Abstract
We report a family with four girls with moderate to severe intellectual disability and epilepsy. Two girls showed regression in adolescence and died of presumed sudden unexpected death in epilepsy at 16 and 22 years. Whole exome sequencing identified a truncating pathogenic variant in IQSEC2 at NM_001111125.2: c.2679_2680insA, p.(D894fs*10), a recently identified cause of epileptic encephalopathy in females (MIM 300522). The IQSEC2 variant was identified in both surviving affected sisters but in neither parent. We describe the phenotypic spectrum associated with IQSEC2 variants, highlighting how IQSEC2 is adding to a growing list of X-linked genes that have a female-specific phenotype typically associated with de novo mutations. This report illustrates the need for careful review of all whole exome data, incorporating all possible modes of inheritance including that suggested by the family history.
Introduction
The first variants that affects function in the IQ motif and Sec7 domain 2 (IQSEC2) gene [NM_001111125.2] (MIM 300522) were described in four separate families with X-linked intellectual disability (XLID).1 Since our first report describing familial cases, high throughput sequencing in intellectual disability or epilepsy cohorts has identified de novo loss-of-function IQSEC2 variants in males and increasingly in females.2, 3, 4, 5, 6, 7, 8, 9, 10, 11 IQSEC2 catalyzes exchange of GDP for GTP in a number of the ARF superfamily of proteins. It has an essential role in modulating the cytoskeleton and vesicle transport at the post-synaptic density and hence is a crucial modifier of synaptic plasticity. All variants were missense and clustered around the sec7 and IQ-like domains, resulting in reduced enzymatic activity.1 Complete abolition of enzymatic activity leads to a more severe phenotype including epileptic encephalopathy (EE) in both males and females. A recent review by Zarem and colleagues12 identified 18 individuals in the literature with IQSEC2 variants and seizures; five of the six females described had a de novo loss-of-function variant.3, 5, 7 IQSEC2 adds to a growing list of genes on the X chromosome, such as USP9X and PHF6, that have distinct phenotypes in males and females depending on the functional severity of the variant.13, 14
Materials and methods
WES identification and molecular analysis
This work was undertaken as part of a study on whole exome sequencing (WES) application to diagnosis in Mendelian disorders at Sydney Children's Hospital and the Kinghorn Centre for Clinical Genomics, approved by the local ethics committee (HREC ref no 13/094). High-quality DNA was extracted from peripheral blood in EDTA. WES was performed on two of the five affected sisters for genetic diagnosis using a NimbleGen VCRome rapid capture expanded exome kit, with libraries analysed on an Illumina HiSeq2500 instrument. Bioinformatics filtering steps and scientific methods are described in Supplementary Methods.
