Abstract
Mutations of the cystatin B gene (CSTB; OMIM 601145) are known to cause Unverricht–Lundborg disease or progressive myoclonic epilepsy-1A (EPM1A, MIM #254800). Most patients are homozygous for an expanded (>30) dodecamer repeat in the promoter region of CSTB, or are compound heterozygotes for the dodecamer repeat and a point mutation. We report two adolescent sisters born to consanguineous parents of Sri Lankan descent who presented with profound global developmental delay, microcephaly, cortical blindness and axial hypotonia with appendicular hypertonia. Neither sibling ever developed head control, independent sitting or ambulation, and never developed speech. The elder sister had a seizure disorder. Both sisters had profound microcephaly and distinct facial features. On serial brain imaging, they had progressive atrophy of the corpus callosum and supratentorial brain, and diffuse hypomyelination with progressive loss of myelin signal. Exome sequencing revealed both siblings to be homozygous for a c.218dupT (p.His75Serfs*2) mutation in exon 3 of CSTB. The neuroimaging features of our patients are consistent with those observed in Cstb-knockout mice, which supports the hypothesis that disease severity is inversely correlated with the amount of residual functional cystatin B protein.
Introduction
Cystatin B, encoded by the CSTB gene (OMIM 601145), is a ubiquitously expressed inhibitor of lysosomal cysteine proteases, also known as cathepsins.1 Mutations of CSTB cause Unverricht–Lundborg disease or progressive myoclonic epilepsy-1A (EPM1A, MIM #254800), the most common form of progressive myoclonic epilepsy, which is characterized by stimulus-sensitive myoclonus, epilepsy and progressive neurological deterioration. Onset is typically between 6 and 16 years of age, usually marked by the appearance of myoclonic jerks and/or tonic–clonic epileptic seizures that increase in frequency over time. Several years after the onset of disease, other features such as intention tremor, dysarthria, ataxia and incoordination can manifest. Overall, cognitive function is preserved, however, cognitive decline has been observed in some patients.2 EPM1A is inherited in an autosomal recessive manner, and most affected individuals are homozygous for an unstable dodecamer expansion repeat (>30) in the promoter region of CSTB, which decreases expression of the transcript without suppressing it completely.3 There are, however, patients who are heterozygous for the expansion repeat and a point mutation creating a null allele (c.202C>T) in CSTB, which have been described to have a more severe phenotype and earlier age of symptom onset.4
The first report of patients homozygous for a nonsense mutation in CSTB was published recently by Mancini et al.5 The patients, homozygous for a c.202C>T mutation, were noted to have severe developmental delay, dyskinesia and microcephaly, as early as 3 months of age.5
We describe the clinical features, serial neuroimaging and long-term follow-up of two adolescent sisters with a severe neurodegenerative phenotype who are homozygous for a novel frameshift variant in CSTB.
Case report
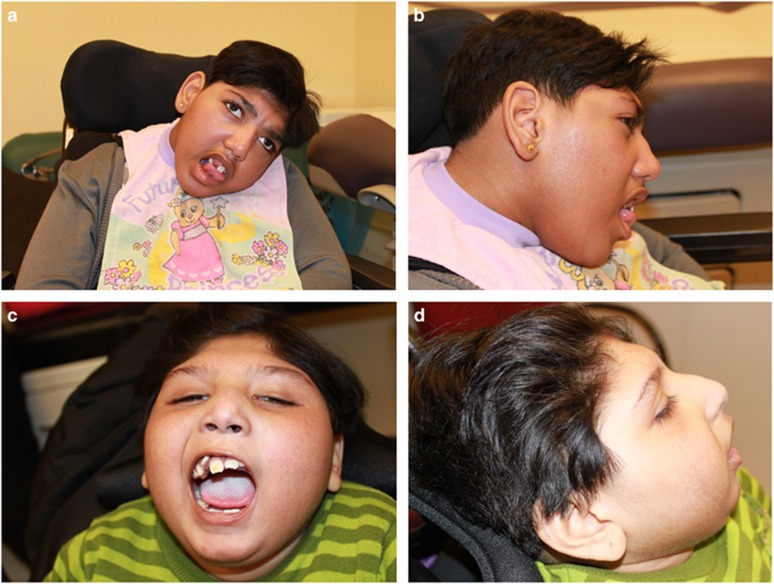
Our patients are two sisters born to distantly consanguineous parents of Sri Lankan descent. Patient 1, the elder sister, was born at term by spontaneous vaginal delivery after an uneventful pregnancy, without perinatal complications. She never achieved any gross motor, fine motor or language milestones. Microcephaly was noted at 3 months, as her head circumference measured 36 cm (<3rd centile for age and 50th centile for 1 month). At 6 months of age, she developed tonic–clonic seizures, but these were well controlled with phenobarbital and clonazepam. EEGs showed multifocal spikes with diffuse background slowing, consistent with seizure activity and encephalopathy. At 3 years of age, she was found to have cortical blindness, with light perception only. Currently, at age 18 years, she is dependent for all activities of daily living, is wheelchair bound and is fed through a gastro-jejunal tube. Physical examination at age 15 years revealed dysmorphic features including a low anterior hair line, prominent nose, short philtrum, high-arched palate, oligodontia with abnormal positioning of the teeth, as well as a short neck (Figures 1a and b). Her head circumference was 48 cm (<<3rd centile for age and 50th centile for 2 years) and her weight was 36 kg (<<3rd centile for age and 50th centile for 10.5 years). She had multiple contractures of her limbs, with significant appendicular rigidity and spasticity, but axial hypotonia. She was hyperreflexic and had bilateral ankle clonus, with extensor plantar response on the left.
