Abstract
Background
Accumulated studies have investigated the prognostic role of insulin-like growth factor II mRNA-binding protein 3 (IMP3) in various cancers, but inconsistent and controversial results were obtained. Therefore, we performed a systematic review and meta-analysis to investigate the potential value of IMP3 in the prognostic prediction of human solid tumors.
Materials and methods
A systematic literature search in the electronic databases PubMed, Embase, Web of Science, and Cochrane library (updated to April 2016) was conducted to identify eligible studies. Pooled hazard ratios (HRs) with 95% confidence intervals (CIs) for survival outcomes were calculated and gathered using STATA 12.0 software.
Results
A total of 53 studies containing 8,937 patients with solid tumors were included in this meta-analysis. High IMP3 expression was significantly associated with worse overall survival (OS) of solid tumors (HR =2.08, 95% CI: 1.80–2.42, P<0.001). Similar results were observed in cancer-specific survival (CSS), disease-free survival (DFS), recurrence-free survival (RFS), progression-free survival (PFS), and metastasis-free survival (MFS). Further subgroup analysis stratified by tumor type showed that elevated IMP3 expression was associated with poor OS in renal cell carcinoma (RCC), lung cancer, oral cancer, urothelial carcinoma, hepatocellular carcinoma (HCC), colorectal cancer, pancreatic cancer, gastric cancer, and intrahepatic cholangiocarcinoma (ICC).
Conclusion
The current evidence suggests that high IMP3 expression is associated with poor prognosis in most solid tumors. IMP3 is a potential valuable prognostic factor and might serve as a promising biomarker to guide clinical decisions in human solid tumors.
Keywords: IMP3, prognosis, solid tumor, biomarker, meta-analysis
Introduction
Insulin-like growth factor II mRNA-binding protein 3 (IMP3 or IGF2BP3) is a member of the RNA-binding protein family, which plays an important role in RNA trafficking and stabilization, cell growth, and cell migration during the early stages of embryogenesis.1 IMP3 was proposed to control the translation or turnover of various candidate target genes, including IGF2, CD44, HMGA2, and MMP9.2–5 This oncofetal protein has been reported to promote tumor cell survival, proliferation, chemoresistance, and tumor cell invasiveness in vitro. In recent years, accumulating studies have shown that IMP3 is specifically expressed in malignant tumors and acts as an important cancer-specific gene involved in many aggressive and advanced cancers.6,7
Numerous studies have reported that upregulated IMP3 expression in tumor tissues is correlated with poor patient survival and can be used as a prognostic factor to guide clinical decisions and distinguish different prognoses in various solid tumors, such as renal cell carcinoma (RCC), lung cancer, oral cancer, bladder cancer, gastrointestinal tumors, and gynecological tumors.8–13 However, some other studies have reported the absence of association between IMP3 expression and cancer prognosis.14,15 Some investigators have also replayed completely opposite results in ovarian cancer. For instance, Kobel et al16 proposed that IMP3 expression is a marker of unfavorable prognosis, whereas Noske et al17 asserted that IMP3 expression is associated with improved survival. Hence, the prognostic role of IMP3 expression in solid tumors remains unclear and controversial.
Therefore, we conducted a systematic review of published studies, with a standard meta-analysis combining available evidence, to evaluate the prognostic value of IMP3 expression in solid tumors.
Materials and methods
This meta-analysis was conducted according to the guideline of Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA)18 (Table S1). Because the data included in this study were retrieved from published articles, ethical approval from ethics committees was not needed.
Literature search
A comprehensive literature search was performed in PubMed, Embase, Web of Science, and Cochrane Library to identify studies evaluating IMP3 expression and clinical prognosis in solid tumors up to April 2016. The search strategy included the following terms through MeSH headings, keywords, and text words: “IMP3” or “Insulin-like growth factor 2 mRNA binding protein 3” or “IGF2BP3” combined with “cancer” or “carcinoma” or “neoplasm”. The references cited in the identified articles were also screened for possible inclusions. The database search and preliminary evaluation of identified studies were performed independently by two investigators (LC and YX). No language limitation existed in the process.
Study selection
The inclusion criteria for selecting articles in our analysis are listed as follows: 1) studies that reported IMP3 expression in cancer tissues, 2) studies analyzing the relationship between IMP3 expression level and clinical cancer outcomes, 3) studies that directly reported survival outcomes with hazard ratio (HR) and corresponding 95% confidence interval (CI) or studies that provided sufficient data for estimating HR and 95% CI by using the methods described by Tierney et al,19 and 4) studies with a median follow-up of at least 6 months. Studies were excluded if they were 1) case reports, letters, conference abstracts, or reviews, 2) non-human research, 3) investigations on the diagnostic role, but not the prognostic role, of IMP3, and 4) studies with insufficient data for calculating the HR and 95% CI. If duplicate publications by the same authors were retrieved, we included only the most informative and recent study. Two independent reviewers (LC and YX) evaluated the full articles for study eligibility, and any disagreement was resolved by consensus.
Data extraction and quality assessment
Two authors (LC and YX) independently extracted data from each eligible study by using predefined item forms. The following information, if available, was recorded: first author’s name, year of publication, study country or region, type of cancer, cancer stage, number of patients, detected method, cutoff definition, percentage of high IMP3 expression, follow-up period, and survival outcomes with their HRs and corresponding 95% CIs. If univariate and multivariate analyses were reported to obtain the HRs, the results of multivariate analysis were preferentially selected. If HRs and 95% CIs were not provided directly, we attempted to estimate these points with Kaplan–Meier curve or other required data in the original study by using Tierney et al’s methods.19 Study quality was scored by two investigators (LC and YX) using the Newcastle–Ottawa Scale, which involves three main categories: selection, comparability, and outcome ascertainment. We defined studies with scores no less than 6 as qualified to be included in the meta-analysis. Discrepancies between investigators were resolved through discussion.
Statistical analysis
Pooled HRs and corresponding 95% CIs were calculated to evaluate the prognostic role of high IMP3 expression in the clinical outcomes of solid tumors. An observed HR greater than 1 implied a worse prognosis in patients with high IMP3 expression, and an HR less than 1 indicated a better prognosis. Statistical heterogeneity of combined HR was assessed using Cochrane Q-test and Higgins I2 metrics. I2>50% was considered a measure of obvious heterogeneity.20 If no evident heterogeneity existed, the fixed-effect model (Mantel–Haenszel method) was used to pool the results.21 Otherwise, the randomeffect model (DerSimonian and Laird method) was selected.22 The potential sources for heterogeneity, if significant, were further explored using a predefined subgroup analysis and meta-regression analysis (based on cancer type, ethnicity, case number, cutoff, cancer stage, HR obtained method, and analysis method). To assess the stability of the pooled results, sensitivity analysis was performed by sequential omission of each single study. Publication bias was also estimated by visually assessing the asymmetry of the funnel plot and then quantitatively evaluated by Begg’s and Egger’s tests.23,24 All the abovementioned analyses were performed using STATA version 12.0 (Stata Corporation, College Station, TX, USA). All statistical tests were two sided, and statistical significance was defined as a P-value less than 0.05.
Results
Search results and study characteristics
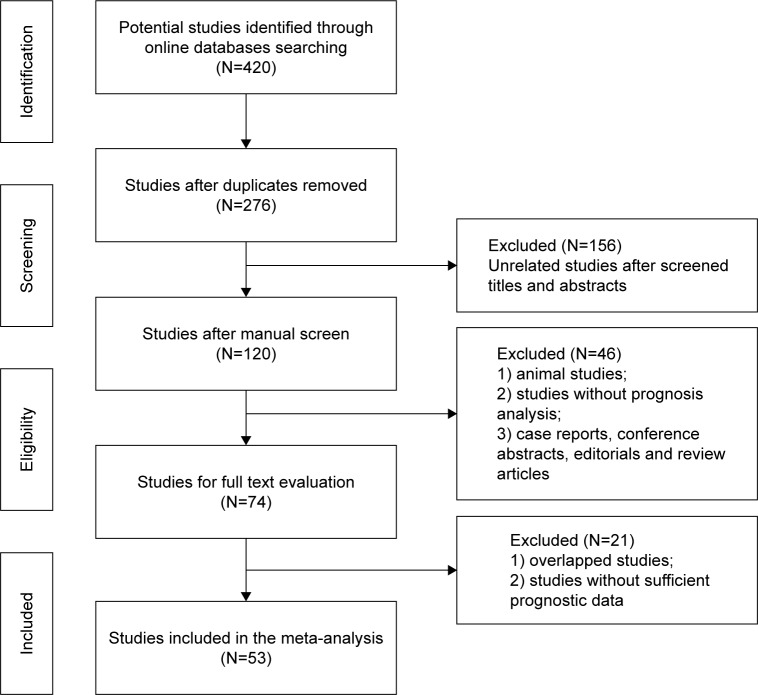
The flowchart of the literature search is shown in Figure 1. A total of 420 potentially relevant studies were retrieved from the initial literature search in the aforementioned electronic databases. A total of 144 duplicated records were excluded by a literature manager software. After carefully screening titles and abstracts of the remaining 120 records, 46 studies were excluded and 74 studies were selected for full-text assessment. Given the inclusion and exclusion criteria, 21 studies that belonged to duplicate publication or failed to offer sufficient prognostic information were excluded. Finally, 53 studies satisfied our eligibility criteria and were included in this meta-analysis.
