Abstract
Pasteurella multocida infection in cattle causes serious epidemic diseases and leads to great economic losses in livestock industry; however, little is known about the interaction between host and P. multocida in the lungs. To explore a fully insight into the host responses in the lungs during P. multocida infection, a mouse model of Pasteurella pneumonia was established by intraperitoneal infection, and then transcriptomic analysis of infected lungs was performed. P. multocida localized and grew in murine lungs, and induced inflammation in the lungs, as well as mice death. With transcriptomic analysis, approximately 107 clean reads were acquired. 4236 differently expressed genes (DEGs) were detected during P. multocida infection, of which 1924 DEGs were up-regulated. By gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) enrichments, 5,303 GO enrichments and 116 KEGG pathways were significantly enriched in the context of P. multocida infection. Interestingly, genes related to immune responses, such as pattern recognition receptors (PRRs), chemokines and inflammatory cytokines, were significantly up-regulated, suggesting the key roles of these genes in P. multocida infection. Transcriptomic data showed that IFN-γ/IL-17-related genes were increased, which were validated by qRT-PCR, ELISA, and immunoblotting. Our study characterized the transcriptomic profile of the lungs in mice upon Pasteurella infection, and our findings could provide valuable information with respect to better understanding the responses in mice during P. multocida infection.
Keywords: P. multocida, lung, IFN-γ, IL-17, transcriptomic analysis
Introduction
Pasteurella multocida is a gram-negative facultative anaerobic bacillus, which infects a wide range of domestic and wild animals, mainly causing hemorrhagic septicemia and respiratory diseases. P. multocida is classified into five capsular serogroups (A, B, D, E, and F) based on specificity of capsular antigens, and 16 serotypes based on lipopolysaccharide (LPS) antigens (Kubera et al., 2017). In China, P. multocida capsular serogroup B induces hemorrhagic septicemia in cattle, while P. multocida capsular serogroup A is mainly associated with pulmonary infection, which causes economic losses in cattle (Dabo et al., 2007). P. multocida serogroup A is one of nasopharyngeal commensal pathogens associated with respiratory diseases in livestock. During the alteration of external environment (e.g., long-distance transport, weather variations) or decrease in host immunity, P. multocida will be extended from the upper respiratory tract to enter the lower respiratory tract, causing pneumonia in cattle. The virulence factors (e.g., the capsule, fimbriae, adhesion proteins, toxins, iron regulation and iron acquisition proteins and outer membrane proteins) of P. multocida are thought to evade immune defenses in cattle, which may result in widespread existence of pneumonic pasteurellosis (Harper et al., 2006; Griffin et al., 2010).
Currently, the vaccine is the main method of preventing pneumonic pasteurellosis. However, complete protection of vaccine against P. multocida infection was not available, which may be due to our inadequate understanding P. multocida-host interaction and limited recognition of immune responses against P. multocida infection in host (Mathy et al., 2002). Recently, it has been reported that the responses in the lungs after P. multocida infection mainly include expression of some inflammatory mediators and cell apoptosis in mice or cattle. For example, P. multocida infection with intratracheal inoculation induces the mRNA expressions of pro-inflammatory cytokines (e.g., Tnf-α, Il-6, Il-1, Il-8, and Il-12), and the increase in the number of neutrophils in the lungs of cattle (Mathy et al., 2002). Although we have the understanding about the expressions of these cytokines and the accumulation of neutrophils in the lungs, a comprehensive immune responses (e.g., Th17 responses) in host against P. multocida remains to be explored. In addition, the alterations of other aspects (e.g., metabolic responses) in the lungs during P. multocida infection are still unclear. Transcriptomic sequencing has been applied to provide the new insights into molecular mechanisms and to explore the interaction between host and pathogens, such as mango-Fusarium mangiferae, tilapia-Streptococcus agalactiae, and epithelial-pneumococcal interaction (Aprianto et al., 2016; Liu et al., 2016; Wang et al., 2016). Thus, to explore the host responses during P. multocida infection, this study firstly established a mouse model of Pasteurella pneumonia using P. multocida serotype A strain CQ2. Subsequently, comprehensive gene expression profiles of infected and sham-infected lungs were explored using RNA-seq analysis. Differently expressed genes (DEGs) were acquired to for functional annotations and classifications, and some DEGs were validated using qRT-PCR, ELISA, and immunoblotting. Mice were selected as models for this study because it is difficult to use the cattle in research considering the high economic cost with the cattle. Also, there are various similarities in pathogenesis between mice and cattle after P. multocida infection, thus the mice are selected to act as substitutable models of cattle in many studies. For example, similar to cattle, P. multocida infection in mice by an intraperitoneal route or an intranasal route also results up-regulated pro-inflammatory cytokines, reactive oxygen species (ROS) and lymphocyte apoptosis (Praveena et al., 2010; Wu et al., 2014). This study revealed transcriptomic profile of murine lungs upon Pasteurella infection and provided a new insight into interaction between mice and P. multocida using transcriptomic analysis.
Materials and methods
Culture of bacteria
This study used P. multocida serotype A strain CQ2 (PmCQ2, GenBank accession number: LIUN00000000), which is a highly virulent strain and isolated from a bovine pneumonic lung, causing severe pulmonary inflammation and high mortality in mice (Li et al., 2016). As previously described, PmCQ2 was routinely cultured in Martin's broth agar containing 5% horse serum (Ren et al., 2013a; Chen et al., 2014).
Mouse infection
All animal studies were performed according to the guidelines of the Laboratory Animal Ethical Commission of the Southwest University, and the procedures used in this study were approved by the Laboratory Animal Ethical Commission. Six to eight week-old, female C57BL/6 mice (weight 18–22 g) were brought from Laboratory Animal Center of Third Military Medical University (Chongqing, China). The mice were housed in the individual ventilated cages (experimental animals IVC factory, Suzhou, China), keeping temperature at 20–30°C relative humidity at 50–60% and lighting cycle at 12 h/day. Total 130 mice were used in this study. In the infected group, the mice were challenged by an intraperitoneal injection of PmCQ2 at the dose of 2.2 × 105 (LD50) colony-forming units (CFU) in 100 μL. In the control group, mice (gender and age matched) were injected intraperitoneally with equal dose of saline. Survival rates (n = 10 in each group) were measured in both groups after injection. Mice were also euthanized to collect lung tissues at 8, 16, and 24 h post-infection. Serum samples were also collected after infection.
Bacterial colonization
To measure the bacterial load, the lung tissues (n = 10 in each group) were collected at 0, 8, 16, and 24 h post-bacterial infection. The tissues were homogenized aseptically and bacterial loads were quantified by 10-fold serial dilution in saline. These different dilutions were plated in triplicate on Martin's broth agar and were incubated at 37°C up to 24 h to count CFU.
Histopathological examination and FISH
For histopathological examination, the lung tissues (n = 6 in each group) were immediately fixed in the 4% paraformaldehyde for 24 h, dehydrated in graded ethanol, and then embedded in paraffin wax. The tissues were sliced at 3 μm thick and then stained with hematoxylin and eosin (H&E). Histopathological scoring was performed by a pathologist blinded to treatment and control groups, and the scoring was mainly based on interstitial inflammation, vascular endothelialitis, bronchitis, edema, serous effusion and thrombus formation (van den Boogaard et al., 2015). All parameters were scored separately from 0 (lesion absent) to 3 (severe lesion) (Praveena et al., 2014; Table 1).
Table 1.
The criteria of histopathological scores.
| Histopathological scores | Inflammatory cells | Alveolar/bronchial epithelium | Blood vessels | Inflammatory exudation |
|---|---|---|---|---|
| 0 (no lesion) | Not detected | Normal | Normal | Not dected |
| 1 (mild) | A small amount of inflammatory cell infiltration (<10 cells in a field of vision) | Slight degeneration and swelling in alveolar/bronchial epithelial cells | Slight congestion | A small amount of serous exudate |
| 2 (moderate) | Moderate inflammatory cell infiltration (10–20 cells in a field of vision) | Moderate degeneration and swelling in alveolar/bronchial epithelial cells | Moderate congestion or hemorrhage; vascular endothelial swelling | Moderate serous exudate |
| 3 (severe) | A lot of inflammatory cell infiltration (>20 cells in a field of vision) | Epithelial cells appear serious degeneration, swelling or even necrosis, shedding | Severe bleeding; plenty of red blood cells in the alveolar cavity; vascular interstitial space widened | A large amount of serous exudate |
For fluorescence in situ hybridization (FISH), the tissues (n = 6 in each group) were fixed in the 4% paraformaldehyde for 1 h. Other steps about paraffin section processing were same as descripted above. Then sections were denatured in degeneration buffer [70% formamide, 30% 2 × saline-sodium citrate (2 × SSC)] at 78°C for 8 min and the probes were denatured in hybridization buffer (0.9 M NaCl, 0.1% SDS, 100 mM Tris PH 7.2, 15% formamide, 10% SDS), followed by hybridization overnight at 42°C. 20 μL hybridization buffer containing 15 μg probes were applied to per section. This study used pmhyb449, 59-CTATTTAACAACATCCCTTC-39 (S-S-Pmul-0449-a-A-20) (Sangon Biotech, China) to detect PmCQ2 (Mbuthia et al., 2001). The probes were labeled with Cy3. After washed by wash buffer (50% formamide, 50% 2 × SSC) for three times with 5 min in each time, cell nuclei were determined by counterstained with 4′, 6-diamidino-2-phenylindole (DAPI, Beyotime Biotech, China) to evaluate cellular morphology. Fluorescence was detected using fluorescence microscope (Olympus). Six mice were included in each group and three slices were conducted in each murine lung. The strength of the fluorescent signals was quantitatively analyzed in three visions per slice.
RNA isolation, cDNA library construction and illumina deep sequencing
Total RNA from approximately 150 mg lung tissues (n = 3 in each group) was extracted using Trizol reagent (Invitrogen, USA) following the manufacturer's protocol. Integrity of RNA was confirmed by 2100 Bioanalyzer (Agilent Technologies). The samples for transcriptomic analysis were prepared using Illumina's kit following manufacturer's recommendations. Briefly, mRNA was purified using oligo (dT) magnetic beads. By using the fragmentation buffer, the mRNA was fragmented into short fragments (about 200 bp), and then the first strand of cDNA was synthesized with random hexamer-primer using the mRNA fragments as templates. Buffer, dNTPs, RNase H, and DNA polymerase I were added to synthesize the second strand. The double-stranded cDNAs were purified with QiaQuick PCR extraction kit (Qiagen) and eluted with elution buffer for end repair and poly (A) addition. Finally, sequencing adapters were ligated to the 5′ and 3′ ends of the fragments. The fragments were purified by agarose gel electrophoresis and enriched by PCR amplification to create a cDNA library. The cDNA library was sequenced on the Illumina sequencing platform (HiSeqTM 2000) and 100 bp paired-end reads were generated.
