Abstract
Bacterial survival requires an intact peptidoglycan layer, a 3-dimensional exoskeleton that encapsulates the cytoplasmic membrane. Historically, the final steps of peptidoglycan synthesis are known to be carried out by d,d-transpeptidases, enzymes that are inhibited by the β-lactams which constitute >50% of all antibacterials in clinical use. Here, we show that the carbapenem subclass of β-lactams is distinctly effective not only because they inhibit d,d-transpeptidases and are poor substrates for β-lactamases, but primarily because they also inhibit non-classical transpeptidases, namely the l,d-transpeptidases, that generate the majority of linkages in the peptidoglycan of mycobacteria. We have characterized the molecular mechanisms responsible for inhibition of l,d-transpeptidases of M. tuberculosis and a range of bacteria, including ESKAPE pathogens, and utilized this information to design, synthesize and test simplified carbapenems with potent antibacterial activity.
INTRODUCTION
Decades-old drugs comprise the standard regimens available today for the treatment of tuberculosis (TB), a disease caused by infection with Mycobacterium tuberculosis. Despite being declared a global health priority by the World Health Organization more than twenty years ago, TB currently causes more deaths worldwide than any other infectious disease 1. The complexity and length of TB treatment, involving the daily administration of multiple drugs for at least six months, precludes its successful implementation in many regions of the world. This failure fuels the development of drug-resistant TB, further exacerbating this already serious global public health problem. Drug regimens that can reduce the complexity and duration of treatment and that also have activity against drug-resistant M. tuberculosis could significantly impact the control of TB and alleviate the suffering of patients with this disease. The development of new anti-TB drugs, including the repurposing and/or improvement of existing drugs, is thus of critical importance.
The β-lactam antibacterials, which target peptidoglycan biosynthesis and are overall the most widely utilized class of antibacterials for the treatment of infections, have traditionally not been effective in TB treatment 2. This lack of activity against M. tuberculosis has primarily been attributed to the presence of the chromosomally-encoded β-lactamase enzyme BlaC, which hydrolyzes the core β-lactam ring to deactivate the drug 3 and also to the potential for limiting penetration of the thick mycolate coat 4. However, recently a number of studies have suggested that a subset of β-lactams, specifically the carbapenems as well as the penem faropenem, may have anti-TB activity 5–8. In addition to being relatively more resistant to β-lactamase activity, these types of β-lactams are unique in that they target more than one enzyme involved in bacterial peptidoglycan synthesis. Most β-lactams inhibit d,d-transpeptidases, commonly known as penicillin-binding proteins; these enzymes catalyze the formation of 4→3 transpeptide linkages in the peptidoglycan network. Emerging evidence suggests that carbapenems target not only the d,d-transpeptidases, but also the non-canonical l,d-transpeptidases, which catalyze the formation of 3→3 transpeptide linkages 5,9,10. This latter function may be key to the anti-TB activity observed with faropenem and carbapenems as M. tuberculosis primarily uses l,d-transpeptidases to cross-link its peptidoglycan 11,12, and mutants lacking the l,d-transpeptidases LdtMt1 and LdtMt2 are attenuated in virulence, have modified peptidoglycan and altered cell physiology and morphology 9,13. A better understanding of how these types of β-lactam antibacterials act against M. tuberculosis may therefore lead to structural improvements for their use as anti-TB drugs.
Here, we have characterized the inhibitory interactions using biochemical and biophysical approaches, including solving multiple crystal structures, of faropenem and carbapenems with the M. tuberculosis l,d-transpeptidases LdtMt1 and LdtMt2 and validated carbapenem anti-TB activity in vivo in a preclinical mouse model of TB treatment. Based on these data, we were able to analyze structure-activity relationships and initiate the synthesis of evolved carbapenems with potent activity against M. tuberculosis. In addition, we identified putative l,d-transpeptidases from other bacteria, including ESKAPE pathogens (organisms associated with severe, difficult-to-treat nosocomial infections), and characterized the inhibitory interactions of faropenem and carbapenems, including our test series of evolved carbapenems, with these enzymes. Thus, our data not only provide critical insights into maximizing the anti-TB activity of carbapenems, but also for wider use to improve their therapeutic potential as antibacterial agents.
RESULTS
Interactions between l,d-transpeptidases and β-lactams
Our group and others have previously demonstrated that faropenem and the carbapenems doripenem, biapenem, and tebipenem exhibit in vitro bactericidal activity against M. tuberculosis, even without co-exposure of the bacteria to a β-lactamase inhibitor 6,7. To identify the β-lactam that is most reactive with l,d-transpeptidases, we incubated the M. tuberculosis LdtMt1 and LdtMt2 enzymes with equimolar mixtures of faropenem, doripenem, biapenem and tebipenem and assessed the identities and abundance of acyl-enzyme adducts (created when the β-lactam ring reacts with the active site cysteine) using ultra performance liquid chromatography-mass spectrometry (UPLC-MS). Although each carbapenem produced a unique adduct when reacted individually with the l,d-transpeptidases, acylation by faropenem was the only adduct detected in the competition assays with the carbapenem mixture, indicating that the LdtMt1 and LdtMt2 enzymes preferentially bound faropenem over the other tested carbapenems (Table 1). As a control, we also reacted these enzymes with equimolar mixtures of faropenem and the non-carbapenem β-lactams amoxicillin, cephalothin, and aztreonam. In the control mixture, the faropenem adduct (+86 Da) was the only acyl-adduct detected, confirming that the M. tuberculosis l,d-transpeptidases preferentially bound faropenem over non-carbapenem β-lactams (Table 1). Finally, the faropenem adduct was again the only species detected when we reacted an equimolar mixture of oxacillin, cephalexin and faropenem with LdtMt2 or a penicillin binding protein DacB2 (Supplementary Results, Supplementary Table 1). Additionally, while carbenicillin failed to bind to LdtMt2 it formed a +379 Da adduct with DacB2. These l,d-transpeptidase/β-lactam interactions were also evaluated by isothermal titration calorimetry (ITC). While enzyme inactivation in these cases is irreversible, we take the absence of ITC response as evidence of no detectable binding and inactivation, whereas a positive response would reflect the sum of binding and subsequent chemical steps. Using this method, while tebipenem and faropenem exhibited single-modality binding to LdtMt1 or LdtMt2 with high affinity, the non-(carba)penem β-lactams either did not bind at all or interacted with reduced affinity to the enzymes (Fig. 1a, Supplementary Fig. 1, Supplementary Table 2).
Table 1.
UPLC-MS analysis of adducts formed between l,d-transpeptidases and β-lactams.
| Enzyme | Molecular weight (Da) of adducts formed with the following β-lactams | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Amoxicillin | Cephalothin | Doripenem | Faropenem | A-C-Z-F | D-B-F-Te | T205 | T206 | T208 | T210 | |
| LdtMt1 | No adduct | No adduct | +376, +421 | +86 | +86 | +86 | +316 | +344 | +385 | +258 |
| LdtMt2 | No adduct | +335 | +123 | +86 | +86 | +86 | +337 | +344 | +383 | +257 |
| LdtMab1 | +366 | +337, +355 | +376, +421 | +86 | +86 | +86 | +337 | +344 | +383 | +258 |
| LdtMab2 | No adduct | No adduct | +68 | +86 | +86 | +86 | No adduct | No adduct | No adduct | No adduct |
| LdtKp | No adduct | No adduct | +376, +421 | +86, +288 | +86 | +86 | +317 | +344 | No adduct | +258 |
| LdtCl | No adduct | +337 | +376, +421 | +86, +172 | +86 | +86 | +316 | +344 | +383 | +258 |
| LdtPa | No adduct | No adduct | No adduct | +86, +288 | +457 | n.d. | +317 | No adduct | +56 | +258 |
Molecular weights (Da) of β-lactams: Amoxicillin (A), 365.4; Cephalothin (C); 396.4; Doripenem (D), 420.1; Faropenem (F), 285.3; Aztreonam (Z), 435.4; Biapenem (B), 350.4; Tebipenem (Te), 383.5; T205, 337.3; T206, 365.4; T208, 405.4; T210, 279.3. A-C-Z-F and D-B-F-Te represent incubation of the enzymes with equimolar mixture of the indicated β-lactams. n.d., not determined.
Figure 1. Interactions of β-lactams with mycobacterial l,d-transpeptidases and activity of biapenem and faropenem in the mouse model of TB.
