Abstract
Background:
Enterococci are indigenous flora of the gastro-intestinal tracts of animals and humans. Recently, interest in two major species, E. faecium and E. faecalis, has heightened because of their ability to cause serious infections and their intrinsic resistance to antimicrobials. This study was aimed at determining the prevalence of E. faecium and E. faecalis in human faecal samples and evaluating the susceptibility of the isolates to antibiotics.
Materials and Methods:
One hundred faecal samples were collected from apparently healthy individuals and analysed using conventionalbacteriological methods. The susceptibility profile of the isolates to nine antibiotics were determined using disk diffusion method.
Results:
Seventy-three (73) Enterococcus were phenotypically identified and 65 of the isolates were differentiated into 36 (55.4%) E. faecium and 29 (44.6%) E. faecalis. Eight (8) isolates could not be identified by the conventional biochemical methods employed. No dual colonization by the E. faecalis and E. faecium was observed and isolation rate was not dependent on sex of the participants. All the isolates were resistant to ceftriaxone, cefuroxime and ceftizoxime. Enterococcus faecium exhibited resistance toerythromycin (88.9%), gentamicin (77.8%), amoxicillin-clavulanate (63.9%), ofloxacin (44.4%), teicoplanin (19.4%) and vancomycin (16.7%). Enterococcus faecalis showed the least resistance to vancomycin (13.8%) and teicoplanin (27.7%). Remarkable multiple antibiotic resistances to the classes of antibiotic tested were observed among the two species.
Conclusion:
The high carriage rate of antibiotic resistant E. faecium and E. faecalis in this study provides information on the local antibiotic patterns of our enterococci isolates thereby suggesting that they could present as important reservoir and vehicle for dissemination of resistant genes in our community.
Keywords: Enterococcus faecium, Human faecal samples, Enterococcus faecalis, Biochemical identification, Antibiotic resistance
Introduction
Members of the genus Enterococcus, are characterized by individual, paired, or short-chain Gram-positive catalase-negative cocci (Ciftci et al., 2009). Although, ubiquitous, they are primarily localized to the human gut and are found in human faeces, where they represent a minority population (up to 1%) within the gut microflora (Song et al., 2005; Bibalan et al., 2015). Once considered as bacteria of minimal clinical impact, enterococci, particularly Enterococcus faecium and Enterococcus faecalis, have now emerged as one of the major causes of human clinical infections (Silva et al., 2011). They have assumed a vital role as etiologic agent of urinary tract infections, hospital-acquired bloodstream infections, endocarditis, abdominal and pelvic abscesses and chronic periodontitis (Sydnor and Perl, 2011; Qian-Qian et al., 2012). Their importance in such infections is reinforced by their intrinsic and acquired resistance to various antimicrobial agents which renders them difficult to treat.
Enterococcal colonization is a known risk factor for developing associated infections and is most commonly caused by the patients’ own commensal flora (Maschieto et al., 2004). Studies have also confirmed that colonising strains of enterococci serve as reservoir for antibiotic resistance genes which can be transferred among enterococci or acquired by other bacteria (Schjørring and Krogfelt, 2011; Boehm and Sassoubre, 2014). High-density colonization by antibiotic-resistant enterococci increases the risk of infections like bacteremia (Lebreton et al., 2014; Tedim et al., 2015). These infections are difficult to treat; chronic, recurrent and sometimes fatal. Thus, there is a need to assess colonisation of multiple antibiotic-resistant enterococci especially in developing countries where the control of antibiotic use is derisory. In Nigeria, most studies on enterococci have been conducted on food, animals and environmental samples (Oladipo et al., 2013; Olawale et al., 2014; 2015; Ayeni et al., 2016) and a few have examined samples from clinical sources (Iregbu et al., 2002, Olawale et al., 2011; Ekuma et al., 2016). We report here the prevalence of multidrug resistant faecal E. faecium and E. faecalis isolates among apparently healthy humans.
