Abstract
Background:
The emergence and spread of extended spectrum β-lactamase (ESBL)-producing Gram- negative bacteria (GNB), particularly in Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa, have increased all over the world. ESBLs are characterized by their ability to hydrolyze β-lactams, early cephalosporins, oxyimino-thiazolyl cephalosporins, and monobactams, but not cephamycins or carbapenems. The rate of nosocomial infections caused by ESBL-producing GNB in Asia Pacific has increased and several studies have identified their prevalence in the region. The aim of this study is to review the prevalence of ESBL-producing GNB in the West Asia and the Middle East with a particular focus on Iran.
Materials and Methods:
The available evidence from various studies (Microbia and clinical studies, retrieved from the PubMed, and Scopus databases) regarding the ESBL producing Gram negative bacteria in Iran were evaluated.
Results:
In almost all parts of the country, high resistance has been observed, especially in the central part of Iran. Up to 89.8% Escherichia coli, 72.1% Klebsiella pneumonia, 84.2% Acinetobacter baumannii, and 83.8% Pseudomonas aeruginosa isolates are ESBL positive.
Conclusion:
The present study showed the increasing prevalence of ESBLs in different regions of Iran, which could be useful to strategic policy towards reducing reduce their prevalence.
Keywords: extended-spectrum β-lactamase, antibiotic resistance, Acinetobacter baumannii, Pseudomonas aeruginosa, Iran
Introduction
Infections due to Gram negative bacteria (GNB) are causing morbidity and mortality worldwide, and the production of β-lactamase remains the most important contributing factor to β-lactam resistance (Leylabadlo et al., 2016). Nowadays, the rapid spread of resistance to β-lactam antibiotics among GNB poses a clinical and public health challenge and increasing rates of drug resistance among them is a major concern worldwide (Little et al., 2012). Monobactams, cephalosporins, penicillins, and carbapenems can be hydrolyzed by the enzymes of the β- lactamase group, which renders them microbiologically ineffective (Bush and Mobashery, 1998). In the early 1980s, new family of beta-lactamases named extended-spectrum β-lactamases (ESBLs) were identified introduced that hydrolyze penicillins and expanded-spectrum cephalosporins have emerged (Kliebe et al., 1985). They are effective against β-lactam antibiotics like ceftazidime, ceftriaxone, cefotaxime, and oxyiminomonobactam, but not cephamycins or carbapenems (Bradford, 2001). ESBL production has been associated with Enterobacteriaceae, mostly Klebsiella pneumoniae isolated from Intensive care unit (ICU) patients; however, it is found also in community setting (Paterson and Bonomo, 2005) and, more recently, other Gram negative bacteria such as Acinetobacter baumannii and Pseudomonas aeruginosa(Oliver et al., 2005).
Nowadays, more than 600 ESBL variants have been described, the majority of which belong to the Cefotaxime hydrolyzing capabilities (CTX-M), Sulfhydryl variable (SHV) and Temoneira (TEM) (http://www.lahey.org/studies/webt.htm). Most of the detected ESBLs are either of SHV or TEM types and associated with nosocomial infections caused by Gram negative bacteria (Paterson and Bonomo 2005). In the recent years, the spread of CTX-M-producing E. coli has been dramatic and they are considered the primary ESBL producers that are always associated with community-acquired infections (Leylabadlo et al., 2015).
Generally, ESBL has been found all over the world; for example, previous studies have reported that the prevalence of ESBL in Europe is higher than the United States, but lower than South America and Asia (Giamarellou, 2005). Asia is almost certainly a part of the world, in which the prevalence of ESBLs is very high among GNBs; however, different countries have different prevalence rates (Giamarellou, 2005). The prevalence of ESBL in GNB has been previously reviewed in the countries of Gulf Cooperation Council (GCC) in the West of Asia and the Middle East (Zowawi et al., 2013), but did not included Iran, a country in both the West of Asia and in the Middle East, which also is another Persian Gulf country. Therefore, this review is focused on obtaining Iranian ESBL epidemiology and population structure of ESBL- producing isolates from hospitals and communities. Also, we have attempted to analyze the nature of mobile genetic elements carrying ESBL genes.
Material and Methods
We conducted a search in the field using the Scopus, PubMed, Google Scholar, and Persian Databases from 2008 to 2016.
Prevalence of ESBLs-producing GNB in Iran
Iran, a country in the western Asia, is the second largest nation in the Middle East. The country is bordered by Azerbaijan and Armenia in the northwest, Russia and Kazakhstan across the Caspian Sea, Turkmenistan in the northeast, Afghanistan and Pakistan in the east, the Gulf of Oman and the Persian Gulf in the south, and Turkey and Iraq in the west. Iran is divided into five regions with thirty-one provinces and various climatic conditions. For convinience of study, we divided it into five hypothetical parts: Northern (Mazandaran, Guilan, and Golestan Provinces), Southern (Kerman, Khuzestan, Yazd, Fars, Hormazgan, Bushehr, as well as Kohgiluyeh and Boyer Ahmad Provinces), Central (Tehran, Isfahan, Makazi, Zanjan, Semnan, Chaharmahal and Bakhtiari, Qom, Qazvin, and Alborz Provinces), Eastern (Khorasan Razavi, Northern and Southern Khorasan, as well as Sistan and Baluchestan Provinces), and Western (Hamedan, Ilam, Kermanshah, Kurdistan, East Azerbaijan, Ardebil, West Azerbaijan, and Lorestan Provinces). There are several studies about the prevalence of ESBL-producing bacteria from most provinces in Iran. Nevertheless, in some provinces (Hormazgan, Kohgiluyeh and Boyer Ahmad, Qom, Northern and Southern Khorasan, as well as Bushehr), no reports are available. Spread and prevalence of ESBLs-producing bacteria are shown in Figure 1.
Figure 1.
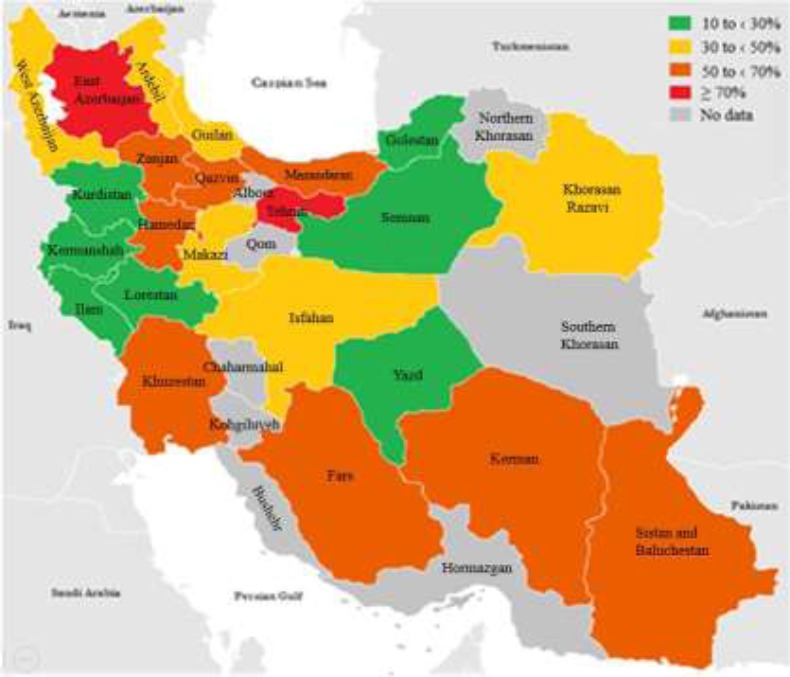
Representation of the frequency of ESBL production (no. of tested isolates) and types of ESBLs produced by GNB isolates from Iranian hospitals and communities. Distribution of ESBLs was not available for all the studies reporting prevalence data.
Northern part of Iran
In the north of Iran, several surveys have been conducted among ESBL-producing GNB isolates from the community and hospitals. A study in Mazandaran Province, which was carried out at Buali Sina Hospital, revealed that 100 out of 327 uropathogenic E. coli (30.5%) were positive for ESBL that were lower in other Asian countries such as India (27%) (Ananthan and Subha 2005). Also, in the same study, TEM gene was the most prevalent one (49%) followed by other genes and about 12% of ESBL-producing isolates were shown to have both TEM and CTX-M genes. In another study from the region, to identify the accompanying Class 1 integrons and ESBLs in the MDR E. coli isolates, urine samples were collected (Moghaddam et al., 2015). The isolates were different in the resistance profiles; however, some others had similar resistance profiles. In a recent study, there was a significant correlation between ESBL production and resistance to all types of antibiotics except for piperacillin and ciprofloxacin, and out of eight ESBL positive isolates, seven (87.5%) carried class 1 integrons (Moghaddam et al., 2015). In another study from pediatric patients for ESBL genes, about 30.5% of isolated E. coli were ESBL-positive and TEM gene was identified in 49% followed by CTX in 28%, SHV in 44%, VEB in 8%, and GES in 0% of isolates (Rezai et al., 2015).