Figure 1.
Flowchart of the study selection process.
The characteristics of these enrolled studies are summarized in Table 1. The 53 studies involved 8,937 patients with different cancer types, including 6 studies of RCC,8,25–29 6 lung cancer,9,30–34 4 oral cancer,10,35–37 4 urothelial carcinoma,38–41 4 ovarian cancer,16,17,42,43 3 hepatocellular carcinoma (HCC),44–46 4 colorectal cancer,12,47–49 3 prostate cancer,14,15,50 3 pancreatic cancer,51–53 2 gastric cancer,11,54 2 intrahepatic cholangiocarcinoma (ICC),55,56 and one study each of tongue cancer,57 thyroid carcinoma,58 sacral chordoma,59 pilocytic astrocytoma and pilomyxoid astrocytoma (PA/PMA),60 neuroblastoma,61 meningioma,62 melanoma,63 breast cancer,64 giant cell tumor,65 bile duct carcinoma,66 esophageal carcinoma,67 and cervical cancer.13 A total of 25 studies involved Caucasians and 28 involved Asians. The survival outcomes in these studies, including overall survival (OS), cancer-specific survival (CSS), disease-free survival (DFS), recurrence-free survival (RFS), progression-free survival (PFS), and metastasis-free survival (MFS), were investigated in 40, 10, 8, 7, 4, and 5 studies, respectively. HRs were reported directly in most of these studies (43/53) and were estimated indirectly in the 10 other studies. Multivariate Cox analysis was performed to evaluate the prognostic role of IMP3 in 38 studies; and univariate analysis was conducted in the other 15 studies. Immunohistochemistry (IHC) staining and quantitative polymerase chain reaction (qPCR) were used to test the IMP3 expression in cancer tissues. Notably, the definition and cutoff of high IMP3 expression were heterogeneous among these studies. The majority of included studies used the percentage of positive staining cells (0%, 10%, 25%, or 50%) as the criteria, whereas in some other studies, staining scores with the percentage and intensity score were obtained as cutoff values for high IMP3 expression. The percentage of high expression in the cohort population varied in different cancer types and ranged from 6.5% to 83.3%. Quality score assessment suggested that the scores of enrolled studies ranged from 6 to 9, which were considered adequate for quantitative meta-analysis.
Table 1.
Characteristics of studies included in the meta-analysis
| Author | Year | Country or region | Cancer type | Case number | Method | Cutoff | High expression | Follow-up | Outcomes | Analysis | HR obtained | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Jiang et al8 | 2006 | USA | RCC | 371 | IHC | Positive vs negative* | 71 (19.1%) | Median 63 months | OS MFS | Multi | Report | 9 |
| Pei et al26 | 2015 | USA | RCC | 346 | IHC | Positive vs negative | 73 (21.1%) | >10 years | OS RFS | Multi | Report | 8 |
| Hoffmann et al25 | 2008 | USA | RCC | 716 | IHC | Positive vs negative | 213 (29.7%) | 9.5 years | CSS MFS | Multi | Report | 8 |
| Park et al27 | 2014 | Korea | RCC | 148 | IHC | >5% of cells stained | 43 (29.1%) | Median 55.5 months | CSS | Multi | Report | 7 |
| Jiang et al28 | 2008 | USA | RCC | 317 | IHC | Positive vs negative | 40 (12.6%) | 8.8 years | OS MFS | Multi | Report | 9 |
| Tantravahi et al29 | 2015 | USA | RCC | 27 | IHC | >20% of cells stained | 14 (51.9%) | >2 years | OS | Multi | Report | 6 |
| Del Gobbo et al34 | 2014 | Italy | Lung cancer | 74 | IHC | Positive vs negative | 24 (32.4%) | Mean 65.6 months | OS DFS | Uni | Report | 7 |
| Sun et al32 | 2015 | China | Lung cancer | 196 | IHC | H-score >100 (0–300) | 83 (42.3%) | Range (16.5–69.0) months | OS DFS | Multi | Report | 8 |
| Yan et al9 | 2016 | China | Lung cancer | 95 | IHC | >25% of cells stained | 39 (41.1%) | >5 years | OS | Multi | Report | 7 |
| Zhang et al33 | 2015 | China | Lung cancer | 186 | IHC | >5% of cells stained | 139 (74.7%) | >5 years | OS | Multi | Report | 8 |
| Lin et al30 | 2015 | China | Lung cancer | 92 | IHC | Positive vs negative | 62 (67.4%) | >5 years | OS | Multi | Report | 8 |
| Beljan Perak et al31 | 2012 | Croatia | Lung cancer | 90 | IHC | >10% of cells stained | 61 (67.8%) | >5 years | OS | Uni | SC | 6 |
| Clauditz et al35 | 2013 | Germany | Oral cancer | 145 | IHC | >10% of cells stained | 79 (54.5%) | Mean 41.3 months | OS | Multi | Report | 8 |
| Lin et al37 | 2011 | Taiwan | Oral cancer | 93 | IHC | >25% of cells stained | 51 (54.8%) | Mean 44.8 months | OS | Multi | Report | 9 |
| Li et al36 | 2010 | Korea | Oral cancer | 96 | IHC | Positive vs negative | 65 (67.7%) | Median 73 months | OS | Multi | Report | 9 |
| Kim and Cha10 | 2011 | Korea | Oral cancer | 95 | IHC | Positive vs negative | 67 (70.5%) | >5 years | OS | Multi | Report | 7 |
| Szarvas et al40 | 2012 | Germany | Urothelial carcinoma | 106 | IHC | Staining index >7 (0–9) | 17 (16.0%) | Median 15 months | OS CSS MFS | Multi | Report | 7 |
| Sitnikova et al39 | 2008 | USA | Urothelial carcinoma | 214 | IHC | Positive vs negative | 42 (19.6%) | Median 35 months | PFS DFS | Multi | Report | 8 |
| Lee et al41 | 2013 | Multicenter | Urothelial carcinoma | 622 | IHC | Positive vs negative | 76 (12.2%) | Median 27 months | OS CSS RFS | Multi | Report | 9 |
| Niedworok et al38 | 2015 | Germany | Urothelial carcinoma | 26 | IHC | H-score >100 (0–300) | 7 (26.9%) | Median 50 months | OS PFS | Uni | Report | 7 |
| Bi et al43 | 2016 | China | Ovarian cancer | 73 | IHC | >10% of cells stained | 46 (63.0%) | >5 years | OS | Uni | SC | 7 |
| Kobel et al16 | 2009 | British and North America | Ovarian cancer | 278 | IHC | >5% of cells stained | 147 (52.9%) | >4.6 years | CSS | Multi | Report | 8 |
| Hus et al42 | 2015 | Taiwan | Ovarian cancer | 140 | IHC | The median value (IRS: 0–9) | NR | Median 39 months | PFS | Multi | Report | 6 |
| Noske et al17 | 2009 | Germany | Ovarian cancer | 68 | IHC | IRS >6 | 32 (47.1%) | Median 37 months | OS | Uni | SC | 7 |
| Hu et al44 | 2014 | China | HCC | 160 | IHC | Staining score (2–7 vs 0–1) | 97 (60.6%) | Median 36 months | OS RFS | Uni | SC | 8 |
| Wachter et al45 | 2011 | Germany | HCC | 365 | IHC | Staining group (2–3 vs 0–1) | 67 (18.4%) | Mean 23.3 months | OS | Multi | Report | 7 |
| Chen et al46 | 2013 | China | HCC | 92 | IHC | Positive vs negative | 65 (70.7%) | >3 years | OS | Multi | Report | 7 |
