Abstract
Pediatric cardiomyopathy is a rare but severe disease with high morbidity and mortality. The causes are poorly understood and can only be established in one-third of cases. Recent advances in genetic technologies, specifically next-generation sequencing, now allow for the detection of genetic causes of cardiomyopathy in a systematic and unbiased manner. This is particularly important given the large clinical variability among pediatric cardiomyopathy patients and the large number of genes (>100) implicated in the disorder. We report on the performance of whole-exome sequencing in members of a consanguineous family with a history of pediatric hypertrophic cardiomyopathy and sudden cardiac death, which led to the identification of a homozygous stop variant in the SLC22A5 gene, implicated in primary carnitine deficiency, as the likely genetic cause. Targeted carnitine tandem mass spectrometry analysis in the patient revealed complete absence of plasma-free carnitine and only trace levels of total carnitine, further supporting the causality of the SLC22A5 variant. l-carnitine supplementation in the proband led to a rapid and marked clinical improvement. This case illustrates the use of exome sequencing as a systematic and unbiased diagnostic tool in pediatric cardiomyopathy, providing an efficient route to the identification of the underlying cause, which lead to appropriate treatment and prevention of premature death.
Introduction
Primary carnitine deficiency (PCD) is an autosomal recessive disorder of carnitine transportation with an estimated incidence between 1/40 000 and 1/120 000.1 It is caused by compound heterozygous or homozygous variants in the SLC22A5 gene, which encodes for the organic cation transporter type 2 (OCTN2).2 OCTN2 has an important role in transporting carnitine across the cell membrane. Defects in OCTN2 lead to impairment of carnitine transportation and result in urinary carnitine wasting, low serum carnitine levels and decreased intracellular carnitine accumulation.3, 4, 5 Newborn screening is available to detect low carnitine levels in infants with PCD and has been implemented in many countries worldwide.6 The clinical presentation varies widely with respect to age of disease onset and organ involvement, where the patient may be asymptomatic or suffer a life-threatening cardiomyopathy.7 In early infancy, patients typically present with metabolic decompensation with episodes of hypoketotic hypoglycemia and hepatomegaly, elevated transaminases, and hepatic encephalopathy. Later in childhood (around the age of 4 years), the presentation is characterized by muscle weakness, dilated or hypertrophic cardiomyopathy and congestive heart failure. If the disease remains unrecognized and no supplementation of l-carnitine is started, heart failure may deteriorate and eventually lead to death. Crucially, early treatment can result in complete resolution of cardiomyopathy.8, 9, 10 We present a family with hypertrophic cardiomyopathy (HCM) and sudden cardiac death (SCD) in multiple siblings in whom genetic testing using whole-exome sequencing (WES), established the diagnosis of PCD leading to treatment with oral l-carnitine supplementation that resulted in a rapid and marked clinical improvement in the sole surviving affected individual.
