and T.W. are supported by European Research Council Starting Investigator Grant AGESPACE (335090). E.D. is supported by the German Research Foundation Collaborative Research Network SFB 776 (TP A07). E.D. and D.B. are part of the JPND working group EUFIND.
Footnotes
Supplementary material to this article can be found online at http://dx.doi.org/10.1016/j.nicl.2017.05.022.
Contributor Information
D. Berron, Email: david.berron@med.ovgu.de.
P. Vieweg, Email: paula.vieweg@dzne.de.
Appendix A. Supplementary material
Supplementary material
References
- Aly M., Ranganath C., Yonelinas A.P. Detecting changes in scenes: the hippocampus is critical for strength-based perception. Neuron. 2013;78:1127–1137. doi: 10.1016/j.neuron.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminoff E.M., Kveraga K., Bar M. The role of the parahippocampal cortex in cognition. Trends Cogn. Sci. 2013;17:379–390. doi: 10.1016/j.tics.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova L.G. Relationship between hippocampal atrophy and neuropathology markers: a 7T MRI validation study of the EADC-ADNI harmonized hippocampal segmentation protocol. Alzheimers Dement. 2015;11:139–150. doi: 10.1016/j.jalz.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S., Vieweg P., Gao F., Gilboa A., Wolbers T., Black S.E., Rosenbaum R.S. The human dentate gyrus plays a necessary role in discriminating new memories. Curr. Biol. 2016;26:2629–2634. doi: 10.1016/j.cub.2016.07.081. [DOI] [PubMed] [Google Scholar]
- Bernasconi N., Bernasconi A., Caramanos Z., Antel S.B., Andermann F., Arnold D.L. Mesial temporal damage in temporal lobe epilepsy: a volumetric MRI study of the hippocampus, amygdala and parahippocampal region. Brain. 2003;126:462–469. doi: 10.1093/brain/awg034. [DOI] [PubMed] [Google Scholar]
- Berron D., Schütze H., Maass A., Cardenas-Blanco A., Kuijf H.J., Kumaran D., Düzel E. Strong evidence for pattern separation in human dentate gyrus. J. Neurosci. 2016;36:7569–7579. doi: 10.1523/JNEUROSCI.0518-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnici H.M., Chadwick M.J., Kumaran D., Hassabis D., Weiskopf N., Maguire E.A. Multi-voxel pattern analysis in human hippocampal subfields. Front. Hum. Neurosci. 2012;6:290. doi: 10.3389/fnhum.2012.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet C., Chupin M., Lehéricy S., Marrakchi-Kacem L., Epelbaum S., Poupon C., Wiggins C.J., Vignaud A., Hasboun D., Defontaines B., Hanon O., Dubois B., Sarazin M., Hertz-Pannier L., Colliot O. Detection of volume loss in hippocampal layers in Alzheimer's disease using 7 T MRI: a feasibility study. NeuroImage Clin. 2014;5:341–348. doi: 10.1016/j.nicl.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H., Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Bronen R.A., Cheung G. Relationship of hippocampus and amygdala to coronal MRI landmarks. Magn. Reson. Imaging. 1991;9:449–457. doi: 10.1016/0730-725x(91)90434-n. [DOI] [PubMed] [Google Scholar]
- Chadwick M.J., Bonnici H.M., Maguire E.A. CA3 size predicts the precision of memory recall. Proc. Natl. Acad. Sci. U. S. A. 2014;2014:10720–10725. doi: 10.1073/pnas.1319641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.R., Pluta J.B., Mancuso L., Kliot D., Yushkevich P.A., Wolk D.A. Anterior and posterior MTL networks in aging and MCI. Neurobiol. Aging. 2015;36:141–150. doi: 10.1016/j.neurobiolaging.2014.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty A.M., Bender A.R., Raz N., Ofen N. Age differences in hippocampal subfield volumes from childhood to late adulthood. Hippocampus. 2015 doi: 10.1002/hipo.22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice L.R. