Abstract
In this study, New Delhi metallo-beta-lactamase (NDM)-producing Enterobacteriaceae were identified in Irish recreational waters and sewage. Indistinguishable NDM-producing Escherichia coli by pulsed-field gel electrophoresis were isolated from sewage, a fresh water stream and a human source. NDM-producing Klebsiella pneumoniae isolated from sewage and seawater in the same area were closely related to each other and to a human isolate. This raises concerns regarding the potential for sewage discharges to contribute to the spread of carbapenemase-producing Enterobacteriaceae.
Keywords: antimicrobial resistance, carbapenemase-producing Enterobacteriaceae, New Delhi metallo-beta-lactamase, recreational waters, wastewater
We report the finding of New Delhi metallo-beta-lactamase (NDM)-producing Enterobacteriaceae in fresh water and seawater samples collected at two beaches located near an untreated human sewage ocean discharge. Isolates of NDM-producing Escherichia coli derived from the sewage collection system, the sewage storage tank and the outflow were 100% identical by pulsed-field gel electrophoresis (PFGE) to those derived from a fresh water stream on one of the beaches, and to a clinical isolate.
Recreational water and sewage sample sites
In 2016, we identified a beach (Beach A) in Ireland, used for bathing and recreation, which is crossed by two fresh water streams (Stream A and Stream B), flowing from the surrounding countryside. These streams were examined for the presence of carbapenemase-producing Enterobacteriaceae (CPE). The detection of NDM-producing E. coli in these waters prompted subsequent additional sampling of the streams. As untreated human sewage was being discharged into the sea in the vicinity of the beach, and the fresh water streams can become immersed in seawater at high tide, sewage was evaluated as a potential source. Sewage samples included samples from the collection system, the storage tank and the outflow. Sampling was performed in the period May to September, 2016. The sewage system is not linked to any hospital or long-term care facility that we are aware of. Further sampling of the fresh water streams and sewage sites was carried out in January 2017. In addition to this, seawater from Beach A and from a second beach (Beach B), ca 950 m in a direct line from Beach A were examined. Figure 1 shows a schematic diagram of the sampling points and their location relative to each other.
Figure 1.
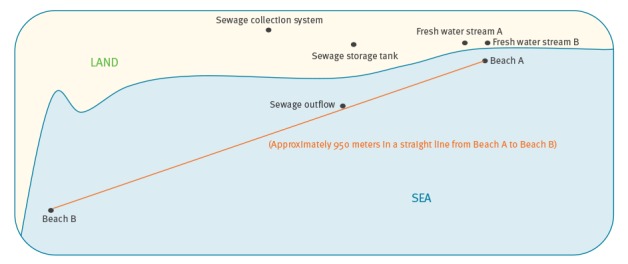
Schematic diagram of water and sewage sampling points and their location relative to each other, Ireland, 2016–2017
Processing of samples
We applied a previously described method (CapE), to examine large volumes of water (30L) from both the fresh water streams and the seawater, for the presence of CPE [1]. Following filtration and overnight enrichment, the samples were sub-cultured onto Brilliance CRE agar (Oxoid). Sewage samples were examined by direct plating onto Brilliance CRE agar. Following purification, presumptive isolates were identified to species level by matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry, and antimicrobial susceptibility testing was performed and interpreted in accordance with European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria [2]. Carbapenemase-encoding genes were detected by real-time PCR, as previously described [3-6]. Typing of NDM-producing Enterobacteriaceae was not performed at the variant level, but PFGE was performed on all isolates, as outlined previously [7]. PFGE profiles of NDM-producing Enterobacteriaceae isolated from recreational water and sewage samples were compared with PFGE profiles of NDM-producing Enterobacteriaceae isolated from clinical specimens.
Findings of New Delhi metallo-beta-lactamase (NDM)-producing Enterobacteriaceae
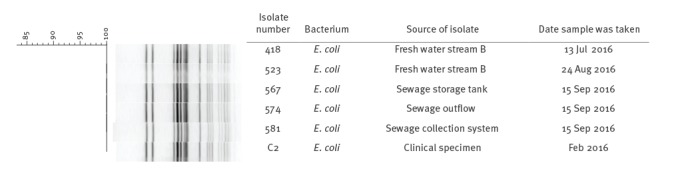
Of eight fresh water samples from Stream B, NDM-producing E. coli were isolated from two samples, which were collected on 13 July and 24 August 2016. NDM-producing E. coli were also isolated in samples collected on 15 September 2016 from the sewage collection system (one of two samples), the storage tank (one of two samples) and the sewage outflow (one of one sample) (Table). All isolates were resistant to ampicillin, cefotaxime, cefoxitin, cefpodoxime, ceftazidime, ciprofloxacin, ertapenem, meropenem and nalidixic acid. The isolates obtained from the fresh water and sewage samples are indistinguishable by PFGE analysis from a human isolate submitted to the National Carbapenemase Producing Enterobacteriaceae Reference Laboratory Service (CPERLS) in early 2016 (Figure 2).