Patient presentation
Initial presentation
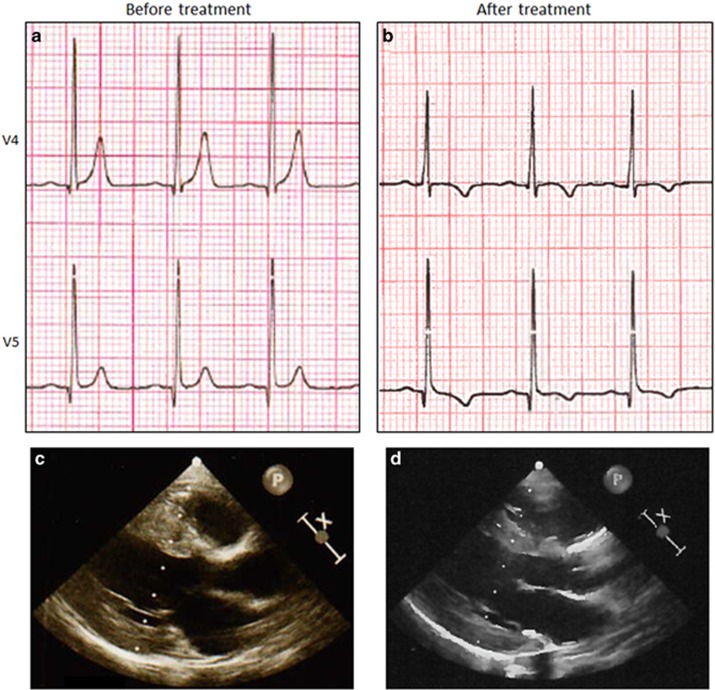
The proband was a 3-year-old girl of Moroccan descent who was born to a 25-year-old female at 39 weeks of gestation via caesarian section. She had normal development until the age of 2 years. In the months prior to admission, the parents noted progressive fatigue, shortness of breath and pallor. The patient had no learning difficulties or mental retardation. She was referred to a pediatric cardiologist and at the time of presentation was in a good overall condition. During physical examination, pallor of skin and conjunctivae were observed. No audible heart murmur was noted and the peripheral pulse was normal. She had normal neurological examination and no dysmorphic features were identified. Her electrocardiogram (ECG) revealed high voltage R and deep S waves in V5 and V2, respectively, suggesting left ventricular hypertrophy, tall peaked T-waves and a short QTc interval, (318 ms; Figure 1a). Trans-thoracic echocardiography revealed situs solitus levocardia with atrioventricular and ventricular arterial concordance. There was good systolic function with a shortening fraction of 36%. Severe concentric hypertrophy was observed without outflow or intra-cavity obstruction (Figure 1c). The left ventricular posterior wall and the interventricular septum were measured 8.3 mm (normal range (NR), 3.2–6.1; z-score,11 3.26; height, 102 cm; weight, 14 kg) and 10.5 mm, respectively (NR, 3.5–7.1; z-score, 3.45; Table 1). There was no mitral regurgitation and the rest of the heart was normal. Her creatine kinase level was 110 U/l (NR, 30–200) and hemoglobin level measured 7 g/dl (NR, 11–13). The patient lived in an isolated area of the south of Morocco with limited access to specialized clinical infrastructure and unfortunately biochemical testing for known treatable causes of pediatric cardiomyopathy was not performed at this stage. In addition, there is, at present, no nationwide newborn screening program in Morocco.
Figure 1.
Electrocardiogram (ECG) and trans-thoracic echocardiography (TTE) before (left) and after (right) treatment with carnitine supplementation. (a) Pretreatment precordial ECG tracings of V4 showing tall peaked T-waves, and high voltage R-wave and a short QTc interval (318 ms). (b) Precordial ECG tracing (lead V4) after 3 months of treatment showing the decreased T-wave amplitude, inversion of the T-wave and QTc interval normalization (400 ms). (c) At pretreatment TTE, LVPWd and the IVSd measured 8.3 and 10.5 mm, respectively. (d) After 3 months of the treatment, both LVPWd and IVSd decreased and measured 6.8 and 7.9 mm, respectively. IVSd, end-diastolic interventricular septal thickness; LVPWd, end-diastolic left ventricular posterior wall thickness.
Table 1. Homozygous variants found in patient II.5.
| Gene | Chromosome | Reference sequence | Allele change | Protein change | Associated disease |
|---|---|---|---|---|---|
| SLC22A5 | Chr5 | NM_003060.3 | c.1188 T>G | p.(Tyr396*) | PCD |
| RNF167 | Chr17 | NM_015528.2 | c.833dup | p.(Pro279Serfs*17) | None |
Abbreviation: PCD, Primary carnitine deficiency.