| Yuan et al48 | 2009 | Taiwan | Colorectal cancer | 186 | IHC | >50% of cells stained | 66 (35.5%) | Median >5 years | OS | Multi | Report | 8 |
| Li et al49 | 2009 | China | Colorectal cancer | 203 | IHC | Staining score (2–7 vs 0–1) | 132 (65.0%) | Median 61 months | OS DFS | Multi | Report | 9 |
| Lochhead et al12 | 2012 | USA | Colorectal cancer | 671 | IHC | Intense or moderate vs weak or absent | 234 (34.9%) | Median 160 months | OS CSS | Multi | Report | 8 |
| Lin et al30 | 2013 | China | Colorectal cancer | 186 | IHC | Positive vs negative | 143 (76.9%) | >2 years | OS | Multi | Report | 7 |
| Ikenberg et al15 | 2010 | Switzerland | Prostate cancer | 425 | IHC | Positive vs negative | 354 (83.3%) | Median 63 months | RFS | Uni | Report | 9 |
| Chromecki et al14 | 2011 | USA | Prostate cancer | 232 | IHC | >10% of cells stained | 42 (18.1%) | Median 69.8 months | RFS | Multi | Report | 9 |
| Szarvas et al50 | 2014 | Germany | Prostate cancer | 124 | IHC | >10% of cells stained | 30 (24.2%) | Median 155 months | OS CSS | Uni | Report | 8 |
| Wang et al52 | 2014 | China | Pancreatic cancer | 50 | qPCR | Cutoff value based on the ROC curve | 30 (60.0%) | >2 years | OS | Multi | Report | 7 |
| Schaeffer et al51 | 2010 | Canada | Pancreatic cancer | 127 | IHC | IHC score >5 | 80 (63.0%) | Mean 13 months | OS | Multi | Report | 8 |
| Morimatsu et al53 | 2012 | Japan | Pancreatic cancer | 32 | IHC | >50% of cells stained | 17 (53.1%) | Median 33.6 months | CSS | Uni | SC | 6 |
| Wang et al54 | 2010 | China | Gastric cancer | 92 | IHC | Positive vs negative | 75 (81.5%) | >2 years | OS | Uni | SC | 7 |
| Okada et al11 | 2011 | Japan | Gastric cancer | 96 | IHC | >10% of cells stained | 71 (74.0%) | Median 5.5 years | OS DFS | Multi | Report | 8 |
| Chen et al46 | 2013 | Taiwan | ICC | 61 | IHC | >10% of cells stained | 25 (41.0%) | Mean 33.5 months | OS DFS | Uni | SC | 7 |
| Gao et al56 | 2014 | China | ICC | 72 | IHC | Positive vs negative | 59 (81.9%) | Median 14.9 months | OS | Multi | Report | 8 |
| Li et al57 | 2011 | China | Tongue carcinoma | 65 | IHC | Positive vs negative | 50 (76.9%) | Median 36 months | CSS | Uni | SC | 8 |
| Asioli et al58 | 2010 | USA | Thyroid carcinoma | 103 | IHC | Final score >2 (0–6) | 61 (59.2%) | >5 years | OS DFS MFS | Multi | Report | 9 |
| Zhou et al59 | 2014 | China | Sacral chordoma | 32 | IHC | Staining score (2–7 vs 0–1) | 20 (62.5%) | Median 110 months | DFS | Uni | SC | 8 |
| Barton et al60 | 2013 | USA | PA/PMA | 77 | IHC | Three groups (1–2 vs 0) | 24 (31.2%) | Mean 8.8 years | PFS | Uni | Report | 7 |
| Chen et al61 | 2011 | Taiwan | Neuroblastoma | 90 | IHC | >10% of cells stained | 52 (57.8%) | Median 39.5 months | OS | Multi | Report | 8 |
| Hao et al62 | 2011 | USA | Meningioma | 107 | IHC | Positive vs negative | 7 (6.5%) | Median 53 months | OS RFS | Multi | Report | 7 |
| Sheen et al63 | 2014 | Taiwan | Melanoma | 97 | IHC | >10% of cells stained | 72 (74.2%) | Median 5.2 years | OS | Multi | Report | 7 |
| Walter et al64 | 2009 | USA | Breast cancer | 138 | IHC | >10% of cells stained | 45 (32.6%) | Median 71.5 months | OS | Multi | Report | 7 |
| Zhang et al33 | 2015 | China | Giant cell tumor | 38 | IHC | Staining score (3–7 vs 0–2) | 13 (34.2%) | Median 88.0 months | RFS | Uni | SC | 6 |
| Riener et al66 | 2009 | Switzerland | Bile duct carcinoma | 115 | IHC | Intense or moderate vs weak or absent | 67 (58.3%) | Median 9 months | CSS | Multi | Report | 8 |
| Takata et al67 | 2014 | Japan | Esophageal carcinoma | 191 | IHC | >10% of cells stained | 113 (59.2%) | Mean 41 months | OS | Multi | Report | 9 |
| Wei et al13 | 2014 | China | Cervical carcinoma | 96 | IHC | >10% of cells stained | 54 (56.3%) | Median 58.1 months | OS | Multi | Report | 8 |
Note:
Positive vs negative: tumor cells with any detectable staining were considered positive.
Abbreviations: CSS, cancer-specific survival; DFS, disease-free survival; HCC, hepatocellular carcinoma; HR, hazard ratio; ICC, intrahepatic cholangiocarcinoma; IHC, immunohistochemistry; IRS, immunoreactivity score; MFS, metastasis-free survival; NOS, Newcastle–Ottawa Scale; NR, not reported; OS, overall survival; PA/PMA, pilocytic astrocytoma and pilomyxoid astrocytoma; PFS, progression-free survival; qPCR, quantitative polymerase chain reaction; RFS, recurrence-free survival; RCC, renal cell carcinoma; SC, survival curve.
Association of IMP3 with OS
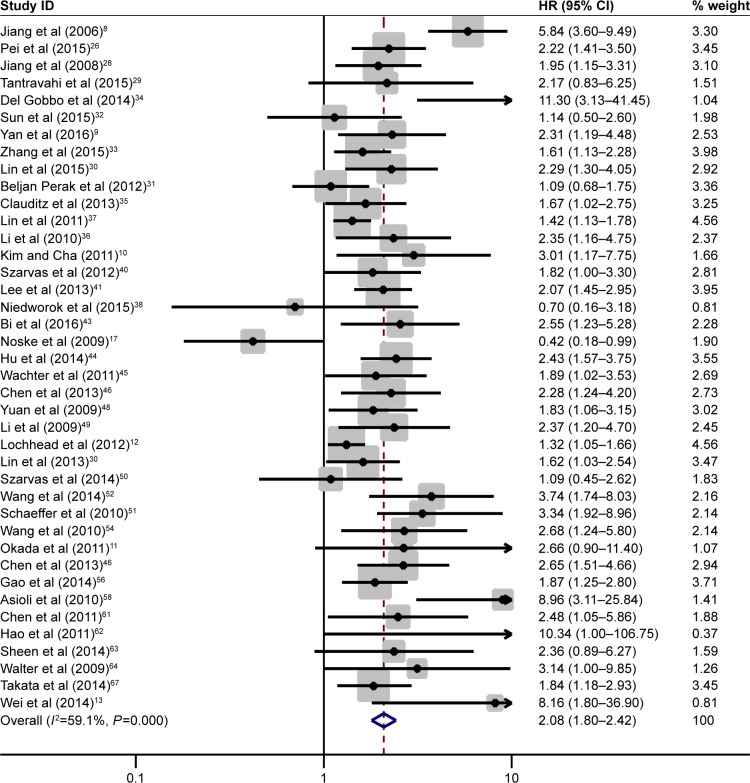
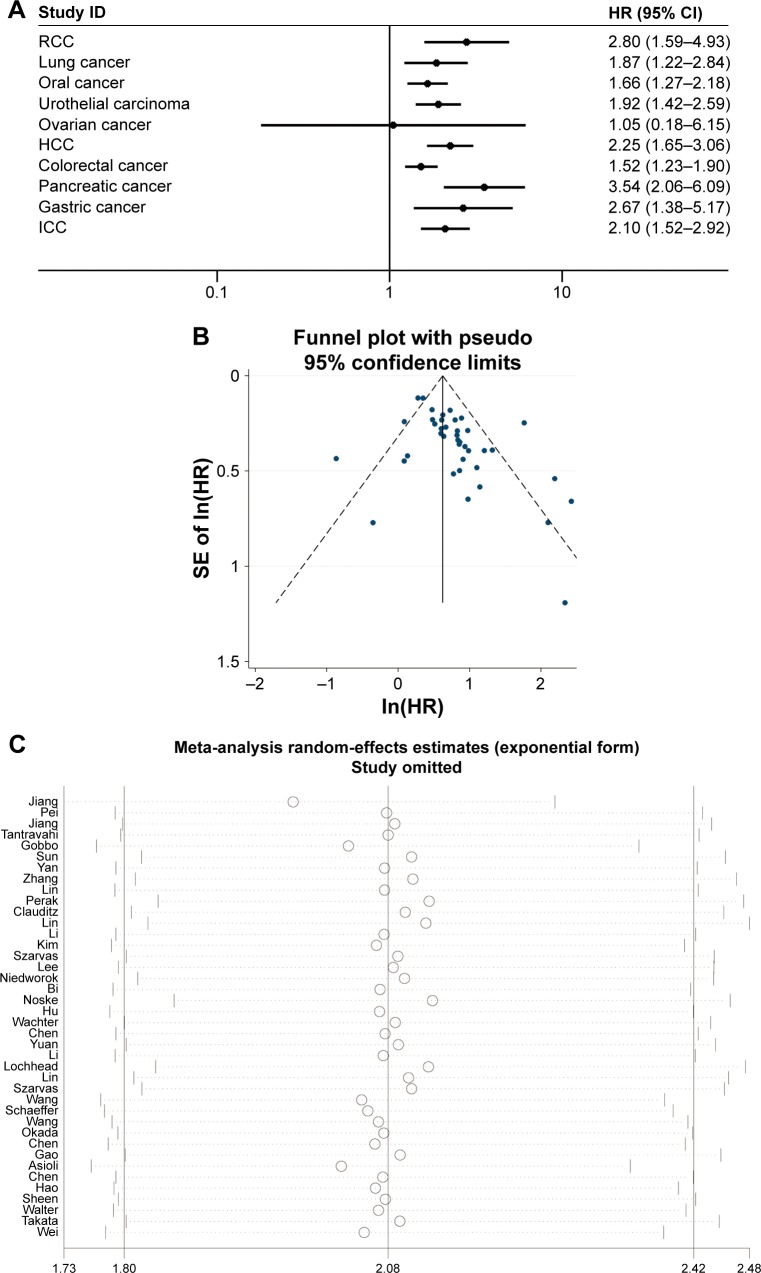
The association of IMP3 expression and OS was investigated in 40 studies containing 6,425 patients with different cancer types. A random-effect model was selected because of the evident interstudy heterogeneity (I2=59.1%, P=0.005). Combined analysis revealed that high IMP3 expression was associated with the worse OS of solid tumors (HR =2.08, 95% CI: 1.80–2.42, P<0.001, Figure 2). The effect of IMP3 expression on OS was further analyzed by tumor types, and the results are presented in Figure 3A. High IMP3 expression was significantly associated with poor OS in RCC (HR =2.80, 95% CI: 1.59–4.93, P<0.001), lung cancer (HR =1.87, 95% CI: 1.22–2.84, P=0.004), oral cancer (HR =1.66, 95% CI: 1.27–2.18, P<0.001), urothelial carcinoma (HR =1.92, 95% CI: 1.42–2.59, P<0.001), HCC (HR =2.25, 95% CI: 1.65–3.06, P<0.001), colorectal cancer (HR =1.52, 95% CI: 1.23–1.90, P<0.001), pancreatic cancer (HR =3.54, 95% CI: 2.06–6.09, P<0.001), gastric cancer (HR =2.67, 95% CI: 1.38–5.17, P=0.003), and ICC (HR =2.10, 95% CI: 1.52–2.92, P<0.001) but not in ovarian cancer (HR =1.05, 95% CI: 0.18–6.15, P=0.957). To explore the source of heterogeneity, subgroup analysis and meta-regression were performed by the following stratification: patient ethnicity, study number, cutoff value, cancer stage, HR obtained method, and analysis style (Table 2). The results indicated that the combined HR estimates for OS in Caucasians and Asians were 2.08 (95% CI: 1.54–2.81, P<0.001) and 1.96 (95% CI: 1.73–2.22, P<0.001), respectively. Differences in the case number, cutoff value, cancer stage, HR obtained method, and analysis method did not influence the effect of IMP3 expression on the OS of solid tumors. Further meta-regression analysis revealed that cancer stage is a potential significant contributor to heterogeneity (P=0.017), unlike other factors (P>0.05).
