Abstract
Dietary restriction (DR) is the most robust environmental manipulation known to increase active and healthy lifespan in many species. Despite differences in the protocols and the way DR is carried out in different organisms, conserved relationships are emerging among multiple species. Elegant studies from numerous model organisms are further defining the importance of various nutrient-signaling pathways including mTOR (mechanistic target of rapamycin), insulin/IGF-1-like signaling and sirtuins in mediating the effects of DR. We here review current advances in our understanding of the molecular mechanisms altered by DR to promote lifespan in three major invertebrate models, the budding yeast Saccharomyces cerevisiae, the nematode Caenorhabditis elegans, and the fruit fly Drosophila melanogaster.
Keywords: Yeast, worms, flies, mitochondrial respiration, sirtuins, autophagy, metabolism
1. Introduction
Reduction of particular or total nutrient intake without causing malnutrition is termed dietary restriction (DR). DR in this broad sense includes caloric restriction (CR), in which total calorie intake is reduced, as well as dietary interventions involving the restriction of specific dietary components (protein, lipid, or carbohydrates). Unraveling the underlying molecular mechanisms by which DR extends lifespan in model organisms like the budding yeast Saccharomyces cerevisiae, the nematode Caenorhabditis elegans and the fruitfly Drosophila melanogaster has played a critical role in shaping the field in part due to the short lifespan, the ease of manipulation, and powerful genetic tools offered by these models. Firstly, it has led to the identification of conserved genetic pathways that mediate the DR response (Fontana et al., 2010; Katewa and Kapahi, 2011; Mair and Dillin, 2008). Secondly, it has helped define the importance of nutrient composition and dispelled the belief that calories alone mediate the effects of DR (Kapahi et al., 2010; Simpson and Raubenheimer, 2009; Tatar, 2011). Finally, it has helped better understand the physiological changes and the ultimate downstream cellular and molecular mechanisms that mediate the effects of DR (Fontana et al., 2010; Kapahi et al., 2010). Below we review insights from S. cerevisiae, C. elegans, and D. melanogaster on the molecular mechanisms of DR.
2. Dietary restriction in S. cerevisiae
2.1. Aging and dietary restriction in S. cerevisiae
As a unicellular model organism, the budding yeast S. cerevisiae allows for the distinct study of both mitotic and post-mitotic aging at the organismal level (Longo et al., 2012). These two types of aging are referred to as replicative and chronological aging, respectively. In the replicative aging model, lifespan is quantified as the number of daughter cells a mother cell produces prior to irreversible cell cycle arrest (Steinkraus et al., 2008a). Chronological lifespan, in contrast, is defined as the length of time that an individual yeast cell can maintain viability in a non-dividing quiescent state and yet retain the ability to re-enter the cell cycle upon appropriate stimulation (Fabrizio and Longo, 2003). Studies of both types of aging in yeast have uncovered novel mechanisms of aging (Kaeberlein, 2010; Longo and Fabrizio, 2012; Mirisola et al., 2014), and genome-wide analyses have been carried out for both replicative (McCormick et al., 2015; Smith et al., 2008b) and chronological (Burtner et al., 2011; Fabrizio et al., 2010; Matecic et al., 2010; Powers et al., 2006; Smith et al., 2016) lifespan, which have identified multiple genetic modifiers of aging in yeast that are shared with multicellular eukaryotes (discussed further below).
Replicative lifespan (RLS) has been most often assessed through manual micro-dissection of yeast daughter cells away from mother cells, as first described by Mortimer and Johnston more than fifty years ago (Mortimer and Johnston, 1959). The vast majority of published studies of RLS in yeast have been performed using variations of this method in which the cells are maintained on a rich nutrient growth medium containing yeast extract, peptone, and glucose (dextrose), referred to as YPD (Steffen et al., 2009). Cells are typically maintained at 30°C during the day and micro-dissected every 2-3 hours, then placed at 4°C overnight to prevent overgrowth. More recently, several microfluidic methods for assessing RLS have been described in which mother cells are physically constrained in a microfluidic device such that fluid flow washes the daughter cells away from the mother cells (Chen et al., 2016). Both YPD and synthetic complete (SC) media types are routinely used for microfluidic RLS experiments.
The methodologies used to measure chronological lifespan (CLS) in yeast are more varied than for RLS. The first method described, and the one still most commonly used, involves culturing cells in SC media until they reach stationary phase, maintaining the cells in their expired media, and periodically plating a subset of the population onto solid YPD media to assess viability based on colony forming units (Fabrizio and Longo, 2003). Similar approaches but with different initial culture media including liquid YPD (Ashrafi et al., 1999) or amino acid restricted solid synthetic media (Madia et al., 2007) have also been used. Higher throughput approaches involving monitoring survival based on outgrowth by optical density in liquid culture using multi-well plate readers have also become common (Jung et al., 2015; Murakami and Kaeberlein, 2009).
DR extends lifespan and has been studied extensively in both yeast-aging paradigms. The most common form of DR used in yeast is a reduction in the starting glucose concentration of the culture medium from 2% to either 0.5% or 0.05% (Longo et al., 2012). Other forms of DR, which have been reported to extend RLS, include growth in the non-fermentable carbon source glycerol and amino-acid restriction (Jiang et al., 2000), but these methods are used much less frequently. DR by growth in glycerol and other non-fermentable carbon sources also extends CLS (Murakami et al., 2011; Smith et al., 2007), as does methionine (Johnson and Johnson, 2014) or tryptophan restriction (He et al., 2014). DR by either a reduction in glucose or growth in a non-fermentable carbon source results in several physiological changes to yeast cells that have been proposed to underlie extension of RLS and CLS. These cellular responses to DR can generally be categorized as changes in metabolism, nutrient signaling, and stress response, as summarized below.
2.2. Effects of dietary restriction on mitochondrial respiration in yeast
The primary metabolic change that yeast cells undergo in response to DR is a shift from non-oxidative glycolysis and ethanol fermentation in the presence of glucose to an induction of mitochondrial respiratory metabolism when glucose becomes limiting or absent. The first model to gain wide attention as a potential mechanism for DR-mediated extension of RLS proposed that this respiratory shift led to activation of the yeast sirtuin protein deacetylase, Sir2, via shifting the coenzyme nicotinamide adenine dinucleotide NAD+:NADH ratio toward NAD+ (Lin et al., 2000; Lin et al., 2002), which is an activator and substrate of Sir2 (Imai et al., 2000). Prior studies had shown that overexpression of Sir2 was sufficient to increase RLS through its pro-silencing and anti-recombinogenic activity in the ribosomal DNA (rDNA) (Kaeberlein et al., 1999). In support of this model, overexpression of the Hap4 transcription factor, which is a limiting factor for expression of mitochondrial respiratory genes (Forsburg and Guarente, 1989), is sufficient to induce respiration and increase lifespan when glucose is present at high levels (Lin et al., 2002), and Hap4 is required for full lifespan extension from DR (Wang et al., 2010). A variant on this model has also been proposed, whereby activation of the NAD+ salvage pathway under conditions of DR results in increased intracellular levels of NAD+ and activation of Sir2 (Anderson et al., 2003).
A direct role for respiration or Sir2 in mediating RLS extension from DR has been weakened by subsequent studies demonstrating that neither respiration (Kaeberlein et al., 2005a) nor Sir2 (Kaeberlein et al., 2004) or other sirtuins (Kaeberlein et al., 2006a; Tsuchiya et al., 2006) are required for RLS extension from DR. Likewise, activation of Sir2 at the rDNA in response to DR was not detected when sensitive measures of rDNA silencing were employed (Smith et al., 2009). It has been argued that Sir2-independent, respiration-independent lifespan extension from DR could simply represent an alternative mechanism by which DR extends lifespan under certain conditions, specifically that the Sir2-mediated pathway is more important under moderate DR (0.5% glucose) while the Sir2-independent pathway is more important under severe DR (0.05% glucose) (Lin and Guarente, 2006). Although sirtuins are dispensible for RLS extension at any glucose level (Tsuchiya et al., 2006), it may be the case that the shift to respiratory metabolism under DR conditions activates Sir2 to promote RLS under certain conditions.
The importance of a metabolic shift toward increased respiration in DR-mediated CLS extension is more straightforward. As mentioned above, cells maintained in standard media with 2% glucose undergo glycolytic and fermentative metabolism. Under standard CLS conditions where cells are grown in liquid culture with a high initial glucose concentration, this metabolic state corresponds to the first several hours of exponential growth until the glucose becomes exhausted and ethanol (from fermentation) accumulates. Once the available glucose is utilized, the cells switch over to respiratory metabolism and begin utilizing the accumulated ethanol as a carbon source to support ATP production. As a byproduct of this metabolic switch, high levels of organic acids, particularly acetic acid, accumulate in the culture medium during the first 24-48 hours of a standard CLS experiment and cause the pH to drop dramatically (Burtner et al., 2009). This acidification of the culture medium induces an apoptotic response that causes a majority of the cell death during the remainder of the CLS experiment. DR, which induces respiration and blocks fermentative metabolism during the early outgrowth phase, increases CLS by preventing culture acidification (Burtner et al., 2009). Buffering the culture medium to prevent acidification under control conditions phenocopies the CLS extension from DR (Burtner et al., 2009), as does overexpression of Hap4 (Piper et al., 2006). Similar to RLS, deletion of Sir2 does not prevent CLS extension from DR (Smith et al., 2007), and instead Sir2 activity appears to limit CLS under both control and DR conditions (Fabrizio et al., 2005).
