SUMMARY
Microbial pattern molecules in the intestine play immunoregulatory roles via diverse pattern recognition receptors. However, the role of the cytosolic DNA sensor AIM2 in the maintenance of intestinal homeostasis is unknown. Here, we show that Aim2−/− mice are highly susceptible to dextran sodium sulfate-induced colitis which is associated with microbial dysbiosis as represented by higher colonic burden of commensal Escherichia coli. Colonization of germ-free mice with Aim2−/− mouse microbiota leads to higher colitis susceptibility. In-depth investigation of AIM2-mediated host defense responses reveals that caspase-1 activation and IL-1β and IL-18 production are compromised in Aim2−/− mouse colons, consistent with defective inflammasome function. Moreover, IL-18 infusion reduces E. coli burden as well as colitis susceptibility in Aim2−/− mice. Altered microbiota in inflammasome-defective mice correlate with reduced expression of several antimicrobial peptides in intestinal epithelial cells. Together, these findings implicate DNA sensing by AIM2 as a regulatory mechanism for maintaining intestinal homeostasis.
Keywords: Pattern Recognition Receptors, AIM2, Inflammasome, DNA Sensor, Colitis, Escherichia coli, Inflammation, Inflammatory Bowel Diseases, Antimicrobial Peptides, Intestinal Epithelium
INTRODUCTION
The intestinal mucosal immune system of mammals evolved to coexist with densely populated microorganisms that reside in the intestinal lumen. The central physiological process for homeostatic immune response in the gut is the recognition of pathogen-associated molecular patterns (PAMPs) by host pattern recognition receptors (PRRs). There are several evolutionary conserved PRRs including Toll-like receptors (TLRs), NOD-like receptors (NLRs), RIG-I-like receptors (RLRs), C-type lectin-like receptors (CLRs) and HIN-200 family receptors. Defects in pathogen sensing systems lead to dysregulated immune responses in the intestine, resulting in the induction of intestinal inflammatory disorders such as inflammatory bowel disease (IBD) and colorectal cancer (Cho, 2008). Clinical and experimental studies have shown that dysfunctions in several TLRs (TLR2, TLR4, TLR9) and NLRs (NOD2, NLRP3, NLRP6, NLRP12) are associated with the pathogenesis of intestinal inflammation and tumorigenesis (Chen et al., 2008; Elinav et al., 2011; Hugot et al., 2001; Katakura et al., 2005; Maeda et al., 2005; Rakoff-Nahoum et al., 2004; Vijay-Kumar et al., 2007; Villani et al., 2009; Zaki et al., 2010a; Zaki et al., 2011b). However, the precise mechanisms of the regulation of intestinal homeostasis by these pathogen sensors are not clearly defined, and the functions of the majority of PRRs in intestinal physiology have yet to be explored.
Absent in melanoma 2 (AIM2), a member of interferon-inducible gene HIN-200 family, has been implicated as a cytosolic sensor of DNA (Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009). Recent studies have underscored the importance of AIM2-mediated sensing of microbial DNA in host defense responses against bacterial and viral infections caused by Listeria monocytogenes, Francisella tularensis, Streptococcus pneumoniae, and murine cytomegalovirus (Fang et al., 2014; Fernandes-Alnemri et al., 2010; Jones et al., 2010; Kim et al., 2010; Park et al., 2014; Rathinam et al., 2010). Structurally, AIM2 contains an N-terminal pyrin domain (PYD) and a C-terminal oligonucleotide binding HIN domain. When cytosolic dsDNA binds to its HIN domain, AIM2 recruits apoptosis speck-like protein containing a caspase recruitment domain (ASC) and caspase-1 to form a molecular platform called the inflammasome. In addition to AIM2, at least three NLR family members - NLRP1, NLRP3, and NLRC4 - are known to activate the inflammasome in biological systems (Lamkanfi and Dixit, 2009). Inflammasome-mediated activation of caspase-1 is required to cleave pro-IL-1β and pro-IL-18 into their active forms. Mounting evidence points to the critical roles caspase-1 and other inflammatory caspases play in regulating intestinal inflammation and tumorigenesis (Allen et al., 2010; Dupaul-Chicoine et al., 2010; Elinav et al., 2011; Salcedo et al., 2010; Zaki et al., 2010a). We and others previously demonstrated that the NLRP3 inflammasome regulates intestinal inflammation and tumorigenesis in mice (Allen et al., 2010; Hirota et al., 2010; Zaki et al., 2010a; Zaki et al., 2010b). Recent studies showed that the inflammasome in the gut can be activated by other PRRs, such as NLRP1 and NLRP6, which also contribute to intestinal homeostasis (Elinav et al., 2011; Williams et al., 2015). However, until now, DNA-dependent activation of the AIM2 inflammasome and its physiological relevance to intestinal homeostasis in the gut has not been reported.
Here, we investigated the physiological function of AIM2 in the gut. Our results show that Aim2−/− mice are highly susceptible to DSS-induced colitis. AIM2-mediated protection against colonic inflammation was associated with its function as an activator of the inflammasome, since caspase-1 activation and IL-18 production were remarkably attenuated in the absence of AIM2. Interestingly, the colons of Aim2−/− and other inflammasome-defective mice, e.g. Nlrp3−/−, caspase-1−/− (caspase1/11−/−) and Il18−/−, contain significantly higher loads of Escherichia coli, an indication of dysbiosis in their gut. Transfer of gut microbiota from Aim2−/− to wild-type (WT) mice confers higher colitis susceptibility in the WT mice, suggesting that the AIM2 inflammasome plays a critical role in the regulation of intestinal inflammation via shaping the intestinal microbial ecology. Further investigation revealed that the AIM2 inflammasome regulates the expression of antimicrobial peptides (AMPs), including C-type lectins (Reg3β and Reg3γ), calprotectin (S100A8 and S100A9) and lipocalin 2 (Lcn2) by intestinal epithelial cells. Overall, our study elucidates a major inflammasome activation pathway in the intestine that involves the DNA sensor AIM2, and offers further insights into the mechanism of inflammasome-mediated protection against intestinal inflammation.
RESULTS
AIM2 Protects Mice from DSS-induced Colitis
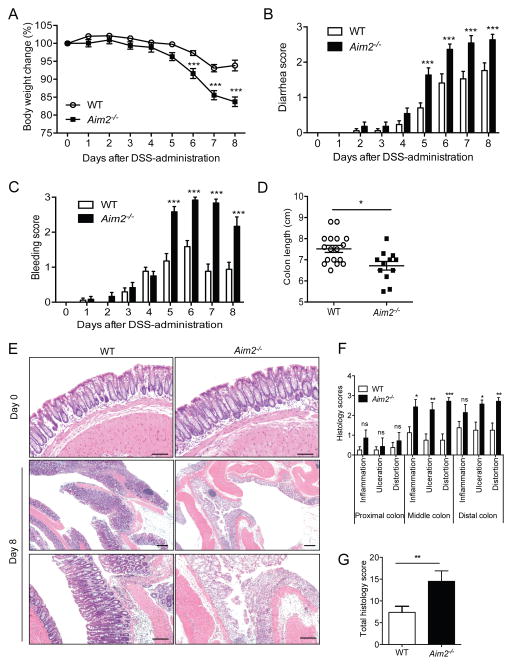
Previous studies have shown that Aim2 is highly expressed in the large intestine (Cridland et al., 2012). Our real-time PCR analyses confirm that Aim2 is ubiquitously expressed in the colon (Figures S1A and S1B). To investigate the physiological function of AIM2 in the colon, we induced colitis in Aim2−/− mice (Rathinam et al., 2010) by feeding them 3% DSS-containing water for 5 days. Colitis susceptibility in Aim2−/− and WT mice was evaluated daily by monitoring body weight and measuring clinical features such as diarrhea and occult stool bleeding. Aim2−/− mice lost significantly more body weight and exhibited increased diarrhea and rectal bleeding compared to WT mice (Figures 1A – 1C). Mice were sacrificed at day 8 and examined macroscopically and microscopically for inflammatory changes. Aim2−/− mouse colons were significantly shorter compared to WT mice (Figure 1D). This was further supported by histological analysis of hematoxylin and eosin (H&E)-stained whole colon tissues sections (Figure 1E). Much of the distal colon in Aim2−/− mice showed complete crypt destruction, ulceration, and submucosal edema, which was far less severe in WT mice (Figures 1E and 1F). Histopathological analyses demonstrated significantly higher scores for inflammation, ulceration, and crypt distortion in the middle and distal part of the Aim2−/− mouse colon at day 8 post DSS administration (Figures 1F and 1G). Notably, Aim2−/− mice exhibited normal colonic histopathology prior to DSS administration (Figure 1E), and the expression of Aim2 was not altered during colitis (Figures S1C – S1E). Taken together, AIM2 is a critical regulator of intestinal homeostasis and Aim2-deficiency leads to higher colitis susceptibility and severity in mice.