Figure 2.
Forest plot of studies evaluating HR of high IMP3 expression in solid tumors for OS.
Notes: A pooled analysis showed that high IMP3 expression was associated with poor OS in solid tumors (HR =2.08, 95% CI: 1.80–2.42, P<0.001). Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; HRs, hazard ratios; IMP3, insulin-like growth factor II mRNA-binding protein 3; OS, overall survival.
Figure 3.
Subgroup analysis of OS stratified by tumor types, funnel plot of OS for publication bias, and sensitive analysis of OS.
Notes: (A) High IMP3 expression was significantly associated with poor OS in RCC, lung cancer, oral cancer, urothelial carcinoma, HCC, colorectal cancer, pancreatic cancer, gastric cancer, and ICC but not in ovarian cancer. (B) The funnel plot for OS was asymmetric, which indicated the probability of publication bias. (C) Sensitivity analysis by sequential omission of individual studies did not alter the significance, which confirmed the credibility of outcomes.
Abbreviations: CI, confidence interval; HCC, hepatocellular carcinoma; HR, hazard ratio; ICC, intrahepatic cholangiocarcinoma; In, natural logarithm; IMP3, insulin-like growth factor II mRNA-binding protein 3; OS, overall survival; RCC, renal cell carcinoma; SE, standard error.
Table 2.
Subgroup analysis and meta-regression of the studies regarding overall survival
| Subgroups | Studies | Patients | Pooled HR and 95% CI | P-value | Heterogeneity (I2) | Meta-regression P-value |
|---|---|---|---|---|---|---|
| Ethnicity | 0.748 | |||||
| Caucasian | 18 | 3,827 | 2.08 (1.54–2.81) | <0.001 | 76.3% | |
| Asian | 22 | 2,598 | 1.96 (1.73–2.22) | <0.001 | 9.6% | |
| No of patients | 0.659 | |||||
| >100 | 20 | 4,850 | 2.08 (1.71–2.53) | <0.001 | 62.3% | |
| <100 | 20 | 1,575 | 2.11 (1.66–2.67) | <0.001 | 57.6% | |
| Cutoff | 0.421 | |||||
| Positive vs negative | 13 | 2,562 | 2.50 (1.96–3.19) | <0.001 | 53.9% | |
| >10% of cells stained | 11 | 1,201 | 1.95 (1.50–2.53) | <0.001 | 29.7% | |
| >25% of cells stained | 2 | 188 | 1.63 (1.06–2.52) | 0.027 | 46.5% | |
| Others | 14 | 2,474 | 1.87 (1.42–2.46) | <0.001 | 65.6% | |
| Cancer stage | 0.017 | |||||
| Nonmetastatic | 14 | 2,918 | 2.01 (1.77–2.29) | <0.001 | 23.4% | |
| Mixed (metastatic and nonmetastatic) | 26 | 3,507 | 1.77 (1.58–1.97) | <0.001 | 16.8% | |
| HR obtain method | 0.326 | |||||
| Reported | 34 | 5,881 | 2.14 (1.84–2.50) | <0.001 | 55.5% | |
| Extracted | 6 | 544 | 1.70 (1.03–2.82) | 0.040 | 76.2% | |
| Analysis | 0.319 | |||||
| Univariable analysis | 9 | 768 | 1.76 (1.09–2.85) | 0.020 | 74.7% | |
| Multivariable analysis | 31 | 5,657 | 2.14 (1.84–2.48) | <0.001 | 52.9% |
Abbreviations: CI, confidence interval; HR, hazard ratio.
To assess the credibility of the pooled outcomes, we performed a sensitivity analysis through the sequential omission of individual studies. The results were not obviously influenced by any single study (Figure 3C). The publication bias of all included studies was evaluated using a vertical funnel plot, Begg’s, and Egger’s tests. However, the funnel plot in Figure 3B appears asymmetrical, and the Begg’s (P=0.015) and Egger’s tests (P=0.002) revealed existing evidence of publication bias, which may be attributed to only seven studies that reported negative results among all the enrolled studies.
Association of IMP3 with CSS, DFS, RFS, PFS, and MFS
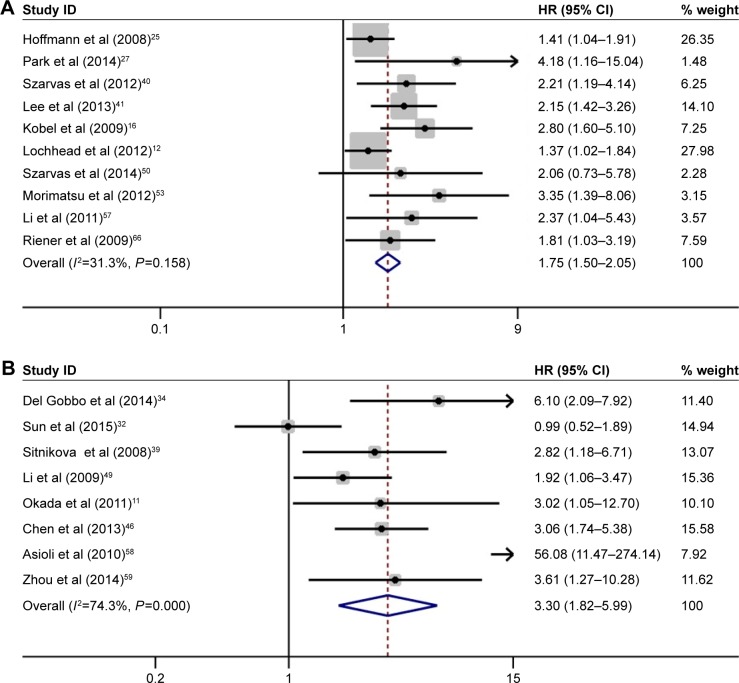
Ten studies that involved a total of 2,877 patients provided sufficient data for CSS analysis. No heterogeneity was observed among these studies (I2=31.3%, P=0.158). Thus, a fixed model was applied to pool the results. The combined HR was 1.75 (95% CI, 1.50–2.05, P<0.001), indicating that high IMP3 expression was associated with worse CSS in the patients with solid tumors (Figure 4A). The subgroup analysis stratified by cancer types showed that high IMP3 expression significantly affected the RCC (HR =1.49, 95% CI: 1.11–2.01, P=0.008) and urothelial carcinoma (HR =2.17, 95% CI: 1.54–3.07, P<0.001). Further sensitivity analysis did not alter the significance of combined HR, which validated the outcome credibility. Eight studies that involved 979 patients reported HRs for DFS, and the effect of high IMP3 expression is presented in Figure 4B. A combined analysis showed that high IMP3 expression was associated with poor DFS in solid tumors (HR =3.30, 95% CI: 1.82–5.99, P<0.001).