Results
Phenotype
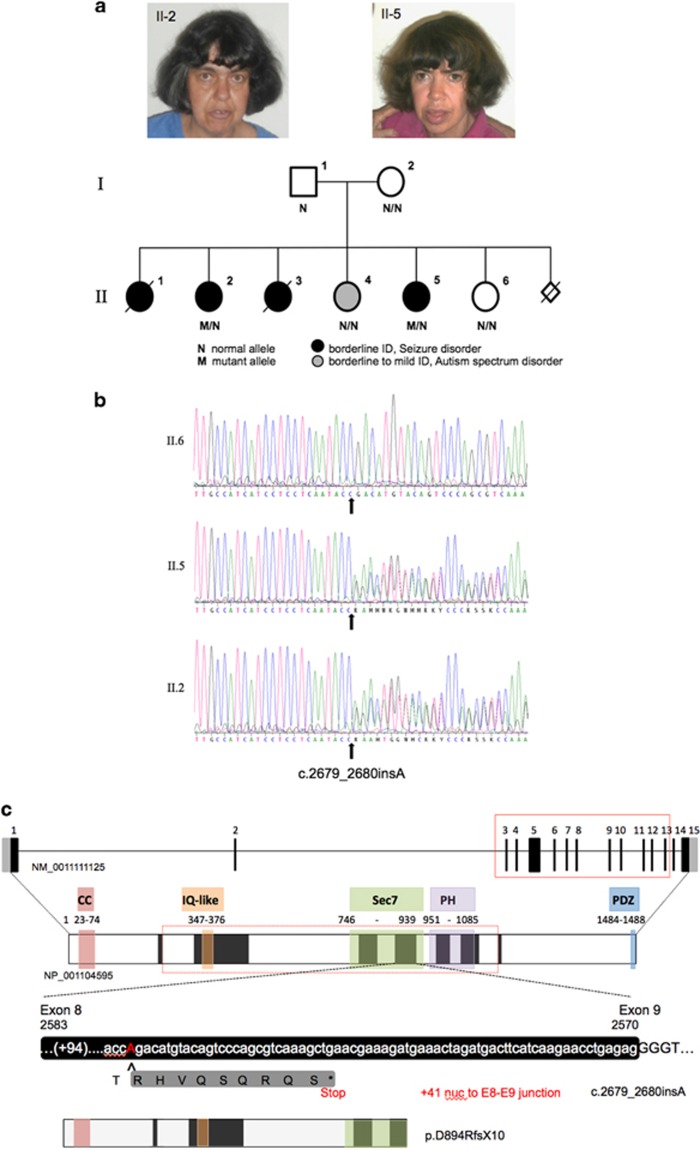
We report a family of six girls born to non-consanguineous Caucasian parents (Figure 1 and Table 1). Four girls had a similar course with developmental delay followed by onset of seizures at 2–5 years of age. Focal seizures were most prominent with seizure triggers including fever, immunisation and the perimenstrual period. They had intellectual disability, which ranged from moderate to severe and aggressive behaviour when young with some autistic features. Two girls showed regression in adolescence, both dying of presumed sudden unexpected death in epilepsy (SUDEP) at 16 and 22 years. The surviving two affected sisters live at home in the care of their parents and are now in their early 40's and have infrequent seizures that often occur around the time of menstruation. II-2 is 159 cm in height (25th centile) with OFC of 57cm (95th centile; Centile based on CDC growth charts). II–5 is 147cm tall (1st centile) with OFC of 55cm (30th centile). There were no dysmorphic features as adults (Figure 1a), and childhood photographs show no evidence of dysmorphic features.
Figure 1.
Identification of a c.2679_2680insA (NM_001111125.2) variant resulting in p.(D894fs*10) (NP_001104595) in IQSEC2. (a) Pedigree of family. Facial photos of II-2 and II-5 shown above the pedigree. Open symbols represent unaffected individuals, filled grey circle represents a female of borderline intellect to mild intellectual disability with ASD, while filled black circles represent females with moderate intellectual disability and epilepsy. Normal (N) and mutant (M) alleles shown for individuals tested. (b) DNA sequence electropherograms for the chrX:g.53276220_53276221insT (GRCh37/hg19 assembly); c.2679_2680insA mutation in exon 8 of 15 of IQSEC2. (c) Predicted impact of the novel variants in IQSEC2. The exon-intron structure of the longest isoform of IQSEC2 gene [NM_0011111125.2] with 15 exons, the ATG, open reading frame and stop codon positions in black and 5' and 3' untranslated regions in light grey. The predicted protein structure (NP_001104595) with known functional domains highlighted; coiled-coiled (CC, red), IQ-like (orange), Sec 7 enzyme domain (Sec 7, green), PH domain (purple) and the PDZ binding motif (blue), corresponding amino acids listed below each domain. The variant c.2679_2680insA causes a frame-shift at the site of insertion with the premature stop codon 29 nucleotides subsequent of the insertion, 41 nucleotides from the exon 8 and 9 junction. This transcript may escape nonsense mediated decay. In addition to a 9 amino acid nonsense peptide encoded prior to the PTC, the C-terminally truncated protein would lack a portion of the Sec7 domain, the PH domain and the PDZ binding domain.
Table 1. Clinical features and investigations of family with novel insertion variant in IQSEC2.