Figure 1. Aim2−/− mice are susceptible to DSS-induced colitis.
(A–E) WT (n=17) and Aim2−/− (n=12) mice were fed with 3% DSS for 5 days, followed by regular drinking water for 3 days. Body weight (A), stool consistency (B) and rectal bleeding (C) were scored daily. (D) Mice were sacrificed on day 8 to measure colon length. (E) Colon tissues collected from healthy mice (day 0) or at day 8 were stained with H&E. Scale bar = 100μm. (F–G) Semi-quantitative scoring of histopathology (n=8–10). Data represent mean ± SEM of a representative experiment; *, p<0.05, **, p<0.01, ***, p<0.001. See also Figure S1.
Colonic Injury Leads to Hyper-inflammation in Aim2−/− Mice
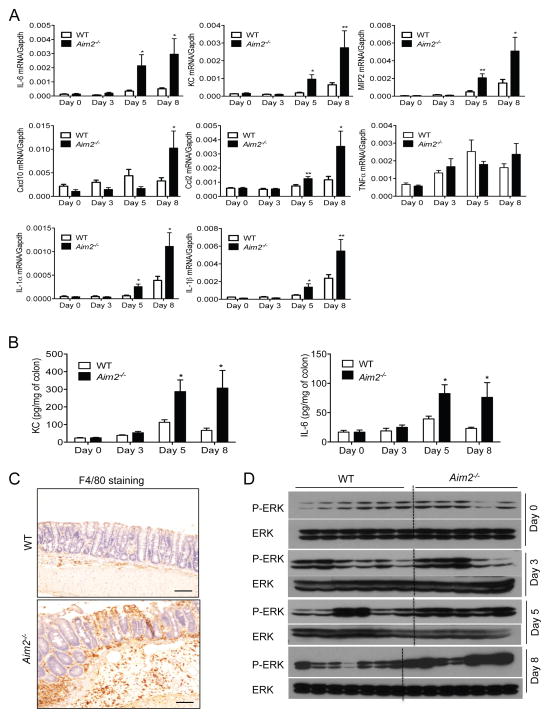
While cytokine production in the intestine is essential for repair and healing of the injured epithelium, overwhelming production of cytokines and chemokines trigger pathologic mucosal tissue injury and inflammatory disorders (Zaki et al., 2011a). Therefore, the types and levels of different mediators determine the fate of colonic inflammation and damage. To understand whether AIM2 signaling regulates cytokine production, we measured the expression of IL-1β, IL-1α, IL-6, KC, TNFα, MIP2, CCL2, CXCL10 in colon tissues collected from wild-type and Aim2−/− mice over the course of colitis (day 0, 3, 5, and 8). Although the initial (day 0) expression of proinflammatory cytokines and chemokines were comparable, IL-1α, IL-1β, IL-6, KC and MIP2 were significantly increased in Aim2−/− mice at day 5 and day 8 (Figure 2A). In addition, CXCL10 and CCL2 were also overexpressed in Aim2−/− mouse colons at day 8 (Figure 2A). Elevated KC and IL-6 levels in Aim2−/− mouse colon homogenates were confirmed by ELISA at day 5 and 8 of colitis (Figure 2B). However, Type I interferons were not regulated by AIM2 (Figure S2A). MIP2, KC and CCL2 are involved in the recruitment and migration of macrophages, neutrophils and lymphocytes. Consistent with the accumulation of these chemokines in Aim2−/− mouse colons, we observed greater macrophage infiltration as measured by immunohistochemistry (Figure 2C). Furthermore, the inflammatory signaling pathway ERK showed elevated activation in Aim2−/− mouse colons collected at day 5 and 8 post DSS as compared to those in WT mice (Figure 2D). However, we did not observe differences in epithelial cell death between WT and Aim2−/− mice during colitis as measured by TUNEL assay (Figures S2B and S2C).
Figure 2. AIM2-deficiency leads to increased inflammation in the colon.
WT and Aim2−/− mice were fed with 3% DSS in drinking water for 5 days, followed by regular drinking water for 3 days. (A) Real-time PCR analyses of IL-1α, IL-1β, IL-6, TNF-α, MIP2, Cxcl10, Ccl2 expression in colons collected at day 0, 3, 5 and 8 after DSS. (B) Colon homogenates were analyzed for the level of KC and IL-6 by ELISA. Data represent mean ± SEM; n=7–10/group; *, p<0.05, **, p<0.01. (C) Colon tissues collected at day 8 was fixed in formalin and immunostained for the macrophage marker F4/80. Brown colors indicate F4/80-positive cells. Scale bar = 100μm (D) Whole colon homogenates were analyzed for the activation of ERK by Western blotting. See also Figure S2.
AIM2 Regulates Intestinal Microbiota
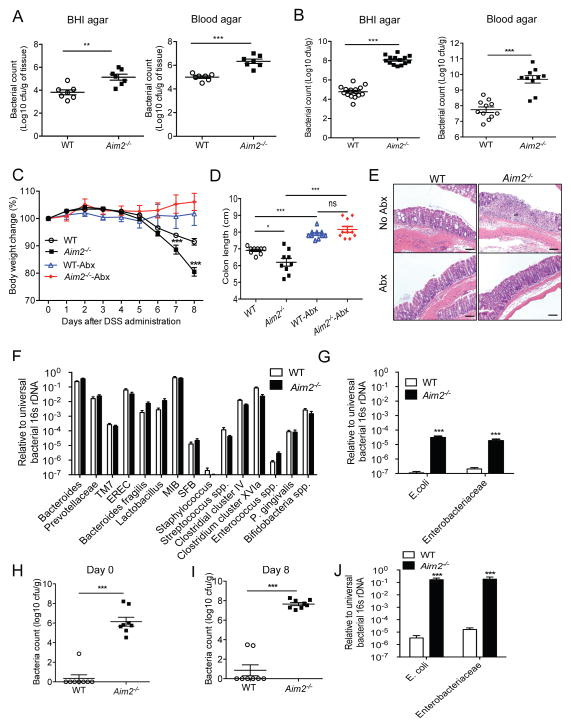
Colitis is initiated by the invasion of gut commensal flora into the lamina propria, driving the activation of immune cells. To investigate the possibility that increased inflammation in Aim2−/− mice is due to increased penetration of the intestinal commensal flora into colonic mucosa during DSS-colitis, we measured the number of culturable bacteria in the colon tissue at day 8 after DSS administration. Colon homogenates were cultured onto brain heart infusion (BHI) and blood agar, under aerobic and anaerobic conditions, respectively. There were significantly greater aerobic and anaerobic/facultative anaerobic bacteria present in the Aim2−/− mouse colon tissue compared to WT mice (Figure 3A). Culture of fecal homogenates revealed a higher bacterial burden in the colonic lumen of Aim2−/− mice compared to those of WT mice (Figure 3B). Notably, no bacteria were detected in WT or Aim2−/− mouse colon tissues (data not shown), and there were no significant differences in total stool bacterial burden prior to colitis induction (Figures S3A and S3B).