(a) Binding of faropenem to M. tuberculosis enzymes LdtMt1 and LdtMt2, and M. abscessus enzymes LdtMab1 and LdtMab2. In each panel, the top plot displays the titration of faropenem to enzyme, and the bottom panel displays the non-linear fit of heat exchange across increasing ligand:enzyme molar ratios. (b) Nitrocefin hydrolyzing activity of LdtMt2 following incubation with various β-lactams. (c) Log10 colony forming units of M. tuberculosis in the lungs of BALB/c mice before, during and after treatment with biapenem and faropenem alone or in combination with rifampin and control regimens. Data represent the mean (n = 5 mice per group per time point) and error bars represent the standard deviation. (d–k) Gross pathology of lungs of M. tuberculosis-infected mice at the end of treatment. One representative lung from each treatment group is shown: (d) no treatment, (e) isoniazid, (f) rifampin, (g) isoniazid + rifampin, (h) biapenem, (i) biapenem + rifampin, (j) faropenem, (k) faropenem + rifampin. Scale bar, 1 cm.
As the M. tuberculosis l,d-transpeptidases clearly demonstrated preferential binding to (carba)penems, we wondered if this drug class could also target l,d-transpeptidases from other bacterial pathogens, thus potentially having wider use as an antibacterial agent. Using a comparative genomics approach, we identified and subsequently cloned two l,d-transpeptidase orthologues in the acid-fast bacillus Mycobacterium abscessus (LdtMab1, LdtMab2) and one orthologue each in the following Gram-negative ESKAPE pathogens: Klebsiella pneumoniae (LdtKp), Enterobacter cloacae (LdtCl), and Pseudomonas aeruginosa (LdtPa) (Supplementary Fig. 2, Supplementary Table 3). UPLC-MS assessment of acyl adduct formation induced by interaction of β-lactams with each of these enzymes revealed that these l,d-transpeptidases also preferentially reacted with faropenem over both the non-carbapenems and carbapenems tested (Table 1). Binding of the l,d-transpeptidases orthologs in M. abscessus, LdtMab1 and LdtMab2, to faropenem and tebipenem, and lack of binding to the non-carbapenems, was also confirmed by ITC (Fig. 1a, Supplementary Table 2). Our data thus suggest that l,d-transpeptidases from diverse bacterial species, including ESKAPE pathogens, can be targeted with select (carba)penems.
As an additional verification, we assessed the activity of LdtMt2 following incubation with penicillins (amoxicillin and ampicillin), cephalosporins (cefoxitin, cefotaxime, cephalexin, cephalothin and cefdinir), and carbapenems (doripenem and biapenem) and the penem faropenem. While doripenem, biapenem and faropenem robustly inhibited the ability of LdtMt2 to hydrolyze nitrocefin, only one cephalosporin, namely cephalothin, showed comparable inhibitory activity and the remaining cephalosporins and penicillins exhibited little or no effect on the enzyme (Fig. 1b). Next, we tested the hypothesis that if carbapenems derive their potency by inhibiting l,d-transpeptidases, then sensitivity of a M. tuberculosis mutant lacking LdtMt2 would not be significantly affected compared to the wild-type strain. While the minimum inhibitory concentration (MIC90) of carbenicillin, methicillin, oxacillin, cefdinir, cefotaxime and cephalexin was 16–>32 fold higher for wild-type M. tuberculosis, MIC90 of meropenem, doripenem, tebipenem and faropenem were only 2–8 fold different between the two strains (Supplementary Table 4).
In vivo anti-TB activity of faropenem and biapenem
The anti-TB activity of faropenem observed in vitro, including potent bactericidal activity against M. tuberculosis 7 and specific inhibitory interactions with the LdtMt1 and LdtMt2 enzymes, prompted us to evaluate the in vivo activity of this drug in a preclinical mouse model of TB treatment. We also evaluated the in vivo anti-TB activity of biapenem, which has a very good in vitro bactericidal activity against M. tuberculosis 7. The antimicrobial activities of faropenem and biapenem were assessed in M. tuberculosis-infected BALB/c mice, and these drugs were administered both alone and in combination with rifampin, a key first-line anti-TB drug. Mice were infected by aerosol with M. tuberculosis H37Rv with an implantation of 3.7 log10 colony-forming units (CFUs) in the lungs; treatment was initiated two days after infection and administered daily for three weeks. As expected, the untreated control mice developed severe disease and either died or were moribund by eighteen days post-infection. Also as expected, the bacterial load in the lungs of mice receiving isoniazid-containing control regimens decreased significantly during the first week of treatment (Fig. 1c). Administration of biapenem alone resulted in bacteriostatic activity as bacterial multiplication in the lungs was inhibited compared to the lungs of untreated control mice. Bacteriostatic activity was also observed in the lungs of mice treated with the faropenem-rifampin regimen. However, potent bactericidal activity was observed in the lungs of mice that received the biapenem-rifampin regimen, which resulted in a greater decline in lung CFUs than was obtained with the isoniazid-rifampin positive control regimen. Furthermore, administration of both faropenem and biapenem, either alone or in combination with rifampin, prevented the development of gross lung lesions in the infected mice (Fig. 1d–k; Supplementary Fig. 3).
Structures of l,d-transpeptidases with (carba)penems
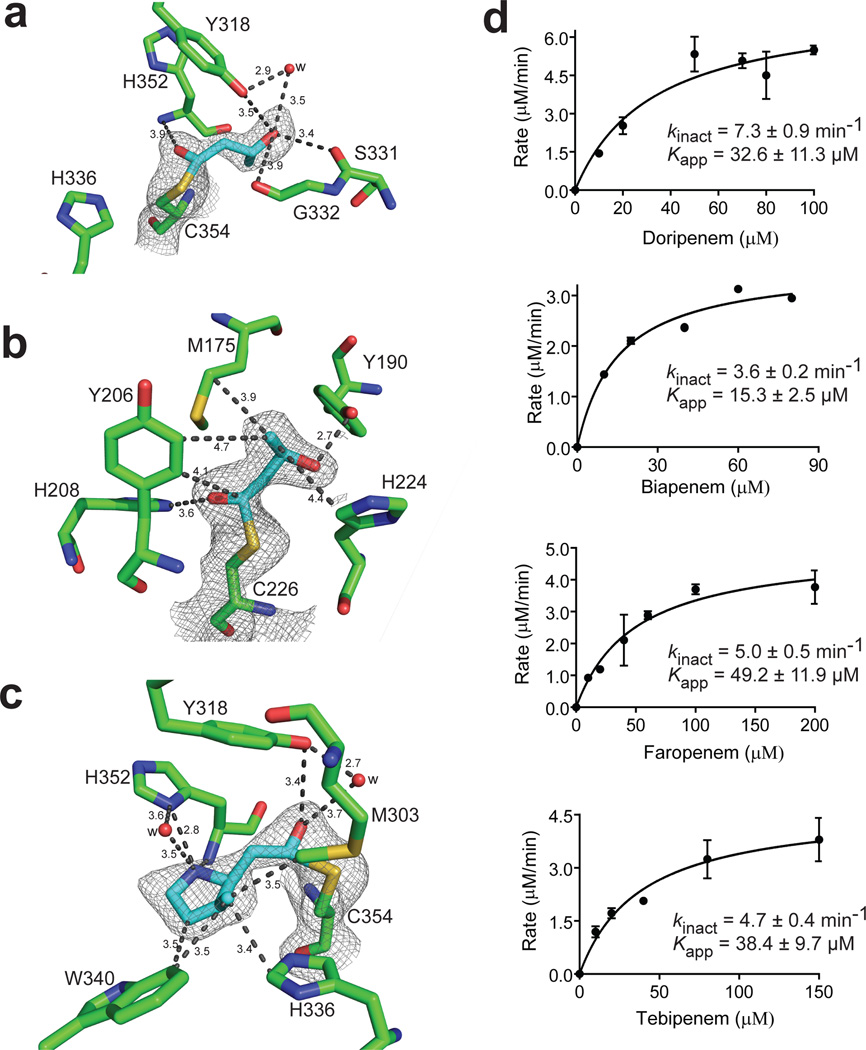
Our in vitro and in vivo data indicated that penems and carbapenems have high potential for use in TB treatment. Understanding the specific molecular interactions responsible for the anti-TB activity of these β-lactams is critical for maximizing their therapeutic capacity. To discern the basis of the preferential binding of the M. tuberculosis l,d-transpeptidases with (carba)penems over other β-lactams, we solved the crystal structures of LdtMt2 in complex with faropenem and doripenem and LdtMt1 with faropenem (Fig. 2a–c, Supplementary Fig. 4a,b; Supplementary Table 5). The catalytic core of LdtMt2 is flanked by two cavities, an inner and outer. The faropenem adduct is bound in the inner cavity of the LdtMt2 active site; this adduct lacked the β-lactam ring, the adjoining 5-membered ring and the R1 group initially present in the drug (Fig. 2a, Supplementary Fig. 4c). Based on these data and the 86 Da adduct detected in UPLC-MS (Table 1), we built a 4-carbon derivative fragment of faropenem into the electron density. The carbonyl carbon, originally C7, was covalently bound to the sulfur of Cys354, and the carbonyl oxygen had electrostatic interactions with the backbone amide nitrogen of His352. The hydroxyethyl substituent (R2) established hydrogen-bonding interactions with the side chain of Tyr318 (both direct and water mediated) and with the backbone carbonyl of Ser331. The carbon adjacent to the hydroxyethyl substituent also displayed hydrophobic packing with the β-carbon of His352. In the crystal structure of LdtMt1 with faropenem, a 86 Da adduct covalently attached to the catalytic Cys226 (equivalent to Cys354 in LdtMt2) at a 110° angle could be modeled into the density (Fig. 2b). Unlike with LdtMt2, the same adduct established different interactions in the active site of LdtMt1. His208 (equivalent to His336 of LdtMt2) did not interact with the sulfur of Cys226 but instead engaged the carbonyl C7 oxygen in the faropenem adduct via electrostatic interactions. The hydroxyethyl side chain and the adjacent carbon of the adduct were stabilized by hydrophobic packing with two methylenes of the Met175 side chain and with the β-carbon of His224 (equivalent to His352 of LdtMt2). In addition, the hydroxyl group of the adduct formed a hydrogen bond with the phenol side chain of Tyr190 (equivalent to Tyr318 of LdtMt2).