The analysis of 100 ESBL phenotypes of A. baumannii from the clinical samples of some hospitals in Mazandaran Province, representing the molecular pattern of Acinetobacter spp., 24% of the isolates were positive, which 79.1%were blaTEM and 31.5% were blaCTX (Ahanjan et al., 2014). The prevalence of blaTEM in this study was the same as reported in China (Jin et al., 2009). In a study from Babol city in Mazandaran during 2011-2012, a high prevalence of β-lactamase-producing E.coli was reported in the hospital, having two or three different types of enzymes in a bacterium, the lowest of which was related to Plasmid-Encoded AmpC (pAmpC) enzyme in 20 (7.7%) isolates (Shahandeh et al., 2015). This value was different from Indian study with the prevalence of 48.5% (Nagdeo et al., 2012).
A study in Guilan Province on urine samples revealed that 55% of E. coli isolates were resistant to the third and fourth cephalosporins. In a similar study on the urine samples of hospitalized patients and outpatients of Valiasr Hospital, the incidence of ESBL was 38.92% and bacterial isolates had the highest levels of resistance to amoxicillin, cephalexin, and tetracycline (Ranjbar et al., 2014). Another province in the northern part of Iran is Golestan Province. In a study carried out on urine samples of the outpatients, 70 isolates (32.1%) were resistant to cefotaxim and 62 (88.6%) were confirmed as ESBL-producing E.coli (NasrolahiOmran et al., 2012). Also, the prevalence of ESBL-producing E.coli in Golestan was in the range of the country’s average and blaCTX-M gene was the most common one (NasrolahiOmran et al., 2012).
In general, class A and D as well as AmpC β-lactamases are the most common in the northern part of Iran. The prevalence of ESBLs was between 24 and 56.85% among the samples of both hospitals and the community. Also, this prevalence was 33.77% in the north of Iran on average. The highest rate belonged to a hospital in Babol city in the north of Iran, which was also mentioned in the present work (Table 1).
Table 1.
Summary of ESBL enzymes and their rates identified in the Iran. All listed studies used phenotypic methods such as double-disk synergy test (DDST) or ESBL Etest
| Section and Province | Producing organism | Setting | ESBL genotypes | (%)among ESBL | Reference |
|---|---|---|---|---|---|
| Northern part of Iran | |||||
| Mazandaran | E. coli | Hospital and Community | TEM, SHV, CTX, VEB | (30.5) | (Rezai et al., 2014) |
| Mazandaran | A. baumannii | Hospital | TEM, CTX | (24) | (Ahanjan et al., 2014) |
| Mazandaran | E. coli | Hospital | AmpC | (56.8) | (Shahandeh et al., 2015) |
| Guilan | E. coli | not specified | CTX-M | (24) | (Hemmati et al., 2015) |
| Guilan | E. coli | Hospital and Community | Not analyzed | (38.9) | (Ranjbar et al., 2014) |
| Golestan | E. coli | Community | CTX-M, TEM, SHV | (28.4) | (Nasrolahi et al., 2012) |
| Southern part of Iran | |||||
| Kerman | P.aeruginosa | Hospital | SHV, PER, TEM | (34) | ( Shakibaie et al., 2008) |
| Kerman | K. pneumoniae E. coli | Hospital | CTX-M, AmpC | (43.8) | (Mansouri et al., 2014) |
| Kerman | P.aeruginosa A. baumannii | Hospital | CTX-M | (59.4) | (Mansouri et al., 2012) |
| Kerman | P.aeruginosa | Hospital | CTX-M, VEB-1, PER-1, GES-1, OXA-4, OXA-10 | (34.1) | (Shacheraghi et al., 2010) |
| Khuzestan | Enterobacteriaceae | Hospital and Community | TEM, SHV | (30.5) | (Moosavian and Deiham 2012) |
| Khuzestan | K. pneumoniae | Hospital | CTX-M1 | (47.72) | (Khosravi et al., 2013) |
| Khuzestan | P.aeruginosa | Hospital | PER-1, OXA-10, CTX-M | (51.9) | (Farshadzadeh et al., 2014) |
| Yazd | P.aeruginosa | Hospital | Not analyzed | (22) | (Tafti et al., 2014) |
| Fars | K. pneumoniae | Hospital | TEM, CTX-M | (60) | (Ghasemi, Archin et al.) |
| Fars | Gram negative bacil | Community | TEM, SHV | (11.75) | (Kargar et al., 2014) |
| Centeral part of Iran | |||||
| Tehran | E. coli | Hospital | TEM, SHV | (28) | (Pakzad et al., 2011) |
| Tehran | K. pneumoniae | Hospital and Community | SHV, CTX-M, TEM, PER | (38.5) | (Leila et al., 2010) |
| Tehran | K. pneumoniae | Hospital and Community | SHV, TEM, CTX-M | (72.1) | (Feizabadi et al., 2010) |
| Tehran | E. coli | Hospital | SHV, TEM | (49) | (Shahcheraghi et al., 2009) |
| Tehran | E. coli | Community | TEM, SHV | (2.7) | (Hosseini et al., 2007) |
| Tehran | K.spp | Hospital | SHV, TEM, CTX-M | (51.6) | (Ghafourian et al., 2010) |
| Tehran | K. pneumoniae | Hospital and Community | SHV, TEM, CTX-M | (27.45) | (Eftekhar et al., 2012) |
| Tehran | K. pneumoniae | Hospital | SHV, TEM-1, CTX-M-15 | (62.85) | (Ashayeri et al., 2014) |
| Tehran | E. coli | Hospital | TEM | (64) | (Yazdi et al., 2013) |
| Tehran | E. coli | Hospital | CTX-M-1 | (70) | (Peerayeh et al., 2013) |
| Tehran | E. coli | Hospital | CTX-M, TEM, SHV | (20) | (Abdi, 2014) |
| Tehran | P.aeruginosa | Hospital | TEM, PER-1 | (39.2) | (Rafiee et al., 2014) |
| Tehran | P.aeruginosa | Hospital (children) | AmpC (Not found PER) | (83.3) | (Fazeli et al., 2014) |
| Tehran | K. pneumoniae | Hospital and Community | CTX-M-15, OXA-48 | (57.5) | (Hashemi et al., 2014) |
| Tehran | K. pneumoniae | Hospital | OXA-48 (first report) | not specified | (Azimi et al., 2014) |
| Tehran | Shigella | Hospital | CTX-M-15, CMY-1 | not specified | (Tajbakhsh et al., 2012) |
| Tehran | Gram negative bacil | Hospital | CTXM | (29.09) | (Vali et al., 2014) |
| Tehran | P.aeruginosa A. baumannii | Hospital | CTX-M-15 | P (31.91) A (21.42) | (Hallajzadeh et al., 2014) |
| Tehran | A. baumannii | Hospital | PER-1, VEB-1 | (84.2) | (Fallah et al., 2014) |
| Tehran | Acineto.spp | Hospital | TEM, SHV | (51) | (Sharif et al., 2014) |
| Tehran | E. coli | Hospital and Community | TEM, CTX-M | not specified | (Goudarzi, 2014) |
| Tehran | E. coli | Hospital | SHV | (89.8) | (Dallal et al., 2013) |
| Esfahan | K. pneumoniae E. coli | Hospital and Community | TEM, SHV | (18.75) | (Gholipour et al., 2014) |
| Esfahan | P.aeruginosa | Hospital | GES-2, SHV-5, CTX –M-1 | (8.1) | (Tavajjohi et al., 2011) |
| Esfahan | P.aeruginosa | Hospital | OXA-10 | not specified | (Golshani and Sharifzadeh 2013) |
| MarkaziQazvin | EnterobacteriaceaeE. cloacae | Hospital and CommunityHospital | CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9 Not analyzed | (44)(55.7) | (Safari et al., 2013)(Peymani et al., 2014) |
| Zanjan | E. coli (EAEC) | Hospital and Community | TEM, CTX-M | (52.7) | (Khoshvaght et al., 2014) |
| Semnan | E. coli K. pneumoniae | Community | Not analyzed | (18.1) | (Moghadas, 2009) |
| Eastern part of Iran | |||||
| Khorasan | E. coli | Hospital | CTX-M, TEM, SHV | (33.3) | (Moghaddam et al., 2012) |
| Khorasan | E. coli Klebsiella | Hospital and Community | SHV, TEM | (35.6) | (Zaniani et al., 2012) |
| Khorasan | Enterobacteriaceae | Hospital and Community | CTX-M-15 | not specified | (Moghaddam et al., 2014) |
| Sistan baluchestan | K. pneumoniae P.aeruginosa | Hospital | TEM, CTX-M | K (66.6) P (13.3) | (Saeidi et al., 2014) |
| Sistan baluchestan | P.aeruginosa | Hospital | TEM, VEB-1, PER-1, SHV | (6.89) | (Bokaeian et al., 2015) |
| Western part of Iran | |||||
| Hamedan | P.aeruginosa | Hospital | PER-1, VEB | (58.3) | (Alikhani et al., 2014) |
| Hamedan | A. baumannii | Hospital | TEM, SHV | (7) | (Safari et al., 2015) |
| Ilam | K.oxytoca | Hospital | Not analyzed | (25) | (Ghafouryan et al., 2010) |
| Kermanshah | E. coli | Community | Not analyzed | (24.5) | (Mohajeri et al., 2014) |
| Kurdistan | Gram negative bacil | Community | TEM, CTX-M, SHV | (12.66) | (Ramazanzadeh et al., 2010) |
| Kurdistan | Gram negative bacil | Community | CTX- M, TEM, SHV | (14.5) | (Ramazanzadeh, 2010) |
| Kurdistan | P.aeruginosa | Hospital | PER-1, OXA-10 | (28) | (Kalantar et al., 2012) |
| Kurdistan | P.aeruginosa | Hospital | SHV | (17.89) | (Bahmani and Ramazanzadeh 2013) |
| East Azerbaijan | K. pneumoniae | Hospital | SHV, TEM, CTX- M | (43.7) | (Ghafourian et al., 2011) |
| East Azerbaijan | A. baumannii | Hospital | PER-1, VEB-1 | (70) | (Farajnia et al., 2013) |
| East Azerbaijan | P.aeruginosa | Hospital | PER-1 | (75.86) | (Akhi et al., 2012) |
| Ardebil | Enterobacteriaceae | Hospital and Community | Not analyzed | (36.75) | (Akbari et al., 2014) |
| West Azerbaijan | E. coli | Hospital | CTX-M-15 | (41.98) | (Parsania and Sohrabpour 2104) |
| Lorestan | Gram negative bacilli | Hospital | Not analyzed | (23.6) | (Zadegan et al., 2009) |
Southern part of Iran
Southern part of Iran has a warm and dry weather. This region mostly includes provinces around the Persian Gulf and the Gulf of Oman. In Kerman Province, in the burn ward of Shafa Hospital during January 2006 to December 2007, 41 (34%) isolated P.aeruginosa were reported to be ESBL producers; however, none of the isolates could produce any MBL enzyme and all ESBL-producing isolate were multidrug resistant (MDR). According to the PCR and multiplex PCR assays, the most prevalent genes among ESBL producers in the recent study were SHV and PER-1; but, no Amp-C gene was detected in any of the isolates (Shakibaie et al., 2008).