Family history
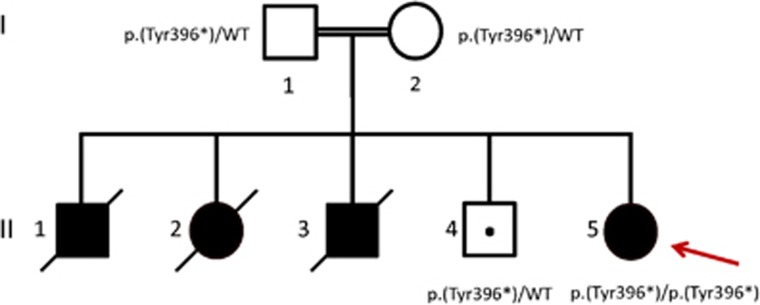
Because of a remarkable family history of two siblings dying suddenly at young age, she was referred for further investigation to the department of medical genetics in Rabat (Morocco). The pedigree of the family is shown in Figure 2. The proband (II-5) is the offspring of consanguineous parents. Her oldest brother (II-1) died suddenly at 6 months of age. Her sister (II-2) had a very similar clinical course as the proband with progressive fatigue and shortness of breath. She was seen by a pediatric cardiologist and diagnosed with severe hypertrophic cardiomyopathy at the age of 3 years. Unfortunately, she died suddenly shortly after the diagnosis was made. The third sibling (a boy, II-3) died at 20 weeks of gestation. His fetal ultrasound showed severe hypertrophic cardiomyopathy. The patient's only living sibling (II-4) is 7 years old and is doing well. Considering the significant family history and heterogeneity of genetic causes of pediatric cardiomyopathy,12, 13 we performed WES to detect the potential underlying genetic cause to assist in the making of a definite diagnosis.
Figure 2.
Pedigree showing segregation of the p.(Tyr396*) variant in SLC22A5. Filled-in symbols indicate affected individuals and symbols with a slash through them indicate deceased individuals. The proband is indicated by an arrow. The dot symbol indicates a carrier; males are indicated by squares and females by circles.
Exome sequencing
The study protocol was approved by the local Institutional Review Board and signed informed consent was obtained from the parents. DNA was extracted from peripheral blood and WES was carried out on the proband (II-5), her unaffected brother (II-4) and her two healthy parents (I-1 and I-2). The coding exons were captured using the Agilent SureSelect Human All Exon v.5 (Agilent Technologies, Santa Clara, CA, USA). The captured fragments were sequenced on the Hiseq2000 sequencer (Illumina, San Diego, CA, USA) to an average depth of >50 reads per target base. The sequence reads were aligned to the human reference genome (UCSC NCBI37.1/hg19) using SOAPaligner (version 2.21, Beijing Genomics Institute, Shenzhen, China), and functional annotation of high-quality variants was performed using SOAPsnp software (Beijing Genomics Institute) for the single-nucleotide variation. For insertion/deletion detection, sequence reads were aligned by BWA and annotated by GATK for break-point identification.
Exome data analysis
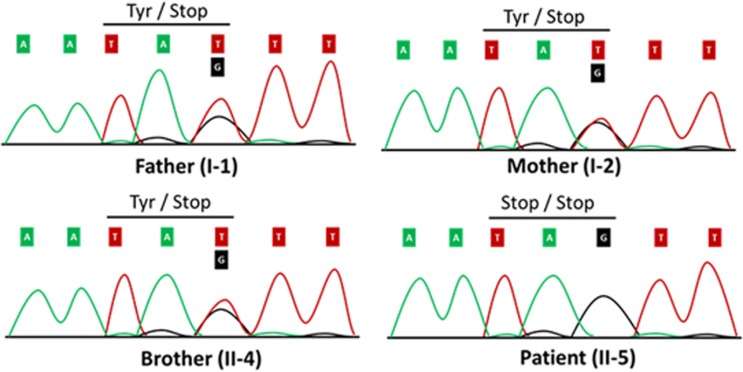
Data analysis was performed in the statistical programming environment R version 3.2.1 (Vienna, Austria). Synonymous variants not located at splice sites and variants with a frequency >0.1% in the following publically available databases were excluded from further analysis: (1) the ExAC Browser,14 (2) the National Heart, Lung, and Blood Institute Exome Sequencing Project,15 (3) Phase 1 version 3 of the 1000 Genomes project (data release October 2012),16 (4) the 69 genomes from Complete Genomics17 and an in-house exome database. Furthermore, only variants compatible with a recessive pattern of inheritance (ie, homozygous in the index patient, heterozygous in both parents and heterozygous or absent in the unaffected sibling) were retained for further consideration. This left only two variants in, respectively, SLC22A5 (p.