Functional annotation of DEGs
Raw data were firstly processed using the NGS QC Toolkit. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. All the downstream analyses were based on clean data with high quality. The clean reads were mapped to reference genome (ftp://ftp.ensembl.org/pub/release-84/fasta/mus_musculus/dna/) and reference transcript (ftp://ftp.ensembl.org/pub/release-84/fasta/mus_musculus/cdna/Mus_musculus.GRCm38.cdna.all.fa.gz) using bowtie2 or Tophat (http://tophat.cbcb.umd.edu/) with default parameters by slightly modified (Langmead and Salzberg, 2012; Kim et al., 2013). Transcriptomic sequencing quality was displayed by mapping ratio to the reference genome and transcriptome. Based on results of clean reads mapping to reference genome, fragments per kilo base of exon per million fragments mapped (FPKM) values of genes were calculated and count value were calculated using eXpress (Mortazavi et al., 2008).
The read counts of each gene were obtained by htseq-count (Anders et al., 2015). Differential expression analysis was performed using estimateSizeFactors in DESeq (2012) R package, and p value and fold-change (FC) value were identified using nbinomTest in DESeq (2012) R package. P < 0.05 was set as the threshold for significantly differential expression. Hierarchical cluster analysis is used to identify differentially expressed genes with certain patterns of expression under two different salinity challenge using R. The settings for the calculations were as follows: similarity was measured by euclidean, and clustering method was complete linkage. GO and KEGG pathway enrichment analysis of the DEGs were performed using R based on hypergeometric distribution.
All the data discussed in this study have been deposited to NCBI's Gene Expression Omnibus (GEO) database and the accession number is SRR5186264.
Quantitative real-time RT-PCR (qRT-PCR)
Total RNA of the lungs (n = 6 in each group) were acquired as described previously (Shi et al., 2015). cDNA was synthesized using a PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). qRT-PCR was performed according to previous study (Ren et al., 2013a). Beta-actin was used as a reference gene to normalize transcript levels of target genes. Specific primers were designed according to the reference sequences in NCBI with Primer 5.0 software and the primer sequences were listed in Supplementary Table 1. The relative expression levels of genes were acted as a ratio of the target gene to the control gene using formula 2-(ΔΔCt), where ΔΔCt = (Ct for a target gene − Ct for the β-actin gene) in a treatment group − (Ct for a target gene − Ct for the β-actin gene) in the control (Ren et al., 2013b).
Immunoblotting analysis
Immunoblotting analysis was performed as described previously (Kamat et al., 2013). Briefly, approximate 100 mg frozen lung tissue powder (n = 5 in each group) was lysed in 1 mL RIPA buffer containing 100 mM PMSF to extract pulmonary total proteins. The protein concentration was determined using a bicinchoninic acid (BCA) protein assay kit (Beyotime, China). Equal amount of proteins (10 μg) were separated by 10% SDS-PAGE and transferred onto PVDF membrane (BioRad). The blots were blocked with 5% (w/v) skimmed milk in TBST (50 mM Tris–HCl, 150 mM NaCl, 0.1% Tween- 20, pH 7.5) for 2 h. Then blots were incubated with primary antibodies overnight at 4°C. Subsequently blots were incubated with the horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at 37°C. After ECL substrates were added, the blots were analyzed using light imaging system (Tanon 5200, China). Before each step, the blots were washed five times for 5 min with TBST.
In this study, β-actin (Proteintech) was determined as an internal control of the western blot. The primary antibodies were activator of transcription 1 (STAT1) (Beyotime Biotech, China), STAT3 (Wanleibio, China), p-STAT1 Y701 (Cell Signaling Technology) and p-STAT3 Y705 (Cell Signaling Technology). All secondary antibodies were peroxidase-conjugated affinipure goat anti-rabbit IgG (Proteintech).
Enzyme linked immunosorbent assay (ELISA)
Lung homogenates (n = 6 in each group) were freezing and thawing (frozen in liquid nitrogen for 5 min and then melted on ice) three times. After centrifugation at 12,000 rpm for 10 min at 4°C, supernatant were acquired. Cytokines (IL-17, IL-6, and IFN-γ) were detected in the supernatant or the serum with ELISA kits in accordance with the manufacturer's protocol. ELISA kit for IFN-γ detection was purchased from Cusabio (Wuhan, China). Other cytokine ELISA kits were purchased from eBioscience, USA.
Statistical analysis
All data were expressed as means ± Standard Error of Mean (SEM). All statistical analyses were performed using GraphPad Prism software. Survival rates of mice were evaluated using Kaplan-Meier analysis (Prism 6.0). Data between groups in H&E experiments were analyzed by Mann-Whitney U tests (Prism 6.0). All the other data between two groups were evaluated using unpaired, two-tailed Student's t-test (Prism 6.0). Data among more than two groups were analyzed by the one-way ANOVA followed by Dunnett multiple comparisons (Prism 6.0). Significant differences were considered at p < 0.05. *means p < 0.05, **means p < 0.01, ***means p < 0.001.
Results
P. multocida infection in mice
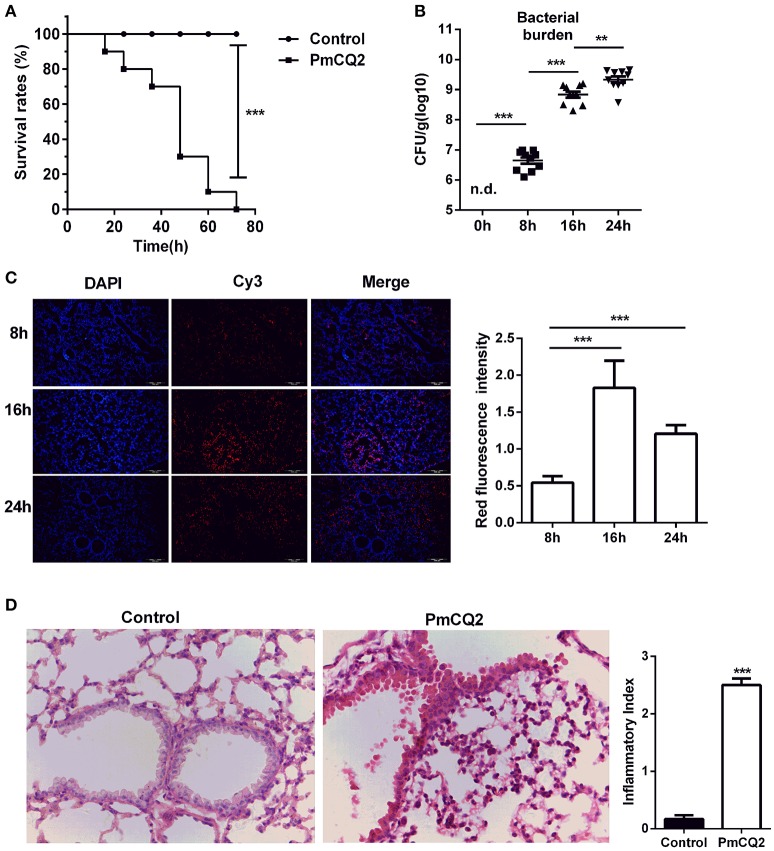
After infection, the survival rates were analyzed. There was a significant difference between infected group and control group (Figure 1A). All mice (n = 10) rapidly approach moribund within 3 days post-infection, whereas, no mortality was observed in control group. Then, bacterial load was counted in murine lungs at 0, 8, 16, 24 h post-infection (hpi). Although, PmCQ2 could not be detected in healthy murine lungs, the bacterial load was approximately 106 CFU/g at 8 hpi, and grew significantly at 16 and 24 hpi (Figure 1B). Also, FISH with specific probe of PmCQ2 (pmhyb449) showed the presence of PmCQ2 in the infected murine lungs (Figure 1C). PmCQ2 was detected widely in the alveolar walls, terminal bronchioles, and interstitial areas. At 16 and 24 hpi, bacterial burden was significantly higher than those at 8 hpi, whereas, there was no significant difference about the bacterial load between 16 and 24 hpi. The pulmonary lesions were also explored by H&E staining. After infection with PmCQ2, there were more severe inflammatory lesions (Figure 1D). The numbers of inflammatory cells in the peribronchial, alveolar and perivascular area of infected group were significantly higher than those in the control group. In addition to inflammatory cell infiltration, respiratory bronchus appears degeneration and necrosis. Capillaries appear dilation, congestion and bleeding in the infected group. Alveoli were suffered from rupture, dissolution and bronchial and alveolar epithelium appeared necrosis and were fell down from the lungs of infected mice. Collectively, similar to previous studies (Ren et al., 2013a; Chen et al., 2014), PmCQ2 infected mice have similar clinical and microscopical characters with what observed in cattle, indicating mice can serve as a surrogate model for studying P. moltocida infection.
Figure 1.
A mouse model of Pasteurella pneumonia caused by PmCQ2. (A) A high mortality rate in mice caused by PmCQ2. (n = 10, Kaplan-Meier analysis). (B) The proliferation of bacteria in the lung tissues (n = 10, one-way ANOVA). (C) The specific distribution of PmCQ2 in the lung tissues via FISH (n = 6, one-way ANOVA). (D) The severe pulmonary inflammatory lesions caused by PmCQ2 with H&E staining (n = 6, Mann-Whitney U tests, magnification 400X). Each point represents an individual mouse in the graph (B) and the data was expressed as means ± SEM. **P < 0.01, ***P < 0.001.
Evaluation of transcriptomic sequencing quality
After Illumina sequencing, there were approximately 5–7 × 107 raw reads in two groups including three mice per group. The average numbers of clean reads in infected and control samples were 71442307 and 56323661, respectively. Thus, there were nearly more raw reads (107) in the infected samples than control samples. The percentages of mapped to reference genome were more than 85% in all samples, in addition to 76.42% in only one infectious sample. Moreover, the percentage of GC content in all these samples was almost 50%. All the assessment criteria of base quality (Q30) were more than 94%. All data about quality of transcriptomic sequencing were shown in Supplementary Table 2.
