Abstract
Soluble N‐ethylmaleimide‐sensitive factor attachment protein receptors (SNAREs) are universal molecular engines that drive membrane fusion. Particularly, synaptic SNAREs mediate fast calcium‐triggered fusion of neurotransmitter‐containing vesicles with plasma membranes for synaptic transmission, the basis of all thought and action. During membrane fusion, complementary SNAREs located on two apposed membranes (often called t‐ and v‐SNAREs) join together to assemble into a parallel four‐helix bundle, releasing the energy to overcome the energy barrier for fusion. A long‐standing hypothesis suggests that SNAREs act like a zipper to draw the two membranes into proximity and thereby force them to fuse. However, a quantitative test of this SNARE zippering hypothesis was hindered by difficulties to determine the energetics and kinetics of SNARE assembly and to identify the relevant folding intermediates. Here, we first review different approaches that have been applied to study SNARE assembly and then focus on high‐resolution optical tweezers. We summarize the folding energies, kinetics, and pathways of both wild‐type and mutant SNARE complexes derived from this new approach. These results show that synaptic SNAREs assemble in four distinct stages with different functions: slow N‐terminal domain association initiates SNARE assembly; a middle domain suspends and controls SNARE assembly; and rapid sequential zippering of the C‐terminal domain and the linker domain directly drive membrane fusion. In addition, the kinetics and pathway of the stagewise assembly are shared by other SNARE complexes. These measurements prove the SNARE zippering hypothesis and suggest new mechanisms for SNARE assembly regulated by other proteins.
Keywords: SNARE assembly, membrane fusion, synaptic exocytosis, optical tweezers, protein folding, energy landscape
Introduction
Soluble N‐ethylmaleimide‐sensitive factor attachment protein receptors (SNAREs) are molecular engines for membrane fusion.1 Similar to molecular motors, SNAREs generate force to perform their biological function. Specifically, the force generated by SNAREs draws two lipid bilayers into close proximity to counteract electrostatic repulsion between the membranes and to dehydrate and deform the bilayers required for membrane fusion.2 The role of force in membrane fusion cannot be overemphasized,1 because this force generation is probably the single most important mission of SNAREs for membrane fusion per se. Supporting this concept, efficient membrane fusion can also be driven by the force produced by DNA hybridization or dimerization of engineered coiled coils.3, 4, 5, 6
However, SNAREs are special in several aspects. First, SNAREs do not contain any ATPase domain and their power strokes are nucleotide‐independent. Instead they produce force by coupled folding and assembly of a pair of cognate SNARE proteins or protein complexes: one on the vesicle membrane (v‐SNARE) and the other typically on the target membrane (t‐SNARE).7 The specific t‐ and v‐SNARE paring also contributes to the specificity of membrane fusion.8 Each eukaryote has 20–50 SNAREs (with 36 in human cells) that mediate membrane fusion involved in various pathways of intracellular transport and trafficking.9 All SNAREs share characteristic SNARE motifs of ∼60 amino acids in ∼16 heptad repeats [Fig. 1(A)].10 Individual SNARE motifs are disordered in solution, but four complementary SNARE motifs fold and assemble into a structurally conserved four‐helix bundle [Fig. 1(B)].11, 12 In the core of the bundle are 15 layers of hydrophobic amino acids and one central ionic layer containing three glutamine residues and one arginine residue (numbered from −7 to +8) [Fig. 1(A)]. After membrane fusion, the fully assembled SNARE complex is disassembled by the AAA+ ATPase NSF (N‐ethylmaleimide‐sensitive factor) with the assistance of SNAPs (Soluble NSF‐attachment proteins) in an ATP‐dependent manner.13, 14, 15, 16 Disassembly of each SNARE complex costs around 10 ATP molecules.17 Therefore, SNARE engines work like battery‐powered motors: they are charged upon disassembly and discharged upon assembly.
Figure 1.
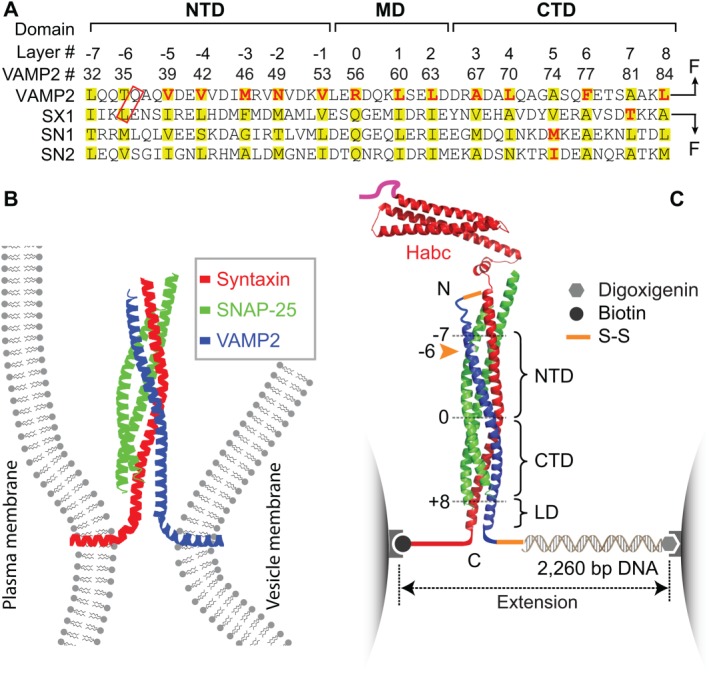
SNARE domain structures, membrane fusion, and the experimental setup to study SNARE assembly. (A) Amino acid sequences and their domain structures of the SNARE motifs in synaptic syntaxin 1 (SX1), VAMP2, and SNAP‐25B (SN1 and SN2). The amino acids in the hydrophobic layers or the central ionic layer are highlighted in yellow. The amino acids colored red are mutated to test their effects on SNARE assembly. Syntaxin and VAMP2 are cross‐linked at their N‐termini, with one of the cross‐linking sites marked by a red rectangle, and pulled from their C‐termini. The extremely fast middle domain (MD) transition and its associated state 3 can only be resolved in some experiments and is often mixed with the CTD transition, as is shown in C. (B) SNARE proteins couple their folding and assembly to draw two membrane into proximity and force them to fuse. An assembled synaptic SNARE complex is depicted to bridge the plasma membrane and the vesicle membrane. (C) Experimental setup to pull a single cytoplasmic domain of the synaptic SNARE complex using optical tweezers (OTs). The assembled SNARE complex consists of an N‐terminal Habc domain in a antiparallel three‐helix bundle, the core four‐helix bundle domain, and a linker domain (LD) in a two‐stranded coiled coil. The four‐helix bundle contains an N‐terminal domain (NTD) and a C‐terminal domain (CTD) that are separated by a central ionic layer (“0” layer). Adapted from Ma et al.53
The synaptic SNAREs have long been a model system to study SNARE assembly and membrane fusion. The synaptic SNARE complex is also the first SNARE complex that was identified.7 The SNAREs consist of t‐SNAREs syntaxin and SNAP‐25 (synaptosome‐associated protein 25) and the v‐SNARE VAMP2 (vesicle‐associated membrane protein 2, also called synaptobrevin), in which SNAP‐25 contains two SNARE motifs (SN1 and SN2) that are connected by a long disordered loop [Fig. 1(A)]. Synaptic SNAREs mediate exocytosis of neurotransmitter‐containing vesicles at synaptic or neuromuscular junctions.18 The exocytosis occurs in several stages.19, 20, 21, 22, 23, 24 First, vesicles are docked onto presynaptic membranes with the assistance of SNARE binding proteins, such as synaptotagmin and Munc18‐1 (mammalian uncoordinated‐18).25, 26 Then, the complementary t‐ and v‐SNAREs associate to form trans‐SNARE complexes that bridge vesicle and plasma membranes. Likely partially assembled, these trans‐SNARE complexes prime the vesicle for calcium‐triggered fusion.27 Finally, the calcium influx induced by the arrival of an action potential triggers synchronous fusion of the primed vesicles, typically within 200 μs in mammalian brains.28, 29 The fusion releases the neurotransmitters into the synaptic or neuromuscular junctions, which bind to receptors on the postsynaptic neurons or muscle cells to activate them.