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- Dickerson B.C., Salat D.H., Bates J.F., Atiya M., Killiany R.J., Greve D.N., Dale A.M., Stern C.E., Blacker D., Albert M.S., Sperling R.A. Vol. 56. 2004. Medial Temporal Lobe Function and Structure in Mild Cognitive Impairment; pp. 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.-L. Comparative anatomy of the prosubiculum, subiculum, presubiculum, postsubiculum, and parasubiculum in human, monkey, and rodent. J. Comp. Neurol. 2013;521:4145–4162. doi: 10.1002/cne.23416. [DOI] [PubMed] [Google Scholar]
- Ding S.-L., Van Hoesen G.W. Borders, extent, and topography of human perirhinal cortex as revealed using multiple modern neuroanatomical and pathological markers. Hum. Brain Mapp. 2010;31:1359–1379. doi: 10.1002/hbm.20940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.-L., Van Hoesen G.W. Organization and detailed parcellation of human hippocampal head and body regions based on a combined analysis of cyto- and chemo-architecture. J. Comp. Neurol. 2015;2253:2233–2253. doi: 10.1002/cne.23786. [DOI] [PubMed] [Google Scholar]
- Ding S.-L., Van Hoesen G.W., Cassell M.D., Poremba A. Parcellation of human temporal polar cortex: a combined analysis of multiple cytoarchitectonic, chemoarchitectonic, and pathological markers. J. Comp. Neurol. 2009;514:595–623. doi: 10.1002/cne.22053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.-L. Comprehensive cellular-resolution atlas of the adult human brain. J. Comp. Neurol. 2016;524:3127–3481. doi: 10.1002/cne.24080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du A.-T., Schuff N., Kramer J.H., Rosen H.J., Maria G.-T., Rankin K., Miller B.L., Weiner M.W. Vol. 130. 2007. Different Regional Patterns of Cortical Thinning in Alzheimer's Disease and Frontotemporal Dementia; pp. 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K., Tompary A., Davachi L. Associative encoding and retrieval are predicted by functional connectivity in distinct hippocampal area CA1 pathways. J. Neurosci. 2014;34:11188–11198. doi: 10.1523/JNEUROSCI.0521-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvernoy H.M., Cattin F., Risold P.-Y., Vannson J.L., Gaudron M. Springer; 2013. The Human Hippocampus: Functional Anatomy, Vascularization, and Serial Sections with MRI. [Google Scholar]
- Eichenbaum H., Yonelinas A.P., Ranganath C. The medial temporal lobe and recognition memory. Annu. Rev. Neurosci. 2007;30:123–152. doi: 10.1146/annurev.neuro.30.051606.094328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrom A.D., Kahana M.J., Caplan J.B., Fields T.A., Isham E.A., Newman E.L., Fried I. Cellular networks underlying human spatial navigation. Nature. 2003;425:184–188. doi: 10.1038/nature01964. [DOI] [PubMed] [Google Scholar]
- Ekstrom A.D., Bazih A.J., Suthana N.A., Al-Hakim R., Ogura K., Zeineh M.M., Burggren A.C., Bookheimer S.Y. Advances in high-resolution imaging and computational unfolding of the human hippocampus. NeuroImage. 2009;47:42–49. doi: 10.1016/j.neuroimage.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein R.A. Parahippocampal and retrosplenial contributions to human spatial navigation. Trends Cogn. Sci. 2008;12:388–396. doi: 10.1016/j.tics.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feczko E., Augustinack J.C., Fischl B., Dickerson B.C. An MRI-based method for measuring volume, thickness and surface area of entorhinal, perirhinal, and posterior parahippocampal cortex. Neurobiol. Aging. 2009;30:420–431. doi: 10.1016/j.neurobiolaging.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Flores R., Joie R., Landeau B., Perrotin A., Mézenge F., de Sayette V., Eustache F., Desgranges B., Chételat G. Vol. 36. 