Summary
Protein synthesis is crucial for the maintenance of long-term-memory-related synaptic plasticity. The prion-like cytoplasmic polyadenylation element-binding protein 3 (CPEB3) regulates the translation of several mRNAs important for long-term synaptic plasticity in the hippocampus. Here, we provide evidence that the prion-like aggregation and activity of CPEB3 is controlled by SUMOylation. In the basal state, CPEB3 is a repressor and is soluble. Under these circumstances, CPEB3 is SUMOylated in hippocampal neurons both in vitro and in vivo. Following neuronal stimulation, CPEB3 is converted into an active form that promotes the translation of target mRNAs, and this is associated with a decrease of SUMOylation and an increase of aggregation. A chimeric CPEB3 protein fused to SUMO cannot form aggregates and cannot activate the translation of target mRNAs. These findings suggest a model whereby SUMO regulates translation of mRNAs and structural synaptic plasticity by modulating the aggregation of the prion-like protein CPEB3.
Graphical abstract
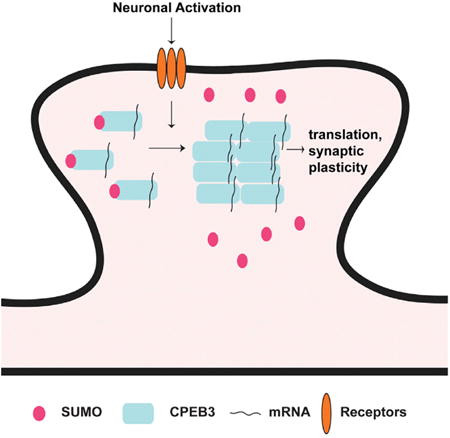
Drisaldi et al. argue that, under basal, unstimulated conditions, CPEB3 is SUMOylated and mostly soluble. After neuronal stimulation, CPEB3 becomes deSUMOylated and more aggregated. DeSUMOylation and aggregation are two crucial steps required for the translation of the mRNA targets of CPEB3 and for dendritic filopodia formation.
Introduction
Persistence of memory is achieved by the growth of new synaptic connections that are maintained by local synthesis at the synapse of key synaptic proteins (Kandel et al., 2014). In Aplysia, local protein synthesis is regulated by the cytoplasmic polyadenylation element-binding protein (ApCPEB), a prion-like protein, which mediates the persistence of long-term synaptic facilitation by its ability to translate dormant mRNAs (Si et al., 2003, 2010). A key feature of Aplysia CPEB is its N-terminal domain, which is enriched in glutamine and asparagine residues (Q/N) resembling the Q/N-rich domain of yeast prions (Si et al., 2003). As is the case with prion proteins, Aplysia CPEB exists in at least two functional states: a soluble, inactive form and an insoluble, aggregated and active prionic form that can self-propagate. An antibody that selectively binds the self-sustaining oligomers of Aplysia CPEB does not interfere with the establishment of long-term facilitation but selectively blocks its maintenance (Si et al., 2010). These results first suggested that the prion-like properties of Aplysia CPEB are functional and regulate persistence of synaptic facilitation. The Drosophila homolog of Aplysia CPEB, Orb2, has similarly been found to be critical for the maintenance of long-term memory through a prion-like mechanism (Majumdar et al., 2012).
Mammals express four CPEB isoforms (CPEB1, CPEB2, CPEB3, and CPEB4; Theis et al., 2003). CPEB3 contains a Q/N-rich domain at its N-terminal and is the mammalian homo-log of Aplysia CPEB. We found that, as with Aplysia CPEB, mouse CPEB3 also exists in two conformational states: a soluble and an insoluble and aggregated form. The presence of the N-terminal domain is required for the aggregation of CPEB3 and for the maintenance of long-term memory (Fioriti et al., 2015). Neuronal stimulation activates CPEB3 and leads to an increase in its ubiquitination by the E3 ubiquitin ligase Neuralized1. The aggregated, active forms of CPEB3 can then initiate the translation of target mRNAs such as GluA1 and GluA2, two crucial components inlong-term synaptic plasticity (Pavlopoulos et al., 2011; Fioriti et al., 2015). Stephan et al. (2015) describe that mouse CPEB3 is also a prion-like protein, which can self-propagate in yeast. However, whereas there is evidence suggesting that aggregation of Drosophila Orb2 is regulated by phosphorylation (White-Grindley et al., 2014), how aggregation of mammalian CPEB3 is regulated is unclear.
In the brain, protein aggregation is highly regulated and, when aberrant, can lead to degenerative brain diseases (Ross and Poirier, 2004). One mechanism that has been proposed to prevent homo-oligomerization in neurons is SUMOylation of the individual moieties of the aggregation-prone proteins (Dorval and Fraser, 2007; Krumova et al., 2011). For example, SUMOylation of α-synuclein inhibits its aggregation in vitro and could prevent the consequent toxicity that contributes to Parkinson's (Krumova et al., 2011). Similarly, Huntingtin, a pathogenic protein that causes Huntington's disease when aggregated, shows less aggregation when SUMOylated (Steffan et al., 2004).
SUMOylation is a post-translational modification that results from the covalent attachment of SUMO-1, SUMO-2, or SUMO-3 to lysine residues of target proteins by SUMO ligases. As the name suggests, small ubiquitin modifier (SUMO) is a post-translational modification similar to ubiquitination. A constellation of proteins are SUMOylated in cells (Gareau and Lima, 2010). In addition to its putative role in preventing protein aggregation, SUMOylation regulates several non-aggregation-related cellular processes and pathways. These include DNA repair, transcription, trafficking of proteins, and synaptic transmission (Gareau and Lima, 2010). Here, we find that SUMOylation of CPEB3 regulates its oligomerization and thereby the activity-dependent translation of some of its targets known to be involvedin synaptic plasticity. When hippocampal neurons are in the basal state, CPEB3 actsasrepressor.Inthis basal state, we find that CPEB3 is SUMOylated. Following stimulation of hippocampal neurons, either in cultures or in vivo, CPEB3 is rapidly de-SUMOylated. This allows the oligomerization and aggregation of CPEB3 necessary for its activation as a regulator of translation. A SUMO-CPEB3 chimeric protein that is uncleavable fails both to aggregate and to induce translation of its targets. We also identify the SUMO-2 mRNA as a target of CPEB3, suggesting a regulatory feedback loop between CPEB3 and SUMO-2. These results suggest that SUMOylation serves as a negative regulator of CPEB3 oligomerization and prevents the aggregation required for the translation of its target mRNAs.
