Abstract
For individual cells in tissues to create the diverse forms of biological organisms, it is necessary that they must reliably sense and generate the correct forces over the correct distances and directions. There is considerable evidence that the mechanical aspects of the cellular microenvironment provide critical physical parameters to be sensed. How proteins sense forces and cellular geometry to create the correct morphology is not understood in detail but protein unfolding appears to be a major component in force and displacement sensing. Thus, the crystallographic structure of a protein domain provides only a starting point to then analyze what will be the effects of physiological forces through domain unfolding or catch‐bond formation. In this review, we will discuss the recent studies of cytoskeletal and adhesion proteins that describe protein domain dynamics. Forces applied to proteins can activate or inhibit enzymes, increase or decrease protein‐protein interactions, activate or inhibit protein substrates, induce catch bonds and regulate interactions with membranes or nucleic acids. Further, the dynamics of stretch‐relaxation can average forces or movements to reliably regulate morphogenic movements. In the few cases where single molecule mechanics are studied under physiological conditions such as titin and talin, there are rapid cycles of stretch‐relaxation that produce mechanosensing signals. Fortunately, the development of new single molecule and super‐resolution imaging methods enable the analysis of single molecule mechanics in physiologically relevant conditions. Thus, we feel that stereotypical changes in cell and tissue shape involve mechanosensing that can be analyzed at the nanometer level to determine the molecular mechanisms involved.
Keywords: mechanobiology, single molecule, protein stretching, mechanosensing, localization microscopy, dSTORM, molecular forces, mechanoenzymatics, bioimaging
Introduction
The growing field of mechanobiology deals with the basic question how the shapes of organisms and all their constituent organs are created by cells through the application of standard functions encoded by the genome. At least in mammalian systems, the organism is dynamically shaped and can change over time with aging, diet, damage, or disease. To create the correct morphology, cells must grow and change their shape in proscribed ways that will influence the development of their neighbors and the surrounding matrix. These processes require highly regulated physical force feedback loops that develop the correct forces on the extracellular matrix (ECM) or neighboring cells as well as the intracellular cytoskeleton, fluid compression, shear stress, and membrane tension. In essence, the cellular complexes that perform these functions are modular machines and they need to have well‐defined input and output parameters to create the proper morphology. Because force is the major input parameter in these functions, the proteins in the machines must be able to create and respond to forces through molecular level changes.
There are potentially many subcellular machines that effectively measure complex parameters such as matrix rigidity, the viability of neighboring cells, curvature of epithelia, or the strength of cell‐cell or cell‐matrix adhesions. When those sensory modules are missing, critical aspects of cell behavior are misregulated as in the case of cancer cells.1, 2 Thus, it is important to understand how those sensory machines work at a molecular level. From many studies, there is clear evidence that mechanical forces directly alter enzyme activities and protein‐protein interactions through either exposing or disrupting binding sites for other proteins or ligands. Because the larger mechanosensory modules are less well understood and involve large protein complexes to generate the emergent properties, they are beyond the scope of this review. We will focus on the basic molecular mechanisms of signaling by forces (see Table 1) that do not tell us about how the modules actually work.
Table 1.
Molecular Changes with Force
| Parameter altered by force | Protein Examples | Force Generated by | Experimental determination |
|---|---|---|---|
| Enzyme Activity | Titin3, FAK4 | Myosin Contraction |
Conventional Biochemistry methods: Cloning, Knock‐down, Enzymatics, etc.5
Live Cell Imaging Techniques(e.g., Single Molecule Imaging, FRET, FRAP)6, 7, 8 Atomic Force Microscopy(AFM)9, 10 Single‐molecule force spectroscopy11, 12 Traction Force Microscopy(TFM)13, 14 PDMS Micropillar Arrays15, 31 Substrate Regidity/Morphology Manupulation16 Laser Ablation17 Molecular Dynamics18 |
| Expose binding sites |
Talin‐vinculin6, 10, 19
Fibronectin20 α‐catenin21, 22 FilaminA23, 24 α‐actinin‐zyxin17 |
Actin flow | |
| Break bonds | Integrin Adhesions25 | Cytoskeletal contractions | |
| Protein Curvature | BAR Proteins26 | Membrane tension | |
| Expose Substrate sites | P130Cas14 | Actin polymerization | |
| Membrane curvature | BAR Proteins27 | Membrane Tension, | |
| Ion Channel | Ca2+Sensitive K+ channe(BK Channel)l16, Piezo1 and Piezo228 | Actin polymerization |
Of the many mechanosensory machines that we imagine are needed to form an organism, the one that is responsible for rigidity sensing is perhaps best understood. Matrix (ECM) rigidity is critical for determining if a cell grows, differentiates, or dies. ECM rigidity or stiffness corresponds to the physical elastic modulus, i.e., the force required per unit contact‐area to cause a unit deformation of the ECM. It assumes the dimension of pressure. ECM rigidity in mammals ranges from hard bone (∼10 GPa) to the soft brain(∼100 kPa).29 Recent studies indicate that a modular machine resembling a muscle sarcomere senses rigidity by displacing the matrix a total of 100 nm and if the force exceeds about 25 pN (the force from about 6 myosin heads), then the surface is considered rigid.30, 31 The rigidity sensing machine is regulated by tyrosine kinases in a variety of ways—the length of the displacement is regulated by the tyrosine kinase AXL, whereas the duration of the displacement is controlled by another tyrosine kinase, ROR2,30 and the complex is thought to require EGFR activity to form (Saxena et al., Nature Materials In Press). For fibroblasts, soft fibronectin‐ coated surfaces cause adhesion disassembly followed by death whereas rigid surfaces stimulate adhesion maturation and growth. Although the force threshold of 25 pN has not been linked to protein unfolding, the displacement limit of 100 nm could reflect the length of protein domains that are being unfolded. There is a lot more work needed to understand how the rigidity sensing contractions and other mechanosensory machines actually work.
At a molecular level, forces on individual proteins have a few clear effects. It is believed that the molecular basis of these mechanisms are changes in protein shape and often protein unfolding directly caused by cellular forces. There is ample experimental evidence that many adhesion and cytoskeleton proteins undergo conformational change in response to cellular forces. Recent studies of protein mechanics both in vivo and in vitro document that mechanical unfolding as well as the distortion of cytoskeletal and adhesion proteins are fundamental to the conversion of mechanical forces into biochemical signals.3, 32, 33 These molecular conformational changes have numerous biochemical consequences. They can activate or deactivate enzymatic functions, expose or hide substrate sites, and regulate cytoskeletal filament dynamics.
Taking the muscle protein titin as an example, titin is a giant sarcomeric protein that stretches from myosin filaments to the z‐band and directly contributes to the passive stiffness and structural integrity of muscles. Titin itself is elastic because it contains a sequence of folded domains that can be progressively unfolded with the force of muscle stretching.34 Energy stored in titin during stretching will aid during the next contraction of the muscle. The elasticity of titin can be modified naturally by isoform splicing and post‐translational modifications at different stages of organism development.35 Mutations that alter the titin stiffness or abnormal expression of certain isoforms are associated with cardiac diseases.36, 37, 38 The adrenergic pathway is also known to alter myocardial stiffness. This is because adrenergic signaling can control protein kinase activity, and the level of titin phosphorylation can then subsequently alter titin's mechanical properties.39, 40
In this example, the titin unfolding is simply used to store mechanical energy for the next contraction; however, there are many other aspects of the stretching of titin that can modify muscles over time—titin exhibits several force‐dependent activities. There is a kinase domain in titin and both the titin and related twitchin kinases change under force.6, 41 Further, the domains of titin unfold in a specific order with the least stable domains unfolding first. Although it is not well documented for titin, there are many titin‐binding proteins whose binding probability to titin increases with the unfolding of their respective binding domain (either released upon unfolding or binding to regions exposed by unfolding). Thus, one protein can be used to mechanically monitor the degree of muscle stretch and send or receive signals that will alter muscle contractility.
For mammalian cells to be able to generate and respond to forces, they assemble a cytoskeleton that forms a continuous mechanical structure across the cell. Unlike macroscopic skeletons, the cell cytoskeleton is very dynamic (undergoing transformation on the timescale of tens of seconds to minutes) and must support tension while the structure is disassembling and reassembling, thus requiring a very robust mechanisms for the cellular proteins to create the proper tension and measure parameters critical for shaping the local as well as the macroscopic tissue over time. Research shows that the accumulation rate and location of MyosinII (the dominant motor protein), actinin (actin cross linker and filamin (actin crosslinker) will be different when the type of mechanical load or the angle of mechanical load are different. In these experiments, the cytoskeleton and its associated proteins adapt to the external mechanical load interactively, in a timely fashion.42, 43
In contrast to the intracellular cytoskeleton, the ECM of most tissues is not turning over although it also experience repeated mechanical stretch‐relaxation cycles.44, 45 For example, collagen in the Achilles tendon that is assembled at birth will remain in situ for the life of the individual.46 The ECM is present in all tissues of multi‐cellular organisms and provides a stable base for maintaining cellular differentiation. In tissue regeneration studies, the currently best way to have stem cells form the desired tissue is to seed them onto a decellularized matrix of that tissue. Although the specific ECM proteins in the matrix will influence cell behavior, the mechanical properties of the matrix are themselves important, since stem cells will differentiate to neurons or osteocytes based primarily upon the rigidity of the collagen matrix (<1 kPa for neurons and >100 kPa for osteocytes). Mutations that cause a cell to fail to react to rigidity differences have been implicated in several cancers.47, 48 As an aside, this emphasizes the importance of the cell mechanosensing systems particularly in the processes of development, regeneration, and disease progression. Matrix mechanical properties are critical for the tissue to be maintained since the outcome of cell testing matrix rigidity strongly influences cell behavior.