Table. Overview of sampling sites, dates and detection of carbapenemase-producing Enterobacteriaceae in a coastal region in Ireland, 2016–2017.
| Sample site | Date of sampling | Carbapenemase-producing Enterobacteriaceae |
|---|---|---|
| Fresh water Stream A | 25 May 2016 | Not detected |
| 22 Jun 2016 | Not detected | |
| 13 Jul 2016 | Not detected | |
| 10 Aug 2016 | Not detected | |
| 24 Aug 2016 | Not detected | |
| 7 Sep 2016 | Not detected | |
| 15 Sep 2016 | Not detected | |
| 18 Jan 2017 | Not detected | |
| Fresh water Stream B | 25 May 2016 | Not detected |
| 22 Jun 2016 | Not detected | |
| 13 Jul 2016 | NDM-producing E. coli | |
| 10 Aug 2016 | Not detected | |
| 24 Aug 2016 | NDM-producing E. coli | |
| 7 Sep 2016 | Not detected | |
| 15 Sep 2016 | Not detected | |
| 18 Jan 2017 | Not detected | |
| Sewage storage tank | 15 Sep 2016 | NDM-producing E. coli |
| NDM-producing K. pneumoniae | ||
| 18 Jan 2017 | NDM-producing K. pneumoniae | |
| Sewage collection system | 15 Sep 2016 | NDM-producing E. coli |
| NDM-producing K. pneumoniae | ||
| 18 Jan 2017 | NDM-producing K. pneumoniae | |
| Sewage outflow | 15 Sep 2016 | NDM-producing E. coli |
| Seawater Beach A | 18 Jan 2017 | NDM-producing K. pneumoniae |
| Seawater Beach B | 18 Jan 2017 | NDM-producing K. pneumoniae |
E. coli: Escherichia coli; K. pneumoniae: Klebsiella pneumoniae; NDM: New Delhi metallo-beta-lactamase.
Figure 2.
PFGE analysis of New Delhi metallo-beta-lactamase-producing Escherichia coli isolated from fresh water, sewage and a clinical source in Ireland, 2016–2017 (n = 6 isolates)
PFGE: pulsed-field gel electrophoresis.
PFGE typing was performed with Xba1. All isolates are 100% similar by PFGE.
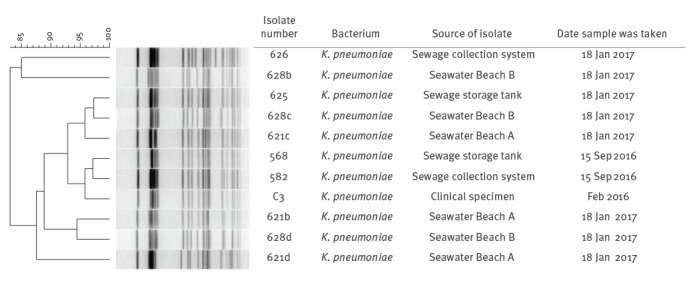
NDM-producing K. pneumoniae were isolated from two of three sewage sampling sites on 15 September 2016 and two of two sewage sampling sites on 18 January 2017. NDM-producing K. pneumoniae was also detected in seawater samples collected at Beach A and Beach B on 18 January 2017 (Table). These isolates were resistant to ampicillin, cefotaxime, cefoxitin, cefpodoxime, ceftazidime, ciprofloxacin, ertapenem, gentamicin, kanamycin, meropenem, nalidixic acid and tetracycline. PFGE analyses of isolates from sewage, seawater and a human isolate (from CPERLS) show isolates to be between 83% and 97% similar (Figure 3).
Figure 3.
PFGE analysis of New Delhi metallo-beta-lactamase-producing Klebsiella pneumoniae isolated from seawater, sewage and a clinical source in Ireland, 2016–2017 (n = 11 isolates)
PFGE: pulsed-field gel electrophoresis.
PFGE typing was performed with Xba1. Isolates are between 83% and 97% similar by PFGE.