Results
CPEB3 Is SUMOylated in Primary Hippocampal Neurons
As a first step in exploring a possible role for SUMOylation in the aggregation of CPEB3, we determined conditions that would allow ready detection of CPEB3 oligomerization. We found that exposure to glycine, a glutamatergic agonist capable of inducing long-term potentiation (LTP) chemically (Lu et al., 2001), increases the level of the CPEB3 and activates the proteins in hippocampal cultured neurons (Fioriti et al., 2015). In so doing, glycine also induces translation of SUMO and at least a subset of known mRNA targets of CPEB3, including GluA1 and GluA2 (Fioriti et al., 2015; Jaafari et al., 2013). Because of its ability to activate CPEB3, we used this protocol for studying the role of SUMO in regulating the activity and aggregation of CPEB3.
To compare the conformation of CPEB3 before and after stimulation, we made use of a biochemical assay that measures the solubility of CPEB3 in a detergent buffer. Depending on their conformations, amyloid and amyloid-like proteins are partially or totally insoluble in these buffers (Drisaldi et al., 2003). We activated hippocampal neurons by exposing them to glycine. Fifteen minutes after stimulation, we harvested the neurons and divided them into a soluble (supernatant) and an insoluble (pellet) fraction by centrifugation. Compared to unstimulated conditions, the treatment causes endogenous CPEB3 to accumulate in the insoluble fraction, suggesting a possible oligomerization of the protein in response to chemical activation of neurons (Figures 1A and 1B).
Figure 1. CPEB3 Is SUMOylated and Aggregates in Cultured Neurons.
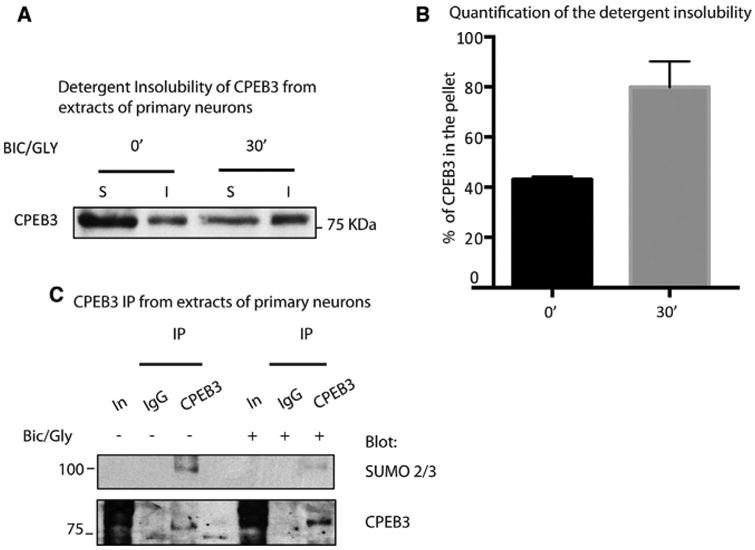
(A) Western blotting (WB) showing detergentinsolubility assay of neuronal cells stimulated for0 and 30min with glycine. Soluble (S) and insoluble(I) fractions were probed with endogenous CPEB3antibody.
(B) Graph showing the percent of CPEB3 in theinsoluble fraction as a mean of five independenttrials ± SE (t- Stud p < 0.0001).
(C) WB showing immunoprecipitation of endogenous CPEB3 from cultured neurons stimulatedwith glycine for 0 and 15 min (CPEB3, immunoprecipitation using an antibody against CPEB3;IgG, IgG negative control; In, Input). The top panelshows WB using an antibody raised againstSUMO-2/3. The bottom panel shows WB using anantibody raised against CPEB3.
To determine whether SUMOylation correlates directly with aggregation of CPEB3, we immunoprecipitated the protein and determined to what degree it was SUMOylated before and after exposure to glycine. Cultured neurons were harvested and lysed in RIPA buffer, and the whole-cell lysates were subjected to immunoprecipitation using a mouse antibody against CPEB3. In unstimulated conditions, an antibody raised specifically against SUMO-2/3 detected a band at approximately 100 KDa, corresponding to the predicted molecular weight of CPEB3 covalently linked to SUMO. This indicated that, in unstimulated neurons, when CPEB3 is mainly soluble and not aggregated, it is SUMOylated (Figure 1C). We also found that CPEB3 can be SUMOylated in 293 cells and in vitro and it interacts with Ubc9 (Figure S2). To study whether SUMOylation affects aggregation, we next stimulated neurons with glycine. Fifteen minutes after stimulation, we found the SUMOylation of CPEB3 to be decreased (Figure 1C). Taken together, these results suggest that chemical activation of CPEB3 can lead to oligomerization and that this oligomerization is correlated with a decrease in SUMOylation of CPEB3.
Learning Decreases SUMOylation of CPEB3 in Hippocampal Extracts
To study further the potential role of SUMO in the aggregation of CPEB3, we determined whether CPEB3 is SUMOylated in vivo and whether SUMOylation is decreased following contextual fear conditioning, a hippocampus-dependent behavioral task that requires CPEB3 (Figure S1).
To first determine whether CPEB3 was SUMOylated in the absence of a stimulus, we immunoprecipitated CPEB3 from hippocampal extracts, and we found that, similar to cultured neurons, a fraction of CPEB3 is SUMOylated in vivo in the basal state. By contrast, 15 min after a foot shock that induces contextual fear conditioning (0.6 mA for 2 s), the fraction of CPEB3 that is SUMOylated decreases compared to control animals exposed to the chamber, but not shocked (Figure 2A). In addition, in earlier studies, Pavlopoulos et al. (2011) found that there was an increase of in vitro monoubiquitination of CPEB3 following neuronal stimulation. We now confirmed these findings in mice by finding that the monoubiquitination of CPEB3 increases following contextual fear conditioning (Figure 2B). We next examined whether the decrease in SUMOylation of CPEB3 following foot shock is associated with increased aggregation. Compared to hippocampal extracts from control animals, we found that, 30 min after foot shock, there is an increase in accumulation of CPEB3 protein in the insoluble fraction (Figures 2C and 2D). Thus, CPEB3 can form oligomers following fear conditioning, mirroring what we found in cultured neurons following neuronal stimulation. Together, these experiments suggest that a reduction in SUMOylation of CPEB3 coincides with its aggregation.