2.3. Effects of dietary restriction on nutrient signaling and stress responses in yeast
Like all eukaryotic cells, yeast have evolved to sense and appropriately respond to nutrient cues from their environment via evolutionarily conserved signaling pathways. These conserved nutrient response signaling factors include three important kinases: the mechanistic target of rapamycin (mTOR, encoded by partially redundant TOR1 and TOR2 genes in yeast), the AMP-activated protein kinase A (PKA, encoded by redundant TPK1, TPK2, and TPK3 genes), and the ribosomal S6 kinase homolog Sch9. These three kinases interact in a still incompletely understood network to coordinately regulate several downstream processes in response to nutritional and other environmental signals. These downstream processes include regulation of the cell cycle, mRNA translation, protein synthesis and degradation, metabolism and mitochondrial function, and a variety of stress response factors (reviewed in Longo et al., 2012).
There is strong evidence supporting a role for these signaling pathways in both CLS and RLS extension. DR reduces the activity of mTOR, PKA and Sch9, and in both aging paradigms, loss-of-function mutations in these kinases are sufficient to extend lifespan (Fabrizio et al., 2004; Fabrizio et al., 2001; Kaeberlein et al., 2005b; Lin et al., 2000; Powers et al., 2006). In the case of mTOR, pharmacological inhibition with the drug rapamycin is also sufficient to extend both RLS (Medvedik et al., 2007) and CLS (Powers et al., 2006). Several of the downstream effectors regulated by these pathways have been proposed to account for lifespan extension in response to DR. Most notably, regulation of mRNA translation and genomic stability at the rDNA appears to be particularly important for RLS (Steffen et al., 2008; Steffen et al., 2012), while induction of stress response transcription factors, autophagy, reduced replication stress and genome instability, as well as altered mitochondrial ROS signaling is important for CLS extension (Alvers et al., 2009a; Alvers et al., 2009b; Bonawitz et al., 2007; Longo, 2003; Madia et al., 2008; Ocampo et al., 2012; Pan et al., 2011; Pan and Shadel, 2009; Wei et al., 2008; Weinberger et al., 2007).
Given the complex physiological changes associated with DR in yeast, it seems likely that multiple overlapping outputs are important for lifespan extension, and the relative importance of individual downstream effectors is dependent on the specific experimental conditions (Figure 1). There is also overlap between the various downstream components of DR. For example, inhibition of mTOR or Sch9 increases mitochondrial respiration and also induces stress response transcription factors that can enhance resistance to acidification-induced cell death (Bonawitz et al., 2007; Fabrizio et al., 2001; Lavoie and Whiteway, 2008; Pedruzzi et al., 2003). In addition, chronological aging leads to reduced RLS (Ashrafi et al., 1999; Delaney et al., 2013; Murakami et al., 2012), suggesting underlying mechanistic similarities between mitotic and post-mitotic lifespan within the same eukaryotic cell.
Figure 1.
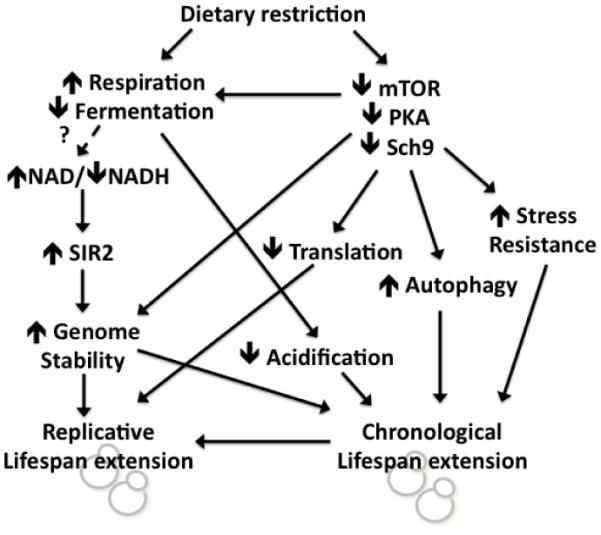
Effector mechanisms of dietary restriction that contribute to lifespan extension in the budding yeast S. cerevisae. See text for details.
Beyond the pathways and mechanisms described above, it is worth nothing that several additional genes and processes have been implicated in RLS or CLS extension from DR. These include chromatin remodeling factors (Dang et al., 2014), maintenance of cellular ATP levels (Choi and Lee, 2013), autophagy and lipid homeostasis (Aris et al., 2013; Lester et al., 2013; Richard et al., 2013), rDNA replication (Kwan et al., 2013), nitric oxide (Li et al., 2011), and glutathione and redox homeostasis (Mannarino et al., 2008; Molin et al., 2011), among others. The relationship of each of these to the established longevity mechanisms and to each other will require additional study. In addition to differences in culture conditions between labs, another complicating factor in developing a unified model for DR in yeast is the use of different strain backgrounds. It is clear that genotype plays a major role in the way that yeast cells respond to DR, with some genotypes showing robust lifespan extension, others showing no response, and some even having their lifespan shortened by DR (Schleit et al., 2013). Thus, studies examining the generality of different mechanisms for lifespan extension across strain backgrounds would be quite valuable and are easily feasible in the yeast-model system.
In summary, DR has been most extensively studied in yeast by reducing the glucose concentration of the culture medium, which extends both RLS and CLS. DR induces a shift in metabolism away from fermentation toward mitochondrial respiration and impacts signaling via a network of nutrient-responsive kinases, including mTOR, PKA, and Sch9. Although further work is needed to determine the precise mechanisms by which DR extends lifespan in both yeast-aging paradigms, many of these mechanisms appear to be conserved in multicellular eukaryotes, as mitochondrial function and nutrient signaling through orthologous factors have been shown to be important for DR-mediated longevity effects in both C. elegans and D. melanogaster (discussed further below), as well as in mice (as reviewed elsewhere in this special issue).
3. Dietary restriction in C. elegans
3.1. Aging and dietary restriction in C. elegans
Similar to the unicellular S. cerevisiae, the microscopic soil nematode C. elegans has played an important role in aging studies due to its short life cycle (3 days for an embryo to become an adult, which lives ~2-3 weeks), relatively complex tissue differentiation (959 cells and multiple differentiated tissues), easy genetic manipulations (RNA interference (RNAi) by feeding, gene overexpression), and highly conserved genome (at least 80% conserved genes). C. elegans has been extremely useful in discovering novel genes important for the beneficial effects of DR and getting insights into their tissue-specific requirements.
Multiple methods of DR has been developed and used in C. elegans (reviewed in Greer and Brunet, 2009; Lan et al., 2015), which normally is maintained in the laboratory on solid media plates on which bacterial lawns of E. coli are growing. A key genetic model is the eat-2 mutant. The eat-2 gene encodes a nicotinic acetyl-choline receptor, which functions postsynaptically in the pharyngeal muscle, the feeding apparatus of the animal, to regulate the rate of feeding. Mutations in this gene render the pharynx non-efficient in grinding the bacteria, and DR is effectively obtained on regular media plates (Raizen et al., 1995). These animals show phenotypes observed in other species subjected to DR, including ~30% longer lifespan (Lakowski and Hekimi, 1998). However, there are important limitations to this model since eat-2 mutants are also subject to DR during development, and the utilized alleles are typically hypomorphic mutations.
Multiple types of food dilutions can also be used to effectively subject C. elegans to DR. These methods include dilution of bacteria in liquid, called bDR (Panowski et al., 2007) or lDR (Bishop and Guarente, 2007). Another method is done by seeding diluted bacterial cultures onto solid media plates, referred to as sDR (Greer et al., 2009), with a variation to also reduce peptone concentration (msDR) (Chen et al., 2009). More extreme regimens include complete dietary deprivation (DD) in which no bacterial is seeded onto the media plates (Kaeberlein et al., 2006b; Lee et al., 2006), or intermittent fasting (IF) where animals are moved between media plates with or without bacteria (Honjoh et al., 2009). Unlike eat-2 mutants, these methods can be initiated at any given point of time, and they all result in significant lifespan extensions (in the range of 30-50%). These DR regimens have been used in C. elegans to identify a number of genes and processes, including several highly conserved transcription factors and signaling pathways, which work in complex tissue-specific fashions, as discussed below and summarized in Figure 2.
Figure 2.
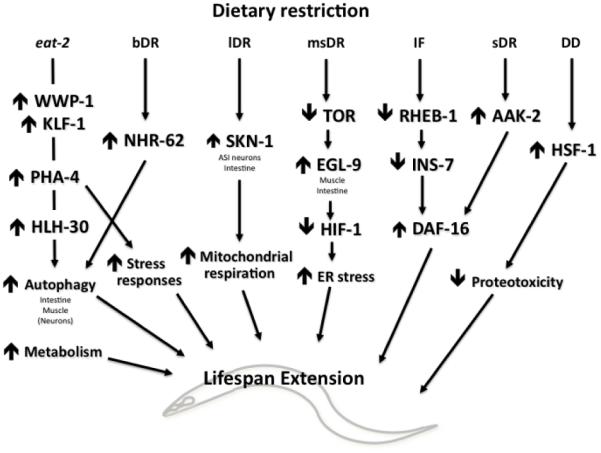
Effector mechanisms of dietary restriction that contribute to lifespan in the nematode C. elegans. Different dietary-restriction regimens are listed, as well as information about the site-of-action of genes and processes where known. See text for details.
3.2. mTOR and autophagy constitute a major effector pathway regulated by dietary restriction in C. elegans
As in yeast, the conserved mTOR and insulin/IGF-1 nutrient-sensing pathways have been linked to longevity and to DR in C. elegans. The nutrient sensor mTOR (called LET-363 in C. elegans) is a kinase with important functions in growth regulation. DR reduces mTOR activity and reduction in mTOR activity extends lifespan in multiple species (Kapahi et al., 2010). Moreover, DR and mTOR inhibition in combination does not further extend lifespan in C. elegans (Hansen et al., 2007), similar to the case in yeast (Kaeberlein et al., 2005b), and in fruit flies (Kapahi et al., 2004).
