Highlights
-
•
Role of the RNA polymerase in the cystoviral life-cycle.
-
•
Spatio-temporal regulation of RNA synthesis in cystoviruses.
-
•
Emerging role of conformational dynamics in polymerase function.
Abbreviations: ATP, adenosine triphosphate; BCS, binding-competent state; BOS, binding-occluded state; BVDV, bovine viral diarrhea virus; CTD, C-terminal domain; DdRP, DNA-directed RNA polymerase; DdDP, DNA-directed DNA polymerase; dsRNA, double-stranded RNA; DTD, dwell-time distribution; FMDV, foot-and-mouth disease virus; GTP, guanosine triphosphate; HCV, hepatitis C virus; HFC, high-fidelity conformation; HIV, human immunodeficiency virus; LFC, low-fidelity conformation; MD, molecular dynamics; NMR, nuclear magnetic resonance; NTP, nucleoside triphosphate; NTPase, nucleoside triphosphatase; PC, procapsid; PX, polymerase complex; RdRP, RNA-directed RNA polymerase; RT, reverse transcriptase; ssRNA, single-stranded RNA; TMC, terminal-base mismatched conformation; TNTase, terminal nucleotidyltransferase
Keywords: Bacteriophage, Cystovirus, Transcription, Replication, RNA-directed RNA polymerase
Abstract
P2, an RNA-directed RNA polymerase (RdRP), is encoded on the largest of the three segments of the double-stranded RNA genome of cystoviruses. P2 performs the dual tasks of replication and transcription de novo on single-stranded RNA templates, and plays a critical role in the viral life-cycle. Work over the last few decades has yielded a wealth of biochemical and structural information on the functional regulation of P2, on its role in the spatiotemporal regulation of RNA synthesis and its variability across the Cystoviridae family. These range from atomic resolution snapshots of P2 trapped in functionally significant states, in complex with catalytic/structural metal ions, polynucleotide templates and substrate nucleoside triphosphates, to P2 in the context of viral capsids providing structural insight into the assembly of supramolecular complexes and regulatory interactions therein. They include in vitro biochemical studies using P2 purified to homogeneity and in vivo studies utilizing infectious core particles. Recent advances in experimental techniques have also allowed access to the temporal dimension and enabled the characterization of dynamics of P2 on the sub-nanosecond to millisecond timescale through measurements of nuclear spin relaxation in solution and single molecule studies of transcription from seconds to minutes. Below we summarize the most significant results that provide critical insight into the role of P2 in regulating RNA synthesis in cystoviruses.
1. Introduction
The Cystoviridae (Φ6 − Φ14, Φ2954 and ΦNN) comprise a family of lipid-enveloped viruses that contain double-stranded RNA (dsRNA) genomes. All members of this family exclusively infect Gram-negative bacteria, notably many strains of the plant pathogen Pseudomonas syringae. Some members of the family are also capable of targeting Escherichia coli or Salmonella (Mindich et al., 1999). The archetypal cystovirus, Φ6, was first isolated from bean straw infested with Pseudomonas syringae pv. phaseolicola in 1973 by Vidaver et al. (Vidaver et al., 1973), following which, eight additional members, Φ7-14, were identified and characterized by Mindich et al. (Mindich et al., 1999). In addition, Φ2954 that showed significant similarities to Φ12 was isolated and characterized by Qiao et al. (2010b). While all members (though not officially classified as such by the International Committee on Taxonomy of Viruses; Φ6 remains the only official member) of the Cystoviridae family share similar overall genetic organization and structural morphology, the level of sequence conservation within their genomes and encoded proteins, are weak at best. Based on the phylogenetic closeness to Φ6, the first discovered and most extensively studied cystovirus, the family may be broadly classified into two sub-groups. The first sub-group, comprising Φ7, Φ9, Φ10 and Φ11, are closely related to Φ6, while the second sub-group comprising, Φ8, Φ12, Φ13, Φ14 and Φ2954 are more distant from Φ6 (Mindich et al., 1999, Qiao et al., 2010b). Recently, a new member of the Cystoviridae family ΦNN with strong genomic similarity to Φ6 was isolated from a Finnish lake (Mäntynen et al., 2015).
Φ6 has been a key player in the development of many aspects of molecular virology including elucidation of detailed mechanisms of capsid assembly (Poranen et al., 2001, Poranen and Tuma, 2004, Poranen et al., 2005, Poranen and Bamford, 2012) and genome packaging (Frilander and Bamford, 1995, Qiao et al., 1997b, Mindich, 1999a, Mindich, 2004, Mindich, 2012). It was also the first RNA virus for which reverse genetics was developed (Olkkonen et al., 1990, Mindich, 1999b). Additionally, Φ6 is still considered to be an ideal model system to study the mechanisms of replication in RNA viruses, most notably the de novo initiation (see below) of viral RNA synthesis (Butcher et al., 2001, Laurila et al., 2002). Since Φ6 remains the prototypical and most thoroughly investigated cystovirus in terms of structure, biochemistry and function, we will use it as our point of reference for most discussions in this review, occasionally referring to other cystoviruses wherever additional information is available.
2. Organization of the cystovirus virion
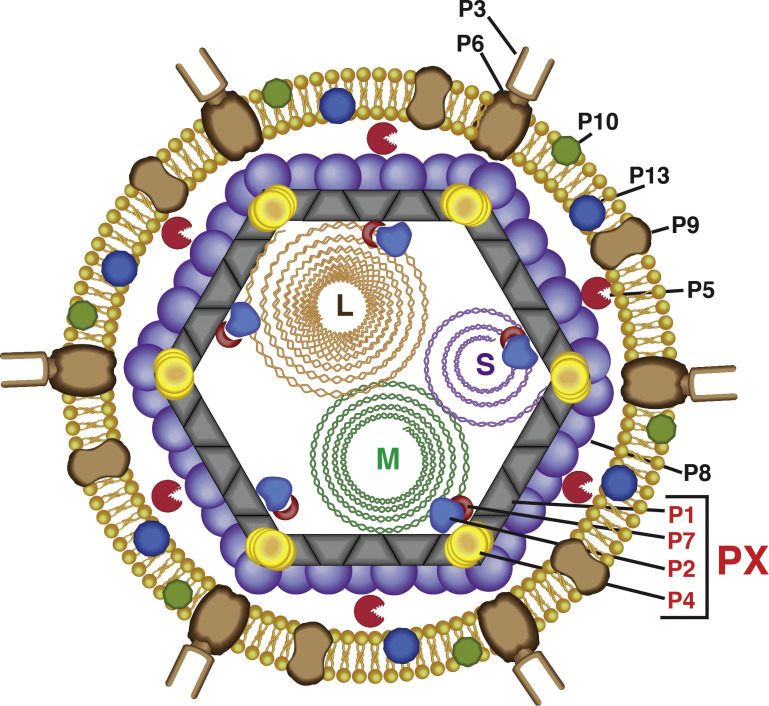
All cystoviruses possess similar overall organization with three structural layers (Fig. 1 ). The outermost layer that is comprised of a combination of phospholipids, extracted from the host (Laurinavičius et al., 2004), houses several integral membrane proteins (P9, P10 and P13) (Gottlieb et al., 1988). This layer also contains the receptor binding spike protein P3, that is critical for interactions with the host and P6, another membrane protein, that mediates fusion of the viral envelope with the bacteria outer membrane (Bamford et al., 1987). Located inside this membrane vesicle, is the first of two protein layers, that in Φ6, is formed by 200 trimers of the capsid protein P8 that assembles on a T = 13 lattice (Butcher et al., 1997, Poranen et al., 2001). However, the P8 shell does not appear to be an invariant feature in cystoviruses and is absent in Φ8 (Jäälinoja et al., 2007). It has been suggested that these differences in organization of the outer protein capsid could explain variations in the mechanisms of infection between the different cystoviral species. Also, present on the outer surface between the lipid and outer protein layers is P5, that is a homolog of the lytic transglycosylases (Caldentey and Bamford, 1992, Dessau et al., 2012) and facilitates the penetration of the host peptidoglycan layer. The second protein layer is composed of 120 copies of the major capsid protein P1 organized as asymmetric dimers on an icosahedral T = 1 lattice (Huiskonen et al., 2006, Jäälinoja et al., 2007, El Omari et al., 2013, Nemecek et al., 2013). Associated with the P1 shell are three other proteins: P4, a hexameric nucleoside triphosphatase (NTPase) packaging motor, 72 copies of which are located on the 5-fold axes (de Haas et al., 1999, Huiskonen et al., 2006); P2, an RNA-directed RNA polymerase (RdRP) that is located inside the P1 shell on the 3-fold axes (Sen et al., 2008, Ilca et al., 2015); and finally an essential protein P7, that appears to be located in close proximity to the 3-fold axes near P2 (Katz et al., 2012, Nemecek et al., 2012, Ilca et al., 2015). The inner protein capsid formed by P1 together with the associated P2, P4 and P7 proteins are collectively known as the polymerase complex (PX). Enclosed within the PX is the three-segmented dsRNA genome. The genomic segments are classified based on overall size and termed large (L, 6.4–7.1 kbp), medium (M, 3.6–4.7 kbp) and small (S, from 2.3 to 3.2 kbp) (McGraw et al., 1986, Gottlieb et al., 1988, Mindich et al., 1988, Hoogstraten et al., 2000, Qiao et al., 2000, Gottlieb et al., 2002, Qiao et al., 2010b, Mäntynen et al., 2015). These genomic segments encode for the various cystoviral proteins; between 14–20 proteins are predicted, some of which, like those that comprise the PX (encoded by the L segment), are present in all cystoviruses, but have a high sequence diversity. Other proteins are predicted to be more specific for a given family member e.g. the P8 protein appears to be absent in Φ8 being replaced by around 60 copies of a putative membrane-associated protein (Jäälinoja et al., 2007).
Fig. 1.
Schematic representation of the structural organization of a mature cystovirus virion. The membrane proteins that include the major envelope protein, P9 (90 residues in Φ6), an envelope protein, P10 (42 residues), and a minor membrane protein, P13 (72 residues) are associated with the outer lipid envelope. Exposed to the extracellular medium, is the receptor-binding spike protein, P3 (648 residues) that forms a complex with the fusogenic membrane protein, P6 (167 residues), is able to interact specifically with the type IV pili of the bacterial host in the case of Φ6 (and Φ2954). In some cystoviruses, e.g. in Φ8, P3 comprises of two separate proteins P3a and P3b. The outer protein capsid is formed by 200 trimers of the major outer capsid protein, P8 (149 residues) assembled on a T = 13 lattice. Not all of all the cystoviruses contain a P8 shell e.g. Φ8. The lytic protein P5 (220 residues) that is involved in the degradation of the peptidoglycan layer during internalization into the host cytoplasm is found in the space between the lipid and outer protein layers. The inner protein capsid is formed from asymmetric dimers of the inner capsid protein, P1 (769 residues) arranged on a T = 1 lattice. Associated with the P1 shell are three other proteins all encoded on the genomic L-segment: an RNA-directed RNA polymerase, P2 (664 residues), a hexameric packaging NTPase, P4 (332 residues) and an accessory protein, P7 (161 residues). The proteins P1, P2, P4 and P7 assemble to form the polymerase complex (PX). The three segments, large (L, 6.4 kbp), medium (M, 4.1 kbp) and small (S, 2.9 kbp) of the double-stranded RNA genome are enclosed within the P1 shell. The L-segment encodes for the P1, P2, P4 and P7 proteins, the M-segment encodes for the P3, P6, P10 and P13 proteins; the S-segment encodes P5, P8 and P9 proteins.
3. Life-cycle of cystoviruses
All cystoviruses infect various species of Pseudomonas but can also target Escherichia coli or Salmonella as in the case of Φ8, Φ12 and Φ13 (Mindich et al., 1999). Infection is initiated through the specific attachment of the P3 spike protein to its receptor (either the type IV pilus in the case of Φ6 or Φ2954, or the lipopolysaccharide coat in the case of Φ8, Φ12 and Φ13) (Romantschuk and Bamford, 1985, Mindich et al., 1999, Qiao et al., 2010b). Following this interaction, the fusion between the viral and bacterial outer membranes is triggered by P6, the fusogenic protein, allowing the intact nucleocapsid (P8 and the PX containing the dsRNA genome) to enter the periplasm (Bamford et al., 1987). P5 on the surface of the capsid then degrades the peptidoglycan layer providing access to the bacterial inner membrane (Caldentey and Bamford, 1992). Subsequently, the nucleocapsid transits through the inner membrane by a process that has been suggested to be facilitated by the interaction of the P8 shell with phospholipid head-groups of the membrane lipids (Poranen et al., 1999, Cvirkaitė-Krupovič et al., 2010). This is followed by the dissociation of the P8 shell and the release of the PX with its genomic contents into the cytoplasm. The PX then starts the process of semi-conservative transcription through the action of P2 (Makeyev and Bamford, 2000a), releasing plus-strand RNA (S+, M+, L+) into the cytoplasm using the P4 protein as a conduit (Kainov et al., 2004). In the host cytoplasm, these segments are utilized as mRNA by the host translational machinery to synthesize the P1, P2, P4 and P7 proteins that self-assemble into empty procapsids (PC). The empty PCs then package the three plus-strand segments in a sequential manner in an NTP-dependent fashion through the action of the packaging NTPase, P4 (Juuti et al., 1998, Kainov et al., 2003, Lísal et al., 2004), using a process that is tightly controlled and requires the presence of pac sequences comprising of a conserved 18 nucleotide sequence at the 5′-end of each segment and an additional ∼200 nucleotide variable sequence further downstream (Gottlieb et al., 1994). S+ is packaged first, then M+, and finally L+ (Frilander and Bamford, 1995, Qiao et al., 1995). The PC particle expands at each stage (Nemecek et al., 2011) and it is hypothesized that this conformational change exposes the binding sites for the following segment (El Omari et al., 2013, Nemecek et al., 2013). Following packaging, P2 then initiates the process of replication or minus-strand RNA synthesis to recapitulate the dsRNA genome to generate fully-expanded PX core particles. The PX then acquires the other viral proteins and the lipid coat from the host membrane to form a mature virion that is released from the host by lysis (Mindich and Bamford, 1988).
The two processes central to the viral life-cycle, transcription and replication, take place within the host and are carried out by a virus-encoded RdRP, P2, that is the focus of this article. In the various sections below, we provide a brief overview of various aspects of the process of RNA polymerization catalyzed by P2, focusing on the enzymatic process and its spatiotemporal regulation in the Cystoviridae family, drawing from a large body of structural, mechanistic and functional data that are available. The assembly, organization and genome packaging of cystoviruses, while also significant areas of interest, will not be reviewed here. For a comprehensive discussion on these subjects the reader is referred to the following excellent treatises (Mindich, 2004, Poranen et al., 2005, Mindich, 2012, Poranen and Bamford, 2012).