The energetics and kinetics of SNARE assembly are keys to understanding the working mechanism of SNARE engines and their biological function. The concept of SNARE engine was inspired by the observation that the synaptic SNARE complex is a parallel helix bundle.11, 30 The observation also suggested a zippering mechanism for SNARE‐mediated fusion because SNARE assembly brings the two membranes into proximity, a prerequisite for fusion.31 Subsequently, SNAREs alone were shown to induce fusion between liposomes with reconstituted t‐ and v‐SNAREs, corroborating that SNAREs are the minimal machine for membrane fusion.32 However, the reconstituted fusion is orders of magnitude slower than synaptic vesicle fusion. The slow fusion seems to correlate with the slow SNARE assembly in vitro, which generally takes from tens of minutes to hours to complete.33, 34, 35 Accordingly, Li et al. have recently measured a small t‐ and v‐SNARE association rate constant of ∼7,300 M−1 s−1 in solution, indicating a large energy barrier for SNARE zippering.34 They have also determined the rate constants of the t‐SNARE complex binding to the Vn peptide (∼530 M−1 s−1) and the Vc peptide (∼6,000 M−1 s−1) corresponding to N‐ and C‐terminal halves of the v‐SNARE, respectively. A comparison of the three rate constants suggests that the association between isolated t‐ and v‐SNAREs is initiated at the C‐terminal half, which is opposite to the N‐to‐C SNARE zippering direction during membrane fusion. The initial association rate constant for membrane‐anchored t‐ and v‐SNAREs should be much smaller than the rate constant for binding between the t‐SNARE complex and the Vn peptide, due to the repulsive force between the apposed membranes. In conclusion, de novo SNARE assembly is extremely slow. In addition, although the SNARE complex has a high melting temperature (>80°C), SNAREs only assemble at a much lower temperature.12 Isothermal titration calorimetry (ITC) measurements reveal an association energy of ∼18 k B T (11 kcal mol−1) between t‐ and v‐SNAREs.34, 36 This energy is much less than the energy barrier of membrane fusion, which is estimated to be >40 k B T.2 Therefore, the energy and rate of SNARE assembly measured from those experiments were inconsistent with the SNARE zippering hypothesis.
Another critical question in SNARE assembly concerns intermediates of SNARE assembly. Jahn et al. considered that SNARE assembly occurs downhill in a steep energy gradient, and thus cannot be suspended midway.37 In this scenario, SNARE assembly would proceed without any intermediates in an all‐or‐none fashion. In contrast, the multiple‐phase kinetics of exocytosis and results of SNARE cleavage by neurotoxins suggest the existence of partially assembled trans‐SNARE complexes prior to membrane fusion.19, 20, 21, 22 Yet, the conformations of these intermediate states and their relation to the different stages of membrane fusion remain unclear.28
In this article, we will focus on the single‐molecule manipulation of SNARE proteins based on optical tweezers (OTs) and the results derived from this approach, with a brief survey of other experimental approaches applied to study SNARE assembly. We will first describe the stagewise SNARE assembly and its associated energies and kinetics. We will then show how SNARE assembly is altered by mutations. Comparing their effects on exocytosis, we will gain insights into distinct functions of different SNARE folding domains in membrane fusion. Finally, we will summarize our improved understanding on t‐SNARE structure and dynamics.
Challenges to Measure the Energy and Kinetics of SNARE Assembly
A variety of biophysical approaches have been applied to investigate SNARE assembly both in solution and on membranes, which greatly contribute to our current understanding of SNARE engines. These include spectroscopic methods based on circular dichroism and fluorescence detection,12, 23, 34, 35, 38, 39, 40 ITC,34, 36 atomic force microscopy (AFM),41, 42, 43, 44, 45 and the surface forces apparatus (SFA).46, 47 The spectroscopic methods consistently revealed a slow t‐ and v‐SNARE association. The reported bimolecular association rate constants vary between 6 × 103 M−1 s−1 and 6 × 105 M−1 s−1.34, 38 Notably, no intermediate states were detected for association between isolated t‐ and v‐SNAREs in solution. The spectroscopic methods and other methods mentioned above have defined the energy of SNARE assembly in the range of 17–35 k B T, which significantly underestimate the stability of synaptic SNARE complexes, as we will discuss later. We have compared AFM, SFA, and OTs in the studies of SNARE assembly elsewhere.48 Here we will discuss more on ITC, which has been widely used to study the folding energy of the SNARE complex, as well as other receptor–ligand interactions. By titrating SNAP‐25 into the syntaxin and VAMP2 mixture, Fasshauer et al. measured an extraordinarily high enthalpy release of 110 kcal/mol (or 185 k B T) upon assembly of the three SNARE proteins, but derived low SNARE folding free energy of ∼18 k B T or a dissociation constant (K d) of 23 nM between t‐ and v‐SNAREs.36 Similarly, by titrating VAMP2 into preformed t‐SNARE complexes, Li et al. again derived a high enthalpy of 40 kcal/mol (67 k B T) for t‐ and v‐SNARE association and an apparent binding free energy of 17 k B T or K d = 132 nM.34 However, as was noted by some of these authors, SNARE assembly was not completely reversible in the titration experiments and the actual folding free energy of the SNARE complex is expected to be much greater than those derived from ITC measurements.36 In addition, we noticed that the experimental condition used in these ITC experiments could only reveal an upper limit of K d ≈ 10 nM for the binding affinity between t‐ and v‐SNAREs. In typical ITC experiments, the heat release per mole of ligand injected into the receptor solution (Q) is measured at different ratios of ligand to receptor concentrations (X). The receptor–ligand binding affinity is obtained by fitting the ITC data with the Wiseman isotherm formula.49 The fitting is accurate when the Wiseman parameter, the ratio of the receptor concentration to K d, is below ∼500. However, when the parameter exceeds 500, the Q–X data approach a step function and the fitting is no longer accurate. In the ITC experiments to study SNARE assembly energy, the concentration of the syntaxin and VAMP2 mixture or the t‐SNARE complex is above micromolar, restricting accurate affinity measurements above 10 nM. Consistent with this prediction, the measured Q–X data are indeed close to a step function,34, 36 indicating that the derived energy for SNARE assembly only represents a lower bound. This conclusion is corroborated by the observations that all ITC measurements of SNARE affinity saturate in the nanomolar range, regardless of SNARE mutations and peptide binding, despite their effects on attenuating SNARE assembly.21, 34, 36 In conclusion, the current ITC measurements underestimate the stability of the SNARE complex and exhibit reduced sensitivity to mutations.