Multiple conserved pathways and genetic factors have been proposed to mediate the lifespan-extending effects of DR and of mTOR inhibition. A major candidate mechanism is macroautophagy (hereafter referred to as autophagy) – a cellular recycling process by which cytosolic components are targeted for degradation in the lysosome that is induced in response to nutrient deprivation (Feng et al., 2014). Studies in C. elegans using dietary-restricted eat-2 mutants have shown that this modulation in autophagy is directly linked to DR-mediated longevity since inhibition of several autophagy-related genes (unc-51/ATG1/Ulk1, bec-1/ATG6/Beclin1, vps-34, atg-18/Wipi, and atg-7) shortens the long lifespan of animals with eat-2 mutations (Gelino et al., 2016; Hansen et al., 2008; Jia and Levine, 2007; Toth et al., 2008). Animals carrying mutations in the mTOR interaction partner, daf-15/Raptor similarly requires autophagy genes for their lifespan extension (Hansen et al., 2008).
Consistently, several transcription factors required for DR- or mTOR-mediated lifespan extension, such as the FOXA ortholog PHA-4 (see below), the nuclear hormone receptor NHR-62 (Heestand et al., 2013), and the TFEB ortholog HLH-30, increase expression of many autophagy genes in eat-2 mutants, and several of these also in C. elegans with reduced mTOR activity (reviewed in Lapierre et al., 2015). Notably, knockdown of autophagy genes atg-18/Wipi, and lgg-1/Atg8 only in the intestine of eat-2 mutants are not only sufficient to shorten their extended lifespan, but also to disrupt an improved intestinal barrier function (i.e., the integrity of the intestine is enhanced by dietary-restriction, as first observed in D. melanogaster (Regan et al., 2016; Rera et al., 2011; Rera et al., 2012), see fly section) as well as to reduce motility, indicating both cell-autonomous and cell non-autonomous roles for autophagy in dietary-restricted animals (Gelino et al., 2016). It is possible that this autophagy-related longevity mechanism involves turnover of mitochondria, as genes involved in mitophagy are similarly required for the long lifespan of eat-2 mutants (Palikaras et al., 2015).
3.3. Other conserved determinants important for diet-restriction-mediated longevity in C. elegans
Additional genetic factors with conserved roles in longevity have been linked to the lifespan extension mediated by DR in C. elegans. Genes acting in the insulin/IGF-1 pathway were the first genes to be directly linked to aging (reviewed in Kenyon, 2010), and some of the genes in this pathway, including the FOXO transcription factor DAF-16 has been linked to DR. Specifically, daf-16 is required for the DR regimens sDR and IF to extend lifespan. In sDR, the nutrient sensor AMP-activated kinase aak-2 activates daf-16 to promote longevity (Greer et al., 2009), whereas IF inhibits RHEB-1, an upstream regulator of mTOR, causing inhibition of the insulin ins-7 that again activates daf-16/Foxo thereby extending C. elegans lifespan (Honjoh et al., 2009). Moreover, DAF-15/Raptor is inhibited by DAF-16/FOXO at the transcriptional levels (Jia et al., 2004). A recent transcriptome analysis further linked rheb-1, daf-16, and aak-2 to the DR response in C. elegans (Hou et al., 2016). The heat-shock inducible transcription factor HSF-1, another transcription factor linked to the lifespan extension observed in long-lived daf-2/insulin/IGF-1 receptor mutants, is also required for the DR regimen DD to promote longevity as well as to protect against protein aggregation (Steinkraus et al., 2008b).
As described above for yeast, the importance of the C. elegans Sir2 homolog, sir-2.1, in DR-mediated lifespan extension is also unclear, as there are conflicting reports on this point. One report showed that the eat-2(ad1113) and eat-2(ad465) alleles require sir-2.1 to promote longevity (Wang and Tissenbaum, 2006), whereas the eat-2(ad1116) allele does not (Hansen et al., 2007). Moreover, several reports have described that sir-2.1 mutants subjected to DR by a variety of bacterial dilution protocols experience full lifespan extension (Kaeberlein et al., 2006b; Lamming et al., 2005; Lee et al., 2006; Mair and Dillin, 2008).
3.4. Additional genetic determinants regulated by dietary restriction in C. elegans
Additional genetic factors including several highly conserved transcription factors have been linked to DR-mediated longevity in C. elegans. It remains to be established to which extent these factors will prove to have conserved effects in DR.
PHA-4/FOXA
The FOXA transcription factor PHA-4, which was first reported to play a key role in the embryonic development of the C. elegans foregut, has been linked specifically to DR in C. elegans (Panowski et al., 2007). PHA-4/FOXA is required during adulthood for the long lifespan of both eat-2 mutants and for animals subjected to DR by bacterial dilution (bDR), but is not required for insulin/IGF-1 receptor mutants, or mitochondrial respiration mutants to live long [it was later showed that pha-4/Foxa is also required for germline-less glp-1 mutants to live long, but these animals may be dietary restricted as they have reduced mTOR levels (Lapierre et al., 2011)]. Moreover, DR increases PHA-4/FOXA levels, and overexpression of PHA-4 is sufficient to extend lifespan and improve stress resistance. In adult animals, PHA-4/FOXA is expressed primarily in the intestine but also in a few head and tail neurons, indicating an important function for the gut and neurons in DR (Panowski et al., 2007). Later work has profiled eat-2 mutants to find a large feed-forward gene regulatory network regulated to a large degree by PHA-4/FOXA. This network is enriched for genes with functions related to cellular signaling and cell-protective processes such as ubiquitin-related degradation and autophagy (Pandit et al., 2014), which play critical roles in DR and longevity. Recent work has also indicated overlap in target genes regulated by PHA-4/FOXA and DAF-16/FOXO, such as the chromatin modifier ZFP-1, which is required for the long lifespan of daf-2/insulin/IGF-1 signaling and the DR regimens of eat-2 and bDR (Singh et al., 2016).
SKN-1
The Nrf2 transcription factor SKN-1, which was first reported to be critical for endodermal development and for regulation of the oxidative stress response, has similarly been linked to DR in C. elegans (Bishop and Guarente, 2007). SKN-1 is also required for other longevity paradigms, including daf-2/insulin/IGF-1 mutants (Tullet et al., 2008). SKN-1 is expressed in two head neuron (ASI neurons) and in the intestine, yet SKN-1 is required only in the ASI neurons to mediate lifespan extension by DR (Bishop and Guarente, 2007). As in yeast, DR increases mitochondrial respiration, and in C. elegans this change in metabolism is mediated by SKN-1 specifically in the ASI neurons (Bishop and Guarente, 2007). Several explanations have been put forward to explain such a neuroendocrine circuit, including the involvement of transient neuronal reactive oxygen species (ROS) signaling (Schmeisser et al., 2013). To this end, it is interesting to note that the thioredoxin trx-1 is important for DR-mediated (eat-2 and DD) lifespan extension in C. elegans via increased expression in ASJ neurons (Fierro-Gonzalez et al., 2011).
Two potential SKN-1 targets, identified in a gene-expression profiling study of genes regulated by SKN-1 under oxidative-stress conditions (Park et al., 2009), have been linked to DR-mediated longevity in C. elegans, namely the neuropeptide NLP-7 and the ligand-gated ion channel CUP-4. nlp-7, the homolog of mammalian ortholog cholecystokinin, is primarily expressed in head neurons, and is required, along with its two putative receptors CKR-1 and CKR-2, for the long lifespan of eat-2 mutants (Park et al., 2010), suggesting a possible signaling mechanism still to be formally tested. The ion-gated channel protein cup-4 is similarly required for the long lifespan of eat-2 as well as in protocols of bacterial dilution on plates (sDR) (Park et al., 2010). CUP-4 is important for endocytosis and is uniquely expressed only in coelomocytes (Patton et al., 2005), six specialized cells with immune and detoxification functions via uptake of material present in the body cavity of the animal. Notably, the coelomocytes are essential for multiple forms of DR (sDR and lDR) to extend lifespan (Cypser et al., 2013), indicating that coelomocyte-mediated endocytosis plays an important role in DR-mediated longevity. Consistent with this notion, two other genes with predicted functions in coelomocyte endocytosis, lgc-26 and cup-5, have been linked to DR (Park et al., 2010).
HIF-1
The conserved hypoxia-inducible transcription factor HIF-1 is a target of the mTOR pathway in mammals, and has been linked to DR in C. elegans (Chen et al., 2009). hif-1 deficiency promotes longevity under nutrient-rich conditions, but fails to show lifespan extension under msDR. HIF-1 is negatively regulated by EGL-9, a proline hydroxylase, and egl-9 mutants, that have increased HIF-1 activity, are not responsive to DR-mediated longevity. EGL-9 works in specific neurons and in muscle to mediate longevity, and functions, as does msDR, via the stress regulator ire-1 to reduce endoplasmatic reticulum (ER) stress (Chen et al., 2009), suggesting tissue-specific functions for HIF-1.
The relationship between HIF-1, DR, and longevity is indeed complex and condition-dependent, as deletion of HIF-1 extends lifespan at high temperature (25°C) but not at low temperature (15°C), while activation of HIF-1 extends lifespan at temperatures ranging from 15°C-25°C (Leiser et al., 2011; Mehta et al., 2009; Muller et al., 2009; Zhang et al., 2009). Recently, neuronal stabilization of HIF-1 was shown to increase lifespan via a cell non-autonomous activation of the flavin-containing monooxygenase FMO-2 in the intestine (Leiser et al., 2015). Interestingly, intestinal FMO-2 is also induced by DD via a HIF-1-independent, but HLH-30-dependent mechanism (Leiser et al., 2015). Collectively, these observations suggest that different forms of DR interact with HIF-1 to modulate expression of downstream longevity factors, but further work is needed to clarify these interactions and to understand the contributions of other environmental factors such as temperature and oxygen availability.