Functional analysis and classification of DEGs
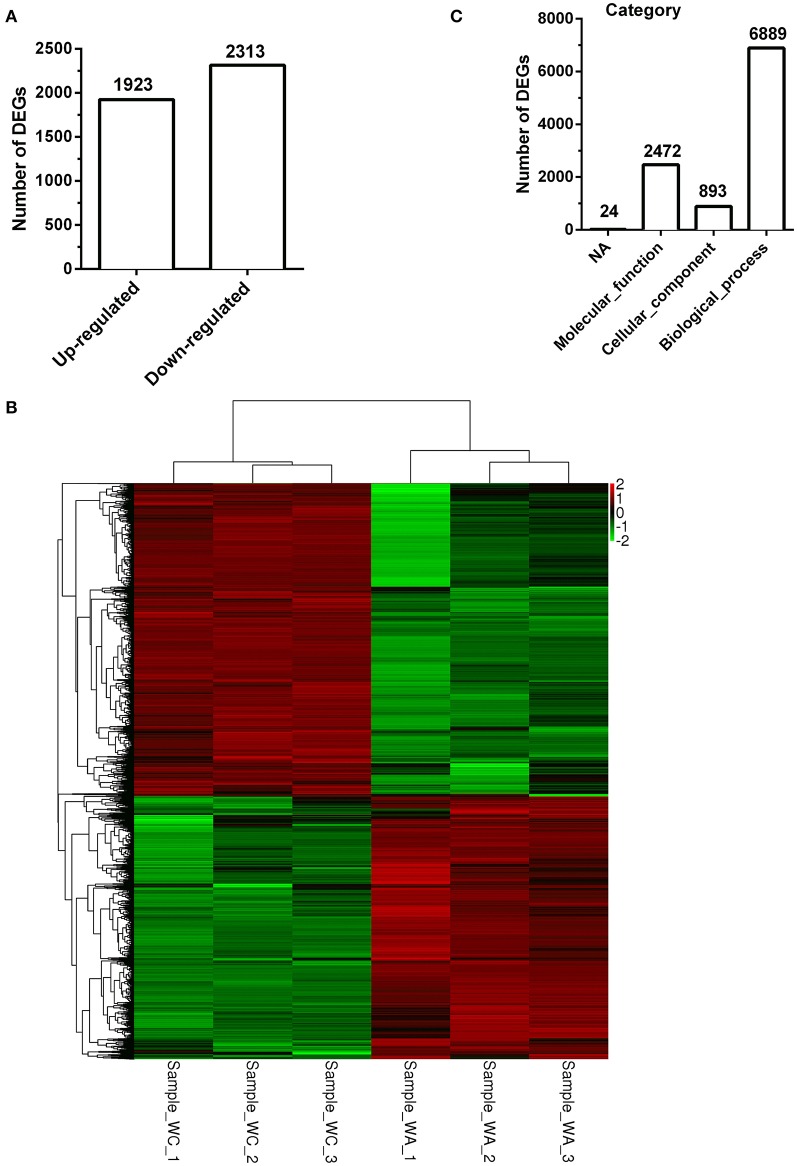
There were total 4236 DEGs between control and infected group with 1924 up-regulated DEGs (Figure 2A). The description of total DEGs was displayed in Supplementary Table 3. DEGs were used to unsupervised clustering hierarchy in this study. Correlation among samples was calculated by expression level of DEGs. In general, the same sample was classified into the same cluster by clustering analysis, and DEGs in the same cluster had lots of similar function, which was displayed with heatmap. Therefore, as shown in Figure 2B, three samples separately in control group or infected group had a tight correlation, whereas, there was almost no correlation between the two groups.
Figure 2.
Clustering and category of DEGs in Illumina sequencing. (A) Numbers of up/down-regulated DEGs (n = 3). (B) The clustering of DEGs in heatmap. Hierarchical cluster analysis is used to identify DEGs with certain patterns of expression. The same sample can appear in the same cluster by the clustering. Sample−WC−1, Sample−WC−2 and Sample−WC−3 represent three duplication of control group, while Sample−WA−1, Sample−WA−2 and Sample−WA−3 represent three duplication of infected group. (C) The category of GO enrichments.
The function of DEGs was annotated by GO enrichments and KEGG enrichments. GO enrichments were divided into biological process (6889), cellular component (893) and molecular function (2,472) (Figure 2C). Total DEGs were annotated by 10278 GO terms of which 5303 GO terms were significant (p < 0.05) (Table 2). Up-regulated DEGs were significantly (p < 0.05) annotated to 4182 GO terms while down-regulated DEGs were 3540 GO terms (Table 2). In addition, DEGs were analyzed using KEGG database. Similarly, there were total 291 KEGG pathways and 116 KEGG pathways were significant (p < 0.05) (Table 3).
Table 2.
GO annotation of DEGs
| Groups | Tested terms | P < 0.05 | P < 0.01 |
|---|---|---|---|
| WC-vs.- WA-Down | 7,063 | 3,540 | 1,667 |
| WC-vs.- WA-Total | 10,278 | 5,303 | 3,376 |
| WC-vs.- WA-Up | 7,198 | 4,182 | 2,739 |
Table 3.
KEGG annotation of DEGs.
| Groups | Tested terms | P < 0.05 | P < 0.01 |
|---|---|---|---|
| WC-vs.- WA-Down | 274 | 74 | 52 |
| WC-vs.- WA-Total | 291 | 116 | 85 |
| WC-vs.- WA-Up | 276 | 97 | 75 |
To further analyze GO/KEGG enrichments of up/down-regulated DEGs, significance of GO/KEGG enrichment score (p < 0.05) and number of DEGs (greater than or equal to 2) were acted as a cutoff. There were 2195 GO enrichments in up-regulated DEGs and 1846 enrichments in down-regulated DEGs (Supplementary Tables 4, 5). Ten top GO enrichments in each functional ontology were showed in Supplementary Figure 1 and Figure 2. In up-regulated DEGs, the dominant GO terms of biological process were “positive regulation of interleukin-23 production” and “response to interferon-beta”; of cellular component were “Ndc80 complex” and “NLRP3 inflammasome complex”; and of molecular function were “C-C chemokine binding” and “interleukin-2 binding” (Supplementary Figure 1). In down-regulated DEGs, they were “positive regulation of integrin biosynthetic process” and “smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation” in biological process; “collagen type V trimer” and “fibrillar collagen trimer” in cellular component; and “hedgehog family protein binding” and “calcium ion binding” in molecular function (Supplementary Figure 2).
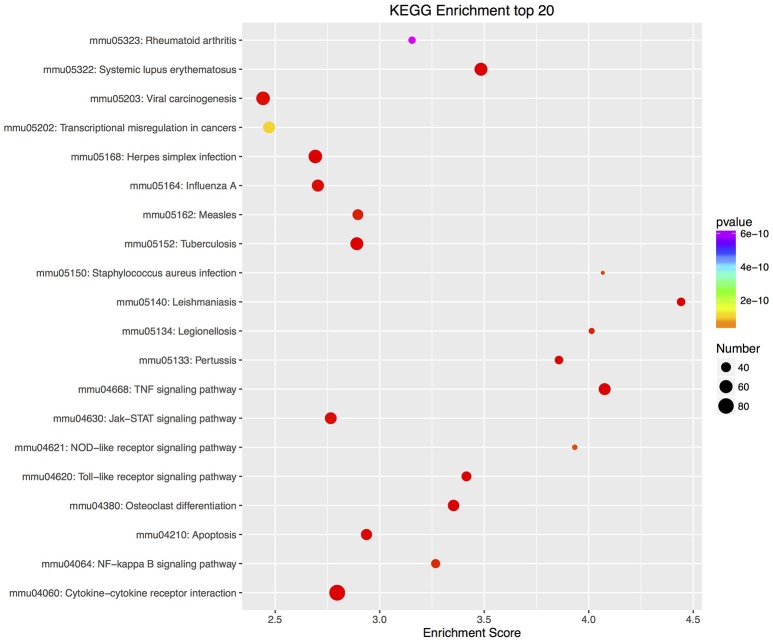
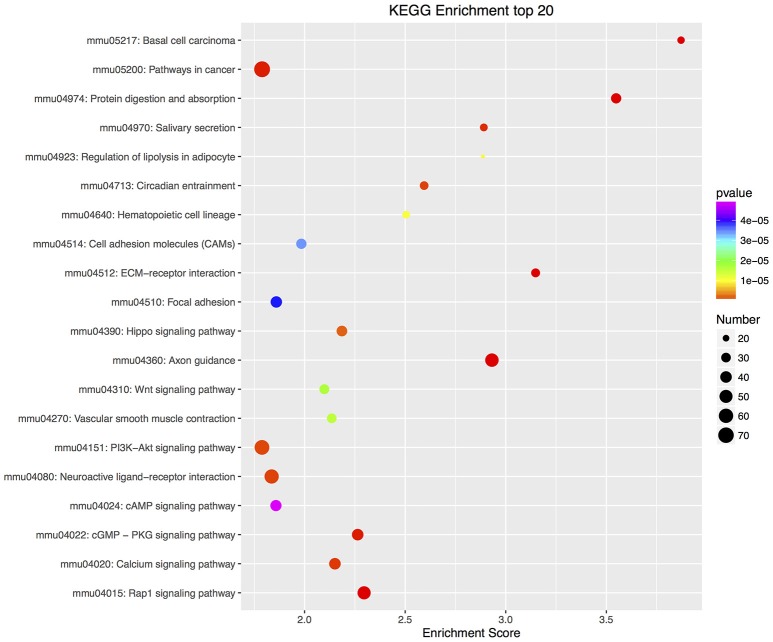
Furthermore, there were 97 significant KEGG pathways in up-regulated DEGs and 74 in down-regulated DEGs (Supplementary Tables 6, 7). Twenty top KEGG enrichments in up/down-regulated DEGs were showed in Figures 3, 4, respectively. In 20 top KEGG enrichments of up-regulated DEGs, there were 7 KEGG pathways which may be involved in host responses against pathogens, namely, “TNF signaling pathway,” “Jak-STAT signaling pathway,” “NOD-like receptor signaling pathway,” “Toll-like receptor signaling pathway,” “Apoptosis,” “NF-kappa B signaling pathway,” and “Cytokine-cytokine receptor interaction” (Figure 3). While, in down-regulated DEGs, there were mainly pathways related to metabolism or autoregulation in 20 top KEGG enrichments (Figure 4), such as, “Protein digestion and absorption,” “Salivary secretion,” “Circadian entrainment,” “Regulation of lipolysis in adipocyte.”
Figure 3.