Materials and Methods
Ethical issues and Collection of samples
Verbal consent in collection of samples was sought and obtained from each person after explaining the procedure. The voiding was done by each participant without any form of coercion. They were provided with wide-mouthed, leak-proof sterile plastic containers. One hundred (100) faecal samples were collected from the participants and their sexes were documented. Other demographic data were not available because most of the participants declined to give the required information. The samples were carefully labelled and processed immediately at the Microbiology and Parasitology Laboratory, University of Lagos, Idi-Araba.
Isolation and Biochemical Identification
Sterile inoculating loop was used to pick a small portion of the sample which was then streaked on Bile-esculin agar (Oxoid, UK) plates and incubated at 37°C for 24 hours. Discrete colonies of suspected Enterococcus species were sub-cultured for purity on MacConkey and blood agar plates which were also incubated at 37°C for 24 hours. Species identification was carried out based on Gram stain, cultural characteristics and various biochemical tests including catalase reaction, growth in 6.5% NaCl broth, pyruvate utilizationtest, growth at 45°C and 60°C for 24 hours in a nutrient broth, haemolytic reaction on blood agar and carbohydrate (mannitol, lactose, glucose, sorbitol, raffinose, fructose, dextrose, xylose, trehalose, galactose, dulcitol, and arabinose) fermentation (Facklam et al., 1999). The carbohydrate fermentation tests were performed in agar containing 1% of each sugar.
Antibiotics susceptibility testing
Testing of susceptibility to ceftizoxime (30µg), cefuroxime (30µg), vancomycin (30µg), erythromycin (25µg), gentamicin (10µg), teicoplanin (25µg), ceftriaxone (30µg), ofloxacin (5µg) and amoxicillin-clavulanate (30µg) was performed by disk diffusion method. The results were interpreted in accordance with the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2009). Isolates were adjudged as multidrug resistant (MDR) if resistance to three or more antibiotics of different antimicrobial classes was demonstrated (Magiorakos et al., 2012). Reference strain Enterococcus faecalis ATCC29212 was used as control.
Results
Of the 100 faecal samples analysed, 73 isolates were identified as Enterococcus. The biochemical scheme differentiated 65 of the 73 isolates into E. faecalis (29; 44.6%) and E. faecium (36; 55.4%). The phenotypic characteristics of eight isolates could not be determined through the conventional methods used and were excluded in the overall analyses. All the Enterococcus faecalis isolates fermented sorbitol, mannitol, glucose and lactose but not arabinose while E. faecium was able to ferment arabinose, mannitol, glucose and lactose but not sorbitol (Table 1). The isolation rate was independent of sex (Table 2) and no individuals exhibited colonisation with dual isolates. The isolated strains of E. faecalis showed resistance to ceftriaxone, cefuroxime and ceftizoxime. The overall antimicrobial susceptibility pattern of the isolates isshown in Table 3. A large proportion (96.6%) of the E. faecalis isolates was resistant to gentamicin and erythromycin. Three percent (10.4%) of the isolates were resistant to vancomycin. Similarly, E. faecium significantly exhibited high degree of resistance to gentamicin (88.9%) and erythromycin (91.7%). Enterococcus faecalis showed the least resistance to vancomycin (13.8%) (Figure 1). Marked multiple antibiotic resistances to the classes of antibiotic tested were observed among the two species (Table 4).
Table 1.
Sugar Utilisation Reaction of Enterococcus faecalis and Enterococcus faecium
| No of Positive Isolates (%) | ||
|---|---|---|
| E. faecalis | E. faecium | |
| Arabinose | 0 (0%) | 36 (100%) |
| Sorbitol | 29 (100%) | 0 (0%) |
| Mannitol | 29 (100%) | 36 (100%) |
| Glucose | 29 (100%) | 36 (100%) |
| Lactose | 29 (100%) | 36 (100%) |
| Trehalose | 29 (100%) | 36 (100%) |
| Raffinose | 29 (100%) | 36 (100%) |
| Fructose | 29 (100%) | 36 (100%) |
| Dextrose | 29 (100%) | 36 (100%) |
| Xylose | 0 (0%) | 0 (0%) |
| Galactose | 29 (100%) | 36 (100%) |
| Dulcitol | 0 (0%) | 0 (0%) |
Table 2.