Another study in the burn ward of Kerman University showed that the ESBL-producing P.aeruginosa was the same as the previous study. However, the frequency of PER-1 gene was very high and molecular results showed that blaVEB-1 in the P.aeruginosa strains harbor (643bp) was the most frequent ESBL gene. These data were in line with other results in the Middle East and Iran (Shakibaie et al., 2008). Surveys on the isolation of MDR non-fermenting GNB of the patients from three hospitals in Kerman indicated that all of the isolates were resistant to cefoxitin, and ESBL production was higher in A. baumannii than other tested bacterial species; but, the difference was not significant. A study from November 2007 to July 2008 on the patients admitted to hospitals in the Kerman showed that, among K. pneumoniae isolates, 44% produced ESBLs and 28% had AmpC (Mansouri et al., 2014). Also, among E. coli isolates, 43.7% isolates had ESBLs and 39.3% produced AmpC. Co-existence of ESBLs and AmpC β-lactamases was observed in 39.3% of isolates. In this study, the prevalence rate of blaCTX-M gene among ESBLs- and AmpC-producing isolates of K. pneumoniae and E. coli was 21.3% and 13.3%, respectively (Mansouri et al., 2014).
Khuzestan is another province in the southern part of Iran. In an 11-month study from August 2011 to July 2012, 500 Enterobacteriaceae isolates were collected from Ahvaz and the prevalence rates of CTX-M-1 genes among ESBLs-positive isolates K. pneumonia were found in 26.92% (Khosravi et al., 2013). In Ganjavian Hospital, Dezful, Khuzestan Province, among 128 ESBL-producing Enterobacteriaceae isolates, 73 (57%) were positive for TEM and/or SHV by multiplex PCR. Also, 14 isolates (19%) had both blaTEM and blaSHV genes; but, blaCTX-M was not detected in ESBL-producing strains (Moosavian and Deiham 2012).
Analysis of P. aeruginosa isolates from the burn ward of a hospital in Khuzestan showed that 176 (95.13%) isolates were MDR, and the prevalence of ESBL was higher than that of a similar study in Yazd Province (Farshadzadeh et al., 2014). During the study period between 2009 and 2010 in Fars Province, K. pneumoniae was collected from Intensive Care Units (ICUs) of Namazi Hospital in Shiraz and high prevalence (60%) of ESBL-producing K. pneumonia were demonstrated. In addition, this study showed a new pattern of blaTEM distribution, in which blaTEM was found to be predominant, followed by blaCTX-M and blaSHV, respectively (Ghasemi et al., 2013). Several Iranian studies have had a similar pattern of ESBLs distribution, such as in Tehran Province (Nasehi et al., 2010). However, this pattern is different from other countries (e.g., Italy, Egypt and Austria) in which CTX-M is the most prevalent β- lactamases (Ahmed et al., 2009). In another study at Namazi Hospital during a 2 year period, 39 Serratia strains were isolated from blood samples; overall, positive ESBL phenotype was identified in 69% (n=27) of the isolates: 70.37% (n=19) for of S. marcescens and 29.62% (n=8) for S.liquefaciens and TEM gene was detected in none of the isolates in this study (Mostatabi et al., 2013). In previous studies from Taiwan and Mexico, it has been reported that blaSHV and blaCTX-M-3 are identified as the most common ESBL genes in Serratia (mostly in S.marcescens) (Cheng et al., 2006). A research on patients with diarrhea conducted in Shiraz among 2010 revealed that the prevalence of ESBLs-producer E. coli isolates was 12.96% (Ghorbani-Dalini et al., 2015). Antimicrobial resistance testing showed a high resistance to cefexime, trimethoprim-sulfamethoxazole, ampicillin and penicillin. In their study, β-lactamase genes were identified in 96.30% isolates which were identified as 83.33% blaTEM, 31.48% blaSHV and 20.37% blaCTX-M. In addition, this study indicated that the blaTEM gene for ESBLs-producer E. coli isolates was widespread in Iran (Ghorbani-Dalini et al., 2015).
Generally, in the southern part of Iran, classes A and D are more common in various samples from the provinces and AmpC is also present in one study. The prevalence of ESBL was between 11.75% and 69% in both hospitals and communities and, on average, it was 42.2%. In addition, the prevalence of ESBL in the community samples was less than in the hospitals (Table-1).
Central part of Iran
Central part of Iran consists of area between Alborz Mountains, Zagros Mountains and the scattered mountains of Khorasan in the east. It includes some provinces that are shown in Table-1. Most of the region has a warm and dry weather with the milder climate in the mountain areas. Tehran, the capital city of Iran, is the largest city of the country and also the largest city in Western Asia. Several surveys have been performed in this area. Analysis of K. pneumoniae isolates in the central parts showed that the rate of ESBL producers was between18.1% and 72% among the sample hospitals and communities. The ESBL-producing K. pneumoniae in the community’s samples was less than that in the hospitals. However, samples from Tehran Hospital were more than other provinces, mostly carrying blaSHV, blaCTX-M, and blaTEM genes predominantly, among which the prevalence of blaSHV was higher than others (26%-77%). An investigation from Motahari and Shariati hospitals revealed that the rates of drug resistance in burn isolates of K. pneumoniae were higher compared to the non-burn strains, while no significant differences were observed in ESBL production between the two groups (Eftekhar and Naseh 2015). In the burn ward of Motahari Hospital, Tehran, in 2011, the first cases of OXA-48-producing K. pneumoniae in Iran were reported and blaOXA-48 gene was detected in96.42% (27/28) of isolates by PCR analysis followed by sequencing (Azimi et al., 2014). Another study in Tehran from October 2011 to May 2012 reported that 83 non-duplicate and non-consecutive K. pneumoniae blaOXA-48 genes were found in two (4.1%) out of the 48 ESBL-producing isolates and 2 isolates harbored both blaOXA-48 and blaCTX-M-15(Hashemi et al., 2014). Reports of blaOXA-48 have been dramatically increased in recent years worldwide and OXA-48-producing K. pneumoniae and other isolates have been reported from various regions such as Turkey, Morocco, Lebanon, Belgium, United Kingdom, Tunisia, Argentina, and India (Cuzon et al., 2011).
A study demonstrates the predominant presence of the gene encoding CTX-M-1 group among ESBLs producing of Salmonella spp. that they can transmit to bacteria of this genus or even other genera of enteric bacteria (Rizi et al., 2015). In the recent study, four Salmonella spp. (3.6%) showed ESBLs phenotype, which all have blaCTX-1 genes group. ESBL-producing E.coli in the central part of Iran has different ranges between hospitals and the community. According to the data, the majority of ESBL-producing isolates were from hospital patients in Tehran. Substantial proportions of E. coli reported from urine samples in the central part of Iran and rate of ESBL-producing E.coli between hospitals in this region were 28%-89.8%, which was higher than that of the communities. Also, CTX-M and TEM type ESBLs appear to be predominating among ESBL-producing E.coli in the region (Pakzad et al., 2011). A study about E.coli isolated from urine samples from patients with urinary tract infection (UTI) referred to two main hospitals in Tehran, during September 2010 to March2011, showed that 20% of E. coli isolates were ESBL-producer and frequency of blaCTX-M, blaTEM, and blaSHV genes was 87%, 82%, and 65%, respectively (Abdi et al., 2014). In a similar study from a Children Medical Center in Tehran between 2012 and 2013, the presence of blaCTX-M and blaTEM were detected in 69 (69%) and 74 (74%) of isolates, respectively, while blaSHV gene was absent (Goudarzi et al., 2013).