4. Cystoviral RNA-dependent RNA polymerases
P2, the cystoviral RdRP (71.6–79.7 kDa, 636–713 residues) performs the dual tasks of replication, synthesis of the minus-strands of the dsRNA genomes using the corresponding plus-stands as template; and transcription, synthesis of the plus-strands utilizing the minus-stands as template. Below, we describe the structure of P2, the mechanism of P2-catalyzed RNA synthesis and its spatiotemporal regulation. We will largely focus on Φ6 P2 (referred to as P2 from here on forward unless explicitly stated) that remains the most comprehensively studied cystoviral RdRP. We will compare and contrast its features with cystoviral RdRPs, for which data are available, and also with other similar, single sub-unit viral RdRPs e.g. those from Flaviviridae (Choi and Rossmann, 2009, Lu and Gong, 2017) or Picornaviridae (Lescar and Canard, 2009, Ferrer-Orta et al., 2015, Peersen, 2017). We will also briefly analyze the possible role of conformational dynamics in modulating P2 function and that is now established in other viral RdRPs (Cameron et al., 2009).
4.1. Structure
The largest amount of high-resolution structural data is available for Φ6 P2 in the apo state, in the presence of variety of divalent metal ions that include the catalytic Mg2+, Mn2+ and inhibitory Ca2+, single-stranded DNA and RNA templates and a variety of nucleoside triphosphates (NTPs) (Butcher et al., 2001, Salgado et al., 2004, Poranen et al., 2008c, Sarin et al., 2012, Wright et al., 2012). High-resolution structural information is also available on Φ12 P2 in the presence of Mg2+ ions (Ren et al., 2013). Table 1 provides a comprehensive list of available high-resolution structures of cystoviral P2 proteins.
Table 1.
List of high-resolution structures of cystoviral RdRPs (P2).
| PDB | Ligands/Variants | Resolution | Species | Reference |
|---|---|---|---|---|
| 1HI0 | 5′-dTdTdTdCdC-3′, GTP, Mg2+, Mn2+ (Initiation Complex) | 3.0 Å | Φ6 | A |
| 1HHS | Mn2+ | 2.0 Å | Φ6 | A |
| 1HHT | 5′-dTdTdTdCdC-3′, Mn2+ | 2.9 Å | Φ6 | A |
| 1HI8 | Mg2+ | 2.5 Å | Φ6 | A |
| 1HI1 | ATP | 3.0 Å | Φ6 | A |
| 1UVI | 5′-UUUUCC-3′, Mn2+ | 2.15 Å | Φ6 | B |
| 1UVJ | 5′-UUCC-3′, Mn2+ | 1.9 Å | Φ6 | B |
| 1UVK | 5′-GG-3′, Mn2+, Mg2+, pyrophosphate (product state) | 2.45 Å | Φ6 | B |
| 1UVL | 5′-UUUCC-3′, Mn2+ | 2.0 Å | Φ6 | B |
| 1UVM | 5′-UUUCC-3′, Mn2+ (alternate conformation) | 2.0 Å | Φ6 | B |
| 1UVN | 5′-UUCC-3′, GTP, Mn2+, Ca2+ | 3.0 Å | Φ6 | B |
| 1WAC | 629-QYKW-632/629-SGT-631 mutant | 3.0 Å | Φ6 | C |
| 2JL9 | E491Q mutant | 3.2 Å | Φ6 | D |
| 2JLF | E491Q mutant, 5′-UUUUCC-3′, Mn2+ | 3.2 Å | Φ6 | D |
| 2JLG | E491Q mutant, 5′-dTdTdTdTdCdC-3′, GTP, Mn2+ | 2.8 Å | Φ6 | D |
| 4A8F | 5′-dAdGdCdG-3′, Mg2+, ATP | 3.3 Å | Φ6 | E |
| 4A8K | 5′-dAdAdTdC-3′, Mg2+, GTP | 3.5 Å | Φ6 | E |
| 4A8M | 5′-dAdAdTdC-3′, Mg2+, ATP, GTP | 2.92 Å | Φ6 | E |
| 4A8O | E634Q mutant, Mn2+ | 2.67 Å | Φ6 | E |
| 4A8Q | E634Q mutant, 5′-dTdTdCdGdCdG-3′, Mg2+, ATP | 3.06 Å | Φ6 | E |
| 4A8S | E634Q mutant, 5′-dTdTdTdTdCdGdCdGdTdAdGdCdG-3′, Mg2+, ATP | 2.9 Å | Φ6 | E |
| 4A8W | E634Q mutant, 5′-dTdTdCdGdCdGdTdAdAdGdCdG-3′, Mg2+, ATP | 3.04 Å | Φ6 | E |
| 4A8Y | E634Q mutant, 5′-dTdTdTdTdCdGdCdGdTdAdAdGdCdG-3′, Mg2+, ATP | 3.41 Å | Φ6 | E |
| 4B02 | Nicked at R607, Mn2+ | 3.3 Å | Φ6 | F |
| 4IEG | Mg2+ (Form A) | 2.1 Å | Φ12 | G |
| 4GZK | Mg2+ (Form B) | 1.69 Å | Φ12 | G |
A (Butcher et al., 2001), B (Salgado et al., 2004), C (Laurila et al., 2005), D (Poranen et al., 2008c), E (Wright et al., 2012), F (Sarin et al., 2012), G (Ren et al., 2013)
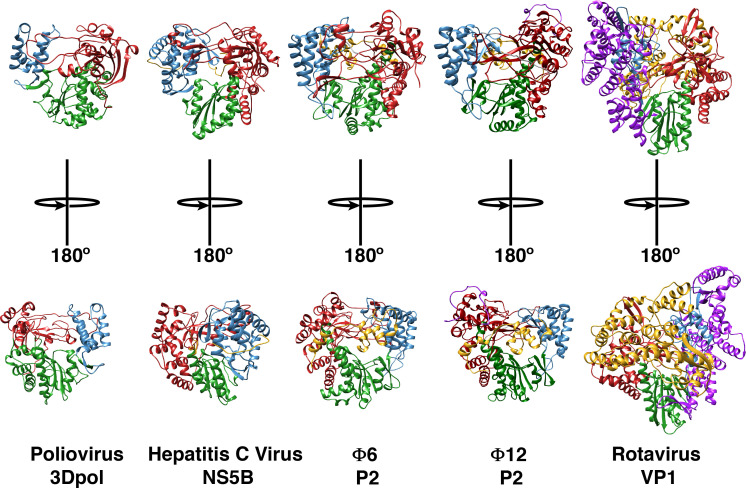
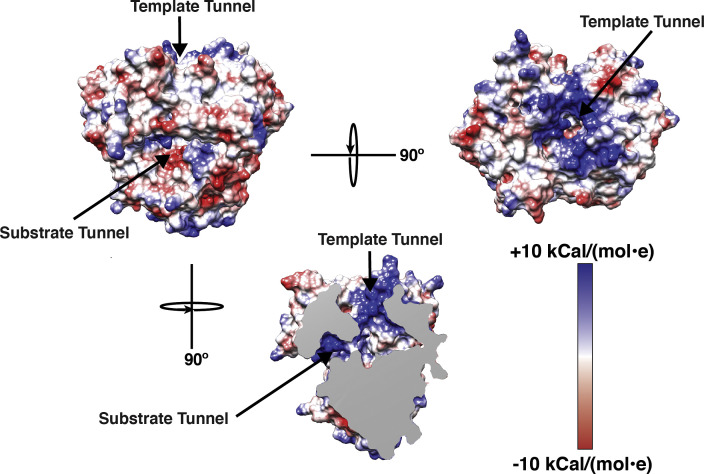
In spite of weak sequence similarity (22% identity, 37% similarity based on the BLOSUM62 matrix), the P2 proteins from Φ6 and Φ12 share a common overall fold (Fig. 2 ). This strong structural similarity with each other, and with other viral RdRps, is not unexpected. The nucleic acid polymerase super-family that includes DNA dependent DNA polymerases (DdDPs), DNA dependent RNA polymerases (DdRPs), reverse transcriptases (RTs), together with the RdRPs share similar overall structural organization in spite of their significant sequence diversity (Ferrer-Orta et al., 2006, Choi, 2012, Mönttinen et al., 2014). The polymerase structure is assembled on a common overall scaffold that resembles a cupped right hand with three conserved domains: “fingers”, “palm” and “thumb”, originally used to describe the structure of the Klenow fragment (Ollis et al., 1985, Steitz, 1998). In viral RdRPs, the fingers and thumb domains are connected either by small linkers (“fingertips”), e.g. in P2, in hepatitis C virus (HCV) NS5B (Lesburg et al., 1999), or in poliovirus 3Dpol (Thompson and Peersen, 2004), or enhanced by additional structural elements e.g. a large N-terminal domain as is the case of the rotavirus VP1 (Lu et al., 2008), forming an encircled active site that limits movement between the fingers and thumb domains. In addition to these conserved domains viral RdRPs contain a variety of additional motifs that facilitate virus specific regulatory activities. Another commonly occurring structural element decorating the basic polymerase fold in viral RdRPs is a C-terminal domain (CTD). In cystoviruses, this is a small segment (∼60 residues) that covers the catalytic cleft formed at the interface of the fingers and the thumb domains and blocks the exit path of the nascent dsRNA, suggesting that a conformational change is necessary to allow egress of the elongating chain (Butcher et al., 2001). In contrast the CTD in the rotavirus VP1, that is expected to play a similar role, is large and comprises about 310 residues (Lu et al., 2008). In cystoviruses, the CTD plays another significant role in conjunction with the catalytic elements (described below) that are located principally on the palm domain, by forming a so-called “initiation platform” or a “priming platform”, that allows the de novo synthesis of RNA on a single-stranded RNA (ssRNA) template without the use of a primer (Butcher et al., 2001, Laurila et al., 2002). This priming platform appears to be a general feature in de novo initiating polymerases (Tao et al., 2002, Lu et al., 2008, Choi and Rossmann, 2009, Appleby et al., 2015). It is notable that this domain is missing in RdRPs that utilize a primer e.g. picornaviruses like foot-and-mouth disease virus (FMDV) or poliovirus 3Dpol proteins, that have larger central cavities (enabled by smaller thumb domains, see Fig. 2) (Ferrer-Orta et al., 2004). The inner surfaces of the fingers and thumb domains in P2 are lined with basic residues forming a template tunnel that provides a positively charged passage for the entry of the ssRNA template into the catalytic site (Fig. 3 ). This channel is however, not wide enough to accommodate dsRNA, and therefore during transcription the dsRNA strands have to be separated to allow the minus-strand to be used as template for the polymerization reaction (semi-conservative transcription). How this strand separation is achieved is unclear though it was previously suggested that a negatively charged “plough” at the edge of the template tunnel could facilitate this separation (Butcher et al., 2001) though recent data seems to indicate otherwise (Sarin et al., 2009). A second channel (the substrate tunnel, Fig. 3) that is almost orthogonal to the template channel is bounded by several positively charged residues and serves as a portal for the entry of the substrate NTPs into the catalytic site where they can be incorporated into the nascent chain during the nucleotidyl-transfer reaction.
Fig. 2.
Structures of viral RNA-directed RNA polymerases. The structures of several viral RdRps are shown in ribbon representation. The structures have been ordered left-to-right with increasing size: poliovirus 3Dpol (PDB: 1RA6, 461 residues, 52.5 kDa) (Thompson and Peersen, 2004), HCV NS5B (PDB: 5CZB, 563 residues, 62.5 kDa) (Pierra Rouvière et al., 2016), Φ6 P2 (PDB: 1HHS, 664 residues, 74.8 kDa) (Butcher et al., 2001); Φ12 P2 (PDB: 4GZK, 659 residues, 75.3 kDa) (Ren et al., 2013) and rotavirus VP1 (PDB: 2R7Q, 1095 residues, 126.1 kDa) (Lu et al., 2008). The fingers, thumb and palm domains are shown in red, blue, and green, respectively. The N- and C-terminal domains, when present, are shown in magenta and yellow, respectively.
Fig. 3.
The template and substrate entry tunnels. The top panel shows two views of P2 with the near orthogonal substrate and template tunnels. The bottom panel shows a third view with the front surface removed to display the basic surfaces of the two tunnels that intersect at the catalytic site. The electrostatic charge surfaces are shown using a red-to-blue gradient.
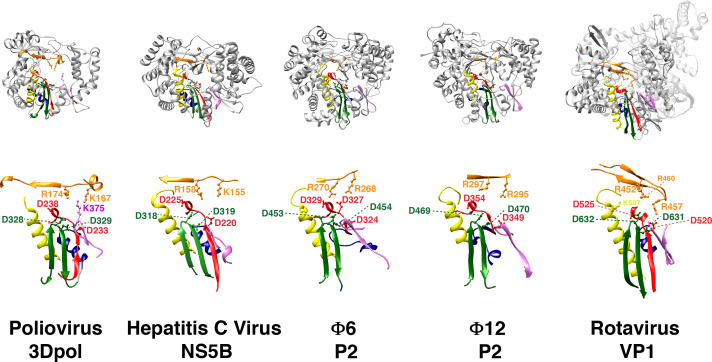
In addition to similarities in their overall fold and the presence of conserved domains, RdRPs contain six conserved sequence motifs that line the central catalytic cavity and play critical catalytic roles (O'Reilly and Kao, 1998, Bruenn, 2003) in both DNA and RNA polymerases. Of these, five of the motifs (A–E) belong to the palm domain and the sixth motif (F) belongs to the fingers domain (Fig. 4 ). These motifs participate in binding the template (A, F), substrate NTPs (B, D, F), catalytic and structural metal ions (C) and in stabilizing the nascent chain (E). An additional fingers domain sequence, termed motif G, was identified in viral RdRPs (Gorbalenya et al., 2002). This motif that plays a role in template binding, shows significant divergence within the RdRP family, and will therefore not be discussed further in this article. As will be discussed below, in addition to their structural roles, the conformational dynamics of these motifs appear to play a critical role in catalysis.
Fig. 4.
Conserved RdRp sequence motifs. The RdRp sequence motifs A–E (palm) and F (fingers) are mapped onto the structures shown in Fig. 2. Motifs A–F are shown in red, yellow, green, blue, pink and orange, respectively. A subset of key conserved charged residues is shown in ball-and-stick representation and labeled.
4.2. Catalytic function
All nucleic acid polymerases create polynucleotide chains by generating a phosphodiester bond between the free 3′-OH group of a 5′-terminal nucleotide of a nascent chain and the α-phosphate of an incoming NTP (Fig. 5 ). In RdRPs this nucleotidyl-transfer reaction can be initiated by one of two broad mechanisms (van Dijk et al., 2004). The first of these requires the use of a primer to initiate the nucleic acid polymerization. This primer could be a short oligonucleotide originating from the host e.g. the cap snatching mechanism in Orthomyxoviridae (e.g. influenza A–C viruses), Bunyaviridae (e.g. Hantavirus) and Arenaviridae (e.g. Lassa virus), where a short sequence cleaved from the 5′-cap of cellular mRNA is used to prime the reaction (Plotch et al., 1981, Ruigrok et al., 2010). The oligonucleotide primer could also be obtained from the virus itself e.g. as a product of so-called abortive synthesis (Nagy et al., 1997) or through back-priming where the template loops back and serves as its own primer (Behrens et al., 1996). The primer could also be a protein attached to the viral genome e.g. the VPg protein in picornaviruses such as FMDV or poliovirus, that is uridylated prior to priming RNA synthesis (Paul and Wimmer, 2015).