Gender-wise Distribution of Enterococcus faecium and Enterococcus faecalis
| Sex | No of Samples Collected | No of Enterococci Isolated | No of : | |
|---|---|---|---|---|
| E. faecium | E. faecalis | |||
| Male | 50 | 38 | 16 | 15 |
| Female | 50 | 35 | 20 | 14 |
| Total | 100 | 73 | 36 | 29 |
Table 3.
Antimicrobial susceptibility of Enterococcus Isolates to antimicrobial agents
| Antibiotics | E. faecalis | E. faecium | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|
| S (%) | I (%) | R (%) | S (%) | I (%) | R (%) | S (%) | I (%) | R (%) | |
| Vancomycin | 25(86.2) | 1(3.5) | 3(10.4) | 30(83.3) | - | 6(16.7) | 55(84.6) | 1(1.5) | 9(13.8) |
| Teicoplanin | 18(62.1) | - | 11(37.9) | 29(80.6) | - | 7(19.4) | 47(72.3) | - | 18(27.7) |
| Amoxicillin-clavulanate | 10(34.5) | 1(3.5) | 18(62.1) | 9(25.0) | 4(11.1) | 23(63.9) | 19(29.2) | 5(7.7) | 41(63.1) |
| Ofloxacin | 13(44.8) | 3(10.4) | 13(44.8) | 17(47.2) | 3(8.3) | 16(44.4) | 30(46.2) | 6(9.2) | 29(44.6) |
| Erythromycin | 1(3.5) | - | 28(96.6) | 1(2.8) | 3(8.3) | 32(88.9) | 2(3.1) | 3(4.6) | 60(92.3) |
| Gentamicin | 1(3.5) | - | 28(96.6) | 8(22.2) | - | 28(77.8) | 9(13.8) | - | 56(86.2) |
| Ceftriaxone | - | - | 29(100) | - | - | 36(100) | - | - | 65(100) |
| Cefuroxine | - | - | 29(100) | - | - | 36(100) | - | - | 65(100) |
| Ceftizoxime | - | - | 29(100) | - | - | 36(100) | - | - | 65(100) |
Abbreviation: S= Susceptible, I= Intermediate, R= Resistant
Figure 1.
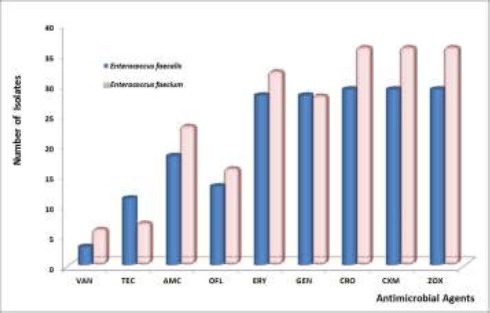
Antimicrobial resistance rates of Enterococcus faecalis and Enterococcus faecium isolates from Apparently Healthy Humans. Abbreviations: VAN-Vancomycin; TEC-Teicoplanin; AMC - Amoxicillin-clavulanate; OFL- Ofloxacin; ERY-Erythromycin; GEN-Gentamicin; CRO - Ceftriaxone; CXM - Cefuroxime; ZOX- Ceftizoxime.
Table 4.