Figure 4.
Forest plot of studies evaluating HRs of high IMP3 expression in solid tumors for CSS and DFS.
Notes: (A) High IMP3 expression was associated with poor CSS in solid tumors (HR =1.75, 95% CI: 1.50–2.05, P<0.001). (B) High IMP3 expression was associated with poor DFS in solid tumors (HR =3.30, 95% CI: 1.82–5.99, P<0.001). Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; CSS, cancer-specific survival; DFS, disease-free survival; HRs, hazard ratios; IMP3, insulin-like growth factor II mRNA-binding protein 3; OS, overall survival.
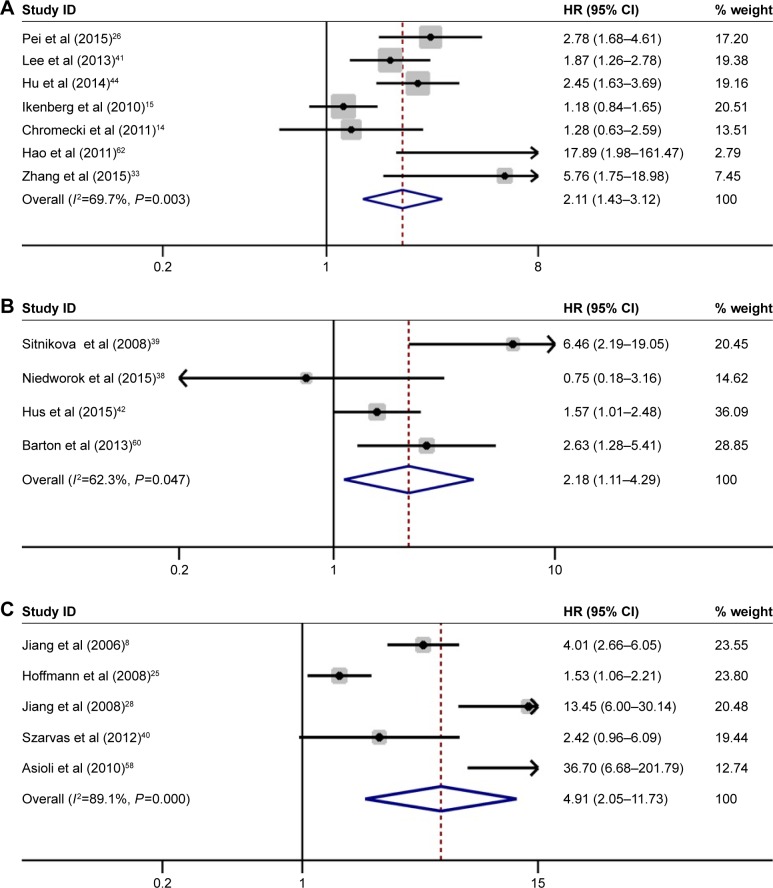
Seven studies with 1,930 patients investigated the prognostic role of IMP3 expression in the RFS of solid tumors. Pooled results demonstrated that high IMP3 adversely influenced the RFS in patients with solid tumors (HR =2.11, 95% CI: 1.43–3.12, P<0.001, Figure 5A). For PFS, four studies with 457 patients were included in the analysis. A forest plot of study-specific HRs for PFS is presented in Figure 5B. The combined results indicated that high IMP3 expression was significantly associated with worse PFS in solid tumors (HR =2.18, 95% CI: 1.11–4.29, P=0.023). In addition, five studies, including 1,613 patients, focused on the influence of IMP3 on solid tumor metastasis. Meta-analysis of these studies suggested that IMP3 expression was also associated with poor MFS (HR =4.91, 95% CI: 2.05–11.73, P<0.001, Figure 5C).
Figure 5.
Forest plot of studies evaluating HRs of high IMP3 expression in solid tumors for RFS, PFS, and MFS.
Notes: (A) High IMP3 expression was associated with poor RFS in solid tumors (HR =2.11, 95% CI: 1.43–3.12, P<0.001). (B) High IMP3 expression was associated with poor PFS in solid tumors (HR =2.18, 95% CI: 1.11–4.29, P=0.023). (C) High IMP3 expression was associated with poor MFS in solid tumors (HR =4.91, 95% CI: 2.05–11.73, P<0.001). Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; HRs, hazard ratios; IMP3, insulin-like growth factor II mRNA-binding protein 3; MFS, metastasis-free survival; PFS, progression-free survival; RFS, recurrence-free survival.
Discussion
Over the past decades, increasing correlative studies describe the elevated IMP3 expression in human cancers, and various functional in vitro or in vivo studies provide strong evidence indicating that this oncofetal protein serves an essential role in modulating tumor cell fate.6 As a molecular biomarker, IMP3 has attracted extensive attention and can be used to distinguish different prognoses, improve prediction accuracy, and better guide clinical decisions in different tumor types.7 Nevertheless, the relationship between IMP3 expression and oncological outcome remains controversial and requires a consensus. Consequently, we attempted to perform a systematic review of published relevant studies and conduct a meta-analysis to clarify the prognostic value of IMP3 expression in patients with solid tumors.
In the present research, given the inclusion criteria, 53 studies involving 8,937 patients were eligible, and the HRs of cumulative survival rates were summarized quantitatively by standard meta-analysis techniques. Our results suggested that high IMP3 expression was associated with worse OS of the solid tumors. Further subgroup analysis stratified by tumor type presented detailed results as follows. The negative prognostic effects of IMP3 on OS were specifically observed in RCC, lung cancer, oral cancer, urothelial carcinoma, HCC, colorectal cancer, pancreatic cancer, gastric cancer, and ICC. Besides OS, we also investigated other frequently used survival outcomes, including CSS, DFS, RFS, PFS, and MFS. Similar influences were found for high IMP3 expression regarding the abovementioned end points, which provide a relatively comprehensive assessment of the value of IMP3 acting as a prognostic biomarker in solid tumors.
Accumulated literature suggests that IMP3 contributes to various aspects of cancer by promoting target genes expression by either preventing mRNA decay or stimulating mRNA translation. IMP3 knockdown in vitro can significantly inhibit the translation of IGF2 mRNA resulting in the marked inhibition of cell proliferation.2 By using solid cancer transcriptome data, IMP3 was also found to be correlated with HMGA2 mRNA expression in a dose-dependent manner. Additional assay for elucidating the mechanism indicated that IMP3 may function as a cytoplasmic safe house and prevents miRNA-directed mRNA decay of HMGA2 during tumor progression.4 Another recent study identified IMP3 as capable of directly binding the mRNAs of cyclins D1, D3, and G1 in vivo and in vitro. The study also found that IMP3 can regulate the expression of these cyclins depending on their protein partner HNRNPM in six human cancer cell lines of different origins.68 In addition, IMP3 promotes tumor cell invasion and migration by targeting the epithelial–mesenchymal transition-associated molecular makers, including E-cadherin, Slug, and vimentin.69 Overall, IMP3 plays an essential and multifaceted role in human cancers. Hence, targeting IMP3 may serve as a potential strategy for anticancer therapy.
To our knowledge, our study is the first meta-analysis that comprehensively evaluated the association between IMP3 expression and prognosis in patients with solid tumors. However, several limitations of our study must be acknowledged. First, we only extracted summarized population-level data rather than individual subject data from published literature. Second, different cutoff values and definitions of high IMP3 expression were used in these included studies. Third, a marked study heterogeneity existed in some analyses. The subgroup analyses and meta-regression revealed that cancer stage might be a significant contributor to heterogeneity. Moreover, several potential factors such as cancer type, cutoff value, baseline characteristics (sample size, sex, age, and pathological subtype), and duration of follow-up may partially contribute to the heterogeneity. Among the enrolled studies, 10 works did not directly report the HRs. The calculated HRs, which were estimated using the methods of Tierney et al, might not be as dependable as those retrieved directly from the reported results. As such, the HRs inevitably introduced some statistical errors and may have influenced the pooled analysis. Furthermore, some studies only provided univariate analysis results, which may have introduced a bias toward overestimation of the prognostic value compared with multivariate analysis. The funnel plot and Egger’s test suggested the probability of publication bias because of fewer studies reporting negative results. However, the greater difficulty in publishing studies with insignificant results than those with significant results may be unavoidable. Finally, despite the well-recognized advantages of systematic review and meta-analysis, the results were based on the quality of the included studies. Thus, further high-quality studies with larger samples and a unified detection method are entailed to achieve a consensus on this matter.
Conclusion
The current evidence suggests that high IMP3 expression in tumor tissues is associated with adverse survival in various cancers. Hence, IMP3 might be a potential and promising biomarker that can be used to improve prognosis stratification and guide decision making in the treatment of solid tumors. Further well-designed studies are needed to confirm our findings and obtain more precise evaluations of the prognostic value of IMP3 in cancers.
Supplementary material
Table S1.