(Tyr396*), c.1188 T>G) and RNF167 c.833dup (p.(Pro279Serfs*17)) (Table 1). The SLC22A5 gene on chromosome 5q31 encodes the high-affinity carnitine transporter OCTN2; biallelic variants in this gene cause autosomal recessive PCD (OMIM: 212140). No associations with disease are known for RNF167. The genetic data in combination with the clinical presentation strongly points to the diagnosis of PCD in our patient. Sanger sequencing was performed confirming that the parents were heterozygous carriers of the SLC22A5 (p.(Tyr396*), c.1188 T>G) variant, whereas the patient carried the variant in homozygous state (Figure 3). The unaffected sibling was a heterozygous carrier. Genetic data were submitted to ClinVar (www.ncbi.nlm.nih.gov/clinvar/) and the following accession numbers were obtained for the SLC22A5 (p.(Tyr396*), c.1188 T>G) and RNF167 c.833dup (p.(Pro279Serfs*17)) variant, respectively: RCV000416283 and RCV000417084.
Figure 3.
The DNA sequence of a segment flanking p.(Tyr396*), c.1188 T>G in SLC22A5 is shown. Both parents and the unaffected brother are heterozygous variant carriers. The patient carries the variant in the homozygous state. The single base-pair substitution leads to a premature stop codon at amino-acid residue 396 of OCTN2.
Biochemical testing, start and evaluation of treatment
To follow-up on our genetic findings, we performed targeted tandem mass spectrometry analysis of plasma carnitine levels in the patient revealing complete absence of free carnitine (0 μmol/l; ref., 30–50 μmol/l) and a trace level of total carnitine (<1 μmol/l; ref., 43–65 μmol/l) further establishing the diagnosis of PCD (Table 2). Because l-carnitine supplementation is highly effective in ameliorating the cardiomyopathy and anemia, high dose oral carnitine supplementation (200 mg/kg per day) was started directly in the patient. The patient was re-evaluated after 3 months of treatment. She displayed significant improvement in her symptoms and the parents noted a major increase in her daily activity. There was a notable increase of free carnitine (5 μmol/l; ref., 30–50 μmol/l) and total carnitine levels (8 μmol/l; ref., 43–65 μmol/l; Table 2). Follow-up trans-thoracic echocardiography showed a reduction of the myocardial hypertrophy. (Table 2; Figure 1d). The patient's hemoglobin increased markedly from 7 to 14 g/dl (NR,11–13). Interestingly, her ECG showed a significant reduction in T-wave amplitude and the QTc interval normalized to 400 ms. Repolarization abnormalities remained in the left-sided leads however (T-wave inversion; Figure 1b). After 8 months of the treatment, her activity and exercise tolerance had normalized completely.
Table 2. Clinical and biochemical characteristics of the patient before and after treatment with carnitine (200 mg/kg per day).
| Plasma-free carnitine levels (μmol/l) | Plasma total carnitine level (μmol/l) | Hemoglobin level (g/dl) | LVPWd (mm) | IVSd (mm) | |
|---|---|---|---|---|---|
| Reference | 30–50 | 43–65 | 11–13 | 3.59–7.31 | 4.38–8.00 |
| Before treatment | 0 | <1 | 7 | 8.3 (z-score 3.26) | 10.5 (z-score 3.45) |
| After 3 months of the treatment | 5 | 8 | 14 | 6.8 (z-score 1.98) | 7.9 (z-score 1.95) |
| After 12 months of the treatment | 5 | 8 | 12 | 5.8 (z-score 1.07) | 6.0 (z-score 0.94) |
Abbreviations: IVSd, end-diastolic interventricular septal thickness; LVPWd, end-diastolic left ventricular posterior wall thickness.