Resistance Rate of the Isolates to Classes of Antimicrobials Tested
| Classes of Antimicrobials | Antimicrobial agent | Species | No. of resistant isolates (%) |
|---|---|---|---|
| Glycopeptide | Vancomycin | E. faecium | 6 (16.67) |
| E. faecalis | 3 (10.34) | ||
| Teicoplanin | E. faecium | 7 (19.44) | |
| E. faecalis | 11 (37.9) | ||
| Beta-lactam inhibitor | Amoxicillin-clavulanate | E. faecium | 23 (63.9) |
| E. faecalis | 18 (62.1) | ||
| Fluoroquinolone | Ofloxacin | E. faecium | 16 (44.4) |
| E. faecalis | 13 (44.8) | ||
| Macrolide | Erythromycin | E. faecium | 32 (88.9) |
| E. faecalis | 28 (96.6) | ||
| Aminoglycoside | Gentamicin | E. faecium | 28 (77.8) |
| E. faecalis | 28 (96.6) | ||
| E. faecium | 36 (100) | ||
| Cephalosporins | Ceftriaxone | E. faecalis | 29 (100) |
| Cefuroxime | E. faecium | 36 (100) | |
| E. faecalis | 29 (100) | ||
| Ceftizoxime | E. faecium | 36 (100) | |
| E. faecalis | 29 (100) |
Discussion
Enterococcal species are found in high concentrations in alimentary tract of humans and animals and may be transferred to other humans through contaminated food animals or the environment (Fisher and Phillips, 2009). Enterococcus faecium and E. faecalis have long been known to be significantly important human pathogens that are specially responsible for nosocomial infections. In this study, E. faecium was the most dominant species with prevalence of 55.4%. This finding is consistent with studies that have established the preponderance of E. faecium in healthy human samples with 44.9% E. faecium and 34.7% E. faecalis (Barreto et al. 2009). A similar distribution of enterococcal species was observed in poultry (Krocko et al., 2007). Enterococcus faecium has also been reported as the most common species in other studies involving chicken, turkey, pork, and beef (Hayes et al., 2003). However, the results presented by authors elsewhere showed a relatively low occurrence of E. faecium compared with E. faecalis among human faecal isolates(Salem-Bekhit et al., 2012). These differences could be attributed to variable host dynamism imposed by gender or sex, diet, age or other environmental conditions. We however found no differences in the frequency of isolation of either E. faecium or E. faecalis between the samples from male and female, suggesting that colonization rate was not sex dependent.
Mostbiochemical identification schemes proposed for enterococci are based on the phenotypic patterns of clinical isolates. Results from previous studies have suggested wide variability in the biochemical profile of E. faecium and E. faecalis isolates from various sources (Facklam and Sahm, 1995, Day et al., 2001). In our case, all the isolates exhibited typical metabolic reaction to key phenotypic characteristics investigated. For instance, the E.faecalis isolates were able to utilize sorbitol unlike the E. faecium strains, contrary to some reports (Teixeira et al., 1995; Castillo-Rojas et al., 2013). Typically, strains of E. faecium would not produce acid from sorbitol but Day et al. (2001) showed that 16 out of 18E. faecium isolates utilized sorbitol. On the otherhand, Facklam and Sahm (1995) speculated that over97% isolates of E. faecium could not utilizesorbitol. Therefore, combined with other biochemical identification criteria, sorbitol fermentation might be a useful marker for the differentiation of E. faecium from E. faecalis isolates in our setting. We, nonetheless, need to confirm this with a larger number of isolates in further study.