Figure 3. AIM2 protects host against colitis via regulation of colonic microbiota.
WT and Aim2−/− mice were fed with 3% DSS in drinking water for 5 days. (A) Colon homogenates collected at day 8 after DSS were plated on BHI and Blood agar and incubated in aerobic (BHI agar) or anaerobic (Blood agar) conditions at 37°C. (B) Feces collected from WT and Aim2−/− mice at day 8 after DSS administration were diluted in PBS and plated on BHI and Blood agar. After overnight culture at 37°C, cfu was counted. (C–E) WT and Aim2−/− mice were treated with a combination of antibiotics or left untreated for 4 weeks and then administered with 3% DSS for 5 days. (C) Body weight changes were monitored daily. (D) Mice were sacrificed at day 8 and colon lengths were measured. (E) Colons collected at day 8 after DSS administration were examined histologically. Scale bar = 100μm. Data represent mean ± SEM; n=7–10 and representative of 3 independent experiment. *, p<0.05, **, p<0.01, ***, p<0.001. (F–G) DNA was isolated from feces of healthy WT and Aim2−/− mice. Bacterial 16S rRNA gene sequences were quantitatively measured by real-time PCR. Data represent mean ± SEM (n=8–10/group); ***p<0.001. (H–I) Stools collected from WT and Aim2−/− mice before or 8 days after DSS administration were diluted in PBS and plated on MacConkey agar plate. Data represent mean ± SEM; ***, p<0.001. (J) 16S rRNA gene sequences of E. coli and Enterobacteriaceae in DNA isolated from stool at day 8 post DSS were quantitatively measured by real-time PCR. See also Figure S3.
These above findings led us to hypothesize that increased colitis susceptibility in Aim2−/− mice is associated with dysregulated host defense response against pre-existing commensal bacteria. Thus, we treated a cohort of WT and Aim2−/− mice with a cocktail of broad-spectrum antibiotics containing vancomycin, neomycin, ampicillin and metronidazole for 4 weeks as described (Rakoff-Nahoum et al., 2004). Antibiotics-treated or untreated WT and Aim2−/− mice were then fed with 3% DSS for 5 days. Interestingly, antibiotic-treated WT and Aim2−/− mice lost minimum body weight as compared to untreated mice (Figure 3C). While untreated Aim2−/− mice lost significantly greater body weight, exhibited shorter colons, and suffered from severe colonic injury and inflammation as compared to those of WT mice, antibiotic treatment completely attenuated the differences between Aim2−/− and WT mice at day 8 after DSS (Figures 3C – 3E). Consistently, the production of proinflammatory cytokines IL-6, KC, MIP2, and CCL2 was reduced to baseline in antibiotic-treated WT and Aim2−/− mouse colons (Figure S3C). These findings support the idea that increased colitis in Aim2−/− mice is due to bacterial overgrowth in the colon during acute colitis.
The composition of bacterial species present in the gut lumen regulates downstream immune responses and affects colitis severity. Accumulating evidence suggests that the presence of certain bacterial species in the gut, such as E. coli, Bacteroides, Clostridia, and Firmicutes, augments experimental colitis in mice (Frank et al., 2007; Li et al., 2014; Martinez-Medina and Garcia-Gil, 2014; Walters et al., 2014). A previous study showed that colitis susceptibility in Nlrp6−/−, ASC−/− and Il18−/− mice was associated with increased number of Prevotellaceae and TM7 (Elinav et al., 2011). To examine the possibility that disease severity in Aim2−/− mice was linked to the colonic load of Prevotellaceae and TM7, we measured the relative abundance of 16S rDNA specific to these bacteria by real-time PCR. Interestingly, there was no significant difference in Prevotellaceae or TM7 between Aim2−/− and WT mice (Figure 3F). In addition, we also measured 16S rDNA levels for other known mouse gut commensal and colitogenic bacteria such as Bacteroides, E. coli, Enterobacteriaceae, Lactobacillus, mouse intestinal Bacteroides (MIB), Eubacterium rectale (EREC), segmented filamentous bacteria (SFB), Staphylococcus, Streptococcus, Enterococcus, Clostridium, Prophyromonas and Bifidobacteria. With the notable exception of E. coli and Enterobacteriaceae, none of those commensal bacteria were significantly different between the two groups of mice (Figures 3F and 3G). However, E. coli and Enterobacteriaceae specific 16S rDNA was several thousand folds higher in Aim2−/− mouse feces compared to those in WT mice (Figure 3G). The overgrowth of E. coli was confirmed by cfu assay of fecal homogenates on MacConkey agar, a selective media for Enterobacteriaceae. While E. coli was barely detectable in WT mouse stool, Aim2−/− mice harbored 106 cfu of E. coli per gram of feces (Figure 3H). DSS administration further enhanced E. coli burden in Aim2−/− mice (Figures 3I and 3J). MacConkey agar colonies were unambiguously identified as E. coli by MALDI-TOF (data not shown). Other known colitogenic bacteria such as Bacteroides, SFB, Clostridium, Bifidobacterium, Prevotellaceae, and TM7 were not significantly different, even after colitis induction, in either set of mice (Figure S3D). However, increased growth of E. coli reflects an alteration of microbial composition in the intestine of Aim2−/− mice.
Dysbiosis Regulates Colitis Susceptibility in Aim2-deficient Mice
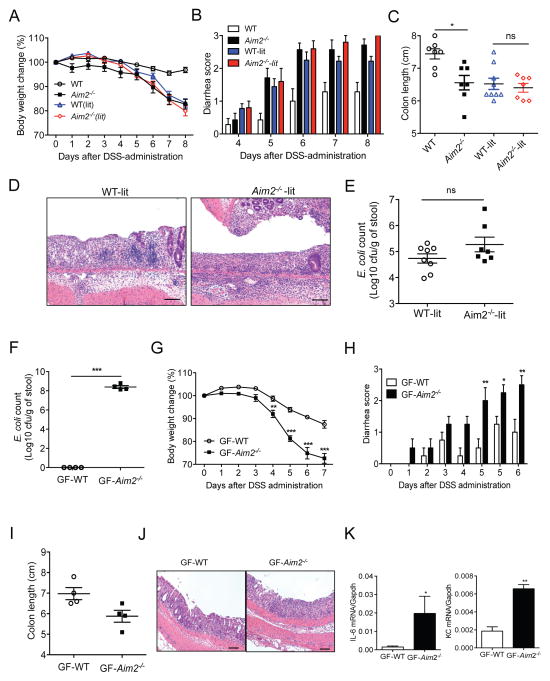
We postulated that dysbiosis, as evidenced by increased growth of E. coli in Aim2−/− mouse colon, exacerbates colitis development. To verify this hypothesis, we generated Aim2 heterozygous mice by breeding of Aim2−/− female and WT male mice. Littermate WT (WT-lit) and Aim2−/− (Aim2−/−-lit) mice obtained from breeding of Aim2 heterozygous mice were then administered with 3% DSS for 5 days. WT-lit and Aim2−/−-lit mice generated in this way should harbor intestinal flora originating from Aim2−/− mice. Interestingly, WT-lit mice developed pronounced colitis which was phenotypically very similar to the colitis in Aim2−/− mice as evidenced by body weight loss, clinical scores for diarrhea and rectal bleeding, and shortening of the colon (Figures 4A – 4C). Histopathological examination revealed that inflammation, ulceration, edema, and crypt destruction in WT-lit mice were morphologically indistinguishable from that seen in Aim2−/−-lit mice (Figure 4D). In agreement with comparable colitis susceptibility in littermate mice, an equal burden of E. coli was detected in WT-lit and Aim2−/−-lit mouse colons (Figure 4E) and similar bacterial growth was observed between WT-lit and Aim2−/−-lit mouse feces (Figures S4A and S4B).