Discussion
The rapid dissemination of carbapenemase-producing Enterobacteriaceae (CPE) in Europe and worldwide is making the delivery of effective healthcare an increasing challenge [8]. The number of CPE confirmed by the national reference laboratory in humans in Ireland has increased every year, rising from 48 in 2013, to 369 in 2016. In 2016, the three most commonly reported carbapenemases in Ireland were K. pneumoniae carbapenemase (KPC), carbapenem-hydrolysing oxacillinase-48 (OXA-48), and NDM [9]. The NDM gene is primarily plasmid-encoded, enabling its easy transfer between bacterial species. The plasmids are diverse and usually harbour a large number of other resistance genes [10]. The NDM gene has been detected extensively in the Indian subcontinent where it has been reported from both environmental and clinical sources [11]. Rapid global spread has been aided greatly by intercontinental travel [12]. However, a recent study in 2016 reported an outbreak of NDM-1-producing Enterobacteriaceae in a number of hospitals in Ireland, where links to foreign travel were not identified [13].
In Europe, a number of studies have reported the presence of CPE in recreational water, including Verona integron-encoded metallo-beta-lactamase (VIM) producing K. pneumoniae in a river in Switzerland in 2013 [14], KPC-producing E. coli in a river in Portugal in 2012 [15], VIM-1, VIM-34, and IMP-type metallo-beta-lactamase (IMP)-8 producing E. coli in the same Portuguese river in 2016 [16], and NDM-1 producing K. pneumoniae in the River Danube in Serbia, in 2016 [17]. Here we identify NDM-producing Enterobacteriaceae in environmental water samples collected at two adjacent beach sites in Ireland. As far as we are aware, this is the first such finding in bathing seawater in Europe.
We consider that contamination of the environment with NDM-producing Enterobacteriaceae from the human sewage outflow is likely to be the source, and that the fresh water streams were contaminated by backwash of sewage onto the beach by tidal currents. The presence of NDM-producing Enterobacteriaceae in the bathing water (seawater) and at a separate bathing site ca 950 m in a direct line indicates the extent of this contamination. It is important to note that by the established regulatory standards, the bathing water quality in the area concerned has been consistently of sufficient quality [18]. Notwithstanding compliance with regulatory standards however, it is reasonable to conclude that those using a beach such as this for recreational purposes might be at least intermittently exposed to NDM-producing Enterobacteriaceae. Although, to date, there is no evidence that NDM-producing Enterobacteriaceae has been acquired as a result of exposure to this beach environment, Leonard et al. have recently reported on the level of risk of exposure to antibiotic resistant bacteria in coastal waters and its relationship to different types of water sports [19].
It appears therefore that there is potential for environmental contamination to contribute to a transition of CPE from largely healthcare-associated organisms, to organisms affecting the general population and the veterinary sector. From a public health perspective, the findings focus attention on the need to accelerate programmes to cease discharge of untreated sewage into the environment. This practice should be unacceptable in the context of discharges in the vicinity of popular bathing and recreation areas where human exposure is highly likely.
We consider that our findings point to potential limitations of the use of E. coli as an indicator bacteria for bathing water quality based on the number of colony forming units (CFU) per 100 mL [20]. In our view, this approach does not reflect the pathogenicity of some variants of E. coli, such as Shiga-toxigenic E. coli for which the infectious dose is very low, (<10 CFU/mL) [21].
Conflict of interest: None declared.
Authors’ contributions: Sampling was carried out by Bláthnaid Mahon, Carina Brehony, Dearbháile Morris, Paul Hickey and Shane Keane. Ann Dolan facilitated gaining access to the sewage supply. Water samples were filtered by Carina Brehony, James Killeen and Bláthnaid Mahon. Screening of samples for CPE was carried out by Bláthnaid Mahon. MALDI-TOF, antimicrobial susceptibility testing and PFGE was completed out by Bláthnaid Mahon, and real-time PCR by Elaine McGrath. Elaine McGrath and Belinda Hanahoe provided access to the clinical isolates. Bláthnaid Mahon, Dearbháile Morris and Martin Cormican were responsible for the study design and coordination, and wrote the manuscript. All authors were involved in reviewing the manuscript and approved the final version.