Fig. 5.
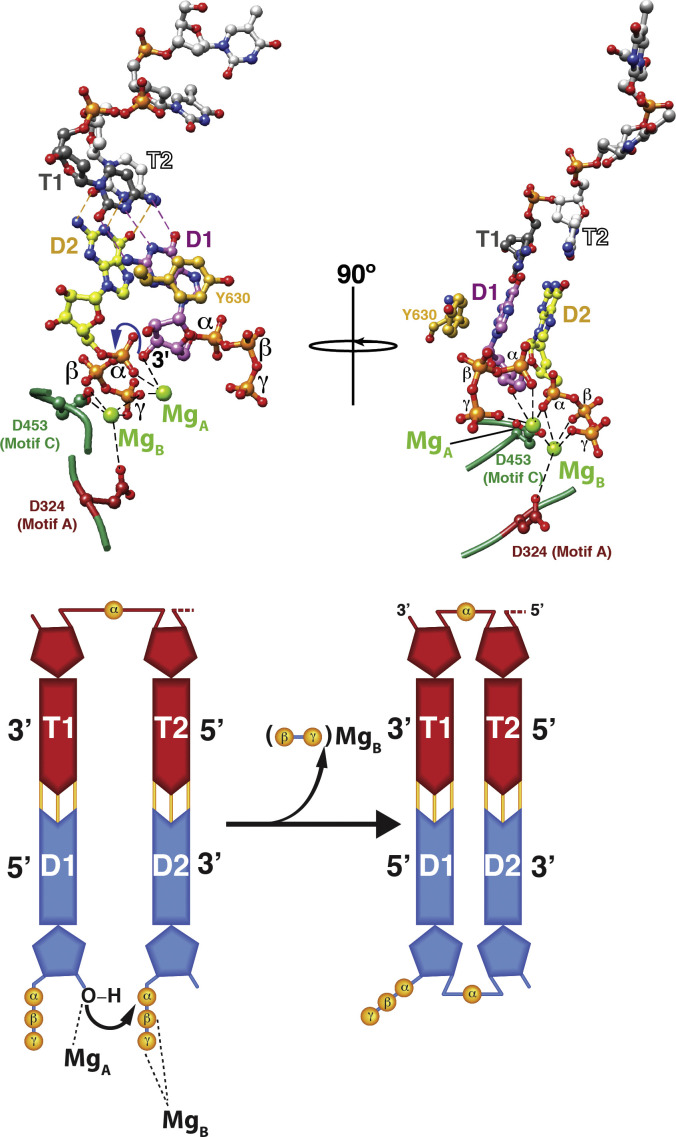
Phosphodiester bond formation. The substrate NTPs (D1 and D2) that would form the first two bases of the daughter chain are paired with the two bases (T1 and T2) at the 3′-end of the template. This priming platform is stabilized by Y630 from the C-terminal domain that base stacks with D1. The nucleotidyl-transfer reaction requires two Mg2+ ions − one (MgA) that coordinates the 3′-OH of NTP D1 and the α-phosphate of the NTP D2; a second (MgB) that coordinates the β- and γ-phosphates of NTP D2 in addition to the sidechains of the conserved motif A (D324) and motif C (D453) aspartates. Coordination with MgA lowers the pKa of the 3′-OH of NTP D1 enabling a nucleophilic attack on the α-phosphate of D2 creating a phosphodiester bond between the two NTPs. The pyrophosphate product is stabilized by MgB. The nucleotidyl-transfer reaction is shown schematically in the lower panel.
The second mechanism, termed de novo synthesis (Kao et al., 2001), is initiated from individual NTPs (these may be considered to be single nucleotide primers) without the use of a polynucleotide or protein primer. This is a common mechanism of RNA synthesis in viral RdRPs and only few families of viruses e.g. the picornaviruses cannot initiate RNA synthesis de novo. Cystoviruses initiate RNA synthesis in de novo fashion, though under certain circumstances, described below, the P2 proteins can be induced to initiate in primed fashion. In the following few sections we describe how P2 enables RNA synthesis (the dual processes of transcription and replication) in cystoviruses as inferred through crystallographic analyses, coupled to mutagenesis and biochemical studies. The process of RNA synthesis may be broadly divided into three distinct stages: initiation, elongation and termination. The various events leading up to the initiation stage i.e. formation of an initiation complex, are collectively termed pre-initiation.
4.2.1. Pre-initiation
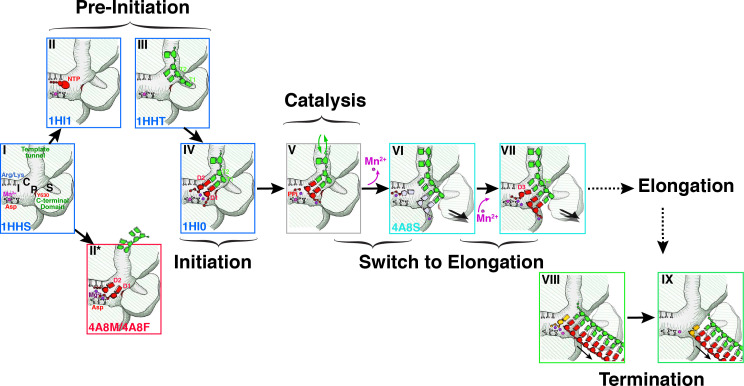
As de novo RNA synthesis does not rely on specific primers to initiate nucleotide polymerization, this step requires tight control, both in terms of structure and chemistry, to ensure that the two nucleotides that form the first phosphodiester bond in the daughter chain are not only properly paired with the complimentary bases at the 3′-end of template but are also in the proper orientation for nucleotidyl-transfer to take place in an optimal fashion (Fig. 5). Additionally, the structural and catalytic metal ions are required to be in place to allow the substrate NTPs to be positioned at the active site, for the activation of the donor —OH to enable an electrophilic attack and to stabilize the product pyrophosphate. Indeed, several structural and biochemical studies on P2 (Makeyev and Bamford, 2000a, Makeyev and Bamford, 2000b, Salgado et al., 2004) have suggested a plausible sequence of events leading up to the initiation of RNA synthesis as detailed below (Fig. 6 ).
Fig. 6.
Proposed sequence of events in P2 mediated RNA synthesis. I, C, P and S indicate the 2nd NTP binding site (I-site or interrogation-site, formed by motif F), the catalytic NTP binding site (C-site or catalytic-site, formed by the motif C G/SDD triplet), the priming platform (P-site, formed by Y630 on the CTD) and the specificity pocket (S-site, also part of the CTD), respectively. In State I, free P2 is complexed with the structural Mn2+ ion located in its non-catalytic binding-site with the CTD in a closed conformation. Under these conditions P2 rapidly samples states II, II* and III corresponding to NTP and template-bound conformations (pre-initiation). In state III, the template bound-state of P2, the 3′-end of the template overshoots the active site and is locked into the specificity pocket (S-site). Metal ions at non-canonical sites facilitate the transit of NTPs through the substrate portal. Step IV: The template ratchets back to allow Watson-Crick base pairing of its 3′-end with the complimentary NTPs (D1 and D2). At this state the catalytic Mg2+ ions are drawn into their appropriate position and the non-catalytic position is occupied by Mn2+. Step V: The nucleotidyl-transfer reaction occurs with the formation of a phosphodiester bond between D2 and D1 with the release of pyrophosphate. Step VI: In order to allow the nascent dsRNA to move forward in register to prime the system for the next nucleotidyl-transfer reaction, the destabilizing effect of the Mn2+ facilitates the opening of the CTD. Mn2+ ions lost at this stage need to be replaced by external Mn2+. The initiation complex is still not stable enough to transit into its processive, elongation stage. Step VII: A new substrate NTP (D3) complementary to the third template base from the 3′-end (T3) now forms a new initiation complex culminating in a repeat of IV–VI and leading to a phosphodiester bond formation between D3 and D2. At this stage (or in cases after a few more cycles) the complex is stable, the CTD is fully open and the Mn2+ is stably bound at the non-catalytic binding site. At this point P2 enters the elongation stage where the entire template can be read through. The alphanumeric codes in the bottom left-hand corner of each box, where present, represent the PDB accession codes of the structures, where available), used to characterize the corresponding step. Modified from Wright et al. (Wright et al., 2012) with permission. Step VII: after the entire daughter strand complimentary to the template has been synthesized, the TNTase activity of P2 attaches a nucleotide to its 5′-end. Step VII: The dsRNA leaves P2. Steps VII and VIII have been proposed to comprise the termination step in RNA synthesis (Poranen et al., 2008a).
The first step in RNA synthesis involves the formation of a binary complex between P2 and the ssRNA template resulting in a so-called a “pre-initiation” state. This complex involves critical but non-specific (i.e. sequence independent) electrostatic interactions between the sugar-phosphate backbone of the ssRNA template and positively charged residues lining the template tunnel. Mutation of some of these basic residues (e.g. R30A or K541L) leads to a reduction in RNA affinity likely resulting from a loss in the ability of P2 to guide the template correctly to the active site to enable the formation of a stable initiation complex (Sarin et al., 2009). In order for initiation to occur, the 3′-terminus of the template has to be accessible and capable of reaching the active site in an unconstrained fashion; the presence of any secondary structure in this region leads to reduced RNA synthesis (Sarin et al., 2009) (see below). Once at the active site, 3′-terminal base of the template (T1) locks into a specificity pocket (the S-site) formed by the CTD. It is notable that at this stage the 3′-terminus of the template overshoots the catalytic site (C-site) that are occupied by NTPs during the nucleotidyl-transfer reaction (Fig. 6). At this point, the penultimate nucleotide (T2) from the 3′-end of the template lies at the priming site (P-site) consisting of a conserved aromatic residue (Y630) that belongs to the CTD.
During pre-initiation, NTP substrates D1 and D2, that will form the first two bases from the 5′-end of the daughter chain, access the catalytic site via the substrate tunnel and occupy a pre-catalytic position (interrogation or I-site), close to the non-catalytic ion-binding site (discussed in detail below). Engagement of a substrate NTP to the I-site involves a patch of positively charged residues on the fingers domain including K223, R225, and the motif F residues, R268 and R270. This set of residues appears to be structurally conserved at least partially, in cystoviral P2 proteins, as evident from the structure of Φ12 P2 (Ren et al., 2013), but also in other viral RdRP e.g. poliovirus 3Dpol (Thompson and Peersen, 2004), HCV NS5B (Lesburg et al., 1999). It has been suggested that the transition of NTPs to the active site during pre-initiation is guided by non-catalytic divalent ions (Wright et al., 2012). Occupancy of sites that are distinct from the canonical non-catalytic site, and indeed the canonical catalytic site, by metal-ions, has been noted both in Φ6 (Wright et al., 2012) and Φ12 (Ren et al., 2013) P2 proteins.
4.2.2. Initiation
Given the simultaneous presence of NTPs at the I-site and the occupancy of the S-site by the 3′-end of the template, the D2 NTP that will form the second base from the 5′-end of the daughter chain base pairs with the T2 base of the template now locked at the P-site. The template ratchets back from the S-site to allow proper base-pairing between the T1 NTP and the D1 base. For Φ6, D1 is a purine NTP since the 3′-ends of all three minus-strands (relevant for transcriptional initiation) are C; they are U for all three plus-strands (relevant for the initiation of replication). At this point, proper Watson-Crick base-pairing between D1 and T1 stabilized by stacking interactions with priming Y630, and between D2 and T2, leads to the formation of the initiation complex (Butcher et al., 2001). Additionally, the shift of the NTPs into base-pairing compatible positions draws their associated Mg2+ ions into the canonical catalytic metal-sites to take part in the nucleotidyl-transfer reaction. The first Mg2+ ion (MgA) coordinates with the 3′-OH of the NTP D1 and with the α-phosphate of the NTP D2 (Fig. 5). Presence of this metal ion lowers the pKa of the 3′-OH of D1 and allows a nucleophilic attack on the α-phosphate of D2 enabling formation of a phosphodiester bond (Steitz, 1998). The second Mg2+ ion (MgB) coordinates to the β- and γ-phosphates of D2, in addition to conserved motif A (D324) and C (D453; a residue that is part of the so-called G/SDD triad conserved in all nucleic acid polymerases) aspartates (Liu and Tsai, 2001, Ng et al., 2008). The MgB serves to stabilize the pyrophosphate product of the nucleotidyl-transfer reaction. This two-ion mechanism is common to all nucleic acid polymerases (Steitz, 1998) although an extension of this mechanism with the requirement of a third divalent metal ion has recently been suggested using time-resolved crystallography in DNA polymerase η (Gao and Yang, 2016, Yang et al., 2016). Whether the requirement for a third metal ion is a general phenomenon for all nucleic acid polymerases remains to be established. It has also recently been demonstrated that the protonation of the pyrophosphate product by a general acid located on motif D (likely to be K487 and K499 in Φ6 and Φ12 P2 proteins, respectively) leads to a 50–1000-fold enhancement in the rate of nucleotide addition (Castro et al., 2009).
While Mg2+ that plays a key role in catalysis, and Mn2+ stimulate P2 activity, Ca2+ ions have a significant inhibitory effect within the context of the PC (Ojala and Bamford, 1995) and in purified P2 (Salgado et al., 2004). The structure of P2 in the context of a stalled initiation complex (Salgado et al., 2004) suggests that this inhibition results largely from the displacement of MgA, rather than MgB (a nearly identical position can be occupied by Ca2+). The presence of Ca2+ rather than Mg2+ at a position displaced from that occupied by MgA in the optimal initiation complex leads to a reorganization of the catalytic site due to the transformation of the octahedral geometry favored by Mg2+ to a pentagonal bi-pyramidal geometry favored by Ca2+. This alteration results in a loss of interactions with the phosphate moieties of D1 and D2. Significantly, the Y630 sidechain now rotates to allow coordination with Ca2+ thus disrupting the priming platform and base-stacking with T1 (as described above) and preventing the formation of an optimal initiation complex. However, the relevance of this Ca2+-induced inhibition in the physiological context is unclear given that the cellular levels of Mg2+ (and Mn2+) exceed that of Ca2+ by several orders of magnitude.
While wild-type P2 initiates RNA synthesis through a de novo mechanism, as described above, P2 variants that can be induced to use a primer have been generated. These variants utilize a mechanism known as back-priming in which the ssRNA template loops back forming a hairpin and is used to prime initiation. It is notable that this mechanism is the dominant initiation mode for wild-type HCV NS5 B in vitro (Behrens et al., 1996). The P2 variant that uses back-priming for initiation is generated by replacing the 630–632 YKW sequence by a much smaller, and more dynamic GSG sequence. This alteration destabilizes the de novo priming platform formed by Y630 (Laurila et al., 2002, Laurila et al., 2005) and leads to back-priming. Clearly, this cannot comprise an optimal method to initiate RNA synthesis in vivo, since the daughter chain remains covalently bound to the template. Nevertheless, this observation emphasizes the critical role of the C-terminal priming platform in stabilizing the de novo initiation complex.