There are many aspects of mechanobiology, in this review though we will focus on the mechanisms of force transduction that are exhibited by a few well‐studied proteins that regularly are part of force bearing elements of cell adhesions or the cytoskeleton. There is not sufficient space to treat the mechanical properties of matrix proteins. We will also discuss the current methods to monitor protein unfolding both in vitro and in vivo that show many proteins are unfolded regularly in cells. This indicates that the functions of many proteins are strongly dependent upon conformations changes induced by mechanical activity.
Mechanosensing by Proteins in Adhesions or the Cytoskeleton
Because cell adhesions are the points where forces are transmitted to the cytoskeleton from the outside, mechanosensing should primarily occur in their vicinity. Perhaps it is not surprising that there are over two hundred cell‐matrix adhesion proteins and about one hundred and fifty verified cell‐cell adhesion proteins.49, 50 The large number of adhesion proteins fits with the idea that adhesions control cell behavior, including growth, death and differentiation. However, for overall cell shape determination and regional specialization, the cytoskeleton, and other factors must be involved. For example, cell polarization is critical for single cell migration, neuronal and epithelial function. In cell shape determination, the mechanical coherence of the cytoskeleton from one side of the cell to the other is important. The coherence is critical for propagating forces across the cell to make a mechanical continuum with the adhesions such that the proper tissue tension can be maintained. In the overall scheme, adhesions can convey forces across the cell membrane to the cytoplasm (through the focal adhesion complex51) and generate biochemical signals (e.g., protein unfolding or force activated ion channels).52 Extracellular forces are propagated through the cytoskeleton by activation of myosin contraction on dynamic actin networks. Remarkably, the system reliably maintains the proper tissue tension through many perturbations including physical activity of the organism, metabolic changes, dehydration, and some diseases.
The cell‐matrix adhesions
Although cell‐matrix adhesions are only one of the many different types of adhesions, they have been extensively studied partially because of the ease of access: cultured cells typically adhere to glass coverslips through matrix adhesions. Further, since fibronectin supports fibroblast growth and is a major component in serum, the integrin‐fibronectin adhesions have been characterized most frequently.
They constitute a paradigm for the further study of other matrix adhesions that are particularly relevant for other cell types and the complexities of fibronectin‐integrin adhesions reveal the types of mechanisms that cells use for mechanosensing.1 The proteins associated with cell‐matrix adhesions are surprisingly dynamic and many are mechanically stretched in vivo. The FA protein turnover rate is typically in the range of tens of seconds to minutes (Table 2). Logically, the effective reaction time (both chemically and or mechanically) for these proteins needs to occur on a comparable time scale. It is important to consider this pace when studying the behavior of mechanosensing proteins.
Table 2.
The Published Dynamic Turnover Times for Popular FA Proteins
| Protein Name | FRAP t1/2(s) | Cell Type |
|---|---|---|
| Vinculin | 1.9/14/50/11/21 | HelaJW53/MEF54, 55/REF5256 |
| Paxillin | 1.5/25/17.7/53.2/20 | HelaJW53/MEF55/Bovine capillary endothelial cells57/MEF, TIRF FRAP58/REF5256 |
| FAK | 2.1/7/6.7/14 | HelaJW53/MEF55/Bovine capillary endothelial cells57/REF5256 |
| Zyxin | 0.5/12/7/10 | HelaJW53/MEF55/REF5256 |
| Talin | 2.2/23/77/18 | HelaJW53/MEF55/Bovine capillary endothelial cells57/REF5256 |
| ILK | 9.2/12 | HelaJW53/REF5256 |
| α‐actinin | 36/14.1/27 | MEF55/Bovine capillary endothelial cells57/REF5256 |
| beta3‐Integrgin | 38.5 | Bovine capillary endothelial cells57 |
| p130Cas | 5.8/14 | MEF, TIRF FRAP58/REF5256 |
| Tensin | 55 | REF5256 |
| Src/CSK | 9 | REF5256 |
| Pinch | 14 | REF5256 |
| VASP | 0.7/15 | HelaJW53/REF5256 |
Notice that FRAP (Fluorescence Recovery After Photobleaching) results may vary between different microscopy systems, cell types, focal adhesion status, and based on other factors. Different FRAP results and cell types have been referenced respectively.
The adhesion proteins exhibit basic paradigms of mechanosensing that differ at the individual protein level. Earlier studies on cytoskeletal signaling proteins have led to a consensus that the mechanical stretching of proteins is a fundamental mechanism of force sensing.59 Force induced conformational change of mechanosensitive proteins can result in the exposure, destruction, or change of (weakening or strengthening) the functional sites, such as phosphorylation sites, binding sites, slip bonds, catch bonds, and ion channels.59, 60 However, we expect that adhesion proteins represent a small fraction of the proteins whose normal functions involve domain unfolding and refolding as a result of mechanical stretch. Specifically, we will consider here the responses of the talin‐vinculin system, FAK, and p‐130Cas to forces.
Formation of cell‐matrix adhesions
Numerous studies of adhesion formation have revealed that the process contains many different steps as well as alternative pathways that serve to make it robust and to enable cell growth under a variety of conditions.61 Initially, cells in suspension contact a matrix‐coated surface and clusters of activated integrins about 100 nm in diameter containing 50 integrins will form and actin polymerization will be activated by the clusters.62 If clusters are close together, myosin will pull actin‐associated clusters together and generate forces that will activate the next step in spreading that involves rapid extension of actin‐based lamellipodia (an alternative mode of spreading involves a series of brief extensions followed by local contractions for rigidity sensing). In the rapid spreading mode, the cell will flatten until it runs out of folded membrane and that will cause a rise in membrane tension thereby activating exocytosis to increase the membrane area plus causing rigidity‐sensing contractions.63 If the surface is rigid, then adhesions will mature and will constantly be under tension due to the flow of actin from the leading edge, contraction of stress fibers, and an actomyosin network that spans the cytoplasm. Within this complex process are many cyclic processes, including assembly and disassembly of actin filaments, stretching and relaxation of talin as well as the rapid turnover of proteins that are part of the cell‐matrix adhesome (Table 2).64 Since tension in the cytoskeleton is needed to maintain adhesions, mechanotransduction is critical.65
The major question that is largely unanswered is how does mechanotransduction occur in the cell. Although the focus is necessarily on the adhesion complex, the integrin adhesome is integrated with the cytoskeleton and many proteins are known to be present in both complexes, for example, vinculin, VASP (Vasodilator‐Stimulated Phosphoprotein), zyxin, and α‐actinin.66 Similarly, super‐resolution 3‐D microscopy reveals the layered transition of protein composition from the adhesions to the cytoskeleton as the observation moves from the membrane to the cytoplasm.67, 68 Two major dynamic processes are occurring at adhesion sites; namely, actin in stress fibers is assembling from adhesion sites and actin filaments assembled at the leading edges of lamellipodia are moving over adhesions. In the first case, actin filament assembly is force dependent and that has been observed in vitro for actin polymerization by formin proteins.69, 70 This is a mechanosensing phenomenon and may extend to other actomyosin networks as well71, 72; however, the major role of this process is to replace actin filaments that are disassembling. When myosin is inhibited, the stress fibers and the actomyosin network in cytoplasm rapidly disassemble by an unknown mechanism. The other mechanotransduction process at adhesions involves the transient binding of adhesion components to the flowing actin filaments. Because the actin flow rates are 10 to 100 nm/s and the lifetime of the adhesions are tens of minutes, the binding of individual proteins to actin can only exist for tens of seconds or less even if the protein is stretching with the actin flow. We will consider the consequences of the interaction of adhesion proteins such as talin with actin below because it appears to be a significant mechanotransduction process that was revealed by recent advances in microscopy.
Talin Mechanosensing
Talin
Among the large number of proteins in FAs, talin is one of the best‐studied FA adaptor proteins. It is large, consisting of a 220kD rod domain and a 50kD head domain and is found in almost all animals and cell types (Fig. 1). In addition to FAs, talin has been reported in podosomes, invasive structures, immunological synapses, cytotoxic synapses and so forth.73, 74 In FAs, Talins work as direct bridges between actin and integrin and as recruiters for other FA proteins.75 There are two isoforms of talin in mammalian cells, Talin1 and Talin2 (with 74% sequence overlap).76 Most cell types express both isoforms, but talin1 is the one that carries out most of the known talin functions, such as integrin beta activation. Talin1 knockout mice die before birth,77 whereas talin2 knockout mice are viable and fertile.78 Recent studies show that talin2 is essential for invadopodia formation and subsequently matrix degradation and cell invasion.79 But since talin2 is not essential for integrin beta activation,80 it is generally considered as redundant for talin1, especially in regular FAs. Although talin has no enzymatic activity, knockdown of both talin isoforms in 2D cultured cells will result in a complete loss of cell adhesion to matrix‐coated surfaces.81
Figure 1.