Checklist of PRISMA 2009
| Section/topic | # | Checklist item | Reported on page # |
|---|---|---|---|
| Title | |||
| Title | 1 | Identify the report as a systematic review, meta-analysis, or both. | 1 |
| Abstract | |||
| Structured summary | 2 | Provide a structured summary including, as applicable: background; objectives; data sources; study eligibility criteria, participants, and interventions; study appraisal and synthesis methods; results; limitations; conclusions and implications of key findings; systematic review registration number. | 2 |
| Introduction | |||
| Rationale | 3 | Describe the rationale for the review in the context of what is already known. | 3 |
| Objectives | 4 | Provide an explicit statement of questions being addressed with reference to participants, interventions, comparisons, outcomes, and study design (PICOS). | 3,4 |
| Methods | |||
| Protocol and registration | 5 | Indicate if a review protocol exists, if and where it can be accessed (eg, Web address), and, if available, provide registration information including registration number. | No |
| Eligibility criteria | 6 | Specify study characteristics (eg, PICOS, length of follow-up) and report characteristics (eg, years considered, language, publication status) used as criteria for eligibility, giving rationale. | 4,5 |
| Information sources | 7 | Describe all information sources (eg, databases with dates of coverage, contact with study authors to identify additional studies) in the search and date last searched. | 4 |
| Search | 8 | Present full electronic search strategy for at least one database, including any limits used, such that it could be repeated. | 4 |
| Study selection | 9 | State the process for selecting studies (ie, screening, eligibility, included in systematic review, and, if applicable, included in the meta-analysis). | 5 |
| Data collection process | 10 | Describe method of data extraction from reports (eg, piloted forms, independently, in duplicate) and any processes for obtaining and confirming data from investigators. | 5 |
| Data items | 11 | List and define all variables for which data were sought (eg, PICOS, funding sources) and any assumptions and simplifications made. | 5,6 |
| Risk of bias in individual studies | 12 | Describe methods used for assessing risk of bias of individual studies (including specification of whether this was done at the study or outcome level), and how this information is to be used in any data synthesis. | 5,6 |
| Summary measures | 13 | State the principal summary measures (eg, risk ratio, difference in means). | 5,6 |
| Synthesis of results | 14 | Describe the methods of handling data and combining results of studies, if done, including measures of consistency (eg, I2) for each meta-analysis. | 6 |
| Risk of bias across studies | 15 | Specify any assessment of risk of bias that may affect the cumulative evidence (eg, publication bias, selective reporting within studies). | 6 |
| Additional analyses | 16 | Describe methods of additional analyses (eg, sensitivity or subgroup analyses, meta-regression), if done, indicating which were pre-specified. | 6 |
| Results | |||
| Study selection | 17 | Give numbers of studies screened, assessed for eligibility, and included in the review, with reasons for exclusions at each stage, ideally with a flow diagram. | 7 |
| Study characteristics | 18 | For each study, present characteristics for which data were extracted (eg, study size, PICOS, follow-up period) and provide the citations. | 7 |
| Risk of bias within studies | 19 | Present data on risk of bias of each study and, if available, any outcome level assessment (see item 12). | 7–14 |
| Results of individual studies | 20 | For all outcomes considered (benefits or harms), present, for each study: (a) simple summary data for each intervention group; (b) effect estimates and confidence intervals, ideally with a forest plot. | 7–14 |
| Synthesis of results | 21 | Present results of each meta-analysis done, including confidence intervals and measures of consistency. | 7–14 |
| Risk of bias across studies | 22 | Present results of any assessment of risk of bias across studies (see item 15). | 7–14 |
| Additional analysis | 23 | Give results of additional analyses, if done (eg, sensitivity or subgroup analyses, meta-regression [see Item 16]). | 7–14 |
| Discussion | |||
| Summary of evidence | 24 | Summarize the main findings including the strength of evidence for each main outcome; consider their relevance to key groups (eg, healthcare providers, users, and policy makers). | 14,15 |
| Limitations | 25 | Discuss limitations at study and outcome level (eg, risk of bias), and at review-level (eg, incomplete retrieval of identified research, reporting bias). | 15,16 |
| Conclusions | 26 | Provide a general interpretation of the results in the context of other evidence, and implications for future research. | 17 |
| Funding | |||
| Funding | 27 | Describe sources of funding for the systematic review and other support (eg, supply of data); role of funders for the systematic review. | None |
Notes: Reproduced from Moher D, Liberati A, Tetzlaff J, et al, Preferred reporting items for systematic reviews and meta-analyses: the PRISMA Statement. PLoS Med. 2009:6(7): e1000097.1
Reference
- 1.Moher D, Liberati A, Tetzlaff J, et al. The PRISMA Group (2009) Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (2014AA020607).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Mueller-Pillasch F, Pohl B, Wilda M, et al. Expression of the highly conserved RNA binding protein KOC in embryogenesis. Mech Dev. 1999;88(1):95–99. doi: 10.1016/s0925-4773(99)00160-4. [DOI] [PubMed] [Google Scholar]
- 2.Liao B, Hu Y, Herrick DJ, Brewer G. The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. J Biol Chem. 2005;280(18):18517–18524. doi: 10.1074/jbc.M500270200. [DOI] [PubMed] [Google Scholar]
- 3.Li W, Liu D, Chang W, et al. Role of IGF2BP3 in trophoblast cell invasion and migration. Cell Death Dis. 2014;5:e1025. doi: 10.1038/cddis.2013.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jønson L, Christiansen J, Hansen TV, Vikeså J, Yamamoto Y, Nielsen FC. IMP3 RNP safe houses prevent miRNA-directed HMGA2 mRNA decay in cancer and development. Cell Rep. 2014;7(2):539–551. doi: 10.1016/j.celrep.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Samanta S, Sharma VM, Khan A, Mercurio AM. Regulation of IMP3 by EGFR signaling and repression by ERbeta: implications for triple-negative breast cancer. Oncogene. 2012;31(44):4689–4697. doi: 10.1038/onc.2011.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lederer M, Bley N, Schleifer C, Huttelmaier S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol. 2014;29:3–12. doi: 10.1016/j.semcancer.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Gong Y, Woda BA, Jiang Z. Oncofetal protein IMP3, a new cancer biomarker. Adv Anat Pathol. 2014;21(3):191–200. doi: 10.1097/PAP.0000000000000021. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Z, Chu PG, Woda BA, et al. Analysis of RNA-binding protein IMP3 to predict metastasis and prognosis of renal-cell carcinoma: a retrospective study. Lancet Oncol. 2006;7(7):556–564. doi: 10.1016/S1470-2045(06)70732-X. [DOI] [PubMed] [Google Scholar]
- 9.Yan J, Wei Q, Jian W, et al. IMP3 predicts invasion and prognosis in human lung adenocarcinoma. Lung. 2016;194(1):137–146. doi: 10.1007/s00408-015-9829-0. [DOI] [PubMed] [Google Scholar]
- 10.Kim KY, Cha IH. Risk stratification of oral cancer patients using a combined prognostic factor including lymph node density and biomarker. J Cancer Res Clin Oncol. 2012;138(3):483–490. doi: 10.1007/s00432-011-1129-3. [DOI] [PubMed] [Google Scholar]
- 11.Okada K, Fujiwara Y, Nakamura Y, et al. Oncofetal protein, IMP-3, a potential marker for prediction of postoperative peritoneal dissemination in gastric adenocarcinoma. J Surg Oncol. 2012;105(8):780–785. doi: 10.1002/jso.22108. [DOI] [PubMed] [Google Scholar]
- 12.Lochhead P, Imamura Y, Morikawa T, et al. Insulin-like growth factor 2 messenger RNA binding protein 3 (IGF2BP3) is a marker of unfavourable prognosis in colorectal cancer. Eur J Cancer. 2012;48(18):3405–3413. doi: 10.1016/j.ejca.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Q, Yan J, Fu B, et al. IMP3 expression is associated with poor survival in cervical squamous cell carcinoma. Hum Pathol. 2014;45(11):2218–2224. doi: 10.1016/j.humpath.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Chromecki TF, Cha EK, Pummer K, et al. Prognostic value of insulin-like growth factor II mRNA binding protein 3 in patients treated with radical prostatectomy. BJU Int. 2012;110(1):63–68. doi: 10.1111/j.1464-410X.2011.10703.x. [DOI] [PubMed] [Google Scholar]
- 15.Ikenberg K, Fritzsche FR, Zuerrer-Haerdi U, et al. Insulin-like growth factor II mRNA binding protein 3 (IMP3) is overexpressed in prostate cancer and correlates with higher Gleason scores. BMC Cancer. 2010;10:341. doi: 10.1186/1471-2407-10-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobel M, Xu H, Bourne PA, et al. IGF2BP3 (IMP3) expression is a marker of unfavorable prognosis in ovarian carcinoma of clear cell subtype. Mod Pathol. 2009;22(3):469–475. doi: 10.1038/modpathol.2008.206. [DOI] [PubMed] [Google Scholar]