Studying SNARE assembly is challenging using traditional experimental approaches based on an ensemble of SNARE proteins. First, assembly of isolated SNARE complexes is irreversible, which makes it impossible to directly measure the energy of SNARE assembly. Second, synaptic SNAREs readily misfold, which complicates studies of functional SNARE assembly and may contribute to the large range of association rates mentioned above. The cognate SNARE motifs can form various antiparallel conformations in vitro.39, 50, 51 In addition, the t‐SNAREs readily form a stable 2:1 t‐SNARE complex with an additional syntaxin molecule.38 Because this syntaxin molecule occupies the binding site for VAMP2, the 2:1 t‐SNARE complex cannot support SNARE zippering. Third, intermediates of SNARE assembly are transient38, 39, 51 and difficult to synchronize in specific d states for traditional kinetics studies. Fourth, the isolated SNAREs show an assembly pathway different from the membrane‐anchored SNAREs, as is discussed above. Trans‐SNARE assembly favors an N‐to‐C assembly pathway due to membrane repulsion and the topological orientation of SNAREs, whereas such constrains are absent in assembly of isolated SNAREs in solution. As a result, SNARE assembly in solution tends to follow a C‐to‐N direction.34 Finally, functional SNARE assembly occurs in the presence of a force load imposed by the apposed membranes, which dramatically alters the energetics and kinetics of SNARE assembly and leads to force‐dependent intermediates.48 These challenges call for new experimental approaches to investigate SNARE assembly. We have established a single‐molecule manipulation method based on high‐resolution OTs to study SNARE assembly, which overcomes the above‐mentioned difficulties and allow us to directly measure the folding intermediates, energies, and kinetics of single SNARE complexes.48, 52, 53, 54, 55, 56
Manipulation of Single SNARE Complexes by OTs
OTs use tightly focused laser beams to trap polystyrene or silica beads (typically 0.5–3 µm in diameter) in a harmonic potential.57, 58 OTs utilize the beads as force and displacement sensors while applying tiny, precisely known forces (0.02–250 pN) to a single molecule attached to the beads [Fig. 1(C)]. An optical interference method detects the bead position with angstrom resolution.59, 60, 61, 62 As a result, OTs measure the extension response of a single molecule to the applied mechanical force with a spatiotemporal resolution of ∼0.3 nm and ∼20 µs.58, 60, 61, 62, 63 The extension and force are used to derive the conformation and energy of the SNARE complex in different folding states in real time. Specifically, for a reversible two‐state transition, the folding energy of the associated protein or protein domain ( ) can be measured from the mechanical work required to unfold the domain ( ), that is, where is the equilibrium force with half unfolding probability of the associated transition, is the corresponding extension change, and is the energy required to stretch the unfolded polypeptide chain to the force .58, 64, 65 This equation shows that the mechanical work is used to unfold the protein and to further pull the unfolded polypeptide to the equilibrium force. For transitions among more than two states under thermodynamic equilibrium, the equilibrium force is not well defined. However, the folding energy of each state at zero force can be similarly derived based on the Boltzmann distribution of state populations at different constant forces.66, 67
Figure 1(C) shows our basic experimental setup and SNARE construct employed to study synaptic SNARE assembly.48, 53, 56 The cytoplasmic SNARE construct contains either full‐length SNAREs [Fig. 1(C)] or truncated SNAREs involved in complex formation. The complex was pulled from the C‐termini of syntaxin and VAMP2 via a long DNA handle. The DNA handle serves as a spacer to facilitate SNARE attachment to the beads and force measurement.68, 69 To help SNARE reassembly and ensure its correct assembly pathway, we cross‐linked syntaxin and VAMP2 at a site toward the N‐terminus of their SNARE motifs. Once a single SNARE complex was tethered between two beads, we used dual‐trap high‐resolution OTs to pull the SNARE complex.
We manipulate a single SNARE complex by either pulling or relaxing the complex or by holding the complex under a constant mean force at a fixed trap separation.70 The force change is controlled by moving one optical trap relative to the other at 10 nm s−1. Both force and extension of the protein–DNA tether are recorded at 10 kHz. We present the data in two formats: plots of force–extension curves (FECs) show global phase diagrams of protein transitions induced by force and extension–time trajectories exhibit detailed transition kinetics of a protein or a protein domain at specific force.48, 58 In these plots, discrete extension increases or decreases represent cooperative protein unfolding or refolding, with sizes of the extension changes roughly proportional to the numbers of amino acids involved in the transitions. In contrast, continuous extension changes in FECs are caused by stretching the DNA and the unfolded polypeptide while the SNARE complex remains in one folding state, which could be fit by the Marko–Siggia formula.71, 72
Stepwise SNARE Assembly and Disassembly
The first two sets of FECs (#1 and #2) shown in Figure 2(A) are obtained by first pulling and then relaxing a single SNARE complex for two consecutive rounds.48 The FECs for the pulling phase show fast extension flickering in two force ranges, one at 10–13 pN (marked by dashed oval) and the other at 15–18 pN (solid oval), followed by an irreversible jump (green arrow). Based on the linear structure of the ternary SNARE complex, these extension changes represent rapid and reversible folding and unfolding transitions in the linker domain (LD) and the C‐terminal domain (CTD) and irreversible unfolding of the N‐terminal domain (NTD) [Fig. (2B)]. When the SNARE complex is relaxed upon NTD unfolding [Fig. (2A), gray curve in FEC #1], the complex takes more than 5 s to reassemble at a low force (red arrow), leading to pronounced hysteresis in SNARE assembly and disassembly. Thus, NTD assembles slowly with a large energy barrier. When the complex is further pulled to a higher force upon NTD unfolding, another unfolding event is found (FEC #2, cyan arrow), corresponding to t‐SNARE unfolding and the accompanying SNAP‐25 dissociation. As a result, the syntaxin‐VAMP2 conjugate fails to reassemble at a low force. However, adding 0.2 µM SNAP‐25 in the solution rescues t‐SNARE folding (FEC#3, blue arrow) and SNARE assembly (red arrow). Therefore, these FECs reveal five distinct states with two intermediates for SNARE zippering [states 2 and 4 in Fig. 2(B)]. Here we have referred to SNARE zippering as SNARE assembly between the partially folded t‐SNARE complex [state 5 in Fig. 2(B) and state i in Fig. 8] and the v‐SNARE. The most important intermediate has a half‐zippered SNARE structure (state 4). Detailed analyses show that folding of its CTD and LD release free energy of ∼36 and ∼8 k B T, respectively, with no or a small energy barrier. Thus, the half‐zippered state folds rapidly and forcefully. The half‐zippered state is also detected by Min et al. using magnetic tweezers.73 However, the CTD unfolds at ∼34 pN and refolds below 11 pN. The difference is caused by a different pulling site used in this work: the SNARE complex is pulled from an artificial site at the junction between CTD and LD, instead of the C‐termini of LD used in our experiment. As a result, a large energy barrier separates the folded and the unfolded CTD states, leading to a large hysteresis of the CTD transitions in the FEC. Finally, the half‐zippered state is supported by many other experiments,35, 40, 74 especially imaging by electron microscopy.75, 76 Combining with earlier evidence,19, 20, 21, 22, 77 these observations corroborate that a half‐zippered trans‐SNARE structure is required for the calcium‐triggered synaptic vesicle fusion. In contrast, de novo SNARE zippering without preassembled NTD is too slow to support rapid synaptic vesicle fusion.78
Figure 2.