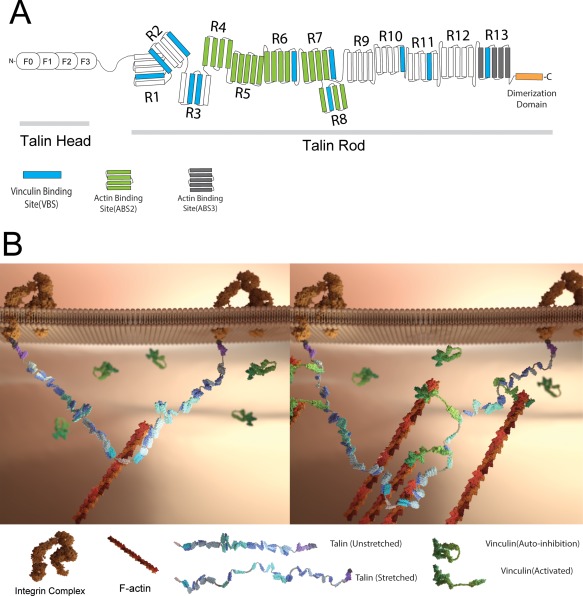
(A) Illustration of the domain structure of full‐length talin. The talin head domain contains a FERM domain (50 kDa), followed by a flexible “neck” (10 kDa) which connects the head domain to its C‐terminal rod domain (220 kDa). The rod domain contains 11 cryptic VBS (drawn in blue). The dimerization domain is a single helix that sits at the end of the rod. (B) Schematics of the talin structure and interaction of the talin dimer with vinculin in cells. A. Illustration of the domain structure of full‐length talin. Talin head domain contains a FERM domain (50 kDa), followed by a flexible “neck” (10 kDa), which connects the head domain to its C‐terminal rod domain (220 kDa). The rod domain contains 11 cryptic VBS (drawn in blue). The dimerization domain is a single helix that sits at the end of the rod domain. B. (left) In the initial stage of FA formation, the talin dimer binds to actin and integrin. At this stage, the cryptic VBSs remain buried among the α‐helical bundles. (right) As the actin filament starts to pull on talin, the formerly buried VBS are revealed to allow vinculin binding, and cause more actin filament recruitment.
Talin and vinculin
Among the many binding partners of talin, vinculin (an FA protein) is the most studied. There are eleven VBSs (Vinculin Binding Sites) on talin. All the VBSs can bind to vinculin independent of each other—talin mutants with different VBS deletions can still recruit vinculin.82 The “co‐localization” between talin and vinculin is extensive and goes beyond matrix adhesions. For example, ARP2/3, vinculin, and talin have been reported in the immunological synaptic complexes initiated by stimulated TCR (T‐Cell Receptor).73
Both vinculin and talin have binding sites for actin. As discussed previously, talin has three active binding sites (ABSes) and talin is commonly considered the primary direct link between integrin and actin. Vinculin has only one ABS, no enzymatic activity, more than fifteen binding partners, but no direct link to integrin. Vinculin knockout results in early lethality in C. elegans and mice.83, 84 Vinculin is usually considered an adaptor protein in FAs, providing additional protein clustering and fine adjustment to the link strength between actin and integrin.85, 86
Several conformation changes regulate talin and vinculin folding and protein interactions. Talin and vinculin each adopt an inactive/auto‐inhibited state in which the N‐terminal head domain binds with the C‐terminal tail domain when in the cytosol.87 Protein activation caused by binding partners stimulates recruitment to the FA.87, 88
The auto‐inhibited form of talin involves the F3 subdomain in the talin head binding to the talin rod R9 helical bundle (F3/R9 interaction). When auto‐inhibited, talin assumes a donut shaped globular form. The globular talin form will change to a more extended conformation under high salt condition, indicating that the F3/R9 interaction that is responsible for talin auto‐inhibition is mediated by hydrophobic and electrostatic interactions.89, 90
Similar to talin, vinculin auto‐inhibition is also described by the interaction between the head and the tail domain, masking the ABS and many other protein binding sites on vinculin. Intra‐molecular FRET (Foerster Resonance Energy Transfer) studies show that only extended/active vinculin is found in FAs, indicating that vinculin activation is necessary for vinculin recruitment into FAs.
Activation of both vinculin and talin occurs by binding to partners that compete with their head and tail interaction. In the case of talin, RIAM is one of its activators as will be discussed below; and for vinculin activation, the talin and actin fibers are both required to overcome the vinculin head and tail interaction.91
All the cryptic VBSs on talin rods are buried in α‐helical bundles and are not accessible for vinculin binding, hence the qualifier “cryptic.” Force dependent talin stretching is required for those sites to become available for vinculin interaction. We have demonstrated that the contractility of the actomyosin cytoskeleton directly causes stretching of talin and recruitment of vinculin.10 In previous studies, talin stretching was monitored by imaging a dually tagged talin (GFP‐talin‐mCherry) using TIRFM. Talin has around 2500 amino acids that could be stretched to a length of around 1000 nm. By tracking the displacement of the GFP and mCherry fluorophores tagged on different ends of talin, our measurements showed that talin was typically 80 to 350nm in length in FAs. This length depended on actomyosin activity (based on Blebbistatin and Y‐27632 treatment studies) and was proportional to the amount of vinculin recruitment (immunostaining).10, 92 Active vinculins should act as instantaneous conformation sensors that bind to any VBS that becomes available. Simulation and in vitro single molecule stretching studies indicate that the R3 helical bundles should be the first domains that open for vinculin binding at small forces that could be generated by a single myosin.22, 93 Thus, the sequence of unfolding of talin domains should dictate the order of vinculin binding.
Talin and RIAM
Magnetic tweezers single molecule studies reveal that the rod domain of talin can unfold in a stepwise fashion (Fig. 2).92 After the R3 α‐helical bundle unfolds at around ∼5pN pulling force, it is postulated that the R2 and R3 bundles will soon unfold thereby revealing two VBS domains. However, the Rap1 small GTPase RIAM binds to talin R2 and R3 α‐helical bundles94 in their closed conformation preventing vinculin binding. This indicates that the binding of RIAM and vinculin onto talin could be mutually exclusive. Indeed, NMR experiments conducted by B. Goult et al demonstrate that vinculin and RIAM compete for talin binding.94
Figure 2.
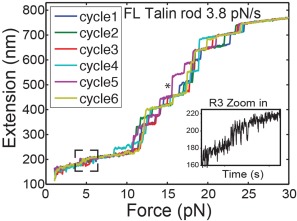
Force extension curves of 13 helical bundles of talin rod domain. Magnetic tweezers were used to develop a constant rate of force increase (3.8 pN/s) to a full length talin rod domain molecule. The R3 domain unfolds first at ∼5 pN and the rest of the talin rod domains unfold at forces between 10 to 25 pN.
Data also shows that the binding of RIAM is necessary for recruiting cytosolic talin to the plasma membrane by connecting Ras GTPases to talin and altering the head domain binding to the rod domain in the cytosol.95 Normally, the integrin‐binding site is masked while the RIAM binding site remains available and RIAM binding to this site frees talin from its auto‐inhibitory form.96 Since talin recruitment to membranes is essential for activation of β‐integrin, RIAM is also important in the process of integrin activation.97 Indeed RIAM activates integrin and its ability to activate integrin is dependent on talin and Rap1 binding. Loss of RIAM causes adhesion deficiency through the talin‐integrin pathway in mice.98 But other studies have shown that this effect is not universally observable, and that RIAM dependent talin or integrin activation may have a significant redundancy, or RIAM might activate only specific talin and/or integrin subpopulations.98, 99
Fluorophore tagged RIAM was found in nascent adhesions but not in mature FAs.94 Taking this and all other available information into consideration, RIAM contributes to talin recruitment to the inner membrane, where it works alongside talin activating and clustering integrins in nascent adhesions. During the maturation of FAs, when the forces exerted on talin start to grow, talin subdomains R2R3 will be unfolded, RIAM will leave and vinculin will bind.
Talin stretching in vivo
Due to talin's unique structure and binding interactions with vinculin, talin is one of the most studied mechanosensing proteins. Talin stretching and its relationship with vinculin binding has been demonstrated by multiple groups under various experimental conditions.13, 92, 93, 100, 101
The cyclic stretching‐relaxation of talin and talin length dependent vinculin binding‐release events in situ have been further characterized by recent single molecule localization microscopy work.9, 10 In these studies, talin length in vivo is determined by tracking the displacement of GFP(talin N‐terminal) and mCherry(talin C‐terminal). A third spectral channel (Atto655) was introduced to tag vinculin with the DHFR‐TMP labeling system.102 These studies reveal that talin undergoes rapid cyclic stretching in mature FAs and vinculin binding occurs in a cooperative fashion not observed before.