Figure 4. Colitis susceptibility in Aim2−/− mice is contributed by altered microbiota.
(A–E) Littermate WT (WT-lit; n=9) and Aim2−/− (Aim2−/−-lit; n=7), non-littermate WT (n=7) and Aim2−/− (n=7) mice were fed with 3% DSS for 5 days, followed by regular drinking water for 3 days. Body weight (A) and stool consistency (B) were scored daily. Mice were sacrificed on day 8 to measure colon length (C) and examined for histopathological changes in colon tissue by H&E staining (D). (E) Fecal pellets collected from healthy littermate WT and Aim2−/− mice were diluted in PBS and cultured on MacConkey agar plate. Cfu of E. coli was counted. (F–L) Germ-free (GF) mice were co-housed with either conventionally raised WT (GF-WT) or Aim2−/− mice (GF-Aim2−/−) for 3 days. (F) 2 weeks after colonization, feces were homogenized in PBS and plated on MacConkey agar plate to count E. coli. (G–J) Microbiota-colonized GF mice (n=4/group) were fed with 3% DSS for 5 days. Body weight changes (G) and clinical scores for diarrhea (H) were monitored daily. (I) Colon lengths were measured at day 7 after DSS administration. (J) Colons were fixed in 10% formalin and processed for H&E staining. (K) A part of the colon was analyzed for IL-6 and KC production by ELISA. Data represent mean ± SEM. *, p<0.05; **, p<0.01. Scale bar = 100μm. See also Figure S4.
The above results suggested that colitis susceptibility of Aim2−/− mice might be transferable. To verify this, we co-housed WT and Aim2−/− mice for 4 weeks. Although adult WT mice were resistant to the maternal transfer of E. coli and failed to develop severe colitis (Figures S4C – S4E), colitis susceptibility of co-housed Aim2−/− mice was more mild than Aim2−/− mice housed without WT mice (Figures S4D and S4E). This finding suggests that unknown microflora transferred from WT mice to co-housed Aim2−/− mice plays a protective role against colitis, and exacerbated colitis in Aim2−/− mice is partly driven by the lack of protective microbiota. To further confirm that altered microbiota is responsible for increased colitis susceptibility in Aim2−/− mice, we colonized germ-free WT (GF) mice with microbiota present in conventionally raised WT or Aim2−/− mice. Two weeks after colonization, E. coli burden in GF mice colonized with fecal microbiota either from WT (GF-WT) or Aim2−/− (GF-Aim2−/−) mice was measured. While GF-Aim2−/− mice harbored huge E. coli burdens, no E. coli was detected in GF-WT mice (Figure 4F). Colitis was then induced in these two groups of mice by feeding them with 2.5% DSS for 5 days. We observed that GF-Aim2−/− mice lost more than 25% of their body weight at day 7 after DSS, while GF-WT mice were relatively resistant to colitis as evaluated by body weight loss, clinical scores, and colon length (Figures 4G – 4I). Consistently, there was significantly increased inflammation, production of proinflammatory cytokines, and bacterial growth in GF-Aim2−/− mouse colons (Figures 4J and 4K; Figures S4F and S4G). These results shed light on the importance of DNA sensor AIM2 in the regulation of commensal E. coli and other uncultured microorganisms that potentially regulate intestinal homeostasis and colitis pathogenesis.
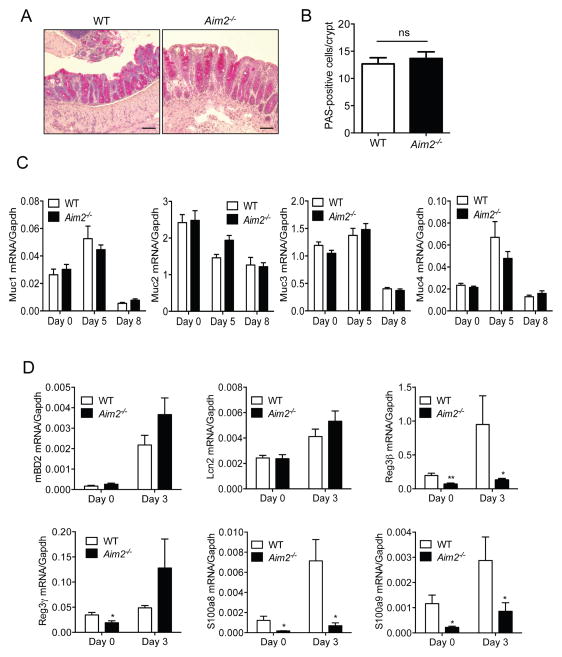
AIM2 Regulates the Expression of Antimicrobial Peptides in the Colon
Intestinal epithelial cells produce antimicrobial effectors that play a central role in the shaping of the gut microbial community and protecting mucosal tissues from colonization and invasion of commensal microorganisms (Gallo and Hooper, 2012). To understand why Aim2−/− mouse colons harbor altered microbiota, we examined whether AIM2 regulates goblet cell function by using periodic acid-schiff (PAS) staining (Figure 5A). We found the number of PAS-positive cells was comparable between WT and Aim2−/− mouse colons collected at day 5 following DSS-induced colitis (Figure 5B). We also did not see any difference in the levels of mucins (Muc1, 2, 3 and 4) present in WT and Aim2−/− mouse colons at baseline or following colitis induction (Figure 5C). Autophagy in intestinal epithelial cells plays an important role in regulating gut microbiota, but the conversion of LC3-I into LC3-II in WT and Aim2−/− was comparable both pre- and post-induction of colitis (Figures S5A and S5B). Intestinal epithelial cells also produce several AMPs, which either directly exert cytotoxicity or inhibit bacterial growth by limiting trace elements (Gallo and Hooper, 2012). We measured the expression of AMPs, including mouse β-defensin 2 (mBD2), Reg3γ and Reg3β, S100A8 and S100A9 and Lcn2 in WT and Aim2−/− mouse colons at day 0 and day 3 following colitis induction. Interestingly, while the expression of mBD2 and Lcn2 in healthy (day 0) WT and Aim2−/− mice was comparable, the level of Reg3γ, Reg3β, S100A8 and S100A9 was significantly reduced in Aim2−/− mouse colons as compared to those of WT mice (Figure 5D). The expression of Reg3β, S100A8 and S100A9 remained low in the early phases of colitis (day 3) (Figure 5D). These results suggest that AIM2 may regulate the bacterial burden of E. coli and Enterobacteriaceae via the production of AMPs. However, expression of Reg3γ, Reg3β, S100A8, S100A9 and Lcn2 were upregulated in Aim2−/− mice during acute colitis (day 8) (Figure S5C). Higher expression of these AMPs in Aim2−/− mice during colitis may be associated with higher bacterial burden since their expression in Aim2−/− and WT mice was reduced to the basal level upon treatment with antibiotics (Figure S5C).
Figure 5. AIM2 regulates production of antimicrobial peptides.
WT and Aim2−/− mice were fed with 3% DSS in drinking water for 5 days. (A) Colons collected at day 5 after DSS were fixed in 10% formalin and cryosections were stained with PAS. Scale bar = 100μm. (B) PAS-positive cells per crypt were counted. (C) Colons were collected at day 0 (n=6/group) and day 5 (n=6/group) after DSS administration. RNA was isolated and expression of Mucins (Muc1, Muc2, Muc3, Muc4) was analyzed by quantitative PCR. (D) Expression of antimicrobial peptides mBD2, Reg3β, Reg3γ, S100a8, S100a9 and Lcn2 in colon tissues collected at day 0 (D) and day 3 (E) after DSS-administration was measured by real-time PCR. Data represent mean ± SEM; n=8–10/group; *, p<0.05, **, p<0.01. See also Figure S5.