Notably, in the other province in the central part of Iran in isolates of Enterobacteriaceae from Markazi province, the frequency of CTX-M-1,-2,-8,-9 among the isolates were as 92.2%, 28.5%, 17.5%, and 38.3%, respectively (Safari et al., 2013). This value was different from other studies in other countries. For example, studies in Sudan, Korea, and Thailand have demonstrated that the prevalence of CTX-M-1 was 45.9%, 16.4%, and14%, respectively (Safari et al., 2013). However, the prevalence of CTX-M-9 in Germany, America, and Canada is in the range of 38.5%-58.5% (Safari et al., 2013). Nevertheless, other studies have shown that the dissemination of CTX-M-8 gene in Iran is higher compared to the rest of the world (Kim et al., 2008). Analysis of non-fermenting GNB in the central part of Iran showed that the lowest ESBL-producing was 8.1% in Isfahan Province and the highest was 83.3% in hospitals of Tehran Province (Kim et al., 2008; Fazeli et al., 2015).
In a study from Tehran, 51 non-duplicate P. aeruginosa isolates were randomly selected from a collection of burn isolates of a hospital in 2011 and it was found that 43 isolates (84.3%) carried β-lactamase genes, out of which 31 (60.8%) harbored blaAmpC, 20 (39.2%) isolates had blaTEM, and 11 (21.6%) carried blaPER-1 genes (Rafiee et al., 2014). But, in another study, during a 3 month period, from November 2011 to January 2012, 72 clinical isolates of P. aeruginosa collected from the samples included blood specimen, urine, wound, tracheal, throat, and other samples from a major pediatric hospital in Tehran reported that none of them had blaPER, blaBEL, and all of the isolates had ampC gene (Fazeli et al., 2015). This study was the first work among the samples from children in which PER was not detected (Fazeli et al., 2015).
Other provinces in the central part of the country have also reported non-fermenting ESBL-producing GNB. In a study from Beheshti Hospital in Kashan that was conducted during 2010 to 2011, it was reported that, from 86 urine, trachea, and stool samples, 19 (20.9%) P. aeruginosa isolates were resistant to all extended-spectrum cephalosporins (Tavajjohi et al., 2011). This study also reported 8.1% ESBL-production among P. aeruginosa isolates and the blaGES-2 gene was detected in all ESBL-producing P. aeruginosa. GES-2 in the same study was different from other results in the Middle East region and Iran (Tavajjohi et al., 2011). It was reported from Kashan that the prevalence of blaCTX-M among Enterobacteriacea isolates was at an alarming rate (Hasan Afzali et al., 2015). It the recent study that conducted in 2013, from 100 Klebsiella spp. isolates, 41% of isolates had resistance ceftazidime or aztreonam and 35% were ESBLproducers, which 80% of the resistant isolates carried the blaCTX-M type genes. Moreover, CTX-M-1, CTX-M-2 and CTX-M-9 were identified in 60%, 42% and 34% of these isolates, respectively (Hasan Afzali et al., 2015). In another research from Kashan, 47% of ESBL-producing isolates were identified by DDST confirmatory test and majority (80.5%) of isolates carried blaCTX-M genes including CTX-M-1 (60%), CTX-M-2 (42.9%), and CTX-M-9 (34.3%) (SHAMS et al., 2015). Also, 77 ESBL-producing K. pneumoniae were positive with PMQR genes, which mostly consisted of aac(6´)-Ib-cr (70.1%) and qnrB (46.0%), followed by qnrS (5.7%). Among these PMQR positive isolates, 35.1% and 1.3% of isolates carried 2 and 3 different PMQR genes, respectively (Shams et al., 2015). Frequency of blaOXA-10 in ESBL-producing P. aeruginosa was 40 (64%) in another study from Isfahan in 2012 (Golshani and Sharifzadeh, 2013). In a study on P. aeruginosa clinical isolates, 76 isolates were collected from Milad hospital in Tehran (Akbariqomi et al., 2015). In their study, 76.3% of isolates classified as MDR and The majority of them were found in ESBL producer P. aeruginosa. Also, the effective antibiotics against ESBL and non-ESBL were meropenem and amikacin, respectively. In the central region of the country, only in the hospitals of Tehran, ESBL-producing Acinetobacters were between 21.4% and 84.2% in three studies. The rate of PER-1, VEB-1, CTX-M-15, TEM, and SHV was 78.03%, 39.5%, 9.3%, 56%, and 63%, respectively (Hakemi et al., 2014; Sharif et al., 2014).
Another study that was conducted in a major pediatric hospital in Tehran showed that 4 out of 55 Shigella isolates, including three S. sonnei and one S. flexneri, had ESBL-positive phenotype (Tajbakhsh et al., 2012). Also one S. sonnei isolate had CMY-59 gene. The ESBL-producing Shigella isolates in this study were higher than previous studies in other countries (Tajbakhsh et al., 2012). Therefore, the prevalence of ESBL-producing GNB in the central part of Iran was between 2.3% and 89.8% and the average prevalence of ESBL-producing GNB in the central part of Iran was 43.55% in hospitals and community samples. Also, class A and D β-lactamases were the most prevalent in the region (Table 1).
Eastern part of Iran
This part of Iran has a warm and dry weather and is near Afghanistan and Pakistan borderlines. In this area, ESBL-producing GNB was reported in Khorasan as well as Sistan and Baluchestan Provinces. Analysis of E. coli isolates from urine samples of hospitalized patients was collected in 2009 at two hospitals in Mashhad city, which showed that 33.3% were ESBL producers and carrying blaCTX in 94.6%, blaTEM in 56.8%, and blaSHV in 13.5% of the isolates (Moghaddam et al., 2012). In this study, blaCTX-M was found to be higher than the developed countries and other studies in Iran (Moghaddam et al., 2012). To determine the frequency of qnr genes in isolates from Imam Reza hospital in Mashhad, 200 clinical isolate of E. coli were investigated (Mood et al., 2015). Results indicated that 43% of isolates were ciprofloxacin-resistant. Also, the phenotypic confirmatory test identified 42.5% isolates as ESBL-producing. Isolates were positive for blaTEM in 76.47%, blaSHV in 27%, qnrA in 31%, qnrB in 17%, and qnrS in 7%. In another research that was carried out in 2010, 82 E. coli and 78 K. pneumoniae were isolated from different clinical samples. In their study, 43.9% of E. coli and 56.1% of K. pneumoniae were ESBL producer, which in the community was lower than the hospital samples and frequencies of SHV and TEM were 14.4% and 20.6%, respectively (Riyahi et al., 2012). Recently, a survey conducted on Enterobacteriaceae isolates from urine samples on inpatients and outpatients who referred to Qaem and 17-Shahrivar Hospitals in Mashhad city in 2012. It was found that 27 (27%) isolates were ESBL-producing organisms with the highest frequency for K. pneumonia (47.4%) and E. coli (17.9%). Also, 26 (26%) Enterobacteriaceae isolates harbored the blaCTX-M gene. It was the first report of blaCTX-M-15 among E. coli isolates in this region. No blaPER was found among the isolates, which was similar to the work conducted by Shahcheraghi et al. in Tehran (Moghaddam et al., 2014).
To evaluate 180 isolates including 30 K. pneumoniae and 150 P. aeruginosa, a study was performed on the urine samples of hospitalized patients in a hospital of Zabol city in Sistan and Baluchestan Province (74). In this work, the prevalence of ESBL-producing isolates was13.3% for P. aeruginosa and 66.6% for K. pneumonia. Also, 75% and 65% of K. pneumoniae harbored the TEM and CTX-M genes, respectively, and 55% of P. aeruginosa isolates harbored the gene TEM; but, none of them demonstrated CTX-M gene. This prevalence of ESBLs-producing organisms was higher than that of other studies (Saeidi et al., 2014). Another study was conducted on non-fermentative GNB isolates during 2012–2013 on 116 P. aeruginosa isolates from Zahedan city, 6.89% isolates were ESBL-positive. Among them, 100% had blaTEM, 13.3% had blaPER-1, 6.6% had blaVEB-1 and 6.6% had blaSHV. blaTEM gene in this study had 100% identity with blaTEM-116(Bokaeian et al., 2015). In characterization of ESBL and AmpC producers among E. coli from three major hospitals in Zahedan, almost half of isolates were resistant to third generation of cephalosporins; of these 62.7% and 5% were ESBL and AmpC producers, respectively. Molecular analysis showed that 49.1% and 5% were positive for blaTEM and blaCMY2, respectively (Shayan and Bokaeian, 2015).
Overall, in this part of Iran, the prevalence of ESBL-producing GNB ranged between 6.89% and 66.6%, the average of which was 31.13%. Among ESBL-producing GNB in this region, class A β-lactamase was the most prevalent one (Table 1).
Western part of Iran
This part of the country with several cities has a cold and rainy weather and is bordered with Turkey and Iraq. The prevalence of ESBL-producing isolates in this region has been also studied and in some provinces and cities. During a 7-month study in 2009, out of 106 isolates of P. aeruginosa from two tertiary hospitals in Hamadan, 88.7% of the isolates were MDR and 58.25% were ESBL producer (Alikhani et al., 2014). Also, among 60 ESBL-producing strains, 16 (26.6%), 9(15%), and 3(5%) strains had blaPER-1, blaVEB-1, blaPER-1, and blaVEB related genes, respectively (Alikhani et al., 2014). The rates of blaVEB and blaPER ESBLs were the same as other studies in Tehran (SHAHCHERAGHI et al., 2009); however, it was higher than those of Turkey and Korea and lower than Thailand and Italy (Shahcheraghi et al., 2009). In another survey, 100 A. baumannii strains were isolated from ICU wards of three educational hospitals in Hamadan (Safari et al., 2015). This study was conducted in 2011 and reported that 7% of isolates were ESBLs producers; this rate was very lower than that of other Iranian studies. Among all the isolates, 58%, and 20% were harboring SHV and CTXM genes, respectively (Safari et al., 2015).