| Clinical Features | II-1 | II-2 | II-3 | II-4 | II-5 |
|---|---|---|---|---|---|
| Mutation | |||||
| Status – Affected female (AF), current age (yrs) | AF Deceased aged 22 yrs | AF 49 yrs | AF Deceased aged 16 yrs | AF 46 yrs | AF 44 yrs |
| p.D894fsX10 in IQSEC2 | Not tested | Yes | Not tested | No (IQSEC2 WT) | Yes |
| Development | |||||
| Early development | Mild delay | Developmental slowing after second immunization (pertussis, diphtheria, tetanus)~4 months | Severe delay (not walking at 33/12) | Mild delay (walking 18/12; single words 24/12) | Minor delay |
| Language development | Intellectual disability | Some language – better receptive language | Non-verbal | Mild intellectual disability | Non-verbal |
| Schooling | Special school, life skills | Special school, life skills | Home schooling | Home schooling, modified curriculum | Home schooling, modified curriculum |
| Independent living and social skills | Dependent | Dependent, some self care skills | Dependent | Semi independent, works in supported environment | Dependent, some self care skills |
| Deceased | 22 years: found dead in bed Presumed SUDEP (no autopsy) | No | 16 years: presumed SUDEP | No | No |
| Epilepsy | Yes | Yes | Yes | No | Yes |
| Age of onset | 5 years | 2 years | 2 years | Nil | 2 years |
| Seizure types | Focal impaired awareness seizures, tonic-clonic seizures~3 / wk | Febrile tonic-clonic after measles. Perimenstrual absence and focal seizures | Focal motor seizure | Nil | Focal impaired awareness seizures Perimenstrual tonic-clonic |
| Anti-epileptic therapy | Phenytoin, carbamazepine; uncontrolled | Phenytoin and carbamazepine | Phenytoin | Nil | Phenytoin and carbamazepine |
| Other features | |||||
| Behaviour | Autistic traits, aggressive behaviour in early childhood | Early childhood; screaming, ADHD | Autistic features | ||
| Regression | 16 yrs: anorexia, food refusal, social withdrawal | Adolescent social withdrawal, concomitant deteriorating condition | ASD | ||
| Neurological examination (childhood) | Limited coordination, stiff movement | Nil | Nil | Nil | Nil |
| Other features | Mild scoliosis | EEG: continual left hemisphere discharges and slowing | True knot in umbilical cord Shorter stature than siblings | Mild scoliosis | |
| Normal Investigations | CDKL5 coding region, Epileptic Encephalopathy gene panel | Karyotype and urine metabolic screen | CDKL5 coding regions; X-tiling array, genome-wide array (0.2 Mb res) | PCDH19 and CDKL5 coding regions; MECP2 MLPA / sequencing; fatty acid, urine metabolic screen | |
Abbreviation: SUPED, Sudden unexpected death in epilepsy.
The fifth daughter, II-4, had autistic features and borderline to mild intellectual disability, without seizures. She had perinatal complications with a true knot in her cord at birth. The sixth daughter, II-6, was unaffected and of normal intellect; she worked in a senior managerial role. There was one single early miscarriage (sex unknown) and no male children. Both parents were health professionals and have no history of seizures or cognitive problems.
Molecular analysis of the IQSEC2 variant
WES analysis identified a heterozygous novel single base pair insertion shared by both sisters at genomic position chrX:g.53276220_53276221insT (GRCh37/hg19 assembly; Supplementary Figure 1). This variant in exon 8 of the NM_001111125.2 isoform inserts a single nucleotide at c.2679_2680insA, p.(D894fs*10) (NP_001104595) in IQSEC2. Variant has been submitted to the gene variant database at www.LOVD.nl/IQSEC2 (patient ID #00081545; DB-ID #000015). Exons are numbered like in NG_021296.1. Sanger sequencing confirmed the presence of this variant in both affected sisters II-2 and II-5 (Figures 1a and b; Supplementary Figure 2). The IQSEC2 single base pair insertion was absent in the father (I-1), mother (I-2) and their unaffected (II-6) and phenotypically distinct (II-4) sisters. Identification of parental origin of the mosacism was not undertaken at the request of the family.
The insertion of this single nucleotide is predicted to cause a frame-shift with a premature stop codon 29 nucleotides downstream (Figure 1c). As expression of IQSEC2 is negligible in lymphoblast cells and fibroblasts, we have been unable to determine if the mutant allele escapes nonsense mediated decay (NMD).