Figure 2. Fear Conditioning Induces CPEB3 Aggregation and a Decrease of SUMOylation in the Hippocampus.
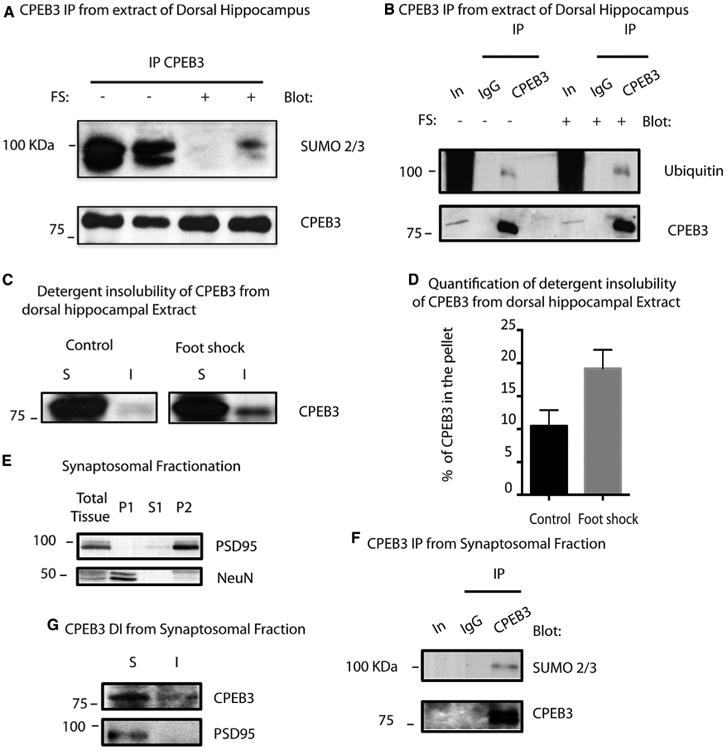
(A) WB showing immunoprecipitation of endogenous CPEB3 from hippocampal extracts 30min after chamber exposure and foot shock (FS).The experiment wasdone in duplicate. An antibody raised against SUMO-2/3 was used in the top panel. The bottom panel shows WB using an antibody raised against CPEB3.
(B) WB showing immunoprecipitation of endogenous CPEB3 from hippocampal extracts 30 min after chamber exposure and FS. An antibody raised againstubiquitin was used in the top panel. CPEB3 was used for the bottom panel.
(C) WB showing detergent insolubility assay of hippocampal extracts 30 min after chamber exposure or foot shock. S and I fractions were probed with antibodyagainst endogenous CPEB3.
(D) Graph showing the percentage of CPEB3 in the insoluble fraction as a mean of three independent trials ± SE (t-Stud p = 0.023).
(E) WB showing the fractionation of cortical and hippocampal tissue in P1 (nuclear fraction), S1 (cytoplasmic fraction), and P2 (synaptosome fraction andmitochondria).
(F) Immunoprecipitation of CPEB3 from synaptosome fraction (P2).
(G) WB showing detergent insolubility of CPEB3 and PSD95 in synaptosomes (P2).
CPEB3 protein is present both in the nucleus and in the cytoplasm of hippocampal neurons. To test whether CPEB3 is also SUMOylated in synapses, we performed a fractionation assay and purified synaptosomes from hippocampal and cortical tissue of adult mice (Figure 2E). We then immunoprecipitated CPEB3 from the synaptosome preparation (Figure 2F) and probed for SUMO-2/3. We found that CPEB3 is also SUMOylated in the synaptosomal fraction, suggesting that SUMOylation of CPEB3 could regulate local protein synthesis at the synapses (Figure 2F). To determine whether CPEB3 protein can form aggregates at the synapse, we subjected the syn-aptosomal fraction to detergent insolubility assay. We found CPEB3 both in the supernatant and in the insoluble fractions, suggesting that it can form amyloid-like aggregates at synapses (Figure 2G).
SUMOylation Affects CPEB3 Aggregation Both In Vitro and In Vivo
To study the role of SUMOylation in the aggregation of CPEB3 more directly, we made a chimeric protein, in which CPEB3 is fused to an uncleavable SUMO-2. Based on our previous results, we predicted that the presence of SUMO at the N terminus of CPEB3 would prevent its aggregation.
We first tested the aggregation properties of CPEB3 and SUMO2-CPEB3 in an in vitro assay. We translated CPEB3, SUMO-1-CPEB3, and SUMO-2-CPEB3 in vitro and performed a detergent insolubility assay using two different concentrations of SDS: 0.1% and 0.2%. We found that, when fused in frame to CPEB3, SUMO-2, but not SUMO-1, increased its solubility at both concentrations, with a stronger effect at higher detergent concentrations (Figure 3A). This result is consistent with previous findings showing that a stronger detergent buffer has more disaggregation capabilities than a milder one (Drisaldi et al., 2003; Lehmann and Grassi, 2004). We next overexpressed CPEB3 and SUMO-2-CPEB3 in 293 cells and performed the detergent insolubility assay. We found CPEB3 to be in both soluble and insoluble fractions, and compared to CPEB3 alone, the presence of SUMO-2 increased the solubility of CPEB3 (Figure 3B).
Figure 3. SUMOylation of CPEB3 Decreases Its Aggregation.