2015. Effects of Age and Alzheimer's Disease on Hippocampal Subfields: Comparison Between Manual and FreeSurfer Volumetry; pp. 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni G.B. The EADC-ADNI harmonized protocol for manual hippocampal segmentation on magnetic resonance: evidence of validity. Alzheimers Dement. 2015;11:111–125. doi: 10.1016/j.jalz.2014.05.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubran M., Rudko D.A., Santyr B., Gati J., Szekeres T., Peters T.M., Khan A.R. Vol. 35. 2014. In vivo normative atlas of the hippocampal subfields using multi-echo susceptibility imaging at 7 tesla; pp. 3588–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.S., Barense M.D., Lee A.C.H. Going beyond LTM in the MTL: a synthesis of neuropsychological and neuroimaging findings on the role of the medial temporal lobe in memory and perception. Neuropsychologia. 2010;48:831–853. doi: 10.1016/j.neuropsychologia.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Huang Y., Coupland N.J., Lebel R.M., Carter R., Seres P., Wilman A.H., Malykhin N.V. Structural changes in hippocampal subfields in major depressive disorder: a high-field magnetic resonance imaging study. Biol. Psychiatry. 2013;74:62–68. doi: 10.1016/j.biopsych.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Iglesias J.E., Augustinack J.C., Nguyen K., Player C.M., Player A., Wright M., Roy N., Frosch M.P., McKee A.C., Wald L.L., Fischl B., Van Leemput K. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: application to adaptive segmentation of in vivo MRI. NeuroImage. 2015;115:117–137. doi: 10.1016/j.neuroimage.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inhoff M.C., Ranganath C. Significance of objects in the perirhinal cortex. Trends Cogn. Sci. 2015;19:302–303. doi: 10.1016/j.tics.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Insausti R., Amaral D.G. Hippocampal formation. In: Mai J.K., Paxinos G., editors. The Human Nervous System. Elsevier Academic Press; 2012. pp. 896–942. [Google Scholar]
- Insausti R., Juottonen K., Soininen H., Insausti A.M., Partanen K., Vainio P., Laakso M.P., Pitkänen A. MR volumetric analysis of the human entorhinal, perirhinal, and temporopolar cortices. Am. J. Neuroradiol. 1998;19:659–671. [PMC free article] [PubMed] [Google Scholar]
- Kerchner G.A., Deutsch G.K., Zeineh M.M., Dougherty R.F., Saranathan M., Rutt B.K. Hippocampal CA1 apical neuropil atrophy and memory performance in Alzheimer's disease. NeuroImage. 2012;63:194–202. doi: 10.1016/j.neuroimage.2012.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisaari S.L., Probst A., Taylor K.I. In: The perirhinal, entorhinal, and parahippocampal cortices and hippocampus: an overview of functional anatomy and protocol for their segmentation in mr images. fMRI, editor. Springer Berlin Heidelberg; Berlin, Heidelberg: 2013. pp. 239–267. [Google Scholar]
- La Joie R., Fouquet M., Mézenge F., Landeau B., Villain N., Mevel K., Pélerin A., Eustache F., Desgranges B., Chételat G. Differential effect of age on hippocampal subfields assessed using a new high-resolution 3T MR sequence. NeuroImage. 2010;53:506–514. doi: 10.1016/j.neuroimage.2010.06.024. [DOI] [PubMed] [Google Scholar]
- La Joie R., Perrotin A., De La Sayette V., Egret S., Doeuvre L., Belliard S., Eustache F., Desgranges B., Chételat G. Hippocampal subfield volumetry in mild cognitive impairment, Alzheimer's disease and semantic dementia. NeuroImage Clin. 2013;3:155–162. doi: 10.1016/j.nicl.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.C.H., Buckley M.J., Pegman S.J., Spiers H.J., Scahill V.L., Gaffan D., Bussey T.J., Davies R.R., Kapur N., Hodges J.R., Graham K.S. Specialization in the medial temporal lobe for processing of objects and scenes. Hippocampus. 2005;15:782–797. doi: 10.1002/hipo.20101. [DOI] [PubMed] [Google Scholar]