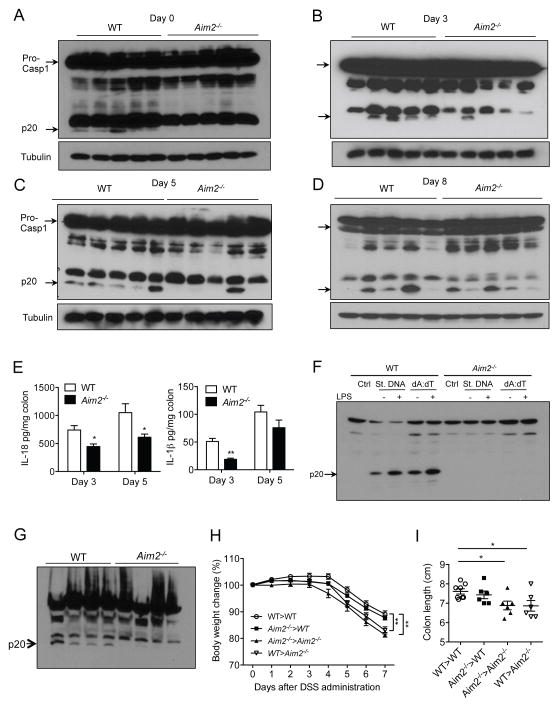
AIM2 is a Major Inflammasome Activator in the Colon
Previous studies have shown that the NLRP3 inflammasome plays a protective role against experimental colitis (Allen et al., 2010; Zaki et al., 2010a). Notably, caspase-1 activation in Nlrp3−/− mice is only partially reduced but not completely abrogated during DSS-colitis (Figure S6A), suggesting that other mechanisms of caspase-1 activation are active in the gut. We hypothesized that the dsDNA from colonizing bacteria contributes to intestinal homeostasis via activation of the AIM2 inflammasome. To test this hypothesis, we measured caspase-1 activation in colon tissues collected from WT and Aim2−/− mice at different days during colitis. Interestingly, caspase-1 activation in Aim2−/− mouse colons at day 0, 3 and day 5 after DSS administration was significantly reduced as compared to WT mouse colons (Figures 6A – 6D). However, a few Aim2−/− mice showed near normal caspase-1 activation in the colon, which may reflect the action of the NLRP3, NLRP6, or other inflammasomes. We consistently observed significantly reduced levels of IL-18, despite enhanced expression of IL-18 mRNA in inflamed colons of Aim2−/− mice at day 3 and 5 after DSS administration (Figure 6E; Figure S6B). Similarly, IL-1β production was suppressed by the absence of AIM2 (Figure 6E). Reduced activation of IL-1β and IL-18 in Aim2−/− mouse colons at healthy and colitic conditions was also seen in ex vivo colon organ culture (Figure S6C). In agreement with the function of IL-18 in epithelial cell proliferation (Dupaul-Chicoine et al., 2010; Zaki et al., 2010a), we observed significantly reduced proliferation of intestinal epithelial cells in Aim2−/− mice during acute colitis (day 5) but not at homeostasis (day 0) or delayed stage of colitis (day 8) (Figures S6D and S6E). These data support the hypothesis that bacterial dsDNA activates an AIM2 dependent inflammasome in the colon. To confirm that bacterial dsDNA present in the colonic lumen can activate the AIM2 inflammasome, we isolated DNA from mouse stool and transfected it into bone marrow-derived macrophages (BMDM). While fecal DNA activated caspase-1 in WT BMDM, cleaved caspase-1 was not detected in Aim2−/− BMDM (Figure 6F). However, activation of the NLRP3 inflammasome induced by LPS plus ATP or Nigericin was completely normal in Aim2−/− BMDM (Figure S6F). To understand which cellular compartment are involved in AIM2-dependent inflammasome activation, we isolated colonic epithelial cells and lamina propria mononuclear cells (LPMN) from WT and Aim2−/− mice at day 5 post DSS. While caspase-1 activation was significantly higher in WT epithelial cells compared to that of Aim2−/− mice (Figure 6G), it was barely detectable in LPMNs of either background of mice (data not shown). The notion that the epithelial compartment is the major site for the activation of the AIM2 inflammasome is further supported by bone marrow chimera studies. Chimeric mice having AIM2 in epithelial compartment, irrespective of the presence of AIM2 in myeloid cells (WT>WT and Aim2−/−>WT), developed less severe colitis as compared to mice deficient in AIM2 in the epithelium (Aim2−/−>Aim2−/− and WT>Aim2−/−) (Figures 6H and 6I). The observed defect in inflammasome activation in Aim2−/− mice was not dependent on altered gut microbiota since inflammasome activation remained defective in Aim2−/− mice co-housed with WT mice (Figure S6G).
Figure 6. AIM2 activates the inflammasome in the colon during colitis.
WT and Aim2−/− mice were fed with 3% DSS in drinking water for 5 days. (A–D) Colons collected at day 0, 3, 5 and 8 after DSS administration were homogenized and lysates were analyzed for caspase-1 activation by Western blotting. Cleaved caspase-1 (p20) is indicated by arrow. (E) Colon homogenates were assayed for IL-1β and IL-18 ELISA. Data represent mean ± SEM; n=7–10/group; *, p<0.05, **, p<0.01. (F) BMDM collected from WT or Aim2−/− mice were stimulated with 500 ng/ml LPS followed by transfected with either stool DNA or poly dA:dT (AIM2 ligand) for 4 h. Cell lysates were analyzed for caspase-1 activation by Western blotting. (G) Epithelial cells were isolated from WT and Aim2−/− mice at day 5 after DSS administration. Cell lysates were analyzed for caspase-1 activation by Western blotting. (H–I) Lethally irradiated WT and Aim2−/− mice were injected with either WT or Aim2−/− bone marrow cells and divided into 4 groups: WT>WT (n=8), WT>Aim2−/− (n=6), Aim2−/−>WT (6), Aim2−/−>Aim2−/− (n=6). Seven weeks after reconstitution, chimeric mice were fed with 3% DSS for 5 days. Body weight changes were monitored daily (H) and colon length was measured at day 7 (I). Data represent mean ± SEM. *, p<0.05; **, p<0.01. See also Figure S6.
The AIM2 Inflammasome Regulates Gut Microbiota and Colitis Susceptibility via Production of AMPs
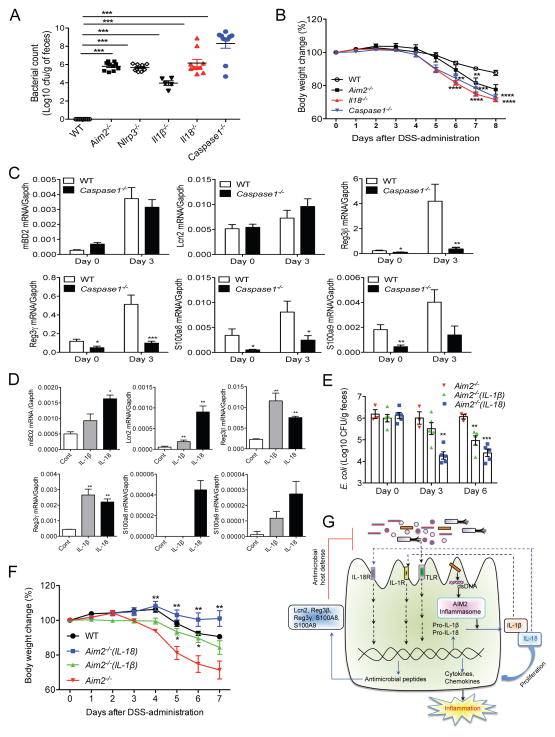
We next sought to determine whether AIM2 controls microbial growth via the inflammasome. Therefore, we measured E. coli burden in healthy WT, Aim2−/−, Nlrp3−/−, caspase-1−/−, Il1β−/− and Il18−/− mice. The E. coli burden was significantly higher than WT in all inflammasome-defective mice (Figure 7A; Figure S7A); however, E. coli burden in Il1β−/− mice was significantly less than other inflammasome-defective mice, suggesting that IL-18 plays a major role in maintaining intestinal microbial homeostasis. Dysregulated microbiota represented by increased growth of E. coli in caspase-1−/− and Il18−/− mice was also associated with increased colitis susceptibility as measured by body weight change, colon shortening, and clinical scores for diarrhea (Figure 7B; Figures S7B – S7D).