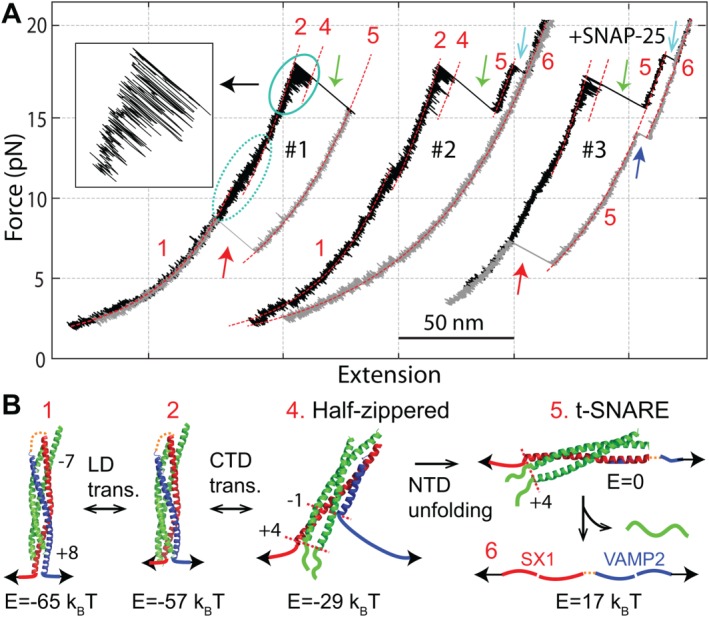
The SNARE complex zippers stepwise. (A) Force‐extension curves (FECs) obtained by first pulling and then relaxing single SNARE complexes in the absence (#1 and #2) and presence (#3) of SNAP‐25 in the solution. Regions of different states (red numbers, see B) are fit by the Marko‐Siggia formula (dashed red lines). Some transitions are marked: the reversible LD transition by a dashed oval, the reversible CTD transition by a solid oval, NTD unfolding by green arrows, t‐SNARE unfolding by cyan arrows, t‐SNARE refolding by the blue arrow, and full SNARE reassembly by red arrows. The inset shows the close‐up view of the CTD transition. (B) Derived SNARE assembly and disassembly states and pathway. The free energies of different states relative to the unzipped state (E) are indicated. Adapted from Gao et al. and Ma et al.48, 53
Figure 8.
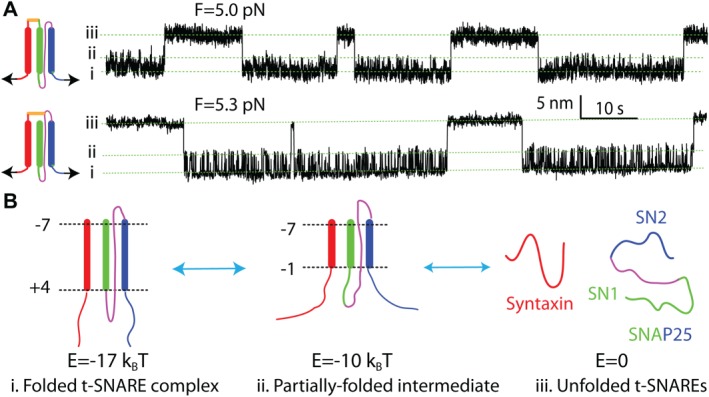
The synaptic t‐SNARE complex folds synchronously via a partially folded intermediate. (A) Extension‐time trajectories obtained by pulling the N‐terminal cross‐linked t‐SNARE complex from different C‐terminal sites (inset on the left). (B) Schematic diagrams of three t‐SNARE folding states (i–iii) and energies (E). Adapted from Zhang et al.55
Energetics, Kinetics, and Pathway of SNARE Assembly
To measure NTD assembly energy, we need to make the NTD transition reversible. To this end, we gradually shift the N‐terminal cross‐linking site toward the SNARE motif. NTD transition becomes reversible only when the cross‐linking site is shifted to the −6 layer [Fig. (1C), yellow arrow].53 This observation suggests that the amino acids N‐terminal to or near the −6 layer significantly limit the rate of NTD assembly. The NTD transition occurs at a slightly higher force range than the CTD transition (Fig. 3, WT). Consequently, both transitions are simultaneously detected in a relatively large force range. The CTD–NTD transitions can be detected at high spatiotemporal resolution at different constant mean forces [Figs. 4(A) and 5]. Interestingly, a new state 3 appears in the CTD transition, which splits the original CTD into a new CTD and a middle domain (MD) that folds and unfolds at submillisecond time scale. Four‐state hidden‐Markov modeling (HMM) fits the extension trajectories well,56, 66, 67 revealing sequential folding and unfolding transitions among the four states 2–5 [Fig. (4A)].53 The HMM also yields state populations, extensions, lifetimes, and transition rates [Fig. (4B)]. Based on its extension, the state 3 has a structure similar to the state 4, but zippers to the +2 layer. Model fitting to the force‐dependent populations and rates yields the energy landscape of SNARE zippering [Fig. (4C)]. In particular, the folding energies of the three domains are derived: 25 (±2, S.D.) k B T for NTD, 13 (±1) k B T for MD, and 22 (±3) k B T for CTD. Thus, the total zippering energy of a single SNARE complex is 68 (±4) k B T. This energy is significantly greater than any previous measurements34, 36, 46 and, to our knowledge, is one of the highest protein folding energies reported. For comparison, we have measured the folding energy of one of the strongest coiled coils that melts above 100°C.79, 80 The two‐stranded coiled coil has an unfolding equilibrium force of 12.4 pN and a folding energy of 24 (±1) k B T.79 Note that the axial length of the coiled coil (33 a.a.) is about half of the size of the SNARE complex. In addition, the duplex DNA of the same length as the SNARE complex (∼12 nm) has an equilibrium unfolding force of 14.5 pN and folding energy of ∼106 k B T.81 Thus, SNARE assembly releases a large amount of energy commensurate with the requirement for membrane fusion. This result is consistent with the observation that a single SNARE complex is sufficient for membrane fusion.82, 83
Figure 3.