Figure 2. Molecular details of interactions of M. tuberculosis l,d-transpeptidases with (carba)penems.
(a) Crystal structures of LdtMt2 bound by faropenem at 2.17 Å, (b) LdtMt1 bound by faropenem at 2.25 Å, and (c) LdtMt2 bound by doripenem at 2.18 Å. Residues that make significant interactions are shown in green and adducts are shown in cyan. The 2Fo-2Fc difference fourier map (gray) is contoured at 1.0 σ. Distances are in Å. (d) Kinetics of inhibition of LdtMt2 by doripenem, biapenem, faropenem and tebipenem. Kinetic constants kinact and Kapp were determined spectrophotometrically. Data represent the mean of three independent experiments and error bars represent standard deviation.
The orientation of doripenem bound to LdtMt2 differed from that of faropenem: the doripenem adduct of 123 Da (Table 1) bound covalently to Cys354 but extended into the outer cavity and interacted with Trp340 (Fig. 2c). Doripenem was highly rearranged, lacking its R1 and R2 substituents and its β-lactam ring opened (C7 bound to Cys354), and could be modeled into the electron density. In the outer cavity, the C2 methyl on the pyrrolidine ring and the adjacent carbon in that ring of the adduct demonstrated van der Waals interactions with Trp340. The side chain of Tyr318 and the backbone amide nitrogen of Cys354 formed hydrogen bonds with the carbonyl C7 oxygen. The amino N4 of the pyrrolidine ring formed a hydrogen bond with the side-chain of His352.
Validation of LdtMt2 binding with (carba)penems
Based on the crystal structure data, as well as sequence conservation among known l,d-transpeptidases, we were able to identify amino acids that are directly involved in molecular interactions with (carba)penems. To build upon this work and further assess the role of these enzyme residues, we generated a series of LdtMt2 constructs each with single amino acid mutations in predicted key residues (Tyr318, His336, His352 and Cys354) and used UPLC-MS to evaluate the interaction of each of these mutants with faropenem, and the carbapenems biapenem and tebipenem (Supplementary Table 6). Stable adducts could be detected for biapenem with wild-type LdtMt2, but no adducts could be detected for any of the mutant proteins, indicating that all four amino acids were involved in binding biapenem. Faropenem and tebipenem failed to produce adducts with mutants H336N, H336A, C354S or C354A but did produce stable adducts with the Y318F, Y318A, and H352A (H352N with faropenem only). Further assessment by ITC indicated loss of binding of both faropenem and tebipenem to C354S, H336N, H352N and Y318F (Supplementary Fig. 5, Supplementary Table 2), thus confirming the involvement of each of these residues in the binding of LdtMt2 with (carba)penems.
Since the UPLC-MS studies indicated irreversible acylation of LdtMt2 by (carba)penems, we performed spectroscopy-based assays to calculate the rate of this irreversible inhibition. The kinact (maximum rate at which an irreversible transformation to enzyme–inhibitor complex occurs) and Kapp (inhibitor concentration required to achieve half maximum velocity) were determined by non-linear regression. These parameters demonstrated variations in the acylation kinetics for faropenem and each of the carbapenems (Fig. 2d). The efficiency of these reactions was assessed by the kinact/Kapp ratios of doripenem (0.22 µM−1 min−1), biapenem (0.23 µM−1 min−1), faropenem (0.10 µM−1 min−1) and tebipenem (0.12 µM−1 min−1). Although all of these β-lactams inactivated LdtMt2 by acylation, there were differences in kinact and Kapp values that may be attributable to variations in the thioether side chain of these carbapenems; these variations may cause changes in the rates of both drug binding and subsequent acylation.
Design, synthesis and assessment of evolved carbapenems
Our structure-activity and mechanistic data regarding the interactions between l,d-transpeptidases and faropenem/carbapenems, as well the previously described crystal structure of meropenem (a carbapenem) bound to LdtMt2 14, provided significant information to guide the design of new carbapenems with improved potential as anti-TB compounds. A key observation from the co-crystal structures was the absence of extensive contacts between the protein and the pyrrolidine-2-carboxamide thioether at C2. We thus hypothesized that the R1 substituent pyrrolidine interacted mainly with the outer cavity and solvent (illustrated in Fig. 3a) and therefore could be used to modify the physiochemical properties to enhance attributes such as permeability and pharmacokinetic profile. We replaced the R1 chain with various groups identified by naive Bayesian machine-learning models as the most frequent sulfur substituents in M. tuberculosis growth inhibitors 15, and initially focused on the 2-substituted analogs of existing carbapenems with an emphasis on R1 substituent to disrupt critical residues in the catalytic site of l,d-transpeptidases. Synthesis of the final carbapenems followed a two-stage process (Supplementary Note) where the enol phosphate and thiol (or immediate precursor) were reacted to afford the desired thioether. The 4-nitrobenzyl ester protecting group was then cleaved to yield the desired carbapenem.
Figure 3. Design and activity of evolved carbapenems.
(a) Model of carbapenem bound to the catalytic core of LdtMt2. While the carbapenem core ring is tightly bound to the catalytic core (gray) of LdtMt2 by covalent bonding to cysteine 354, its R1 group protrudes and makes extensive contacts with either the inner cavity (pocket colored purple, ligand colored beige) or the outer cavity (pocket colored yellow, ligand colored green), depending on the ligand binding mode. (b) Chemical structures of evolved carbapenems T123, T206, T208 and T210. (c) Kinetics of acylation of LdtMt2 by T208 and T210. Kinetic constants kinact and Kapp were determined spectrophotometrically. Data represent the mean of three independent experiments and error bars represent standard deviation.
Of the thirteen fully synthetic carbapenems illustrated in Fig. 3b and Supplementary Note (1–13), twelve exhibited in vitro activity against M. tuberculosis, having minimum inhibitory concentration (MIC) values at the low or sub µg/mL level (Supplementary Table 7); most of which were lower than the MIC values of meropenem, doripenem, biapenem, faropenem and tebipenem against M. tuberculosis 7. For four of the synthesized carbapenems with the lowest MICs for M. tuberculosis, namely T205 (4), T206 (5), T208 (7) (all MICs 1–2 µg/mL) and T210 (9) (MIC 0.25–0.5 µg/mL), we evaluated their specific binding interactions with LdtMt1 and LdtMt2 by UPLC-MS (Table 1). Each of these evolved carbapenems reacted with these enzymes and formed covalent adducts, indicating retention of the carbapenem skeleton/framework by the enzymes. Next, to assess the stability of the evolved carbapenems we reacted T208 and T210 with LdtMt2 and quantified the adducts at 1 and 24 hrs. We found no observable adduct degradation even at 24 hrs (Supplementary Table 8). The kinact/Kapp ratios of the LdtMt2 interactions with T208 and T210 were 0.56 and 0.07 min−1 µM−1, respectively (Fig. 3c), indicating that these experimental carbapenems were effective acylators of LdtMt2 and at least as efficient as meropenem 14, doripenem, biapenem, faropenem and tebipenem (Fig. 2d) in inactivating the enzyme. The LdtMt2 mutants Y318F, H336N, H352N and C354S all exhibited poor binding to T210 (Supplementary Table 2), again indicating that these residues are key for productive binding to carbapenems.