Figure 7. The inflammasome regulates intestinal microbiota via production of AMPs.
(A) Feces collected from healthy WT, Aim2−/−, Nlrp3−/−, Il1β−/−, caspase-1−/− and Il18−/− mice were diluted in PBS and plated on MacConkey agar. Colonies were counted after 24 h culture at 37°C. (B) Colitis was induced in WT, Aim2−/−, caspase-1−/− and Il18−/− (n=8–10) mice with 3% DSS. Body weights were monitored daily. Data represent mean ± SEM of a representative experiment; **, p<0.01, *** p<0.001. (C) Expression of mBD2, Reg3β, Reg3γ, S100a8, S100a9 and Lcn2 in colons collected at day 0 and 3 was measured by real-time PCR. Data represent mean ± SEM; n=7–10/group; *, p<0.05, **, p<0.01. (D) Colonic crypts were cultured and treated with IL-1β or IL-18 for 4 h. Expression of mBD2, Reg3β, Reg3γ, S100a8, S100a9 and Lcn2 was measured by RT-PCR. Data represent mean ± SD of triplicate wells; *, p<0.05, **, p<0.01. (E–F) Aim2−/− mice were treated with either IL-18 (0.5μg/mouse) or IL-1β (0.3μg/mouse) for 5 consecutive days. Feces collected at day 0, 3 and 6 after cytokine infusion were cultured on MacConkey agar to count E. coli burden. (F) WT (n=5), Aim2−/− (n=3), Aim2−/−(1L-1β) (n=5) and Aim2−/−(IL-18) (n=5) mice were fed with 3% DSS for 5 days. IL-1β- and IL-18-treated mice further received IL-1β (0.3μg/mouse) and IL-18 (0.5μg/mouse) respectively at day 1, 3 and 5 following DSS administration. Body weight changes were monitored daily. Data represent mean ± SEM. *, p<0.05, **, p<0.01. (G) The proposed pathway for the AIM2 inflammasome-mediated regulation of intestinal inflammation and microbiota. See also Figure S7.
To understand whether dysbiosis in caspase-1−/− mice is also due to the defects in AMP production, we measured the expression of mBD2, Reg3γ, Reg3β, S100A8, S100A9 and Lcn2 at homeostasis (day 0) or day 3 after DSS administration. Interestingly, similar to Aim2−/− mice (Figure 5D), the expression of Reg3γ, Reg3β, S100A8 and S100A9, was significantly reduced in caspase-1−/− mouse colon as compared to those in WT mice (Figure 7C). However, the level of mBD2 and Lcn2 was not different between caspase-1−/− and WT mouse colons (Figure 7C). This observation is further supported by in vitro studies showing that Reg3γ, Reg3β, S100A9 and Lcn2 were induced by both IL-1β and IL-18, and S100A8 was induced by IL-18 in colonic epithelial cells (Figure 7D; Figures S7E and S7F). Similarly, intestinal organoids expressed Reg3γ, Reg3β, S100A9 and Lcn2 in response to IL-1β and IL-18 (Figure S7G). Culture supernatants of IL-1β- and IL-18-stimulated colon organ culture effectively killed E. coli (Figure S7H). In vivo infusion of IL-18 led to a dramatic reduction of colonic E. coli growth in Aim2−/− mice, and protected them against DSS-induced colitis (Figures 7E and 7F). IL-1β infusion also demonstrated a suppressive effect on E. coli growth, but its effect was much less than that of IL-18 (Figures 7E and 7F). These results suggest that IL-18 produced by the AIM2 inflammasome binds to the IL-18R on epithelial and immune cells (Figures S7I and S7J), leading to the induction of antimicrobial peptides that play a central role in regulating the intestinal microbial ecology and inflammation (Figure 7G).
DISCUSSION
The surveillance of PAMPs by diverse PRRs expressed in intestinal epithelial cells, as well as immune cells, triggers a homeostatic immune response that allows for the peaceful coexistence of the intestinal mucosal tissue and gut microbiota. Microbial DNA is known to be sensed by TLR9, AIM2 and cGAS, leading to the activation of NF-κB, inflammasome, and interferon signaling pathways respectively (Hornung et al., 2009; Roberts et al., 2009; Sun et al., 2013; Tsujimura et al., 2004). TLR9-mediated recognition of DNA in the intestine exerts a protective effect against experimental colitis (Katakura et al., 2005; Rachmilewitz et al., 2002). Although the physiological relevance of DNA sensing by cGAS during intestinal inflammation has not been investigated to date, type-I interferon signaling has been shown to be critical in regulating Paneth and T-cell functions (Lee et al., 2012; Tschurtschenthaler et al., 2014). However, little is known about the role of AIM2 as a DNA sensor in intestinal homeostasis. This study demonstrates the involvement of AIM2 in DNA-dependent activation of the intestinal mucosal immune system, which critically provides protection against intestinal inflammation.
Pathogenesis of inflammatory bowel diseases (IBD) such as Crohn’s disease (CD) and ulcerative colitis involves both host genetic susceptibility and altered gut microbiota. Several bacterial species, including Escherichia coli, Bacteroides, Clostridia, Bifidobacteria and Firmicutes have been identified in clinical samples from IBD patients, suggesting a role for these bacteria in IBD pathogenesis (Frank et al., 2007; Li et al., 2014; Martinez-Medina and Garcia-Gil, 2014; Walters et al., 2014). Therefore, how host immune system regulates the growth of these pathobionts is a long sought after goal in IBD research. A major limitation of studies investigating the regulation of gut microbiota in experimental models is the diversity of microflora in animals from different laboratories. This may explain why we found no remarkable difference in Prevotellaceae and TM7 in inflammasome-deficient mice, unlike what had been previously described (Elinav et al., 2011). WT and all mutant mice included in this study were obtained from Jackson Laboratory and they were housed in the same animal facility at UT Southwestern. Thus, altered microbiota as represented by increased growth of E. coli in Aim2−/−, Nlrp3−/−, caspase-1−/− and Il18−/− mice is less likely due to the difference in source, location and other environmental factors. Notably, we did not see similar level of E. coli as of Aim2−/− and caspase-1−/− mice in Nod2−/− and Nlrp12−/− mice, which are also susceptible to experimental colitis and housed in the same animal facility (data not shown). However, this study warrants that microbiota-related disease phenotypes in experimental studies should be re-evaluated in different laboratory settings.