WWP-1 and KLF-1
The conserved HECT E3 ubiquitin ligase WWP-1 has been linked specifically to the DR in C. elegans (Carrano et al., 2009). wwp-1 is required for the long lifespan of both eat-2 mutants and in response to bDR, and overexpression of WWP-1 is sufficient to extend lifespan in a pha-4-dependent manner (Carrano et al., 2009). The Kruppel- like transcription factor KLF-1 is a substrate for ubiquitylation by WWP-1, and, consistently, klf-1 is similarly required for DR-mediated lifespan, and intestinal overexpression is sufficient to extend C. elegans lifespan (Carrano et al., 2014), highlighting a DR-specific longevity pathway that remains to be investigated in other organisms.
SAMS-1, RAB-10, DRR-1, and DRR-2
A genome-wide RNAi screen to discover new longevity genes identified several new genes with specific effects in DR-mediated longevity. These include the S-adenosyl methionine synthetase sams-1, the small GTPase rab-10, and two proteins with no clear homologs, named drr-1 and drr-2 for dietary-restriction response. Inhibition of these genes extended the lifespan of wild-type animals, but not of eat-2 mutants; consistently, the expression of these genes is reduced in eat-2 mutants (Hansen et al., 2005). Later investigations of drr-2 have shown that it is a functional ortholog of human eukaryotic translation initiation factor 4H (eIF4H), and it is partially required for lifespan extension by sDR or by reduced DAF-15/Raptor levels (Ching et al., 2010). Further epistasis analysis has suggested that drr-2/eIF4H might act downstream of both sams-1 and rab-10 to mediate longevity effects of DR via reduced mTOR signaling (Ching et al., 2010).
Additional genes with roles in metabolism
A recent study systematically examined eat-2 mutants by quantitative proteomics, immunochemistry, and metabolic quantification to identify effects on energy metabolism and their contribution to DR-mediated responses, including lifespan (Yuan et al., 2012). These studies showed a switch to fatty acid metabolism as an energy source, and an enhanced rate of energy metabolism via complex alterations in the tricarboxylic acid (TCA) cycle in eat-2 mutants compared to wild-type animals. Consistently, several metabolic genes were required for DR-mediated longevity, including the acetyl-CoA carboxylase pod-2 (Yuan et al., 2012), as observed in flies (Katewa et al., 2012).
In conclusion, C. elegans has been successfully used to identify many new genes and biological processes with critical roles in DR-mediated longevity (Figure 2). Many of these genes and processes are appearing to act in complex, tissue-specific and cell non-autonomous manners. The availability of many different experimental regimens to model DR in C. elegans has been a strength but also a challenge to the field. These different DR models provide opportunities to identify a variety of potential mechanisms of action for DR; however, they have also to some extent prevented the field from achieving consensus about which mechanisms are most important. Inhibition of mTOR signaling and enhanced autophagy, in particular, appear to be necessary for enhanced longevity across a variety of different DR paradigms in C. elegans, as observed in other organisms. A major future challenge for the C. elegans DR field will be to better understand how the numerous genes implicated in different forms of DR interact within the longevity network, are coordinately regulated between tissues, and also how they have affect the healthspan of the organism.
4. Dietary restriction in D. melanogaster
4.1. Aging and dietary restriction in D. melanogaster
Drosophila melanogaster was one of the earliest invertebrate models to be adopted for aging research and has contributed significantly both in terms of identifying environmental factors and genes that modulate lifespan. In addition to the strength of genetic tools, which have been established over the last century, the complex behaviors, the short lifespan of two months and a life cycle around 2 weeks has contributed to the success of D. melanogaster as a popular model in the aging field. More recently, the use of GAL4-UAS system (Brand and Perrimon, 1993) and the availability of RNAi strains (Dietzl et al., 2007) have allowed for tissue-specific manipulation which has had a significant influence in discovering novel genes and their tissue-specific effects that affect DR-dependent responses on lifespan.
Research in D. melanogaster has been critical in demonstrating the role of individual nutrients in mediating the effects of DR rather than total calories. In flies, reduction of total calories by changing the concentration of media components (termed caloric restriction, CR) has been used by several laboratories to model nutrient restriction (Mair et al., 2003; Pletcher et al., 2002; Rogina and Helfand, 2004). A number of groups have also shown that reducing yeast or yeast extract, the main source of protein in the fly diet, alone is sufficient to extend lifespan in D. melanogaster and is referred to as DR (Bruce et al., 2013; Carvalho et al., 2006; Chippindale, 1993; Kapahi et al., 2004; Lee et al., 2008; Mair et al., 2005; Skorupa et al., 2008). Consistent with the idea of restricting protein alone (De Marte and Enesco, 1986; Miller et al., 2005) or lowering the protein-to-carbohydrate ratio is sufficient for lifespan extension in rodents (Solon-Biet et al., 2014), suggesting that these findings are likely to be conserved across species. These studies have had a major influence in dispelling the long-held notion that total calories but not individual nutrients are critical for lifespan extension (Masoro, 2003).
Given the importance of nutrient composition on lifespan, the behavioral choices that influence macronutrient ingestion and nutrient balance are likely to be relevant to aging. Though the majority of laboratory experiments are undertaken under fixed food composition, most organisms including humans face choices for various nutrients and their smells in real life. Thus dietary choices and nutrient perception are likely to play important roles in determining their aging rates. The importance of olfaction in DR-mediated lifespan extension was first demonstrated in D. melanogaster by the shortening of lifespan simply from exposure to nutrient-derived odors (Libert et al., 2007). Interestingly, a similar phenomenon also occurs in C. elegans subjected to DD but exposed to soluble or volatile components of the bacterial diet (Smith et al., 2008a).
Studies in female D. melanogaster have investigated the mechanisms by which a dietary switch upon mating leads to increased preference for yeast, a major source of protein in the fly diet (Ribeiro and Dickson, 2010; Vargas et al., 2010). Furthermore, these studies also showed that mated female flies have a much stronger preference towards yeast to sustain egg production. This increased plasticity in behavior in response to changes in yeast concentration is likely an important reason that mated female flies are much more responsive than males to the physiological effects of DR (Katewa et al., 2012; Vargas et al., 2010). The switch towards enhanced feeding (Carvalho et al., 2006) and increased preference to yeast (Ribeiro and Dickson, 2010; Vargas et al., 2010) upon mating in D. melanogaster is likely to influence aging, and may in part explain the observed lifespan extension in virgin female flies compared to mated controls (Chapman et al., 1995). The molecular bases of these phenomena are still largely undefined. Simple invertebrates serve as excellent model systems to study the mechanisms of nutrient balance and homeostasis and their impact on aging.
4.2. Physiological and behavioral effects of dietary restriction in D. melanogaster
Research in D. melanogaster has yielded important insights into the physiological and behavioral changes that accompany or manifest lifespan extension upon DR (Figure 3). DR in D. melanogaster leads to a reduction in egg production but an increase in resistance to certain stresses. Increase in starvation resistance upon DR has been associated with a metabolic adaptation that increases lipid content in the flies, allowing for prolonged survival in the absence of nutrients (Djawdan et al., 1998; Katewa et al., 2012; Skorupa et al., 2008). Further insight into these physiological changes has resulted from experiments, which have varied the ratio of carbohydrate to protein in the diet. A consistent finding from these studies in the fly is that increased lifespan and triglyceride accumulation is favored by diets that have reduced yeast to carbohydrate ratio, and vice versa (Djawdan et al., 1998; Katewa et al., 2012; Skorupa et al., 2008). On the surface, these results are at odds with those observed in mice where body fat decreases under CR, which robustly extends lifespan (Bruss et al., 2010). In mice, there is also no clear correlation between steady-state fat levels and lifespan, as mice that are fed on a diet with reduced methionine content are long-lived, yet show an increase in body fat (Miller et al., 2005).
Figure 3.
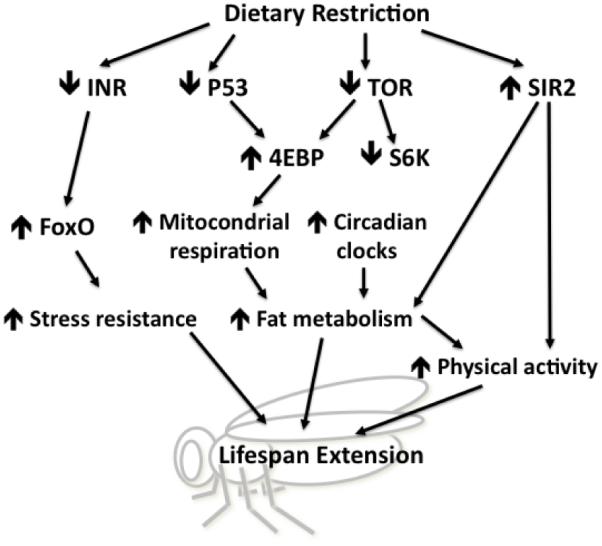
Effector mechanisms of dietary restriction that contribute to lifespan in the fruit fly D. melanogaster. See text for details.
One reason for a lack of apparent connection between fat and longevity is that the fat metabolism is assessed based on steady-state levels of triglycerides. One cannot deduce from steady-state levels of triglycerides whether a change in triglyceride levels is due to alterations in synthesis, storage, or breakdown of triglycerides. Interestingly, studies from mice using CR and from flies using restriction of yeast in the diet both demonstrate an increase in both triglyceride synthesis and breakdown, suggesting an increased turnover of triglycerides upon DR (Katewa et al., 2012). The enhanced turnover of triglycerides is a critical metabolic adaptation for lifespan extension as inhibition of triglyceride synthesis or breakdown in D. melanogaster prevents the maximal lifespan extension upon DR (Katewa et al., 2012). Interestingly, upon CR, where mice were fed 70% of the calories consumed ad libitum, show increased turnover of triglycerides despite the lowering of steady-state levels of triglycerides (Bruss et al., 2010). This increase in fat turnover is presumably required to maintain the increase in spontaneous activity, which is known to take place in mice, flies and other species upon conditions of nutrient restriction, presumably to support the animal’s need to search more actively for food. The increase in physical activity has been causally associated with lifespan extension upon DR as flies whose movement is restricted fail to maximize the lifespan extension effects of DR in D. melanogaster (Katewa et al., 2012). These results argue for the need of a behavioral adaptation that increases physical activity in addition to nutrient restriction to attain maximal lifespan extension by DR.