Figure 1.
Photographs of the two patients showing their dysmorphic features. Patient 1 (a, b) at 15 years old and Patient 2 (c, d) at 10 years old. Both have a low anterior hair line, prominent nose, short philtrum, high-arched palate, abnormal dentition with oligodontia and abnormal positioning of the teeth, and a short neck.
Patient 2, the younger sister, was born at term following an uneventful pregnancy. She never achieved developmental milestones and was also found to be microcephalic at 3 months of age, with a head circumference of 36.5 cm (2nd centile for age and 50th centile for 1.5 months). Although she had occasional episodes of stiffening and opisthotonic posturing, she never developed a seizure disorder, and multiple EEGs ruled out seizures, but did reveal an encephalopathic background. She was also shown to be cortically blind. At 10 years, she was wheelchair bound, fed by gastro-jejunal tube and completely dependent for all activities of daily living. Physical examination at age 10 years revealed the same dysmorphic features as her sister (Figures 1c and d). She had a head circumference of 41 cm (<<3rd centile and 50th centile for 6 months) and a weight of 20 kg (<3rd centile and 50th centile for 6 years). She also had similar findings on neurological examination, including axial hypotonia with appendicular hypertonia, hyperreflexia and extensor plantar response bilaterally. At 10 years and 9 months of age, she was admitted to the hospital for abdominal pain and irritability of unknown cause, despite extensive investigations. She passed away in hospital shortly after admission.
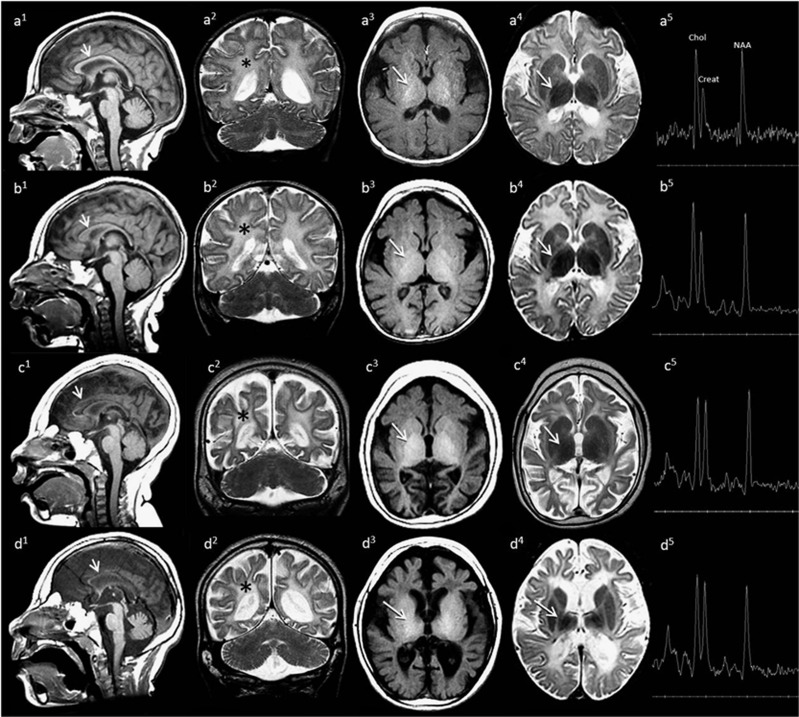
Brain MRIs carried out in the first and third year of life for Patient 1 showed progressive cerebral volume loss involving the basal ganglia and thalami, atrophy of the corpus callosum and supratentorial brain, and diffuse hypomyelination with progressive loss of myelin signal (Figures 2a and d). There was relative sparing of brainstem and cerebellum on early imaging for Patient 1 and on later imaging in Patient 2. Subtle brainstem volume loss, widening of the interfoliate fissures, moderate loss of vermian height and increasing prominence of the infravermian cistern are identified in Patient 1 at 33 months of age. The neuroimaging findings for Patient 2 were similar (Figures 2b and c).