To examine the potential broader antibacterial activity of these experimental carbapenems, we also characterized their activity (i.e., determined the MICs) against M. abscessus and a full panel of ESKAPE pathogens: E. faecalis, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa and E. cloacae (Supplementary Table 7). While none of the experimental carbapenems inhibited the growth of M. abscessus, several of the compounds (T221 (10), T222 (11), T223 (12) and T224 (13)) inhibited the growth of all pathogens except P. aeruginosa, which was inhibited by T123 (1). Furthermore, compounds T205, T206, T208 and T210 reacted and formed acyl adducts with the l,d-transpeptidases from M. abscessus (LdtMab1 only), K. pneumoniae, E. cloacae and P. aeruginosa (Table 1) and the acyl-adducts formed are similar to those observed with LdtMt2 indicating these evolved carbapenems acylate varying l,d-transpeptidases from unrelated bacteria by a similar mechanism (LdtPa is an exception as a smaller adduct, +56 Da, is formed with T208). Thus, although the experimental carbapenems were designed for improved binding to the M. tuberculosis l,d-transpeptidases, several display potential for use as broader spectrum antibacterial agents.
Structures of LdtMt2 in complex with evolved carbapenems
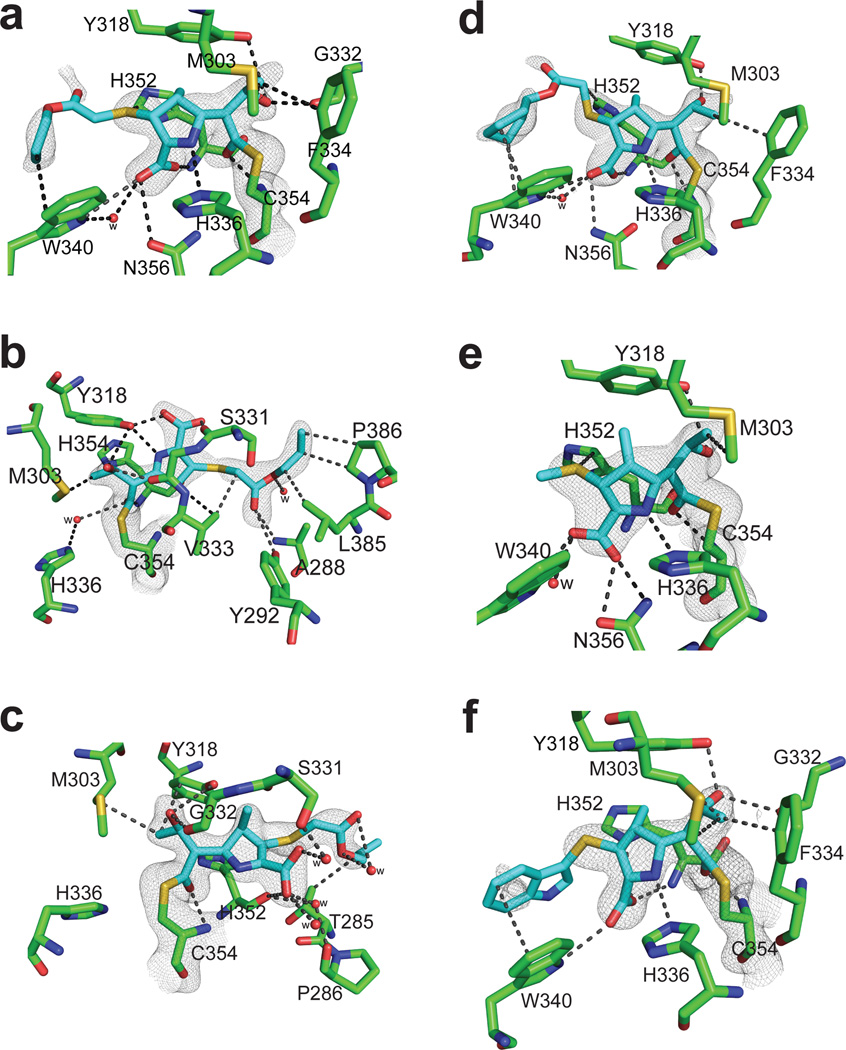
The evolved carbapenems also provided new insights to improve our ability to maximize the antimicrobial, and specifically the anti-TB activity, of this class of drugs. We thus solved the crystal structures of LdtMt2 in complex with T206, T208, T210 and T224 (Fig. 4, Supplementary Fig. 6,7, Supplementary Table 9). T206 binds in three different orientations in the active site of the enzyme, via the outer cavity (conformation A1) or via the inner cavity (conformation A2 and B), from two different crystal structure forms, LdtMt2-T206-A and LdtMt2-T206-B. In LdtMt2-T206-A, the electron density map of T206 in conformation A1 in chain A shows this carbapenem interacting with the outer cavity and covalently attached to the catalytic Cys354 (Fig 4a). The electron density of carbapenem T206 is contiguous to the Sγ atom of Cys354, but it is missing for much of the R1 group beyond the thioether sulfur. The hydroxyl of the R2 group hydrogen bonds with the side chain of Tyr318 and displayed both a direct and a water-mediated hydrogen bond with the backbone carbonyl of Gly332. The carboxyl group at C3 of the pyrrolidine ring forms a hydrogen bond with the side chain of Trp340 and with the backbone amide of His352. Trp340 also has hydrophobic interactions with the alkyl tail of the R1 group of T206. In conformation A2, chain B of LdtMt2-T206-A, T206 is bound in the inner cavity resembling the meropenem-bound structure (PDB ID 3VYP) 16, but the R2 group tail makes additional extensive electrostatic and hydrophobic interactions with Ala288, Tyr292, Leu385 and Pro386 (Fig 4b). The C3 carboxyl group accepts a hydrogen bond from Tyr318, and Met303 has a hydrophobic interaction with the terminal carbon of the C6 hydroxyethyl side chain. From the crystal structure form LdtMt2-T206-B, in conformation B of chain A, the carbonyl C7 (of what was the β-lactam ring) is covalently bound to Cys354, with the rest of the molecule extending into the inner cavity of the active site (Fig. 4c), similar to previous crystal structures of the meropenem:LdtMt2 complexes 14. The pyrrolidine ring and its carboxylate substituent have a conformation similar to the corresponding substructure of meropenem in chain A (PDB ID: 4GSU) 14 and in conformation A of chain A (PDB ID: 3VYP) 16, but this ring was rotated with respect to the orientation observed in the other chains of 4GSU and 3VYP. Interestingly, T206 is fully ordered in conformation B, with its R1 group tail stabilized by hydrophobic interactions with residue Thr285 and by a network of water-mediated hydrogen bonds between the carboxylate of T206 and the side-chain Ser331, the backbone amide nitrogen of Ala288, and the backbone carbonyl oxygen of Pro286.
Figure 4. Crystal structures of LdtMt2 with evolved carbapenems.
a–c) Structure with T206 at the catalytic site of LdtMt2a) Conformation A1, b) conformation A2 and c) conformation B showing interactions of T206. Structure of adducts (cyan) of (d) T208, (e) T210 and (f) T224 at the catalytic site of LdtMt2. For each panel, the 2Fo-2Fc difference fourier map (gray) is contoured at 1.0 σ. Distances are in Å.
In the crystal structure of LdtMt2 with T208, T210 and T224, the maps showed electron density for the respective inhibitors (and specifically the carbonyl carbon of what was the β-lactam moiety) within the distance necessary for covalent bonding to C354 (Fig. 4d–f). T208, T210 and T224 extended into the outer cavity in a manner similar to conformation A of T206 and that of imipenem with LdtMt1 (PDB ID: 4JMX) 17. The conserved region of T208, T210 and T224 forms the same quaternary interactions with the outer cavity of LdtMt2, and their C3 carboxyl accepts hydrogen bonds from Trp340 and Asn356 side chains thereby mimicking those found in T206 conformation A1. In all of the evolved carbapenems bound in the outer cavity, electron densities for the methyl group of the pyrrolidine ring at C1 position and the R1 group tail beyond the thioether sulfur are missing.
DISCUSSION
The traditionally understood targets of β-lactams are d,d-transpeptidases. Likewise, resistance to β-lactams has traditionally been attributed to β-lactamases, most of which have evolved as soluble β-lactam hydrolases from their membrane-bound d,d-transpeptidase ancestors 18. However, l,d-transpeptidases are biochemically and structurally distinct from d,d-transpeptidases and use cysteine to catalyze the formation of non-classical 3→3 linkages in the peptidoglycan and thereby provide an alternative mechanism by which bacteria can survive in the presence of traditional d,d-transpeptidase-targeting β-lactams 19. Therefore, molecules that can also target the l,d-transpeptidases have significant potential as agents with bactericidal activity against organisms that utilize both classical and non-classical transpeptidases when causing human disease.