Z-scores were calculated from reference.11
Discussion
Pediatric cardiomyopathy is a rare but serious disease with high morbidity and mortality. Nearly 40% of children with symptomatic cardiomyopathy receive a heart transplant or die within 2 years of diagnosis.18 In children, cardiomyopathy is poorly understood and a specific cause can only be identified in one-third of cases, whereas the remaining two-third are classified as idiopathic.18 Without the knowledge of a specific cause, specialized therapies cannot be started, which may contribute to the poor outcomes observed. The myriad of genetic and acquired causes of cardiomyopathy, the limited availability of diagnostics tests at some clinical centers (eg, availability of metabolic tests only at specialized centers) and the lack of a systematic genetic and clinical approach, all contribute to the low diagnostic yield in pediatric cardiomyopathy. Recent advances in genetic techniques, specifically next-generation sequencing (NGS), now allow for the detection of genetic causes of cardiomyopathy in a systematic and unbiased manner.19, 20 This is an important advantage given the extensive clinical variability and the large number of genes (>100) in which variants leading to pediatric cardiomyopathy may occur.12, 13
In this case, we used WES in a family with HCM and SCD, and identified PCD as the underlying cause. The homozygous truncating SLC22A5 variant p.(Tyr396*) found in our patient has been identified previously in a compound heterozygous state in another patient diagnosed with PCD.21 PCD is an autosomal recessive disorder of carnitine transportation that results in defective fatty acid oxidation.1, 22 The disease is caused by biallelic variants in the SLC22A5 gene, encoding the organic cation transporter type 2.2 This defect results in urinary carnitine wasting, low serum carnitine levels and decreased intracellular carnitine accumulation, leading to impaired oxidation of long-chain fatty acids.3, 4, 5 This leads to reduced energy generation, ketogenesis and the accumulation of long-chain fatty acids in affected organs.23 OCTN2 is highly expressed in the myocardium, skeletal muscle, fibroblasts, renal tubules, placental tissue and the intestine. Patients with PCD can present in early infancy with metabolic decompensation and later in childhood with cardiomyopathy as the prominent feature. Less common features such as anemia, muscular weakness and developmental delay, respiratory distress, arrhythmia and an abnormal ECG have also been described.1 Measurement of plasma carnitine will reveal extremely low plasma carnitine levels (<5 μm, normal 25–50 μm).24 One study of 61 literature-reported PCD cases showed an exclusive cardiac manifestation in 62% of PCD cases in addition to a very low likelihood of both cardiac and metabolic manifestations being present.25 Our patient presented with tall T-waves and QTc interval shortening, which resolved after treatment with carnitine. Characteristic ECG features, similar to our case, have indeed been described in PCD patients and these include high peaked T-waves, shortening or prolongation of the QTc interval and ventricular ectopy alongside evidence for left ventricular hypertrophy.7, 8, 9, 26, 27 Interestingly, mice with induced carnitine deficiency display shortening of the QTc interval and spontaneous sustained ventricular tachycardia, which was not observed in control mice.28 These data suggest that the mechanism of sudden death from arrhythmia in PCD patients might involve abnormalities of the QTc interval. An interaction between long-chain acylcarnitines and potential depolarization abnormalities by regulating the rapidly activated delayed rectifier potassium channel (Ikr/HERG) has been suggested.7
Supplementation with l-carnitine led to an increase in blood carnitine levels. The patient showed a reduction in the degree of cardiac hypertrophy and her exercise tolerance improved markedly after l-carnitine supplementation. Her quality of life and life expectancy have been drastically improved, and her risk of sudden death is expected to be significantly reduced by the identification of the cause, and treatment of her illness. Diagnosis in this family was followed by reproductive counseling and provided the parents the reassurance that the oldest living son (Figure 2, II-4) is unaffected.
