Abstract
Glucose transporter type 1 deficiency syndrome (GLUT1DS) is a neurometabolic disorder with a complex phenotypic spectrum but simple biomarkers in cerebrospinal fluid. The disorder is caused by impaired glucose transport into the brain resulting from variants in SCL2A1. In 10% of GLUT1DS patients, a genetic diagnosis can not be made. Using whole-genome sequencing, we identified a de novo 5′-UTR variant in SLC2A1, generating a novel translation initiation codon, severely compromising SLC2A1 function. This finding expands our understanding of the disease mechanisms underlying GLUT1DS and encourages further in-depth analysis of SLC2A1 non-coding regions in patients without variants in the coding region.
Introduction
Glucose transporter type 1 deficiency syndrome (GLUT1DS) is a treatable disorder caused by heterozygous SLC2A1 variants leading to defective glucose transport into the brain.1, 2, 3, 4, 5 Most patients suffer from intellectual disability (ID), early-onset epilepsy, and a movement disorder.3, 4, 5 Cerebrospinal fluid (CSF) analysis reveals the biochemical hallmark of GLUT1DS being decreased concentrations of glucose and lactate.6
Despite extensive efforts, including whole-exome sequencing (WES), a molecular diagnosis remains elusive in ~10% of patients with clinical features supporting GLUT1DS and a typical CSF profile.3, 5, 7, 8, 9 Here, we report the use of whole-genome sequencing (WGS) to identify a de novo non-coding variant disrupting SLC2A1 function in a patient with GLUT1DS, who was previously tested negative by targeted sequencing of SLC2A1 and WES.
Subjects and methods
Clinical report
Patient 1 is a 14-year-old girl who was born after an uneventful pregnancy and delivery. From the age of 3 months, her parents considered her ‘jittery', which was recognized as myoclonic seizures at the age of 5 months and successfully treated with lamotrigine. She had a severe ID (IQ ~50). From the age of 9 years, she was able to walk a few steps independently, however, she generally used a wheelchair. She spoke short sentences. Her parents noticed that her movements were very ‘clumsy' in the mornings, with clear improvement after breakfast, but never noticed episodes that might fit with a diagnosis of paroxysmal dyskinesia. Initiation of the ketogenic diet improved her early morning ‘clumsiness' and her general well-being.
On neurological examination at age 10, she is a friendly, dysarthric girl with a normal head circumference, spastic–atactic movements of arms and legs, some dystonic posturing of the hands, and symmetric increased deep tendon reflexes of all extremities with a bilateral Babinski sign. MRI revealed mild, non-specific hypomyelination. On EEG, there were sporadic, multifocal epileptiform discharges. CSF analysis showed low glucose (1.6 mm; age-specific reference range (10th–90th percentile): 2.6–3.8 mm6) and lactate (1.0 mm; age-specific reference range (10th–90th percentile): 1.2–1.9 mm6) concentrations, in the context of normoglycemia (serum glucose 4.1 mm).
Patient 1 had no SLC2A1 variant after Sanger sequencing of the coding sequence and the intron/exon boundaries (up to ±20 bp), and analysis of copy-number variation by multiplex ligation probe amplification. Because subsequent WES did not reveal clinically relevant variants,10 she was included in a study to assess the diagnostic potential of WGS.11 Notably, three de novo single nucleotide variants were detected in CNGA3, RTP5, and LPPR3, respectively, but neither of these variants was considered to cause the patients' disorder.11
Replication cohort of ‘SLC2A1-negative' GLUT1DS patients
Twenty-two ‘SLC2A1-negative' GLUT1DS patients from the Radboud University Medical Center (Nijmegen, the Netherlands) and the Children's Hospital Aschaffenburg Alzenau (Aschaffenburg, Germany) were additionally included in this study. All patients were clinically suspected of GLUT1DS and had low CSF glucose concentrations. Routine molecular analysis of SLC2A1 (in all 22 patients) and WES (in 12 patients) had not identified clinically relevant variants in SLC2A1 or any other gene.