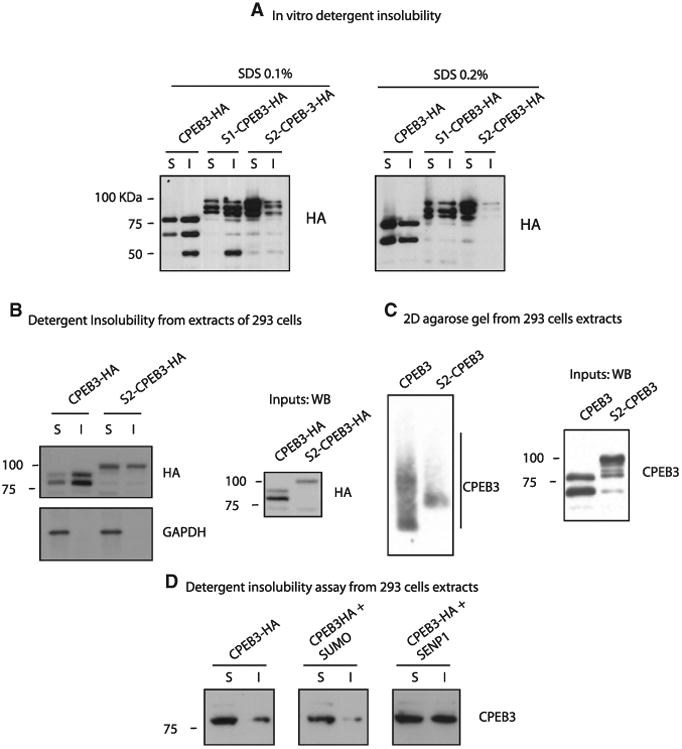
(A) WB showing detergent insolubility assay of in-vitro-translated CPEB3-HA, SUMO-1-CPEB3-HA,and SUMO-2-CPEB3-HA fusion proteins in thepresence of two different concentrations of SDS(0.1% and 0.2%).
(B) WB showing detergent insolubility assay ofHEK293 cells extracts overexpressing CPEB3-HAand SUMO-2-CPEB3-HA fusion proteins. An HAantibody was used to detect the constructs.
(C) Left panel shows a WB of a 2D agarose gelshowing aggregates formation for CPEB3-HA andSUMO-2-CPEB3-HA in HEK293 cells. Right panelshows a WB of the input material.
(D) WB showing detergent insolubility assay ofHeLa cells extracts overexpressing CPEB3 alone,CPEB3-HA and SUMO-2, and CPEB3-HA andSENP1.
These findings prompted us to characterize further the differences in oligomeric state between CPEB3 and SUMO-2-CPEB3 by performing a native agarose gel analysis, a technique that allows one to study the prion-like properties of proteins. In contrast to a detergent insolubility assay, an agarose gel allows the discrimination of different species of insoluble aggregates with dissimilar molecular weight, thus allowing a better resolution of the aggregates formed by CPEB3 and by SUMO-2-CPEB3 (Bagriantsev et al., 2006). We found that, whereas overex-pressed CPEB3 formed a smear throughout the lane, suggesting that the protein exists in different aggregated forms, SUMO-2-CPEB3 formed a much more restricted smear, indicating the inability of the protein to form heavy-molecular-weight oligomers (Figure 3C). These findings suggest that SUMO-2-CPEB3 does not block oligomerization of CPEB3 completely but only inhibits the formation of the largest oligomers. Taken together, these experiments suggest that CPEB3 forms amyloid-like aggregates both in vitro and in cells and that SUMO-2 prevents the formation of the highest-molecular-weight subset of them.
To study further the role of SUMO-2 in negatively regulating the aggregation of CPEB3, we cotransfected CPEB3 and SUMO-2 in HeLa cells. We found that overexpression of SUMO decreases the amount of CPEB3 in the insoluble fraction, whereas overexpression of the SUMO-specific protease SENP2 increases the amount of CPEB3 in the insoluble fraction (Figure 3D). These experiments support the idea that inducing SUMOylation of CPEB3 increases its solubility, whereas decreasing its SUMOylation enhances its insolubility. Together, these experiments illustrate that the SUMO machinery regulates CPEB3 aggregation.
SUMOylation of CPEB3 Affects Its Target Expression
To test whether aggregation and SUMOylation regulate CPEB3 mRNA targets, we decided to overexpress either CPEB3 or SUMO-2-CPEB3, and we tested the expression of CPEB3 targets after neuronal stimulation. We overexpressed CPEB3 and SUMO-2-CPEB3 together with the actin 3′ UTR reporterplasmids in hippocampal neurons (Stephan et al., 2015). Following stimulation withglycine,wefound that,incontrasttothe activation that occurs when CPEB3 itself is expressed, SUMO-2-CPEB3 inhibited induction of the target luciferase mRNA (Figure 4A). These findings indicate that SUMOylation not only prevents CPEB3 aggregation but also prevents the translation of the target mRNAsofCPEB3.Wenextexaminedtheeffectofoverexpressing CPEB3 and SUMO-2-CPEB3 on the formation of filopodia, following stimulation (Pavlopoulos et al., 2011). We found that glycine rapidly increases the number of dendritic filopodia, the precursors of dendritic spines (Alvarez and Sabatini, 2007; Segal, 2005). Cells overexpressing CPEB3 showed an increase in the number of filopodia compared to untreated control 30 min after stimulation. However, neurons expressing SUMO-2-CPEB3 failed to show an increase in filopodia, suggesting that aggregation of CPEB3 following stimulus is not only important for expression of the mRNA targets ofCPEB3 but also for the morphological changes that occur at the level of the synapses that underlie synaptic plasticity (Figures 4B and 4C).
Figure 4. SUMOylation Prevents Translation of CPEB3 mRNA Targets.
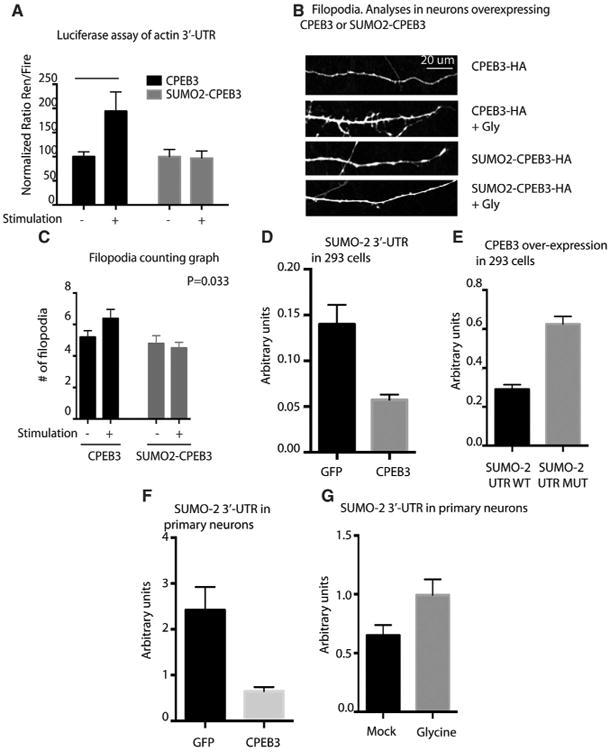
(A) Luciferase assay of actin UTR in 293 with overexpression of CPEB3 or SUMO-2-CPEB3. n = 3 experiments; n = 8 replicates. ANOVA shows a significant interaction between the four conditions (p = 0.0003).