This case illustrates the use of exome sequencing as a systematic and unbiased diagnostic tool in a pediatric case with HCM, resulting in the appropriate treatment of the patient and the prevention of further deterioration and premature sudden death. A major obstacle to the systematic implementation of NGS in clinical practice has been time to result. Recent technological advances now allow rapid NGS to be applied in clinical practice. This approach can aid in the diagnosis, prognosis and treatment of critically ill pediatric patients, especially when a high degree of clinical and genetic heterogeneity exists, as suggested by recent guidelines.29 In a recent study, rapid NGS in severe early onset pediatric disease showed the potential of NGS to provide a definitive or likely molecular diagnosis in ~50 h. It is important to note that despite our proof-of-concept that WES can be of great importance in the diagnostic work-up of pediatric cardiomyopathy patients, biochemical testing to identify treatable causes of cardiomyopathy (such as PCD) should be performed at an early stage and should not be delayed by genetic testing.
Acknowledgments
We thank the patient and her family for their participation in this study. We thank L Beekman and M Klerk for helpful discussion. We thank Professor FA Wijburg for clinical discussions. We acknowledge support from the Netherlands CardioVascular Research Initiative (CVON-PREDICT project, CVON 2012-10).
Author contributions
Dr Lahrouchi and Dr Lodder carried out the whole-exome sequencing analyses, drafted the initial manuscript, reviewed and revised the manuscript, and approved the final manuscript as submitted. Dr Mansouri, Dr Adadi and Dr Ratbi recruited the patient and her family, gathered clinical information, prepared DNA samples, and approved the final manuscript as submitted. Dr Zniber provided clinical care for the patient, gathered clinical information and approved the final manuscript as submitted. Dr Postma and Dr Tadros supervised data analysis, critically reviewed the manuscript and approved the final manuscript as submitted. Dr van Spaendonck-Zwarts (Clinical Geneticist) and Dr Sally-Ann Clur (Pediatric Cardiologist) critically assessed the manuscript and approved the final manuscript as submitted. Professor Bezzina and Professor Sefiani coordinated and supervised data analysis, critically reviewed the manuscript, and approved the final manuscript as submitted.
Footnotes
The authors declare no conflict of interest.
References
- Magoulas PL, El-Hattab AW: Systemic primary carnitine deficiency: an overview of clinical manifestations, diagnosis, and management. Orphanet J Rare Dis 2012; 7: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezu J, Tamai I, Oku A et al: Primary systemic carnitine deficiency is caused by mutations in a gene encoding sodium ion-dependent carnitine transporter. Nat Genet 1999; 21: 91–94. [DOI] [PubMed] [Google Scholar]
- Tamai I, Ohashi R, Nezu J-i et al: Molecular and functional identification of sodium ion-dependent, high affinity human carnitine transporter OCTN2. J Biol Chem 1998; 273: 20378–20382. [DOI] [PubMed] [Google Scholar]
- Wu X, Prasad PD, Leibach FH, Ganapathy V: cDNA sequence, transport function, and genomic organization of human OCTN2, a new member of the organic cation transporter family. Biochem Biophys Res Commun 1998; 246: 589–595. [DOI] [PubMed] [Google Scholar]
- Scaglia F, Wang Y, Singh RH et al: Defective urinary carnitine transport in heterozygotes for primary carnitine deficiency. Genet Med 1998; 1: 34–39. [DOI] [PubMed] [Google Scholar]
- Wilcken B, Wiley V, Sim KG, Carpenter K: Carnitine transporter defect diagnosed by newborn screening with electrospray tandem mass spectrometry. J Pediatr 2001; 138: 581–584. [DOI] [PubMed] [Google Scholar]
- Fu L, Huang M, Chen S: Primary carnitine deficiency and cardiomyopathy. Korean Circ J 2013; 43: 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp ME, Katcher ML, Peters HA et al: Systemic carnitine deficiency presenting as familial endocardial fibroelastosis: a treatable cardiomyopathy. N Engl J Med 1981; 305: 385–390. [DOI] [PubMed] [Google Scholar]
- Waber LJ, Valle D, Neill C, DiMauro S, Shug A: Carnitine deficiency presenting as familial cardiomyopathy: a treatable defect in carnitine transport. J Pediatr 1982; 101: 700–705. [DOI] [PubMed] [Google Scholar]
- Pierpont ME, Breningstall GN, Stanley CA, Singh A: Familial carnitine transporter defect: a treatable cause of cardiomyopathy in children. Am Heart J 2000; 139 (2 Pt 3): S96–S106. [DOI] [PubMed] [Google Scholar]
- Pettersen MD, Du W, Skeens ME, Humes RA: Regression equations for calculation of Z scores of cardiac structures in a large cohort of healthy infants, children, and adolescents: an echocardiographic study. J Am Soc Echocardiogr 2008; 21: 922–934. [DOI] [PubMed] [Google Scholar]
- Towbin JA, Lowe AM, Colan SD et al: Incidence, causes, and outcomes of dilated cardiomyopathy in children. JAMA 2006; 296: 1867–1876. [DOI] [PubMed] [Google Scholar]
- Colan SD, Lipshultz SE, Lowe AM et al: Epidemiology and cause-specific outcome of hypertrophic cardiomyopathy in children: findings from the Pediatric Cardiomyopathy Registry. Circulation 2007; 115: 773–781. [DOI] [PubMed] [Google Scholar]
- Exome Aggregation Consortium (ExAC). Available at: http://exac.broadinstitute.org/terms (accessed on 30 October 2014).