SLC2A1 mRNA expression analysis in fibroblasts
Fibroblasts were cultured in the presence and absence of cycloheximide for RNA isolation. Total RNA was isolated using RNeasy mini columns (QIAgen, Venlo, the Netherlands), and processed for cDNA generation using the iScript cDNA Synthesis Kit (Bio-Rad, Veenendaal, the Netherlands). Primers for qPCR primers were designed to span the exon 1–exon 2 boundary (forward: GAGCCTGAGCGGGAGAGC; reverse: GACCCGTCAGCTTCTTGC). For qPCR normalization, GUSB was used (forward: AGAGTGGTGCTGAGGATTGG; reverse: CCCTCATGCTCTAGCGTGTC). Experiments were performed in triplicate. ΔΔCt values were calculated using SDS software according to standard procedures (Applied Biosystems, Bleiswijk, The Netherlands).
Expression constructs
Genomic DNA (Chr1:g.43424305-43424844) of exon 1 of the SLC2A1 gene (NG_008232.1, almost the entire 5′-UTR and the entire coding region of exon 1; NM_006516.2:c.-522_c.18) was amplified from patient DNA using primers ACCATTTTGCTAGAGAAGGC (FW) and CTTGCTGCTGGGCTCCAT (RV) with gateway compatible linkers (Attb1). pDONR201 was used as donor vector and pcDNA-DEST47, containing a GFP reporter protein, as expression vector (Invitrogen, Waltham, MA, USA). The start codon of GFP in pcDNA-DEST47 was mutated to prevent translation of GFP alone. Site-directed mutagenesis was performed using standard thermal cycling procedures. Sanger sequencing was used to verify the fidelity of the constructs.
Transfection, imaging, and western blotting
Constructs were transfected in HEK293T cells using FuGENE (Promega, Leiden, The Netherlands). Imaging was performed by fluorescent microscopy using the ZOE fluorescent cell imager system (Bio-Rad, Veenendaal, The Netherlands). Settings were as follows: Zoom 1, Gain 40, Exposure 1000, LED intensity 50, and contrast 0. Protein isolates were subjected to immunoblotting using primary antibodies directed against GFP (ab290, rabbit, polyclonal, Abcam, Cambridge, UK) and alpha-tubulin (ab7291, mouse, monoclonal, Abcam), and secondary antibodies Goat-α-Mouse IRDye800 (926-32210; LI-Cor biosciences, Leusden, the Netherlands) and Goat-α-Rabbit Alexa Fluor 680 (A21076, Molecular Probes, Breda, The Netherlands).
Data deposition
The gene variant was deposited to the LOVD database at http://databases.lovd.nl/shared/ (variant ID 0000132189 and patient ID 00081421).
Results
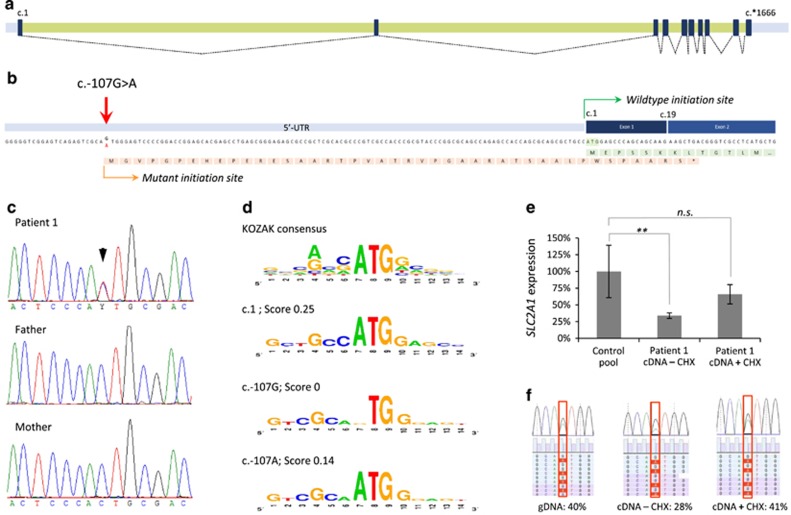
The previously generated WGS data of patient 111 were reanalyzed aiming to identify potential de novo variants in the non-coding sequences of SLC2A1. This analysis revealed one potential de novo variant in the 5′-UTR of SLC2A1 (Chr1(GRCh37):g.43424429C>T; NM_006516.2:c.-107G>A), which was validated by Sanger sequencing (Figures 1a–c). Modeling12 indicated that the variant introduces a putative start codon and KOZAK sequence with a reliability score of 0.14 (Figure 1d). The use of this novel translation initiation site was predicted to result in a premature termination codon in exon 2, thereby targeting this transcript for nonsense-mediated mRNA decay (NMD), and hence in SLC2A1 haploinsufficiency. SLC2A1 mRNA expression analysis in patient fibroblasts indeed showed a ~60% reduction of expression compared to normal controls, and a partial restoration of the expression levels after addition of cycloheximide (CHX), an inhibitor of NMD (Figure 1e). In addition, sequencing of the 5′UTR using the patient's cDNA showed NMD of the mutant allele (Figure 1f).