4.2.3. Elongation
Elongation represents the processive state of the RdRP, where the template is read through completely and the full sequence complimentary to the template is synthesized. However, the transition from initiation to elongation does not necessarily occur automatically since the ternary complex involving P2, NTPs and the template is not stable enough after formation of the first few phosphodiester bonds to sustain multiple nucleotidyl-transfer reactions without dissociation. This leads to abortive initiation i.e. formation of daughter chains that are no more than a few nucleotides long (2-nt appears to be most common in P2 under standard in vitro conditions) (Sarin et al., 2012). This behavior is not unusual in de novo initiating RdRPs and it has been suggested that multiple initiation attempts may be required before a productive initiation complex, that can successfully transition to the elongation stage, can be assembled (Farsetta et al., 2000). One reason for these difficulties is the presence of steric barriers that prevent the formation of a stable elongation complex. In P2 this barrier is provided by the CTD that, while contributing Y630 to create the priming platform to enable the formation of a stable initiation complex, also blocks the exit path of the double-stranded nascent chain. It was recently suggested that a disordered hinge (603–609), that is part of the CTD, performs the task of gatekeeper in controlling the passage from initiation to elongation (Sarin et al., 2012) by reducing the association of the CTD with the main body P2 and allowing an outward motion to allow the egress of the newly synthesized dsRNA. Indeed, detachment of this region (by formation of a nick at R607) from the rest of the CTD leads to a significant increase in the ratio of abortive to productive initiation products compared to wild-type P2 (Sarin et al., 2012). Additionally, there is mounting evidence that suggests that a non-catalytic, structural Mn2+ ion, that is coordinated by E491, D454 (the second aspartate of the conserved G/SDD triad on Motif C) and A495, serves to enhance the flexibility of P2 (Poranen et al., 2008c) and to ease the transition into elongation. It has been shown that transition from initiation to elongation is the only step in the P2 RNA synthesis pathway that is dependent on the presence of external Mn2+ ions (Wright et al., 2012). Thus, based on detailed biochemical and structural analyses, the following model was proposed (Fig. 6): After formation of the first phosphodiester bond, an outward motion of the CTD enables the nascent dsRNA and the template to move forward in register, and to bring T3 into position to allow proper base-pairing with the D3 NTP. Following this, motional rearrangements near the non-catalytic metal site reduce the affinity for the bound Mn2+ that is now released and must be replaced by external Mn2+ to allow the catalytic elements to re-establish their active conformations to enable the next nucleotidyl-transfer reaction (linking D3 to D2). This cycling continues for a few additional steps until the nascent dsRNA chain is large enough to lock the CTD into its fully open position. At this stage, the Mn2+ can no longer be displaced from its binding site and the P2-RNA-NTP ternary complex is stable enough to enter the processive state (Wright et al., 2012). It has been noted that this structural Mn2+ ion appears to be a general feature in RNA polymerases from both ssRNA and dsRNA viruses and in HIV RT (Mönttinen et al., 2012). Therefore, it is possible that this non-structural metal ion fulfills a function that is conserved in viral nucleic acid polymerases.
4.2.4. Termination
As mentioned above, elongation comprises a non-abortive stage, where the entire ssRNA template is read-through producing a complete complimentary dsRNA product at the end of the polymerase reaction. But, in the case of linear genomes, as for the cystoviruses, it is still unclear how the 5′ termini of the template are processed. Analyses of cystovirus genomes do not reveal the presence of 5′-terminal sequences that could be classified as canonical termination signals e.g. the A- and U-tracts and GC dyads in bacterial RNA polymerases (Ray-Soni et al., 2016). However, a possible mechanism for chain termination in cystoviruses was suggested by Poranen et al. (2008a) through the discovery of non-templated terminal nucleotidyltransferase (TNTase) activity in P2. This reaction, that takes place not only in purified P2 but also in viral core particles, allows the addition of nucleotides with little apparent specificity at the 3′-end of the nascent RNA chain. TNTase activity has been reported in other viral RdRPs e.g. in the HCV (Behrens et al., 1996), bovine viral diarrhea virus (BVDV) (Zhong et al., 1998) and poliovirus (Neufeld et al., 1994), to name a few. It is therefore possible that after the synthesis of the daughter strand on the template is complete, the TNTase activity adds the final nucleotide to the 3′-end of the daughter chain and the terminated dsRNA leaves through the CTD that is now in its fully-open position characteristic of the elongation phase. A small wrinkle in this hypothesis is the fact that this activity would yield a single-base overhang on both ends of the genome (assuming that both the transcription and replication are affected) but no such overhangs have been detected in the sequencing of cystoviral genomic RNA. A plausible explanation for this could lie in the TNTase activities of the intact core particles. Since P2 adds the terminal nucleotide to the 3′-end of the plus-strand produced from the last round of transcription as it leaves its catalytic site, the initiation complex templated by this strand would form in a new host and consequently the overhang would not be detected in the mature virus particles. However, this would require RNA synthesis to originate not from the last nucleotide from the 3′-end of the template (T1) but rather from the penultimate nucleotide (T2). Such a mechanism has been proposed for replication in the bacteriophage Qβ (Rensing and August, 1969). This scenario would require the mechanism for initiation discussed above to be refined.
5. Regulation of RNA synthesis
5.1. Imbalance in plus-strand and minus-strand synthesis
While P2 can utilize a large number of ssRNA templates (Makeyev and Bamford, 2000b), the efficiency of synthesis depends on the nature of the 3′-end, with a net preference for pyrimidines rather purines with a C-3′ being preferred in most cases (Makeyev and Bamford, 2001). Similar trends were also seen for Φ8 and Φ13 P2 proteins (Yang et al., 2001). However in vitro, when using native Φ6 dsRNA templates and purified P2, or in the context of core particles, plus-strand RNA is preferentially synthesized i.e. reactions appear to be more efficiently templated by the minus-strands (Makeyev and Bamford, 2000a, Makeyev and Bamford, 2000b). A similar behavior is also seen for P2 from Φ8 and Φ13 (in the absence of external Mn2+) (Yang et al., 2003b), suggesting that this asymmetry in RNA synthesis is a common characteristic of the cystoviruses. For Φ13 P2, the M− template ending in a CC-3′ allows more efficient initiation than M+ that is terminated by a CU-3′. This observation is in agreement with the higher transcriptional efficiency for the M segment i.e. the synthesis of M+. However, the efficiency of synthesis does not appear to be dependent simply on the nature of the template 3′-end given that while S− and S+ both terminate in a -CC-3′ the transcription of S+ is higher than that of S−. An analysis of the termini of the cystoviral genomic sequences suggests a lower GC content at the transcription initiation sites (i.e. the 3′-ends of the minus-strands) than the replication initiation sites (the 3′-ends of the plus-strands) (Yang et al., 2003b). Therefore, the latter sites are more likely to form stable duplexes (preventing efficient strand separation) limiting their availability as ssRNA templates. Alternatively, the presence of stable secondary structures near the 3′-ends of the plus-strands would also make them less available for use as linear templates during replication given the narrow size of the template entry tunnel in the cystoviral RdRPs. The latter scenario i.e. the loss of linearity due to secondary structure formation is likely to be more relevant while using purified RdRPs though both mechanisms could, in principle, contribute to asymmetric RNA synthesis in the context of the viral particles (Yang et al., 2003b).
5.2. Temporal control of RNA synthesis
In Φ6, the synthesis of the minus- and plus-strands of the three segments (S, M and L) of the dsRNA genome, occurs in a tightly controlled fashion. Using purified PC particles, it was suggested (Frilander et al., 1995) that replication i.e. minus-strand synthesis initiates only when all three plus-strands are fully packaged (S+ then M+ and finally L+, as mentioned above); the complete packaging of the L+ segment, that is packaged last, serves as a signal for initiation of minus-strand synthesis of all segments (replication). Additionally, the required signal for the initiation of plus-strand synthesis (transcription) was suggested to be the synthesis of the L− strand. Thus, it was suggested that the synthesis of minus-strands and of plus-strands relied on the presence of the complimentary L segment, L+ for replication, L− for transcription. However, it was later demonstrated that the initiation of both the plus- and minus-strand syntheses were not dependent on the presence of the L-segment or fragments thereof, but rather the size of the RNA packaged that initiates replication and the size of the dsRNA produced that initiates transcription (Qiao et al., 1997b). Thus, it is highly likely that RNA synthesis is regulated by conformational changes resulting from the filling and the consequent expansion of the PC particles.
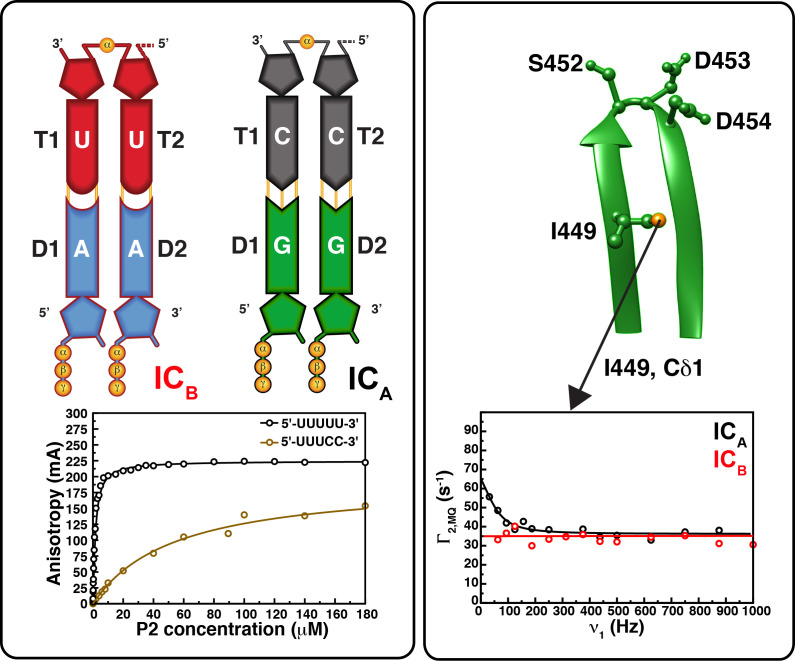
An additional level of control is obtained through the differential synthesis of L+ (that has a 5′-GU i.e. templated by a AC-3′) compared to M+ and S+ that both have 5′-GG (i.e. use CC-3′ templates). However, the synthesis of L+ can be achieved by simply changing its penultimate 5′-base to G (i.e. using a C as template). This phenomenon may be partially explained by the lower affinity of P2 for the large A compared to smaller C at the penultimate 3′-position. Indeed, P2 has a ∼5-fold higher affinity for 5′-UUUCC-3′ than for 5′-UUUAC-3′ (Ren and Ghose, 2011). It is notable that while only the M+ and S+ segments are transcribed in the absence of Mn2+, the presence of which enables the synthesis of L+, suggesting that factors beyond affinity for the template are at play. Indeed, 5′-UUUUU-3′ has a ∼50-fold higher affinity for P2 than 5′-UUUCC-3′.
The differences in template utilization in vitro can explain the variations in the relative levels of transcripts in the latter stages of infection i.e. high levels of M+ and S+ and low levels of L+. However, the overall levels of all three transcripts were found to be similar at the early stages of infection (Coplin et al., 1975). This suggests that the transcription/replication cycle is also regulated by an alternate mechanism during the early stages of infection. One possible mode of regulating RNA synthesis within the host cell could be through the influence of host factors. One of these regulatory factors has since been identified as YajQ (Qiao et al., 2008), a protein of unknown function that is found in many Gram-negative bacteria. The presence of YajQ enables a change in the template specificity of P2 and enables the synthesis of the L+. Indeed, Φ6 is unable to replicate in yajQ knockout strains. Additionally, the requirement for YajQ is specific to the transcription of the L-segment and a variant containing a modified L+ segment where 5′-GU is replaced by a 5′-GG is YajQ-independent. Interestingly, specific mutations on P1 (I632V or E369G and G405S) or on P2 (K34N on the fingers or I642V on the CTD) switch the Φ6 cystovirus to a YajQ-independent infection mode (Qiao et al., 2008). Other residues of the CTD located in 603–612 region, that overlaps with the hinge discussed above, are also able to alter this YajQ-dependent transcription (Qiao and Mindich, 2013). It was previously demonstrated that YajQ could interact directly with the viral core particles (Qiao et al., 2010c) through P1 (Qiao et al., 2008). Therefore, it is likely that the mutations in P1 enable conformational changes similar to those resulting from the interaction with YajQ to facilitate the synthesis of L+. However, there is no evidence of a direct interaction between P2 and YajQ, and indeed addition of the latter does not affect the in vitro transcriptional behavior of purified P2. This suggests that the effect of YajQ in altering the enzymatic function of P2 is indirect, likely transmitted through P1. Given that there are low levels of YajQ in the cell and most are bound to infectious particles, there is little free YajQ available to interact with the newly assembled particles to activate significant levels of L+ synthesis at the latter stages of infection (Qiao et al., 2010c).
In Φ2954, transcription of the L+ segment (5′-AC compared to 5′-GC for the M+ and S+ segments) (Qiao et al., 2010b) is weak compared to that of the M+ and S+ segments. In a manner similar to Φ6, changing the first base to G (rather the second base in Φ6) increases the levels of L+ to those of M+ and S+ (Qiao et al., 2010a). In this case, however, the transcription of the L+ segment is not activated through interactions with YajQ but rather through the removal of the P8 shell by a host glutaredoxin (GrxC) (Qiao et al., 2010a). In contrast, for Φ8 core particles, all genomic segments (5′-GA in all cases) are well-transcribed in vitro without the need of a host protein. However, behavior similar to in Φ6 and Φ2954 is seen for Φ8 in the context of an infected host cell, where the levels of L+ are significantly reduced late in infection. In contrast to Φ6 and Φ2954, this appears to be achieved in Φ8 through differential degradation of the transcripts through a process controlled by the expression of specific gene products (Qiao et al., 2009). Therefore, while all the cystovirus species maintain temporal control over transcription, they do so using distinct mechanisms.
5.3. Regulatory interactions with PX components
In addition to the roles of distinct host proteins in affecting the ability of P2 for polynucleotide synthesis through indirect means e.g. conformational effects transmitted through P1, there is mounting evidence that other components of the PX are also able to directly or indirectly affect P2 function. The accessory protein P7, is known to be essential for the regulation of the transcription with minimal effects on replication (Juuti and Bamford, 1997). Indeed, incomplete core particles lacking P7 (P1-P2-P4 particles) are capable of the normal production of minus-strand RNA while producing transcripts of the incorrect size. This effect of P7 in influencing plus-strand synthesis is in addition to its roles in accelerating both the assembly of the PX and the packaging of the genomic segments into empty PC particles (Poranen et al., 2008b, Sun et al., 2012). P7 is a small accessory protein (between 156 and 177 residues) that shows little overall sequence conservation within the cystoviruses. However, all the cystoviral P7 proteins share a common feature − a highly acidic C-terminal tail that has been shown to be highly flexible (Eryilmaz et al., 2008, Poranen et al., 2008b). This tail is indispensable for the formation of infectious virions (Poranen et al., 2008b). NMR analyses using purified P2 and P7 proteins from Φ12 suggests that the dynamic C-terminal tail of P7 can interact with P2 and in doing so mimic the negatively charged sugar-phosphate backbone of a linear ssRNA template (Alphonse et al., 2014) by binding at the template tunnel. Follow-up biochemical analyses suggest that this interaction decreases the affinity of P2 for RNA thereby inhibiting RNA synthesis. While the degree of in trans inhibition is a modest 3-fold reduction in P2 activity (Alphonse et al., 2014), it is possible that this modest in trans effect could be substantially amplified in the context of an assembled PX. Multiple cryo-EM studies on Φ6 have suggested that P2 and P7 occupy overlapping (Nemecek et al., 2012) or proximal positions (Katz et al., 2012) and this is supported by biochemical data (Sun et al., 2012). It was even suggested than P7 could “hold” P2 to the P1 shell (Katz et al., 2012). Thus, the close proximity of P7 to P2 on the assembled PX and the consequent increase in the local concentration of P7 would greatly enhance the weak interaction (KD ∼75 μM) seen in trans and perhaps allow it to regulate template entry. The plausibility of this hypothesis is yet to be assessed in the context of the intact core particles.