Increased growth of E. coli in Aim2−/− and other inflammasome-deficient mice is of great clinical significance. A growing number of clinical studies demonstrate that E. coli resides in the colonic and celiac mucosa of CD patients (Burke and Axon, 1988; Darfeuille-Michaud et al., 2004; Martinez-Medina and Garcia-Gil, 2014; Tabaqchali et al., 1978). Our understanding of the genetic link between increased E. coli colonization and CD susceptibility is poor. More than 100 loci associated with CD susceptibility have been identified including NOD2, ATG16L, IRGM, IL-23R, IL-10 and IL-18RAP (Cleynen et al., 2013). However, a common mechanism of controlling E. coli growth by these functionally diverse genes is less clear. Perhaps defects in innate and adaptive immune system lead to an altered host defense in the gut allowing increased growth of E. coli and other Enterobacteriaceae. Supporting this view, increased growth of Enterobacteriaceae was noted in T-bet−/−Rag2−/− mice, a mice model for spontaneous colitis (Garrett et al., 2010). An imbalance in the host immune response during inflammation has also been shown to boost E. coli growth in the colon (Winter et al., 2013). Our study demonstrates that the AIM2 inflammasome is a part of intestinal innate host defense against an overgrowth of commensal E. coli. Notably, the colitis phenotype in Aim2−/− mice is not solely dependent on E. coli since co-housed Aim2−/− mice developed attenuated colitis despite higher E. coli burden, suggesting that other unidentified bacteria that are transferred from WT to Aim2−/− mice during co-housing may have protected Aim2−/− mice against exacerbated colitis.
Although the inflammasome has emerged as a critical regulator of intestinal homeostasis, our knowledge of how it regulates the intestinal microbial community is limited. This study reveals a pathway for the regulation of gut microbiota by inflammasome-mediated production of AMPs, including Reg3β, Reg3γ, S100A8, S100A9 and Lcn2. AMPs produced by intestinal epithelial cells play critical roles in regulating the growth of the intestinal microbial community (Lai and Gallo, 2009; Ostaff et al., 2013). Previous studies have elucidated a role for Reg3β in host defense against Gram-negative bacteria (Stelter et al., 2011; van Ampting et al., 2012). S100A8 and S100A9 act as metal chelators, thus they limit the growth of pathogens by reducing the availability of essential trace elements such as Zn2+ and Mn2+ (Gallo and Hooper, 2012). Similarly, Lcn2 was shown to inhibit bacterial growth, including E. coli, by sequestering iron (Berger et al., 2006; Flo et al., 2004). Although the level of Lcn2 in healthy Aim2−/− and caspase-1−/− mouse colons were seen comparable to WT mice, its expression was strikingly induced by cytokines IL-1β and IL-18, suggesting that Lcn2 may be induced by the inflammasome and contribute to antimicrobial host defense in the gut. Since Lcn2 is also induced by MyD88-dependent pathways (Layoun et al., 2012), it is possible that an increase in the E. coli burden in inflammasome-defective mice may induce Lcn2 in the gut through activation of the TLR-MyD88 pathways, making the level of Lcn2 indistinguishable in comparison to WT mice. Taken together, these data suggest that inflammasome-associated cytokines regulate the growth E. coli and other gut microbiota through induction of multiple AMPs.
Our study shows that AIM2 is expressed in different cell population including myeloid, lymphocyte, and epithelial cells in the colon. However, how microbial DNA is delivered in the host cells is unknown. Since the AIM2 inflammasome is activated by diverse bacterial infections, it might be possible that gut bacteria are taken up by intestinal epithelial and myeloid cells leading to the release of bacterial DNA into the cytosol. A possibility of natural transfection of microbial DNA into intestinal immune and epithelial cell should be investigated in future studies.
In summary, this study underscores the critical role the DNA sensor AIM2 plays in regulating intestinal microbial community and inflammation. We show that AIM2 activates the inflammasome via sensing bacterial DNA in the gut. Although a protective role for the inflammasome in intestinal inflammatory disorders has been known for last five years, the underlying mechanism of inflammasome-mediated protection remains elusive. Therefore, the findings of this study, showing regulation of gut microbiota by the inflammasome via production of antimicrobial effector molecules, offer an insight into the pathogenesis of inflammatory disorders of the gut. Moreover, the AIM2 inflammasome may serve as a therapeutic target for the regulation of intestinal microbiota and inflammation.
EXPERIMENTAL PROCEDURES
Mice
Aim2−/−, Nlrp3−/−, Il18−/−, Caspase-1−/− (Caspase-1/11−/−) and WT (C57Bl6/J) mice were purchased from Jackson Laboratory. Il1β−/− mice (Shornick et al., 1996) were kindly shared by Dr. Chandrashekhar Pasare at UT Southwestern. All mice are maintained in a specific pathogen free (SPF) facility. All studies were approved by the Institutional Animal Care and Use Committee (IACUC) of UT Southwestern Medical Center and were conducted in accordance with the IACUC guidelines and the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All experiments were conducted with sex and age-matched mice and both male and female mice were included.
Colitis
Mice were treated with 3% (w/v) DSS (Molecular mass 36–40 kDa; TdB Consultancy) dissolved in sterile, distilled water ad libitum for the experimental days 1–5 followed by normal drinking water until the end of the experiment. Body weight, stool consistency and the presence of occult blood were determined daily up to day 8. Scoring system for diarrhea and rectal bleeding is described in Supplemental Information.
Gene Expression
For gene expression analysis, tissues were homogenized in Trizol reagent (Invitrogen). RNA was extracted according to the manufacturer’s instructions and reverse transcripted into cDNA. Quantitative PCR assays were performed with an CFX Connect real-time PCR detection system (Bio-rad) using iTaq Universal SYBR Green Supermix (Bio-rad). Expression of GAPDH was used as an endogenous control. Primers used for real-time PCR are listed in Supplementary Table 1.
Western Blot Analyses
For Western blot analysis, total proteins from tissues or cells were homogenized in RIPA lysis buffer containing protease and phosphatase inhibitor (Roche), resolved by SDS-PAGE, and transferred onto a PVDF membrane. Membranes were immunoblotted with rabbit antibodies against ERK (4695, Cell Signaling), Phospho-ERK (4370, Cell Signaling) and LC3A (4599, Cell Signaling), caspase-1 (AG-20B-0042, Adipogen) and AIM2 (13095,Cell Signaling). β-actin or tubulin was used as a loading control.
Bacterial Genomic DNA Isolation and Microbiota Analysis
Frozen fecal samples were processed for DNA isolation using QIAamp Fast DNA Stool Mini Kit (Qiagen). To analysis the relative number of specific intestinal bacterial groups, extracted bacterial genomic DNA was amplified for 16S rDNA with iTaq Universal SYBR Green Supermix (Bio-rad). Specific 16S rDNA primer sequences for real-time PCR are listed in Supplementary Table 2.
Statistical Analysis
Data are represented as mean ± SEM. Data were analyzed by Prism5 (GraphPad Software) and statistical significance was determined by two-tailed student t-test. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank John Shelton and Angie Mobley for their assistance in histopathology and flow cytometry respectively. This work was supported by Crohn’s and Colitis Foundation Career Development Award (3711), a New Investigator Award from the American Cancer Society (IRG-02-196-10) and the Harold C. Simmons Comprehensive Cancer Center (National Cancer Institute/National Institutes of Health, P30 CA142543), and UT Southwestern startup funding given to M.H.Z.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, two tables and Supplemental References.
AUTHOR CONTRIBUTIONS
S.H., Y.T.K. and M.H.Z. designed and performed experiments and analyzed data. L.P. performed histopathological examination of colon tissues. E.M.T. identified bacterial species isolated from feces by MALDI-TOF system. J.S.M., C.P. and L.V.H. provided reagents and helped editing the manuscript. M.H.Z conceived this study and wrote the manuscript.