KEGG annotation of up-regulated DEGs. The numbers of DEGs in each pathway are counted and KEGG enrichment scores and P-values are acquired by hypergeometric distribution test. Top 20 KEGG enrichments with up-regulated DEGs larger than 2 were showed. The x-axis represents KEGG enrichment scores and the y –axis represents pathway terms. The colors of circle indicate P-values and the size of circle indicates the numbers of DEGs. The circle with redder and larger indicating that the enrichment of the pathway is higher and DEG number is larger in the pathway.
Figure 4.
KEGG annotation of down-regulated DEGs. Similar to up-regulated DEGs, all down-regulated DEGs were mapped to signal pathway by KEGG database. DEGs numbers in each pathway were counted and KEGG enrichment scores and P-value were acquired by hypergeometric distribution test. Top 20 KEGG enrichments with down-regulated DEGs larger than 2 were showed. The x-axis represents KEGG enrichment scores and the y –axis represents pathway terms. The colors of circle indicate P-values and the size of circle indicates the numbers of DEGs. The circle with redder and larger indicating that the enrichment of the pathway is higher and DEG number is larger in the pathway.
DEGs related to host immune responses against bacterial infection
DEGs related to immune responses were summarized via GO enrichments (P < 0.05) and listed in the Supplementary Table 8. There were 13 GO terms correlated with immune responses, including, “interferon-gamma-mediated signaling pathway,” “positive regulation of interleukin-23 production,” “response to interferon-beta,” “Bcl-2 family protein complex,” “NLRP3 inflammasome complex,” “Toll-like receptor 2-Toll-like receptor 6 protein complex,” “interleukin-2 binding,” “Fc-gamma receptor signaling pathway,” “positive regulation of natural killer cell chemotaxis,” “chemokine activity,” “cytokine activity,” “response to molecule of bacterial origin,” and “T cell differentiation involved in immune response.” The 13 GO terms included 158 DEGs in total and in which there were only 26 down-regulated DEGs (16%). These GO terms were mainly involved in pattern recognition receptor (PRR), apoptosis, chemokine, inflammatory cytokines and T cell differentiation, which were involved in resistant to bacterial infection.
In addition, biosynthetic or signal transduction pathways about defense response via KEGG annotation were significantly enriched. There were 20 KEGG pathways in up-regulated DEGs, while only 7 KEGG pathways in down-regulated DEGs involved in immune responses (Supplementary Tables 9, 10). By using DEGs number with greater than 50 in these KEGG pathways as a cutoff, up-regulated DEGs were mapped to 4 KEGG pathways, including “Cytokine-cytokine receptor interaction,” “PI3K-Akt signaling pathway,” “TNF signaling pathway,” and “Jak-STAT signaling pathway,” while down-regulated DEGs were mapped to “PI3K-Akt signaling pathway” and “Rap1 signaling pathway.”
Experimental verification of DEGs
By comprehensive analysis of GO and KEGG annotation of DEGs, numerous DEGs in IL-12/IFN-γ axis and IL-23/IL-17 axis were found (Supplementary Table 11). The previous study reported that in pulmonary inflammation, IL-23 or IL-6 and transforming growth factor β (TGF-β) synergistically induced differentiation of naïve CD4+ cells into Th17 cells, favoring proliferation of memory T-cells and production of IL-17 (IL-17A) and IL-17F, associating with the STAT3 (signal transducers and activators of transcription-3) signaling (Iwakura and Ishigame, 2006; Mangan et al., 2006; Aujla et al., 2007; Wilson et al., 2007; Vanaudenaerde et al., 2008). IL12 is an important regulatory cytokine in Th1 responses through inducing the differentiation of Th1 cells to produce interferon (IFN-γ), associating with STAT1 signaling (Trinchieri, 2003; Qing and Stark, 2004; Iwakura and Ishigame, 2006). Interestingly, the key components of IL-23/IL-17 axis, including IL-23, IL-6, TGF-β, IL-17, IL-17F, IL-17 receptor, and STAT3, were significantly increased in transcriptional level and the key components of IL-12/IFN-γ axis, including IL12a/b (p35/p40), IFN-γ, IFN-γ receptor, JAK2, and STAT, were also all significantly increased (Supplementary Table 11).
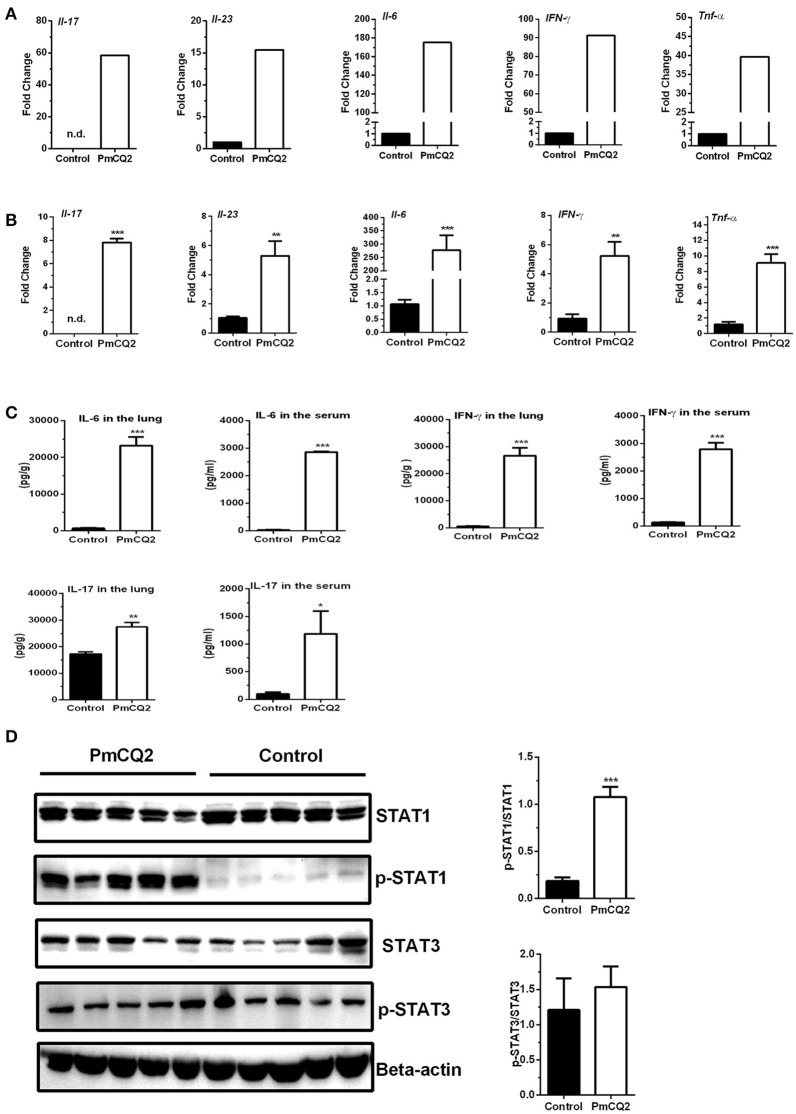
To validate the reliability of transcriptomic sequencing, inflammatory cytokines in IL-12/IFN-γ axis and IL-23/IL-17 axis were selected to detect by qRT-PCR. Figures 5A,B showed expression level of DEGs (Il-6, Il-23, Il-17, IFN-γ, and Tnf-α) by RNA-seq and qRT-PCR, respectively. Similar to that observed with RNA-seq analysis, they are found to significantly increase by qRT-PCR analysis (Figures 5A,B). To further confirm post-translational level of these DEGs in vivo, levels of IL-6, IL-17, and IFN-γ in the lungs or serum were determined using ELISA kits, and activation of STAT1 and STAT3 signaling were analyzed with immunoblotting. We found that levels of IL-6, IL-17, and IFN-γ in the lungs or serum in infectious mice were significantly higher than that in control mice (Figure 5C). There was no difference in the abundance of total STAT1 in the lungs between infectious mice and control mice, whereas, abundance of p-STAT1 in the lungs of infectious mice were significantly increased, compared to control group (Figure 5D). While the abundance of total STAT3 and p-STAT3 were similar between two groups (Figure 5D).
Figure 5.
Up-regulation of genes related to Ifn-γ/Il-17 at 16 hpi. (A) The high expression of Ifn-γ and Il-17 after P. multocida infection by Illumina sequencing (n = 3). The fold change indicated the ratio of the experimental group basemean (sample-WA average expression level) to the control group basemean (sample-WC average expression level) by Illumina sequencing. (B) The high expression of Ifn-γ and Il-17 after P. multocida infection by qRT-PCR (n = 6, unpaired, two-tailed Student's t-test). (C) The levels of IFN-γ and IL-17 in the lungs or serum using ELISA analysis (n = 6, unpaired, two-tailed Student's t-test). (D) The abundance of p-STAT1Y701 and p-STAT3Y705 in the murine lungs by immunoblotting analysis (n = 5, unpaired, two-tailed Student's t-test). All data were expressed as means ±SEM. Because Il17 was not detected in control mice, the fold changes of Il17 in treatment mice were showed via presuming expression quantity = 1 of control mice in RNA-Seq (A) and Ct = 30 in qRT-PCR (B), respectively. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
P. multocida serogroup A is an opportunistic pathogen causing severe respiratory diseases in cattle (Rerat et al., 2012; Li et al., 2016). In this study, a mouse model of Pasteurella pneumonia caused by PmCQ2 was established via intraperitoneal infection. It is well known that the natural infectious route for P. multocida serotype A strain is through intranasal infection, but we found that intranasal infection results in a poor reproducibility among mice. Moreover, there are some similarities between intranasal infection and intraperitoneal infection, including the pathological alterations and expressions of inflammatory cytokines (Praveena et al., 2010; Hodgson et al., 2013). Thus, intraperitoneal infection was used in this study. The mice died within 3 days post-infected by PmCQ2. This result indicates that PmCQ2 is a virulent strain for mice as reported previously (Li et al., 2016). Likewise, this study indicates that PmCQ2 also proliferates significantly in the lungs of mice after inoculation, but the bacterial burden tends to be stable after 16 h post-infection from previous observation (Li et al., 2016). However, in current study, the bacterial load increases after 16 h post-infection by bacterial counting analysis. Traditional counting bacteria by gradient dilution cannot display detailed distribution of bacteria (Amann et al., 1995). Thus, the bacterial load in the lungs was also detected with FISH using an oligonucleotide probe pmhyb449, which recognizes specifically 16S rRNA of P. multocida (Mbuthia et al., 2001). The conclusion from FISH analysis is similar with the previous investigation (Li et al., 2016). It is worth noting that bacterial load after 24 h post-infection from FISH is not consistent to that from gradient dilution. We suspect this difference may be caused by limitations of the number of lung sections in FISH. Pasteurella pneumonia in cattle, rabbits, fowl and pigs has been reported and is mainly caused by P. multocida serogroup A (Blackall et al., 1995; Al-Haddawi et al., 2000; Pors et al., 2011; Praveena et al., 2014). A previous study reported that P. multocida causes inflammation because of neutrophil and macrophage infiltration in the lungs of cattle by intratracheal route (Praveena et al., 2014). In this study, like other P. multocida serogroup A, PmCQ2 causes serious lung lesions in mice characterized by inflammation in murine lungs. This suggests that there is a similar inflammatory response in murine lungs after PmCQ2 infection by intraperitoneal routes. However, the cell type in the lungs was not further confirmed by other methods, such as flow cytometry and IHC. In addition, the infected tissue section shows that there are different degrees of bronchitis, alveolar rupture and some hemorrhage, which are consistent to pathological changes in cattle infected by P. multocida (Mathy et al., 2002; Dabo et al., 2007). However, the underlying mechanism of host inflammatory responses to this bacterial infection is still elusive. Thus, the DEGs related to P. multocida infection are explored in this study with transcriptomic sequencing because it can provide a wide range of changes in transcription level. In this study, we mainly studied the immune responses in mice against P. multocida, such as, PRRs, downstream pathways of PRRs and effector molecules (e.g., cytokines and chemokines).