Kurdistan is another province located in the west of the country. Studies have shown that the rate of ESBL-producing strains in communities is lower than hospitals. For example, out of158 GNB isolated from the outpatients referring to hospital in Sanandaj, 20 (12.66%) isolates were ESBL producer and the majority of community acquired ESBL types belonging to CTX-M and SHV were 10.76% and TEM was 9.49% (Ramazanzadeh et al., 2010). In another study from Kurdistan that was conducted on 188 urinary tract infection outpatients in 2008, it was revealed that 27 (14.5%) isolates were ESBL-producing GNB and CTX-M type was the most prevalent one (12.7%). Also, the prevalence of SHV, TEM was 11.7% and 10.1%, respectively (Ramazanzadeh et al., 2010). The analysis of P. aeruginosa isolates from the patients who were hospitalized in Sanandaj showed that 17.89% of the isolates were ESBL producer and the prevalence of blaSHV was 10.57%; this rate was lower than that of other Iranian studies (Bahmani and Ramazanzadeh, 2013). Another work on the burn patients of Tohid Hospital in Kurdistan showed that, out of 100 P. aeroginusa isolates, 28% were positive for ESBL production and 48% and 52% had PER-1 and OXA-10 producers, respectively (Kalantar et al., 2012).
East Azerbaijan and West Azerbaijan are other provinces in this part of the country which have the highest prevalence of β-lactamases. The prevalence of ESBLs rate in East Azerbaijan was reported 43.3% in a survey (Ghafourian et al., 2011). In a study that was conducted in 2008, blaSHV, blaTEM, and blaCTX-M were identified as 86.7%, 15.6%, and 15.6%, respectively (Ghafourian et al., 2011). ESBLs-producing E. coli at hospitals of West Azerbaijan Province was relatively similar to that of the previous study (41.98%) that carried 23.63% blaCTX-M-15 gene (Parsania and Sohrabpour, 2014). Analysis of ESBL-producing non-fermentative bacteria in East Azerbaijan showed that 70% and 75.68% of the isolates were A. baumannii and P.aeruginosa, respectively; this rate was the maximum in Iran. Also, analysis of ESBL genes in this bacteria showed that, among 100 A. baumannii isolates, 51% were positive for PER-1 and 10% positive for VEB1, whereas none of the isolates were positive for PER2 type gene. In this study, the prevalence of blaPER-1 among 56 P. aeruginosa isolates was 27.5% (Farajnia et al., 2013). In a study from the Tabriz region, K. pneumoniae isolates had the same ESBL prevalence as E.coli isolates (SHAMS et al., 2015). In a same study, the presence of integron class 1 was associated with presence of ciprofloxacin resistance in both organisms and co-presence of class 1 and 2 and the presence of ESBL production. However, nalidixic acid resistance was related significantly with only integron class 1 and to the presence of ESBL production. Class 1 and 2 integrons were found in 73.5% of MDR isolates with 13.2% of them possessing both intI1 and intI2 genes (Shams et al., 2015).
The frequency of ESBL-producing GNB is reported in other provinces in this region; however, ESBL genes have not been analyzed. For example, in a study performed in Ilam hospitals during March 2007 to April 2008, among the samples taken from the wards of surgery as well as lesion and infection of the respiratory tract, 25% of K. oxytoca were reported to be ESBL-positive (Ghafouryan et al., 2010). In another study from Kermanshah Province, 200 isolates of Uropathogenic Escherichia coli (UPEC) were collected among urine samples in the community from February 2012 to February 2013 and 24.5% (n = 49) of isolates were found to be positive for ESBL production (Mohajeri et al., 2014). The prevalence of ESBL-producing GNB in the urine samples of hospitalized patients in Imam Khomeini Hospital of Ardabil Province in 2012 showed that, out of 400 Enterobacteriaceae, 147 (36.75%) isolates were positive for ESBL (Akbari, Amir Mozaffari et al., 2014). In another study from Shohada-ye Ashayer Hospital in Lorestan Province, from225 isolates, 53 (23.6%) were positive in terms of ESBL production (Zadegan et al., 2009).
Finally, in the western part of Iran, the prevalence of ESBL-producing GNB ranged from 7% to 75.86% in the samples of communities and hospitals and, on average, it was 34.26%. Also, Class A, D Ambler was the most prevalent one (Table 1).
Conclusion
Iran, like many other parts of the world, has experienced a significant increase in the numberof ESBL-producing GNB in the hospitals and communities. Thus, this review indicates that spread of ESBL-producing GNB is a great problem in healthcare centers. In the community setting, ESBL-producing GNB mostly have a lower prevalence than the hospital. However, it should be considered that the prevalence of ESBL genes varies in different geographical areas. The current rates in some parts of the country are very high, especially in the central part of Iran, such as Tehran Province. So, the highest rate of ESBL-producing E. coli, K. pneumonia, A. baumannii, and P.aeruginosa has been reported in the hospitals of Tehran in the recent years. Other parts of the country also have high prevalence of ESBL-producing GNB in both hospitals and communities. It is clear that ESBL-producing organisms are widely distributed globally; but, this rate is lower in several parts of the world than Iran. Most frequent types of ESBL enzymes in Iran include TEM, CTX-M, SHV, and OXA; but, there are other ESBL enzymes with different frequencies among GNB such as PER, VEB, and GES. In many parts of the world such as European Countries, Latin America, East and West Asia in Gulf Cooperation Council (GCC), and Africa, CTX-M variants become dominant β- lactamases in compare to TEM and SHV enzymes in GNB. In most reports, TEM, CTX-M, and SHV are the predominant and OXA-type ESBLs have been found mainly in P. aeruginosa and A. baumannii isolates in this region. Therefore, the presence of ESBLs genes is a risk factor for the future use of antimicrobial treatment in Iran. ESBLs distribution and the facilitation of their spread in different regions may be caused by factors such as “mobility” of ESBL genes, strong selective pressure of antibiotic use, purchase antibiotics without prescriptions, lack of observing hand hygiene, use of antibiotics in animals, travel, and different weather conditions. Future research should be focused on the areas in which no reports of ESBL have been made. Also, the areas with less prevalence of ESBL-producing GNBs and ESBL genes must be considered in order to take specific measures and increase supervision in the hospitals and the community in case any changes or increase occur in terms of prevalence.
Acknowledgements
This study was supported by Drug Applied Research Centre and the authors thank all staff of Microbiology laboratory for their collaborations and help.