References
- Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, Herfarth HH, Jobin C, Ting JP. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger T, Togawa A, Duncan GS, Elia AJ, You-Ten A, Wakeham A, Fong HE, Cheung CC, Mak TW. Lipocalin 2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia-reperfusion injury. Proc Nat Acad Sci of U S A. 2006;103:1834–1839. doi: 10.1073/pnas.0510847103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Burke DA, Axon AT. Adhesive Escherichia coli in inflammatory bowel disease and infective diarrhoea. BMJ. 1988;297:102–104. doi: 10.1136/bmj.297.6641.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Shaw MH, Redondo G, Nunez G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Can Res. 2008;68:10060–10067. doi: 10.1158/0008-5472.CAN-08-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- Cleynen I, Gonzalez JR, Figueroa C, Franke A, McGovern D, Bortlik M, Crusius BJ, Vecchi M, Artieda M, Szczypiorska M, et al. Genetic factors conferring an increased susceptibility to develop Crohn’s disease also influence disease phenotype: results from the IBDchip European Project. Gut. 2013;62:1556–1565. doi: 10.1136/gutjnl-2011-300777. [DOI] [PubMed] [Google Scholar]
- Cridland JA, Curley EZ, Wykes MN, Schroder K, Sweet MJ, Roberts TL, Ragan MA, Kassahn KS, Stacey KJ. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol Biol. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, Meunier C, Turbide C, Gros P, Beauchemin N, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–378. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. NLRP6 Inflammasome Regulates Colonic Microbial Ecology and Risk for Colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Hara H, Sakai S, Hernandez-Cuellar E, Mitsuyama M, Kawamura I, Tsuchiya K. Type I interferon signaling regulates activation of the absent in melanoma 2 inflammasome during Streptococcus pneumoniae infection. Infect Immun. 2014;82:2310–2317. doi: 10.1128/IAI.01572-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Nat Acad Sci of U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota SA, Ng J, Lueng A, Khajah M, Parhar K, Li Y, Lam V, Potentier MS, Ng K, Bawa M, et al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis. 2011;17:1359–72. doi: 10.1002/ibd.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O’Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Nat Acad Sci of U S A. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katakura K, Lee J, Rachmilewitz D, Li G, Eckmann L, Raz E. Toll-like receptor 9-induced type I IFN protects mice from experimental colitis. J Clin Invest. 2005;115:695–702. doi: 10.1172/JCI22996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Bauernfeind F, Ablasser A, Hartmann G, Fitzgerald KA, Latz E, Hornung V. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur J Immunol. 2010;40:1545–1551. doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Inflammasomes: guardians of cytosolic sanctity. Immunol Rev. 2009;227:95–105. doi: 10.1111/j.1600-065X.2008.00730.x. [DOI] [PubMed] [Google Scholar]
- Layoun A, Huang H, Calve A, Santos MM. Toll-like receptor signal adaptor protein MyD88 is required for sustained endotoxin-induced acute hypoferremic response in mice. Am J Pathol. 2012;180:2340–2350. doi: 10.1016/j.ajpath.2012.01.046. [DOI] [PubMed] [Google Scholar]
- Lee SE, Li X, Kim JC, Lee J, Gonzalez-Navajas JM, Hong SH, Park IK, Rhee JH, Raz E. Type I interferons maintain Foxp3 expression and T-regulatory cell functions under inflammatory conditions in mice. Gastroenterology. 2012;143:145–154. doi: 10.1053/j.gastro.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Butcher J, Mack D, Stintzi A. Functional Impacts of the Intestinal Microbiome in the Pathogenesis of Inflammatory Bowel Disease. Inflamm Bowel Dis. 2014;21:139–53. doi: 10.1097/MIB.0000000000000215. [DOI] [PubMed] [Google Scholar]
- Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, Kagnoff MF, Eckmann L, Karin M. Nod2 mutation in Crohn’s disease potentiates NF-kappaB activity and IL-1beta processing. Science. 2005;307:734–738. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- Martinez-Medina M, Garcia-Gil LJ. Escherichia coli in chronic inflammatory bowel diseases: An update on adherent invasive Escherichia coli pathogenicity. World J Gastrointest Pathophysiol. 2014;5:213–227. doi: 10.4291/wjgp.v5.i3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostaff MJ, Stange EF, Wehkamp J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol Med. 2013;5:1465–1483. doi: 10.1002/emmm.201201773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Na HS, Song YR, Shin SY, Kim YM, Chung J. Activation of NLRP3 and AIM2 inflammasomes by Porphyromonas gingivalis infection. Infect Immun. 2014;82:112–123. doi: 10.1128/IAI.00862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmilewitz D, Karmeli F, Takabayashi K, Hayashi T, Leider-Trejo L, Lee J, Leoni LM, Raz E. Immunostimulatory DNA ameliorates experimental and spontaneous murine colitis. Gastroenterology. 2002;122:1428–1441. doi: 10.1053/gast.2002.32994. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Salcedo R, Worschech A, Cardone M, Jones Y, Gyulai Z, Dai RM, Wang E, Ma W, Haines D, O’HUigin C, et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J Exp Med. 2010;207:1625–1636. doi: 10.1084/jem.20100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shornick LP, De Togni P, Mariathasan S, Goellner J, Strauss-Schoenberger J, Karr RW, Ferguson TA, Chaplin DD. Mice deficient in IL-1beta manifest impaired contact hypersensitivity to trinitrochlorobenzone. J Exp Med. 1996;183:1427–1436. doi: 10.1084/jem.183.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter C, Kappeli R, Konig C, Krah A, Hardt WD, Stecher B, Bumann D. Salmonella-induced mucosal lectin RegIIIbeta kills competing gut microbiota. PloS one. 2011;6:e20749. doi: 10.1371/journal.pone.0020749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabaqchali S, O’Donoghue DP, Bettelheim KA. Escherichia coli antibodies in patients with inflammatory bowel disease. Gut. 1978;19:108–113. doi: 10.1136/gut.19.2.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschurtschenthaler M, Wang J, Fricke C, Fritz TM, Niederreiter L, Adolph TE, Sarcevic E, Kunzel S, Offner FA, Kalinke U, et al. Type I interferon signalling in the intestinal epithelium affects Paneth cells, microbial ecology and epithelial regeneration. Gut. 2014;63:1921–31. doi: 10.1136/gutjnl-2013-305863. [DOI] [PubMed] [Google Scholar]
- Tsujimura H, Tamura T, Kong HJ, Nishiyama A, Ishii KJ, Klinman DM, Ozato K. Toll-like receptor 9 signaling activates NF-kappaB through IFN regulatory factor-8/IFN consensus sequence binding protein in dendritic cells. J Immunol. 2004;172:6820–6827. doi: 10.4049/jimmunol.172.11.6820. [DOI] [PubMed] [Google Scholar]
- van Ampting MT, Loonen LM, Schonewille AJ, Konings I, Vink C, Iovanna J, Chamaillard M, Dekker J, van der Meer R, Wells JM, et al. Intestinally secreted C-type lectin Reg3b attenuates salmonellosis but not listeriosis in mice. Infect Immun. 2012;80:1115–1120. doi: 10.1128/IAI.06165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Sanders CJ, Taylor RT, Kumar A, Aitken JD, Sitaraman SV, Neish AS, Uematsu S, Akira S, Williams IR, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007;117:3909–3921. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, Baba N, Libioulle C, Belaiche J, Bitton A, et al. Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat Genet. 2009;41:71–76. doi: 10.1038/ng285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS lett. 2014;588:4223–33. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TM, Leeth RA, Rothschild DE, Coutermarsh-Ott SL, McDaniel DK, Simmons AE, Heid B, Cecere TE, Allen IC. The NLRP1 inflammasome attenuates colitis and colitis-associated tumorigenesis. J Immunol. 2015;194:3369–3380. doi: 10.4049/jimmunol.1402098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010a;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Lamkanfi M, Kanneganti TD. The Nlrp3 inflammasome: contributions to intestinal homeostasis. Trends Immunol. 2011a;32:171–179. doi: 10.1016/j.it.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti TD. IL-18 Production Downstream of the Nlrp3 Inflammasome Confers Protection against Colorectal Tumor Formation. J Immunol. 2010b;185:4912–20. doi: 10.4049/jimmunol.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Vogel P, Malireddi RK, Body-Malapel M, Anand PK, Bertin J, Green DR, Lamkanfi M, Kanneganti TD. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 2011b;20:649–660. doi: 10.1016/j.ccr.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.