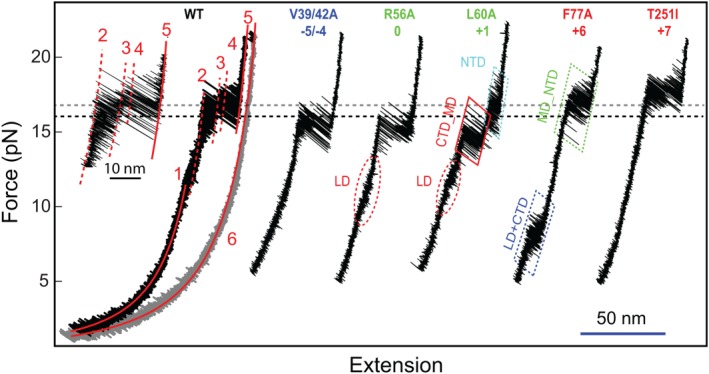
FECs of wild‐type (WT) and mutant SNARE complexes showing effects of layer mutations on SNARE assembly. The combined or mixed domain transitions are indicated by their domain names connected by “_” and “+,” respectively (also see legend for Figure 5). Adapted from Ma et al.53
Figure 4.
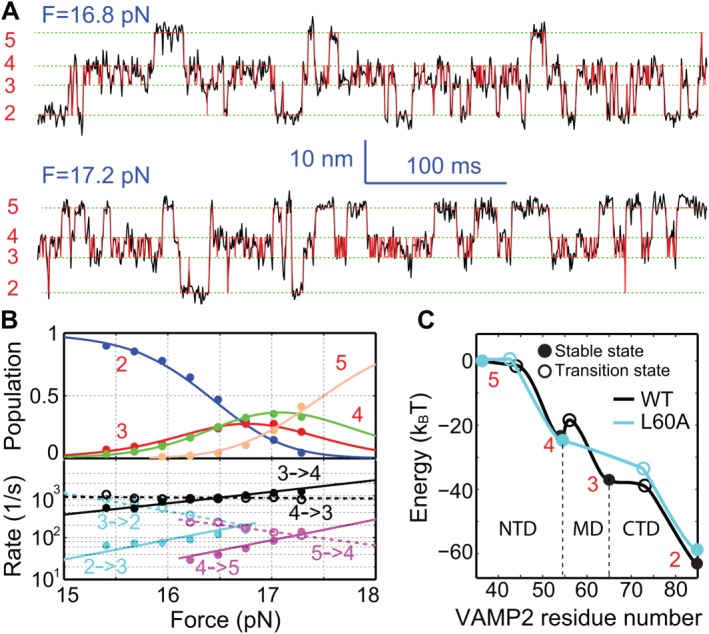
Force‐dependent reversible folding and unfolding of the WT SNARE complex among four states. (A) Extension‐time trajectories (black) of a single SNARE complex at two indicated mean forces (F). The red trajectories are idealized state transitions derived from hidden‐Markov modeling (HMM). (B) Force‐dependent state populations (top, symbols) and transition rates (bottom) and their best model fits (solid or dashed curves). (C) Simplified energy landscapes66 of the wild‐type (WT) and mutant (L60A) SNARE complexes. The reaction coordinate is the number of the amino acid in VAMP2 located at the C‐terminal border of the zippered region that starts from the amino acid number 36 at the N‐terminal cross‐linking site [Fig. 1(A)]. From Ma et al.53
Figure 5.
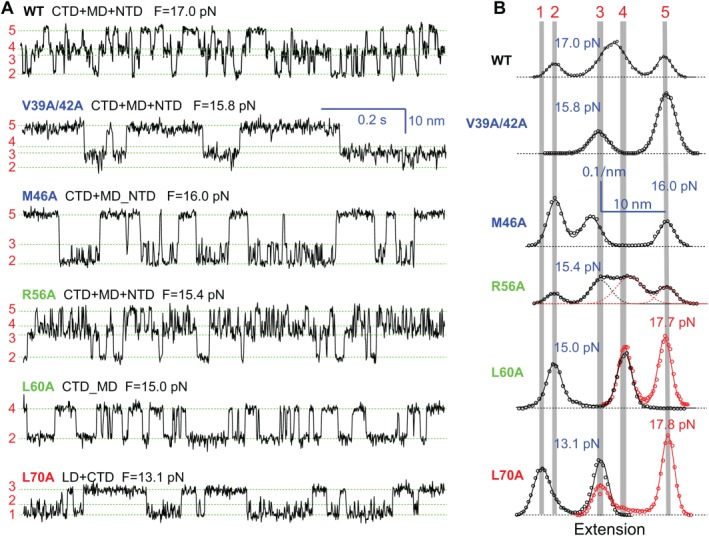
Position‐dependent effects of layer mutations on SNARE assembly. (A) Extension‐time trajectories of the WT and mutant SNARE complexes at the indicated constant mean forces (F). Domains that are involved in sequential folding and unfolding transitions are indicated by the domain names connected by “+” signs. However, some mutations cause two neighboring domains to cooperatively fold and unfold as single extended domains, which are indicated by two individual domains connected by “_” signs. (B) Probability‐density distributions of the extensions shown in A (symbols) and their best‐fits by sums of two to four Gaussian functions (curves). The red symbols and curves for L60A and L70A show the distributions of the extensions for NTD transitions (not shown) at high forces. From Ma et al.53
The NTD zippering energy measured by us depends on the cross‐linking at the −6 layer. On one hand, the cross‐linking may prevent the amino acids at or N‐terminal to the −6 layer from unzipping [Fig. (1A)], which approximately underestimates the NTD zippering energy by 14%. On the other hand, the cross‐linking tethers the t‐ and v‐SNAREs together, leading to a high effective concentration of the v‐SNARE around the t‐SNARE complex. The concentration is estimated to be >1 molar, leading to an overestimation of the bimolecular NTD association energy.55 Therefore, the measured NTD zippering energy may not significantly deviate from the actual bimolecular NTD association energy.81, 84 It will be interesting to test more cross‐linking sites around the −7 layer for potential reversible NTD transition and to confirm the prediction. It must be noted that the N‐terminal cross‐linking does not affect the folding of MD, CTD, and LD.53
Position‐Dependent Effects of Point Mutations and Distinct Functions of Different Domains
Functional studies of SNAREs make use of numerous SNARE mutations.18, 21 However, data interpretation has been hindered by lack of clear understanding on the effects of these mutations on SNARE assembly. To correlate the energetics and kinetics of SNARE assembly to its biological function, we have examined 16 SNARE mutations in all layers except −7, mainly single alanine substitutions in VAMP2.53 The FECs and extension–time trajectories of representative mutations are shown in Figures 3 and 5, respectively. We have found that effects of these mutations are strongly position‐dependent and may alter the energetics, kinetics, or pathway of SNARE assembly, indicating distinct functions of different SNARE folding domains (Fig. 6). Note that the folding energy and the folding pathway of the SNARE complex are correlated. The four folding domains of the wild‐type SNARE complex are partly maintained by their distinct mechanical stabilities. Most mutations specifically destabilize the domains in which the mutations are located, thus altering their mechanical stabilities relative to other domains. Consequently, two domains may merge into a single domain to cooperatively fold and unfold, changing the folding pathway.