- 17.Noske A, Faggad A, Wirtz R, et al. IMP3 expression in human ovarian cancer is associated with improved survival. Int J Gynecol Pathol. 2009;28(3):203–210. doi: 10.1097/PGP.0b013e318189a724. [DOI] [PubMed] [Google Scholar]
- 18.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010;8(5):336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darroch JN. The Mantel-Haenszel test and tests of marginal symmetry; fixed-effects and mixed models for a categorical response, correspondent paper. Int Stat Rev. 1981;49(3):285–307. [Google Scholar]
- 22.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 23.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 24.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann NE, Sheinin Y, Lohse CM, et al. External validation of IMP3 expression as an independent prognostic marker for metastatic progression and death for patients with clear cell renal cell carcinoma. Cancer. 2008;112(7):1471–1479. doi: 10.1002/cncr.23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei X, Li M, Zhan J, et al. Enhanced IMP3 expression activates NF-small ka, CyrillicB pathway and promotes renal cell carcinoma progression. PLoS One. 2015;10(4):e0124338. doi: 10.1371/journal.pone.0124338. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Park JY, Choe M, Kang Y, Lee SS. IMP3, a promising prognostic marker in clear cell renal cell carcinoma. Korean J Pathol. 2014;48(2):108–116. doi: 10.4132/KoreanJPathol.2014.48.2.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Z, Lohse CM, Chu PG, et al. Oncofetal protein IMP3: a novel molecular marker that predicts metastasis of papillary and chromophobe renal cell carcinomas. Cancer. 2008;112(12):2676–2682. doi: 10.1002/cncr.23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tantravahi SK, Albertson D, Agarwal AM, et al. Survival outcomes and tumor IMP3 expression in patients with sarcomatoid metastatic renal cell carcinoma. J Oncol. 2015;2015:181926. doi: 10.1155/2015/181926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin L, Zhang J, Wang Y, et al. Expression of insulin-like growth factor 2 mRNA-binding protein 3 expression and analysis of prognosis in the patients with lung squamous cell carcinoma. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2013;29(7):694–697. [PubMed] [Google Scholar]
- 31.Beljan Perak R, Durdov MG, Capkun V, et al. IMP3 can predict aggressive behaviour of lung adenocarcinoma. Diagn Pathol. 2012;7:165. doi: 10.1186/1746-1596-7-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun X, Wei P, Shen C, et al. Prognostic value of the IASLC/ATS/ERS classification and IMP3 expression in lung adenocarcinoma of Chinese cases. Am J Cancer Res. 2015;5(7):2266–2276. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Ou Y, Ma Y, et al. Clinical implications of insulin-like growth factor II mRNA-binding protein 3 expression in non-small cell lung carcinoma. Oncol Lett. 2015;9(4):1927–1933. doi: 10.3892/ol.2015.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Del Gobbo A, Vaira V, Guerini Rocco E, et al. The oncofetal protein IMP3: a useful marker to predict poor clinical outcome in neuroendocrine tumors of the lung. J Thorac Oncol. 2014;9(11):1656–1661. doi: 10.1097/JTO.0000000000000316. [DOI] [PubMed] [Google Scholar]
- 35.Clauditz TS, Wang CJ, Gontarewicz A, et al. Expression of insulin-like growth factor II mRNA-binding protein 3 in squamous cell carcinomas of the head and neck. J Oral Pathol Med. 2013;42(2):125–132. doi: 10.1111/j.1600-0714.2012.01178.x. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Cha J, Kim J, et al. Insulin-like growth factor II mRNA-binding protein 3: a novel prognostic biomarker for oral squamous cell carcinoma. Head Neck. 2011;33(3):368–374. doi: 10.1002/hed.21457. [DOI] [PubMed] [Google Scholar]
- 37.Lin CY, Chen ST, Jeng YM, et al. Insulin-like growth factor II mRNA-binding protein 3 expression promotes tumor formation and invasion and predicts poor prognosis in oral squamous cell carcinoma. J Oral Pathol Med. 2011;40(9):699–705. doi: 10.1111/j.1600-0714.2011.01019.x. [DOI] [PubMed] [Google Scholar]
- 38.Niedworok C, Panitz M, Szarvas T, et al. Urachal carcinoma of the bladder: impact of clinical and immunohistochemical parameters on prognosis. J Urol. 2016;195(6):1690–1696. doi: 10.1016/j.juro.2015.11.067. [DOI] [PubMed] [Google Scholar]
- 39.Sitnikova L, Mendese G, Liu Q, et al. IMP3 predicts aggressive superficial urothelial carcinoma of the bladder. Clin Cancer Res. 2008;14(6):1701–1706. doi: 10.1158/1078-0432.CCR-07-2039. [DOI] [PubMed] [Google Scholar]
- 40.Szarvas T, vom Dorp F, Niedworok C, et al. High insulin-like growth factor mRNA-binding protein 3 (IMP3) protein expression is associated with poor survival in muscle-invasive bladder cancer. BJU Int. 2012;110(6 pt B):E308–E317. doi: 10.1111/j.1464-410X.2012.11149.x. [DOI] [PubMed] [Google Scholar]
- 41.Lee DJ, Xylinas E, Rieken M, et al. Insulin-like growth factor messenger RNA-binding protein 3 expression helps prognostication in patients with upper tract urothelial carcinoma. Eur Urol. 2014;66(2):379–385. doi: 10.1016/j.eururo.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 42.Hsu KF, Shen MR, Huang YF, et al. Overexpression of the RNA-binding proteins Lin28B and IGF2BP3 (IMP3) is associated with chemoresistance and poor disease outcome in ovarian cancer. Br J Cancer. 2015;113(3):414–424. doi: 10.1038/bjc.2015.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bi R, Shen X, Zhang W, et al. Clear cell carcinomas of the ovary: a mono-institutional study of 73 cases in China with an analysis of the prognostic significance of clinicopathological parameters and IMP3 expression. Diagn Pathol. 2016;11:17. doi: 10.1186/s13000-016-0467-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu S, Wu X, Zhou B, et al. IMP3 combined with CD44s, a novel predictor for prognosis of patients with hepatocellular carcinoma. J Cancer Res Clin Oncol. 2014;140(6):883–893. doi: 10.1007/s00432-014-1639-x. [DOI] [PubMed] [Google Scholar]
- 45.Wachter DL, Kristiansen G, Soll C, et al. Insulin-like growth factor II mRNA-binding protein 3 (IMP3) expression in hepatocellular carcinoma. A clinicopathological analysis with emphasis on diagnostic value. Histopathology. 2012;60(2):278–286. doi: 10.1111/j.1365-2559.2011.04091.x. [DOI] [PubMed] [Google Scholar]
- 46.Chen LT, Lin LJ, Zheng LL. The correlation between insulin-like growth factor II mRNA binding protein 3 expression in hepatocellular carcinoma and prognosis. Hepatogastroenterology. 2013;60(123):553–556. doi: 10.5754/hge12855. [DOI] [PubMed] [Google Scholar]
- 47.Lin L, Zhang J, Wang Y, et al. Insulin-like growth factor-II mRNA-binding protein 3 predicts a poor prognosis for colorectal adenocarcinoma. Oncol Lett. 2013;6(3):740–744. doi: 10.3892/ol.2013.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan RH, Wang CC, Chou CC, Chang KJ, Lee PH, Jeng YM. Diffuse expression of RNA-binding protein IMP3 predicts high-stage lymph node metastasis and poor prognosis in colorectal adenocarcinoma. Ann Surg Oncol. 2009;16(6):1711–1719. doi: 10.1245/s10434-009-0446-0. [DOI] [PubMed] [Google Scholar]
- 49.Li D, Yan D, Tang H, et al. IMP3 is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Ann Surg Oncol. 2009;16(12):3499–3506. doi: 10.1245/s10434-009-0648-5. [DOI] [PubMed] [Google Scholar]
- 50.Szarvas T, Tschirdewahn S, Niedworok C, et al. Prognostic value of tissue and circulating levels of IMP3 in prostate cancer. Int J Cancer. 2014;135(7):1596–1604. doi: 10.1002/ijc.28808. [DOI] [PubMed] [Google Scholar]
- 51.Schaeffer DF, Owen DR, Lim HJ, et al. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) overexpression in pancreatic ductal adenocarcinoma correlates with poor survival. BMC Cancer. 2010;10:59. doi: 10.1186/1471-2407-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang BJ, Wang L, Yang SY, Liu ZJ. Expression and clinical significance of IMP3 in microdissected premalignant and malignant pancreatic lesions. Clin Transl Oncol. 2015;17(3):215–222. doi: 10.1007/s12094-014-1216-4. [DOI] [PubMed] [Google Scholar]
- 53.Morimatsu K, Aishima S, Yamamoto H, et al. Insulin-like growth factor II messenger RNA-binding protein-3 is a valuable diagnostic and prognostic marker of intraductal papillary mucinous neoplasm. Hum Pathol. 2013;44(9):1714–1721. doi: 10.1016/j.humpath.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 54.Wang L, Li HG, Xia ZS, Lu J, Peng TS. IMP3 is a novel biomarker to predict metastasis and prognosis of gastric adenocarcinoma: a retrospective study. Chin Med J (Engl) 2010;123(24):3554–3558. [PubMed] [Google Scholar]