PRRs are crucial for the initiation of innate immunity against bacterial infection, via recognizing pathogen-associated molecular patterns (PAMPs) (Jang et al., 2015). Retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), toll-like receptors (TLRs), C-type lectin receptors (CLRs), AIM2-like receptors (ALRs) and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) are from PRR families (Kawai and Akira, 2009; Jang et al., 2015). Activation of TLRs, except for TLR3, recruits the adaptor protein myeloid differentiation factor 88 (MyD88) to form receptor complex (Kawai and Akira, 2005). NOD1, NOD2, and NLRP3 (NOD-like receptor family, pyrin domain containing, NLRP) are important components of NLR families. NOD1, NOD2, TLRs/MyD88 complex stimulate nuclear factor kappa B (NF-κB) signaling and mitogen activated protein kinase (MAPK) signaling against bacteria. NLRP3 inflammasome consists of NLRP3, the adaptor apoptosis-associated speck-like protein containing a CARD (ASC) and the effector caspase-1 (Levy et al., 2015). Activation of NLRP3 inflammasome leads to secretion of mature IL-18/IL-1β (Kanneganti et al., 2007).
In this study, there were various DEGs involved in “Toll-like receptor signaling pathway,” “RIG-I-like receptor signaling pathway,” and “NOD-like receptor signaling pathway” by KEGG annotation (Supplementary Table 12). In “Toll-like receptor signaling pathway,” more than 90.9% of DEGs are up-regulated, including Tlr1, Tlr2, Tlr6, Tlr9, and MyD88. TLR2 functions as a homodimer or as a heterodimer with TLR1 or TLR6 to recognize a variety of PAMPs (Li and Sun, 2016). TLR2/TLR1 recognizes triacylated lipoproteins/lipopeptides while TLR2/TLR6 recognizes diacylated lipoproteins/lipopeptides (Schenk et al., 2009). TLR9 recognizes CpG DNA in bacterial genomes (Medzhitov, 2007). To date, various investigations have reported the involvement of TLRs during infections, such as Sporothrix schenckii (TLR2), Staphylococcus aureus (TLR2), Salmonella enterica (TLR1/TLR2 complex, TLR9), Vibrio cholerae cytolysin (TLR2/TLR6 complex, TLR9) (Totemeyer et al., 2005; de C Negrini et al., 2014; Khilwani et al., 2015; Nandi and Bishayi, 2015). However, as far as we known, the association of TLRs during P. multocida infection is mainly about TLR4 to recognize P. multocida toxin (PMT) and LPS (Hildebrand et al., 2012; Kubatzky, 2012). Our results indicate that other TLRs, especially, TLR2, mediate P. multocida infection. Nod1, Nod2, and Nlrp3 significantly up-regulate from transcriptional level. NOD1 recognizes γ-D-glutamyl-meso-diaminopimelic acid (iE-DAP) mainly from gram-negative bacteria, while NOD2 is a general sensor of peptidoglycan through the recognition of muramyl dipeptide (MDP) (Girardin et al., 2003). Like TLR induction, Shigella flexneri, entero-invasive Escherichia coli, Chlamydophila pneumoniae, Campylobacter jejuni, and Listeria monocytogenes are sensed by NOD1, while Streptococcus pneumonia and Mycobacterium tuberculosis are recognized by NOD2 (Kanneganti et al., 2007). Therefore, we presume that NLRs are also involved in the P. multocida infection.
As downstream pathways of PRRs, NF-κB, and MAPK signaling pathways are enriched by KEGG annotation. There are total 82 DEGs involved in MAPK signal pathway and 45 DEGs in NF-κB signaling. NF-κB and MAPK signaling play important roles in defense responses against bacterial infection by regulating production of cytokines, such as Enterococcus faecalis, S. aureus, and M. tuberculosis (Zhu et al., 2013; Deng et al., 2014; Zou and Shankar, 2015). Thus, the NF-κB and MAPK signaling may also have the critical importance during P. multocida infection. Nlrp3, caspase-1, and Pycard up-regulate significantly, which are involved in assembling of NLRP3 inflammasome to promote maturation of pro-IL18/IL-1β. Its activation contributes to host defense during a variety of bacterial infections, suggesting that it is involved in defense against P. multocida infection. Indeed, we have found that Nlrp3 is significantly up-regulated in RAW264.7 cells after P. multocida infection (data unpublished).
During the development of inflammation, cytokines play important roles, mainly embodying as restriction and balance between proinflammatory and anti-inflammatory cytokines. Cytokines have also key functions in host defense responses against bacteria. Some cytokines (e.g., IL-1) are acted as potential useful biomarkers of community-acquired bacterial infection (Holub et al., 2013). There are various DEGs involved in cytokine mediating signal pathway by KEGG enrichments during P. multocida infection (Supplementary Table 13). 117 DEGs are included in cytokine-cytokine receptor interaction, and in which 84 DEGs are up-regulated. The type of up-regulated cytokines (e.g., Ifn-γ, Tnf-α, Il-1β, Il-6) in this study has some similarity to that of bovine lung infected by P. multocida (Mathy et al., 2002), which suggests that in addition to similar pathological changes, immunological changes in mice may be also similar to those in cattle caused by P. multocida. However, a further study is needed to further explore the similarity and difference in responses to P. multocida in mice and cattle.
Interestingly, various up-regulated cytokines is involved in Th17 responses. In this study, although p-STAT3 is not significantly increased, Th17 related cytokines (Il-17, Il-17ra, Il-23a, Il-6, and Il-6rα) are significantly increased using transcriptomic sequencing. Moreover, increased expressions of Il-6, Il-17, and Il-23 are validated by qRT-PCR. Also, the levels of IL-17 and IL-6 are significantly increased in the pulmonary homogenate and serum at 16 h post-infection. It has been reported that host immune responses are mainly Th17 responses in pulmonary inflammation caused by bacteria (Caucheteux et al., 2016; Hua et al., 2016; Wong et al., 2016). For example, in pulmonary inflammation, IL-23 is secreted mainly by alveolar dendritic cells (DCs) and macrophages to induce the differentiation of naïve CD4 cells into Th17 cells for production of IL-17 (Iwakura and Ishigame, 2006; Aujla et al., 2007; Caucheteux et al., 2016). In addition, IL-6 and TGF-β synergistically stimulates the production of IL-17 (Mangan et al., 2006; Wilson et al., 2007). This process is regulated mainly by transcription factors Foxp3, RORγt (retinoid-related orphan receptor), STAT3 and IRF4 (interferon regulatory factor-4) (Vanaudenaerde et al., 2008; Voo et al., 2009). IL-17 could recruit neutrophils into lung to eliminate the invasive bacteria through granulopoiesis and CXC chemokines induction (Kolls and Linden, 2004). Release of IL-17 also induces antimicrobial proteins and proinflammatory cytokines, including granulocyte colony-stimulating factor (G-CSF), IL-6, IL-1β, and TNF-α and hematopoietic growth factor (Jovanovic et al., 1998; Happel et al., 2005; Dubin and Kolls, 2007). Thus, IL-23/IL-17 axis plays a critical role in anti-bacterial immunity, such as Pseudomonas aeruginosa, Citrobacter rodentium, E. coli, Porphyromonas gingivalis (Mangan et al., 2006; Dubin and Kolls, 2007; Shibata et al., 2007; Yu et al., 2008; Ren et al., 2017). The results from current study indicate like other reported bacteria, Th17 responses is also involved in P. multocida infection by an intraperitoneal route. In fact, it has been reported that PMT induces the differentiation of Th17 cells through the activation of STAT transcription factors to induce IL-17 production (Hildebrand et al., 2015), which suggest IL-17 is involved in P. multocida infection. Notably, intranasal immunization in mice protects mice against multiple bacterial pneumonia (e.g., P. aeruginosa, S. pneumonia) in an antibody-independent but IL-17–dependent manner (Wu et al., 2012; Wang et al., 2017), which suggest the feasibility of development a broadly protective vaccine against bacterial pneumonia by targeting lung Th17 cells. Thus, it is interesting to know whether the manipulation of lung Th17 responses could regulate P. multocida infection or not.