- Lee J.K., Ekstrom A.D., Ghetti S. Volume of hippocampal subfields and episodic memory in childhood and adolescence. NeuroImage. 2014;94:162–171. doi: 10.1016/j.neuroimage.2014.03.019. [DOI] [PubMed] [Google Scholar]
- Lim C., Mufson E.J., Kordower J.H., Blume H.W., Madsen J.R., Saper C.B. Connections of the hippocampal formation in humans: II. The endfolial fiber pathway. J. Comp. Neurol. 1997;385 [PubMed] [Google Scholar]
- Maass A., Schütze H., Speck O., Yonelinas A.P., Tempelmann C., Heinze H.-J., Berron D., Cardenas-Blanco A., Brodersen K.H., Enno Stephan K., Düzel E. Laminar activity in the hippocampus and entorhinal cortex related to novelty and episodic encoding. Nat. Commun. 2014;5:5547. doi: 10.1038/ncomms6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass A., Berron D., Libby L.A., Ranganath C., Düzel E. Functional subregions of the human entorhinal cortex. elife. 2015;4:1–20. doi: 10.7554/eLife.06426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai J.K., Majtanik M., Paxinos G. Elsevier; Oxford: 2015. Atlas of the Human Brain. [Google Scholar]
- Malykhin N.V., Lebel R.M., Coupland N.J., Wilman A.H., Carter R. In vivo quantification of hippocampal subfields using 4.7 T fast spin echo imaging. NeuroImage. 2010;49:1224–1230. doi: 10.1016/j.neuroimage.2009.09.042. [DOI] [PubMed] [Google Scholar]
- Mueller S.G., Stables L., Du A.T., Schuff N., Truran D., Cashdollar N., Weiner M.W. Measurement of hippocampal subfields and age-related changes with high resolution MRI at 4 T. Neurobiol. Aging. 2007;28:719–726. doi: 10.1016/j.neurobiolaging.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neunuebel J.P., Knierim J.J. CA3 retrieves coherent representations from degraded input: direct evidence for CA3 pattern completion and dentate gyrus pattern separation. Neuron. 2014;81:416–427. doi: 10.1016/j.neuron.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R.K., Palombo D.J., Rabin J.S., Levine B., Ryan J.D., Rosenbaum R.S. Volumetric analysis of medial temporal lobe subregions in developmental amnesia using high-resolution magnetic resonance imaging. Hippocampus. 2013;23:855–860. doi: 10.1002/hipo.22153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombo D.J., Amaral R.S.C., Olsen R.K., Muller D.J., Todd R.M., Anderson A.K., Levine B. KIBRA polymorphism is associated with individual differences in hippocampal Subregions: evidence from anatomical segmentation using high-resolution MRI. J. Neurosci. 2013;33:13088–13093. doi: 10.1523/JNEUROSCI.1406-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh M.B., Rutt B.K., Purcell R., Chen Y., Zeineh M.M. Ultra-high resolution in-vivo 7.0T structural imaging of the human hippocampus reveals the endfolial pathway. NeuroImage. 2015;112:1–6. doi: 10.1016/j.neuroimage.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenk J.L., Evensmoen H.R., Moscovitch M., Nadel L. Long-axis specialization of the human hippocampus. Trends Cogn. Sci. 2013;17:230–240. doi: 10.1016/j.tics.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Prasad K.M.R., Patel A.R., Muddasani S., Sweeney J., Keshavan M.S. The entorhinal cortex in first-episode psychotic disorders: a structural magnetic resonance imaging study. Am. J. Psychiatry. 2004;161:1612–1619. doi: 10.1176/appi.ajp.161.9.1612. [DOI] [PubMed] [Google Scholar]