- 55.Chen YL, Jeng YM, Hsu HC, et al. Expression of insulin-like growth factor II mRNA-binding protein 3 predicts early recurrence and poor prognosis in intrahepatic cholangiocarcinoma. Int J Surg. 2013;11(1):85–91. doi: 10.1016/j.ijsu.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 56.Gao Y, Yang M, Jiang Z, et al. IMP3 expression is associated with poor outcome and epigenetic deregulation in intrahepatic cholangiocarcinoma. Hum Pathol. 2014;45(6):1184–1191. doi: 10.1016/j.humpath.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 57.Li HG, Han JJ, Huang ZQ, Wang L, Chen WL, Shen XM. IMP3 is a novel biomarker to predict metastasis and prognosis of tongue squamous cell carcinoma. J Craniofac Surg. 2011;22(6):2022–2025. doi: 10.1097/SCS.0b013e3182319750. [DOI] [PubMed] [Google Scholar]
- 58.Asioli S, Erickson LA, Righi A, et al. Poorly differentiated carcinoma of the thyroid: validation of the Turin proposal and analysis of IMP3 expression. Mod Pathol. 2010;23(9):1269–1278. doi: 10.1038/modpathol.2010.117. [DOI] [PubMed] [Google Scholar]
- 59.Zhou M, Chen K, Yang H, et al. Expression of insulin-like growth factor II mRNA-binding protein 3 (IMP3) in sacral chordoma. J Neurooncol. 2014;116(1):77–82. doi: 10.1007/s11060-013-1274-4. [DOI] [PubMed] [Google Scholar]
- 60.Barton VN, Donson AM, Birks DK, et al. Insulin-like growth factor 2 mRNA binding protein 3 expression is an independent prognostic factor in pediatric pilocytic and pilomyxoid astrocytoma. J Neuropathol Exp Neurol. 2013;72(5):442–449. doi: 10.1097/NEN.0b013e31829023dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen ST, Jeng YM, Chang CC, et al. Insulin-like growth factor II mRNA-binding protein 3 expression predicts unfavorable prognosis in patients with neuroblastoma. Cancer Sci. 2011;102(12):2191–2198. doi: 10.1111/j.1349-7006.2011.02100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hao S, Smith TW, Chu PG, et al. The oncofetal protein IMP3: a novel molecular marker to predict aggressive meningioma. Arch Pathol Lab Med. 2011;135(8):1032–1036. doi: 10.5858/2009-0652-OAR2. [DOI] [PubMed] [Google Scholar]
- 63.Sheen YS, Liao YH, Lin MH, et al. IMP-3 promotes migration and invasion of melanoma cells by modulating the expression of HMGA2 and predicts poor prognosis in melanoma. J Invest Dermatol. 2015;135(4):1065–1073. doi: 10.1038/jid.2014.480. [DOI] [PubMed] [Google Scholar]
- 64.Walter O, Prasad M, Lu S, Quinlan RM, Edmiston KL, Khan A. IMP3 is a novel biomarker for triple negative invasive mammary carcinoma associated with a more aggressive phenotype. Hum Pathol. 2009;40(11):1528–1533. doi: 10.1016/j.humpath.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 65.Zhang K, Zhou M, Chen H, Wu G, Chen K, Yang H. Expression of IMP3 and IGF2 in giant cell tumor of spine is associated with tumor recurrence and angiogenesis. Clin Transl Oncol. 2015;17(7):570–575. doi: 10.1007/s12094-015-1280-4. [DOI] [PubMed] [Google Scholar]
- 66.Riener MO, Fritzsche FR, Clavien PA, et al. IMP3 expression in lesions of the biliary tract: a marker for high-grade dysplasia and an independent prognostic factor in bile duct carcinomas. Hum Pathol. 2009;40(10):1377–1383. doi: 10.1016/j.humpath.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 67.Takata A, Takiguchi S, Okada K, et al. Expression of insulin-like growth factor-II mRNA-binding protein-3 as a marker for predicting clinical outcome in patients with esophageal squamous cell carcinoma. Oncol Lett. 2014;8(5):2027–2031. doi: 10.3892/ol.2014.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rivera Vargas T, Boudoukha S, Simon A, et al. Post-transcriptional regulation of cyclins D1, D3 and G1 and proliferation of human cancer cells depend on IMP-3 nuclear localization. Oncogene. 2014;33(22):2866–2875. doi: 10.1038/onc.2013.252. [DOI] [PubMed] [Google Scholar]
- 69.Su P, Hu J, Zhang H, et al. IMP3 expression is associated with epithelial-mesenchymal transition in breast cancer. Int J Clin Exp Pathol. 2014;7(6):3008–3017. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Checklist of PRISMA 2009
| Section/topic | # | Checklist item | Reported on page # |
|---|---|---|---|
| Title | |||
| Title | 1 | Identify the report as a systematic review, meta-analysis, or both. | 1 |
| Abstract | |||
| Structured summary | 2 | Provide a structured summary including, as applicable: background; objectives; data sources; study eligibility criteria, participants, and interventions; study appraisal and synthesis methods; results; limitations; conclusions and implications of key findings; systematic review registration number. | 2 |
| Introduction | |||
| Rationale | 3 | Describe the rationale for the review in the context of what is already known. | 3 |
| Objectives | 4 | Provide an explicit statement of questions being addressed with reference to participants, interventions, comparisons, outcomes, and study design (PICOS). | 3,4 |
| Methods | |||
| Protocol and registration | 5 | Indicate if a review protocol exists, if and where it can be accessed (eg, Web address), and, if available, provide registration information including registration number. | No |
| Eligibility criteria | 6 | Specify study characteristics (eg, PICOS, length of follow-up) and report characteristics (eg, years considered, language, publication status) used as criteria for eligibility, giving rationale. | 4,5 |
| Information sources | 7 | Describe all information sources (eg, databases with dates of coverage, contact with study authors to identify additional studies) in the search and date last searched. | 4 |
| Search | 8 | Present full electronic search strategy for at least one database, including any limits used, such that it could be repeated. | 4 |
| Study selection | 9 | State the process for selecting studies (ie, screening, eligibility, included in systematic review, and, if applicable, included in the meta-analysis). | 5 |
| Data collection process | 10 | Describe method of data extraction from reports (eg, piloted forms, independently, in duplicate) and any processes for obtaining and confirming data from investigators. | 5 |
| Data items | 11 | List and define all variables for which data were sought (eg, PICOS, funding sources) and any assumptions and simplifications made. | 5,6 |
| Risk of bias in individual studies | 12 | Describe methods used for assessing risk of bias of individual studies (including specification of whether this was done at the study or outcome level), and how this information is to be used in any data synthesis. | 5,6 |
| Summary measures | 13 | State the principal summary measures (eg, risk ratio, difference in means). | 5,6 |
| Synthesis of results | 14 | Describe the methods of handling data and combining results of studies, if done, including measures of consistency (eg, I2) for each meta-analysis. | 6 |
| Risk of bias across studies | 15 | Specify any assessment of risk of bias that may affect the cumulative evidence (eg, publication bias, selective reporting within studies). | 6 |
| Additional analyses | 16 | Describe methods of additional analyses (eg, sensitivity or subgroup analyses, meta-regression), if done, indicating which were pre-specified. | 6 |
| Results | |||
| Study selection | 17 | Give numbers of studies screened, assessed for eligibility, and included in the review, with reasons for exclusions at each stage, ideally with a flow diagram. | 7 |
| Study characteristics | 18 | For each study, present characteristics for which data were extracted (eg, study size, PICOS, follow-up period) and provide the citations. | 7 |
| Risk of bias within studies | 19 | Present data on risk of bias of each study and, if available, any outcome level assessment (see item 12). | 7–14 |
| Results of individual studies | 20 | For all outcomes considered (benefits or harms), present, for each study: (a) simple summary data for each intervention group; (b) effect estimates and confidence intervals, ideally with a forest plot. | 7–14 |
| Synthesis of results | 21 | Present results of each meta-analysis done, including confidence intervals and measures of consistency. | 7–14 |
| Risk of bias across studies | 22 | Present results of any assessment of risk of bias across studies (see item 15). | 7–14 |
| Additional analysis | 23 | Give results of additional analyses, if done (eg, sensitivity or subgroup analyses, meta-regression [see Item 16]). | 7–14 |
| Discussion | |||
| Summary of evidence | 24 | Summarize the main findings including the strength of evidence for each main outcome; consider their relevance to key groups (eg, healthcare providers, users, and policy makers). | 14,15 |
| Limitations | 25 | Discuss limitations at study and outcome level (eg, risk of bias), and at review-level (eg, incomplete retrieval of identified research, reporting bias). | 15,16 |
| Conclusions | 26 | Provide a general interpretation of the results in the context of other evidence, and implications for future research. | 17 |
| Funding | |||
| Funding | 27 | Describe sources of funding for the systematic review and other support (eg, supply of data); role of funders for the systematic review. | None |
Notes: Reproduced from Moher D, Liberati A, Tetzlaff J, et al, Preferred reporting items for systematic reviews and meta-analyses: the PRISMA Statement. PLoS Med. 2009:6(7): e1000097.1