Figure 1.
Molecular characterization and expression analysis of the SLC2A1 c.-107G>A variant. (a) Schematic depiction of the SLC2A1 exons (black boxes), introns (green bars), and UTRs (gray bars) with splicing information (dotted lines) based on NM_006516.2. (b) The c.-107G>A variant results in a potential novel translation initiation site. The predicted protein effect is indicated in orange, whereas the wild-type protein (partial) is shown in green. The mutant protein is predicted to result in a premature termination codon in exon 2 (*). (c) Sanger sequencing validation of the c.-107G>A variant in patient 1 and her parents, indicating de novo occurrence of the variant. (d) Evaluation of the sequence context surrounding c.107G for the presence of a KOZAK sequence. Top panel shows the consensus sequence, with the relative height of the nucleotides representing the frequency observed at the specified position. Second panel shows the wild-type SLC2A1 c.1 KOZAK sequence. Third panel shows the predication for the wild-type c.107G sequence and its absence of a KOZAK sequence. Lower panel shows the prediction for c.107 A with the presence of a KOZAK sequence. Scores represent reliability of the KOZAK prediction. (e) Relative SLC2A1 mRNA expression in patient 1, indicating a significantly reduced expression (cDNA – CHX) that is partially increased by blocking of nonsense-mediated decay with cycloheximide (cDNA+CHX); **P<0.01. (f) Nonsense-mediated decay of the SLC2A1 transcript bearing the c.-107G>A variant. Patient's fibroblasts were cultured in the absence (−CHX) or presence (+CHX) of cycloheximide, and both genomic (gDNA) and mRNA were isolated. Copy-DNA (cDNA) was made from the mRNA samples, and both gDNA and cDNA were subjected to semi-quantitative ion-torrent sequencing (>40 000 reads per sample) after PCR. The percentage of reads containing the c.-107G>A variant is given. NS, not significant.
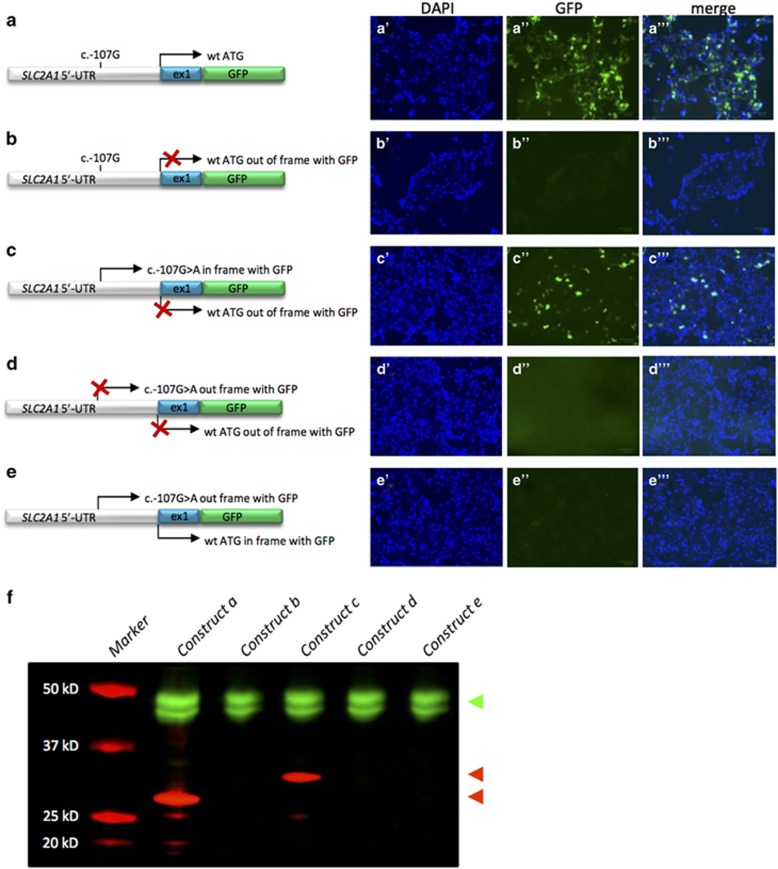
To analyze whether the de novo variant in the 5′-UTR introduces a functional translation initiation codon (ATG), we transfected HEK293T cells with constructs of the 5′-UTR and exon 1 of SLC2A1 fused with the coding sequence of GFP (Figure 2). Since the new ATG formed by the variant would result in the use of another reading frame, constructs were adapted accordingly. Cells containing this construct of wild-type SLC2A1 (wild-type ATG) in frame with GFP (Figure 2a) or the construct with the mutant SLC2A1 (mutant ATG) in frame with GFP (Figure 2c) showed green fluorescent signals suggestive for the use of both