6. Fidelity of nucleotide incorporation
RNA viruses are characterized by their significantly higher mutation rates, in general, compared to DNA viruses (Sanjuán and Domingo-Calap, 2016). Mutation rates for DNA viruses are estimated to be between 10−8 and 10−6 substitutions per nucleotide per cell infection (s/n/c) while the corresponding range for RNA viruses lies between 10−6 and 10−4 s/n/c (10−6 for Φ6 (Chao et al., 2002)). Most polymerases in RNA viruses, but not all e.g. the plus-strand coronaviruses (Denison et al., 2011), lack 3′-exonuclease proofreading activity, that is more common in their DNA virus counterparts (Steinhauer et al., 1992). Factors that contribute to the fidelity of nucleotide incorporation include the stability of the Watson-Crick base-pairing between the template and the NTP to be incorporated (Sloane et al., 1988), the availability of the template in linear form i.e. its tendency to assume stable secondary-structures (as discussed above), and the presence of a post-incorporation repair function. No structure exists for an elongating complex involving P2; therefore, to analyze the structural determinants of fidelity, one has to rely on the structures of elongation complexes involving other viral RdRPs that include poliovirus 3Dpol (Gong and Peersen, 2010) or HCV NS5B (Appleby et al., 2015). Based on these structures, it has been suggested that fidelity control is established by a sequence of events that follows the initial base-pairing to form appropriate Watson-Crick pairs between the NTP and template bases. A conformational change (that has been termed the ribose-Asp switch (Shu and Gong, 2016)) that occurs in the presence of the appropriate NTP involves a conserved aspartate on motif A (likely D329 on P2) and a serine on motif B (likely S393 on P2). This change is facilitated by the breaking of a hydrogen-bond between the motif A aspartate (D238 for poliovirus 3Dpol, D225 for NS5B) with an asparagine residue (N297 for 3Dpol, N291 for NS5B) on motif B. This reorientation allows the aspartate sidechain to establish interactions with a motif B serine (S288 on 3Dpol, S282 for NS5B) that also interacts with the 2′-OH of the NTP and the asparagine. For 3Dpol, mutation of either D238 or N297 to alanine leads to altered fidelity (Gohara et al., 2004). In P2, a somewhat similar mode involving motif A D329 (fully conserved in cystoviral P2 proteins, e.g. D354 and D347 in Φ12 and Φ2954, respectively) and the motif B S393 (also fully conserved e.g. S420 and S414 in Φ12 and Φ2954, respectively) is possible, but the details of the conformational change are likely to be different. While the sidechain of D329 does rotate slightly in the presence of NTPs, the change is nowhere near as drastic as the ones seen for 3Dpol and NS5B. A comparison of the P2 structures in the presence and absence of NTP also does not suggest the possibility of the aspartate-serine sidechain interactions reminiscent of the D238-S288 interactions in the NTP-bound state of the 3Dpol elongation complex. A caveat is that for P2, we infer this change from the structure of the initiation complex since no structure of the elongation complex, where conformations could be different, is available. Another indication that the details of the fidelity-determining conformational changes could be somewhat different for P2 is the fact that the motif B asparagine (or equivalent residue) appears to be absent. The fidelity determination process has been suggested to be completed by the coupling of these transitions to the closure of motif D to enable the optimal orientation of the lysine sidechain (K359 in 3Dpol, K487 in P2) to protonate the pyrophosphate leaving group (Moustafa et al., 2014). This scenario is explored in greater detail in the following section.
7. Role of dynamics in function
While a variety of structural studies have provided significant mechanistic insights into the function of viral RdRPs including the cystoviral P2 proteins, others have strongly indicated that conformational dynamics occurring on multiple timescales ranging from nanoseconds to milliseconds also play a significant role in determining the functional behavior of viral RdRPs (Cameron et al., 2009). Molecular dynamics (MD) simulations (Moustafa et al., 2011) and NMR relaxation studies (Alphonse et al., 2015) have revealed viral RdRPs to be highly connected dynamic enzymes where remote regions are well networked with the catalytic site. NMR relaxation studies on Φ6 and Φ12 P2 proteins in the unliganded states have suggested that motifs A, B and to some extent C, are largely ordered, while motifs D-F are disordered on the nanosecond timescale (Alphonse et al., 2015), in line with that suggested by MD studies on several picornavirus RdRPs (Moustafa et al., 2011) (Fig. 7 ). Further, it was shown for Φ12 P2, that local perturbations through metal-ion or NTP binding or though the introduction of a functional mutation (T425I on motif B to generate a ts-mutant with a significantly altered temperature-dependence of activity) (Yang et al., 2003a) are not restricted to the site of binding/mutation but are transmitted to remote regions of the protein (Alphonse et al., 2015). Further, in Φ6 P2, slow dynamics on the same millisecond timescale appears to link the template entry tunnel and the CTD suggesting that these dynamics were the result of the same transient conformational changes transmitted through the protein core (Ren et al., 2010). It was speculated that this dynamic mode (the sum of the forward and reverse rates for an exchange between two conformations: kex = 1200–1500 s−1) coupled the motion of the template to that of the CTD to allow the exit of the newly synthesized dsRNA chain, as described above. Additionally, another slower motional mode (kex = 500–800 s−1) was detected and largely involved regions near the catalytic site including several of the conserved sequence motifs. These motions were predicted to be related to those that enable the chemical step (Ren et al., 2010) and the flanking conformational changes described in greater detail below. This was supported by the fact that this dynamic mode coupled all the observable RdRP sequence motifs only upon formation of a physiologically relevant initiation complex that included a template with the correct genomic 3′-ends (CC-3′, corresponding to S− and M−, as discussed above) and the appropriate NTP (a non-hydrolyzable GTP analog) but not with a higher affinity (∼50-fold) non-genomic ssRNA template (UU-3′) in spite of the formation of the correct Watson-Crick base-pairs with the added ATP analog (Ren and Ghose, 2011) (Fig. 8 ).
Fig. 7.
Fast timescale dynamics. Flexibility measured from spin-relaxation utilizing the methyl positions of leucine, valine, methionine and isoleucine (δ1 only) residues in Φ12 P2 and isoleucine (δ1) for Φ6 P2. Σ values represent flexibility on the fast, nanoseconds timescale with residues with Σ = 0.5 having average flexibility for a given residue type and Σ > 0.5 and Σ < 0.5 being less and more flexible than average, respectively (Alphonse et al., 2015). The inset on the top panel shows the distribution of the least (largely distributed around the template and substrate tunnels) and most flexible positions. The bottom panel shows the flexibility of measured positions in the various conserved RdRp sequence motifs for Φ12 and Φ6 P2. Positions for motifs D and F are overall the most dynamic. The Σ values are represented using a red-to-blue gradient ranging from the most-to-least dynamic elements.
Fig. 8.
Slow timescale dynamics in P2. Dynamics measurements on two separate stalled initiation complexes one 5′-UUUCC-3′ with GMPCPP (ICA) and a second with a 5′-UUUUU-3′ template with AMPCPP (ICB). The 5′-UUUUU-3′ and 5′-UUUCC-3′ templates have affinities that are ∼50-fold different (fluorescence anisotropy traces with increasing concentration shown on the bottom left panel) but only the latter for which the last two nucleotides correspond to the S− and M− 3′-ends, shows dynamics on a catalytically relevant timescale in its catalytic elements. Slow timescale dynamics for ICA (black trace) using the δ1 position of the Motif C residue I449 (also shown is the SDD triad) as probe is reflected in a change in the relaxation rate with field. No such dynamics is seen for ICB as reflected by the flat trace (in red) (Ren and Ghose, 2011).
While biochemical experiments are required to confirm the functionality of the dynamics measured by NMR in P2, extensive NMR (Yang et al., 2010, Yang et al., 2012) and molecular dynamics (MD) studies (Moustafa et al., 2011, Moustafa et al., 2014) have been performed on poliovirus 3Dpol and these are backed up by biochemical assays that indicate the role of dynamics on the fidelity of nucleotide incorporation. The most critical “functional” dynamics in this regard are those involving motif D and the conserved basic residue that acts as a general acid in protonating the pyrophosphate product, as discussed above. A conservative mutation of this motif D lysine (K359R) leads to a 5-fold increase in incorporation fidelity (Castro et al., 2009). NMR studies have indicated that key structural rearrangements of motif D seen in the presence of the correct nucleotide are absent in the presence of the incorrect nucleotide (Yang et al., 2010, Yang et al., 2012). Recent MD studies on high (G64S) and low-fidelity (H273R) 3Dpol variants (Moustafa et al., 2014) have suggested a hierarchical connection of dynamical timescales that has been noted in other systems (Henzler-Wildman et al., 2007). Data for the wild-type enzyme and the fidelity variants suggest the presence of two distinct states of the enzyme, one that is capable of binding nucleotide (binding-competent state, BCS) and the other that is not (binding-occluded state, BOS). The interconversion between these states occurs on the nanosecond timescale and these dynamics appear to drive a slower conformational transition involving motif D (Arnold and Cameron, 2004). This transition between the BOS and BCS requires a conformational change driven by the breaking of a hydrogen-bond involving R174 on motif F and D238 on motif A (that is involved in the ribose-Asp switch discussed above). These simulations further suggest that nature of these dynamics are significantly different between wild-type 3Dpol and its fidelity variants. Solution NMR studies on Φ12 P2 have suggested significant dynamics on the nanosecond timescale involving motif F (Fig. 7) that in principle could reflect dynamics like those seen in 3Dpol (Alphonse et al., 2015). These data are backed up by MD simulations on Φ12 P2 that also suggest occupancy of similar state involving the motif F residue R297 (R270 in Φ6 P2) and D354 on motif A (D329 in Φ6 P2) (unpublished). Moustafa et al. (Moustafa et al., 2014) proposed a model in which the transition between the binding-occluded and binding-competent states drives a slower transition that involves the closing of the motif D to allow the K359 on 3Dpol (discussed above; K487 and K499 in Φ6 and Φ12 P2, respectively) to protonate the pyrophosphate leaving group. It was further suggested that the fidelity checkpoint involved a conformational transition involving motif B that couples these two timescales. Indeed, the end-points of motions involving motif B appear to be crystallographically visible (Gong and Peersen, 2010) and involve S288 on 3Dpol (the equivalent positions are S393 in Φ6 P2 and S420 in Φ12 P2, as described above). While similar structural information is lacking in P2, NMR relaxation analyses demonstrate the presence of millisecond timescale motion in Φ12 P2 involving motif B and motif D (unpublished). Motif D dynamics on the millisecond timescale was also seen in Φ6 P2 in studies that utilized a smaller number of reporters (Ren et al., 2010, Ren and Ghose, 2011). However, in order to unequivocally confirm the functional relevance of these NMR-detected conformational dynamics, additional studies, including pre-steady-state kinetic analyses on wild-type and functional variants of P2 similar to those performed for poliovirus 3Dpol (Arnold and Cameron, 2004, Arnold et al., 2004, Gohara et al., 2004, Yang et al., 2012), are necessary.
8. Mechanism of nucleic acid polymerization
A necessary step in probing the process of nucleotide incorporation is the elucidation of the complete kinetic mechanism of elongation. Extensive pre-steady state kinetic analyses have been carried out on a variety of DNA and RNA polymerases (Dahlberg and Benkovic, 1991, Patel et al., 1991, Kati et al., 1992, Smidansky et al., 2011) including viral RdRPs e.g. poliovirus 3Dpol (Arnold and Cameron, 2004, Arnold et al., 2004, Gohara et al., 2004) and HCV NS5B (Jin et al., 2012, Jin et al., 2013). Nucleic acid polymerases appear to follow a general mechanism for nucleotide incorporation as shown in Fig. 9 (Arnold and Cameron, 2004) in which the central chemistry step is sandwiched between two sets of conformational changes. The first of these conformational steps involves active site closure to generate a state that is competent to perform chemistry. The second conformational step involves a change in the structure of the post-chemistry state to facilitate the dissociation of the pyrophosphate product and likely coincides with the movement on the template forward in register to reset the polymerase for the next nucleotidyl-transfer reaction. This is followed by the rapid release of the pyrophosphate product. Any or all of these events starting from the selection for the correct over the incorrect nucleotide to the last conformational step could, in principle, be used as a fidelity checkpoint. However, several studies have suggested that the pre-chemistry conformational step (discussed at length above) constitutes the fidelity checkpoint for a variety of viral RdRPs e.g. poliovirus 3Dpol (Arnold et al., 2005). While there is no reason to suspect that an overall kinetic mechanism different from that depicted in Fig. 9 is operative in P2, determination of the mechanism and of the corresponding kinetic rate constants should provide clear insight into the fidelity of nucleotide incorporation in P2 and indeed enable the elucidation of rate-limiting step (or steps) of the reaction.
Fig. 9.
Mechanism for the nucleotide addition cycle. A kinetic scheme that is applicable to nucleic acids polymerases is shown. The first step involves binding of the substrate NTP to the enzyme in complex with RNA at the nth stage of elongation i.e. the daughter chain is n nucleotides long. The second step involves a conformation change (shown by the red arrows) of the ERn•NTP complex into a “closed” state that is capable of catalysis. The third step is the chemistry step (shown by the blue arrows) extending the daughter chain by one nucleotide (n + 1). The fourth step (shown by the red arrows) is a second conformational change that enables the release of the pyrophosphate product. The fifth and the final step involves release of pyrophosphate that is likely coupled to movement of the template forward in register to prime it for the next nucleotidyl-transfer reaction.