Footnotes
Source of support: This study was supported by Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
References
- 1.Abdi S, Ranjbar R, Vala M. H, Jonaidi N, Bejestany O. B, Bejestany F. B. Frequency of bla TEM, bla SHV, bla CTX-M, and qnrA Among Escherichia coli Isolated From Urinary Tract Infection. Archives of Clinical Infectious Diseases. 2014;9(1) [Google Scholar]
- 2.Ahanjan M, Kholdi S, Rafiei A. Antibiotic-resistance Patterns and Frequency of TEM and CTX Type Extended-spectrum β-lactamases in Acinetobacter Clinical Isolates. Journal of Mazandaran University of Medical Sciences. 2014;24(116):32–40. [Google Scholar]
- 3.Ahmed S. H, Daef E. A, Badary M. S, Mahmoud M. A, Abd-Elsayed A. A. Nosocomial blood stream infection in intensive care units at Assiut University Hospitals (Upper Egypt) with special reference to extended spectrum β-lactamase producing organisms. BMC Research Notes. 2009;2(1):76. doi: 10.1186/1756-0500-2-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akbari M, Amir Mozaffari N, Peeri Dogaheh H. Prevalence of Extended-Spectrum Beta Lactamase in Entrobacteriaceae Isolated from Urinary Tract Infections in Ardabil, Iran. Journal of Ardabil University of Medical Sciences. 2014;14(3):285–291. [Google Scholar]
- 5.Akbariqomi M, Ghafourian S, Taherikalani M, Mohammadi S, Pakzad I, Sadeghifard N. Antibiotic Susceptibility Patterns of Extended Spectrum Beta-Lactamase and Non Extended Spectrum Beta-Lactamase Pseudomonas aeruginosa Clinical Isolates. Recent patents on anti-infective drug discovery. 2015;10(2):128–133. doi: 10.2174/1574891x10666150901111312. [DOI] [PubMed] [Google Scholar]
- 6.Akhi M. T, Khalili Y, Ghottaslou R, Aghazadeh M. Prevalence of PER-1-type extended-spectrum beta-lactamaes in clinical strains of Pseudomonas aeruginosa isolated from Tabriz, Iran. Iranian journal of basic medical sciences. 2012;15(1):678. [PMC free article] [PubMed] [Google Scholar]
- 7.Alikhani M. Y, Tabar Z. K, Mihani F, Kalantar E, Karami P, Sadeghi M, Farajnia S. Antimicrobial resistance patterns and prevalence of blaPER-1 and blaVEB-1 genes among ESBL-producing Pseudomonas aeruginosa isolates in West of Iran. Jundishapur Journal of Microbiology. 2014;7(1) doi: 10.5812/jjm.8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ananthan S, Subha A. Cefoxitin resistance mediated by loss of a porin in clinical strains of Klebsiella pneumoniae and Escherichia coli. Indian journal of medical microbiology. 2005;23(1):20. doi: 10.4103/0255-0857.13867. [DOI] [PubMed] [Google Scholar]
- 9.Ashayeri-Panah M, Feizabadi M, Eftekhar F. Correlation of multi-drug resistance, integron and blaESBL gene carriage with genetic fingerprints of extended-spectrum β-lactamase producing Klebsiella pneumoniae. Jundishapur Journal of Microbiology. 2014;7(2) doi: 10.5812/jjm.8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azimi L, Nordmann P, Lari A. R, Bonnin R. A. First report of OXA-48-producing Klebsiella pneumoniae strains in Iran. GMS Hygiene & Infection Control. 2014;9(1) doi: 10.3205/dgkh000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahmani N, amazanzadeh R. Detection of SHV type extended-spectrum B-lactamase and risk factors in pseudomonas aeruginosa clinical isolates. Pakistan journal of medical sciences. 2013;29(3):788. doi: 10.12669/pjms.293.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bokaeian M, Zahedani S. S, Bajgiran M. S, Moghaddam A. A. Frequency of PER, VEB, SHV, TEM and CTX-M Genes in Resistant Strains of Pseudomonas aeruginosa Producing Extended Spectrum β-Lactamases. Jundishapur journal of microbiology. 2015;8(1) doi: 10.5812/jjm.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradford P. A. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clinical microbiology reviews. 2001;14(4):933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bush K, Mobashery S. In Resolving the Antibiotic Paradox. Springer US: 1998. How β-lactamases have driven pharmaceutical drug discovery; pp. 71–98. [PubMed] [Google Scholar]
- 15.Cheng K, Chuang Y, Wu L, Huang G, Yu W. Clinical experiences of the infections caused by extended-spectrum beta-lactamase-producing Serratia marcescens at a medical center in Taiwan. Japanese journal of infectious diseases. 2006;59(3):147. [PubMed] [Google Scholar]
- 16.Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrobial agents and chemotherapy. 2011;55(5):2420–2423. doi: 10.1128/AAC.01452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dallal M. S, Sabbaghi A, Aghamirzaeie H. M, Lari A. R, Eshraghian M. R, Mehrabad J. F, Rajabi Z. Prevalence of AmpC and SHV β-lactamases in clinical isolates of Escherichia coli from Tehran Hospitals. Jundishapur Journal of Microbiology. 2013;6(2):176–180. [Google Scholar]
- 18.Eftekhar F, Naseh Z. Extended-spectrum β-lactamase and carbapenemase production among burn and non-burn clinical isolates of Klebsiella pneumoniae. Iranian journal of microbiology. 2015;7(3):144. [PMC free article] [PubMed] [Google Scholar]
- 19.Eftekhar F, Rastegar M, Golalipoor M, Mansoursamaei N. Detection of Extended Spectrum B-Lactamases in Urinary Isolates of Klebsiella pneumoniae in Relation to blaSHV, blaTEM and blaCTX-M gene carriage. Iranian journal of public health. 2012;41(3):127. [PMC free article] [PubMed] [Google Scholar]
- 20.Fallah F, Noori M, Hashemi A, Goudarzi H, Karimi A, Erfanimanesh S, Alimehr S. Prevalence of blaNDM, blaPER, blaVEB, blaIMP, and blaVIM genes among Acinetobacter baumannii isolated from two hospitals of Tehran, Iran. Scientifica; 2014. 2014 doi: 10.1155/2014/245162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farajnia S, Azhari F, Alikhani M. Y, Hosseini M. K, Peymani A, Sohrabi N. Prevalence of PER and VEB type extended spectrum betalactamases among multidrug resistant Acinetobacter baumannii isolates in North-West of Iran. Iranian journal of basic medical sciences. 2013;16(6):751–755. [PMC free article] [PubMed] [Google Scholar]
- 22.Farshadzadeh Z, Khosravi A. D, Alavi S. M, Parhizgari N, Hoveizavi H. Spread of extended-spectrum β-lactamase genes of bla OXA-10, bla PER-1 and bla CTX-M in Pseudomonas aeruginosa strains isolated from burn patients. Burns. 2014;40(8):1575–1580. doi: 10.1016/j.burns.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Fazeli H, Sadighian H, Nasr-Esfahani B, Pourmand M. Identification of class-1 integron and various β-lactamase classes among clinical isolates of Pseudomonas aeruginosa at children’s medical center hospital. Journal of Medical Bacteriology. 2015;1(3-4):25–36. [Google Scholar]
- 24.Feizabadi M. M, Mahamadi-Yeganeh S, Mirsalehian A, Mirafshar S. M, Mahboobi M, Nili F, Yadegarinia D. Genetic characterization of ESBL producing strains of Klebsiella pneumoniae from Tehran hospitals. The Journal of Infection in Developing Countries. 2010;4(10):609–615. doi: 10.3855/jidc.1059. [DOI] [PubMed] [Google Scholar]
- 25.Ghafourian S, Sadeghifard N, bin Sekawi Z, Neela V. K, Shamsudin M. N, Mohebi R, Raftari M. Antimicrobial pattern and clonal dissemination of extended-spectrum beta-lactamase producing Klebsiella Spp isolates. American Journal of Infectious Diseases. 2010;6(4):110. [Google Scholar]
- 26.Ghafourian S, Sadeghifard N, Sekawi Z, Neela V. K, Shamsudin M. N, Pakzad I, Rahbar M. Phenotypic and genotypic assay for detection of extended spectrum B-lactamases production by Klebsiella pnemoniae isolates in Emam Reza Hospital in Tabriz, Iran. Journal of Pure and Applied Microbiology. 2011;5(1):1–10. [Google Scholar]
- 27.Ghafouryan S, Sadeghifard N, Neela V, Mariana N. S, Mohebi R, Rahbar M, Pakzad I. Occurrence of Klebsiella oxytoca producing extended-spectrum beta-lactamases in different seasons in Ilam Hospitals, Iran. African Journal of Microbiology Research. 2010;4(22):2381–2387. [Google Scholar]
- 28.Ghasemi Y, Archin T, Kargar M, Mohkam M. A simple multiplex PCR for assessing prevalence of extended-spectrum B-lactamases producing Klebsiella pneumoniae in Intensive Care Units of a referral hospital in Shiraz, Iran. Asian Pacific Journal of Tropical Medicine. 2013:703–708. doi: 10.1016/S1995-7645(13)60122-4. [DOI] [PubMed] [Google Scholar]
- 29.Gholipour A, Soleimani N, Shokri D, Mobasherizadeh S, Kardi M, Baradaran A. Phenotypic and molecular characterization of extended-Spectrum β-lactamase produced by Escherichia coli, and Klebsiella pneumoniae isolates in an educational hospital. Jundishapur journal of microbiology. 2014;7(10) doi: 10.5812/jjm.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghorbani-Dalini S, Kargar M, Doosti A, Abbasi P, Sarshar M. Molecular Epidemiology of ESBL Genes and Multi-Drug Resistance in Diarrheagenic Escherichia coli strains Isolated from Adults in Iran. Iranian journal of pharmaceutical research: IJPR. 2015;14(4):1257. [PMC free article] [PubMed] [Google Scholar]
- 31.Giamarellou H. Multidrug resistance in Gram-negative bacteria that produce extended-spectrum β-lactamases (ESBLs) Clinical Microbiology and infection. 2005;11:1–16. doi: 10.1111/j.1469-0691.2005.01160.x. [DOI] [PubMed] [Google Scholar]