An interesting question is how DR increases both the synthesis and breakdown of triglycerides in both flies and mice. A likely possibility is that these processes are temporally separated, and regulated by circadian clocks. Circadian clocks are responsible for oscillations in various cellular processes including metabolic functions that are critical for maintaining organismal homeostasis. These rhythms are controlled by endogenous clocks located in the brain and in many peripheral tissues (Schibler and Sassone-Corsi, 2002). A recent study using D. melanogaster supports the idea that circadian-clock genes play a critical role in the improved fat turnover observed upon DR (Katewa et al., 2016). Specifically, both tim and per mutant flies showed reduced starvation resistance under DR conditions, and this was associated with reduced fat synthesis and reduced breakdown. Furthermore, metabolomic analysis of cycling triglycerides identified various medium chain triglycerides (MCTs). MCTs are triglycerides contain medium chain fatty acids (MCFA, C8 to C12 length). Feeding of MCTs to humans also increases energy expenditure (Mumme and Stonehouse, 2015), and ameliorates insulin resistance and inflammation in high fat diet-induced obese mice (Geng et al., 2016).
Disruption of circadian rhythms is associated with cancer and metabolic disorders that accelerate aging and age-related diseases (Kondratov, 2007; Turek et al., 2005). Furthermore, rhythms of rest:activity or sleep:wake break down with age in both flies and mice (Koh et al., 2006; Turek et al., 2005). Consistently, two studies in flies (Katewa et al., 2016) and in mice (Patel et al., 2016), have identified a key role for circadian clocks in lifespan extension upon DR. Disruption of clocks in flies, by keeping flies kept under constant light or using the clock mutants timeless or period, diminished the maximal lifespan extension upon DR (Katewa et al., 2016). Consistently, in mice knockout of the clock gene Bmal1 is also required for the beneficial effects of CR (30% restriction) on lifespan extension (Patel et al., 2016). Interestingly, both the fly and mouse studies observed that nutrient deprivation increases the amplitude of circadian clock genes, especially in peripheral tissues. As aging is known to attenuate various circadian behaviors so conversely (Kondratova and Kondratov, 2012), improving oscillations upon nutrient deprivation might slow down the age-related decline in function. Though it is not clear how a reduction in dietary protein or yeast (in case of flies) leads to regulation of circadian clocks, it is likely that either mTOR, insulin-like signaling or sirtuins, which are known nutrient-responsive pathways, may lie upstream to modulate circadian clocks upon DR (Katewa et al., 2016).
DR also affects other physiological parameters in flies, including intestinal integrity or permeability, a key contributing factor of mortality in flies (Rera et al., 2012), that also may explain the increase in infection observed in old flies (Ren et al., 2007; Rera et al., 2012). Interestingly, larger lifespan extensions are observed in females upon DR has been attributed to sexual dimorphism in diet dependent changes in intestinal pathology (Regan et al., 2016). Feminization of intestinal regions using genetic approaches induced similar pathology in male flies. Thus the intestine is likely to play an important role in mediating the DR-dependent changes in lifespan extension, as also observed in worms. In addition to the influence of the fly intestinal tissue on gut permeability and immune function, it may also influence lifespan through modulation of fat metabolism. The IRE1/XBP1 endoplasmic reticulum (ER) stress-signaling pathway mediates both the changes in fat metabolism and lifespan extension upon DR in flies (Luis et al., 2016). Furthermore, sugarbabe, a Gli-like zinc-finger transcription factor that was previously shown to be diet-responsive in flies (Zinke et al., 2002) acts downstream of IRE1/XBP1 to mediate the intestinal changes in fat metabolism upon DR. These studies argue for the importance of studying tissue-specific mediators of the DR response to better understand the protective effects of DR on healthspan and lifespan.
4.3. Genetic pathways modulating lifespan extension upon dietary restriction in D. melanogaster
Similar to DR in yeast and nematodes, the three major nutrient-signaling pathways, mTOR, insulin, and Sir2 have been linked with the lifespan extension upon DR in flies (Figure 3). It is becoming evident that multiple nutrient signals modulate various signaling pathways to mediate the lifespan extension upon DR. Thus modulation of one pathway is not sufficient to completely abrogate the maximal lifespan extension upon DR.
The conserved nutrient-sensing mTOR pathway, was first shown in D. melanogaster to mediate the effects of DR (Kapahi et al., 2004). Since then at least 60 studies have demonstrated that inhibition of various components of the mTOR pathway extends lifespan in multiple species (Kapahi et al., 2010; Lamming, 2016). Protein synthesis, a key downstream output of the mTOR pathway takes place at multiple levels, which include modulation of the ribosomal S6 kinase (S6K), 4E-BP, autophagy, and ribosomal biogenesis. Modulation of mRNA translation by these outputs modulates lifespan in multiple species, including flies (Kapahi et al., 2010). mTOR phosphorylates translation initiation factor 4E-binding protein 4E-BP, relieving its inhibition of eIF4E and enhancing overall mRNA translation (Sonenberg and Hinnebusch, 2009). It was previously shown that 4E-BP mediates lifespan extension by DR in D. melanogaster (Zid et al., 2009). Furthermore, DR induces d4E-BP protein levels and overexpression of a gain of function form of d4E-BP extends lifespan under nutrient-rich conditions, but not during DR (Zid et al., 2009). Overexpression of d4E-BP just in the muscle leads to lifespan extension and organism-wide improvement of proteostasis in D. melanogaster (Demontis and Perrimon, 2010). d4E-BP has also been shown to be required for the lifespan extension observed due to overexpression of a dominant-negative version of P53, which overlaps with the lifespan extension by CR in flies (Bauer et al., 2010). Downstream of d4E-BP, it has been shown that its overexpression or DR increased the ribosomal binding of some nuclear-encoded mitochondrial genes, including those encoding the electron transport chain (ETC) complexes I and IV and mitochondrial ribosomal proteins. Consistent with enhanced ribosomal loading increasing mRNA translation, DR was found to increase mitochondrial density and respiratory function (Zid et al., 2009). Furthermore, different studies have shown that in D. melanogaster, inhibition of the mitochondrial ETC abrogates the benefits of DR on lifespan extension (Bahadorani et al., 2010; Zid et al., 2009). Thus, the enhanced translation of mitochondrial ETC genes may serve as a protective mechanism not only by increasing mitochondrial efficiency but also by maintaining the function of the ETC and, hence, ATP production. Consistent with the genetic manipulation of mTOR extending lifespan, rapamycin has also ben shown to extend lifespan in flies through modulation of autophagy and mRNA translation (Bjedov et al., 2010).
The insulin-like signaling pathway is activated in response to the amount of yeast in the diet (Britton et al., 2002), and a number of studies have shown that mutants in this pathway regulate growth and development in D. melanogaster (Edgar, 1999; Oldham and Hafen, 2003) The interaction of the insulin-like-signaling pathway with diet has been examined in different studies upon manipulation of genes that modulate the insulin signaling pathway at different steps. Using a mutant for the insulin-receptor substrate, chico, or Foxo, a downstream transcription factor in this pathway, it was shown that the lifespan extension, using a specific paradigm for DR, surprisingly, may be independent of the insulin-like signaling pathway in flies (Gems and Partridge, 2001; Tatar, 2007). However, other experiments using a similar line of approach suggest that the insulin-signaling pathway in flies may exert its effects on lifespan both independently of DR and in a protein-dependent manner (Tatar, 2007).
The role of Sir2, which has been shown to mediate the effects of CR in yeast, has also been examined in D. melanogaster. Recent studies using overexpression of Sir2 in flies has shown it to be important for enhancing fat metabolism (Banerjee et al., 2012) and physical activity (Parashar and Rogina, 2009), whereas loss of function of Sir2 has been shown to abrogate the lifespan extension by CR (i.e., limitation of total food) in D. melanogaster (Rogina and Helfand, 2004). However, the role of Sir2 in longevity remains controversial, and these earlier findings have been disputed to be independent of Sir2 (Astrom et al., 2003; Burnett et al., 2011; Newman et al., 2002). Some of these discrepancies are also likely due to strain differences and DR protocols used by the different groups, which play a critical role in influencing lifespan (Piper and Partridge, 2007). Thus the role of Sir2 in mediating nutrient-dependent changes in lifespan extension remains unclear and further evidence is warranted to improve this understanding.
One of the strengths of the fly models is the ability to undertake tissue-specific analysis of genetic perturbations. Future analysis is likely to provide additional genetic pathways and information about their tissue-specificities important for mediating the effects of DR in D. melanogaster. Furthermore, these experiments will also help dissect the distinction between the protective effects of DR on various healthspan measures in addition to lifespan. In addition to the manipulation of nutrient-responsive pathways, a number of gene expression and metabolomics studies have implicated various cellular processes and signaling pathways to be altered upon DR in flies (Ding et al., 2014; Katewa et al., 2012; Laye et al., 2015; Pletcher et al., 2002; Whitaker et al., 2014). Thus, the field is well poised to ultimately provide several other candidate targets that can be manipulated to mimic the effects of DR.
5. Concluding remarks
The last decade has seen a burst of studies in invertebrate model organism establishing that several highly regulated nutrient-sensing pathways play conserved roles in mediating the effects of DR on lifespan. The most prominent example is the nutrient-sensor mTOR, which subsequently has been shown to also play conserved roles in longevity in rodents, and possibly in humans (Johnson et al., 2015; Kennedy and Lamming, 2016). The identified genes and pathways modulate lifespan by distinct as well as overlapping mechanisms. However, the downstream biological processes and the tissue-specificity by which such genes mediate the protective effects of DR remain largely unknown. Such information will be critical to guide experiments that could ultimately identify and test drugs for the targeting of nutrient-sensitive genes and pathways. Candidate drugs that may target either the upstream nutrient-responsive pathways (e.g., mTOR, insulin, or sirtuins) or downstream biological processes (e.g., antioxidants, glycolytic inhibitors, autophagy stimulators) could prove to be promising DR mimetics. Invertebrate model systems will likely be important for careful investigations of such DR mimetics before these agents are ready to, ultimately, be tested in more complex organisms, including in human subjects.