Integrin
Another well‐established mechanosensing protein is integrin. The well‐characterized integrin activation events involve a conformation change that can be readily introduced by stretch in a catch‐bond dependent manner.2, 103 Unlike talin, the conformation change that activates integrins is the switch between the bent (inactivate) and extended (activate) states. The activation of integrin α5β1 is directly associated with the actomyosin contraction. Chemical crosslinking assays demonstrate that treatment with a myosin II inhibitor significantly reduces activated integrin α5β1 compared to non‐treated cells.104 Another study employing a bead micromanipulation assay shows that Integrin αLβ2 undergoes a force‐dependent conformational change/activation. This live cell single molecule experiment indicates that integrin αLβ2 cycles between bent and extended conformations, and that both the switching frequency and the ratio between the two states are affected by the application of force.60 Thus, force on integrin‐ligand bonds may stabilize the bound states through a catch‐bond like mechanism.
Mechanosensing in the cytoskeleton
Filamin a
Filamin A is a large actin cross‐linking protein that is believed to have mechanosensing activity. It forms homodimers through the C‐terminal IgG‐like domain. The dimerization domain is capable of crosslinking actin fibers at a relatively large angle (orthogonal crosslinking).105 Similar to talin, the rod 2 domain of filamin A contains cryptic binding sites that required force induced strain to activate.106 An in vitro FLAC(fluorescence loss after photoconversion) assay confirms this by showing that applying force on filamin crosslinked actin fiber changes the binding affinity of binding partners of filamin A. Increased integrin binding and reduced filGAP binding was observed on the filaminA‐actin junction after strain.24 Thus, it appears that tension in the cytoskeleton can be transduced through the alteration of filamin A conformation that causes altered binding to other proteins and changes in cell behavior.
Mechanoenzymatics
Besides altering protein‐protein interactions through domain unfolding, catch bonds or conformational changes, there are a number of enzymatic activities that are linked to mechanosensing. Of particular interest are the FAK and titin kinases3 that are activated by mechanical domain separation. In addition to kinase activation by stretch, there is substrate activation by domain unfolding. The p130Cas substrate domain can be spread opened to catalyze its phosphorylation. In addition, a number of enzymatic signaling pathways have been implicated as mechanosensing pathways, these include the twitchin kinase, Src,8 RhoA,107, 108 Rac1,109 Cdc42,107 and FAK.110
Titin kinase
Titin has over 100 distinct domains in its micrometer length and one of those has kinase activity. Single‐molecule force spectroscopy studies confirm that titin unfolding involves the kinase domain.111, 112 Molecular dynamics and enzymatic studies further indicate that the strain induced conformation changes of titin activate its C‐terminal kinase domain as well as the ATP binding site, in a stepwise fashion.3 As an integral part of the muscle contractile unit, titin is responsible for the assembly of sarcomere, preventing overstretching and maintaining muscle elasticity. It can also act as a strain sensor and relay the mechanical signal into a chemical signal induced by strain‐dependent alterations in ATP binding affinity and kinase activity.
FAK
Focal Adhesion Kinase(FAK) is a non‐receptor protein tyrosine kinase that is important in cellular mechanosensing and is activated through a myosin‐dependent process. FAK‐/‐ cell lines fail to initiate the bone formation process triggered by load. Reintroducing wild type FAK but not mutated FAK (point mutations in Tyr‐397 or Tyr‐925) will restore bone formation.110 Recent molecular dynamics (MD) and force‐probe molecular dynamics (FPMD) simulation studies further support FAK's role as a mechano‐sensor.4 FAK consist of three main domain structures, a FERM domain, a kinase domain and a FAT(focal adhesion targeting) domain. According to this in silico study, the FERM domain of FAK needs to be pulled away from the kinase domain to allow auto phorsophorylation and subsequent activation of FAK. Thus it is likely that force is needed to activate FAK.
P130Cas
Another example is P130Cas. P130Cas itself is not a kinase, but it is ubiquitously expressed and is considered as the major substrate for Src (a non‐receptor tyrosine kinase).113 After stretching triton treated cytoskeletons, Sawada et al reported that P130Cas was phosphorylated in focal adhesions and the extension dependence of P130Cas phosphorylation was confirmed in vitro.114, 115
Mechanosensing at cell‐cell adhesions
The other adhesions that are involved in mechanosensing are the cell‐cell adhesions, of which there are several types. Most is known about the role of α‐catenin in cellular mechanotransduction, where both the interaction with actin and the binding of vinculin are force dependent. This is part of cadherin‐dependent cell‐cell junctions and there are many additional proteins that bind to those adhesions to create the cadherin adhesome or cadhesome50, 116 that has a variety of mechanosensing functions. Further, it is likely that there are mechanosensing modules in cells that rely upon cell‐cell adhesion machinery but we know relatively little about those at this time. An important component of tissue mechanics is provided by the intermediate filaments that have the interesting property of telescoping under force and this could provide passive tension for epithelia once an intermediate filament network is assembled and stretched. Again, we have few examples of mechanotransduction but they also appear to involve stretching of proteins or filaments to alter binding activities.
α‐Catenin
α‐Catenin is an adaptor protein that connects cadherin to actin cytoskeleton. Vinculin can bind to α‐Catenin and modulate the binding affinity of cadherin junctions to the cytoskeleton.117 An X‐ray crystallography study reveals that it prefers an auto‐inhibitory status, which renders its vinculin binding site buried. In addition, it normally binds only weakly to actin filaments but force on actin filaments will activate the α‐catenin‐actin bond formation, again a catch‐bond type of behavior.12 Magnetic tweezer single molecule stretching experiments demonstrate that α‐Catenin can unfold under physiological relevant force levels(5∼15 pN), and the unfolding of α‐Catenin promotes vinculin binding in vitro.22
Imaging the effects of molecular forces
Imaging techniques have been extensively used in the field of mechanobiology. Both traction force microscopy and pillar tracking imaging systems rely on localization techniques of either beads or pillar to trace the substrate deformation and therefore calculate the actual force that's being apply to the ECM by the cell. Forster‐Resonance‐Energy‐Transfer(FRET) probes can measure the mechanical load on individual molecules and studies using those indicate that vinculin is under higher tension when located at the leading edge of the cell.118 In the second session of this review, we are going to discuss the application of localization based super‐resolution imaging technique in the study of mechanobiology.
Molecular microscopy
Fluorescent tags enable the in‐vivo study of individual or aggregates of proteins. They can be expressed within the proteins119 or linked to them (Table 3). The imaging modality used depends on the molecular scale required126 and can extend down to the few‐nanometer scale.
Table 3.
Summary of In Vivo Chemical Dye Labeling Systems
| Name | Protein tagging | Emission | Staining | Comments |
|---|---|---|---|---|
| FlAsH/ReAsH120 | TetraCystein sequence(TC), 6 aa | Green,Red | Incubation | Covalent, fluorogenic, commericialized |
| HaloTag121, 122 | Bacterial enzyme haloalkane dehalogenase, 30kDa | Haloalkane conjugated dyes | Incubation, electroporation | Covalent, commercialized, enzyme and dye raion 1:1. |
| SNAP‐Tag121 | Human enzyme AGT, 182aa, 19.4kDa | O6‐benzylguanine conjugated dye123s | Incubation, electroporation | Covalent, commercialized |
| CLIP‐Tag121 | Human enzyme, AGT | O6‐benzylcytosine conjugated dyes | Incubation, electroporation | Covalent, modified from SNAP‐tag, commercialize |
| eDHFR‐TMP123 | Bacterial enzyme, eDHFR, 18kDa | TMP conjugated dyes | Incubation, electroporation | Covalent, fluorogenic version available, commercialized |
| Metal Chelators124, 125 | 2∼3 kDa | Nitrilotriacetate conjugated dyes | Incubation | Low affinity and low stability, not membrane permeable |
FRET, the electromagnetic coupling of one optical dipole (donor) to a nearby one (acceptor) covers the shortest scale (1–10nm). The acceptor will be excited more frequently with proximity to and alignment with the donor.127 The method enables measurements of (un)folding of a single site, recruitment, or binding events on a sub‐molecular scale.126 Further, it generally does not provide an image of the proteins of interest.
Superresolution microscopy is a form of Far field optical microscopy that considerably breaks the diffraction limit and involves techniques such as PALM and STORM. We won't consider methods like STED that work best on pools of molecules and NSOM which are not contact free and hence disruptive.128
An isolated sub‐resolution particle will form a diffraction blurred imaged aptly termed Point Spread Function (PSF) whose center can be localized much more finely than the width of the blur: used in astronomy,129 particle microscopy,130 and for single proteins.131 The FWHM of the PSF narrows with the square root of observations.132 Thus the localization of a single fluorophore can be better than the resolution limit by 1–3 orders of magnitude, enabling single molecule tracking if fluorophores are sparse.6, 133 Measuring molecular length can be done with different wavelength tags,7 blinking tags or through analysis of the deformed PSFs that occur when two fluorophores have overlapping PSFs.6
Using dense assemblies of fluorophores that are active at different times and hence allow imaging by localization, emerged in 2006. At first with photoactivation localization microscopy (PALM),134 then stochastic switching and coupling (STORM),135, 136 and finally by fast optical pumping into the fluorescent dark state,137, 138 direct‐STORM. These methods share the signal processing methods but differ in hardware needs.139 In general, they have not been exploited to measure molecular lengths dynamically.