Figure 6.
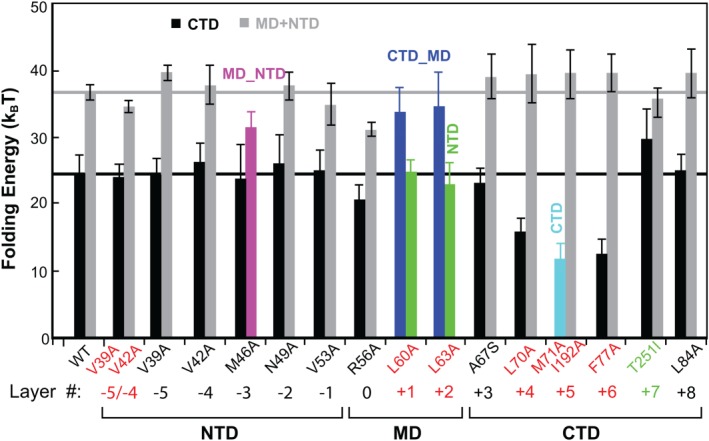
Effects of layer mutations on SNARE folding energy. Gray bars indicate the sum of the MD and NTD folding energies. Colored bars indicate energies of the combined domains denoted. All mutations are layer mutations in VAMP2, except the double mutations M71A and I192A in SNAP‐25 and T251I in syntaxin. Mutations highlighted in red correspond to impaired exocytosis. Reproduced from Ma et al.53
The NTD is generally insensitive to single alanine substitutions and is responsible for vesicle priming. Except M46A (−3 layer), all single alanine substitutions in NTD barely change SNARE assembly, indicating robust NTD folding (Fig. 6). Correspondingly, many of these mutations do not significantly affect exocytosis.21, 22 However, double mutations V39A/V42A decrease the rate and stability of NTD assembly [Figs. 3, 5(A), and 6)], which slows down vesicle priming and reduces the number of primed vesicles and the amplitude of calcium‐triggered neurotransmitter release.21 The comparison corroborates that the NTD is responsible for vesicle priming. M46A is special, because it destabilizes NTD, leading to comparable mechanical stabilities for NTD and MD. As a result, the two domains merge into an extended domain (designated as MD_NTD) to cooperatively fold and unfold (Fig. 5). The effect of this mutation on exocytosis remains to be tested. However, the −3 layer appears to play an important role in stabilizing NTD. A mutation in the same layer in the human SNARE protein membrin causes epilepsy.85
The CTD is sensitive to layer mutations and directly drives membrane fusion. CTD mutations generally reduce CTD stability (Fig. 6). In particular, L70A and F77A in VAMP2 and M71A/I192A in SNAP‐25 dramatically destabilize the CTD. Consequently, these mutations abolish exocytosis.21, 86 This comparison demonstrates that CTD catalyzes membrane fusion by lowering the energy barrier for membrane fusion. In contrast, L84A (+8) alters neither SNARE assembly nor exocytosis.21 Among all mutations tested, syntaxin T251I is the only mutation that enhances SNARE assembly. Electrophysiological measurements show that this mutation in flies enhances synaptic exocytosis.87 Therefore, CTD assembly energy dictates the speed of exocytosis, corroborating that the CTD directly drives membrane fusion.
The three layer mutations in the MD change the reaction pathway of SNARE assembly or globally reduce the stability of the SNARE complex, thereby impairing membrane fusion.88, 89 L60A and L63A unite CTD and MD into a single domain (MD_CTD), leading to an extension change equal to the sum of the extension changes for individual CTD and MD transitions [Figs. 4(C) and 5)]. In contrast, the ionic layer mutation R56A clearly shows MD as an independent domain, as is seen from both the extension–time trajectory and the corresponding histogram distribution (Fig. 5). However, R56A is unique among all mutations tested because it globally reduces the folding energies of CTD, MD, and NTD by 3.2, 0.8, and 4.2 k B T, respectively, compared to the WT.53, 67 This observation suggests that R56A induces a subtle long‐range conformational change that spreads to both NTD and CTD, an unusual property likely shared by all the hydrophilic amino acids in the ionic layer.55 More data demonstrate that MD is important for the accuracy of SNARE assembly, as MD mutations cause heterogeneity in SNARE transition kinetics.53
The stepwise SNARE zippering continues to the LD and the transmembrane domain (TMD), contributing to additional SNARE zippering energy.90 Yet, the functions of LD and TMD zippering remain a matter of debate.91, 92, 93, 94 Highly positively charged, LD interacts strongly with membranes and may constitute a membrane anchor with the TMD.95, 96, 97 In addition, TMD may destabilize membranes or help form fusion pores during membrane fusion.98, 99, 100, 101, 102 In conclusion, different SNARE folding domains play distinct functions in SNARE zippering and membrane fusion.
Conserved SNARE Assembly Pathway and Kinetics
The partially assembled intermediates are crucial for the fast calcium‐triggered synaptic SNARE assembly and vesicle fusion. Do they exist in folding of other SNARE complexes, especially those mediate slower membrane fusion? To address this question, we have tested three SNARE complexes responsible for translocation of glucose transporter type 4 (designated as GLUT4) in adipocytes or muscle cells, constitutive fusion of endocytic vesicles to early endosome in mammals (Endosome), and fusion of post‐Golgi vesicles to plasma membranes in yeast (Yeast).52 We have discovered that these SNARE complexes share the same assembly pathway and kinetics as the neuronal SNARE complex (Neuron), that is, slow NTD association and fast CTD zippering (Fig. 7). This demonstrates that the SNARE assembly pathway and kinetics are highly conserved,103 likely caused by the highly conserved primary sequences of SNARE motifs and structures of assembled SNARE complexes. Thus, the half‐zippered intermediate in SNARE assembly likely plays an essential role in SNARE assembly and membrane fusion, not specific to synaptic SNAREs for fast fusion. We speculate that the half‐zippered intermediate is required for accurate and regulated SNARE assembly and may serve as a substrate for Sec/Munc18 (SM) proteins that are essential for SNARE‐mediated membrane fusion.104, 105 Consistent with this view, we found that Munc18‐1 stabilizes the synaptic SNARE complex in a half‐zippered state.53 However, the CTD folding energies significantly differ among these SNARE complexes, as is indicated by their different equilibrium forces (Fig. 7). Correspondingly, the GLUT4, endosomal, and yeast SNARE complexes have CTD folding energies of 23, 16, and 13 k B T, respectively, which may partly account for the different speed of membrane fusion mediated by these SNARE complexes.
Figure 7.