- Preston A.R., Bornstein A.M., Hutchinson J.B., Gaare M.E., Glover G.H., Wagner A.D. High-resolution fMRI of content-sensitive subsequent memory responses in human medial temporal lobe. J. Cogn. Neurosci. 2010;22:156–173. doi: 10.1162/jocn.2009.21195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruessner J.C., Köhler S., Crane J., Pruessner M., Lord C., Byrne A., Kabani N., Collins D.L., Evans A.C. Volumetry of temporopolar, perirhinal, entorhinal and parahippocampal cortex from high-resolution MR images: considering the variability of the collateral sulcus. Cereb. Cortex. 2002;12:1342–1353. doi: 10.1093/cercor/12.12.1342. [DOI] [PubMed] [Google Scholar]
- Ranganath C., Ritchey M. Two cortical systems for memory-guided behaviour. Nat. Rev. Neurosci. 2012;13:713–726. doi: 10.1038/nrn3338. [DOI] [PubMed] [Google Scholar]
- Reagh Z.M., Yassa M.A. Object and spatial mnemonic interference differentially engage lateral and medial entorhinal cortex in humans. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4264–E4273. doi: 10.1073/pnas.1411250111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinek H., Endo Y., Santi D.S., Frid D., Tsui -H.W., Segal S., Convit A., de Leon M.J. Vol. 63. 2004. Atrophy Rate in Medial Temporal Lobe During Progression of Alzheimer Disease; pp. 2354–2359. [DOI] [PubMed] [Google Scholar]
- Sahay A., Hen R. Adult hippocampal neurogenesis in depression. Nat. Neurosci. 2007;10:1110–1115. doi: 10.1038/nn1969. [DOI] [PubMed] [Google Scholar]
- Schlichting M.L., Zeithamova D., Preston A.R. CA1 subfield contributions to memory integration and inference. Hippocampus. 2014;24:1248–1260. doi: 10.1002/hipo.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shing Y.L., Rodrigue K.M., Kennedy K.M., Fandakova Y., Bodammer N.C., Werkle-Bergner M., Lindenberger U., Raz N. Hippocampal subfield volumes: age, vascular risk, and correlation with associative memory. Front. Aging Neurosci. 2011;3:2. doi: 10.3389/fnagi.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout P., Fleiss J. Vol. 86. 1979. Intraclass correlations: uses in assessing rater reliability; pp. 420–428. [DOI] [PubMed] [Google Scholar]
- Simić G., Kostović I., Winblad B., Bogdanović N. Volume and number of neurons of the human hippocampal formation in normal aging and Alzheimer's disease. J. Comp. Neurol. 1997;379:482–494. doi: 10.1002/(sici)1096-9861(19970324)379:4<482::aid-cne2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Suthana N.A., Donix M., Wozny D.R., Bazih A.J., Jones M., Heidemann R.M., Trampel R., Ekstrom A.D., Scharf M., Knowlton B.J., Turner R., Bookheimer S.Y. High-resolution 7T fMRI of human hippocampal subfields during associative learning. J. Cogn. Neurosci. 2015;27:1194–1206. doi: 10.1162/jocn_a_00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis S.G., Huang Y., Fujiwara E., Radomski A., Olsen F., Carter R., Seres P., Malykhin N.V. High field structural MRI reveals specific episodic memory correlates in the subfields of the hippocampus. Neuropsychologia. 2014;53:233–245. doi: 10.1016/j.neuropsychologia.2013.11.016. [DOI] [PubMed] [Google Scholar]
- van Veluw S.J., Wisse L.E.M., Kuijf H.J., Spliet W.G.M., Hendrikse J., Luijten P.R., Geerlings M.I., Biessels G.J. Hippocampal T2 hyperintensities on 7 tesla MRI. NeuroImage Clin. 2013;3:196–201. doi: 10.1016/j.nicl.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Neylan T.C., Mueller S.G., Lenoci M., Truran D., Marmar C.R., Weiner M.W., Schuff N. Magnetic resonance imaging of hippocampal subfields in posttraumatic stress disorder. Arch. Gen. Psychiatry. 2010;67:296. doi: 10.1001/archgenpsychiatry.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterburn J.L., Pruessner J.C., Chavez S., Schira M.M., Lobaugh N.J., Voineskos A.N., Chakravarty M.M. A novel in vivo atlas of human hippocampal subfields using high-resolution 3 T magnetic resonance imaging. NeuroImage. 