For the more in‐depth in vivo analysis of the weakly parallel dimer Talin (which are linked at the C‐terminus), no proven tracking technique is ideal as different spectrum proteins will only report a length corresponding to the geometric center of both N‐termini. To overcome this, we exploited the deformation of the overlapping PSFs of both green tagged N‐termini to estimate their relative separation. Unlike more general localization methods that function in dense assemblies of fluorophores that either rely on blinking,140, 141 or estimate complex clusters of few fluorophores,142 having two known identical tags within one diffraction limited spot allows for continuous observation and orientation estimation at the same time.6 The approach is less potent than pure localization microscopy and needs separations in excess of 35nm to reliably detect a separation. It relies on the fact that any spatial separation of the tags will project a spot that is the sum of two non‐concentric PSFs and the dislocation is solved by matching two PSFs to the observed spot.
Single molecule counting
In fixed specimen, PALM provides accurate molecular counting and hence densitometry. Artifacts only arise from multiple events within one optical blob and the error can be capped by low activation power. In living specimens however, protein turnover and recovery compete with the destruction process and the count may become meaningless as number of proteins at one location grows over time when the cell stays active. Still oriented samples can provide length information.
dSTORM is a less suitable candidate for fluorometry as dye off‐times are mere stochastic decay properties. The timing cannot be controlled universally, and many dyes sport multiple dark state populations.143 Currently, the permitted densities for potentially analyzing single moleculaes are capped by the tolerable laser power to the order of below thousand molecules per m2.
Alternative techniques
Combining tagged Talin with its non‐tagged equivalent which then would cleanly report the dynamics of one strand while staying viable, reliably failed—the early dimerization of the proteins heavily favors dual tagged dimers. Speckle promotion reduces the density of optically active dimers but generates few optical monomers. Bleaching one of the strands of a dimer also proved to be both inefficient and detrimental to the life span of the other strand. It does however provide a welcome reference within the same experiment to assess the quality of the multiple‐tags‐per‐PSF methods employed for these experiments. Upon one of two identical tags bleaching, the optical centroid travels half the tags' separation along the molecules' orientation—the very last measurement of orientation and separation is hence an accurate one. The earlier estimates provide inferior accuracy than dSTORM but without impacting the viability and activity of the proteins and hence enabling long term in‐vivo observations.
Discussion
Proteins have different stages of life and many of those involve changes in the folding pattern particularly when the protein is part of a larger mechanically important complex. For cells to assume the proper morphology and contribute to the shaping of the tissue, those unfolding events must be transduced into biochemical signals that can be distinguished from the normal thermal unfolding events. Through force dependent conformational changes, cells convert mechanical cues experienced at signaling hubs such as cell‐matrix and cell‐cell adhesions into biochemical signals. The magnitude of the forces needed for domain unfolding are often large enough that it is highly improbable that unfolding could occur by thermal diffusion. These mechanosensing events can lead to regulation of cell behavior and fate decisions in development and tissue homeostasis through the mechanosensory machines like the rigidity sensing complex. As noted in the introduction, there are few cases where the mechanoregulatory pathways are even roughly understood; however, malfunctions in those pathways are linked to a wide range of diseases such as cancer and muscular dystrophies.
Direct consequences of force dependent conformation changes of proteins include enhanced phosphorylation of p130Cas, enhanced kinase activity of titin and enhanced binding of vinculin to stretched talin and α‐catenin. In all of these cases, there is evidence of stretch in vivo that is correlated with cell function; although that correlation is weak in some cases. In the future, the combination of in vivo measurements of mechanical tension and conformational dynamics can be combined with in vitro biophysical characterization of these mechanical proteins to give good quantitative understanding of what possible mechanisms can transduce the signal into relevant cellular responses. For example, in the case of talin stretching in vivo, there are rapid changes in length and stretch‐relaxation cycles of 4–20 s are generally observed with vinculin binding during stretch and presumably releasing during relaxation. One possible model for how a mechanosensory signal is developed assumes that vinculin is modified (e.g., phosphorylated) when bound to talin. After many stretch‐relaxation cycles, the concentration of phosphorylated vinculin could reach a threshold to activate growth or other pathways. Such a hypothetical mechanism would be robust to transient stretching of the cell since it is the average number of stretch‐relaxation cycles per unit time that will determine the level of the phosphorylated vinculin (assuming a soluble phosphatase will slowly decrease the level). Now that we know that the mechanical unfolding of important proteins is occurring in vivo, it will be important to link those unfolding events to cellular functions through logical pathways.
In addition, at the cellular level, these mechanosensing pathways can create subpopulations of altered proteins that change cell behavior. The subpopulations of the same protein are usually defined by location and we have studied the FAK‐dependent movement of a LIM domain transcription factor, FHL2, from adhesions to the nucleus.144 Most dramatically, the inhibition of myosin II activity causes FHL2 to leave fibronectin adhesions where it is phosphorylated by mechanically activated FAK and move to the nucleus where it activates p21 transcription that inhibits cell cycle progression. The details of this pathway are not known but we can see from this example that multiple steps in different cell compartments are triggered by the relaxation of tension on adhesions. What complicates our model building is that these force‐sensing proteins like FAK show very fast dynamics (less than 10s) according to FRAP (Table 2) as well as fluorescence localization after photobleaching (FLAP).145 With super‐resolution microscopy, it is also evident that structures like matrix adhesions are actually loose aggregates of much smaller protein clusters that seem to behave independently.146
It is important to emphasize the mechanosensing systems need to be robust to a wide variety of mechanical perturbations including most of those experienced in normal physical activity. In other words the shape of the organs needs to depend upon long‐term forces and not transient events like a football game. Consistent with that, the unfolding of protein domains depends both on the rate of force increase and the level of the force, the dynamics of the pulling is important. When a protein is being stretched with AFM or Magnetic Tweezers, the force required to unfold the same domain varies exponentially with the loading rate.33 Cytoskeletal tensions in tissues are relatively uniform but physical activity of the organism can cause very rapid increases in force. However, static tensions will have a greater effect than sudden changes in tension. Obviously, the integration of the tension over time is the important parameter and the rapid exchange of many adhesion proteins will mean that there can be integration of force‐sensing over time.
The dynamics of the stretching and binding events in mechanosensing proteins underlie the fundamental steps that result in the mechanical signaling processes. We have emphasized the dynamic processes and the need to integrate mechanical signals over time to get reliable responses. At the molecular level, however, individual domain unfolding and refolding events appear to initiate force‐sensing processes. Thus, the surprisingly rapid stretch‐dependent binding and relaxation events in vivo are difficult to mechanistically link with cellular functions because the simultaneous measurement of the critical events in situ has yet to be achieved. Enabling technologies that measure molecular mechanics in situ can shed light on important aspects of molecular sensing mechanisms.