- NHLBI GO Exome Sequencing Project (ESP). Available at: http://evs.gs.washington.edu/EVS (accessed on March 2016).
- Abecasis GR, Altshuler D, Auton A et al: A map of human genome variation from population-scale sequencing. Nature 2010; 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ et al: Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 2010; 327: 78–81. [DOI] [PubMed] [Google Scholar]
- Cox GF, Sleeper LA, Lowe AM et al: Factors associated with establishing a causal diagnosis for children with cardiomyopathy. Pediatrics 2006; 118: 1519–1531. [DOI] [PubMed] [Google Scholar]
- Goodwin S, McPherson JD, McCombie WR: Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016; 17: 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Ng SB, Bigham AW et al: Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 2011; 12: 745–755. [DOI] [PubMed] [Google Scholar]
- Lee N-C, NL-S Tang, Chien Y-H et al: Diagnoses of newborns and mothers with carnitine uptake defects through newborn screening. Mol Genet Metab 2010; 100: 46–50. [DOI] [PubMed] [Google Scholar]
- Stanley CA: Carnitine deficiency disorders in children. Ann NY Acad Sci 2004; 1033: 42–51. [DOI] [PubMed] [Google Scholar]
- Longo N, Amat di San Filippo C, Pasquali M: Disorders of carnitine transport and the carnitine cycle. Am J Med Genet C Semin Med Genet 2006; 142C: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hattab AW, Li F-Y, Shen J et al: Maternal systemic primary carnitine deficiency uncovered by newborn screening: clinical, biochemical, and molecular aspects. Genet Med 2010; 12: 19–24. [DOI] [PubMed] [Google Scholar]
- Shibbani K, Fahed AC, Al-Shaar L et al: Primary carnitine deficiency: novel mutations and insights into the cardiac phenotype. Clin Genet 2014; 85: 127–137. [DOI] [PubMed] [Google Scholar]
- Matsuishi T, Hirata K, Terasawa K et al: Successful carnitine treatment in two siblings having lipid storage myopathy with hypertrophic cardiomyopathy. Neuropediatrics 1985; 16: 6–12. [DOI] [PubMed] [Google Scholar]
- Tein I, De Vivo DC, Bierman F et al: Impaired skin fibroblast carnitine uptake in primary systemic carnitine deficiency manifested by childhood carnitine-responsive cardiomyopathy. Pediatr Res 1990; 28: 247–255. [DOI] [PubMed] [Google Scholar]
- Roussel J, Labarthe F, Thireau J et al: Carnitine deficiency induces a short QT syndrome. Heart Rhythm 2016; 13: 165–174. [DOI] [PubMed] [Google Scholar]
- ACMG Board of Directors: Points to consider in the clinical application of genomic sequencing. Genet Med 2012; 14: 759–761. [DOI] [PubMed] [Google Scholar]