2013;74:254–265. doi: 10.1016/j.neuroimage.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Wisse L.E.M., Gerritsen L., Zwanenburg J.J.M., Kuijf H.J., Luijten P.R., Biessels G.J., Geerlings M.I. Subfields of the hippocampal formation at 7 T MRI: in vivo volumetric assessment. NeuroImage. 2012;61:1043–1049. doi: 10.1016/j.neuroimage.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Wisse L.E.M. A harmonized segmentation protocol for hippocampal and parahippocampal subregions: why do we need one and what are the key goals? Hippocampus. 2016;27:3–11. doi: 10.1002/hipo.22671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse L.E.M., Adler D.H., Ittyerah R., Pluta J.B., Robinson J.L., Schuck T., Trojanowski J.Q., Grossman M., Detre J.A., Elliott M.A., Toledo J.B., Liu W., Pickup S., Das S.R., Wolk D.A., Yushkevich P.A. Comparison of in vivo and ex vivo MRI of the human hippocampal formation in the same subjects. Cereb. Cortex. 2016:1–12. doi: 10.1093/cercor/bhw299. (Epub) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse L.E.M., Kuijf H.J., Honingh A.M., Wang H., Pluta J.B., Das S.R., Wolk D.A., Zwanenburg J.J.M., Yushkevich P.A., Geerlings M.I. Automated hippocampal subfield segmentation at 7 T MRI. Am. J. Neuroradiol. 2016;37:1050–1057. doi: 10.3174/ajnr.A4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbers T., Büchel C. Dissociable Retrosplenial and hippocampal contributions to successful formation of survey representations. J. Neurosci. 2005;25:3333–3340. doi: 10.1523/JNEUROSCI.4705-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk D.A., Das S.R., Mueller S.G., Weiner M.W., Yushkevich P.A. Medial temporal lobe subregional morphometry using high resolution MRI in Alzheimer's disease. Neurobiol. Aging. 2017;49:204–213. doi: 10.1016/j.neurobiolaging.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Wisse L.E.M., Das S.R., Wang H., Wolk D.A., Manjón J.V., Yushkevich P.A. 2016. Accounting for the Confound of Meninges in Segmenting Entorhinal and Perirhinal Cortices in T1-Weighted MRI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Pluta J.B., Das S.R., Wisse L.E.M., Wang H., Mancuso L., Kliot D., Avants B.B., Ding S.-L., Manjón J.V., Wolk D.A., Yushkevich P.A. Multi-template analysis of human perirhinal cortex in brain MRI: explicitly accounting for anatomical variability. NeuroImage. 2017;144:183–202. doi: 10.1016/j.neuroimage.2016.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich P.A., Piven J., Hazlett H.C., Smith R.G., Ho S., Gee J.C., Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Yushkevich P.A., Wang H., Pluta J.B., Das S.R., Craige C., Avants B.B., Weiner M.W., Mueller S.G. Nearly automatic segmentation of hippocampal subfields in in vivo focal T2-weighted MRI. NeuroImage. 2010;53:1208–1224. doi: 10.1016/j.neuroimage.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich P.A. Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: towards a harmonized segmentation protocol. NeuroImage. 2015;111:526–541. doi: 10.1016/j.neuroimage.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich P.A., Pluta J.B., Wang H., Xie L., Ding S.-L., Gertje E.C., Mancuso L., Kliot D., Das S.R., Wolk D.A. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum. Brain Mapp. 2015;36:258–287. doi: 10.1002/hbm.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineh M.M., Engel S.A., Thompson P.M., Bookheimer S.Y. Vol. 265. 2001. Unfolding the human hippocampus with high resolution structural and functional MRI; pp. 111–120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material