9. Elongation at the single-molecule level
9.1. Error incorporation
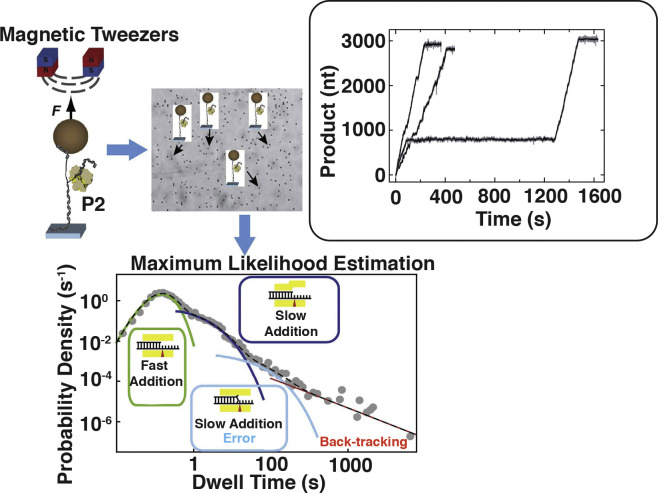
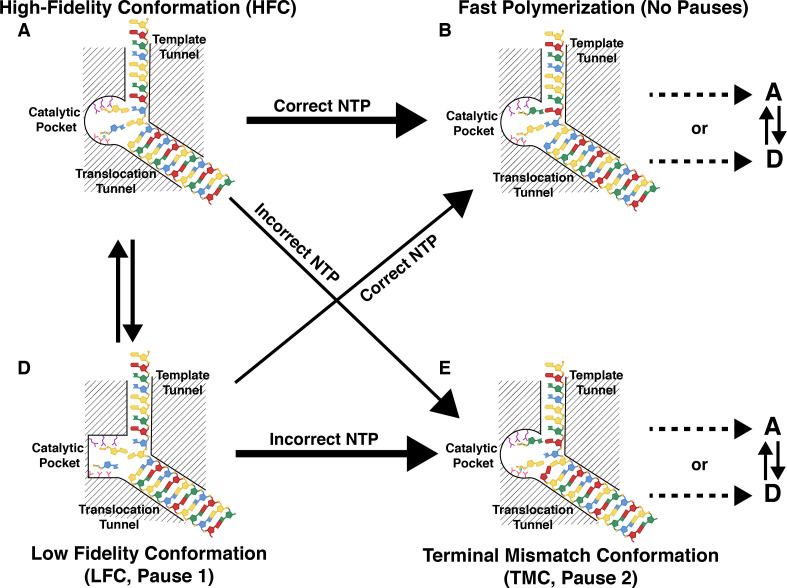
Recent developments in single molecule approaches (Dulin et al., 2013) have yielded the ability to follow nucleotide incorporation from timescales that range from seconds to hours providing access to unique features and rare events that are hidden in bulk measurements while allowing the robust statistical analysis of these events. These studies complement those described earlier in that they reveal dynamic features that occur on timescales much slower than the chemical or conformational steps described above. They also reveal dynamics on longer length scales and provide insight into the variety of scenarios that the RdRP may encounter while moving on the template ssRNA during elongation. While bulk measurements of kinetics are unavailable for P2, Dulin et al. (2015b) have studied elongation at the single molecule level using a clever setup utilizing magnetic tweezers (Fig. 10 ). A length of dsRNA is tethered to a surface at one end to a magnetic bead on the other. A constant force is applied to the magnetic bead using a pair of permanent magnets that are placed above a flow cell. One strand of the dsRNA has a short 3′ overhang that allows initiation by P2. RNA synthesis is carried out on the overhang by addition of NTPs that generates dsRNA while creating ssRNA on the tethered construct. The extension on the tethered strand is monitored and converted in the number of nucleotide incorporation steps. Independent measurements on several such assemblies at the single-molecule level allow a highly multiplexed analysis of individual chain elongation traces and their detailed analyses in a statistically robust fashion that is not possible when using bulk measurements. Using this approach coupled to a maximum-likelihood analysis of a large set of traces recorded under different conditions (e.g. variation of NTP concentrations, presence of Mn2+ ions etc.) provides several interesting observations. Plotting the probability distribution of the times that it takes P2 to travel on the template through two consecutive 10nt windows against time (dwell-time distribution, DTD) reveals a convolution of four distinct behaviors (Dulin et al., 2015c): (1) An nth order Γ-distribution for n-steps with the same rate constant corresponding to the pause-free nucleotide incorporation through a 10nt window. (2) An off-pathway pause (Pause 1) suggested to be due to a large-scale conformation change at the active site that leads to a transition from a state of high catalytic activity (high-fidelity conformation, HFC) to a catalytically deficient state (low fidelity conformation, LFC). A similar transition has been previously suggested for E. coli RNA polymerase (Neuman et al., 2003). (3) A second pause (Pause 2) due to an on-pathway slowdown in polymerase activity after base mismatch (Johnson, 2008) representing a terminal-base mismatched conformation (TMC). (4) A power-law distribution (best described by an exponent of −3/2) over a large range of dwell-times due to backtracking (described below) corresponding to diffusive reversal of the RdRP (Fig. 10). Using this formalism, one can derive a model for fidelity control as follows: P2 may be considered to be in equilibrium between two conformations of disparate activity (Fig. 11 ), HFC and LFC, each of which can incorporate a correct or an incorrect nucleotide though the catalytic efficiency is much higher in the HFC state. In the presence of a nucleotide mismatch the system enters the TMC state (that is not populated in the presence of the correct NTP) leading to Pause 2 and a slow nucleotide incorporation rate.
Fig. 10.
Measurement of elongation at the single molecule level. Schematic depiction of the magnetic tweezers setup to P2 mediated transcription (top left panel). Array of multiple tweezers setup in a homogeneous magnetic field to allow data collection in a highly multiplexed fashion (top middle panel). The setup comprises of an RNA strand that this largely in duplex form tethered to a magnetic bead at one extremity and to a surface on the other. The RNA contains a short 15-nt overhang at the 3′-end to allow transcriptional initiation in the presence of NTPs. The inset on the top right depicts three typical traces (displaying three distinct types of dynamic behaviors − no pauses, a few short pauses, fast transcription interrupted by a very long pause) that represent the increase in product length versus time due to transcription under a constant force (F) in the presence of an optimal NTP concentration. The lower panel depicts a dwell time distribution (DTD) plot displaying four regions: rapid addition without pause (a Γ-distribution), two sets of short pauses (Pause 1 and Pause 2 described by exponential distributions) and a power-law distribution representing long pauses due to back-tracking. Modified from (Dulin et al., 2015b) with permission.
Fig. 11.
Kinetic view of the elongation cycle. The P2 elongation complex samples two specific conformations, one with an optimal conformation of the catalytic elements (A, a High-Fidelity Conformation or HFC) that allows rapid NTP incorporation (with the correct NTP) leading to B; and an unfavorable conformation of the catalytic pocket (D, a Low Fidelity Conformation or LFC) which has a lower rate of NTP incorporation leading to pauses (Pause 1). In the presence of an incorrect nucleotide, the system enters a Terminal Mismatch Conformation (TMC) that leads to the longest pauses (Pause 2) with the slowest rate of incorporation (misincorporation). The arrows symbolize the transition between the different states with the thick arrows representing the preferred path. Adapted from Dulin et al. (2015b).
9.2. Backtracking and recombination
As mentioned above, the power-law distribution at longer dwell-times corresponds to back-tracking events which in the experimental setup for single molecule measurements lead to reannealing of the tethered dsRNA (Dulin et al., 2015b, Dulin et al., 2015a). This reversal of the direction of motion has also seen in bacterial and eukaryotic RNA polymerases (Shaevitz et al., 2003). For example, in E. coli RNA polymerase, short backtracking motions play a critical role in the proof-reading mechanism, leading to the high-fidelity characteristic of this enzyme in vivo (Shaevitz et al., 2003). However, the precise role of this back-tracking is still unclear in the case of P2. Given the high misincorporation rate in P2, it is unlikely to encode any kind of proofreading activity. The backtracking is also unlikely to be the result of NTP-mediated 3′-terminal nucleotide excision activity as demonstrated for HCV NS5B given that the catalytic turnover is much faster during the back-tracking motions in P2 than that seen for NS5B (Jin et al., 2013). These events are also unlikely to result from nucleotide excision by pyrophosphorolysis as seen in BVDV 3Dpol since very high pyrophosphate concentrations would be required to accomplish this process (D'Abramo et al., 2004). However, an intriguing proposal is that back-tracking could be utilized for a rudimentary repair mechanism. The reversal of the motion of the RdRP through the newly synthesized dsRNA would unwind it and release a new 3′-end that could be used as template by a second RdRP (Onodera et al., 2001). This, in principle could be utilized to create short RNA segments for use in recombination events to repair damaged RNA. Indeed both homologous (Onodera et al., 2001) and heterologous (Mindich et al., 1992, Qiao et al., 1997a) recombination has been known to occur in cystoviruses and involve the 3′-end of genomic segments. Recombination in Φ6 was shown to be capable of repairing damaged 3′-ends or to remove non-native insertions and hence to rescue replication in vitro (Onodera et al., 1993).
10. Conclusions and future perspectives
In spite of a large body of structural, biochemical and functional studies on P2 mediated transcription and replication in cystoviruses, there are several open questions. A high-resolution structure of the elongation complex for P2 is still not available and it is possible that certain structural differences may emerge that distinguish it from HCV NS5B (Appleby et al., 2015) and poliovirus 3Dpol (Gong and Peersen, 2010). A key mechanistic feature that could, in principle, be different for P2, involves the conformational events leading up to closure of the active site and events that modulate the fidelity of nucleotide incorporation. This structural data together with an elucidation of the complete mechanism of the nucleotide addition cycle through pre-steady state kinetics analysis should enable the identification and design of mutator and antimutator variants and provide insight into diversity and adaptability in cystoviral quasispecies (Vignuzzi et al., 2006, Vignuzzi and Andino, 2012, Korboukh et al., 2014).
While crystal structures provide static snapshots of conformational states that are stable under crystal packing forces, some conformations that are transiently sampled or are unstable under crystallographic conditions may nevertheless be functionally relevant. While NMR in general and the measurement of spin-relaxation, in particular, is ideally suited to characterize these conformations, the large size of RdRps including P2 had previously prevented these studies. Recently however, NMR studies of the dynamics of viral RdRps using a variety of sparse labeling schemes are beginning to appear in the literature (Ren et al., 2010, Yang et al., 2010, Ren and Ghose, 2011, Alphonse et al., 2015) and hold promise, post-validation through kinetic and functional studies, to yield the missing-link (Cameron et al., 2009) between structure and function. In addition to bulk measurements of conformational dynamics in solution, the recent single-molecule studies over a large timescale range have yielded a variety of unexpected features, the functional role of which remain to be ascertained (Dulin et al., 2015a).
Acknowledgements
Research support from the National Science Foundation (NSF) MCB 1412007 is acknowledged. Award G12 MD007603 from the National Institutes of Health (NIH) towards partial support of research facilities is also acknowledged. This article was written when RG was a Program Director at the NSF in the Molecular and Cellular Biosciences (MCB) division. His IR/D activities were supported by award MCB 1657192. The authors thank Drs. Leonard Mindich (The Public Health Research Institute), Reza Khayat (The City College of New York) and Craig Cameron (Pennsylvania State University) for critical reading of this manuscript.