In this present communication, very high percentage of resistance to almost all antimicrobial tested were demonstrated by the Enterococcus isolates. The multiple antibiotic resistances of our isolates agree with those found in other studies (Rams et al., 2013;Komiyama et al., 2016). The recovery of MDR colonising strains of E. faecium and E. faecalis highlights the ability to these organisms to develop resistance to an array of antimicrobial drugs. E. faecium were more resistant to most of the antibiotics than E. faecalis, particularly, gentamicin and erythromycin. This is similar to those reported byMengeloğlu and colleagues (Mengeloğlu et al., 2011). However, resistance to erythromycin and gentamycin varies substantially in other studies depending on the setting. Frequencies of 100% and 93.6% erythromycin resistance had been documented for E. faecalis and E. faecium respectively in a trial conducted in Spain (Aarestrup et al., 2002). In another instance, Peters and others observed that 74% of their E. faecalis strains and all their E. faecium strains were moderately or completely resistant to erythromycin (Peters et al., 2003). They also noted that the isolates showed low gentamicin resistance with 1% among E. faecalis isolates and 5% among E. faecium (Peters et al., 2003). The high resistance of our isolates to gentamicin is consistent with findings of some researchers who examined the carriage rate of vancomycin resistant enterococci among patients on prolonged hospitalization in Lagos University Teaching Hospital in Lagos, Nigeria (Ekuma et al., 2016). It has however been suggested that gentamicin resistance in enterococci is caused by the difficulty of penetration of this agent through the cell membrane and the secreting of the enzymes modifying gentamicin as a result of genes acquired by plasmids and transposons (Shepard and Gilmore, 2002).
In this study, the sensitivity pattern observed for ofloxacin (a quinolone) was close to that reported by Mengeloglu et al. (2011). The workers observed that all E. faecium isolates investigated were resistant to ciprofloxacin, but the resistance rate in E. faecalis group was 65%. Both ofloxacin and ciprofloxacin belong to fluoroquinolone class of antimicrobial agents. Fluoroquinolones have been the preferred antibiotics for treatment of Enterococcus and resistance to the agent could have arisen principally from its misuse since antibiotics are readily available over–the–counter in Nigeria. Resistance to several antibiotics has also been reported in other Nigerian studies (Olawale et al., 2011, Ayeni et al., 2016).
Glycopeptides (teicoplanin and vancomycin) are drugs that are rarely sold across the counter. Surprisingly, resistance to vancomycin was exhibited by some of our isolates. The resistance was however, higher in E. faecium (16.7%) than E. faecalis (10.4%). This was in contrast to the reported rate by Aarestrup et al. (2002) where 100% susceptibility to vancomycin in all E. faecalis isolates was recorded. Nevertheless, there was an agreement between our findings and that of Metiner et al. (2013) who observed that 26.6% of the E.faecalis strains isolated from pig faecal samples in Istanbul, Turkey were resistant to vancomycin while 6.4% of E. faecium were resistant to the antibiotic. Similar report from Northwest Ethiopia (Abebe et al., 2014) showed that prior antibiotic treatment was associated with vancomycin resistant enterococci colonization among clients with and without HIV. Although, we are not able to provide information on the use of antibiotics of the participants but expression of resistance is usually favoured by antibiotic misuse which conceivably can cause the emergence of vancomycin resistant isolates. As proposed by Teymournejad and others, expression of resistance genes and selection of strains already expressing these genes may alter the competing microbial flora in the GI tract, thereby increasing vancomycin resistant enterococcal concentration in the stools (Teymournejad et al., 2015). Based on this point, it is apparent that over-use or inappropriate use of this antibioticmay seriously compromise the treatment options for these organisms.
Conclusion
Our data indicate that while gender was not a determinant in Enterococcus carriage, high prevalence of resistant E. faecium and E. faecalis was identified among the individuals investigated. Resistance to gentamicin, erythromycin, ceftriaxone, ceftizoxime and cefuroxime were the most commonly detected among the two species. This infers that these antibiotics may not be suitable candidates for the treatment of enterococcal infections in our environment. We therefore stress the need for development of strategies to stop the sales of antibiotics across the counter and create awareness among the populace by Government and Health Agencies on the consequences of unregulated antibiotic use. More detailed microbiological studies using genotypic methods are warranted.