- 32.Golshani Z, Sharifzadeh A. Prevalence of blaOxa10 Type Beta-lactamase Gene in Carbapenemase Producing Pseudomonas aeruginosa Strains Isolated From Patients in Isfahan. Jundishapur Journal of Microbiology. 2013;6(5) [Google Scholar]
- 33.Goudarzi H, Aghamohammad S, Hashemi A, Nikmanesh B, Noori M. Distribution of bla TEM, bla SHV and bla CTX-M genes among Escherichia coli isolates causing urinary tract infection in children. Archives of Clinical Infectious Diseases. 2013;8(3) [Google Scholar]
- 34.Hakemi Vala M, Hallajzadeh M, Hashemi A, Goudarzi H, Tarhani M, Sattarzadeh Tabrizi M, Bazmi F. Detection of Ambler class A, B and D ß-lactamases among Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates from burn patients. Ann Burns Fire Disasters. 2014;27(1):8–13. [PMC free article] [PubMed] [Google Scholar]
- 35.Hasan Afzali F. F, Amiri A, Moniri R, Zibaei M. Characterization of CTX-M-Type Extend-Spectrum β-Lactamase Producing Klebsiella spp. in Kashan, Iran. Jundishapur journal of microbiology. 2015;8(10) doi: 10.5812/jjm.27967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hashemi A, Fallah F, Erfanimanesh S, Hamedani P, Alimehr S, Goudarzi H. Detection of β-lactamases and outer membrane porins among Klebsiella pneumoniae strains isolated in Iran. Scientifica. 2014;2014(6) doi: 10.1155/2014/726179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hemmati T. B, Mehdipour Moghaddam M. J, Salehi Z, Habibzadeh S. M. Prevalence of CTX-M-Type β-Lactamases in Multi-Drug Resistant Escherichia coli Isolates from North of Iran, Rasht. Biological Journal of Microorganism. 2015;3(12) doi: 10.7508/ibj.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosseini-Mazinani S. M, Eftekhar F, Milani M, Ghandili S. Characterization of β-Lactamases from Urinary Isolates of Escherichia coli in Tehran. Iranian Biomedical Journal. 2007;11(2):95–99. [PubMed] [Google Scholar]
- 39.Jin H, Xu X. M, Mi Z. H, Mou Y, Liu P. Drug-resistant gene based genotyping for Acinetobacter bauinannii in tracing epidemiological events and for clinical treatment within nosocomial settings. Chinese Medical Journal (English Edition) 2009;122(3):301. [PubMed] [Google Scholar]
- 40.Kalantar E, Taherzadeh S, Ghadimi T, Soheili F, Salimizand H, Hedayatnejad A. Pseudomonas aeruginosa, an emerging pathogen among burn patients in Kurdistan Province, Iran. Southeast Asian Journal of Tropical Medicine and Public Health. 2012;43(3):712. [PubMed] [Google Scholar]
- 41.Kargar M, Kargar M, Jahromi M. Z, Najafi A, Ghorbani-Dalini S. Molecular detection of ESBLs production and antibiotic resistance patterns in Gram negative bacilli isolated from urinary tract infections. Indian Journal of Pathology and Microbiology. 2014;57(2):244. doi: 10.4103/0377-4929.134688. [DOI] [PubMed] [Google Scholar]
- 42.Khoshvaght H, Haghi F, Zeighami H. Extended spectrum betalactamase producing Enteroaggregative Escherichia coli from young children in Iran. Gastroenterology and Hepatology from bed to bench. 2014;7(2):131. [PMC free article] [PubMed] [Google Scholar]
- 43.Khosravi A. D, Hoveizavi H, Mehdinejad M. Prevalence of Klebsiella pneumoniae encoding genes for CTX-M-1, TEM-1 and SHV-1 extended-spectrum beta lactamases (ESBL) enzymes in clinical specimens. Jundishapur Journal of Microbiology. 2013;6(10) [Google Scholar]
- 44.Kim J. Y, Kim S. H, Jeon S. M, Park M. S, Rhie H. G, Lee B. K. Resistance to fluoroquinolones by the combination of target site mutations and enhanced expression of genes for efflux pumps in Shigella flexneri and Shigella sonnei strains isolated in Korea. Clinical Microbiology and Infection. 2008;14(8):760–765. doi: 10.1111/j.1469-0691.2008.02033.x. [DOI] [PubMed] [Google Scholar]
- 45.Kliebe C, Nies B. A, Meyer J. F, Tolxdorff-Neutzling R. M, Wiedemann B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrobial Agents and Chemotherapy. 1985;28(2):302–307. doi: 10.1128/aac.28.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lasehi K, Shahcheraghi F, Nikbin V. S, Nematzadeh S. PER, CTX-M, TEM and SHV Beta-lactamases in clinical isolates of Klebsiella pneumoniae isolated from Tehran, Iran. Iranian Journal of Basic Medical Sciences. 2010;13(3):111–118. [Google Scholar]
- 47.Leylabadlo H. E, Asgharzadeh M, Aghazadeh M. Dissemination of carbapenemases producing Gram negative bacteria in the Middle East. Iranian journal of microbiology. 2015;7(5):226. [PMC free article] [PubMed] [Google Scholar]
- 48.Leylabadlo H. E, Kafil H. S, Yousefi M, Aghazadeh M, Asgharzadeh M. Persistent infection with metallo-beta-lactamase and extended spectrum β-lactamase producer Morganella morganii in a patient with urinary tract infection after kidney transplantation. Journal of Natural Science, Biology, and Medicine. 2016;7(2):179. doi: 10.4103/0976-9668.184707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Little M. L, Qin X, Zerr D. M, Weissman S. J. Molecular diversity in mechanisms of carbapenem resistance in paediatric Enterobacteriaceae. International journal of antimicrobial agents. 2012;39(1):52–57. doi: 10.1016/j.ijantimicag.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mansouri S, Neyestanaki D. K, Shokoohi M, Halimi S, Beigverdi R, Rezagholezadeh F, Hashemi A. Characterization of AmpC, CTX-M and MBLs types of β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli producing Extended Spectrum β-lactamases in Kerman, Iran. Jundishapur Journal of Microbiology. 2014;7(2) doi: 10.5812/jjm.8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mansouri S, Razavi M, Norouzi F, Gholamhoseinian N. S. Prevalence of β-Lactamase Production and Antimicrobial Susceptibility of Multidrug Resistant Clinical Isolates of Non-Fermenting Gram Negative Bacteria From Hospitalized Patients in Kerman/Iran. Jundishapur J Microbiol. 2012;5(2):405–10. [Google Scholar]
- 52.Mozhdeh R, Fatemeh N, Sasan G. N. Prevalence of β-Lactamase production and antimicrobial susceptibility of multidrug resistant clinical isolates of non-fermenting Gram negative bacteria from hospitalized patients in Kerman/Iran. Jundishapur Journal of Microbiology. 2012;2012(2, Spring):405–410. [Google Scholar]
- 53.Moghadas A. J, Irajian G, Beheshti A. Prevalence of extended-spectrum beta lactamase positive and multidrug resistance pattern of Escherichia coli and Klebsiella pneumoniae isolates, Semnan, Iran. Iranian Journal of Microbiology. 2009;1(1):49–53. [Google Scholar]
- 54.Moghaddam M. J. M, Mirbagheri A. A, Salehi Z, Habibzade S. M. Prevalence of class 1 integrons and extended spectrum beta lactamases among multi-drug resistant Escherichia coli isolates from North of Iran. Iranian biomedical journal. 2015;19(4):233. doi: 10.7508/ibj.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moghaddam M. N, Beidokhti M. H, Jamehdar S. A, Ghahraman M. Genetic properties of blaCTX-M and blaPER β-lactamase genes in clinical isolates of Enterobacteriaceae by polymerase chain reaction. Iranian journal of basic medical sciences. 2014;17(5):378. [PMC free article] [PubMed] [Google Scholar]
- 56.Moghaddam M. N, Forghanifard M. M, Moshrefi S. Prevalence and Molecular Characterization of Plasmid-mediated Extended-Spectrum β-Lactamase Genes (balaTEM, blaCTX and blASHV) Among Urinary Escherichia coli Clinical Isolates in Mashhad, Iran. Iranian journal of basic medical sciences. 2012;15(3):833–839. [PMC free article] [PubMed] [Google Scholar]
- 57.Mohajeri P, Darfarin G, Farahani A. Genotyping of ESBL producing Uropathogenic Escherichia coli in west of Iran. International journal of microbiology. 2014;2014 doi: 10.1155/2014/276941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mood E. H, Meshkat Z, Izadi N, Rezaei M, Jamehdar S. A, Nasab M. N. Prevalence of Quinolone Resistance Genes Among Extended-Spectrum B-Lactamase-Producing Escherichia coli in Mashhad, Iran. Jundishapur journal of microbiology. 2015;8(12) doi: 10.5812/jjm.16217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moosavian M, Deiham B. Distribution of TEM, SHV and CTX-M Genes among ESBL-producing Enterobacteriaceae isolates in Iran. African Journal of Microbiology Research. 2012;6(26):5433–5439. [Google Scholar]
- 60.Mostatabi N, Farshad S, Ranjbar R. Molecular evaluations of extended spectrum β-lactamase producing strains of Serratia isolated from blood samples of the patients in Namazi Hospital, Shiraz, Southern Iran. Iranian journal of microbiology. 2013;5(4):328. [PMC free article] [PubMed] [Google Scholar]
- 61.Nagdeo N. V, Kaore N. M, Thombare V. R. Phenotypic methods for detection of various β-lactamases in Gram-negative clinical isolates: Need of the hour. Chronicles of Young Scientists. 2012;3(4):292. [Google Scholar]
- 62.Nasehi L, Shahcheraghi F, Nikbin V. S, Nematzadeh S. PER, CTX-M, TEM and SHV Beta-lactamases in clinical isolates of Klebsiella pneumoniae isolated from Tehran, Iran. Iranian Journal of Basic Medical Sciences. 2010;13(3):111–118. [Google Scholar]