Figure 2.
Longitudinal changes in brain magnetic resonance imaging for the two patients. Images are arranged chronologically. Row 1: Patient 1 at 7 months. Row 2: Patient 2 at 7.4 months of age. Row 3: Patient 2 at 28.8 months of age. Row 4: Patient 1 at 33 months of age. Delayed myelin maturation with later progressive loss of myelin signal, atrophy and calvarial thickening are seen in both siblings. Sequential T1-weighted sagittal images (a1–d1) demonstrate progressive thinning and loss of myelin signal in the corpus callosum (arrows). Coronal T2-weighted images (a2–d2) reveal progressive white matter (*) signal increase and volume loss. Axial T1- (a3–d3) and T2- (a4–d4) weighted images demonstrate early delay in myelin maturation with loss of myelin signal on later images. Only the posterior limb of the internal capsule (arrows) is myelinated on early scans (a3, a4; b3, b4). Loss of myelin signal is noted on later scans (c3, c4; d3, d4). Intermediate echo TE 144 MRS in the basal ganglia demonstrates altered choline:creatine ratios and decreased NAA peaks at all ages. Note the relative sparing of brainstem and cerebellum on early imaging in both (a1, b1) and on later imaging in Patient 2 (c1). Subtle brainstem volume loss, widening of the interfoliate fissures, moderate loss of vermian height and increasing prominence of the infravermian cistern are identified in Patient 1 (d1, d2) at 33 months of age.
Both patients had extensive investigations for genetic diseases, including inborn errors of metabolism, which were all normal.
Molecular methods and results
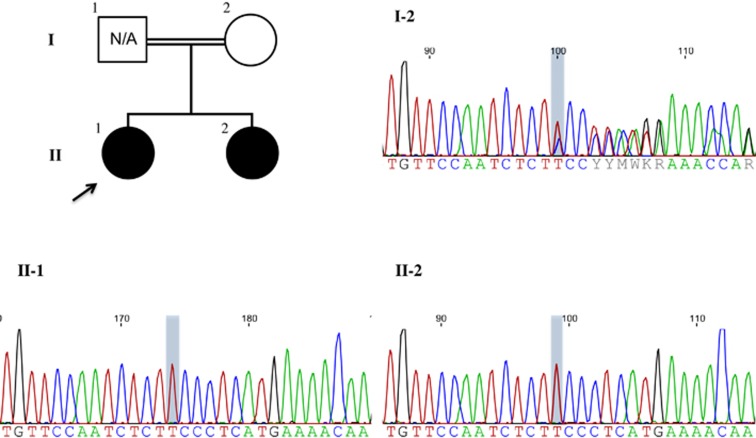
Exome capture was carried out using the Agilent SureSelect Clinical Research Exome target enrichment kit from 3 μg of genomic DNA using the Bravo Automated Liquid Handling Platform (Agilent Technologies, Santa Clara, CA, USA). Sequencing was carried out on Illumina HiSeq2500 (San Diego, CA, USA) in rapid mode and V1 sequencing chemistry following the manufacturer's instructions. Base calling was performed using CASAVA v1.8.2 (Illumina, San Diego, CA, USA) and reads were mapped to the NCBI Build 37/hg19 genome reference sequence with decoy using BWA-MEM v0.7.8 (http://bio-bwa.sourceforge.net). Duplicate reads were removed using MarkDuplicates from Picard v1.108 (http://picard.sourceforge.net). Local read realignment around indels, base quality score recalibration and variant calling with UnifiedGenotyper were accomplished using GATK v2.8.1 (https://software.broadinstitute.org/gatk). SNP calls were subjected to variant quality score recalibration and hard filters by variant call annotations QualByDepth (QD≥2.0), Phred-scaled P-value using Fisher's exact test to detect strand bias (FS≤60.0), RMS Mapping quality (MQ≥40.0), Mapping Quality RankSum (MQRS≥12.5), Haplotype Score (HS≤13.0) and ReadPosRankSum (ReadPosRankSum≥−8.0). Indel calls were hard-filtered by variant call annotations QualByDepth (QD≥2.0), ReadPosRankSumTest (ReadPosRankSum<−20.0) and Phred-scaled P-value using Fisher's exact test to detect strand bias (FS<=200.0). Resulting variant calls were annotated with population frequency information, variant effect on protein and in silico prediction program using a custom pipeline based on ANNOVAR (http://annovar.openbioinformatics.org; downloaded on 14 July 2014) and SnpEff 4.0 (http://snpeffect.switchlab.org). Whole-exome sequencing was initially carried out for Patient 2 and revealed a homozygous c.218dupT (p.His75Serfs*2) variant in exon 3 of CSTB (exon numbering according to transcript NM_000100.3 and LRG_485 (http://ftp.ebi.ac.uk/pub/databases/lrgex/LRG_485.xml)). Sanger sequencing subsequently confirmed the homozygous variant in Patients 1 and 2. The mother is a heterozygous carrier of the variant; the father was not available to be tested (Figure 3). Our patients' variants were submitted to the Leiden Open Variation Database (patient IDs 87233 and 87236), accessible at www.lovd.nl/cstb.