(B and C) Filopodia counts. Primary hippocampal neurons express CPEB3 or SUMO-2-CPEB3. ANOVA shows a significant interaction between the four conditions (p = 0.0335).
(D and E) Luciferase assay of SUMO-2 UTR or SUMO-2 CPE mutant UTR in 293 with over-expression ofCPEB3.n=3 replicates (p<0.0001).
(F and G) Luciferase assay of SUMO-2 UTR in primary hippocampal neurons with over-expression of CPEB3 and stimulation with glycine. n = 3 replicates (p < 0.0001).
CPEB3 Regulates Translation of SUMO-2 mRNA in Cells and Neurons
The 3′ UTR of the SUMO-1 mRNA is thought to contain cytoplasmic polyadenylation elements (CPEs) that can be recognized by the CPEB protein (Jaafari et al., 2013). We noticed that the mRNA of SUMO-2 contains four putative CPEs in its 3′ UTR (Figure S3A). We therefore asked whether CPEB3 could regulate the translation of the SUMO-2 mRNA. We first cloned the full-length 3′ UTR of SUMO-2 in a luciferase plasmid and then assessed whether overexpression of CPEB3 in HeLa (Figure S3B) or 293 cells can modulate its translation. In 293 cells, compared to GFP alone, overexpression of CPEB3 reduced the translation of the luciferase construct. Moreover, the mutation of the CPE elements of the SUMO-2 UTR rescued the translation of the luciferase construct in the presence of overexpression of CPEB3 (Figures 4D and 4E). We next determined whether CPEB3 also regulates SUMO-2 mRNA translation in neurons. We found that, in neurons, as in immortalized human cell lines, SUMO-2 mRNA is a target of CPEB3. Interestingly, in hippocampal neurons, CPEB3 suppresses SUMO-2 translation in unstimulated conditions, whereas, following glycine treatment, CPEB3 induces the translation of SUMO-2 mRNA (Figures 4F and 4G). These results suggest that CPEB3 and SUMO-2 might regulate one another.
We summarize this analysis in a model illustrated in Figure 5. According to this model, SUMOylation is a dynamic post-translational modification that negatively regulates the aggregation of CPEB3. In basal, unstimulated conditions, CPEB3 is SUMOylated and mostly soluble. After neuronal stimulation, CPEB3 becomes deSUMOylated and more aggregated. DeSUMOylation and aggregation are two crucial steps required for the translation of the mRNA targets of CPEB3 and for dendritic filopodia formation. In addition, once active, CPEB3 can induce translation of the SUMO-2 mRNA, suggesting a negative feedback regulatory loop between CPEB3 and SUMO-2 presumably designed to limit the aggregation of CPEB3 in both time and amount.
Figure 5. CPEB3 and SUMO-2 Regulate Each Other.
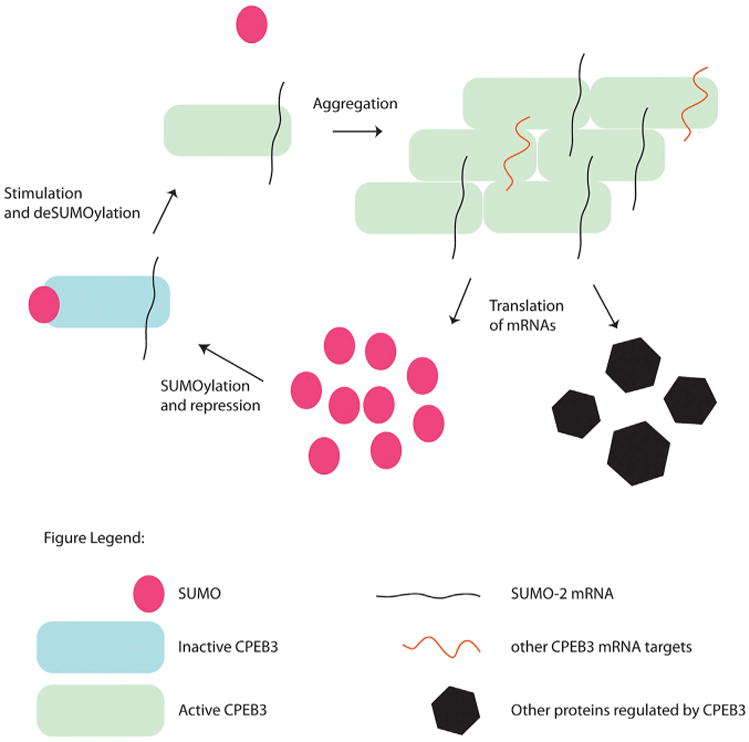
SUMO-2 prevents aggregation of CPEB3 molecules before treatment. DeSUMOylation of the protein facilitates aggregation and mRNA translation. The SUMO-2 mRNA is a target of CPEB3, and it is induced upon stimulation of neurons and deSUMOylation of CPEB3.
Discussion
Because CPEB3 can form aggregates in yeast resembling prion-like molecules (Stephan et al., 2015), it seemed likely that the aggregation process is highly regulated in vivo. Using primary hippocampal neurons, we found that, in unstimulated conditions, CPEB3 is both soluble and SUMOylated (by SUMO-2). Following stimulation of hippocampal neurons, SUMOylation of CPEB3 decreases, and this is accompanied by an enhancement of both the levels of CPEB3 and its aggregation. A similar mechanism is operative in vivo. Hippocampal extracts taken from control animals reveal CPEB3 to be SUMOylated. By contrast, 30 min after fear conditioning, SUMOylation of CPEB3 decreases and the protein becomes more aggregated, consistent with the idea that the inhibition of aggregation correlates with the degree of SUMO-2 conjugation. To determine whether this mechanism is causal, we explored whether a chimeric CPEB3 fused in frame to an uncleavable SUMO-2 fails to aggregate to the same extent as the wild-type version of the protein. Indeed, we find that this inhibition of the aggregation prevents the translation of target mRNAs. These experiments define SUMO as a central inhibitory constraint of the physiological formation of the prion-like form of CPEB3, which is essential for the maintenance of long-term memories (Fioriti et al., 2015).