- 63.NasrolahiOmran A, Javid N, Shakeri F, Yazdi M, Ghaemi E. A. Extended spectrum beta lactamase producing E. coli isolated from Gorgan, North of Iran. Medical Laboratory Journal. 2012;6(1):51–58. [Google Scholar]
- 64.Oliver A, Coque T. M, Alonso D, Valverde A, Baquero F, Cantón R. CTX-M-10 linked to a phage-related element is widely disseminated among Enterobacteriaceae in a Spanish hospital. Antimicrobial agents and chemotherapy. 2005;49(4):1567–1571. doi: 10.1128/AAC.49.4.1567-1571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pakzad I, Ghafourian S, Taherikalani M, Abtahi H, Rahbar M, Mansory Jamshidi N. Qnr prevalence in extended spectrum beta-lactamases (ESBLs) and none-ESBLs producing Escherichia coli isolated from urinary tract infections in central of Iran. Iranian journal of basic medical sciences. 2011;14(5):458–464. [PMC free article] [PubMed] [Google Scholar]
- 66.Parsania S, Sohrabpour M. Prevalence of CTX-M-15β-lactamase Genes in Escherichia coli Strains Using PCR. Journal of Mazandaran University of Medical Sciences. 2014;24(116):58–62. [Google Scholar]
- 67.Paterson D. L, Bonomo R. A. Extended-spectrum β-lactamases: a clinical update. Clinical microbiology reviews. 2005;18(4):657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peerayeh S. N, Eslami M, Memariani M, Siadat S. D. High prevalence of blaCTX-M-1 group extended-spectrum β-lactamase genes in Escherichia coli isolates from Tehran. Jundishapur Journal of Microbiology. 2013;6(7) [Google Scholar]
- 69.Peymani A, Marandi M, Najafipour R. Prevalence of OXA-1 type ESBLs amonge clinical isolates of Entrobacter cloacae collected from Qazvin Hospitals, Iran Iranian. Journal of Public Health. 2014;43(2):84. [Google Scholar]
- 70.Rafiee R, Eftekhar F, Tabatabaei S. A. Prevalence of Extended-Spectrum and Metallo β-Lactamase Production in AmpC β-Lactamase Producing Pseudomonas aeruginosa Isolates From Burns. Jundishapur journal of microbiology. 2014;7(9):e16436–e16436. doi: 10.5812/jjm.16436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramazanzadeh R. Etiologic agents and extended-spectrum beta-lactamase production in urinary tract infections in Sanandaj, Iran. Eastern Journal of Medicine. 2010;15(2):57. [Google Scholar]
- 72.Ranjbar R. E. Z. A, Ahmadnezhad B. A. H. M. A. N, Jonaidi N. E. M. A. T. O. L. L. A. H. The Prevalance of Beta Lactamase Producing Escherichia coli Strains Isolated from the Urine Samples in Valiasr Hospital. Biomed & Pharmacol J. 2014;7:425–431. [Google Scholar]
- 73.Rezai M. S, Salehifar E, Rafiei A, Langaee T, Rafati M, Shafahi K, Eslami G. Characterization of multidrug resistant extended-spectrum beta-lactamase-producing Escherichia coli among uropathogens of pediatrics in North of Iran. BioMed research international. 2015;2015 doi: 10.1155/2015/309478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riyahi Zaniani F, Meshkat Z, Naderi Nasab M, Khaje-Karamadini M, Ghazvini K, Rezaee A, Darban Hoseini M. The prevalence of TEM and SHV genes among extended-spectrum beta-lactamases producing Escherichia coli and Klebsiella pneumoniae. Iranian journal of basic medical sciences. 2012;15(1):654–660. [PMC free article] [PubMed] [Google Scholar]
- 75.Rizi K. S, Peerayeh S. N, Bakhshi B, Rahbar M. Prevalence of the blaCTX-M-1 group and their transferability in resistant clinical isolates of Salmonella serogroups from several hospitals of Tehran. Iranian journal of microbiology. 2015;7(4):203. [PMC free article] [PubMed] [Google Scholar]
- 76.Saeidi S, Alavi-Naini R, Shayan S. Antimicrobial susceptibility and distribution of tem and ctx-m genes among esbl-producing Klebsiella pneumoniae and Pseudomonas aeruginosa causing urinary tract infections. Zahedan Journal of Research in Medical Sciences. 2014;16(4):1–5. [Google Scholar]
- 77.Safari M, Nejad A. S. M, Bahador A, Jafari R, Alikhani M. Y. Prevalence of ESBL and MBL encoding genes in Acinetobacter baumannii strains isolated from patients of intensive care units (ICU) Saudi journal of biological sciences. 2015;22(4):424–429. doi: 10.1016/j.sjbs.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Safari M, Shojapour M, Akbari M, Pourbabaee A, Abtahi H. Dissemination of CTX-M-type beta-lactamase among clinical isolates of Enterobacteriaceae in Markazi province, Iran. Jundishapur Journal of Microbiology. 2013;6(8) [Google Scholar]
- 79.Shacheraghi F, Shakibaie M. R, Noveiri H. Molecular identification of ESBL Genes blaGES-blaVEB-blaCTX-M blaOXA-blaOXA-4, blaOXA-10 andblaPER-in Pseudomonas aeruginosa strains isolated from burn patients by PCR, RFLP and sequencing techniques. Int J Biol life Sci. 2010;3(6):138–42. [Google Scholar]
- 80.Shahandeh Z, Sadighian F, Rekabpou K. B. Phenotypic study of Extended-spectrum beta-lactamase, AmpC and Carbapenemase among E. coli clinical isolates in affiliated hospitals of Babol University of Medical Sciences. International Journal of Health System and Disaster Management. 2015;3(2):74. [Google Scholar]
- 81.Shahcheraghi F, Nasiri S, Noveiri H. Detection of extended-spectrum β-lactamases (ESBLs) Archives of Clinical Infectious Diseases. 2009;4(2):65–70. [Google Scholar]
- 82.Shahcheraghi F, Nikbin V. S, Feizabadi M. M. Prevalence of ESBLs genes among multidrug-resistant isolates of Pseudomonas aeruginosa isolated from patients in Tehran. Microbial Drug Resistance. 2009;15(1):37–39. doi: 10.1089/mdr.2009.0880. [DOI] [PubMed] [Google Scholar]
- 83.Shams E, Firoozeh F, Moniri R, Zibaei M. Prevalence of plasmid-mediated quinolone resistance genes among extended-spectrum β-Lactamase-producing Klebsiella pneumoniae human isolates in Iran. Journal of pathogens. 2015;2015 doi: 10.1155/2015/434391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shams F, Hasani A, Rezaee M. A, Nahaie M. R, Hasani A, Haghi M. H. S. B, Arbatan A. E. Carriage of Class 1 and 2 Integrons in Quinolone, Extended-Spectrum-β-Lactamase-Producing and Multi Drug Resistant E. coli and K. pneumoniae: High Burden of Antibiotic Resistance. Advanced pharmaceutical bulletin. 2015;5(3):335. doi: 10.15171/apb.2015.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sharif M, Mirnejad R, Amirmozafari N. Molecular identification of TEM and SHV extended spectrum β-lactamase in clinical isolates of Acinetobacter baumannii from Tehran hospitals. J Gen Microb Immun. 2014;2:1–9. [Google Scholar]
- 86.Shayan S, Bokaeian M. Detection of ESBL-and AmpC-producing E. coli isolates from urinary tract infections. Advanced biomedical research. 2015:4. doi: 10.4103/2277-9175.166643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tafti A, Zandi H, Vakli M, Mousavi S. M, Zarei M. Frequency of β-lactamase and metallo-β-lactamase in Pseudomonas aeruginosa strains isolated from burn wounds in Yazd burn hospital during 2011-2012. KAUMS Journal (FEYZ) 2014;18(2):167–174. [Google Scholar]
- 88.Tajbakhsh M, Migura L. G, Rahbar M, Svendsen C. A, Mohammadzadeh M, Zali M. R, Hendriksen R. S. Antimicrobial-resistant Shigella infections from Iran: an overlooked problem? Journal of antimicrobial chemotherapy. 2012;67(5):1128–1133. doi: 10.1093/jac/dks023. [DOI] [PubMed] [Google Scholar]
- 89.Tavajjohi Z, Moniri R, Khorshidi A. Detection and characterization of multidrug resistance and extended-spectrum-beta-lactamase-producing (ESBLS) Pseudomonas aeruginosa isolates in teaching hospital. African Journal of Microbiology Research. 2011;5(20):3223–3228. [Google Scholar]
- 90.Vali P, Shahcheraghi F, Seyfipour M, Zamani M. A, Allahyar M. R, Feizabadi M. M. Phenotypic and genetic characterization of carbapenemase and ESBLs producing gram-negative bacteria (GNB) isolated from patients with cystic fibrosis (CF) in Tehran hospitals. Journal of clinical and diagnostic research: JCDR. 2014;8(1):26. doi: 10.7860/JCDR/2014/6877.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yazdi M. K. S, Dallal M. M. S, Mirzael H. M. A, Sabbaghi A, Mehrabadi J. F, Lari A. R, Bakhtiari R. Molecular detection of TEM broad spectrum β-lactamase in clinical isolates of Escherichia coli. African Journal of Biotechnology. 2011;10(46):9454–9458. [Google Scholar]
- 92.Zadegan H. H, Ramazanzadeh R, Hasany A. Cross-sectional study of extended spectrum beta-lactamase producing gram-negative bacilli from clinical cases in Khorramabad, Iran. Iranian Journal of Microbiology. 2009;1(3):16–19. [Google Scholar]
- 93.Zaniani F, Meshkat Z, Naderi Nasab M, Khaje-Karamadini M, Ghazvini K, Rezaee A, Darban Hoseini M. The prevalence of TEM and SHV genes among extended-spectrum beta-lactamases producing Escherichia coli and Klebsiella pneumoniae. Iranian journal of basic medical sciences. 2012;15(1):654–660. [PMC free article] [PubMed] [Google Scholar]
- 94.Zowawi H. M, Balkhy H. H, Walsh T. R, Paterson D. L. β-Lactamase production in key gram-negative pathogen isolates from the Arabian Peninsula. Clinical microbiology reviews. 2013;26(3):361–380. doi: 10.1128/CMR.00096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