Figure 3.
Pedigree showing the two affected sisters (II-1 and II-2) and the unaffected consanguineous parents of Sri Lankan descent. Accompanying chromatograms demonstrate the homozygous CSTB c.218dupT (p.His75Serfs*2) variant in the two sisters (reference transcript: NM_000100.3). The mother (I-2) is heterozygous for the variant. The father was not available to be tested.
Discussion
Brain anomalies have been described in patients with EPM1A, including volume loss of the brainstem and cerebellum,6 or atrophy of the cortical motor areas and thalamus.7 The cerebral atrophy and loss of white matter seen in our patients are much more severe, and in keeping with that described in Cstb-knockout mice.8 The clinical features of our patients are also much more severe and of earlier onset than those described in EPM1A patients. In the latter, onset is generally around 6 years of age, is preceded by normal development and seizures are the most striking aspect of the clinical picture. In contrast, our patients had neonatal onset of symptoms with complete absence of development, microcephaly and dysmorphic features. Although seizures were present, they were superseded by profound developmental delays as the most striking aspect of the phenotype.
Among patients with mutations in CSTB, those homozygous for the dodecamer repeat expansion, resulting in decreased but still detectable protein expression, have the mildest phenotype (EPM1A). Patients who are compound heterozygous for the dodecamer repeat and a loss-of-function mutation, have less protein expression than typical EPM1A patients, and have a more severe phenotype with earlier age of onset. Finally, patients homozygous for loss-of-function alleles of CSTB, as reported recently by Mancini et al5 and in this report, appear to have a much more severe phenotype. The c.218dupT (p.His75Serfs*2) variant we report in CSTB is located in exon 3 and causes a frameshift, and thus most likely results in a loss-of-function allele.
This is the second report of patients homozygous for a point mutation in CSTB and represents the most extensive longitudinal clinical follow-up described for such patients, highlighting the severity of the associated phenotype, which until recently was unknown. The neurological findings in our patients suggest that cerebral white matter development and maintenance is dependent on proper function of cystatin B. Our observations further support the hypothesis that clinical severity is inversely proportional to residual cystatin B production.
Acknowledgments
We would like to thank the family for participating in this study. This study was supported by the University of Toronto McLaughlin Centre: Next Generation Diagnostics: Advancing the Clinical Application of Genomic Analysis, grant ID MC-2012-13A.
Footnotes
The authors declare no conflict of interest.
References
- Stoka V, Turk B, Turk V: Lysosomal cysteine proteases: structural features and their roles in apoptosis. IUBMB Life 2005; 57: 347–353. [DOI] [PubMed] [Google Scholar]
- Kälviäinen R, Khyuppenen J, Koskenkorva P, Eriksson K, Vanninen R, Mervaala E: Clinical picture of EPM1-Unverricht-Lundbord disease. Epilepsia 2008; 49: 549–556. [DOI] [PubMed] [Google Scholar]
- Joensuu T, Lehesjoki AE, Kopra O: Molecular background of EMP1-Unverricht-Lundborg disease. Epilepsia 2008; 49: 557–563. [DOI] [PubMed] [Google Scholar]
- Koskenkorva P, Hyppönen J, Aikiä M et al: Severer phenotype in Unverricht-Lundborg disease (EPM1) patients compound heterozygous for the dodecamer repeat expansion and the c.202C>T mutation in the CSTB gene. Neurodegener Dis 2011; 8: 515–522. [DOI] [PubMed] [Google Scholar]
- Mancini GM, Schot R, de Wit MC et al: CSTB null mutation associated with microcephaly, early developmental delay and severe dyskinesia. Neurology 2016; 86: 877–878. [DOI] [PubMed] [Google Scholar]
- Mascalchi M, Michelucci R, Cosottini M et al: Brainstem involvement in Unverricht-Lundborg disease (EPM1): an MRI and 1-H MRS study. Neurology 2002; 58: 1686–1689. [DOI] [PubMed] [Google Scholar]
- Koskenkorva P, Khyuppenen J, Niskanen E et al: Motor cortex and thalamic atrophy in Unverricht-Lundborg disease: voxel-based morphometric study. Neurology 2009; 73: 606–611. [DOI] [PubMed] [Google Scholar]
- Manninen O, Laitinen T, Lehtimaki KK et al: Progressive volume loss and white matter degeneration in cstb-deficient mice: a diffusion tensor and longitudinal volumetry MRI study. PLoS One 2014; 9: e90709. [DOI] [PMC free article] [PubMed] [Google Scholar]