Here, we demonstrated that penem and carbapenem β-lactam antibacterials have potent anti-TB activity using a preclinical mouse model of TB treatment. The bioavailability of many carbapenems is sub-optimal due to metabolism by dehydropepetidase-1 (DHP-1) in the renal proximal tubules 20. Faropenem and early carbapenems meropenem and imipenem exhibit limited activity in mice infected with M. tuberculosis 6,21–23. A recent clinical trial (NCT02349841) demonstrated a promising early bactericidal activity of meropenem in TB patients 24. Compared to meropenem, biapenem is more potent against M. tuberculosis 7 and is resistant to inactivation by DHP-1 20. The anti-tubercular activity seen in the mouse model in this work suggests that new and evolved (carba)penems have promise in treatment of TB. In pursuit of developing new carbapenems, we conducted an in-depth biochemical and structural analyses of (carba)penem interactions with M. tuberculosis l,d-transpeptidases allowing for the development of evolved carbapenems with improved anti-TB activity. We have also characterized the activities of our evolved compounds with other bacterial pathogens, including difficult-to-treat ESKAPE pathogens, demonstrating their broad-spectrum antibacterial potential.
Experiments characterizing the interactions between faropenem and carbapenems with the M. tuberculosis l,d-transpeptidases LdtMt1 and LdtMt2, including analysis of co-crystal structures of drug-enzyme interactions, have provided data that allowed us to propose molecular mechanisms responsible for enzyme inhibition. The reaction of faropenem with LdtMt2 and LdtMt1, proposed in Fig. 5a, was rapid (Fig. 2d) and yielded the inactivated enzymes bearing only (R)-3-hydroxybutyryl covalently linked to the active site cysteine, Cys354 in LdtMt2 (Fig. 2a) and Cys226 in LdtMt1 (Fig. 2b). Nucleophilic attack by this cysteine on the reactive β-lactam is proposed to open the 4-membered ring followed by scission of the remaining thiazoline ring. Such a cleavage process is known in the analogous masked enol opening of clavulanic acid by seryl β-lactamases 25. The resulting imine/iminium species, again paralleling clavulanate cleavage, is proposed to hydrate to an aminal, whose facile retro-aldol scission affords the observed inactivated enzyme adduct.
Figure 5. Proposed mechanism of acylation of l,d-transpeptidases by (carba)penems.
(a) Reaction of faropenem with LdtMt1 or LdtMt2, and reaction of LdtMt2 with (b) doripenem and (c) T208.
In contrast, reaction of the carbapenem doripenem at the LdtM2 catalytic center (proposed in Fig. 5b) takes a different course, but not one driven by the formation of a stable (aromatic) pyrrole ring of unchanged oxidation state as expected. Instead, we observed formation of the net two-electron reduced pyrrolinium adduct hydrogen-bonded to the nearby His352. Specifically, attack of the active site cysteine on the carbapenem carbonyl is proposed to form an imine/iminium species, which may decarboxylate to a stabilized ylid, analogous to the active form of the cofactor thiamin. After protonation, the resulting pyrrolinium may be captured by the adjacent thioether and the resulting bridged sulfonium species, either by attack of a nucleophile or by reversible release of the enamine and scavenge of the sulfonium ion, achieves both the required two-electron reduction and preservation of the C1 and C5 stereocenters evident in the crystal structure. Subsequent to these events, or earlier, precedented retro-aldol reaction can ensue to expel the hydroxyethyl side chain as acetaldehyde 3,26. While these proposals are in agreement with a mechanism of carbapenem-induced inhibition of LdtMt2 derived from a theoretical free energy study 27, our data provide direct evidence of additional cleavage and rearrangement mechanisms beyond the loss of the C6 hydroxymethyl group thought to be common to carbapenems 14. This information expands design options for carbapenems having alternative modes of transpeptidase inactivation. It remains to be seen how these fragmentation pathways for doripenem and faropenem are implicit in their whole-cell efficacy against M. tuberculosis.
Evaluation of molecular interactions of (carba)penems with l,d-transpeptidases facilitated the development of evolved carbapenems. Although a few of these compounds have been synthesized previously 28–31, their ability to target M. tuberculosis transpeptidases has not been described. Of note was the prevalence of active molecules with –SCH2CO2R (R = CH3, CH2CH3, (CH2)2CH3, CH(CH3)2, C(CH3)3, (CH2)3CH3, c-C6H12), as well as –S(2-pyridyl) moieties (Fig. 3b; Supplementary Table 7, Supplementary Note). Consistent detection of the entire drug molecule bound to enzymes from five different organisms (Table 1) suggests that acylation occurs through a similar mechanism of inactivation for each carbapenem (T205, T206, T208 and T210), which is further supported by crystal structure data of T206, T208, T210 and T224 with LdtMt2 (Fig. 4, 5c). Superposition of existing carbapenems and T206 in all conformations at the active site of LdtMt2 (Supplementary Fig. 6b) shows that the R1 sidechain of T206 establishes additional interactions with residues in the outer and inner cavities. In the inner cavity, the R1 sidechain is either stabilized by Pro386 or Thr285 accounting for why the R1 tail is ordered in the crystal structure (Fig. 4b,c). In the outer cavity, the R1 tail makes weak interactions with Trp340 as evidenced by the partial electron density (Fig. 4a).
Understanding how (carba)penems target l,d-transpeptidases is of critical importance for maximizing their potential for use in TB treatment, as these enzymes are not targeted by other anti-TB drugs, and therefore could play significant roles in the treatment of drug-resistant TB where new treatment options are greatly needed. A few clinical reports exist regarding the use of meropenem in combination with the β-lactamase inhibitor clavulanate in patients with multidrug-/extensively drug-resistant TB 32–34, although the direct impact of the carbapenem on treatment outcome could not be clearly assessed from these reports. Finally, this work has shown promising activity of carbapenems against each category of ESKAPE organisms, which represent difficult-to-treat, often nosocomially-transmitted bacterial pathogens for which new treatment options are also immediately needed. Thus, advancing knowledge of (carba)penem-based inhibition of bacterial transpeptidases impacts not only TB treatment, but also provide a rich source of new antibacterial agents effective against resistant organisms evolved to use 3→3 crosslinks in their cell walls. Bioinformatics/comparative genomics show that orthologs of the five l,d-transpeptidase genes in M. tuberculosis are widely distributed among bacterial pathogens and thus pose the threat of a genetic reservoir of antibiotic resistance that can now be recognized and blunted. The presence of multiple genes encoding d,d-transpeptidases, l,d-transpeptidases and β-lactamase in M. tuberculosis genome underscores their importance in peptidoglycan metabolism in this organism. Enzymes in each class, while similar in sequence, have distinct structures. Therefore, there is a continuum of structural variations around a similar function/theme. This is directly reflected in the varying affinities for different β-lactam structures in each sub-class. We hypothesize that a combination of β-lactams that are effective against each enzyme class will be necessary to bring about a simultaneous inhibition of the enzymes and subsequently a total inhibition of peptidoglycan metabolism.
MATERIALS AND METHODS
Experimental Design
The main objectives of this study were (i) not only to characterize, from the molecular to the pre-clinical (animal model) level, the inhibitory mechanisms of penem and carbapenem β-lactams against bacterial l,d-transpeptidases (specifically for M. tuberculosis, but also other bacterial pathogens) from the molecular to the pre-clinical level; and (ii) to utilize our data to design, synthesis and test evolved carbapenems with improved anti-TB activity. Sample sizes for the mouse experiment were determined based the ability to detect statistically significant differences between control and test groups, considering that inbred BALB/c mice were used, which represent a standard, well-established model organism for TB chemotherapy. Replication numbers for each experiment are provided either in the figure legends or below in the detailed methods.
Bacterial Strains, Growth Media and Drugs
The following bacterial strains were used in this study: M. tuberculosis H37Rv, M. abscessus (ATCC 19977), A. baumannii (strain 6M-1b, Clinical Microbiology, Johns Hopkins University), K. pneumoniae (ATCC 35657), E. cloacae (ATCC 13047), P. aeruginosa (PA14), E. faecalis (ATCC 19433) and methicillin-sensitive S. aureus (ATCC 29213). M. tuberculosis and M. abscessus were grown and assessed as previously described 7. Cation-adjusted Mueller-Hinton broth was used to grow A. baumannii, K. pneumoniae, E. cloacae, P. aeruginosa, E. faecalis and S. aureus at 35 °C according to Clinical and Laboratory Standard Institute guidelines 35. Rifampin, isoniazid and all β-lactams except evolved carbapenems were obtained from commercial vendors. Compounds were 95%–99% pure when random samples were analyzed using LC-MS.