References
- 1.Aarestrup F. M, Hasman H, Jensen L. B, Moreno M, Herrero I. A, Dominguez L, Finn M, Franklin A. Antimicrobial resistance among enterococci from pigs in three European countries. Appl. Environ. Microbiol. 2002;68:4127–4129. doi: 10.1128/AEM.68.8.4127-4129.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abebe W, Endris M, Tiruneh M, Moges F. Prevalence of vancomycin resistant Enterococci and associated risk factors among clients with and without HIV in Northwest Ethiopia: a cross-sectional study. BMC Public Health 2014. 2014;14:185. doi: 10.1186/1471-2458-14-185. http://www.biomedcentral.com/1471-2458/14/185 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayeni F. A, Odumosu B. T, Oluseyi A. E, Ruppitsch W. Identification and prevalence of tetracycline resistance in enterococci isolated from poultry in IIishan, Ogun State, Nigeria. J. Pharm. Bioall. Sci. 2016;8:69–73. doi: 10.4103/0975-7406.171729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barreto A, Guimaraes B, Radhouani H, Araujo C, Goncalves A, Gaspar E, Rodrigues J, Igrejas G, Poeta P. Detection of antibiotic resistant E. coli and Enterococcus spp. in stool of healthy growing children in Portugal. J. Basic Microbiol. 2009;49:503–512. doi: 10.1002/jobm.200900124. [DOI] [PubMed] [Google Scholar]
- 5.Bibalan M. H, Eshaghi M, Sadeghi J, Asadian M, Narimani T, Talebi M. Clonal Diversity in Multi Drug Resistant (MDR) Enterococci Isolated from Fecal Normal Flora. Int. J. Mol. Cell. Med. 2015;4:240–244. [PMC free article] [PubMed] [Google Scholar]
- 6.Boehm A. B, Sassoubre L. M. Enterococci as indicators of environmental fecal contamination. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: from commensals to leading causes of drug resistant infection. Boston: Massachusetts Eye and Ear Infirmary; 2014. p. 1e17. [PubMed] [Google Scholar]
- 7.Castillo-Rojas G, Mazari-Hirı´art M, Ponce de Leo´n S, Amieva-Ferna´ndez R. I, Agis-Jua´rez R. A, Huebner J, Lo´pez-Vidal Y. Comparison of Enterococcus faecium and Enterococcus faecalis Strains Isolated from Water and Clinical Samples: Antimicrobial Susceptibility and Genetic Relationships. PLoS ONE. 2013;8(4):e59491. doi: 10.1371/journal.pone.0059491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciftci A, Findik A, Ica T, Bas B, Onuk E. E, Gungordu S. Slime production and antibiotic resistance of Enterococcus faecalis isolated from arthritis in chickens. J. Vet. Med. Sci. 2009;71:849–853. doi: 10.1292/jvms.71.849. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. CLSI, 19th International Supplement, M100-S19 2009 [Google Scholar]
- 10.Day A. M, Sandoe J. A, Cove J. H, Phillips-Jones M. K. Evaluation of a biochemical test scheme for identifying clinical isolates of Enterococcus faecalis and Enterococcus faecium. Lett Appl Microbiol. 2001;33(5):392–6. doi: 10.1046/j.1472-765x.2001.01017.x. [DOI] [PubMed] [Google Scholar]
- 11.Ekuma E. A, Oduyebo O. O, Efunshile A. M, Konig B. Surveillance for Vancomycin Resistant Enterococci in a Tertiary Institution in South Western Nigeria. Afr. J. Infect. Dis. 2016;10(2):121–126. doi: 10.21010/ajid.v10i2.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Facklam R. R, Sahm D, Teixeira L. M. Enterococcus. In: Murray P, Baron EJ, Pfaller EA, Tenover FC, Yolke RH, editors. Manual of Clinical Microbiology. Washington, DC: American Society for Microbiology; 1999. pp. 297–305. [Google Scholar]