Discussion
IQSEC2 is a gene on the X-chromosome initially implicated in XLID through identification of variants in affected males in large X-linked families.1 Subsequent population-based screening approaches in intellectual disability and epilepsy cohorts using next generation sequencing (NGS) strategies have identified variants predicted to affect function in IQSEC2 in both sexes. This study identified a novel frameshift variant in IQSEC2, NM_001111125.2: c.2679_2680insA, p.(D894fs*10) in affected females that was absent in DNA extracted from both blood and saliva samples in their unaffected parents. Parental gonadal mosaicism is therefore the most likely mechanism, with the parent of origin not clarified. Gonadal mosaicism as a cause of sibling recurrence of genetic disorders is an increasingly recognised molecular mechanism, with studies suggesting as many as 3.8–10% of apparently de novo variants are mosaic in the parental germline.15, 16
The IQSEC2 variant identified in this family truncates the protein within the Sec7 domain, but distal to the critical residue for enzymic activity at p.E849. The premature stop codon is encoded just 41 nucleotides from the exon 8 and 9 junction, less than the required 50 nucleotides required to trigger NMD. As such, we cannot rule out that the RNA may escape NMD to some extent. Despite this, even if a C-terminal truncated protein was produced and potentially retain residual Sec7 enzyme activity, it would lack both the PH and PDZ domains. The PH domain is predicted to mediate membrane localisation and play a regulatory role in binding to phosphoinositides, with the type 1 PDZ domain at the C-terminal end of the IQSEC protein required for interaction with the PSD complex.17 Removal of last 74 amino acids of the protein results in a dramatic mis-localisation of the truncated protein to the dendritic shaft of neurons instead of the usual location within the dendritic spines.18 Even if the C-terminal protein was to escape NMD to some degree, the impact on the functional capacity of the mutant IQSEC2 protein is predicted to be consistent with a loss-of-function and subsequent severe phenotype in affected females.
We assessed all the reported missense and nonsense variants associated with disease reported in IQSEC2, listing them as they occur across the length of the coding region1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 19 (Table 2), and several notable correlations were identified. The missense variants that either impact the Sec7 domain directly or interfere with Sec7 activity are all inherited in a familial manner (pale grey in Table 2). As highlighted by the recent report in the MRX78 family, these families present with affected males having a more severe phenotype than affected females.9 The remaining cases (dark grey in Table 2) present with a spectrum of seizure and behavioural phenotypes due to de novo nonsense variants or variants that lead to splicing defects.
Table 2. Summary of IQSEC2 variants reported in affected males and females.
| Mutation cDNA | Ex | Mutation Protein AA | Domain affected | Mutation type | Phenotype | Family | Inheritance | Sex | GRCh37/hg19 coordinate | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| c.273_282del | 1 | p.N91Kfs*112 | – | nonsense | Rett like | P7 | De novo | F | 53350040-53350049 | 6 |
| c.804delC | 3 | p.Y269Tfs*3 | – | nonsense | Seizures | 48 | De novo | F | 53285177 | 9 |
| c.928G>T | 3 | p.E310* | – | nonsense | FE, mild-mod ID | P16 | De novo | F | 53285053 | 12 |
| c.1049C>T | 4 | p.A350V | IQ-like | missense | EE, mod-severe ID | P1 | De novo | M | 53284064 | 12 |
| c.1075C>T | 4 | p.R359C | IQ-like | missense | NS-XLID | AU128 | Familial | M | 53284038 | 1 |
| c.1618C>T | 5 | p.Q540* | – | nonsense | EE, DD | P14 | De novo | M | 53280140 | 12 |
| c.1688G>A | 5 | p.R563Q | – | missense | ASD | S0090 | Familial | M | 53280070 | 19(supp) |
| c.2052_2053delCG | 5 | p.C684* | – | nonsense | DD, Seizures | P1 | De novo | M | 53279705-53279706 | 5 |
| Seizures | 47 | De novo | F | 9 | ||||||
| c.2203C>T | 5 | p.Q735* | – | nonsense | SGE | T17563 | De novo | F | 53279555 | 11 |