ATGs. Moreover, cells expressing the construct with the wild-type ATG out of frame and no mutant ATG present (Figure 2b), or both ATG's out of frame (Figure 2d) did not reveal any fluorescent signals. Finally, cells expressing a construct with wild-type ATG in frame, but mutant ATG out of frame (Figure 2e) showed no fluorescent signals, suggesting a preferential use of the mutant ATG over the wild-type site. A western blot of cell lysates showed the anticipated ~3 kD larger protein when using the mutant ATG compared to the wild-type ATG (Figure 2f), further underscoring the functionality of the new translation initiation site.
Figure 2.
Functional characterization of SLC2A1 c.-107G>A. HEK293T cells were transfected with constructs of the 5′-UTR and exon 1 of SLC2A1 fused to GFP. Left: cartoons of the constructs used. The positions and possible use (arrow) of the wild-type initiation codon (a–e) and the novel initiation codon introduced by the c.-107G>A variant (c–e) are indicated. If no expression is anticipated, the arrow is crossed out (X). Right: representative images of cells transfected with the respective constructs with signals for nuclei by DAPI staining (blue) or green fluorescent protein (green). (f) Western blot of the SLC2A1-GFP fusion proteins created by the constructs in HEK293T cells. An antibody directed against tubulin was used as control (in green, top arrow head), whereas an antibody against GFP was used to detect the SLC2A1-GFP fusion (in red). The fusion proteins resulting from usage of the novel initiation site or the wild-type initiation codon are 32 kD (upper red arrow head) and 29 kD (lower red arrow head) in size, respectively.
We next sequenced the entire 5′-UTR of SLC2A1 (NM_006516.2:c.-525_-1) in a cohort of 22 ‘SLC2A1-negative' GLUT1DS patients. These efforts, however, did not identify any further relevant variants.
Discussion
Since the first description of GLUT1DS,1 many patients have been diagnosed on clinical and biochemical grounds, and benefit from a rational therapeutic approach. In ~90% of these patients, GLUT1DS is molecularly confirmed by demonstration of an SLC2A1 variant that likely affects function. Remarkably, in the remaining patients, no (other) genetic cause can be identified.3, 5, 7, 8, 9 Deep-intronic variants were recently reported to cause GLUT1DS, but detailed functional and genetic evidence for pathogenicity of the variants was lacking.5 Taken together, SLC2A1 may be the only gene responsible for GLUT1DS and ‘SLC2A1-negative' patients may harbor so far undetected, non-coding SLC2A1 variants.