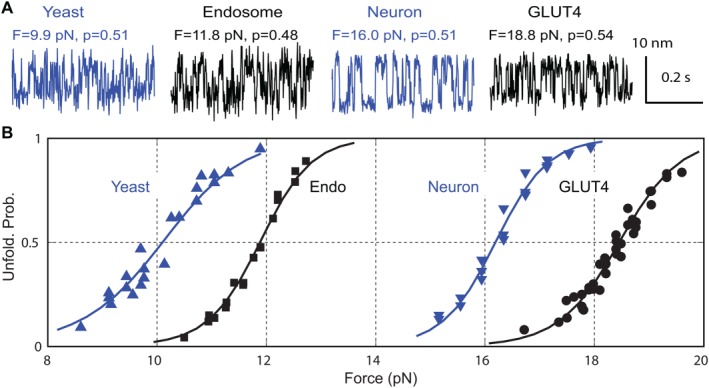
Different SNARE complexes share fast CTD folding, but exhibit different equilibrium forces and folding energies. (A) Extension‐time trajectories of two‐state CTD transitions in different SNARE complexes at different constant mean forces (F) near half unfolding probabilities (p). (B) CTD unfolding probability (symbol) as a function of force and its best model fit (solid curve). To compare folding dynamics of these SNARE complexes, we have used chimeric SNARE constructs in which the four SNARE motifs are joined into one polypeptide, which minimally interferes with CTD transitions and NTD unfolding. Adapted from Zorman et al.52
T‐SNARE conformation and its two‐step folding
The experiments on synaptic SNARE zippering demonstrate that the t‐SNARE complex is mechanically stable with significantly folded syntaxin even after the SNARE complex is unzipped [Fig. (2B)]. To elucidate the overall structure, stability, and folding dynamics of the t‐SNARE complex, we pulled the t‐SNARE complex from the C‐termini of syntaxin and SN1 or SN2.55 From both pulling sites, the t‐SNARE complex folds and unfolds similarly via an obligate half‐assembled intermediate [Fig. (8A)]. The large extension changes associated with t‐SNARE folding and results of SNARE mutations show that the folded t‐SNARE complex is a three‐helix bundle with a small frayed C‐terminus in layers +5 to +8 [Fig. (8B)]. This folded t‐SNARE conformation confirms the one derived from our SNARE zippering assay [Fig. (2B)]. The unfolding energy of NTD and CTD are 5 and 7 k B T, respectively. Accounting for the effect of the N‐terminal cross‐linking, we obtained a total bimolecular association energy of 17 k B T between syntaxin and SNAP‐25,55 consistent with the ensemble measurement of 18 k B T.33, 36 Combining this result with the ternary SNARE zippering energy yields a total unfolding energy of ∼80 k B T for a single SNARE complex. In comparison, NSF consumes ∼10 ATP molecules or ∼200 k B T energy to disassemble a single SNARE complex,17 leading to an energy efficiency of ∼40% for NSF‐dependent SNARE disassembly.
Perspectives
OTs make it possible to accurately measure the energy and kinetics of SNARE assembly for the first time. These measurements verify the SNARE zippering mechanism and reveal stepwise SNARE assembly via intermediates and domains with distinct functions. In particular, the observed half‐zippered intermediate is conserved across four different SNARE complexes and likely constitutes an essential intermediate for all SNARE‐mediated membrane fusion. The new methodology and improved understanding of SNARE engines will help elucidate the regulatory mechanisms of SNARE assembly, membrane fusion, and associated diseases. Many proteins directly bind SNAREs to regulate SNARE assembly, enabling calcium‐triggered exocytosis to occur at right time and location.1, 104, 106 Using OTs, we have obtained interesting effects of several key regulatory proteins on SNARE assembly, including Munc18‐1, α‐SNAP, and various Vn and Vc peptides.53, 54, 55 It will be interesting to apply the approach to test other regulatory proteins, such as synaptotagmin, complexin, and NSF. In addition, membranes directly modulate SNARE assembly and are bound by key regulatory proteins including synaptotagmin,107, 108 complexin,109 and α‐SNAP.110, 111 Investigating SNARE assembly in a natural membrane context will better recapitulate the energetics and kinetics SNARE assembly and its regulatory mechanisms in vivo.112, 113 In this case, model membranes can be incorporated into the SNARE zippering assay as bilayers supported on bead surfaces or nanodiscs.83, 114 Finally, numerous mutations of SNAREs or their regulatory proteins have been identified in a variety of human diseases.85, 115, 116, 117 OTs will be an important tool to understand how these mutations alter the energetics, kinetics, and pathway of regulated SNARE assembly, leading to dysfunction of SNARE assembly and membrane fusion.
Acknowledgments
The author thanks Aleksander A. Rebane for proofreading the manuscript.
References
- 1. Sudhof TC, Rothman JE (2009) Membrane fusion: grappling with SNARE and SM proteins. Science 323:474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ryham RJ, Klotz TS, Yao LH, Cohen FS (2016) Calculating transition energy barriers and characterizing activation states for steps of fusion. Biophys J 110:1110–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan YHM, van Lengerich B, Boxer SG (2009) Effects of linker sequences on vesicle fusion mediated by lipid‐anchored DNA oligonucleotides. Proc Natl Acad Sci USA 106:979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stengel G, Zahn R, Hook F (2007) DNA‐induced programmable fusion of phospholipid vesicles. J Am Chem Soc 129:9584–9585. [DOI] [PubMed] [Google Scholar]
- 5. Rabe M, Schwieger C, Zope HR, Versluis F, Kros A (2014) Membrane interactions of fusogenic coiled‐coil peptides: implications for lipopeptide mediated vesicle fusion. Langmuir 30:7724–7735. [DOI] [PubMed] [Google Scholar]
- 6. Kumar P, Guha S, Diederichsen U (2015) SNARE protein analog‐mediated membrane fusion. J Pept Sci 21:621–629. [DOI] [PubMed] [Google Scholar]
- 7. Sollner T, Whiteheart SW, Brunner M, Erdjument‐Bromage H, Geromanos S, Tempst P, Rothman JE (1993) SNAP receptors implicated in vesicle targeting and fusion. Nature 362:318–324. [DOI] [PubMed] [Google Scholar]
- 8. Rothman JE (2014) The principle of membrane fusion in the cell (Nobel Lecture). Angew Chem Int Ed 53:12676–12694. [DOI] [PubMed] [Google Scholar]
- 9. Jahn R, Scheller RH (2006) SNAREs ‐ engines for membrane fusion. Nat Rev Mol Cell Biol 7:631–643. [DOI] [PubMed] [Google Scholar]
- 10. Fasshauer D, Sutton RB, Brunger AT, Jahn R (1998) Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q‐ and R‐SNAREs. Proc Natl Acad Sci USA 95:15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sutton RB, Fasshauer D, Jahn R, Brunger AT (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 angstrom resolution. Nature 395:347–353. [DOI] [PubMed] [Google Scholar]
- 12. Fasshauer D, Antonin W, Subramaniam V, Jahn R (2002) SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat Struct Biol 9:144–151. [DOI] [PubMed] [Google Scholar]
- 13. Malhotra V, Orci L, Glick BS, Block MR, Rothman JE (1988) Role of an N‐ethylmaleimide‐sensitive transport component in promoting fusion of transport vesicles with cisternae of the golgi stack. Cell 54:221–227. [DOI] [PubMed] [Google Scholar]