| c.2273G>A | 5 | p.R758Q | Sec7 | missense | NS-XLID | US166 | Familial | M | 53279485 | 1 |
| c.2366C>T | 6 | p.A789V | Sec7 | missense | Severe-ID, Seizures, Ag | MRX78 | Familial | M | 53277996 | 8 |
| Mild-ID | F | |||||||||
| c.2402 A>C | 6 | p.Q801P | Sec7 | missense | NS-XLID | MRX18 | Familial | M | 53277960 | 1 |
| c.2563C>T | 7 | p.R855* | Sec7 | nonsense | NS-XLID | De novo | M | 53277315 | 2 | |
| c.2582G>C | 7 | p.S861T | Sec7 | splicing | DD, Seizures | P2 | De novo | M | 53277296 | 4 |
| Seizures | 46 | De novo | F | 9 | ||||||
| c.2587C>T | 8 | p.R863W | Sec7 | missense | NS-XLID | MRX1 | Familial | M | 53276313 | 1 |
| c.2662dup | 8 | p.I888Nfs*16 | Sec7 | nonsense | XLID | P7 | De novo | M | 53276237 | 7 |
| c.2679_2680insA | 8 | p.(D894fs*10) | Sec7 | nonsense | ID and epilepsy | K2 | Familial | F | 53276220 | This report |
| c.2776C>T | 9 | p.R926* | Sec7 | nonsense | Rett like | P3 | F | 3272627 | 10 | |
| c.3097C>T | 11 | p.Q1033* | PH | nonsense | Severe ID, Seizures | APN-68 | De novo | M | 53268395 | 5 |
| c.3163C>T | 12 | p.R1055* | PH | nonsense | Severe ID, | Fam 19 | F | 53267441 | 7 | |
| c.3322C>T | 13 | p.Q1108* | – | nonsense | LGS | KO | De novo | F | 53265633 | 3 |
Abbreviations: Ag, Aggression; ASD-F, autistic features; DevR, developmental regression; DD, Developmental delay; EE, Epileptic encephalopathy; FE, Focal epilepsy; ID, intellectual disability; LGS, Lennox-Gastaut syndrome; NS-XLID, non-syndromic X-linked intellectual disability; SGE, symptomatic generalised epilepsy.
Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence for the IQSEC2 gene [GenBank: NM_ 001111125.2].
Affected females with de novo variants present with epileptic encephalopathy including Rett-like phenotypes and Lennox-Gastaut syndrome. Our affected sisters presented with epilepsy and intellectual disability. They may have had an epileptic encephalopathy (EE) but there was only scant EEG data to show that developmental slowing in infancy or the adolescent regression in two sisters had an epileptic basis. Interestingly these two sisters died of presumed SUDEP in adolescence and young adult life. This is the first report of SUDEP in IQSEC2 diseases suggesting that IQSEC2 adds to the number of EE genes that may increase risk for SUDEP.20
This gene region is also interesting from an epigenetic perspective. IQSEC2 escapes X-inactivation in females.21 Genes that escape X-inactivation are potential candidates for dosage-mediated phenotypic disruptions. This has been shown recently for microduplications in Xp11.2 involving the IQSEC2 gene.22 Even though X inactivation is not apparent at this loci, expression of IQSEC2 is similar in males and females.22 We speculate that if a female has a heterozygous variant in IQSEC2 that results in a significant loss-of-function of IQSEC2 activity, the as-yet unknown mechanism that limits the gene dosage of IQSEC2 to mirror that in males does not allow for compensation for the loss of IQSEC2 function in females. The IQSEC2 region contributes to a growing body of evidence that the extent of escape from X-inactivation may be underappreciated as a substantial number of genes are subject to some degree of escape.23
The phenotypic spectrum associated with these cases demonstrates that IQSEC2 should be considered as an X-linked gene extending to a female-specific phenotype, including intellectual disability, epilepsy and SUDEP. In addition we suggest careful analysis of NGS data with consideration of all modes of inheritance, even those that seem unlikely based on family history.
Acknowledgments
We thank the patients, their families and physicians for their participation in this study, in particular Dr Michael Partington for his work with this family over many years. We would like to thank Kevin Ying, Jiang Tao, Paula Morris and Kerith-Rae Dias from the Kinghorn Centre for Clinical Genomics for sequencing these patients. Whole exome sequencing and salaries for TR and MED were funded through a donation by the Kinghorn Foundation to the Garvan Institute of Medical Research. MJC is supported by Cancer Institute NSW (13/ECF/1-46). CS is supported by the Australian Research Council (Future Fellowship FT120100086).