SUMO and Synaptic Plasticity
Biochemical and imaging experiments have defined SUMO as one of many regulators of synaptic plasticity, contributing, for example, to the regulation of receptors trafficking during the induction of chemical LTP (Luo et al., 2013). Indeed, induction of chemical LTP in cultured neurons with glycine induces the expression of SUMO-1 and Ubc9 (Jaafari et al., 2013). Here, we describe a new role for SUMO in negatively regulating synaptic plasticity. In cultured neurons, SUMOylation of CPEB3 prevents the formation of new filopodia following stimulation, inhibiting the translation of CPEB3 mRNA targets. Interestingly, we also found that CPEB3 regulates the translation of the SUMO-2 mRNA. Upon stimulation, CPEB3 is deSUMOylated and induces the translation not only of other transcripts such as those of GluA1 and GluA2 but also of SUMO-2. It is therefore possible that an increase of SUMO-2 protein levels is required not only to regulate synaptic plasticity but also to regulate the degree to which CPEB3 itself is SUMOylated, thereby providing a negative feedback mechanism regulating the activity of CPEB3 after the initial stimulus.
The Modulation of Prion-like Proteins by SUMOylation Suggests a Connection between Functional and Pathological Prions
We have analyzed the formation of CPEB3 oligomers in the mouse under two different in vivo conditions: (1) in hippocampal neuronal cells following chemical activation with glycine and (2) in brain homogenates of the dorsal hippocampus following training for learned fear (foot shock). In both cases, the stimuli that trigger a transient deSUMOylation of CPEB3 precede the formation of oligomers. Conversely, fusing SUMO in frame to CPEB3 prevents its aggregation.
A similar role for SUMO-2 in the inhibition of aggregation has been attributed to α-synuclein in vitro, where SUMOylation of just 10% of the proteins dramatically affects the aggregation of α-synuclein. Insoluble aggregates of this protein are implicated in Parkinson's disease, and SUMOylation is thought to prevent its aggregation in cells (Krumova et al., 2011). Thus, the disease is thought to represent, at least in part, a failure of the inhibitory constraint on aggregation mediated by SUMOylation.
SUMO has also been proposed to regulate aggregation of β-amyloid and tau, two proteins found aberrantly aggregated in Alzheimer's disease, by affecting their expression and stability (Lee et al., 2013). We here described the role of SUMO in regulating the aggregation of the physiological prion-like protein CPEB3. Although we find that physiological amyloids share similar biophysical properties to pathological ones and both are SUMOylated in vivo, physiological and pathological amyloids differ in the consequence of their SUMOylation. SUMOylation of CPEB3 is a constitutive, reversible process in the cell, mediating a physiological function. By contrast, SUMOylation of α-synuclein, amyloid-β, and tau might prevent an aberrant aggregation, as it serves as a self-protective mechanism in these neurons.
SUMO increases the solubility of other proteins, and it is often used to tag proteins expressed in bacteria that are insoluble and prone to aggregation and thus difficult to purify (Peroutka III et al., 2011). Although we still do not know how SUMO prevents aggregation, it has been proposed that the 3D structure of SUMO prevents the aggregation of proteins directly. It may also affect the aggregation properties of target proteins. An alternative mechanism that has been proposed is that SUMO directly affects inter- or intramolecular interactions of target proteins (Graña-Montes et al., 2014; Sabate et al., 2012). As mentioned above, our bioinformatics analysis has not as yet located a putative SUMO-interacting motif in the protein sequence of CPEB3, thereby excluding a direct interaction of SUMO and CPEB3. It is possible, however, that SUMO prevents the binding of CPEB3 with itself or other molecules, thus preventing the formation of amyloid-like aggregates. Nevertheless, we cannot exclude the possibility that, in addition to SUMO, other post-translational modifications, such as phosphory-lation, can participate in the regulation of CPEB3 aggregates as has been suggested for the regulation of protein aggregation in RNA granules (Han et al., 2012; Kato et al., 2012). There is also the question of how CPEB3 aggregates are disaggregated. Whereas SUMO might have a role in disaggregation, alternatively, it is also possible that SUMO can only act on the soluble CPEB3 molecules, preventing them from forming aggregates.
A Model for the Regulation of Physiological Prions Involved in Memory
Long-term memory is mediated by an increase in synaptic strength that persists over time (Kandel et al., 2014; Si et al., 2010). This persistence is synapse-specific and regulates local protein synthesis. In previous work, we and others found that, in Aplysia, Drosophila, and mice, learning leads to the activation of the aggregation-prone molecules ApCPEB, ORB2, and CPEB3, respectively. These proteins are translational regulators at the synapse necessary for the maintenance of long-term memory. Intriguingly, in each case, their activation is associated with homo-oligomerization (Bailey et al., 2004; Si et al., 2003; Stephan et al., 2015). Both recombinant ApCPEB and CPEB3 are soluble in urea-containing buffers, but they promptly form aggregates in more permissive solutions with a concentration of urea lower than 2 M. This suggests that, like other prions, the ApCPEB and CPEB3 proteins can exist in multiple stable conformations in the absence of any post-translational modifications. We here find that the aggregation of CPEB3 in vivo and in vitro is restrained by SUMOylation. SUMO inhibits CPEB3 aggregation and consequently impairs its activation and translation of targets and could have a detrimental effect on memory formation when misregulated. Thus, we provide further evidence that, following a stimulus, the aggregation of the prion-like molecule CPEB3 is a fundamental step in the function of protein synthesis required in memory maintenance.