Cloning, Site-Directed Mutagenesis, Expression and Purification of Proteins
l,d- and d,d-transpeptidases were cloned (excluding the N-terminal transmembrane anchoring domains) (Supplementary Table 3), expressed and purified to enhance chances of obtaining apo and co-crystals with drugs. Desired gene fragments were cloned into the multiple cloning site in pET28a+ to afford a N-terminal His6 tagged protein cleavable by TEV 10. Single amino acid substitutions of LdtMt2 (fragment ΔN55) were constructed by site directed mutagenesis as follows. Primers were designed such that mutations resulting in amino acid substitution were placed at the center of the oligo (Supplementary Table 3). For each mutagenesis, two separate PCR reactions, each with forward or reverse primer, using NEB high-fidelity buffer was used to amplify the pET28a+ vector carrying wild-type sequence for LdtMt2 (fragment ΔN55). DNA from the two reactions was combined, denatured at 95 °C, slowly renatured to 37 °C and digested with Dpn I as described. E. coli DH5α (C2987H, NEB Labs) was used for cloning and manipulation of plasmids. E. coli BL21δɛ3 (C2527H, NEB Labs) was used to overproduce proteins as described 36.
Mass Spectrometry
UPLC-MS analysis was carried out with a Waters Acquity H-Class system utilizing a Waters Acquity BEH-300 UPLC column packed with a C4 stationary phase (2.1 × 50 mm, 1.7 µm) in conjunction with HRMS analysis by a Waters Xevo-G2 quadropole-TOF electrospray mass spectrometer to detect and analyze protein-drug adducts. Each enzyme (2 µM) in 25 mM Tris buffer, pH 8, was incubated in the absence or presence of drug (50 µM) at room temperature for 5 hrs or for 1 hr and 24 hrs to assess stability of the adducts. Trifluoroacetic acid (0.1% final) was used to quench the reactions, samples were filtered through a 0.22 um filter and analyzed using UPLC-MS as follows: Mobile phase: 0–1 min 90% water + 10% ACN + 0.1% formic acid, 1–7.5 min gradient up to 20% water + 80% ACN + 0.1% formic acid, 7.5–8.4 min 20% water + 80% ACN + 0.1% formic acid, 8.4–8.5 min gradient up to 90% water + 10% ACN + 0.1% formic acid, 8.5–10 min 90% water + 10% ACN + 0.1% formic acid. Flow rate = 0.3 mL min−1. T = 60 °C.
Isothermal Titration Calorimetry
Varying concentrations of ligands and proteins (100 µM) were solubilized in 50mM Tris pH 8.0, 150mM NaCl, 0.5mM TCEP. Calorimetry experiments were performed using a microcalorimeter (iTC200, MicroCal) at 24 °C, by titration of the ligand (1 × 0.5 µL + 16 × 2.5 µL injections at 280 s intervals; stirring speed of 1000 rpm). Titrations of ligands into buffer were performed as a control, and the resulting heats of ligand dilution were subtracted from the experimental data prior to curve fitting. MicroCal Data Analysis software, Auto-iTC200 was used to monitor ligand binding behavior using non-linear least-squares fitting assuming a single-site model.
Inhibition of Nitrocefin Hydrolysis
To demonstrate inhibition of LdtMt2 by β-lactams, nitrocefin (Calbiochem) was used as a substrate for LdtMt2 as previously described 10. 10 µM LdtMt2 in 25mM HEPES/MES/Diethanolamine, 300 mM NaCl, pH 7.5, was incubated with 50 µM of each β-lactam at 25 °C for 2 hrs. Nitrocefin at a range of concentrations was added to the enzyme/inhibitor mixture and incubated for 50 min at 25 °C and its hydrolysis was measured at 496 nm. Absorbance data were converted to µM/minute using Beer’s Law (e= 20,500 M−1 cm−1 for nitrocefin; L = 0.5 cm).
Determination of Catalytic Constants
The acylation kinetics of LdtMt2 were determined by measuring the reduction in absorbance of the carbapenems following β-lactam ring opening using UV/Vis spectrophotometry (Shimadzu UV1800). The kinetics of acyl-enzyme complex formation were determined by incubating 10 µM LdtMt2 with increasing concentrations of drugs in a buffer containing MES (100 mM; pH 6.5), 150 mM NaCl and 0.05 mM TCEP. The assays were performed at 10 °C to reduce the rate of reactions, similar to previous investigations via stopped-flow/fluorescence approach 5. The reduction in carbapenem absorbance was used to compute the rate of acyl-enzyme complex formation, which was then plotted as a function of the concentration of various carbapenems assayed at their λmax (Supplementary Fig. 8). The absorption coefficients (molar absorptivities) in buffer containing 100 mM MES, pH 6.5 were determined to be 7500 M−1cm−1 for doripenem , 6980 M−1cm−1 for faropenem, 9846 M−1cm−1 for biapenem, 6652 M−1cm−1 for tebipenem, 9900 M−1cm−1 for T208 and 9880 M−1cm−1 for T210. The kinact, and Kapp. The kinact and Kapp values were calculated using non-linear regression analysis 37.
Protein Crystallization
Purified LdtMt2 (fragment ΔN55) was dialyzed against 50mM Tris pH 8.0, 150mM NaCl, 0.5mM TCEP and concentrated to 21 mg/mL. The crystallization screens Hampton Crystal Screen HT, JBScreen JCSG ++ HTS and JBScreen PEG /Salt HTS were used to identify crystallization conditions using the sitting drop vapor diffusion method. Plate-like crystals appeared in 20% 5000MME and 200 mM ammonium sulfate within a week and these crystals were used as seeds to produce better quality crystals by the hanging drop vapor diffusion method. For co-crystallization of LdtMt2 with faropenem and doripenem, enzyme was incubated with 5 mM of each drug for 2 hrs at room temperature and crystallization trays were set up using the vapor diffusion method. For crystallization of LdtMt2 with evolved drugs T206, T208, T210 and T224, LdtMt2 crystals were soaked overnight with 1–2 mM of each drug.
Purified LdtMt1 (fragment ΔN31) at a concentration of 16 mg/mL was crystallized using the sitting drop vapor diffusion method with 10% PEG6000 and 100mM Bicine pH 9.0. Crystals from a preliminary screen were used as seeds for further production of good quality crystals by the hanging drop vapor diffusion method. For co-crystallization of LdtMt1 with faropenem, 10 mM of drug was mixed with enzyme and incubated for 2 hrs at room temperature before setting up crystallization condition using the hanging drop vapor diffusion method.
Crystal Diffraction and Data Collection
LdtMt2-apo, co-crystals as well as crystals soaked with carbapenem drugs were cryo-protected in 30% glycerol, 20% 5000 MME and 120 mM ammonium sulfate buffer and flash cooled in liquid nitrogen. For LdtMt2-apo and co-crystals with faropenem and doripenem, X-ray diffraction data were collected at a wavelength of 1.54 Å using an in-house CuKα X-ray source (Rigaku FR-E+ SuperBright generator with a Saturn 944+ CCD Detector; Rigaku). for LdtMt2 apo and crystals soaked with evolved the carbapenems T206, T208 and T210, diffraction data were collected at 100K at a wavelength of 0.98 Å on beamline 19-ID at the Advanced Photon Source (Argonne National Laboratory) and the diffraction data were recorded on an ADSC Quantum 315r CCD detector, and processed with HKL3000 38.
LdtMt1-apo and co-crystal with faropenem were cryoprotected with 30% PEG8000, 10% PEG6000 and 100 mM Bicine pH 9.0 buffer and flash cooled in liquid nitrogen. The X-ray diffraction data were collected and analyzed as described below.
Structure Determination
The LdtMt2-apo crystals belong to the primitive triclinic space group P1 with four molecules in the asymmetric cell. The crystal structure of LdtMt2 was solved by molecular replacement method using the program, PHASER-MR from the CCP4 suite of programs 39,40 using the coordinates of LdtMt2 (PDB ID: 3VYN) as a search model. The initial solution was subjected to multiple rounds of crystallographic refinement with REFMAC 5.8.0103 41 from the CCP4 suite of programs and rebuilt to fit the electron density with COOT 42. The R values of refined structure (Supplementary Table 5) are well within the range of typical resolution. Ramachandran analysis with MolProbity 43 indicated that 96.46% residues are under favorable region and 3.54% in additional allowed region. The LdtMt2 crystals with different (carba)penems (faropenem, doripenem, T206, T208, T210 and T224) belong to the primitive monoclinic space group P21 with two molecules in the asymmetric unit. The crystal structures of LdtMt2 with carbapenem drugs were solved by the molecular replacement method using the program, PHASER-MR, either in Phenix or the CCP4 suite of programs 39,40. The initial solutions of LdtMt2 with different carbapenem drugs were subjected to multiple rounds of crystallographic refinement with program, REFMAC 5.8.0103 from the CCP4 suite of programs or with program phenix.refine from the Phenix suite of programs, and rebuilt to fit the electron density with COOT. The R values of refined structures (Supplementary Tables 5 and 9) are well within the range that is typical at corresponding resolution. Ramachandran analysis with MolProbity indicated that, in the structure of LdtMt2-faropenem, 96.69% residues are under favorable region and 3.02% in additional allowed region. In the structure of LdtMt2-doripenem, 97.56% residues are under favorable region and 2.44% in additional allowed region. In the structure of LdtMt2-T206-A, 96.12% residues are under favorable region and 3.74% in additional allowed region. In the structure of LdtMt2-T206-B, 96.12% residues are under favorable region and 3.59% in additional allowed region. In the structure of LdtMt2-T208, 96.55% residues are under favorable region and 3.16% in additional allowed region. In the structure of LdtMt2-T210, 96.56% residues are under favorable region and 3.16% in additional allowed region. For LdtMt2-T224, 97.27% residues are under favorable region and 2.73% in additional allowed region. The crystallographic parameters and final refinement statistics are summarized in Supplementary Tables 5 and 9. 2Fo-Fc difference Fourier maps were calculated for all different carbapenems bound to the active site of LdtMt2 using calculate maps in Phenix Suite and figures were prepared using PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.