We now present a patient with GLUT1DS, resulting from a de novo variant in the 5′-UTR of SLC2A1. The variant introduces a novel translation initiation site, consequently leading to an out of frame transcript degraded by NMD. Whereas the wild-type initiation codon can be used in principle, the 5′-UTR variant is preferentially used, or alternatively, disrupts proper recognition of the wild-type initiation codon, and thus results in an effective null allele. This mechanism, in which a 5′-UTR variant leads to an upstream open reading frame is a rare mechanism described in disease.13 Although we were unable to identify additional patients with non-coding variants in the 5′-UTR that likely affect function, we have provided compelling evidence that such variants can cause GLUT1DS.
In summary, we present a de novo non-coding SLC2A1 variant, leading to SLC2A1 haploinsufficiency and GLUT1DS. Our results provide a window of opportunity for identification of other non-coding variants in ‘SLC2A1-negative' patients with GLUT1DS.
Acknowledgments
We thank the patients and their parents, as well as the referring physicians for their contributions to this study.
Footnotes
JK has received speakers honoraria and travel costs from Nutricia GmbH, Eerlangen, Germany and Vitaflo Pharma GmbH, Bad Homburg, Germany. The remaining authors declare no conflict of interest.
References
- De Vivo DC, Trifiletti RR, Jacobson RI et al: Defective glucose transport across the blood-brain barrier as a cause of persistent hypoglycorrhachia, seizures, and developmental delay. N Engl J Med 1991; 325: 703–709. [DOI] [PubMed] [Google Scholar]
- Seidner G, Alvarez MG, Yeh JI et al: GLUT-1 deficiency syndrome caused by haploinsufficiency of the blood-brain barrier hexose carrier. Nat Genet 1998; 18: 188–191. [DOI] [PubMed] [Google Scholar]
- Leen WG, Klepper J, Verbeek MM et al: Glucose transporter-1 deficiency syndrome: the expanding clinical and genetic spectrum of a treatable disorder. Brain 2010; 133: 655–670. [DOI] [PubMed] [Google Scholar]
- Hully M, Vuillaumier-Barrot S, Le Bizec C et al: From splitting GLUT1 deficiency syndromes to overlapping phenotypes. Eur J Med Genet 2015; 58: 443–454. [DOI] [PubMed] [Google Scholar]
- Liu YC, Lee JW, Bellows ST et al: Evaluation of non-coding variation in GLUT1 deficiency. Dev Med Child Neurol 2016; 58: 1295–1302. [DOI] [PubMed] [Google Scholar]
- Leen WG, Wevers RA, Kamsteeg EJ et al: Cerebrospinal fluid analysis in the workup of GLUT1 deficiency syndrome: a systematic review. JAMA Neurol 2013; 70: 1440–1444. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang D, Engelstad K et al: Glut1 deficiency syndrome and erythrocyte glucose uptake assay. Ann Neurol 2011; 70: 996–1005. [DOI] [PubMed] [Google Scholar]
- Hashimoto N, Kagitani-Shimono K, Sakai N et al: SLC2A1 gene analysis of Japanese patients with glucose transporter 1 deficiency syndrome. J Hum Genet 2011; 56: 846–851. [DOI] [PubMed] [Google Scholar]
- Klepper J: Absence of SLC2A1 mutations does not exclude Glut1 deficiency syndrome. Neuropediatrics 2013; 44: 235–236. [DOI] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BW et al: Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012; 367: 1921–1929. [DOI] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT et al: Genome sequencing identifies major causes of severe intellectual disability. Nature 2014; 511: 344–347. [DOI] [PubMed] [Google Scholar]
- Salamov AA, Nishikawa T, Swindells MB: Assessing protein coding region integrity in cDNA sequencing projects. Bioinformatics 1998; 14: 384–390. [DOI] [PubMed] [Google Scholar]
- Hornig NC, de Beaufort C, Denzer F et al: A recurrent germline mutation in the 5′UTR of the androgen receptor causes complete androgen insensitivity by activating aberrant uORF translation. PLoS One 2016; 11: e0154158. [DOI] [PMC free article] [PubMed] [Google Scholar]