Many questions remain unanswered, however. For example, how are CPEB3 aggregates regulated? Does SUMO also have a role in disaggregation? Or is its role limited to preventing soluble CPEB3 to become part of aggregates? Moreover, is CPEB3 the only functional prion-like protein regulated by SUMOylation? Or is this a general mechanism for limiting the aggregation of functional prion-like proteins and perhaps even prions in general, be they functional or pathogenic? Conversely, are there other types of regulating mechanisms that inhibit the aggregation of prion-like proteins besides SUMOylation? Because prion-like proteins with a physiological function have recently also been found in the immune system (Hou et al., 2011), it now becomes of general interest to determine whether the capability of SUMOylation to inhibit aggregation is unique for CPEB3 or whether it is serving as a more general mechanism for restraining prion-like molecules from aggregating.
Experimental Procedures
Neuronal Stimulation
Chem-LTP was induced as described previously (Jaafari et al., 2013). Briefly, neuronal cultures were transferred from Neurobasal growth medium to extracellular solution (ECS) containing 150 mM NaCl, 2 mM CaCl2, 5 mM KCl, 10 mM HEPES (pH 7.4), 30 mM glucose, 0.5 mM TTX, and 20 mM bicuculline methiodide. After 5 min in ECS, neuronal cultures were treated with glycine (200 mM) for 3 min in ECS and then incubated in ECS without glycine for different time points.
Detergent Insolubility Assay
We used a modified procedure described by Tatzelt et al. (1996). Cells and brain homogenates were lysed for 30 min at 4°C in the following buffer: 0.5% Triton X-100; 0.5% NP-40; 0.5% sodium deoxycholate; and 50 mM Tris-HCl (pH 7.5). After debris was removed by centrifugation at 16,000 × g for 10 min, the supernatant was centrifuged in a TLA 55 rotor at 65,000 rpm for 40 min. Proteins in the supernatant were precipitated with methanol, and CPEB3 in the supernatant and insoluble fractions was analyzed by western blotting (Chiesa et al., 1998; Drisaldi et al., 2003). For the in vitro detergent insolubility assay, CPEB3-HA, SUMO-1-CPEB3-HA, and SUMO-2-CPEB3-HA were in vitro translated in the TNT rabbit reticulocyte lysate system (Promega) and incubated for 1 hr in the detergent insolubility buffer.
2D Agarose Gel
Cells were washed in ice-cold PBS and re-suspended in the protein extraction buffer (50 mM Tris-HCl [pH7.5], 50 mM KCl, and 10mM MgCl2) supplemented with 10 mM phenylmethylsulfonyl fluoride and Complete protease inhibitor cocktail (Roche Applied Science) to prevent protein degradation. SDS-AGE gel electrophoresis of whole-cell lysates was performed as previously described (Bagriantsev and Liebman, 2004). Priorto loading on the gel, lysates (∼40 mg of total protein) were incubated in the sample buffer (25 mMTris, 200 mM glycine [pH 8.3], 2% SDS, 5% glycerol, and 0.025% bromophenol blue) at room temperature (∼25°C) for 7 min. To dismantle aggregates, samples were incubated in boiling water bath instead of the room temperature incubation.
Synaptosomes Preparation
Synaptosome preparation was done according to Munton et al. (2007). In brief, hippocampi and cortex were dissected from 10- to 12-week-old mice and homogenized in synaptoneurosome buffer (10 mM Tris [pH 7.5], 2.2 mM CaCl2, 0.5 mM Na2HPO4,0.4 mM Kh2PO4,4 mM NaHCO3, and 80 mM NaCl) using a Teflon-glass mechanical tissue grinder. The homogenate was centrifuged at 1,000 g for 5 min at 4°C. The supernatant was further centrifuged at 10,000 g for 10 min at 4°C. The synaptoneurosome and mitochondrial pellet was next re-suspended and lysed in RIPA buffer for immunoprecipitation or detergent insolubility buffer to perform a detergent insolubility assay.
Luciferase Assay
Luciferase assays were performed using the Dual Luciferase Promega Kit following the instructions of the manufacturer. Briefly, primary hippocampal neurons at 8/9 DIV were transfected with CPEB3 orSUMO-2-CPEB3 plasmids with a mixture of Renilla luciferase harboring actin or SUMO UTR and Firefly luciferase containing no UTR. Twenty-four hours after transfection, cells were stimulated with glycine for 30 min, lysed, and dual luciferase activity was measured. Cells were lysed in 100 ml of 1 × passive lysis buffer. Twenty microliters of the lysates were used for the quantification of Firefly and Renilla luciferase activity using a Luminometer (Turner Design).
Filopodia Analysis
Primary hippocampal neurons at 9 DIV were transfected with CPEB3-HA or SUMO-2-CPEB3 bi-cistronic plasmids expressing also GFP. Twenty-four hours after transfection, cells were stimulated with glycine for 30 min and fixed and GFP-positive cells were analyzed for filopodia. Forty-micrometer branches per neuron were analyzed, and filopodia were counted by eye. For each group, we counted eight neurons and four branches each.
Highlights.
CPEB3 is SUMOylated in hippocampal neurons
CPEB3 regulates SUMO-2 translation
SUMO-2 inhibits aggregation of CPEB3
Following neuronal activity, CPEB3 SUMOylation is decreased
Acknowledgments
We would like to thank the members of the E.R.K. laboratory for their technical help, comments, and criticism. We would like to also thank M.J.K. Jones for the gift of HeLa and 293 cells and for helpful comments on the manuscript, J.D. Richter for the gift of the CPEB3 antibody, and D.B. Beck for helpful comments on the manuscript. The work was supported by HHMI (to E.R.K.). B.D. and L.C. were partially supported by The Italian Academy for Advanced Studies in America and by the Alexander Bodini Foundation. S.H. is affiliated with Inception 3, Inc.
Footnotes
Supplemental Information: Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.04.061.
Author Contributions: B.D., L.C., and L.F. designed, performed, and analyzed the experiments. With all authors' comments, B.D., L.C., and L.F. wrote the manuscript.