References
- 1. Lim CT, Bershadsky A, Sheetz MP (2010) Mechanobiology. J R Soc Interface 7:S291–S293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Geiger B, Spatz JP, Bershadsky A (2009) Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol 10:21–33. [DOI] [PubMed] [Google Scholar]
- 3. Puchner EM, Alexandrovich A, Kho AL, Hensen U, Schafer LV, Brandmeier B, Grater F, Grubmuller H, Gaub HE, Gautel M (2008) Mechanoenzymatics of titin kinase. Proc Natl Acad Sci USA 105:13385–13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou J, Aponte‐Santamaria C, Sturm S, Bullerjahn JT, Bronowska A, Grater F (2015) Mechanism of focal adhesion kinase mechanosensing. PLoS Comput Biol 11:e1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gautel M (2011) Cytoskeletal protein kinases: titin and its relations in mechanosensing. Pflugers Arch 462:119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu X, Jing C, Xu X, Nakazawa N, Cornish VW, Margadant FM, Sheetz MP (2016) Cooperative vinculin binding to talin mapped by time‐resolved super resolution microscopy. Nano Lett 16:4062–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Margadant F, Chew LL, Hu X, Yu H, Bate N, Zhang X, Sheetz M (2011) Mechanotransduction in vivo by repeated talin stretch‐relaxation events depends upon vinculin. PLoS Biol 9:e1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S (2005) Visualizing the mechanical activation of Src. Nature 434:1040–1045. [DOI] [PubMed] [Google Scholar]
- 9. Rief M, Gautel M, Schemmel A, Gaub HE (1998) The mechanical stability of immunoglobulin and fibronectin III domains in the muscle protein titin measured by atomic force microscopy. Biophys J 75:3008–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. del Rio A, Perez‐Jimenez R, Liu R, Roca‐Cusachs P, Fernandez JM, Sheetz MP (2009) Stretching single talin rod molecules activates vinculin binding. Science 323:638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen H, Fu H, Zhu X, Cong P, Nakamura F, Yan J (2011) Improved high‐force magnetic tweezers for stretching and refolding of proteins and short DNA. Biophys J 100:517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR (2014) Cell adhesion. The minimal cadherin‐catenin complex binds to actin filaments under force. Science 346:1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dembo M, Wang YL (1999) Stresses at the cell‐to‐substrate interface during locomotion of fibroblasts. Biophys J 76:2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Branis J, Pataki C, Sporrer M, Gerum RC, Mainka A, Cermak V, Goldmann WH, Fabry B, Brabek J, Rosel D (2017) The role of focal adhesion anchoring domains of CAS in mechanotransduction. Sci Rep 7:46233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ghassemi S, Meacci G, Liu S, Gondarenko AA, Mathur A, Roca‐Cusachs P, Sheetz MP, Hone J (2012) Cells test substrate rigidity by local contractions on submicrometer pillars. Proc Natl Acad Sci USA 109:5328–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao H, Yu Y, Wu X, Liu S, Liu B, Du J, Li B, Jiang L, Feng X (2017) A role of BK channel in regulation of Ca2+ channel in ventricular myocytes by substrate stiffness. Biophys J 112:1406–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Colombelli J, Besser A, Kress H, Reynaud EG, Girard P, Caussinus E, Haselmann U, Small JV, Schwarz US, Stelzer EH (2009) Mechanosensing in actin stress fibers revealed by a close correlation between force and protein localization. J Cell Sci 122:1665–1679. [DOI] [PubMed] [Google Scholar]
- 18. Hytonen VP, Vogel V (2008) How force might activate talin's vinculin binding sites: SMD reveals a structural mechanism. PLoS Comput Biol 4:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yao M, Goult BT, Klapholz B, Hu X, Toseland CP, Guo Y, Cong P, Sheetz MP, Yan J (2016) The mechanical response of talin. Nat Commun 7:11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klotzsch E, Smith ML, Kubow KE, Muntwyler S, Little WC, Beyeler F, Gourdon D, Nelson BJ, Vogel V (2009) Fibronectin forms the most extensible biological fibers displaying switchable force‐exposed cryptic binding sites. Proc Natl Acad Sci USA 106:18267–18272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M (2010) alpha‐Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol 12:533–542. [DOI] [PubMed] [Google Scholar]
- 22. Yao M, Qiu W, Liu R, Efremov AK, Cong P, Seddiki R, Payre M, Lim CT, Ladoux B, Mege RM, et al. (2014) Force‐dependent conformational switch of alpha‐catenin controls vinculin binding. Nat Commun 5:4525. [DOI] [PubMed] [Google Scholar]
- 23. Chen H, Chandrasekar S, Sheetz MP, Stossel TP, Nakamura F, Yan J (2013) Mechanical perturbation of filamin A immunoglobulin repeats 20–21 reveals potential non‐equilibrium mechanochemical partner binding function. Sci Rep 3:1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP (2011) Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 478:260–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C (2009) Demonstration of catch bonds between an integrin and its ligand. J Cell Biol 185:1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Galic M, Jeong S, Tsai FC, Joubert LM, Wu YI, Hahn KM, Cui Y, Meyer T (2012) External push and internal pull forces recruit curvature‐sensing N‐BAR domain proteins to the plasma membrane. Nat Cell Biol 14:874–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi Z, Baumgart T (2015) Membrane tension and peripheral protein density mediate membrane shape transitions. Nat Commun 6:5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coste B, Xiao B, Santos JS, Syeda R, Grandl J, Spencer KS, Kim SE, Schmidt M, Mathur J, Dubin AE, et al. (2012) Piezo proteins are pore‐forming subunits of mechanically activated channels. Nature 483:176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim HN, Kang DH, Kim MS, Jiao A, Kim DH, Suh KY (2012) Patterning methods for polymers in cell and tissue engineering. Ann Biomed Eng 40:1339–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang B, Lieu ZZ, Wolfenson H, Hameed FM, Bershadsky AD, Sheetz MP (2016) Mechanosensing controlled directly by tyrosine kinases. Nano Lett 16:5951–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wolfenson H, Meacci G, Liu S, Stachowiak MR, Iskratsch T, Ghassemi S, Roca‐Cusachs P, O'Shaughnessy B, Hone J, Sheetz MP (2016) Tropomyosin controls sarcomere‐like contractions for rigidity sensing and suppressing growth on soft matrices. Nat Cell Biol 18:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tee YH, Shemesh T, Thiagarajan V, Hariadi RF, Anderson KL, Page C, Volkmann N, Hanein D, Sivaramakrishnan S, Kozlov MM, et al. (2015) Cellular chirality arising from the self‐organization of the actin cytoskeleton. Nat Cell Biol 17:445–457. [DOI] [PubMed] [Google Scholar]
- 33. Yao M, Chen H, Yan J (2015) Thermodynamics of force‐dependent folding and unfolding of small protein and nucleic acid structures. Integr Biol 7:1154–1160. [DOI] [PubMed] [Google Scholar]
- 34. Bianco P, Martonfalvi Z, Naftz K, Koszegi D, Kellermayer M (2015) Titin domains progressively unfolded by force are homogenously distributed along the molecule. Biophys J 109:340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lahmers S, Wu Y, Call DR, Labeit S, Granzier H (2004) Developmental control of titin isoform expression and passive stiffness in fetal and neonatal myocardium. Circ Res 94:505–513. [DOI] [PubMed] [Google Scholar]
- 36. Anderson BR, Granzier HL (2012) Titin‐based tension in the cardiac sarcomere: molecular origin and physiological adaptations. Prog Biophys Mol Biol 110:204–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nagueh SF, Shah G, Wu Y, Torre‐Amione G, King NM, Lahmers S, Witt CC, Becker K, Labeit S, Granzier HL (2004) Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation 110:155–162. [DOI] [PubMed] [Google Scholar]
- 38. Warren CM, Jordan MC, Roos KP, Krzesinski PR, Greaser ML (2003) Titin isoform expression in normal and hypertensive myocardium. Cardiovasc Res 59:86–94. [DOI] [PubMed] [Google Scholar]
- 39. Hidalgo C, Hudson B, Bogomolovas J, Zhu Y, Anderson B, Greaser M, Labeit S, Granzier H (2009) PKC phosphorylation of titin's PEVK element: a novel and conserved pathway for modulating myocardial stiffness. Circ Res 105:631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kruger M, Kotter S, Grutzner A, Lang P, Andresen C, Redfield MM, Butt E, dos Remedios CG, Linke WA (2009) Protein kinase G modulates human myocardial passive stiffness by phosphorylation of the titin springs. Circ Res 104:87–94. [DOI] [PubMed] [Google Scholar]
- 41. von Castelmur E, Strumpfer J, Franke B, Bogomolovas J, Barbieri S, Qadota H, Konarev PV, Svergun DI, Labeit S, Benian GM, et al. (2012) Identification of an N‐terminal inhibitory extension as the primary mechanosensory regulator of twitchin kinase. Proc Natl Acad Sci USA 109:13608–13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bausch AR, Schwarz US (2013) Cellular mechanosensing: Sharing the force. Nat Mater 12:948–949. [DOI] [PubMed] [Google Scholar]
- 43. Luo T, Mohan K, Iglesias PA, Robinson DN (2013) Molecular mechanisms of cellular mechanosensing. Nat Mater 12:1064–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Humphrey JD, Dufresne ER, Schwartz MA (2014) Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Biol 15:802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kubow KE, Vukmirovic R, Zhe L, Klotzsch E, Smith ML, Gourdon D, Luna S, Vogel V (2015) Mechanical forces regulate the interactions of fibronectin and collagen I in extracellular matrix. Nat Commun 6:8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Freedman BR, Gordon JA, Soslowsky LJ (2014) The Achilles tendon: fundamental properties and mechanisms governing healing. Muscles Ligaments Tendons J 4:245–255. [PMC free article] [PubMed] [Google Scholar]
- 47. Jaalouk DE, Lammerding J (2009) Mechanotransduction gone awry. Nat Rev Mol Cell Biol 10:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Giannone G, Sheetz MP (2006) Substrate rigidity and force define form through tyrosine phosphatase and kinase pathways. Trends Cell Biol 16:213–223. [DOI] [PubMed] [Google Scholar]