The LdtMt1-apo and co-crystal structures with faropenem belong to the primitive trigonal space group P31 with four molecules in the asymmetric unit. Since the two axis a and b are equal in the space group, analysis of data quality with Xtriage from the Phenix suite of programs 44 suggested three 2-fold merohedral twin operators (−h,−k,l; h,−h–k,−l; and –k,−h,−l). The crystal structure of LdtMt1 was solved by molecular replacement using PHASER-MR from the Phenix suite of programs. The coordinates of LdtMt1 (PDB ID: 4JMN) were used as search models. The initial solution was further built using Autobuild (Automated model building and refinement) 45 from the Phenix suite of programs. The output model from Autobuild was further subjected to multiple rounds of NCS-based (non-crystallographic symmetry) crystallographic refinement using the program phenix.refine 46 from the Phenix suite of programs and the twin law (−h,−k,l) was applied during the refinement cycles. The structure was rebuilt to fit the electron density with COOT. The R values of refined structures (Supplementary Table 5) are well within the range that is typical at corresponding resolution. Ramachandran analysis with MolProbity indicated that, in the structure of LdtMt1-apo, 94.69% residues are under favorable region and 4.77% in additional allowed region and in the structure of LdtMt1-faropenem, 94.01% residues are under favorable region and 5.45% in additional allowed region. The crystallographic parameters and final refinement statistics are summarized in Supplementary Table 5.
Design and Synthesis of Evolved Carbapenems
All reagents were purchased from commercial suppliers and used without further purification unless otherwise noted. All chemical reactions occurring solely in an organic solvent were carried out under an inert atmosphere of argon or nitrogen. Analytical TLC was performed with Merck silica gel 60 F254 plates. Silica gel column chromatography was conducted with Teledyne Isco CombiFlash Companion or Rf+ systems. 1H- and 13C-NMR spectra were acquired on Varian Inova 400, 500 and 600 MHz instruments and are listed in parts per million downfield from TMS. LC-MS was performed on an Agilent 1260 HPLC coupled to an Agilent 6120 MS. All synthesized compounds were at least 95% pure as judged by their HPLC trace at 250 nm and were characterized by the expected parent ion(s) in the MS trace. The Bayesian models were constructed previously using a commercial software Discovery Studio 4.0 (Biovia) and trained with publicly available M. tuberculosis growth inhibition high-throughput screening data to distinguish actives from inactives given a specified efficacy cutoff 47,48. Methods for synthesis of evolved carbapenems are described in Supplementary Notes.
T205, T206, T208 and T210 were tested for cytotoxicity using the lactate dehydrogenase release assay (Pierce™ LDH Cytotoxicity Assay Kit, Thermo Fisher). We used J774A.1 macrophages as this cell line is most relevant to TB: intracellular M. tuberculosis reside in macrophages and, therefore, the drug is likely to accumulate more in these cells. Drug concentrations ranged from 8–64 µg/mL. None of the carbapenems exhibited toxicity (Supplementary Fig. 9).
Minimum Inhibitory Concentration
The standard broth dilution method was used to determine the MIC of drugs and experimental carbapenems 35,49. Briefly, each bacterial strain was grown in appropriate medium under aforementioned conditions to exponential phase and the cultures were used to inoculate 105 colony forming units (CFU) into microtiter wells containing a drug at two fold dilutions ranging from 64 µg/mL to 0.06 µg/mL. Medium inoculated with bacteria but without drug was used as a positive control for growth. The negative control was medium alone. As per CLSI guidelines cultures were incubated at 37 °C and evaluated for growth by visual inspection at 14 days for M. tuberculosis and at 3 days for M. abscessus at 30 °C 35. Similarly, MIC assessments for A. baumannii, K. pneumoniae, E. cloacae, P. aeruginosa, E. faecalis and S. aureus were performed as per CLSI guidelines. Experiments were repeated to verify MIC results. MIC was defined as the lowest concentration that inhibited growth by 90% compared to the no drug control.
Evaluation of Carbapenems in mice
Female BALB/c mice, 4–5 weeks old (Charles River Labs), were used according to the protocol approved by the Johns Hopkins University Animal Care & Use Committee (MO15M25). We used an acute model of active M. tuberculosis infection in mice to assess the activities of faropenem and biapenem. Using M. tuberculosis culture at exponential phase of growth, we prepared a suspension with an optical density (A600nm) of 0.2 and infected mice by aerosolizing 10 ml of this suspension in a Glas-Col chamber. This high dose infection resulted in implantation of 3.7 log10 CFU in the lungs of each mouse. Treatment was initiated two days after infection, a time when M. tuberculosis bacilli are rapidly proliferating. While isoniazid (10 mg/kg) and rifampicin (10 mg/kg) were administered by oral gavage once daily (7 days per week), biapenem (200 mg/kg) and faropenem (200 mg/kg) were administered twice daily by subcutaneous injection. Five mice were sacrificed one day after infection, lungs homogenized and plated on Middlebrook 7H11 selective medium to determine the actual infection burden. Next, five mice were sacrificed on the day of treatment initiation and at three weeks following initiation of treatment, the final time point, to determine bacterial burden in the lungs. Gross pathology of lungs and mouse body weights were recorded at each time point.
Supplementary Material
Acknowledgments
Assistance of Varsha Chauhan for protein expression is appreciated. We thank Leighanne Basta for critical discussions. P. aeruginosa PA14 was a kind gift from Dr. Stephen Lory, Harvard University. Discovery Studio was kindly provided to SE and JSF by Biovia. This study was supported by NIH awards R21AI111739 and DP2OD008459 to GL. Structural results shown in this report are derived in part from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Footnotes
AUTHOR CONTRIBUTIONS: PK (protein/ligand, crystallization and structure studies), AK (antibacterial potency, animal studies), EPL (antibacterial synthesis and mass-spectrometry, mechanism), SL (antibacterial synthesis), RM, DTB (enzyme kinetics), TAZ and ALP (structure data analysis), SE (antibacterial chemiiformatics), SLG (crystal data collection/analysis), CAT (antibacterial design/synthesis, mass-spectrometry, data analysis, mechanism), JSF (antibacterial design/synthesis and data analysis), GL (overall study conception, cloning, animal studies, data analysis). PK, NCA and GL prepared the manuscript and all authors contributed to the final draft.
COMPETING FINANCIAL INTERESTS: Authors declare no competing financial interests.
Accession codes
Coordinates and structure factors have been deposited in the PDB with the following accession codes: 5DU7 (LdtMt2-apo), 5DUJ (LdtMt2-faropenem), 5DVP (LdtMt2-doripenem), 5E5L (LdtMt1-apo), 5E51 (LdtMt1-faropenem), 5DZJ (LdtMt2-T206-conformation A1&A2), 5DZP (LdtMt2- T206-conformation B), 5E1G (LdtMt2-T208), 5E1I (LdtMt2-T210) and 5K69 (LdtMt2-T224).