References
- Alphonse S., Arnold J.J., Bhattacharya S., Wang H., Kloss B., Cameron C.E., Ghose R. Cystoviral polymerase complex protein P7 uses its acidic C-terminal tail to regulate the RNA-directed RNA polymerase P2. J. Mol. Biol. 2014;426:2580–2593. doi: 10.1016/j.jmb.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alphonse S., Bhattacharya S., Wang H., Ghose R. Methyl relaxation measurements reveal patterns of fast dynamics in a viral RNA-directed RNA polymerase. Biochemistry. 2015;54:5828–5838. doi: 10.1021/acs.biochem.5b00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleby T.C., Perry J.K., Murakami E., Barauskas O., Feng J., Cho A., Fox D., 3rd., Wetmore D.R., McGrath M.E., Ray A.S., Sofia M.J., Swaminathan S., Edwards T.E. Viral replication: structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J.J., Gohara D.W., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+ Biochemistry. 2004;43:5138–5148. doi: 10.1021/bi035213q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J.J., Vignuzzi M., Stone J.K., Andino R., Cameron C.E. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J. Biol. Chem. 2005;280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford D.H., Romantschuk M., Somerharju P.J. Membrane fusion in prokaryotes: bacteriophage Φ6 membrane fuses with the Pseudomonas syringae outer membrane. EMBO J. 1987;6:1467–1473. doi: 10.1002/j.1460-2075.1987.tb02388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S.E., Tomei L., De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- Bruenn J.A. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003;31:1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher S.J., Dokland T., Ojala P.M., Bamford D.H., Fuller S.D. Intermediates in the assembly pathway of the double-stranded RNA virus Φ6. EMBO J. 1997;16:4477–4487. doi: 10.1093/emboj/16.14.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher S.J., Grimes J.M., Makeyev E.V., Bamford D.H., Stuart D.I. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- Caldentey J., Bamford D.H. The lytic enzyme of the Pseudomonas phage Φ6: purification and biochemical characterization. Biochim. Biophys. Acta. 1992;1159:44–50. doi: 10.1016/0167-4838(92)90073-m. [DOI] [PubMed] [Google Scholar]
- Cameron C.E., Moustafa I.M., Arnold J.J. Dynamics: the missing link between structure and function of the viral RNA-dependent RNA polymerase? Curr. Opin. Struct. Biol. 2009;19:768–774. doi: 10.1016/j.sbi.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C., Smidansky E.D., Arnold J.J., Maksimchuk K.R., Moustafa I., Uchida A., Götte M., Konigsberg W., Cameron C.E. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nature Struct. Mol. Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L., Rang C.U., Wong L.E. Distribution of spontaneous mutants and inferences about the replication mode of the RNA bacteriophage Φ6. J. Virol. 2002;76:3276–3281. doi: 10.1128/JVI.76.7.3276-3281.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.H., Rossmann M.G. RNA-dependent RNA polymerases from Flaviviridae. Curr. Opin. Struct. Biol. 2009;19:746–751. doi: 10.1016/j.sbi.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Choi K.H. Viral polymerases. Adv. Exp. Med. Biol. 2012;726:267–304. doi: 10.1007/978-1-4614-0980-9_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coplin D.L., Van Etten J.L., Koski R.K., Vidaver A.K. Intermediates in the biosynthesis of double-stranded ribonucleic acids of bacteriophage Φ6. Proc. Natl. Acad. Sci. U. S. A. 1975;72:849–853. doi: 10.1073/pnas.72.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvirkaitė-Krupovič V., Poranen M.M., Bamford D.H. Phospholipids act as secondary receptor during the entry of the enveloped, double-stranded RNA bacteriophage Φ6. J. Gen. Virol. 2010;91:2116–2120. doi: 10.1099/vir.0.020305-0. [DOI] [PubMed] [Google Scholar]
- D'Abramo C.M., Cellai L., Götte M. Excision of incorporated nucleotide analogue chain-terminators can diminish their inhibitory effects on viral RNA-dependent RNA polymerases. J. Mol. Biol. 2004;337:1–14. doi: 10.1016/j.jmb.2004.01.030. [DOI] [PubMed] [Google Scholar]
- Dahlberg M.E., Benkovic S.J. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- de Haas F., Paatero A.O., Mindich L., Bamford D.H., Fuller S.D. A symmetry mismatch at the site of RNA packaging in the polymerase complex of dsRNA bacteriophage Φ6. J. Mol. Biol. 1999;294:357–372. doi: 10.1006/jmbi.1999.3260. [DOI] [PubMed] [Google Scholar]
- Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessau M., Goldhill D., McBride R., Turner P.E., Modis Y. Selective pressure causes an RNA virus to trade reproductive fitness for increased structural and thermal stability of a viral enzyme. PLoS Genet. 2012;8:e1003102. doi: 10.1371/journal.pgen.1003102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulin D., Lipfert J., Moolman M.C., Dekker N.H. Studying genomic processes at the single-molecule level: introducing the tools and applications. Nature Rev. Genet. 2013;14:9–22. doi: 10.1038/nrg3316. [DOI] [PubMed] [Google Scholar]
- Dulin D., Vilfan I.D., Berghuis B.A., Poranen M.M., Depken M., Dekker N.H. Backtracking behavior in viral RNA-dependent RNA polymerase provides the basis for a second initiation site. Nucleic Acids Res. 2015;43:10421–10429. doi: 10.1093/nar/gkv1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulin D., Vilfan I.D., Berghuis B.A., Hage S., Bamford D.H., Poranen M.M., Depken M., Dekker N.H. Elongation-competent pauses govern the fidelity of a viral RNA-dependent RNA polymerase. Cell Rep. 2015;10:983–992. doi: 10.1016/j.celrep.2015.01.031. [DOI] [PubMed] [Google Scholar]
- Dulin D., Berghuis B.A., Depken M., Dekker N.H. Untangling reaction pathways through modern approaches to high-throughput single-molecule force-spectroscopy experiments. Curr. Opin. Struct. Biol. 2015;34:116–122. doi: 10.1016/j.sbi.2015.08.007. [DOI] [PubMed] [Google Scholar]
- El Omari K., Sutton G., Ravantti J.J., Zhang H., Walter T.S., Grimes J.M., Bamford D.H., Stuart D.I., Mancini E.J. Plate tectonics of virus shell assembly and reorganization in phage Φ8, a distant relative of mammalian reoviruses. Structure. 2013;21:1384–1395. doi: 10.1016/j.str.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eryilmaz E., Benach J., Su M., Seetharaman J., Dutta K., Wei H., Gottlieb P., Hunt J.F., Ghose R. Structure and dynamics of the P7 protein from the bacteriophage Φ12. J. Mol. Biol. 2008;382:402–422. doi: 10.1016/j.jmb.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsetta D.L., Chandran K., Nibert M.L. Transcriptional activities of reovirus RNA polymerase in recoated cores. Initiation and elongation are regulated by separate mechanisms. J. Biol. Chem. 2000;275:39693–39701. doi: 10.1074/jbc.M004562200. [DOI] [PubMed] [Google Scholar]
- Ferrer-Orta C., Arias A., Perez-Luque R., Escarmís C., Domingo E., Verdaguer N. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 2004;279:47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- Ferrer-Orta C., Arias A., Escarmís C., Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 2006;16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Ferrer-Orta C., Ferrero D., Verdaguer N. RNA-Dependent RNA polymerases of picornaviruses: from the structure to regulatory mechanisms. Viruses. 2015;7:4438–4460. doi: 10.3390/v7082829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frilander M., Bamford D.H. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage Φ6: the three segments modulate each other's packaging efficiency. J. Mol. Biol. 1995;246:418–428. doi: 10.1006/jmbi.1994.0096. [DOI] [PubMed] [Google Scholar]
- Frilander M., Poranen M., Bamford D.H. The large genome segment of dsRNA bacteriophage Φ6 is the key regulator in the in vitro minus and plus strand synthesis. RNA. 1995;1:510–518. [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yang W. Capture of a third Mg2+ is essential for catalyzing DNA synthesis. Science. 2016;352:1334–1337. doi: 10.1126/science.aad9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohara D.W., Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry. 2004;43:5149–5158. doi: 10.1021/bi035429s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P., Peersen O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Pringle F.M., Zeddam J.L., Luke B.T., Cameron C.E., Kalmakoff J., Hanzlik T.N., Gordon K.H., Ward V.K. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 2002;324:47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb P., Metzger S., Romantschuk M., Carton J., Strassman J., Bamford D.H., Kalkkinen N., Mindich L. Nucleotide sequence of the middle dsRNA segment of bacteriophage Φ6: placement of the genes of membrane-associated proteins. Virology. 1988;163:183–190. doi: 10.1016/0042-6822(88)90245-0. [DOI] [PubMed] [Google Scholar]
- Gottlieb P., Qiao X., Strassman J., Frilander M., Mindich L. Identification of the packaging regions within the genomic RNA segments of bacteriophage Φ6. Virology. 1994;200:42–47. doi: 10.1006/viro.1994.1160. [DOI] [PubMed] [Google Scholar]
- Gottlieb P., Potgieter C., Wei H., Toporovsky I. Characterization of phi12, a bacteriophage related to Φ6: nucleotide sequence of the large double-stranded RNA. Virology. 2002;295:266–271. doi: 10.1006/viro.2002.1436. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K.A., Lei M., Thai V., Kerns S.J., Karplus M., Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- Hoogstraten D., Qiao X., Sun Y., Hu A., Onodera S., Mindich L. Characterization of Φ8, a bacteriophage containing three double-stranded RNA genomic segments and distantly related to Φ6. Virology. 2000;272:218–224. doi: 10.1006/viro.2000.0374. [DOI] [PubMed] [Google Scholar]
- Huiskonen J.T., de Haas F., Bubeck D., Bamford D.H., Fuller S.D., Butcher S.J. Structure of the bacteriophage Φ6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure. 2006;14:1039–1048. doi: 10.1016/j.str.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Ilca S.L., Kotecha A., Sun X., Poranen M.M., Stuart D.I., Huiskonen J.T. Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 2015;6:8843. doi: 10.1038/ncomms9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäälinoja H.T., Huiskonen J.T., Butcher S.J. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages Φ6 and Φ8. Structure. 2007;15:157–167. doi: 10.1016/j.str.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Jin Z., Leveque V., Ma H., Johnson K.A., Klumpp K. Assembly, purification, and pre-steady-state kinetic analysis of active RNA-dependent RNA polymerase elongation complex. J. Biol. Chem. 2012;287:10674–10683. doi: 10.1074/jbc.M111.325530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Leveque V., Ma H., Johnson K.A., Klumpp K. NTP-mediated nucleotide excision activity of hepatitis C virus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E348–E357. doi: 10.1073/pnas.1214924110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.A. Role of induced fit in enzyme specificity: a molecular forward/reverse switch. J. Biol. Chem. 2008;283:26297–26301. doi: 10.1074/jbc.R800034200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juuti J.T., Bamford D.H. Protein P7 of phage phi6 RNA polymerase complex, acquiring of RNA packaging activity by in vitro assembly of the purified protein onto deficient particles. J. Mol. Biol. 1997;266:891–900. doi: 10.1006/jmbi.1996.0817. [DOI] [PubMed] [Google Scholar]
- Juuti J.T., Bamford D.H., Tuma R., Thomas G.J., Jr. Structure and NTPase activity of the RNA-translocating protein (P4) of bacteriophage Φ6. J. Mol. Biol. 1998;279:347–359. doi: 10.1006/jmbi.1998.1772. [DOI] [PubMed] [Google Scholar]
- Kainov D.E., Pirttimaa M., Tuma R., Butcher S.J., Thomas G.J., Jr., Bamford D.H., Makeyev E.V. RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein. J. Biol. Chem. 2003;278:48084–48091. doi: 10.1074/jbc.M306928200. [DOI] [PubMed] [Google Scholar]
- Kainov D.E., Lísal J., Bamford D.H., Tuma R. Packaging motor from double-stranded RNA bacteriophage Φ12 acts as an obligatory passive conduit during transcription. Nucleic Acids Res. 2004;32:3515–3521. doi: 10.1093/nar/gkh680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C.C., Singh P., Ecker D.J. De novo initiation of viral RNA-dependent RNA synthesis. Virology. 2001;287:251–260. doi: 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- Kati W.M., Johnson K.A., Jerva L.F., Anderson K.S. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- Katz G., Wei H., Alimova A., Katz A., Morgan D.G., Gottlieb P. Protein P7 of the cystovirus Φ6 is located at the three-fold axis of the unexpanded procapsid. PLoS One. 2012;7:e47489. doi: 10.1371/journal.pone.0047489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korboukh V.K., Lee C.A., Acevedo A., Vignuzzi M., Xiao Y., Arnold J.J., Hemperly S., Graci J.D., August A., Andino R., Cameron C.E. RNA virus population diversity, an optimum for maximal fitness and virulence. J. Biol. Chem. 2014;289:29531–29544. doi: 10.1074/jbc.M114.592303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lísal J., Kainov D.E., Bamford D.H., Thomas G.J., Jr., Tuma R. Enzymatic mechanism of RNA translocation in double-stranded RNA bacteriophages. J. Biol. Chem. 2004;279:1343–1350. doi: 10.1074/jbc.M309587200. [DOI] [PubMed] [Google Scholar]
- Laurila M.R., Makeyev E.V., Bamford D.H. Bacteriophage Φ6 RNA-dependent RNA polymerase: molecular details of initiating nucleic acid synthesis without primer. J. Biol. Chem. 2002;277:17117–17124. doi: 10.1074/jbc.M111220200. [DOI] [PubMed] [Google Scholar]
- Laurila M.R., Salgado P.S., Stuart D.I., Grimes J.M., Bamford D.H. Back-priming mode of phi6 RNA-dependent RNA polymerase. J. Gen. Virol. 2005;86:521–526. doi: 10.1099/vir.0.80492-0. [DOI] [PubMed] [Google Scholar]
- Laurinavičius S., Käkelä R., Bamford D.H., Somerharju P. The origin of phospholipids of the enveloped bacteriophage Φ6. Virology. 2004;326:182–190. doi: 10.1016/j.virol.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Lesburg C.A., Cable M.B., Ferrari E., Hong Z., Mannarino A.F., Weber P.C. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 1999;6:937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- Lescar J., Canard B. RNA-dependent RNA polymerases from flaviviruses and Picornaviridae. Curr. Opin. Struct. Biol. 2009;19:759–767. doi: 10.1016/j.sbi.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Liu J., Tsai M.D. DNA polymerase beta: pre-steady-state kinetic analyses of dATPαS stereoselectivity and alteration of the stereoselectivity by various metal ions and by site-directed mutagenesis. Biochemistry. 2001;40:9014–9022. doi: 10.1021/bi010646j. [DOI] [PubMed] [Google Scholar]
- Lu X., McDonald S.M., Tortorici M.A., Tao Y.J., Vasquez-Del Carpio R., Nibert M.L., Patton J.T., Harrison S.C. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008;16:1678–1688. doi: 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Gong P. A structural view of the RNA-dependent RNA polymerases from the Flavivirus genus. Virus Res. 2017;234:34–43. doi: 10.1016/j.virusres.2017.01.020. [DOI] [PubMed] [Google Scholar]
- Mäntynen S., Laanto E., Kohvakka A., Poranen M.M., Bamford J.K., Ravantti J.J. New enveloped dsRNA phage from freshwater habitat. J. Gen. Virol. 2015;96:1180–1189. doi: 10.1099/vir.0.000063. [DOI] [PubMed] [Google Scholar]
- Mönttinen H.A., Ravantti J.J., Poranen M.M. Evidence for a non-catalytic ion-binding site in multiple RNA-dependent RNA polymerases. PLoS One. 2012;7:e40581. doi: 10.1371/journal.pone.0040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönttinen H.A., Ravantti J.J., Stuart D.I., Poranen M.M. Automated structural comparisons clarify the phylogeny of the right-hand-shaped polymerases. Mol. Biol. Evol. 2014;31:2741–2752. doi: 10.1093/molbev/msu219. [DOI] [PubMed] [Google Scholar]
- Makeyev E.V., Bamford D.H. The polymerase subunit of a dsRNA virus plays a central role in the regulation of viral RNA metabolism. EMBO J. 2000;19:6275–6284. doi: 10.1093/emboj/19.22.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev E.V., Bamford D.H. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage Φ6. EMBO J. 2000;19:124–133. doi: 10.1093/emboj/19.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev E.V., Bamford D.H. Primer-independent RNA sequencing with bacteriophage Φ6 RNA polymerase and chain terminators. RNA. 2001;7:774–781. doi: 10.1017/s1355838201002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw T., Mindich L., Frangione B. Nucleotide sequence of the small double-stranded RNA segment of bacteriophage Φ6: novel mechanism of natural translational control. J. Virol. 1986;58:142–151. doi: 10.1128/jvi.58.1.142-151.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L., Bamford D.H. Lipid-containing bacteriophages. In: Calendar R., editor. The Bacteriophages. Plenum Publishing Corporation; New York: 1988. pp. 475–520. [Google Scholar]