- 13.Facklam R. R, Sahm D. F. In Manual of Clinical Microbiology. 6th ed. Washington D.C: American Society for Microbiology Press; 1995. Enterococcus; pp. 308–314. [Google Scholar]
- 14.Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiol. 2009;155:1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 15.Hayes J. R, English L. L, Carter P. J. Prevalence and antimicrobial resistance of Enterococcus species isolated from retail meats. Appl. Environ. Microbiol. 2003;12:7153–7160. doi: 10.1128/AEM.69.12.7153-7160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iregbu K. C, Ogunsola F. T, Odugbemi T. O. Susceptibility profile of Enterococcus faecalis isolated at the Lagos University Teaching Hospital, Nigeria. Nig. Postgrad. Med. J. 2002;9:125–128. [PubMed] [Google Scholar]
- 17.Komiyama E. Y, Lepesqueur L. S. S, Yassuda C. G, Samaranayake L. P, Parahitiyawa N. B, Balducci I, Koga-Ito C. Y. Enterococcus Species in the Oral Cavity: Prevalence, Virulence Factors and Antimicrobial Susceptibility. PLoS ONE. 2016;11(9):e0163001. doi: 10.1371/journal.pone.0163001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krocko M, Canigova M, Duckova V. Occurrence, isolation and antibiotic resistance of Enterococcus species isolated from raw pork, beef and poultry. J. Food Nutr. Res. 2007;46:91–95. [Google Scholar]
- 19.Lebreton F, Willems R. J. L, Gilmore M. S. Enterococcus diversity, origins in nature, and gut colonization. In: InGilmore M. S, Clewell D. B, Ike Y, Shankar N, editors. Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary. Boston, MA: 2014. Available at http://www.ncbi.nlm.nih.gov/books/NBK190427/ [PubMed] [Google Scholar]
- 20.Magiorakos A. P, Srinivasan A, Carey R. B, Carmeli Y, Falagas M. E, Giske C. G, Harbarth S, Hindler J. F, Kahlmeter G, Olsson-Liljequist B, Paterson D. L, Rice L. B, Stelling J, Struelens M. J, Vatopoulos A, Weber J. T, Monnet D. L. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18(3):268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 21.Maschieto A, Martinez R, Palazzo I. C. Z, Darini A. Antimicrobial Resistance of Enterococcus sp. Isolated from the Intestinal Tract of Patients from a University Hospital in Brazil. Mem. Inst. Oswaldo Cruz, Rio de Janeiro. 2004;99(7):763–767. doi: 10.1590/s0074-02762004000700018. [DOI] [PubMed] [Google Scholar]
- 22.Mengeloğlu F. Z, Çakır D, Terzi H. A. Comparison of resistance in isolates of Enterococcus faecalis and Enterococcus faecium. J. Microbiol. Infect. Dis. 2011;1(1):10–1. [Google Scholar]
- 23.Metiner K, Küçüker M. A, Boral O. Z, Ang O. First isolation of Enterococcus strains in pig faeces in Turkey and determination of antibiotic susceptibilities. Acta Vet. Brno. 2013;82:231–235. [Google Scholar]
- 24.Oladipo I. C, Sanni A, Swarnakar S. Phenotypic and Genomic Characterization of Enterococcus Species from Some Nigerian Fermented Foods. Food Biotechnol. 2013;27(1):39–53. [Google Scholar]
- 25.Olawale A. K, David O. M, Oluyege A. O, Osuntoyinbo R. T, Laleye S. A, Famurewa O. Histopathological changes induced in an animal model by potentially pathogenic Enterococcus faecalis strains recovered from ready-to-eat food outlets in Osun State, Nigeria. Infect. Drug Resist. 2015;8:181–187. doi: 10.2147/IDR.S61381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olawale A. K, Onasanya A, Oyelakin O. O, David O. M, Famurewa O. Enterococcus faecalis isolates of food origin and detection of their virulence determinant factors and genes in Osun State, Nigeria. Microbiol. Res. Int. 2014;2(2):18–27. [Google Scholar]