References
- 1. Morris D, Kavanagh S, Carney K, MacDomhnaill B, Cormican M. CapE (capture, amplify, extract): A rapid method for detection of low level contamination of water with Verocytotoxigenic Escherichia coli (VTEC). Sci Total Environ. 2016;563-564:267-72. 10.1016/j.scitotenv.2016.04.075 [DOI] [PubMed] [Google Scholar]
- 2.The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Växjö: EUCAST. Version 7.0; 2017. Available from: http://www.eucast.org
- 3. Swayne RL, Ludlam HA, Shet VG, Woodford N, Curran MD. Real-time TaqMan PCR for rapid detection of genes encoding five types of non-metallo- (class A and D) carbapenemases in Enterobacteriaceae. Int J Antimicrob Agents. 2011;38(1):35-8. 10.1016/j.ijantimicag.2011.03.010 [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC). Multiplex real-time PCR detection of klebsiella pneumoniae carbapenemase (KPC) and new delhi metallo-β-lactamase (NDM-1) genes. Atlanta: CDC. 2011;6-7. [Google Scholar]
- 5. Birkett CI, Ludlam HA, Woodford N, Brown DFJ, Brown NM, Roberts MTM, et al. Real-time TaqMan PCR for rapid detection and typing of genes encoding CTX-M extended-spectrum β-lactamases. J Med Microbiol. 2007;56(Pt 1):52-5. 10.1099/jmm.0.46909-0 [DOI] [PubMed] [Google Scholar]
- 6. Geyer CN, Reisbig MD, Hanson ND. Development of a TaqMan multiplex PCR assay for detection of plasmid-mediated ampC β-lactamase genes. J Clin Microbiol. 2012;50(11):3722-5. 10.1128/JCM.02038-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV, CDC PulseNet Task Force PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis. 2001;7(3):382-9. 10.3201/eid0703.017303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Albiger B, Glasner C, Struelens MJ, Grundmann H, Monnet DL, European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) working group Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro Surveill. 2015;20(45):30062. 10.2807/1560-7917.ES.2015.20.45.30062 [DOI] [PubMed] [Google Scholar]
- 9.McGrath E, Cormican M. CPEaRLS ANNUAL REPORT 2015, National Carbapenemase Producing Enterobacteriaceae (CPE) Reference Laboratory Service; 2015.1-26. Available from: https://www.researchgate.net/publication/307601382_Annual_Report_2015_Carbapenemase_Producing_Enterobacteriaceae_Reference_Laboratory_of_Ireland
- 10. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791-8. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bushnell G, Mitrani-Gold F, Mundy LM. Emergence of New Delhi metallo-β-lactamase type 1-producing enterobacteriaceae and non-enterobacteriaceae: global case detection and bacterial surveillance. Int J Infect Dis. 2013;17(5):e325-33. 10.1016/j.ijid.2012.11.025 [DOI] [PubMed] [Google Scholar]
- 12. Berrazeg M, Diene S, Medjahed L, Parola P, Drissi M, Raoult D, et al. New Delhi Metallo-beta-lactamase around the world: an eReview using Google Maps. Euro Surveill. 2014;19(20):20809. 10.2807/1560-7917.ES2014.19.20.20809 [DOI] [PubMed] [Google Scholar]
- 13. O’Connor C, Cormican M, Boo TW, McGrath E, Slevin B, O’Gorman A, et al. An Irish outbreak of New Delhi metallo-β-lactamase (NDM)-1 carbapenemase-producing Enterobacteriaceae: increasing but unrecognized prevalence. J Hosp Infect. 2016;94(4):351-7. 10.1016/j.jhin.2016.08.005 [DOI] [PubMed] [Google Scholar]
- 14. Zurfluh K, Hächler H, Nüesch-Inderbinen M, Stephan R. Characteristics of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae Isolates from rivers and lakes in Switzerland. Appl Environ Microbiol. 2013;79(9):3021-6. 10.1128/AEM.00054-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poirel L, Barbosa-Vasconcelos A, Simões RR, Da Costa PM, Liu W, Nordmann P. Environmental KPC-producing Escherichia coli isolates in Portugal. Antimicrob Agents Chemother. 2012;56(3):1662-3. 10.1128/AAC.05850-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kieffer N, Poirel L, Bessa LJ, Barbosa-Vasconcelos A, da Costa PM, Nordmann P. VIM-1, VIM-34, and IMP-8 Carbapenemase-Producing Escherichia coli Strains Recovered from a Portuguese River. Antimicrob Agents Chemother. 2016;60(4):2585-6. 10.1128/AAC.02632-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kittinger C, Lipp M, Folli B, Kirschner A, Baumert R, Galler H, et al. Enterobacteriaceae isolated from the River danube: Antibiotic resistances, with a focus on the presence of ESBL and carbapenemases. PLoS One. 2016;11(11):e0165820. 10.1371/journal.pone.0165820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Environmental Protection Agency (EPA). Bathing water quality in Ireland 2015. EPA; 2016. Available from: https://www.epa.ie/pubs/reports/water/bathing/BW_Report_2015.pdf
- 19. Leonard AFC, Zhang L, Balfour AJ, Garside R, Gaze WH. Human recreational exposure to antibiotic resistant bacteria in coastal bathing waters. Environ Int. 2015;82:92-100. 10.1016/j.envint.2015.02.013 [DOI] [PubMed] [Google Scholar]
- 20. Edberg SC, Rice EW, Karlin RJ, Allen MJ. Escherichia coli: the best biological drinking water indicator for public health protection. Symp Ser Soc Appl Microbiol. 2000;88(29):106S-16S. 10.1111/j.1365-2672.2000.tb05338.x [DOI] [PubMed] [Google Scholar]
- 21. Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26(4):822-80. 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]