- Mindich L., Nemhauser I., Gottlieb P., Romantschuk M., Carton J., Frucht S., Strassman J., Bamford D.H., Kalkkinen N. Nucleotide sequence of the large double-stranded RNA segment of bacteriophage Φ6: genes specifying the viral replicase and transcriptase. J. Virol. 1988;62:1180–1185. doi: 10.1128/jvi.62.4.1180-1185.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L., Qiao X., Onodera S., Gottlieb P., Strassman J. Heterologous recombination in the double-stranded RNA bacteriophage Φ6. J. Virol. 1992;66:2605–2610. doi: 10.1128/jvi.66.5.2605-2610.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L., Qiao X., Qiao J., Onodera S., Romantschuk M., Hoogstraten D. Isolation of additional bacteriophages with genomes of segmented double-stranded RNA. J. Bacteriol. 1999;181:4505–4508. doi: 10.1128/jb.181.15.4505-4508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage Φ6. Microbiol. Mol. Biol. Rev. 1999;63:149–160. doi: 10.1128/mmbr.63.1.149-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L. Reverse genetics of dsRNA bacteriophage Φ6. Adv. Virus Res. 1999;53:341–353. doi: 10.1016/s0065-3527(08)60355-3. [DOI] [PubMed] [Google Scholar]
- Mindich L. Packaging, replication and recombination of the segmented genome of bacteriophage Φ6 and its relatives. Virus Res. 2004;101:83–92. doi: 10.1016/j.virusres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Mindich L. Packaging in dsRNA viruses. Adv. Exp. Med. Biol. 2012;726:601–608. doi: 10.1007/978-1-4614-0980-9_26. [DOI] [PubMed] [Google Scholar]
- Moustafa I.M., Shen H., Morton B., Colina C.M., Cameron C.E. Molecular dynamics simulations of viral RNA polymerases link conserved and correlated motions of functional elements to fidelity. J. Mol. Biol. 2011;410:159–181. doi: 10.1016/j.jmb.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa I.M., Korboukh V.K., Arnold J.J., Smidansky E.D., Marcotte L.L., Gohara D.W., Yang X., Sánchez-Farrán M.A., Filman D., Maranas J.K., Boehr D.D., Hogle J.M., Colina C.M., Cameron C.E. Structural dynamics as a contributor to error-prone replication by an RNA-dependent RNA polymerase. J. Biol. Chem. 2014;289:36229–36248. doi: 10.1074/jbc.M114.616193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Carpenter C.D., Simon A.E. A novel 3'-end repair mechanism in an RNA virus. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1113–1118. doi: 10.1073/pnas.94.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D., Cheng N., Qiao J., Mindich L., Steven A.C., Heymann J.B. Stepwise expansion of the bacteriophage varphi6 procapsid: possible packaging intermediates. J. Mol. Biol. 2011;414:260–271. doi: 10.1016/j.jmb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D., Qiao J., Mindich L., Steven A.C., Heymann J.B. Packaging accessory protein P7 and polymerase P2 have mutually occluding binding sites inside the bacteriophage Φ6 procapsid. J. Virol. 2012;86:11616–11624. doi: 10.1128/JVI.01347-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D., Boura E., Wu W., Cheng N., Plevka P., Qiao J., Mindich L., Heymann J.B., Hurley J.H., Steven A.C. Subunit folds and maturation pathway of a dsRNA virus capsid. Structure. 2013;21:1374–1383. doi: 10.1016/j.str.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld K.L., Galarza J.M., Richards O.C., Summers D.F., Ehrenfeld E. Identification of terminal adenylyl transferase activity of the poliovirus polymerase 3Dpol. J. Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman K.C., Abbondanzieri E.A., Landick R., Gelles J., Block S.M. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- Ng K.K., Arnold J.J., Cameron C.E. Structure-function relationships among RNA-dependent RNA polymerases. Curr. Top. Microbiol. Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly E.K., Kao C.C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- Ojala P.M., Bamford D.H. In vitro transcription of the double-stranded RNA bacteriophage phi 6 is influenced by purine NTPs and calcium. Virology. 1995;207:400–408. doi: 10.1006/viro.1995.1099. [DOI] [PubMed] [Google Scholar]
- Olkkonen V.M., Gottlieb P., Strassman J., Qiao X.Y., Bamford D.H., Mindich L. In vitro assembly of infectious nucleocapsids of bacteriophage Φ6: formation of a recombinant double-stranded RNA virus. Proc. Natl. Acad. Sci. U. S. A. 1990;87:9173–9177. doi: 10.1073/pnas.87.23.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D.L., Brick P., Hamlin R., Xuong N.G., Steitz T.A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Onodera S., Qiao X., Gottlieb P., Strassman J., Frilander M., Mindich L. RNA structure and heterologous recombination in the double-stranded RNA bacteriophage Φ6. J. Virol. 1993;67:4914–4922. doi: 10.1128/jvi.67.8.4914-4922.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera S., Sun Y., Mindich L. Reverse genetics and recombination in Φ8, a dsRNA bacteriophage. Virology. 2001;286:113–118. doi: 10.1006/viro.2001.0995. [DOI] [PubMed] [Google Scholar]
- Patel S.S., Wong I., Johnson K.A. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- Paul A.V., Wimmer E. Initiation of protein-primed picornavirus RNA synthesis. Virus Res. 2015;206:12–26. doi: 10.1016/j.virusres.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peersen O.B. Picornaviral polymerase structure, function and fidelity modulation. Virus Res. 2017;234:4–20. doi: 10.1016/j.virusres.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierra Rouvière C., Amador A., Badaroux E., Convard T., Da Costa D., Dukhan D., Griffe L., Griffon J.F., LaColla M., Leroy F., Liuzzi M., Loi A.G., McCarville J., Mascia V., Milhau J., Onidi L., Paparin J.L., Rahali R., Sais E., Seifer M., Surleraux D., Standring D., Dousson C. Synthesis of potent and broad genotypically active NS5B HCV non-nucleoside inhibitors binding to the thumb domain allosteric site 2 of the viral polymerase. Bioorg. Med. Chem. Lett. 2016;26:4536–4541. doi: 10.1016/j.bmcl.2016.01.042. [DOI] [PubMed] [Google Scholar]
- Plotch S.J., Bouloy M., Ulmanen I., Krug R.M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- Poranen M.M., Bamford D.H. Assembly of large icosahedral double-stranded RNA viruses. Adv. Exp. Med. Biol. 2012;726:379–402. doi: 10.1007/978-1-4614-0980-9_17. [DOI] [PubMed] [Google Scholar]
- Poranen M.M., Tuma R. Self-assembly of double-stranded RNA bacteriophages. Virus Res. 2004;101:93–100. doi: 10.1016/j.virusres.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Poranen M.M., Daugelavicius R., Ojala P.M., Hess M.W., Bamford D.H. A novel virus-host cell membrane interaction: membrane voltage-dependent endocytic-like entry of bacteriophage straight Φ6 nucleocapsid. J. Cell Biol. 1999;147:671–682. doi: 10.1083/jcb.147.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poranen M.M., Paatero A.O., Tuma R., Bamford D.H. Self-assembly of a viral molecular machine from purified protein and RNA constituents. Mol. Cell. 2001;7:845–854. doi: 10.1016/s1097-2765(01)00228-3. [DOI] [PubMed] [Google Scholar]
- Poranen M.M., Tuma R., Bamford D.H. Assembly of double-stranded RNA bacteriophages. Adv. Virus Res. 2005;64:15–43. doi: 10.1016/S0065-3527(05)64002-X. [DOI] [PubMed] [Google Scholar]
- Poranen M.M., Koivunen M.R., Bamford D.H. Nontemplated terminal nucleotidyltransferase activity of double-stranded RNA bacteriophage Φ6 RNA-dependent RNA polymerase. J. Virol. 2008;82:9254–9264. doi: 10.1128/JVI.01044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poranen M.M., Butcher S.J., Simonov V.M., Laurinmäki P., Bamford D.H. Roles of the minor capsid protein P7 in the assembly and replication of double-stranded RNA bacteriophage Φ6. J. Mol. Biol. 2008;383:529–538. doi: 10.1016/j.jmb.2008.08.082. [DOI] [PubMed] [Google Scholar]
- Poranen M.M., Salgado P.S., Koivunen M.R., Wright S., Bamford D.H., Stuart D.I., Grimes J.M. Structural explanation for the role of Mn2+ in the activity of phi6 RNA-dependent RNA polymerase. Nucleic Acids Res. 2008;36:6633–6644. doi: 10.1093/nar/gkn632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J., Mindich L. The template specificity of bacteriophage Φ6 RNA polymerase. J. Virol. 2013;87:10190–10194. doi: 10.1128/JVI.01467-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Qiao J., Mindich L. Interference with bacteriophage Φ6 genomic RNA packaging by hairpin structures. J. Virol. 1995;69:5502–5505. doi: 10.1128/jvi.69.9.5502-5505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Qiao J., Mindich L. An in vitro system for the investigation of heterologous RNA recombination. Virology. 1997;227:103–110. doi: 10.1006/viro.1996.8311. [DOI] [PubMed] [Google Scholar]
- Qiao X., Qiao J., Mindich L. Stoichiometric packaging of the three genomic segments of double-stranded RNA bacteriophage Φ6. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4074–4079. doi: 10.1073/pnas.94.8.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Qiao J., Onodera S., Mindich L. Characterization of phi 13, a bacteriophage related to Φ6 and containing three dsRNA genomic segments. Virology. 2000;275:218–224. doi: 10.1006/viro.2000.0501. [DOI] [PubMed] [Google Scholar]
- Qiao X., Sun Y., Qiao J., Mindich L. The role of host protein YajQ in the temporal control of transcription in bacteriophage Φ6. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15956–15960. doi: 10.1073/pnas.0807489105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Sun Y., Qiao J., Mindich L. Temporal control of message stability in the life cycle of double-stranded RNA bacteriophage Φ8. J. Virol. 2009;83:633–639. doi: 10.1128/JVI.01766-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J., Qiao X., Sun Y., Mindich L. Role of host protein glutaredoxin 3 in the control of transcription during bacteriophage Φ2954 infection. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6000–6004. doi: 10.1073/pnas.1000383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Sun Y., Qiao J., Di Sanzo F., Mindich L. Characterization of Φ2954, a newly isolated bacteriophage containing three dsRNA genomic segments. BMC Microbiol. 2010;10:55. doi: 10.1186/1471-2180-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Sun Y., Qiao J., Mindich L. Interaction of a host protein with core complexes of bacteriophage Φ6 to control transcription. J. Virol. 2010;84:4821–4825. doi: 10.1128/JVI.00026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Soni A., Bellecourt M.J., Landick R. Mechanisms of bacterial transcription termination: all good things must end. Annu. Rev. Biochem. 2016;85:319–347. doi: 10.1146/annurev-biochem-060815-014844. [DOI] [PubMed] [Google Scholar]
- Ren Z., Ghose R. Slow conformational dynamics in the cystoviral RNA-directed RNA polymerase P2: influence of substrate nucleotides and template RNA. Biochemistry. 2011;50:1875–1884. doi: 10.1021/bi101863g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z., Wang H., Ghose R. Dynamics on multiple timescales in the RNA-directed RNA polymerase from the cystovirus Φ6. Nucleic Acids Res. 2010;38:5105–5118. doi: 10.1093/nar/gkq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z., Franklin M C., Ghose R. Structure of the RNA-directed RNA polymerase from the cystovirus Φ12. Proteins. 2013;81:1479–1484. doi: 10.1002/prot.24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing U., August J.T. The 3'-terminus and the replication of phage RNA. Nature. 1969;224:853–856. doi: 10.1038/224853a0. [DOI] [PubMed] [Google Scholar]
- Romantschuk M., Bamford D.H. Function of pili in bacteriophage Φ6 penetration. J. Gen. Virol. 1985;66:2461–2469. doi: 10.1099/0022-1317-66-11-2461. [DOI] [PubMed] [Google Scholar]
- Ruigrok R.W., Crépin T., Hart D.J., Cusack S. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr. Opin. Struct. Biol. 2010;20:104–113. doi: 10.1016/j.sbi.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Salgado P.S., Makeyev E.V., Butcher S.J., Bamford D.H., Stuart D.I., Grimes J.M. The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure. 2004;12:307–316. doi: 10.1016/j.str.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Sanjuán R., Domingo-Calap P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016;73:4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin L.P., Poranen M.M., Lehti N.M., Ravantti J.J., Koivunen M.R., Aalto A.P., van Dijk A.A., Stuart D.I., Grimes J.M., Bamford D.H. Insights into the pre-initiation events of bacteriophage Φ6 RNA-dependent RNA polymerase: towards the assembly of a productive binary complex. Nucleic Acids Res. 2009;37:1182–1192. doi: 10.1093/nar/gkn1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin L.P., Wright S., Chen Q., Degerth L.H., Stuart D.I., Grimes J.M., Bamford D.H., Poranen M.M. The C-terminal priming domain is strongly associated with the main body of bacteriophage Φ6 RNA-dependent RNA polymerase. Virology. 2012;432:184–193. doi: 10.1016/j.virol.2012.05.035. [DOI] [PubMed] [Google Scholar]
- Sen A., Heymann J.B., Cheng N., Qiao J., Mindich L., Steven A.C. Initial location of the RNA-dependent RNA polymerase in the bacteriophage Φ6 procapsid determined by cryo-electron microscopy. J. Biol. Chem. 2008;283:12227–12231. doi: 10.1074/jbc.M710508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaevitz J.W., Abbondanzieri E.A., Landick R., Block S.M. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu B., Gong P. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E4005–4014. doi: 10.1073/pnas.1602591113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloane D.L., Goodman M.F., Echols H. The fidelity of base selection by the polymerase subunit of DNA polymerase III holoenzyme. Nucleic Acids Res. 1988;16:6465–6475. doi: 10.1093/nar/16.14.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidansky E.D., Arnold J.J., Reynolds S.L., Cameron C.E. Human mitochondrial RNA polymerase: evaluation of the single-nucleotide-addition cycle on synthetic RNA/DNA scaffolds. Biochemistry. 2011;50:5016–5032. doi: 10.1021/bi200350d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer D.A., Domingo E., Holland J.J. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122:281–288. doi: 10.1016/0378-1119(92)90216-c. [DOI] [PubMed] [Google Scholar]
- Steitz T.A. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- Sun X., Bamford D.H., Poranen M.M. Probing, by self-assembly, the number of potential binding sites for minor protein subunits in the procapsid of double-stranded RNA bacteriophage Φ6. J. Virol. 2012;86:12208–12216. doi: 10.1128/JVI.01505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Farsetta D.L., Nibert M.L., Harrison S.C. RNA synthesis in a cage–structural studies of reovirus polymerase λ3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- Thompson A.A., Peersen O.B. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23:3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk A.A., Makeyev E.V., Bamford D.H. Initiation of viral RNA-dependent RNA polymerization. J. Gen. Virol. 2004;85:1077–1093. doi: 10.1099/vir.0.19731-0. [DOI] [PubMed] [Google Scholar]
- Vidaver A.K., Koski R.K., Van Etten J.L. Bacteriophage Φ6: a lipid-Containing virus of Pseudomonas phaseolicola. J. Virol. 1973;11:799–805. doi: 10.1128/jvi.11.5.799-805.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M., Andino R. Closing the gap: the challenges in converging theoretical, computational, experimental and real-life studies in virus evolution. Curr. Opin. Virol. 2012;2:515–518. doi: 10.1016/j.coviro.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M., Stone J.K., Arnold J.J., Cameron C.E., Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., Poranen M.M., Bamford D.H., Stuart D.I., Grimes J.M. Noncatalytic ions direct the RNA-dependent RNA polymerase of bacterial double-stranded RNA virus Φ6 from de novo initiation to elongation. J. Virol. 2012;86:2837–2849. doi: 10.1128/JVI.05168-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Makeyev E.V., Bamford D.H. Comparison of polymerase subunits from double-stranded RNA bacteriophages. J. Virol. 2001;75:11088–11095. doi: 10.1128/JVI.75.22.11088-11095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Gottlieb P., Wei H., Bamford D.H., Makeyev E.V. Temperature requirements for initiation of RNA-dependent RNA polymerization. Virology. 2003;314:706–715. doi: 10.1016/s0042-6822(03)00460-4. [DOI] [PubMed] [Google Scholar]
- Yang H., Makeyev E.V., Butcher S.J., Gaidelyte A., Bamford D.H. Two distinct mechanisms ensure transcriptional polarity in double-stranded RNA bacteriophages. J. Virol. 2003;77:1195–1203. doi: 10.1128/JVI.77.2.1195-1203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Welch J.L., Arnold J.J., Boehr D.D. Long-range interaction networks in the function and fidelity of poliovirus RNA-dependent RNA polymerase studied by nuclear magnetic resonance. Biochemistry. 2010;49:9361–9371. doi: 10.1021/bi100833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Smidansky E.D., Maksimchuk K.R., Lum D., Welch J.L., Arnold J.J., Cameron C.E., Boehr D.D. Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure. 2012;20:1519–1527. doi: 10.1016/j.str.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Weng P.J., Gao Y. A new paradigm of DNA synthesis: three-metal-ion catalysis. Cell Biosci. 2016;6:51. doi: 10.1186/s13578-016-0118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W., Gutshall L.L., Del Vecchio A.M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J. Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]