- 49. Zaidel‐Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B (2007) Functional atlas of the integrin adhesome. Nat Cell Biol 9:858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zaidel‐Bar R (2013) Cadherin adhesome at a glance. J Cell Sci 126:373–378. [DOI] [PubMed] [Google Scholar]
- 51. Maniotis AJ, Chen CS, Ingber DE (1997) Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA 94:849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martinac B (2004) Mechanosensitive ion channels: molecules of mechanotransduction. J Cell Sci 117:2449–2460. [DOI] [PubMed] [Google Scholar]
- 53. Lavelin I, Wolfenson H, Patla I, Henis YI, Medalia O, Volberg T, Livne A, Kam Z, Geiger B (2013) Differential effect of actomyosin relaxation on the dynamic properties of focal adhesion proteins. PLoS One 8:e73549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Janostiak R, Brabek J, Auernheimer V, Tatarova Z, Lautscham LA, Dey T, Gemperle J, Merkel R, Goldmann WH, Fabry B, et al. (2014) CAS directly interacts with vinculin to control mechanosensing and focal adhesion dynamics. Cell Mol Life Sci 71:727–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM (2010) Myosin II activity regulates vinculin recruitment to focal adhesions through FAK‐mediated paxillin phosphorylation. J Cell Biol 188:877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hoffmann JE, Fermin Y, Stricker RL, Ickstadt K, Zamir E (2014) Symmetric exchange of multi‐protein building blocks between stationary focal adhesions and the cytosol. Elife 3:e02257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lele TP, Thodeti CK, Pendse J, Ingber DE (2008) Investigating complexity of protein‐protein interactions in focal adhesions. Biochem Biophys Res Commun 369:929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Donato DM, Ryzhova LM, Meenderink LM, Kaverina I, Hanks SK (2010) Dynamics and mechanism of p130Cas localization to focal adhesions. J Biol Chem 285:20769–20779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vogel V, Sheetz MP (2009) Cell fate regulation by coupling mechanical cycles to biochemical signaling pathways. Curr Opin Cell Biol 21:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen W, Lou J, Evans EA, Zhu C (2012) Observing force‐regulated conformational changes and ligand dissociation from a single integrin on cells. J Cell Biol 199:497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wolfenson H, Iskratsch T, Sheetz MP (2014) Early events in cell spreading as a model for quantitative analysis of biomechanical events. Biophys J 107:2508–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Changede R, Xu X, Margadant F, Sheetz MP (2015) Nascent integrin adhesions form on all matrix rigidities after integrin activation. Dev Cell 35:614–621. [DOI] [PubMed] [Google Scholar]
- 63. Gauthier NC, Fardin MA, Roca‐Cusachs P, Sheetz MP (2011) Temporary increase in plasma membrane tension coordinates the activation of exocytosis and contraction during cell spreading. Proc Natl Acad Sci USA 108:14467–14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Whittaker CA, Bergeron KF, Whittle J, Brandhorst BP, Burke RD, Hynes RO (2006) The echinoderm adhesome. Dev Biol 300:252–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Na S, Collin O, Chowdhury F, Tay B, Ouyang M, Wang Y, Wang N (2008) Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc Natl Acad Sci USA 105:6626–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Han MK, de Rooij J (2016) Converging and unique mechanisms of mechanotransduction at adhesion sites. Trends Cell Biol. [DOI] [PubMed] [Google Scholar]
- 67. Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM (2010) Nanoscale architecture of integrin‐based cell adhesions. Nature 468:580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shtengel G, Wang Y, Zhang Z, Goh WI, Hess HF, Kanchanawong P (2014) Imaging cellular ultrastructure by PALM, iPALM, and correlative iPALM‐EM. Methods Cell Biol 123:273–294. [DOI] [PubMed] [Google Scholar]
- 69. Aspenstrom P (2010) Formin‐binding proteins: modulators of formin‐dependent actin polymerization. Biochim Biophys Acta 1803:174–182. [DOI] [PubMed] [Google Scholar]
- 70. Smith BA, Gelles J, Goode BL (2014) Single‐molecule studies of actin assembly and disassembly factors. Methods Enzymol 540:95–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rossier OM, Gauthier N, Biais N, Vonnegut W, Fardin MA, Avigan P, Heller ER, Mathur A, Ghassemi S, Koeckert MS, et al. (2010) Force generated by actomyosin contraction builds bridges between adhesive contacts. EMBO J 29:1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Luo W, Lieu ZZ, Manser E, Bershadsky AD, Sheetz MP (2016) Formin DAAM1 organizes actin filaments in the cytoplasmic nodal actin network. PLoS One 11:e0163915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nolz JC, Medeiros RB, Mitchell JS, Zhu P, Freedman BD, Shimizu Y, Billadeau DD (2007) WAVE2 regulates high‐affinity integrin binding by recruiting vinculin and talin to the immunological synapse. Mol Cell Biol 27:5986–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ham H, Guerrier S, Kim J, Schoon RA, Anderson EL, Hamann MJ, Lou Z, Billadeau DD (2013) Dedicator of cytokinesis 8 interacts with talin and Wiskott‐Aldrich syndrome protein to regulate NK cell cytotoxicity. J Immunol 190:3661–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP (2003) Two‐piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424:334–337. [DOI] [PubMed] [Google Scholar]
- 76. Debrand E, El Jai Y, Spence L, Bate N, Praekelt U, Pritchard CA, Monkley SJ, Critchley DR (2009) Talin 2 is a large and complex gene encoding multiple transcripts and protein isoforms. FEBS J 276:1610–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Monkley SJ, Zhou XH, Kinston SJ, Giblett SM, Hemmings L, Priddle H, Brown JE, Pritchard CA, Critchley DR, Fassler R (2000) Disruption of the talin gene arrests mouse development at the gastrulation stage. Dev Dyn 219:560–574. [DOI] [PubMed] [Google Scholar]
- 78. Debrand E, Conti FJ, Bate N, Spence L, Mazzeo D, Pritchard CA, Monkley SJ, Critchley DR (2012) Mice carrying a complete deletion of the talin2 coding sequence are viable and fertile. Biochem Biophys Res Commun 426:190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Qi L, Jafari N, Li X, Chen Z, Li L, Hytonen VP, Goult BT, Zhan CG, Huang C (2016) Talin2‐mediated traction force drives matrix degradation and cell invasion. J Cell Sci 129:3661–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jin JK, Tien PC, Cheng CJ, Song JH, Huang C, Lin SH, Gallick GE (2015) Talin1 phosphorylation activates beta1 integrins: a novel mechanism to promote prostate cancer bone metastasis. Oncogene 34:1811–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang X, Jiang G, Cai Y, Monkley SJ, Critchley DR, Sheetz MP (2008) Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat Cell Biol 10:1062–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Atherton P, Stutchbury B, Wang DY, Jethwa D, Tsang R, Meiler‐Rodriguez E, Wang P, Bate N, Zent R, Barsukov IL, et al. (2015) Vinculin controls talin engagement with the actomyosin machinery. Nat Commun 6:10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Barstead RJ, Waterston RH (1989) The basal component of the nematode dense‐body is vinculin. J Biol Chem 264:10177–10185. [PubMed] [Google Scholar]
- 84. Xu W, Baribault H, Adamson ED (1998) Vinculin knockout results in heart and brain defects during embryonic development. Development 125:327–337. [DOI] [PubMed] [Google Scholar]
- 85. Das M, Subbayya Ithychanda S, Qin J, Plow EF (2014) Mechanisms of talin‐dependent integrin signaling and crosstalk. Biochim Biophys Acta 1838:579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Carisey A, Ballestrem C (2011) Vinculin, an adapter protein in control of cell adhesion signalling. Eur J Cell Biol 90:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chen H, Cohen DM, Choudhury DM, Kioka N, Craig SW (2005) Spatial distribution and functional significance of activated vinculin in living cells. J Cell Biol 169:459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Johnson RP, Craig SW (1994) An intramolecular association between the head and tail domains of vinculin modulates talin binding. J Biol Chem 269:12611–12619. [PubMed] [Google Scholar]
- 89. Goult BT, Xu XP, Gingras AR, Swift M, Patel B, Bate N, Kopp PM, Barsukov IL, Critchley DR, Volkmann N, et al. (2013) Structural studies on full‐length talin1 reveal a compact auto‐inhibited dimer: implications for talin activation. J Struct Biol 184:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Goult BT, Bate N, Anthis NJ, Wegener KL, Gingras AR, Patel B, Barsukov IL, Campbell ID, Roberts GC, Critchley DR (2009) The structure of an interdomain complex that regulates talin activity. J Biol Chem 284:15097–15106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ciobanasu C, Faivre B, Le Clainche C (2014) Actomyosin‐dependent formation of the mechanosensitive talin‐vinculin complex reinforces actin anchoring. Nat Commun 5:3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yao M, Goult BT, Chen H, Cong P, Sheetz MP, Yan J (2014) Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci Rep 4:4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yan J, Yao M, Goult BT, Sheetz MP (2015) Talin dependent mechanosensitivity of cell focal adhesions. Cell Mol Bioeng 8:151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Goult BT, Zacharchenko T, Bate N, Tsang R, Hey F, Gingras AR, Elliott PR, Roberts GC, Ballestrem C, Critchley DR, et al. (2013) RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. J Biol Chem 288:8238–8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Banno A, Goult BT, Lee H, Bate N, Critchley DR, Ginsberg MH (2012) Subcellular localization of talin is regulated by inter‐domain interactions. J Biol Chem 287:13799–13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yang J, Zhu L, Zhang H, Hirbawi J, Fukuda K, Dwivedi P, Liu J, Byzova T, Plow EF, Wu J, et al. (2014) Conformational activation of talin by RIAM triggers integrin‐mediated cell adhesion. Nat Commun 5:5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lee HS, Lim CJ, Puzon‐McLaughlin W, Shattil SJ, Ginsberg MH (2009) RIAM activates integrins by linking talin to ras GTPase membrane‐targeting sequences. J Biol Chem 284:5119–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Klapproth S, Sperandio M, Pinheiro EM, Prunster M, Soehnlein O, Gertler FB, Fassler R, Moser M (2015) Loss of the Rap1 effector RIAM results in leukocyte adhesion deficiency due to impaired beta2 integrin function in mice. Blood 126:2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Su W, Wynne J, Pinheiro EM, Strazza M, Mor A, Montenont E, Berger J, Paul DS, Bergmeier W, Gertler FB, et al. (2015) Rap1 and its effector RIAM are required for lymphocyte trafficking. Blood 126:2695–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hirata H, Tatsumi H, Lim CT, Sokabe M (2014) Force‐dependent vinculin binding to talin in live cells: a crucial step in anchoring the actin cytoskeleton to focal adhesions. Am J Physiol Cell Physiol 306:C607–C620. [DOI] [PubMed] [Google Scholar]