In this study, Th1 related cytokines (Il-12a, Il-12b, Il-12rβ1, Il-12rβ2, Ifn-γ, Tnf-α, Il-2rα, Il-2rγ, Il-2rβ) are up-regulated from transcriptomic sequencing, qRT-PCR, and ELISA analysis. Moreover, increased activation of p-STAT1, the regulator of Th1, is also observed. These results indicate that there are strong Th1 immune responses in mice during P. multocida infection. IL-12 is structural similar to IL-23 with the same IL-12p40 subunit and unique IL-12p35, and is an important regulatory cytokine in Th1 responses by inducing the differentiation of naïve CD4 cells into Th1 cells and activating natural killer cells for the production of IFN-γ (Trinchieri, 2003; Happel et al., 2005; Iwakura and Ishigame, 2006). IFN-γ activates both neutrophils and macrophages for intracellular killing of bacteria or fungi (Nguyen et al., 2015). The process of IFN-γ or IFN regulatory factor-1 (IRF-1) production is regulated by STAT1 (Qing and Stark, 2004; Iwakura and Ishigame, 2006). In return, recognition of IFN-γ by IFN-γ receptor (IFNGR1/2) activates Janus kinase (JAK, mainly JAK1 and JAK2)/STAT1 pathway to activate transcription factor T-bet, resulting in extensive production of IFN-γ to eliminate virus or bacteria (Rawlings et al., 2004; Hu and Ivashkiv, 2009). Thus, IL-12/IFN-γ axis has critical importance in lung infection. IL-12 is important in the initial phase of bacterial, parasitic, and viral infections (Tekkanat et al., 2001). Recently, a study indicated that IL-12 plays a critical role in protection from pulmonary methicillin-resistant S. aureus (MRSA) via activating IL-12/ IFN-γ axis (Nguyen et al., 2015). In addition, IL-12 promotes IFN-γ-dependent neutrophil recruitment into the lungs of mice after intranasal infection with S. pneumonia (Sun et al., 2007). Specifically, IL-12 and IFN-γ induce synergistically bacterial clearance from the lungs and pulmonary cellular infiltration during extracellular and intracellular bacterial infection, such as L. monocytogenes, S. pneumonia, Chlamydia muridarum, M. tuberculosis (Haring et al., 2005; Mpiga and Ravaoarinoro, 2006; Sun et al., 2007; Cooper and Khader, 2008; Jupelli et al., 2010; Hashiguchi et al., 2014). More importantly, Il-12a, Il-12b, Il-2, Ifn-γ are up-regulated in bovine lungs by an intratracheal P. multocida inoculation (Mathy et al., 2002), which suggest that Th1 responses are induced by P. multocida and there are some similarities between mice and cattle during P. multocida infection by an intratracheal or intraperitoneal route. However, Th1 responses in host during P. multocida infection with an intranasal route still need to further investigation.
In addition to cytokine mediating signal pathway, chemokine signaling is also a significant signal pathway from KEGG annotation. Based on cysteine residues position, chemokines are classified into two major chemokine sub-families, CXC chemokines and CC chemokines. The former is mainly chemotactic for neutrophils and the latter mainly hemotactic for monocytes and sub-set of lymphocytes (Palomino and Marti, 2015). In this study, there are 14 CC DEGs, including, Ccl3, Ccl4, Ccl9, Ccl11, Ccl22, Ccl17, Ccl5, Ccl12, Ccl7, Ccl2, Ccl19, Ccl28, Ccl27a, and Ccl21a. Except for the last two, others are up regulated. In addition, 8 CXC DEGs are up regulated, including Cxcl16, Cxcl14, Cxcl13, Cxcl5, Cxcl19, Cxcl9, Cxcl10, and Cxcl2. Only Cxcl12 decreases significantly in transcriptional level. Tagawa et al. showed CC chemokines induces macrophage trafficking to the site of infection during E. coli infection (Tagawa et al., 2004). Abundant macrophage inflammatory protein 1α (MIP-1α, namely CCL3) is detected in the liver during L. monocytogenes (Bubonja et al., 2006). Similarly, MIP-1α, MIP-1β, and RANTES (namely CCL3, CCL4, and CCL5) play key roles in Helicobacter pylori (Raghavan et al., 2003). Moreover, CXC chemokines are involved in various bacterial infections by impacting neutrophils, for example, P. aeruginosa, M. tuberculosis (Almeida et al., 2009; Gregson et al., 2013). In this study, we found infiltrated cells in the lungs are mainly the neutrophils, with lower number of macrophages in pulmonary lesion, suggesting that CXC chemokines have critical roles during P. multocida infection. However, the involvement of neutrophils, macrophages or CXC chemokines in P. multocida infection needs further investigation.
In conclusion, a mouse model of Pasteurella pneumonia is established successfully by P. multocida serogroup A strain PmCQ2 in this study. This study firstly showed that there is significant alteration in expression profile in the lungs during P. multocida infection in vivo using transcriptomic sequencing. Most of altered genes are correlated to PRRs, PRR downstream signaling, chemokines and cytokines, indicating the complex immune responses in the lungs during P. multocida infection. This study provides a wide range of new insights to defense responses during P. multocida infection.
Author contributions
YP, NL, and WR conceived the experiments. CW and XQ conducted the experiments. XQ, PL, and TP contributed to data analysis. CW and WR wrote the paper. YP, NL, and WR supervised the project and revised the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study is supported by the earmarked fund for China Agriculture Research System (Beef/Yak Cattle, CARS-38), Chongqing Science and Technology Commission (cstc2015shmszx80022, cstc2016shmszx80085) and Doctoral Funds of Southwest University (SWU112006).
Glossary
Abbreviations
- ALRs
AIM2-like receptor
- ASC
apoptosis-associated speck-like protein containing a CARD
- BCA
bicinchoninic acid
- CLRs
C-type lectin receptors
- CFU
colony-forming units
- DEGs
differently expressed genes
- DAPI, 4′
6-diamidino-2-phenylindole
- ELISA
enzyme linked immunosorbent assay
- FPKM
per million fragments mapped
- FC
fold-change
- FISH
fluorescence in situ hybridization
- GO
gene ontology
- G-CSF
granulocyte colony-stimulating factor
- H&E
hematoxylin and eosin
- iE-DAP
γ-D-glutamyl-meso-diaminopimelic acid
- IFN-γ
interferon
- IRF4
interferonregulatoryfactor-4
- LPS
lipopolysaccharide
- MAPK
mitogen activated protein kinase
- MDP
muramyl dipeptide
- MIP-1α
macrophage inflammatory protein 1α
- MyD88
myeloid differentiation factor 88
- NLRP
NOD-like receptor family, pyrin domain containing
- NLRs
nucleotide-binding oligomerization domain (NOD)-like receptors
- NF-κB
nuclear factor kappa B
- PMT
pasteurella multocida toxin
- PAMPs
pathogen-associated molecular patterns
- qRT-PCR
quantitative real-time RT-PCR
- KEGG
kyoto encyclopedia of genes and genomes
- RLRs
retinoic acid-inducible gene-I (RIG-I)-like receptors
- RORγt
retinoid-related orphan receptor
- ROS
reactive oxygen species
- STAT3
signal transducers and activators of transcription-3
- SSC
saline-sodium citrate
- SEM
standard error of mean
- TLRs
toll-like receptors
- TGF-β
transforming growth factor β.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00251/full#supplementary-material
GO analysis of up-regulated DEGs. All up-regulated DEGs were analyzed by GO enrichments and numbers of DEGs in each GO term were counted. Significance of GO enrichments was described as P < 0.05. In this figure, top 10 GO enrichments with up-regulated DEGs larger than 2 in each functional ontologies were showed.
GO analysis of down-regulated DEGs. All down-regulated DEGs were analyzed by GO enrichments and numbers of DEGs in each GO term were counted. Significance of GO enrichments was described as P < 0.05. In this figure, top 10 GO enrichments with down-regulated DEGs larger than 2 in each functional ontologies were showed.
Primers used in qRT-PCR analysis.
RNA-seq sequencing evaluation.
Total DEGs in this study.
Significant GO enrichment analysis of up-regulated DEGs.
Significant GO enrichment analysis of down-regulated DEGs.
Significant KEGG pathways of up-regulated DEGs.
Significant KEGG pathway of down-regulated DEGs.
DEGs related to immune response.
KEGG pathway of up-regulated DEGs related to immune response.
KEGG pathway of down-regulated DEGs related to immune response.
Gene list of significant DEGs in IL-12/IFN-γ axis and IL23/IL17 axis.
Expression level of DEGs related to PRR and PRR downstream signaling.
Expression level of DEGs related to cytokine and chemokine signaling.