Acknowledgements
The authors apologize to all the colleagues whose work has not been cited as a consequence of space constraint. PK was supported by NIH grants R01AG038688 and AG045835 and the Julie Martin Mid-Career Award in Aging Research supported by The Ellison Medical Foundation and AFAR. MK was supported by NIH grants AG038518, AG031108, AG005136, and AG013280. MH was supported by NIH/NIA grants AG038664 and AG039756, and a Julie Martin Mid-Career Award in Aging Research supported by The Ellison Medical Foundation and AFAR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interests
The authors declare that there is no conflict of interests regarding the publication of this manuscript.
References
- Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA, Jr., Aris JP. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009a;8:353–369. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvers AL, Wood MS, Hu D, Kaywell AC, Dunn WA, Jr., Aris JP. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy. 2009b;5:847–849. doi: 10.4161/auto.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris JP, Alvers AL, Ferraiuolo RA, Fishwick LK, Hanvivatpong A, Hu D, Kirlew C, Leonard MT, Losin KJ, Marraffini M, et al. Autophagy and leucine promote chronological longevity and respiration proficiency during calorie restriction in yeast. Exp Gerontol. 2013;48:1107–1119. doi: 10.1016/j.exger.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Sinclair D, Gordon JI, Guarente L. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96:9100–9105. doi: 10.1073/pnas.96.16.9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrom SU, Cline TW, Rine J. The Drosophila melanogaster sir2+ gene is nonessential and has only minor effects on position-effect variegation. Genetics. 2003;163:931–937. doi: 10.1093/genetics/163.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahadorani S, Hur JH, Lo T, Jr., Vu K, Walker DW. Perturbation of mitochondrial complex V alters the response to dietary restriction in Drosophila. Aging Cell. 2010;9:100–103. doi: 10.1111/j.1474-9726.2009.00537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee KK, Ayyub C, Ali SZ, Mandot V, Prasad NG, Kolthur-Seetharam U. dSir2 in the adult fat body, but not in muscles, regulates life span in a diet-dependent manner. Cell Rep. 2012;2:1485–1491. doi: 10.1016/j.celrep.2012.11.013. [DOI] [PubMed] [Google Scholar]
- Bauer JH, Chang C, Bae G, Morris SN, Helfand SL. Dominant-negative Dmp53 extends life span through the dTOR pathway in D. melanogaster. Mech Ageing Dev. 2010;131:193–201. doi: 10.1016/j.mad.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- Bruce KD, Hoxha S, Carvalho GB, Yamada R, Wang HD, Karayan P, He S, Brummel T, Kapahi P, Ja WW. High carbohydrate-low protein consumption maximizes Drosophila lifespan. Exp Gerontol. 2013;48:1129–1135. doi: 10.1016/j.exger.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruss MD, Khambatta CF, Ruby MA, Aggarwal I, Hellerstein MK. Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. Am J Physiol Endocrinol Metab. 2010;298:E108–116. doi: 10.1152/ajpendo.00524.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8:1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Olsen B, Kennedy BK, Kaeberlein M. A genomic analysis of chronological longevity factors in budding yeast. Cell Cycle. 2011;10:1385–1396. doi: 10.4161/cc.10.9.15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano AC, Dillin A, Hunter T. A Kruppel-like factor downstream of the E3 ligase WWP-1 mediates dietary-restriction-induced longevity in Caenorhabditis elegans. Nat Commun. 2014;5:3772. doi: 10.1038/ncomms4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano AC, Liu Z, Dillin A, Hunter T. A conserved ubiquitination pathway determines longevity in response to diet restriction. Nature. 2009;460:396–399. doi: 10.1038/nature08130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho GB, Kapahi P, Anderson DJ, Benzer S. Allocrine modulation of feeding behavior by the Sex Peptide of Drosophila. Curr Biol. 2006;16:692–696. doi: 10.1016/j.cub.2006.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Liddle LF, Kalb JM, Wolfner MF, Partridge L. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature. 1995;373:241–244. doi: 10.1038/373241a0. [DOI] [PubMed] [Google Scholar]
- Chen D, Thomas EL, Kapahi P. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000486. doi: 10.1371/journal.pgen.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KL, Crane MM, Kaeberlein M. Microfluidic technologies for yeast replicative lifespan studies. Mech Ageing Dev. 2016 doi: 10.1016/j.mad.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching TT, Paal AB, Mehta A, Zhong L, Hsu AL. drr-2 encodes an eIF4H that acts downstream of TOR in diet-restriction-induced longevity of C. elegans. Aging Cell. 2010;9:545–557. doi: 10.1111/j.1474-9726.2010.00580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippindale AK, Leroi AM, Kim SB, Rose MR. Phenotypic plasticity and selection in Drosophila life•history evolution. I. Nutrition and the cost of reproduction. Journal of Evolutionary Biology. 1993;6:171–193. [Google Scholar]
- Choi JS, Lee CK. Maintenance of cellular ATP level by caloric restriction correlates chronological survival of budding yeast. Biochem Biophys Res Commun. 2013;439:126–131. doi: 10.1016/j.bbrc.2013.08.014. [DOI] [PubMed] [Google Scholar]
- Cypser JR, Kitzenberg D, Park SK. Dietary restriction in C. elegans: recent advances. Exp Gerontol. 2013;48:1014–1017. doi: 10.1016/j.exger.2013.02.018. [DOI] [PubMed] [Google Scholar]
- Dang W, Sutphin GL, Dorsey JA, Otte GL, Cao K, Perry RM, Wanat JJ, Saviolaki D, Murakami CJ, Tsuchiyama S, et al. Inactivation of yeast Isw2 chromatin remodeling enzyme mimics longevity effect of calorie restriction via induction of genotoxic stress response. Cell Metab. 2014;19:952–966. doi: 10.1016/j.cmet.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marte ML, Enesco HE. Influence of low tryptophan diet on survival and organ growth in mice. Mech Ageing Dev. 1986;36:161–171. doi: 10.1016/0047-6374(86)90017-5. [DOI] [PubMed] [Google Scholar]
- Delaney JR, Murakami C, Chou A, Carr D, Schleit J, Sutphin GL, An EH, Castanza AS, Fletcher M, Goswami S, et al. Dietary restriction and mitochondrial function link replicative and chronological aging in Saccharomyces cerevisiae. Exp Gerontol. 2013;48:1006–1013. doi: 10.1016/j.exger.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Ding F, Gil MP, Franklin M, Ferreira J, Tatar M, Helfand SL, Neretti N. Transcriptional response to dietary restriction in Drosophila melanogaster. J Insect Physiol. 2014;69:101–106. doi: 10.1016/j.jinsphys.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djawdan M, Chippindale AK, Rose MR, Bradley TJ. Metabolic reserves and evolved stress resistance in Drosophila melanogaster. Physiol Zool. 1998;71:584–594. doi: 10.1086/515963. [DOI] [PubMed] [Google Scholar]
- Edgar BA. From small flies come big discoveries about size control. Nat Cell Biol. 1999;1:E191–193. doi: 10.1038/70217. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Hoon S, Shamalnasab M, Galbani A, Wei M, Giaever G, Nislow C, Longo VD. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet. 2010;6:e1001024. doi: 10.1371/journal.pgen.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pletcher SD, Minois N, Vaupel JW, Longo VD. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS Lett. 2004;557:136–142. doi: 10.1016/s0014-5793(03)01462-5. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro-Gonzalez JC, Gonzalez-Barrios M, Miranda-Vizuete A, Swoboda P. The thioredoxin TRX-1 regulates adult lifespan extension induced by dietary restriction in Caenorhabditis elegans. Biochem Biophys Res Commun. 2011;406:478–482. doi: 10.1016/j.bbrc.2011.02.079. [DOI] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Gelino S, Chang JT, Kumsta C, She X, Davis A, Nguyen C, Panowski S, Hansen M. Intestinal Autophagy Improves Healthspan and Longevity in C. elegans during Dietary Restriction. PLoS Genet. 2016;12:e1006135. doi: 10.1371/journal.pgen.1006135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Partridge L. Insulin/IGF signalling and ageing: seeing the bigger picture. Curr Opin Genet Dev. 2001;11:287–292. doi: 10.1016/s0959-437x(00)00192-1. [DOI] [PubMed] [Google Scholar]
- Geng S, Zhu W, Xie C, Li X, Wu J, Liang Z, Xie W, Zhu J, Huang C, Zhu M, et al. Medium-chain triglyceride ameliorates insulin resistance and inflammation in high fat diet-induced obese mice. Eur J Nutr. 2016;55:931–940. doi: 10.1007/s00394-015-0907-0. [DOI] [PubMed] [Google Scholar]
- Greer EL, Banko MR, Brunet A. AMP-activated protein kinase and FoxO transcription factors in dietary restriction-induced longevity. Ann N Y Acad Sci. 2009;1170:688–692. doi: 10.1111/j.1749-6632.2009.04019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- He C, Tsuchiyama SK, Nguyen QT, Plyusnina EN, Terrill SR, Sahibzada S, Patel B, Faulkner AR, Shaposhnikov MV, Tian R, et al. Enhanced longevity by ibuprofen, conserved in multiple species, occurs in yeast through inhibition of tryptophan import. PLoS Genet. 2014;10:e1004860. doi: 10.1371/journal.pgen.1004860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heestand BN, Shen Y, Liu W, Magner DB, Storm N, Meharg C, Habermann B, Antebi A. Dietary restriction induced longevity is mediated by nuclear receptor NHR-62 in Caenorhabditis elegans. PLoS Genet. 2013;9:e1003651. doi: 10.1371/journal.pgen.1003651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjoh S, Yamamoto T, Uno M, Nishida E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457:726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- Hou L, Wang D, Chen D, Liu Y, Zhang Y, Cheng H, Xu C, Sun N, McDermott J, Mair WB, et al. A Systems Approach to Reverse Engineer Lifespan Extension by Dietary Restriction. Cell Metab. 2016;23:529–540. doi: 10.1016/j.cmet.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 2000;14:2135–2137. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Johnson FB. Methionine restriction activates the retrograde response and confers both stress tolerance and lifespan extension to yeast, mouse and human cells. PLoS One. 2014;9:e97729. doi: 10.1371/journal.pone.0097729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Sangesland M, Kaeberlein M, Rabinovitch PS. Modulating mTOR in aging and health. Interdiscip Top Gerontol. 2015;40:107–127. doi: 10.1159/000364974. [DOI] [PubMed] [Google Scholar]