- 101. Golji J, Mofrad MR (2010) A molecular dynamics investigation of vinculin activation. Biophys J 99:1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chen Z, Jing C, Gallagher SS, Sheetz MP, Cornish VW (2012) Second‐generation covalent TMP‐tag for live cell imaging. J Am Chem Soc 134:13692–13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Askari JA, Buckley PA, Mould AP, Humphries MJ (2009) Linking integrin conformation to function. J Cell Sci 122:165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Friedland JC, Lee MH, Boettiger D (2009) Mechanically activated integrin switch controls alpha5beta1 function. Science 323:642–644. [DOI] [PubMed] [Google Scholar]
- 105. Lynch CD, Gauthier NC, Biais N, Lazar AM, Roca‐Cusachs P, Yu CH, Sheetz MP (2011) Filamin depletion blocks endoplasmic spreading and destabilizes force‐bearing adhesions. Mol Biol Cell 22:1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Pentikainen U, Ylanne J (2009) The regulation mechanism for the auto‐inhibition of binding of human filamin A to integrin. J Mol Biol 393:644–657. [DOI] [PubMed] [Google Scholar]
- 107. Wojciak‐Stothard B, Ridley AJ (2003) Shear stress‐induced endothelial cell polarization is mediated by Rho and Rac but not Cdc42 or PI 3‐kinases. J Cell Biol 161:429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Goldyn AM, Rioja BA, Spatz JP, Ballestrem C, Kemkemer R (2009) Force‐induced cell polarisation is linked to RhoA‐driven microtubule‐independent focal‐adhesion sliding. J Cell Sci 122:3644–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Tzima E, Del Pozo MA, Kiosses WB, Mohamed SA, Li S, Chien S, Schwartz MA (2002) Activation of Rac1 by shear stress in endothelial cells mediates both cytoskeletal reorganization and effects on gene expression. EMBO J 21:6791–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Castillo AB, Blundo JT, Chen JC, Lee KL, Yereddi NR, Jang E, Kumar S, Tang WJ, Zarrin S, Kim JB, et al. (2012) Focal adhesion kinase plays a role in osteoblast mechanotransduction in vitro but does not affect load‐induced bone formation in vivo. PLoS One 7:e43291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Puchner EM, Franzen G, Gautel M, Gaub HE (2008) Comparing proteins by their unfolding pattern. Biophys J 95:426–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Greene DN, Garcia T, Sutton RB, Gernert KM, Benian GM, Oberhauser AF (2008) Single‐molecule force spectroscopy reveals a stepwise unfolding of Caenorhabditis elegans giant protein kinase domains. Biophys J 95:1360–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Matsui H, Harada I, Sawada Y (2012) Src, p130Cas, and mechanotransduction in cancer cells. Genes Cancer 3:394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Sawada Y, Hirata H (2016) p130Cas‐mediated regulation of mechanical functions of cells. Clin Calcium 26:1743–1749. [PubMed] [Google Scholar]
- 115. Sawada Y, Tamada M, Dubin‐Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP (2006) Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell 127:1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ladoux B, Nelson WJ, Yan J, Mege RM (2015) The mechanotransduction machinery at work at adherens junctions. Integr Biol 7:1109–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Mege RM, Gavard J, Lambert M (2006) Regulation of cell‐cell junctions by the cytoskeleton. Curr Opin Cell Biol 18:541–548. [DOI] [PubMed] [Google Scholar]
- 118. Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, et al. (2010) Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466:263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Heim R, Cubitt AB, Tsien RY (1995) Improved green fluorescence. Nature 373:663–664. [DOI] [PubMed] [Google Scholar]
- 120. Griffin BA, Adams SR, Tsien RY (1998) Specific covalent labeling of recombinant protein molecules inside live cells. Science 281:269–272. [DOI] [PubMed] [Google Scholar]
- 121. Stagge F, Mitronova GY, Belov VN, Wurm CA, Jakobs S (2013) SNAP‐, CLIP‐ and Halo‐tag labelling of budding yeast cells. PLoS One 8:e78745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, et al. (2008) HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol 3:373–382. [DOI] [PubMed] [Google Scholar]
- 123. Miller LW, Cai Y, Sheetz MP, Cornish VW (2005) In vivo protein labeling with trimethoprim conjugates: a flexible chemical tag. Nat Methods 2:255–257. [DOI] [PubMed] [Google Scholar]
- 124. Hauser CT, Tsien RY (2007) A hexahistidine‐Zn2+‐dye label reveals STIM1 surface exposure. Proc Natl Acad Sci USA 104:3693–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Hoffmann C, Gaietta G, Zurn A, Adams SR, Terrillon S, Ellisman MH, Tsien RY, Lohse MJ (2010) Fluorescent labeling of tetracysteine‐tagged proteins in intact cells. Nat Protoc 5:1666–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Pawley JB (2006) Handbook of biological confocal microscopy. Springer. [Google Scholar]
- 127. Stryer L, Haugland RP (1967) Energy transfer: a spectroscopic ruler. Proc Natl Acad Sci USA 58:719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hell SW, Wichmann J (1994) Breaking the diffraction resolution limit by stimulated emission: stimulated‐emission‐depletion fluorescence microscopy. Opt Lett 19:780–782. [DOI] [PubMed] [Google Scholar]
- 129. Bessel FW (1838) On the parallax of 61 Cygni. Monthly Notices Roy Astron Soc 4:152–161. [Google Scholar]
- 130. De Brabander M, Geerts H, Nuydens R, Nuyens R (1989) Detection of gold probes with video‐enhanced contrast microscopy: nanovid microscopy. Am J Anat 185:282–295. [DOI] [PubMed] [Google Scholar]
- 131. Saxton MJ, Jacobson K (1997) Single‐particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct 26:373–399. [DOI] [PubMed] [Google Scholar]
- 132. Thompson RE, Larson DR, Webb WW (2002) Precise nanometer localization analysis for individual fluorescent probes. Biophys J 82:2775–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Waterman‐Storer CM, Desai A, Bulinski JC, Salmon ED (1998) Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol 8:1227–1230. [DOI] [PubMed] [Google Scholar]
- 134. Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott‐Schwartz J, Hess HF (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642–1645. [DOI] [PubMed] [Google Scholar]
- 135. Klein T, Loschberger A, Proppert S, Wolter S, van de Linde S, Sauer M (2011) Live‐cell dSTORM with SNAP‐tag fusion proteins. Nat Methods 8:7–9. [DOI] [PubMed] [Google Scholar]
- 136. Rust MJ, Bates M, Zhuang X (2006) Sub‐diffraction‐limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 3:793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. de Linde SV (2008) Photoswitching microscopy with standard fluorophores. Appl Phys B. [Google Scholar]
- 138. Siegel AP, Baird MA, Davidson MW, Day RN (2013) Strengths and weaknesses of recently engineered red fluorescent proteins evaluated in live cells using fluorescence correlation spectroscopy. Int J Mol Sci 14:20340–20358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Smith AM, Mancini MC, Nie S (2009) Bioimaging: second window for in vivo imaging. Nat Nanotechnol 4:710–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Dertinger T, Colyer R, Vogel R, Enderlein J, Weiss S (2010) Achieving increased resolution and more pixels with Superresolution Optical Fluctuation Imaging (SOFI). Opt Express 18:18875–18885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Cox S, Rosten E, Monypenny J, Jovanovic‐Talisman T, Burnette DT, Lippincott‐Schwartz J, Jones GE, Heintzmann R (2011) Bayesian localization microscopy reveals nanoscale podosome dynamics. Nat Methods 9:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Holden SJ, Uphoff S, Kapanidis AN (2011) DAOSTORM: an algorithm for high‐ density super‐resolution microscopy. Nat Methods 8:279–280. [DOI] [PubMed] [Google Scholar]
- 143. Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A (2011) Quantitative photo activated localization microscopy: unraveling the effects of photoblinking. PLoS One 6:e22678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Nakazawa N, Sathe AR, Shivashankar GV, Sheetz MP (2016) Matrix mechanics controls FHL2 movement to the nucleus to activate p21 expression. Proc Natl Acad Sci USA 113:E6813–E6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Dunn GA, Dobbie IM, Monypenny J, Holt MR, Zicha D (2002) Fluorescence localization after photobleaching (FLAP): a new method for studying protein dynamics in living cells. J Microsc 205:109–112. [DOI] [PubMed] [Google Scholar]
- 146. Changede R, Sheetz M (2017) Integrin and cadherin clusters: A robust way to organize adhesions for cell mechanics. Bioessays 39:1–12. [DOI] [PubMed] [Google Scholar]