REFERENCES
- 1.WHO. Global Tuberculosis Report. World Health Organization; 2015. [Google Scholar]
- 2.Walsh C. Antibiotics: Actions, Origins, Resistance. Vol. 3. ASM Press; 2003. [Google Scholar]
- 3.Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE, 3rd, Blanchard JS. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science (New York, N.Y) 2009;323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wivagg CN, Bhattacharyya RP, Hung DT. Mechanisms of β-lactam killing and resistance in the context of Mycobacterium tuberculosis. The Journal of antibiotics. 2014;67:645–654. doi: 10.1038/ja.2014.94. [DOI] [PubMed] [Google Scholar]
- 5.Cordillot M, et al. In vitro cross-linking of peptidoglycan by Mycobacterium tuberculosis L,D-transpeptidases and inactivation of these enzymes by carbapenems. Antimicrobial agents and chemotherapy. 2013 doi: 10.1128/AAC.01663-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhar N, et al. Rapid Cytolysis of Mycobacterium tuberculosis by Faropenem, an Orally Bioavailable β-Lactam Antibiotic. Antimicrobial agents and chemotherapy. 2015;59:1308–1319. doi: 10.1128/AAC.03461-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaushik A, et al. Carbapenems and Rifampin Exhibit Synergy against Mycobacterium tuberculosis and Mycobacterium abscessus. Antimicrobial agents and chemotherapy. 2015;59:6561–6567. doi: 10.1128/AAC.01158-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rullas J, et al. Combinations of β-lactam antibiotics currently in clinical trials are efficacious in a DHP-I deficient mice model of TB infection. Antimicrobial agents and chemotherapy. 2015 doi: 10.1128/AAC.01063-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta R, et al. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nature medicine. 2010;16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdemli SB, et al. Targeting the cell wall of Mycobacterium tuberculosis: structure and mechanism of L,D-transpeptidase 2. Structure. 2012;20:2103–2115. doi: 10.1016/j.str.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavollay M, et al. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. Journal of bacteriology. 2008;190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar P, et al. Meropenem inhibits D,D-carboxypeptidase activity in Mycobacterium tuberculosis. Molecular microbiology. 2012;86:367–381. doi: 10.1111/j.1365-2958.2012.08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoonmaker MK, Bishai WR, Lamichhane G. Nonclassical transpeptidases of Mycobacterium tuberculosis alter cell size, morphology, the cytosolic matrix, protein localization, virulence, and resistance to β-lactams. Journal of bacteriology. 2014;196:1394–1402. doi: 10.1128/JB.01396-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HS, et al. Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains. Acta crystallographica. Section D, Biological crystallography. 2013;69:420–431. doi: 10.1107/S0907444912048998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekins S, et al. Enhancing Hit Identification in Mycobacterium tuberculosis Drug Discovery Using Validated Dual-Event Bayesian Models. PloS one. 2013;8:e63240. doi: 10.1371/journal.pone.0063240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li WJ, et al. Crystal structure of L,D-transpeptidase LdtMt2 in complex with meropenem reveals the mechanism of carbapenem against Mycobacterium tuberculosis. Cell research. 2013;23:728–731. doi: 10.1038/cr.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Correale S, Ruggiero A, Capparelli R, Pedone E, Berisio R. Structures of free and inhibited forms of the L,D-transpeptidase LdtMt1 from Mycobacterium tuberculosis. Acta crystallographica. Section D, Biological crystallography. 2013;69:1697–1706. doi: 10.1107/S0907444913013085. [DOI] [PubMed] [Google Scholar]
- 18.Meroueh SO, Minasov G, Lee W, Shoichet BK, Mobashery S. Structural aspects for evolution of β-lactamases from penicillin-binding proteins. Journal of the American Chemical Society. 2003;125:9612–9618. doi: 10.1021/ja034861u. [DOI] [PubMed] [Google Scholar]
- 19.Mainardi JL, et al. A novel peptidoglycan cross-linking enzyme for a β-lactam-resistant transpeptidation pathway. The Journal of biological chemistry. 2005;280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- 20.Hikida M, Kawashima K, Yoshida M, Mitsuhashi S. Inactivation of new carbapenem antibiotics by dehydropeptidase-I from porcine and human renal cortex. The Journal of antimicrobial chemotherapy. 1992;30:129–134. doi: 10.1093/jac/30.2.129. [DOI] [PubMed] [Google Scholar]
- 21.Chambers HF, Turner J, Schecter GF, Kawamura M, Hopewell PC. Imipenem for treatment of tuberculosis in mice and humans. Antimicrobial agents and chemotherapy. 2005;49:2816–2821. doi: 10.1128/AAC.49.7.2816-2821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veziris N, Truffot C, Mainardi JL, Jarlier V. Activity of carbapenems combined with clavulanate against murine tuberculosis. Antimicrobial agents and chemotherapy. 2011;55:2597–2600. doi: 10.1128/AAC.01824-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.England K, et al. Meropenem-clavulanic acid shows activity against Mycobacterium tuberculosis in vivo. Antimicrobial agents and chemotherapy. 2012;56:3384–3387. doi: 10.1128/AAC.05690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diacon AH, et al. β-Lactams against Tuberculosis--New Trick for an Old Dog? The New England journal of medicine. 2016;375:393–394. doi: 10.1056/NEJMc1513236. [DOI] [PubMed] [Google Scholar]
- 25.Charnas RL, Fisher J, Knowles JR. Chemical studies on the inactivation of Escherichia coli RTEM β-lactamase by clavulanic acid. Biochemistry. 1978;17:2185–2189. doi: 10.1021/bi00604a025. [DOI] [PubMed] [Google Scholar]
- 26.Drawz SM, et al. Inhibition of the class C β-lactamase from Acinetobacter spp.: insights into effective inhibitor design. Biochemistry. 2010;49:329–340. doi: 10.1021/bi9015988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva JR, et al. Targeting the cell wall of Mycobacterium tuberculosis: A molecular modelling investigation of the interaction of imipenem and meropenem with L,D-transpeptidase 2. Journal of biomolecular structure & dynamics. 2015;1-34 doi: 10.1080/07391102.2015.1029000. [DOI] [PubMed] [Google Scholar]
- 28.Fung-Tomc JC, et al. Structure-activity relationships of carbapenems that determine their dependence on porin protein D2 for activity against Pseudomonas aeruginosa. Antimicrobial agents and chemotherapy. 1995;39:394–399. doi: 10.1128/aac.39.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Natsugari H, Matsushita Y, Yoshioka K, Matsui S. 5,6-cis-carbapenem-3-carboxylic acid derivatives and process for their preparation. Japan patent. 1983 [Google Scholar]
- 30.Abe T, Hayashi K, Isoda T, Kumagai T. 2-((pyridyl-substituted)thio)-carbapenem derivative. Japan patent JP19940170499 19940630. 1996 [Google Scholar]
- 31.Seki M, Kondo K, Iwasaki T. Efficient synthesis of 1β-methylcarbapenems based on the counter-attack strategy. Journal of the Chemical Society, Perkin Transactions. 1996;1:2851–2856. [Google Scholar]
- 32.Palmero D, et al. First series of patients with XDR and pre-XDR TB treated with regimens that included meropenen-clavulanate in Argentina. Archivos de bronconeumologia. 2015;51:e49–e52. doi: 10.1016/j.arbres.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 33.Payen MC, et al. Clinical use of the meropenem-clavulanate combination for extensively drug-resistant tuberculosis. Int J Tuberc Lung Dis. 2012;16:558–560. doi: 10.5588/ijtld.11.0414. [DOI] [PubMed] [Google Scholar]
- 34.De Lorenzo S, et al. Efficacy and safety of meropenem-clavulanate added to linezolid-containing regimens in the treatment of MDR-/XDR-TB. The European respiratory journal. 2013;41:1386–1392. doi: 10.1183/09031936.00124312. [DOI] [PubMed] [Google Scholar]
- 35.Desmond E. Susceptibility Testing of Mycobacteria, Nocardiae and Other Aerobic Actinomycetes. Clinical Laboratory Standard Institute. 2011:M24–A2. [PubMed] [Google Scholar]
- 36.Brammer Basta LA, et al. Loss of a Functionally and Structurally Distinct L,D-Transpeptidase, LdtMt5, Compromises Cell Wall Integrity in Mycobacterium tuberculosis. The Journal of biological chemistry. 2015;290:25670–25685. doi: 10.1074/jbc.M115.660753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Triboulet S, et al. Inactivation kinetics of a new target of β-lactam antibiotics. The Journal of biological chemistry. 2011;286:22777–22784. doi: 10.1074/jbc.M111.239988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta crystallographica. Section D, Biological crystallography. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta crystallographica. Section D, Biological crystallography. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.CCP4. The CCP4 suite: programs for protein crystallography. Acta crystallographica. Section D, Biological crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. Section D, Biological crystallography. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zwart PH, et al. Automated structure solution with the PHENIX suite. Methods in molecular biology (Clifton, N.J) 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 45.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta crystallographica. Section D, Biological crystallography. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta crystallographica. Section D, Biological crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maddry JA, et al. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis (Edinburgh, Scotland) 2009;89:354–363. doi: 10.1016/j.tube.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reynolds RC, et al. High throughput screening of a library based on kinase inhibitor scaffolds against Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinburgh, Scotland) 2012;92:72–83. doi: 10.1016/j.tube.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gavan TL, Town MA. A microdilution method for antibiotic susceptibility testing: an evaluation. American journal of clinical pathology. 1970;53:880–885. doi: 10.1093/ajcp/53.6.880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.