- 27.Olawale K. O, Fadiora S. O, Taiwo S. S. Prevalence of hospital-acquired Enterococci infections in two primary-care hospitals in Osogbo, Southwestern Nigeria. Afr. J. Infect. Dis. 2011;5(2):40–46. doi: 10.4314/ajid.v5i2.66513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters J, Mac K, Wichmann-Schauer H, Klein G, Ellerbroek L. Species distribution and antibiotic resistance patterns of enterococci isolated from food of animal origin in Germany. Int. J. Food Microbiol. 2003;1:311–4. doi: 10.1016/s0168-1605(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 29.Qian-Qian W, Zhang C, Chu C, Zhu X. Prevalence of Enterococcus faecalis in saliva and filled root canals of teeth associated with apical periodontitis. Int. J. Oral Sci. 2012;4:19–23. doi: 10.1038/ijos.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rams T. E, Feik D, Mortensen J. E, Degener J. E, van Winkelhoff A. J. Antibiotic Susceptibility of Periodontal Enterococcus faecalis. J. Periodontol. 2013;84(7):1026–1033. doi: 10.1902/jop.2012.120050. [DOI] [PubMed] [Google Scholar]
- 31.Salem-Bekhit M. M, Moussa I. M, Muharram M. M, Alanazy F. K, Hefni H. M. Prevalence and antimicrobial resistance pattern of multidrug-resistant enterococci isolated from clinical specimens. Indian J Med Microbiol. 2012;30(1):44–51. doi: 10.4103/0255-0857.93032. [DOI] [PubMed] [Google Scholar]
- 32.Schjørring S, Krogfelt K. A. Assessment of Bacterial Antibiotic Resistance Transfer in the Gut. Int. J. Microbiol. 2011;2011 doi: 10.1155/2011/312956. ID 312956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shepard B. D, Gilmore MS. Antibiotic-resistant enterococci: the mechanisms and dynamics of drug introduction and resistance. Microbes Infect. 2002;4:215–224. doi: 10.1016/s1286-4579(01)01530-1. [DOI] [PubMed] [Google Scholar]
- 34.Silva N, Igrejas G, Gonçalves A, Poeta P. Commensal gut bacteria: distribution of Enterococcus species and prevalence of Escherichia coli phylogenetic groups in animals and humans in Portugal. Annals Microbiol. 2011;62:449–459. [Google Scholar]
- 35.Song J. Y. I, Hwang S, Eom J. S, Cheong H. J, Bae W. K, Park Y. H, Kim W. J. Prevalence and molecular epidemiology of vancomycin-resistant enterococci (VRE) strains isolated from animals and humans in Korea. Korean J. Intern. Med. 2005;20:55–62. doi: 10.3904/kjim.2005.20.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sydnor E. R, Perl T. M. Hospital epidemiology and infection control in acute-care settings. Clin. Microbiol. Rev. 2011;24:141–173. doi: 10.1128/CMR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tedim A. P, Garbajosa P. R, Corander J, Rodríguez C. M, Cantón R, Willems R. J, Baquero F, Coque T. M. Population biology of intestinal Enterococcus isolates from hospitalized and non-hospitalized individuals in different age groups. Appl. Environ. Microbiol. 2015;81(5):1820–1831. doi: 10.1128/AEM.03661-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teixeira L. M, Facklam R. R, Steigerwalt A. G, Pigott N. E, Merquior V. L. C, Brenner D. J. Correlation between phenotypic characteristics and DNA relatedness within Enterococcus faecium strains. J. Clin. Microbiol. 1995;33:1520–1523. doi: 10.1128/jcm.33.6.1520-1523.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teymournejad O, Mobarez A. M, Doust R. H, Yaslianifard S. Prevalence of VanA and B Genotype among Vancomycin Low Resistant Enterococcus in Fecal Normal Flora and Clinical Samples Isolated from Tehran’s Hospitals. Int. J. Enteric Pathog. 2015;3(1):e22254. [Google Scholar]