References
- Al-Haddawi M. H., Jasni S., Zamri-Saad M., Mutalib A. R., Zulkifli I., Son R., et al. (2000). In vitro study of Pasteurella multocida adhesion to trachea, lung and aorta of rabbits. Vet J. 159, 274–281. 10.1053/tvjl.1999.0418 [DOI] [PubMed] [Google Scholar]
- Almeida C. S., Abramo C., Alves C. C., Mazzoccoli L., Ferreira A. P., Teixeira H. C. (2009). Anti-mycobacterial treatment reduces high plasma levels of CXC-chemokines detected in active tuberculosis by cytometric bead array. Mem. Inst. Oswaldo Cruz. 104, 1039–1041. 10.1590/S0074-02762009000700018 [DOI] [PubMed] [Google Scholar]
- Amann R. I., Ludwig W., Schleifer K. H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprianto R., Slager J., Holsappel S., Veening J. W. (2016). Time-resolved dual RNA-seq reveals extensive rewiring of lung epithelial and pneumococcal transcriptomes during early infection. Genome Biol. 17:198. 10.1186/s13059-016-1054-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aujla S. J., Dubin P. J., Kolls J. K. (2007). Th17 cells and mucosal host defense. Semin. Immunol. 19, 377–382. 10.1016/j.smim.2007.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackall P. J., Pahoff J. L., Marks D., Fegan N., Morrow C. J. (1995). Characterisation of Pasteurella multocida isolated from fowl cholera outbreaks on turkey farms. Aust. Vet. J. 72, 135–138. 10.1111/j.1751-0813.1995.tb15033.x [DOI] [PubMed] [Google Scholar]
- Bubonja M., Wraber B., Brumini G., Gobin I., Veljkovic D., Abram M. (2006). Systemic and local CC chemokines production in a murine model of Listeria monocytogenes infection. Mediators Inflamm. 2006:54202. 10.1155/MI/2006/54202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caucheteux S. M., Hu-Li J., Mohammed R. N., Ager A., Paul W. E. (2016). Cytokine regulation of lung Th17 response to airway immunization using LPS adjuvant. Mucosal Immunol. 10, 361–372. 10.1038/mi.2016.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Liu S., Zhang F., Ren W., Li N., Yin J., et al. (2014). Effects of dietary L-glutamine supplementation on specific and general defense responses in mice immunized with inactivated Pasteurella multocida vaccine. Amino Acids. 46, 2365–2375. 10.1007/s00726-014-1789-9 [DOI] [PubMed] [Google Scholar]
- Cooper A. M., Khader S. A. (2008). The role of cytokines in the initiation, expansion, and control of cellular immunity to tuberculosis. Immunol. Rev. 226, 191–204. 10.1111/j.1600-065X.2008.00702.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabo S. M., Taylor J. D., Confer A. W. (2007). Pasteurella multocida and bovine respiratory disease. Anim. Health Res. Rev. 8, 129–150. 10.1017/S1466252307001399 [DOI] [PubMed] [Google Scholar]
- de C Negrini T., Ferreira L. S., Arthur R. A., Alegranci P., Placeres M. C., Spolidorio L. C., et al. (2014). Influence of TLR-2 in the immune response in the infection induced by fungus Sporothrix schenckii. Immunol. Invest. 43, 370–390. 10.3109/08820139.2013.879174 [DOI] [PubMed] [Google Scholar]
- Deng W., Li W., Zeng J., Zhao Q., Li C., Zhao Y., et al. (2014). Mycobacterium tuberculosis PPE family protein Rv1808 manipulates cytokines profile via co-activation of MAPK and NF-kappaB signaling pathways. Cell. Physiol. Biochem. 33, 273–288. 10.1159/000356668 [DOI] [PubMed] [Google Scholar]
- Dubin P. J., Kolls J. K. (2007). IL-23 mediates inflammatory responses to mucoid Pseudomonas aeruginosa lung infection in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 292, L519–L528. 10.1152/ajplung.00312.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin S. E., Boneca I. G., Viala J., Chamaillard M., Labigne A., Thomas G., et al. (2003). Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278, 8869–8872. 10.1074/jbc.C200651200 [DOI] [PubMed] [Google Scholar]
- Gregson A. L., Wang X., Weigt S. S., Palchevskiy V., Lynch J. R., Ross D. J., et al. (2013). Interaction between Pseudomonas and CXC chemokines increases risk of bronchiolitis obliterans syndrome and death in lung transplantation. Am. J. Respir. Crit. Care Med. 187, 518–526. 10.1164/rccm.201207-1228OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D., Chengappa M. M., Kuszak J., McVey D. S. (2010). Bacterial pathogens of the bovine respiratory disease complex. Vet. Clin. North Am. Food Anim. Pract. 26, 381–394. 10.1016/j.cvfa.2010.04.004 [DOI] [PubMed] [Google Scholar]
- Happel K. I., Dubin P. J., Zheng M., Ghilardi N., Lockhart C., Quinton L. J., et al. (2005). Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J. Exp. Med. 202, 761–769. 10.1084/jem.20050193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring J. S., Corbin G. A., Harty J. T. (2005). Dynamic regulation of IFN-gamma signaling in antigen-specific CD8+ T cells responding to infection. J. Immunol. 174, 6791–6802. 10.4049/jimmunol.174.11.6791 [DOI] [PubMed] [Google Scholar]
- Harper M., Boyce J. D., Adler B. (2006). Pasteurella multocida pathogenesis: 125 years after Pasteur. FEMS Microbiol. Lett. 265, 1–10. 10.1111/j.1574-6968.2006.00442.x [DOI] [PubMed] [Google Scholar]
- Hashiguchi T., Oyamada A., Sakuraba K., Shimoda K., Nakayama K. I., Iwamoto Y., et al. (2014). Tyk2-dependent bystander activation of conventional and nonconventional Th1 cell subsets contributes to innate host defense against Listeria monocytogenes infection. J. Immunol. 192, 4739–4747. 10.4049/jimmunol.1303067 [DOI] [PubMed] [Google Scholar]
- Hildebrand D., Heeg K., Kubatzky K. F. (2015). Pasteurella multocida toxin manipulates T cell differentiation. Front. Microbiol. 6:1273. 10.3389/fmicb.2015.01273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand D., Sahr A., Wolfle S. J., Heeg K., Kubatzky K. F. (2012). Regulation of Toll-like receptor 4-mediated immune responses through Pasteurella multocida toxin-induced G protein signalling. Cell Commun. Signal. 10:22. 10.1186/1478-811X-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson J. C., Dagleish M. P., Gibbard L., Bayne C. W., Finlayson J., Moon G. M., et al. (2013). Seven strains of mice as potential models of bovine pasteurellosis following intranasal challenge with a bovine pneumonic strain of Pasteurella multocida A:3; comparisons of disease and pathological outcomes. Res. Vet. Sci. 94, 634–640. 10.1016/j.rvsc.2013.01.015 [DOI] [PubMed] [Google Scholar]
- Holub M., Lawrence D. A., Andersen N., Davidova A., Beran O., Maresova V., et al. (2013). Cytokines and chemokines as biomarkers of community-acquired bacterial infection. Mediators Inflamm. 2013:190145. 10.1155/2013/190145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Ivashkiv L. B. (2009). Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 31, 539–550. 10.1016/j.immuni.2009.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Jiao Y. Y., Ma Y., Peng X. L., Fu Y. H., Zheng Y. P., et al. (2016). DNA vaccine encoding central conserved region of G protein induces Th1 predominant immune response and protection from RSV infection in mice. Immunol. Lett. 179, 95–101. 10.1016/j.imlet.2016.09.011 [DOI] [PubMed] [Google Scholar]
- Iwakura Y., Ishigame H. (2006). The IL-23/IL-17 axis in inflammation. J. Clin. Invest. 116, 1218–1222. 10.1172/JCI28508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. H., Shin H. W., Lee J. M., Lee H. W., Kim E. C., Park S. H. (2015). An overview of pathogen recognition receptors for innate immunity in dental pulp. Mediators Inflamm. 2015:794143. 10.1155/2015/794143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic D. V., Di Battista J. A., Martel-Pelletier J., Jolicoeur F. C., He Y., Zhang M., et al. (1998). IL-17 stimulates the production and expression of proinflammatory cytokines, IL-beta and TNF-alpha, by human macrophages. J. Immunol. 160, 3513–3521. [PubMed] [Google Scholar]
- Jupelli M., Selby D. M., Guentzel M. N., Chambers J. P., Forsthuber T. G., Zhong G., et al. (2010). The contribution of interleukin-12/interferon-gamma axis in protection against neonatal pulmonary Chlamydia muridarum challenge. J. Interferon Cytokine Res. 30, 407–415. 10.1089/jir.2009.0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat P. K., Kalani A., Givvimani S., Sathnur P. B., Tyagi S. C., Tyagi N. (2013). Hydrogen sulfide attenuates neurodegeneration and neurovascular dysfunction induced by intracerebral-administered homocysteine in mice. Neuroscience 252, 302–319. 10.1016/j.neuroscience.2013.07.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti T. D., Lamkanfi M., Nunez G. (2007). Intracellular NOD-like receptors in host defense and disease. Immunity 27, 549–559. 10.1016/j.immuni.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. (2005). Toll-like receptor downstream signaling. Arthritis Res. Ther. 7, 12–19. 10.1186/ar1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Akira S. (2009). The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 21, 317–337. 10.1093/intimm/dxp017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khilwani B., Mukhopadhaya A., Chattopadhyay K. (2015). Transmembrane oligomeric form of Vibrio cholerae cytolysin triggers TLR2/TLR6-dependent proinflammatory responses in monocytes and macrophages. Biochem. J. 466, 147–161. 10.1042/BJ20140718 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolls J. K., Linden A. (2004). Interleukin-17 family members and inflammation. Immunity 21, 467–476. 10.1016/j.immuni.2004.08.018 [DOI] [PubMed] [Google Scholar]
- Kubatzky K. F. (2012). Pasteurella multocida and immune cells. Curr. Top. Microbiol. Immunol. 361, 53–72. 10.1007/82_2012_204 [DOI] [PubMed] [Google Scholar]
- Kubera A., Thamchaipenet A., Shoham M. (2017). Biofilm inhibitors targeting the outer membrane protein A of Pasteurella multocida in swine. Biofouling 33, 14–23. 10.1080/08927014.2016.1259415 [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M., Thaiss C. A., Zeevi D., Dohnalova L., Zilberman-Schapira G., Mahdi J. A., et al. (2015). Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163, 1428–1443. 10.1016/j.cell.2015.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Long Q., Du H., Zhang J., Pan T., Wu C., et al. (2016). High and low-virulent bovine Pasteurella multocida capsular type A isolates exhibit different virulence gene expression patterns in vitro and in vivo. Vet. Microbiol. 196, 44–49. 10.1016/j.vetmic.2016.10.017 [DOI] [PubMed] [Google Scholar]
- Li X. P., Sun L. (2016). Toll-like receptor 2 of tongue sole Cynoglossus semilaevis: signaling pathway and involvement in bacterial infection. Fish Shellfish Immunol. 51, 321–328. 10.1016/j.fsi.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Liu F., Wu J. B., Zhan R. L., Ou X. C. (2016). Transcription profiling analysis of mango-Fusarium mangiferae interaction. Front. Microbiol. 7:1443. 10.3389/fmicb.2016.01443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan P. R., Harrington L. E., O'Quinn D. B., Helms W. S., Bullard D. C., Elson C. O., et al. (2006). Transforming growth factor-beta induces development of the T(H)17 lineage. Nature 441, 231–234. 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- Mathy N. L., Mathy J. P., Lee R. P., Walker J., Lofthouse S., Meeusen E. N. (2002). Pathological and immunological changes after challenge infection with Pasteurella multocida in naive and immunized calves. Vet. Immunol. Immunopathol. 85, 179–188. 10.1016/S0165-2427(01)00427-5 [DOI] [PubMed] [Google Scholar]