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Shoubridge C, Tarpey PS, Abidi F et al: Mutations in the guanine nucleotide exchange factor gene IQSEC2 cause nonsyndromic intellectual disability. Nat Genet 2010; 42: 486–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E et al: Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012; 380: 1674–1682. [DOI] [PubMed] [Google Scholar]
- Epi KC Epilepsy Phenome/Genome PAllen AS et al: De novo mutations in epileptic encephalopathies. Nature 2013; 501: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandomi SK, Farwell Gonzalez KD, Parra M et al: Diagnostic exome sequencing identifies two novel IQSEC2 mutations associated with X-linked intellectual disability with seizures: implications for genetic counseling and clinical diagnosis. J Genet Counsel 2014; 23: 289–298. [DOI] [PubMed] [Google Scholar]
- Redin C, Gerard B, Lauer J et al: Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J Med Genet 2014; 51: 724–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson HE, Tambunan D, LaCoursiere C et al: Mutations in epilepsy and intellectual disability genes in patients with features of Rett syndrome. Am J Med Genet A 2015; 167A: 2017–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzschach A, Grasshoff U, Beck-Woedl S et al: Next-generation sequencing in X-linked intellectual disability. Eur J Hum Genet 2015; 23: 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer VM, James VM, Himelright ML et al: Novel missense mutation A789V in IQSEC2 underlies X-Linked intellectual disability in the MRX78 family. Front Mol Neurosci 2015; 8: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig KL, Farwell Hagman KD, Shinde DN et al: Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet Med: Off J Am College Med Genet 2016; 18: 898–905. [DOI] [PubMed] [Google Scholar]
- Allou L, Julia S, Amsallem D et al: Rett-like phenotypes: expanding the genetic heterogeneity to the KCNA2 gene and first familial case of CDKL5-related disease. Clin Genet 2016, e-pub ahead of print 7 April 2016; doi:10.1111/cge.12784. [DOI] [PubMed]
- Epi KCEaekce, Epi KC: De novo mutations in SLC1A2 and CACNA1A are important causes of epileptic encephalopathies. Am J Hum Genet 2016; 99: 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerem A, Haginoya K, Lev D: The molecular and phenotypic spectrum of IQSEC2 related epilepsy. Epilepsia 2016; 57: 1858–1869. [DOI] [PubMed] [Google Scholar]
- Zweier C, Kraus C, Brueton L et al: A new face of Borjeson-Forssman-Lehmann syndrome? De novo mutations in PHF6 in seven females with a distinct phenotype. J Med Genet 2013; 50: 838–847. [DOI] [PubMed] [Google Scholar]
- Reijnders MR, Zachariadis V, Latour B et al: De novo loss-of-function mutations in USP9X cause a female-specific recognizable syndrome with developmental delay and congenital malformations. Am J Hum Genet 2016; 98: 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbari R, Wuster A, Lindsay SJ et al: Timing, rates and spectra of human germline mutation. Nat Genet 2016; 48: 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yang X, Wu Q et al: Amplicon resequencing identified parental mosaicism for approximately 10% of ‘de novo' SCN1A mutations in children with dravet syndrome. Hum Mutat 2015; 36: 861–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakagami H, Sanda M, Fukaya M et al: IQ-ArfGEF/BRAG1 is a guanine nucleotide exchange factor for Arf6 that interacts with PSD-95 at postsynaptic density of excitatory synapses. Neurosci Res 2008; 60: 199–212. [DOI] [PubMed] [Google Scholar]
- Sanda M, Kamata A, Katsumata O et al: The postsynaptic density protein, IQ-ArfGEF/BRAG1, can interact with IRSp53 through its proline-rich sequence. Brain Res 2009; 1251: 7–15. [DOI] [PubMed] [Google Scholar]
- Piton A, Gauthier J, Hamdan FF et al: Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry 2011; 16: 867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall RD, Crompton DE, Petrovski S et al: Exome-based analysis of cardiac arrhythmia, respiratory control, and epilepsy genes in sudden unexpected death in epilepsy. Ann Neurol 2016; 79: 522–534. [DOI] [PubMed] [Google Scholar]
- Tsuchiya KD, Greally JM, Yi Y, Noel KP, Truong JP, Disteche CM: Comparative sequence and x-inactivation analyses of a domain of escape in human xp11.2 and the conserved segment in mouse. Genome Res 2004; 14: 1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moey C, Hinze SJ, Brueton L et al: Xp11.2 microduplications including IQSEC2, TSPYL2 and KDM5C genes in patients with neurodevelopmental disorders. Eur J Hum Genet 2016; 24: 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton AM, Price EM, Jones MJ, Balaton BP, Kobor MS, Brown CJ: Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum Mol Genet 2015; 24: 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.