- Jung PP, Christian N, Kay DP, Skupin A, Linster CL. Protocols and programs for high-throughput growth and aging phenotyping in yeast. PLoS One. 2015;10:e0119807. doi: 10.1371/journal.pone.0119807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Hu D, Kerr EO, Tsuchiya M, Westman EA, Dang N, Fields S, Kennedy BK. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2005a;1:e69. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005b;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Steffen KK, Hu D, Dang N, Kerr EO, Tsuchiya M, Fields S, Kennedy BK. Comment on "HST2 mediates SIR2-independent life-span extension by calorie restriction". Science. 2006a;312:1312. doi: 10.1126/science.1124608. author reply 1312. [DOI] [PubMed] [Google Scholar]
- Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, Fields S, Kennedy BK, Kaeberlein M. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006b;5:487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa SD, Akagi K, Bose N, Rakshit K, Camarella T, Zheng X, Hall D, Davis S, Nelson CS, Brem RB, et al. Peripheral Circadian Clocks Mediate Dietary Restriction-Dependent Changes in Lifespan and Fat Metabolism in Drosophila. Cell Metab. 2016;23:143–154. doi: 10.1016/j.cmet.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa SD, Demontis F, Kolipinski M, Hubbard A, Gill MS, Perrimon N, Melov S, Kapahi P. Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell Metab. 2012;16:97–103. doi: 10.1016/j.cmet.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa SD, Kapahi P. Role of TOR signaling in aging and related biological processes in Drosophila melanogaster. Exp Gerontol. 2011;46:382–390. doi: 10.1016/j.exger.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Lamming DW. The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging. Cell Metab. 2016;23:990–1003. doi: 10.1016/j.cmet.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Koh K, Evans JM, Hendricks JC, Sehgal A. A Drosophila model for age-associated changes in sleep:wake cycles. Proc Natl Acad Sci U S A. 2006;103:13843–13847. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV. A role of the circadian system and circadian proteins in aging. Ageing Res Rev. 2007;6:12–27. doi: 10.1016/j.arr.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nat Rev Neurosci. 2012;13:325–335. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan EX, Foss EJ, Tsuchiyama S, Alvino GM, Kruglyak L, Kaeberlein M, Raghuraman MK, Brewer BJ, Kennedy BK, Bedalov A. A natural polymorphism in rDNA replication origins links origin activation with calorie restriction and lifespan. PLoS Genet. 2013;9:e1003329. doi: 10.1371/journal.pgen.1003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW. Inhibition of the Mechanistic Target of Rapamycin (mTOR)-Rapamycin and Beyond. Cold Spring Harb Perspect Med. 2016:6. doi: 10.1101/cshperspect.a025924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Lan J, Zhang X, Chen D. Molecular mechanisms of dietary restriction in aging-insights from Caenorhabditis elegans research. Sci China Life Sci. 2015;58:352–358. doi: 10.1007/s11427-015-4824-5. [DOI] [PubMed] [Google Scholar]
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun. 2013;4:2267. doi: 10.1038/ncomms3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, Gelino S, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, Kumsta C, Sandri M, Ballabio A, Hansen M. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy. 2015;11:867–880. doi: 10.1080/15548627.2015.1034410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H, Whiteway M. Increased respiration in the sch9Delta mutant is required for increasing chronological life span but not replicative life span. Eukaryot Cell. 2008;7:1127–1135. doi: 10.1128/EC.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laye MJ, Tran V, Jones DP, Kapahi P, Promislow DE. The effects of age and dietary restriction on the tissue-specific metabolome of Drosophila. Aging Cell. 2015;14:797–808. doi: 10.1111/acel.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, Ingram DK, Zou S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Simpson SJ, Clissold FJ, Brooks R, Ballard JW, Taylor PW, Soran N, Raubenheimer D. Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proc Natl Acad Sci U S A. 2008;105:2498–2503. doi: 10.1073/pnas.0710787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiser SF, Begun A, Kaeberlein M. HIF-1 modulates longevity and healthspan in a temperature-dependent manner. Aging Cell. 2011;10:318–326. doi: 10.1111/j.1474-9726.2011.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiser SF, Miller H, Rossner R, Fletcher M, Leonard A, Primitivo M, Rintala N, Ramos FJ, Miller DL, Kaeberlein M. Cell nonautonomous activation of flavin-containing monooxygenase promotes longevity and health span. Science. 2015;350:1375–1378. doi: 10.1126/science.aac9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester RL, Withers BR, Schultz MA, Dickson RC. Iron, glucose and intrinsic factors alter sphingolipid composition as yeast cells enter stationary phase. Biochim Biophys Acta. 2013;1831:726–736. doi: 10.1016/j.bbalip.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Skinner C, Castello PR, Kato M, Easlon E, Xie L, Li T, Lu SP, Wang C, Tsang F, et al. Identification of potential calorie restriction-mimicking yeast mutants with increased mitochondrial respiratory chain and nitric oxide levels. J Aging Res. 2011;2011:673185. doi: 10.4061/2011/673185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert S, Zwiener J, Chu X, Vanvoorhies W, Roman G, Pletcher SD. Regulation of Drosophila life span by olfaction and food-derived odors. Science. 2007;315:1133–1137. doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Guarente L. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2006;2:e33. doi: 10.1371/journal.pgen.0020033. author reply e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Longo VD. The Ras and Sch9 pathways regulate stress resistance and longevity. Exp Gerontol. 2003;38:807–811. doi: 10.1016/s0531-5565(03)00113-x. [DOI] [PubMed] [Google Scholar]
- Longo VD, Fabrizio P. Chronological aging in Saccharomyces cerevisiae. Subcell Biochem. 2012;57:101–121. doi: 10.1007/978-94-007-2561-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Shadel GS, Kaeberlein M, Kennedy B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012;16:18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis NM, Wang L, Ortega M, Deng H, Katewa SD, Li PW, Karpac J, Jasper H, Kapahi P. Intestinal IRE1 Is Required for Increased Triglyceride Metabolism and Longer Lifespan under Dietary Restriction. Cell Rep. 2016;17:1207–1216. doi: 10.1016/j.celrep.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madia F, Gattazzo C, Fabrizio P, Longo VD. A simple model system for age-dependent DNA damage and cancer. Mech Ageing Dev. 2007;128:45–49. doi: 10.1016/j.mad.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madia F, Gattazzo C, Wei M, Fabrizio P, Burhans WC, Weinberger M, Galbani A, Smith JR, Nguyen C, Huey S, et al. Longevity mutation in SCH9 prevents recombination errors and premature genomic instability in a Werner/Bloom model system. J Cell Biol. 2008;180:67–81. doi: 10.1083/jcb.200707154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- Mair W, Goymer P, Pletcher SD, Partridge L. Demography of dietary restriction and death in Drosophila. Science. 2003;301:1731–1733. doi: 10.1126/science.1086016. [DOI] [PubMed] [Google Scholar]
- Mair W, Piper MD, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 2005;3:e223. doi: 10.1371/journal.pbio.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannarino SC, Amorim MA, Pereira MD, Moradas-Ferreira P, Panek AD, Costa V, Eleutherio EC. Glutathione is necessary to ensure benefits of calorie restriction during ageing in Saccharomyces cerevisiae. Mech Ageing Dev. 2008;129:700–705. doi: 10.1016/j.mad.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Subfield history: caloric restriction, slowing aging, and extending life. Sci Aging Knowledge Environ. 2003;2003:RE2. doi: 10.1126/sageke.2003.8.re2. [DOI] [PubMed] [Google Scholar]
- Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, Boeke JD, Smith JS. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet. 2010;6:e1000921. doi: 10.1371/journal.pgen.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A, Sim S, Chou AC, Ahmed U, Carr D, Murakami CJ, et al. A Comprehensive Analysis of Replicative Lifespan in 4,698 Single-Gene Deletion Strains Uncovers Conserved Mechanisms of Aging. Cell Metab. 2015;22:895–906. doi: 10.1016/j.cmet.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta R, Steinkraus KA, Sutphin GL, Ramos FJ, Shamieh LS, Huh A, Davis C, Chandler-Brown D, Kaeberlein M. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science. 2009;324:1196–1198. doi: 10.1126/science.1173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, Smith-Wheelock M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4:119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirisola MG, Braun RJ, Petranovic D. Approaches to study yeast cell aging and death. FEMS Yeast Res. 2014;14:109–118. doi: 10.1111/1567-1364.12112. [DOI] [PubMed] [Google Scholar]