- Mbuthia P. G., Christensen H., Boye M., Petersen K. M., Bisgaard M., Nyaga P. N., et al. (2001). Specific detection of Pasteurella multocida in chickens with fowl cholera and in pig lung tissues using fluorescent rRNA in situ hybridization. J. Clin. Microbiol. 39, 2627–2633. 10.1128/JCM.39.7.2627-2633.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. (2007). Recognition of microorganisms and activation of the immune response. Nature 449, 819–826. 10.1038/nature06246 [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 5, 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Mpiga P., Ravaoarinoro M. (2006). Chlamydia trachomatis persistence: an update. Microbiol. Res. 161, 9–19. 10.1016/j.micres.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Nandi A., Bishayi B. (2015). Host antioxidant enzymes and TLR-2 neutralization modulate intracellular survival of Staphylococcus aureus: evidence of the effect of redox balance on host pathogen relationship during acute staphylococcal infection. Microb. Pathog. 89, 114–127. 10.1016/j.micpath.2015.09.007 [DOI] [PubMed] [Google Scholar]
- Nguyen Q. T., Furuya Y., Roberts S., Metzger D. W. (2015). Role of Interleukin-12 in Protection against Pulmonary Infection with Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 59, 6308–6316. 10.1128/AAC.00968-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomino D. C., Marti L. C. (2015). Chemokines and immunity. Einstein (Sao. Paulo). 13, 469–473. 10.1590/S1679-45082015RB3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pors S. E., Hansen M. S., Bisgaard M., Jensen H. E. (2011). Occurrence and associated lesions of Pasteurella multocida in porcine bronchopneumonia. Vet. Microbiol. 150, 160–166. 10.1016/j.vetmic.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Praveena P. E., Periasamy S., Kumar A. A., Singh N. (2010). Cytokine profiles, apoptosis and pathology of experimental Pasteurella multocida serotype A1 infection in mice. Res. Vet. Sci. 89, 332–339. 10.1016/j.rvsc.2010.04.012 [DOI] [PubMed] [Google Scholar]
- Praveena P. E., Periasamy S., Kumar A. A., Singh N. (2014). Pathology of experimental infection by Pasteurella multocida serotype A: 1 in buffalo calves. Vet. Pathol. 51, 1109–1112. 10.1177/0300985813516647 [DOI] [PubMed] [Google Scholar]
- Qing Y., Stark G. R. (2004). Alternative activation of STAT1 and STAT3 in response to interferon-gamma. J. Biol. Chem. 279, 41679–41685. 10.1074/jbc.M406413200 [DOI] [PubMed] [Google Scholar]
- Raghavan S., Nystrom J., Fredriksson M., Holmgren J., Harandi A. M. (2003). Orally administered CpG oligodeoxynucleotide induces production of CXC and CC chemokines in the gastric mucosa and suppresses bacterial colonization in a mouse model of Helicobacter pylori infection. Infect. Immun. 71, 7014–7022. 10.1128/IAI.71.12.7014-7022.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings J. S., Rosler K. M., Harrison D. A. (2004). The JAK/STAT signaling pathway. J. Cell Sci. 117(Pt 8), 1281–1283. 10.1242/jcs.00963 [DOI] [PubMed] [Google Scholar]
- Ren W., Liu S., Chen S., Zhang F., Li N., Yin J., et al. (2013a). Dietary L-glutamine supplementation increases Pasteurella multocida burden and the expression of its major virulence factors in mice. Amino Acids. 45, 947–955. 10.1007/s00726-013-1551-8 [DOI] [PubMed] [Google Scholar]
- Ren W., Yin J., Xiao H., Chen S., Liu G., Tan B., et al. (2017). Intestinal microbiota-derived gaba mediates interleukin-17 expression during enterotoxigenic Escherichia coli infection. Front. Immunol. 7:685. 10.3389/fimmu.2016.00685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Zou L., Ruan Z., Li N., Wang Y., Peng Y., et al. (2013b). Dietary L-proline supplementation confers immunostimulatory effects on inactivated Pasteurella multocida vaccine immunized mice. Amino Acids. 45, 555–561. 10.1007/s00726-013-1490-4 [DOI] [PubMed] [Google Scholar]
- Rerat M., Albini S., Jaquier V., Hussy D. (2012). Bovine respiratory disease: efficacy of different prophylactic treatments in veal calves and antimicrobial resistance of isolated Pasteurellaceae. Prev. Vet. Med. 103, 265–273. 10.1016/j.prevetmed.2011.09.003 [DOI] [PubMed] [Google Scholar]
- Schenk M., Belisle J. T., Modlin R. L. (2009). TLR2 looks at lipoproteins. Immunity 31, 847–849. 10.1016/j.immuni.2009.11.008 [DOI] [PubMed] [Google Scholar]
- Shi B., Huang Z., Xiang X., Huang M., Wang W. X., Ke C. (2015). Transcriptome analysis of the key role of GAT2 gene in the hyper-accumulation of copper in the oyster Crassostrea angulata. Sci. Rep. 5:17751. 10.1038/srep17751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata K., Yamada H., Hara H., Kishihara K., Yoshikai Y. (2007). Resident Vdelta1+ gammadelta T cells control early infiltration of neutrophils after Escherichia coli infection via IL-17 production. J. Immunol. 178, 4466–4472. 10.4049/jimmunol.178.7.4466 [DOI] [PubMed] [Google Scholar]
- Sun K., Salmon S. L., Lotz S. A., Metzger D. W. (2007). Interleukin-12 promotes gamma interferon-dependent neutrophil recruitment in the lung and improves protection against respiratory Streptococcus pneumoniae infection. Infect. Immun. 75, 1196–1202. 10.1128/IAI.01403-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa T., Nishimura H., Yajima T., Hara H., Kishihara K., Matsuzaki G., et al. (2004). Vdelta1+ gammadelta T cells producing CC chemokines may bridge a gap between neutrophils and macrophages in innate immunity during Escherichia coli infection in mice. J. Immunol. 173, 5156–5164. 10.4049/jimmunol.173.8.5156 [DOI] [PubMed] [Google Scholar]
- Tekkanat K. K., Maassab H., Berlin A. A., Lincoln P. M., Evanoff H. L., Kaplan M. H., et al. (2001). Role of interleukin-12 and stat-4 in the regulation of airway inflammation and hyperreactivity in respiratory syncytial virus infection. Am. J. Pathol. 159, 631–638. 10.1016/S0002-9440(10)61734-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totemeyer S., Kaiser P., Maskell D. J., Bryant C. E. (2005). Sublethal infection of C57BL/6 mice with Salmonella enterica Serovar Typhimurium leads to an increase in levels of Toll-like receptor 1 (TLR1), TLR2, and TLR9 mRNA as well as a decrease in levels of TLR6 mRNA in infected organs. Infect. Immun. 73, 1873–1878. 10.1128/IAI.73.3.1873-1878.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G. (2003). Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3, 133–146. 10.1038/nri1001 [DOI] [PubMed] [Google Scholar]
- Vanaudenaerde B. M., De Vleeschauwer S. I., Vos R., Meyts I., Bullens D. M., Reynders V., et al. (2008). The role of the IL23/IL17 axis in bronchiolitis obliterans syndrome after lung transplantation. Am. J. Transplant. 8, 1911–1920. 10.1111/j.1600-6143.2008.02321.x [DOI] [PubMed] [Google Scholar]
- van den Boogaard F. E., van 't Veer C., Roelofs J. J. T. H., Meijers J. C., Schultz M. J., Broze G. J., et al. (2015). Endogenous tissue factor pathway inhibitor has a limited effect on host defence in murine pneumococcal pneumonia. Thromb. Haemost. 114, 115–122. 10.1160/TH14-12-1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voo K. S., Wang Y. H., Santori F. R., Boggiano C., Wang Y. H., Arima K., et al. (2009). Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc. Natl. Acad. Sci. U.S.A. 106, 4793–4798. 10.1073/pnas.0900408106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu P., Wan Z. Y., Huang S. Q., Wen Y. F., Lin G., et al. (2016). RNA-Seq revealed the impairment of immune defence of tilapia against the infection of Streptococcus agalactiae with simulated climate warming. Fish Shellfish Immunol. 55, 679–689. 10.1016/j.fsi.2016.06.058 [DOI] [PubMed] [Google Scholar]
- Wang Y., Jiang B., Guo Y., Li W., Tian Y., Sonnenberg G. F., et al. (2017). Cross-protective mucosal immunity mediated by memory Th17 cells against Streptococcus pneumoniae lung infection. Mucosal Immunol. 10, 250–259. 10.1038/mi.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N. J., Boniface K., Chan J. R., McKenzie B. S., Blumenschein W. M., Mattson J. D., et al. (2007). Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat. Immunol. 8, 950–957. 10.1038/ni1497 [DOI] [PubMed] [Google Scholar]
- Wong T. M., Petrovsky N., Bissel S. J., Wiley C. A., Ross T. M. (2016). Delta inulin-derived adjuvants that elicit Th1 phenotype following vaccination reduces respiratory syncytial virus lung titers without a reduction in lung immunopathology. Hum. Vaccin. Immunother. 12, 2096–2105. 10.1080/21645515.2016.1162931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Yu L., Qiu J., Shen B., Wang D., Soromou L. W., et al. (2014). Linalool attenuates lung inflammation induced by Pasteurella multocida via activating Nrf-2 signaling pathway. Int. Immunopharmacol. 21, 456–463. 10.1016/j.intimp.2014.05.030 [DOI] [PubMed] [Google Scholar]
- Wu W., Huang J., Duan B., Traficante D. C., Hong H., Risech M., et al. (2012). Th17-stimulating protein vaccines confer protection against Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 186, 420–427. 10.1164/rccm.201202-0182OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. J., Ruddy M. J., Conti H. R., Boonanantanasarn K., Gaffen S. L. (2008). The interleukin-17 receptor plays a gender-dependent role in host protection against Porphyromonas gingivalis-induced periodontal bone loss. Infect. Immun. 76, 4206–4213. 10.1128/IAI.01209-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Qin H., Cheng T., Tan H. L., Guo Y. Y., Shi S. F., et al. (2013). Staphylococcus aureus supernatant induces the release of mouse beta-defensin-14 from osteoblasts via the p38 MAPK and NF-kappaB pathways. Int. J. Mol. Med. 31, 1484–1494. 10.3892/ijmm.2013.1346 [DOI] [PubMed] [Google Scholar]
- Zou J., Shankar N. (2015). Roles of TLR/MyD88/MAPK/NF-kappaB signaling pathways in the regulation of phagocytosis and proinflammatory cytokine expression in response to E. faecalis infection. PLoS ONE 10:e0136947. 10.1371/journal.pone.0136947 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GO analysis of up-regulated DEGs. All up-regulated DEGs were analyzed by GO enrichments and numbers of DEGs in each GO term were counted. Significance of GO enrichments was described as P < 0.05. In this figure, top 10 GO enrichments with up-regulated DEGs larger than 2 in each functional ontologies were showed.
GO analysis of down-regulated DEGs. All down-regulated DEGs were analyzed by GO enrichments and numbers of DEGs in each GO term were counted. Significance of GO enrichments was described as P < 0.05. In this figure, top 10 GO enrichments with down-regulated DEGs larger than 2 in each functional ontologies were showed.
Primers used in qRT-PCR analysis.
RNA-seq sequencing evaluation.
Total DEGs in this study.
Significant GO enrichment analysis of up-regulated DEGs.
Significant GO enrichment analysis of down-regulated DEGs.
Significant KEGG pathways of up-regulated DEGs.
Significant KEGG pathway of down-regulated DEGs.
DEGs related to immune response.
KEGG pathway of up-regulated DEGs related to immune response.
KEGG pathway of down-regulated DEGs related to immune response.
Gene list of significant DEGs in IL-12/IFN-γ axis and IL23/IL17 axis.
Expression level of DEGs related to PRR and PRR downstream signaling.
Expression level of DEGs related to cytokine and chemokine signaling.