References
- Alvarez VA, Sabatini BL. Anatomical and physiological plasticity of dendritic spines. Annu Rev Neurosci. 2007;30:79–97. doi: 10.1146/annurev.neuro.30.051606.094222. [DOI] [PubMed] [Google Scholar]
- Bagriantsev S, Liebman SW. Specificity of prion assembly n vivo [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem. 2004;279:51042–51048. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- Bailey CH, Kandel ER, Si K. The persistence of long-term memory: a molecular approach to self-sustaining changes in learning-induced synaptic growth. Neuron. 2004;44:49–57. doi: 10.1016/j.neuron.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Chiesa R, Piccardo P, Ghetti B, Harris DA. Neurological illness n transgenic mice expressing a prion protein with an insertional mutation. Neuron. 1998;21:1339–1351. doi: 10.1016/s0896-6273(00)80653-4. [DOI] [PubMed] [Google Scholar]
- Dorval V, Fraser PE. SUMO on the road to neurodegeneration. Biochim Biophys Acta. 2007;1773:694–706. doi: 10.1016/j.bbamcr.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Drisaldi B, Stewart RS, Adles C, Stewart LR, Quaglio E, Biasini E, Fior-iti L, Chiesa R, Harris DA. Mutant PrP is delayed in its exit from the endoplasmic reticulum, but neither wild-type nor mutant PrP undergoes retrotranslocation prior to proteasomal degradation. J Biol Chem. 2003;278:21732–21743. doi: 10.1074/jbc.M213247200. [DOI] [PubMed] [Google Scholar]
- Fioriti L, Myers C, Huang YY, Li X, Stephan JS, Trifilieff P, Colnaghi L, Kosmidis S, Drisaldi B, Pavlopoulos E, Kandel ER. The persistence of hippocampal-based memory requires protein synthesis mediated by the prion-like protein CPEB3. Neuron. 2015;86:1433–1448. doi: 10.1016/j.neuron.2015.05.021. [DOI] [PubMed] [Google Scholar]
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graña-Montes R, Marinelli P, Reverter D, Ventura S. N-terminal protein tails act as aggregation protective entropic bristles: the SUMO case. Biomacromolecules. 2014;15:1194–1203. doi: 10.1021/bm401776z. [DOI] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, McKnight SL. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafari N, Konopacki FA, Owen TF, Kantamneni S, Rubin P, Craig TJ, Wilkinson KA, Henley JM. SUMOylation is required for glycine-induced increases in AMPA receptor surface expression (ChemLTP) in hippo-campal neurons. PLoS ONE. 2013;8:e52345. doi: 10.1371/journal.pone.0052345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER, Dudai Y, Mayford MR. The molecular and systems biology of memory. Cell. 2014;157:163–186. doi: 10.1016/j.cell.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumova P, Meulmeester E, Garrido M, Tirard M, Hsiao HH, Bossis G, Urlaub H, Zweckstetter M, Kügler S, Melchior F, et al. Sumoylation inhibits alpha-synuclein aggregation and toxicity. J Cell Biol. 2011;194:49–60. doi: 10.1083/jcb.201010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L, Sakurai M, Matsuzaki S, Arancio O, Fraser P. SUMO and Alzheimer's disease. Neuromolecular Med. 2013;15:720–736. doi: 10.1007/s12017-013-8257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S, Grassi J. Techniques in Prion Research (Birkhä user) 2004 [Google Scholar]
- Lu W, Man H, Ju W, Trimble WS, MacDonald JF, Wang YT. Activation of synaptic NMDA receptors induces membrane insertion of new AMPA receptors and LTP in cultured hippocampal neurons. Neuron. 2001;29:243–254. doi: 10.1016/s0896-6273(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Luo J, Ashikaga E, Rubin PP, Heimann MJ, Hildick KL, Bishop P, Gir-ach F, Josa-Prado F, Tang LT, Carmichael RE, et al. Receptor trafficking and the regulation of synaptic plasticity by SUMO. Neuromolecular Med. 2013;15:692–706. doi: 10.1007/s12017-013-8253-y. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Cesario WC, White-Grindley E, Jiang H, Ren F, Khan MR, Li L, Choi EM, Kannan K, Guo F, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148:515–529. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Munton RP, Tweedie-Cullen R, Livingstone-Zatchej M, Weinandy F, Waidelich M, Longo D, Gehrig P, Potthast F, Rutishauser D, Gerrits B, et al. Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol Cell Proteomics. 2007;6:283–293. doi: 10.1074/mcp.M600046-MCP200. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Trifilieff P, Chevaleyre V, Fioriti L, Zairis S, Pagano A, Malleret G, Kandel ER. Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell. 2011;147:1369–1383. doi: 10.1016/j.cell.2011.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peroutka RJ, III, Orcutt SJ, Strickler JE, Butt TR. SUMO fusion technology for enhanced protein expression and purification inprokaryotes and eukaryotes. Methods Mol Biol. 2011;705:15–30. doi: 10.1007/978-1-61737-967-3_2. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Sabate R, Espargaro A, Gran˜ a-Montes R, Reverter D, Ventura S. Native structure protects SUMO proteins from aggregation into amyloid fibrils. Biomacromolecules. 2012;13:1916–1926. doi: 10.1021/bm3004385. [DOI] [PubMed] [Google Scholar]
- Segal M. Dendritic spines and long-term plasticity. Nat Rev Neurosci. 2005;6:277–284. doi: 10.1038/nrn1649. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers insensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, et al. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- Stephan JS, Fioriti L, Lamba N, Colnaghi L, Karl K, Derkatch I, Kandel ER. The CPEB3 protein is a functional prion that interacts with actin cytoskeleton. Cell Rep. 2015;11:1772–1785. doi: 10.1016/j.celrep.2015.04.060. this issue. [DOI] [PubMed] [Google Scholar]
- Tatzelt J, Maeda N, Pekny M, Yang SL, Betsholtz C, Eliasson C, Cayetano J, Camerino AP, DeArmond SJ, Prusiner SB. Scrapie in mice deficient in apolipoprotein E or glial fibrillary acidic protein. Neurology. 1996;47:449–453. doi: 10.1212/wnl.47.2.449. [DOI] [PubMed] [Google Scholar]
- Theis M, Si K, Kandel ER. Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc Natl Acad Sci USA. 2003;100:9602–9607. doi: 10.1073/pnas.1133424100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Grindley E, Li L, Mohammad Khan R, Ren F, Saraf A, Florens L, Si K. Contribution of Orb2A stability in regulated amyloid-like oligomerization of Drosophila Orb2. PLoS Biol. 2014;12:e1001786. doi: 10.1371/journal.pbio.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]