- Molin M, Yang J, Hanzen S, Toledano MB, Labarre J, Nystrom T. Life span extension and H(2)O(2) resistance elicited by caloric restriction require the peroxiredoxin Tsa1 in Saccharomyces cerevisiae. Mol Cell. 2011;43:823–833. doi: 10.1016/j.molcel.2011.07.027. [DOI] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Muller RU, Fabretti F, Zank S, Burst V, Benzing T, Schermer B. The von Hippel Lindau tumor suppressor limits longevity. J Am Soc Nephrol. 2009;20:2513–2517. doi: 10.1681/ASN.2009050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumme K, Stonehouse W. Effects of medium-chain triglycerides on weight loss and body composition: a meta-analysis of randomized controlled trials. J Acad Nutr Diet. 2015;115:249–263. doi: 10.1016/j.jand.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Murakami C, Delaney JR, Chou A, Carr D, Schleit J, Sutphin GL, An EH, Castanza AS, Fletcher M, Goswami S, et al. pH neutralization protects against reduction in replicative lifespan following chronological aging in yeast. Cell Cycle. 2012;11:3087–3096. doi: 10.4161/cc.21465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami C, Kaeberlein M. Quantifying yeast chronological life span by outgrowth of aged cells. J Vis Exp. 2009 doi: 10.3791/1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami CJ, Wall V, Basisty N, Kaeberlein M. Composition and acidification of the culture medium influences chronological aging similarly in vineyard and laboratory yeast. PLoS One. 2011;6:e24530. doi: 10.1371/journal.pone.0024530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman BL, Lundblad JR, Chen Y, Smolik SM. A Drosophila homologue of Sir2 modifies position-effect variegation but does not affect life span. Genetics. 2002;162:1675–1685. doi: 10.1093/genetics/162.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo A, Liu J, Schroeder EA, Shadel GS, Barrientos A. Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab. 2012;16:55–67. doi: 10.1016/j.cmet.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Hafen E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 2003;13:79–85. doi: 10.1016/s0962-8924(02)00042-9. [DOI] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521:525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Shadel GS. Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density. Aging (Albany NY) 2009;1:131–145. doi: 10.18632/aging.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit A, Jain V, Kumar N, Mukhopadhyay A. PHA-4/FOXA-regulated microRNA feed forward loops during Caenorhabditis elegans dietary restriction. Aging (Albany NY) 2014;6:835–855. doi: 10.18632/aging.100697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Parashar V, Rogina B. dSir2 mediates the increased spontaneous physical activity in flies on calorie restriction. Aging (Albany NY) 2009;1:529–541. doi: 10.18632/aging.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Link CD, Johnson TE. Life-span extension by dietary restriction is mediated by NLP-7 signaling and coelomocyte endocytosis in C. elegans. FASEB J. 2010;24:383–392. doi: 10.1096/fj.09-142984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Tedesco PM, Johnson TE. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. 2009;8:258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SA, Chaudhari A, Gupta R, Velingkaar N, Kondratov RV. Circadian clocks govern calorie restriction-mediated life span extension through BMAL1- and IGF-1-dependent mechanisms. FASEB J. 2016;30:1634–1642. doi: 10.1096/fj.15-282475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton A, Knuth S, Schaheen B, Dang H, Greenwald I, Fares H. Endocytosis function of a ligand-gated ion channel homolog in Caenorhabditis elegans. Curr Biol. 2005;15:1045–1050. doi: 10.1016/j.cub.2005.04.057. [DOI] [PubMed] [Google Scholar]
- Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, Winderickx J, De Virgilio C. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell. 2003;12:1607–1613. doi: 10.1016/s1097-2765(03)00485-4. [DOI] [PubMed] [Google Scholar]
- Piper MD, Partridge L. Dietary restriction in Drosophila: delayed aging or experimental artefact? PLoS Genet. 2007;3:e57. doi: 10.1371/journal.pgen.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper PW, Harris NL, MacLean M. Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast. Mech Ageing Dev. 2006;127:733–740. doi: 10.1016/j.mad.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, Partridge L. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol. 2002;12:712–723. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Lee RY, Avery L. Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics. 1995;141:1365–1382. doi: 10.1093/genetics/141.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan JC, Khericha M, Dobson AJ, Bolukbasi E, Rattanavirotkul N, Partridge L. Sex difference in pathology of the ageing gut mediates the greater response of female lifespan to dietary restriction. Elife. 2016;5:e10956. doi: 10.7554/eLife.10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jr., Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Clark RI, Walker DW. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci U S A. 2012;109:21528–21533. doi: 10.1073/pnas.1215849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C, Dickson BJ. Sex peptide receptor and neuronal TOR/S6K signaling modulate nutrient balancing in Drosophila. Curr Biol. 2010;20:1000–1005. doi: 10.1016/j.cub.2010.03.061. [DOI] [PubMed] [Google Scholar]
- Richard VR, Leonov A, Beach A, Burstein MT, Koupaki O, Gomez-Perez A, Levy S, Pluska L, Mattie S, Rafesh R, et al. Macromitophagy is a longevity assurance process that in chronologically aging yeast limited in calorie supply sustains functional mitochondria and maintains cellular lipid homeostasis. Aging (Albany NY) 2013;5:234–269. doi: 10.18632/aging.100547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- Schleit J, Johnson SC, Bennett CF, Simko M, Trongtham N, Castanza A, Hsieh EJ, Moller RM, Wasko BM, Delaney JR, et al. Molecular mechanisms underlying genotype-dependent responses to dietary restriction. Aging Cell. 2013;12:1050–1061. doi: 10.1111/acel.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser S, Priebe S, Groth M, Monajembashi S, Hemmerich P, Guthke R, Platzer M, Ristow M. Neuronal ROS signaling rather than AMPK/sirtuin-mediated energy sensing links dietary restriction to lifespan extension. Mol Metab. 2013;2:92–102. doi: 10.1016/j.molmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson SJ, Raubenheimer D. Macronutrient balance and lifespan. Aging (Albany NY) 2009;1:875–880. doi: 10.18632/aging.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Kumar N, Matai L, Jain V, Garg A, Mukhopadhyay A. A chromatin modifier integrates insulin/IGF-1 signalling and dietary restriction to regulate longevity. Aging Cell. 2016;15:694–705. doi: 10.1111/acel.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorupa DA, Dervisefendic A, Zwiener J, Pletcher SD. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell. 2008;7:478–490. doi: 10.1111/j.1474-9726.2008.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Jr., Li C, Matecic M, Maqani N, Bryk M, Smith JS. Calorie restriction effects on silencing and recombination at the yeast rDNA. Aging Cell. 2009;8:633–642. doi: 10.1111/j.1474-9726.2009.00516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Jr., Maharrey CH, Carey CR, White RA, Hartman J.L.t. Gene-nutrient interaction markedly influences yeast chronological lifespan. Exp Gerontol. 2016 doi: 10.1016/j.exger.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Jr., McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell. 2007;6:649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- Smith ED, Kaeberlein TL, Lydum BT, Sager J, Welton KL, Kennedy BK, Kaeberlein M. Age- and calorie-independent life span extension from dietary restriction by bacterial deprivation in Caenorhabditis elegans. BMC Dev Biol. 2008a;8:49. doi: 10.1186/1471-213X-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, Johnston ED, Tchao BN, Pak DN, Welton KL, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008b;18:564–570. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, McMahon AC, Ballard JW, Ruohonen K, Wu LE, Cogger VC, Warren A, Huang X, Pichaud N, Melvin RG, et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014;19:418–430. doi: 10.1016/j.cmet.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, Kennedy BK, Kaeberlein M. Measuring replicative life span in the budding yeast. J Vis Exp. 2009 doi: 10.3791/1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, McCormick MA, Pham KM, MacKay VL, Delaney JR, Murakami CJ, Kaeberlein M, Kennedy BK. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics. 2012;191:107–118. doi: 10.1534/genetics.111.136549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol. 2008a;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkraus KA, Smith ED, Davis C, Carr D, Pendergrass WR, Sutphin GL, Kennedy BK, Kaeberlein M. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell. 2008b;7:394–404. doi: 10.1111/j.1474-9726.2008.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M. Diet restriction in Drosophila melanogaster. Design and analysis. Interdiscip Top Gerontol. 2007;35:115–136. doi: 10.1159/000096559. [DOI] [PubMed] [Google Scholar]
- Tatar M. The plate half-full: status of research on the mechanisms of dietary restriction in Drosophila melanogaster. Exp Gerontol. 2011;46:363–368. doi: 10.1016/j.exger.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- Tsuchiya M, Dang N, Kerr EO, Hu D, Steffen KK, Oakes JA, Kennedy BK, Kaeberlein M. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging Cell. 2006;5:505–514. doi: 10.1111/j.1474-9726.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas MA, Luo N, Yamaguchi A, Kapahi P. A role for S6 kinase and serotonin in postmating dietary switch and balance of nutrients in D. melanogaster. Curr Biol. 2010;20:1006–1011. doi: 10.1016/j.cub.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jiang JC, Jazwinski SM. Gene regulatory changes in yeast during life extension by nutrient limitation. Exp Gerontol. 2010;45:621–631. doi: 10.1016/j.exger.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, Longo VD. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008;4:e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M, Feng L, Paul A, Smith DL, Jr., Hontz RD, Smith JS, Vujcic M, Singh KK, Huberman JA, Burhans WC. DNA replication stress is a determinant of chronological lifespan in budding yeast. PLoS One. 2007;2:e748. doi: 10.1371/journal.pone.0000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker R, Gil MP, Ding F, Tatar M, Helfand SL, Neretti N. Dietary switch reveals fast coordinated gene expression changes in Drosophila melanogaster. Aging (Albany NY) 2014;6:355–368. doi: 10.18632/aging.100662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Kadiyala CS, Ching TT, Hakimi P, Saha S, Xu H, Yuan C, Mullangi V, Wang L, Fivenson E, et al. Enhanced energy metabolism contributes to the extended life span of calorie-restricted Caenorhabditis elegans. J Biol Chem. 2012;287:31414–31426. doi: 10.1074/jbc.M112.377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shao Z, Zhai Z, Shen C, Powell-Coffman JA. The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS One. 2009;4:e6348. doi: 10.1371/journal.pone.0006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]