- 14. Block MR, Glick BS, Wilcox CA, Wieland FT, Rothman JE (1988) Purification of an N‐ethylmaleimide‐sensitive protein catalyzing vesicular transport. Proc Natl Acad Sci USA 85:7852–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao M, Wu S, Zhou Q, Vivona S, Cipriano DJ, Cheng Y, Brunger AT (2015) Mechanistic insights into the recycling machine of the SNARE complex. Nature 518:61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ryu JK, D Min, SH Rah, SJ Kim, Y Park, H Kim, C Hyeon, HM Kim, R Jahn, TY Yoon (2015) Spring‐loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover. Science 347:1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shah N, Colbert KN, Enos MD, Herschlag D, Weis WI (2015) Three alpha SNAP and 10 ATP molecules are used in SNARE complex disassembly by N‐ethylmaleimide‐sensitive Factor (NSF). J Biol Chem 290:2175–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen YA, Scales SJ, Patel SM, Doung YC, Scheller RH (1999) SNARE complex formation is triggered by Ca2+ and drives membrane fusion. Cell 97:165–174. [DOI] [PubMed] [Google Scholar]
- 19. Xu T, Rammner B, Margittai M, Artalejo AR, Neher E, Jahn R (1999) Inhibition of SNARE complex assembly differentially affects kinetic components of exocytosis. Cell 99:713–722. [DOI] [PubMed] [Google Scholar]
- 20. Hua SY, Charlton MP (1999) Activity‐dependent changes in partial VAMP complexes during neurotransmitter release. Nat Neurosci 2:1078–1083. [DOI] [PubMed] [Google Scholar]
- 21. Walter AM, Wiederhold K, Bruns D, Fasshauer D, Sorensen JB (2010) Synaptobrevin N‐terminally bound to syntaxin‐SNAP‐25 defines the primed vesicle state in regulated exocytosis. J Cell Biol 188:401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sorensen JB (2009) Conflicting views on the membrane fusion machinery and the fusion pore. Annu Rev Cell Dev Biol 25:513–537. 2009). [DOI] [PubMed] [Google Scholar]
- 23. Yoon TY, Okumus B, Zhang F, Shin YK, Ha T (2006) Multiple intermediates in SNARE‐induced membrane fusion. Proc Natl Acad Sci USA 103:19731–19736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hernandez JM, Stein A, Behrmann E, Riedel D, Cypionka A, Farsi Z, Walla PJ, Raunser S, Jahn R (2012) Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science 336:1581–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Wit H, Walter AM, Milosevic I, Gulyas‐Kovacs A, Riedel D, Sorensen JB, Verhage M (2009) Synaptotagmin‐1 docks secretory vesicles to syntaxin‐1/SNAP‐25 acceptor complexes. Cell 138:935–946. [DOI] [PubMed] [Google Scholar]
- 26. Baker RW, Hughson FM (2016) Chaperoning SNARE assembly and disassembly. Nat Rev Mol Cell Biol 17:465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Munch AS, et al. (2016) Extension of Helix 12 in Munc18‐1 induces vesicle priming. J Neurosci 36:6881–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaeser PS, Regehr WG (2014) Molecular mechanisms for synchronous, asynchronous, and spontaneous neurotransmitter release. Annu Rev Physiol 76:333–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sabatini BL, Regehr WG (1996) Timing of neurotransmission at fast synapses in the mammalian brain. Nature 384:170–172. [DOI] [PubMed] [Google Scholar]
- 30. Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE (1997) Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick‐freeze/deep‐etch electron microscopy. Cell 90:523–535. [DOI] [PubMed] [Google Scholar]
- 31. Hanson PI, Heuser JE, Jahn R (1997) Neurotransmitter release ‐ four years of SNARE complexes. Curr Opin Neurobiol 7:310–315. [DOI] [PubMed] [Google Scholar]
- 32. Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE (1998) SNAREpins: minimal machinery for membrane fusion. Cell 92:759–772. [DOI] [PubMed] [Google Scholar]
- 33. Fasshauer D, Margittai M (2004) A transient N‐terminal interaction of SNAP‐25 and syntaxin nucleates SNARE assembly. J Biol Chem 279:7613–7621. [DOI] [PubMed] [Google Scholar]
- 34. Li F, Tiwari N, Rothman JE, Pincet F (2016) Kinetic barriers to SNAREpin assembly in the regulation of membrane docking/priming and fusion. Proc Natl Acad Sci USA 113:10536–10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li F, Kummel D, Coleman J, Reinisch KM, Rothman JE, Pincet F (2014) A half‐zippered snare complex represents a functional intermediate in membrane fusion. J Am Chem Soc 136:3456–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wiederhold K, Fasshauer D (2009) Is assembly of the SNARE complex enough to fuel membrane fusion? J Biol Chem 284:13143–13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jahn R, Fasshauer D (2012) Molecular machines governing exocytosis of synaptic vesicles. Nature 490:201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pobbati AV, Stein A, Fasshauer D (2006) N‐ to C‐terminal SNARE complex assembly promotes rapid membrane fusion. Science 313:673–676. [DOI] [PubMed] [Google Scholar]
- 39. Weninger K, Bowen ME, Chu S, Brunger AT (2003) Single‐molecule studies of SNARE complex assembly reveal parallel and antiparallel configurations. Proc Natl Acad Sci USA 100:14800–14805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kyoung M, Srivastava A, Zhang YX, Diao JJ, Vrljic M, Grob P, Nogales E, Chu S, Brunger AT (2011) In vitro system capable of differentiating fast Ca2+‐triggered content mixing from lipid exchange for mechanistic studies of neurotransmitter release. Proc Natl Acad Sci USA 108:E304–E313. (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yersin A, et al. (2003) Interactions between synaptic vesicle fusion proteins explored by atomic force microscopy. Proc Natl Acad Sci USA 100:8736–8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu W, Montana V, Bai JH, Chapman ER, Parpura V (2006) Single molecule mechanical probing of the SNARE protein interactions. Biophys J 91:744–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parpura V, Mohideen U (2008) Molecular form follows function: (un)snaring the SNAREs. Trends Neurosci 31:435–443. [DOI] [PubMed] [Google Scholar]
- 44. Liu W, Montana V, Parpura V, Mohideen U (2009) Single molecule measurements of interaction free energies between the proteins within binary and ternary SNARE complexes. J Nanoneurosci 1:120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Abdulreda MH, Bhalla A, Rico F, Berggren PO, Chapman ER, Moy VT (2009) Pulling force generated by interacting SNAREs facilitates membrane hemifusion. Integr Biol 1:301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li F, Pincet F, Perez E, Eng WS, Melia TJ, Rothman JE, Tareste D (2007) Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat Struct Mol Biol 14:890–896. [DOI] [PubMed] [Google Scholar]
- 47. Wang YJ, Li F, Rodriguez N, Lafosse X, Gourier C, Perez E, Pincet F (2016) Snapshot of sequential SNARE assembling states between membranes shows that N‐terminal transient assembly initializes fusion. Proc Natl Acad Sci USA 113:3533–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao Y, Zorman S, Gundersen G, Xi ZQ, Ma L, Sirinakis G, Rothman JE, Zhang YL (2012) Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science 337:1340–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Turnbull WB, Daranas AH (2003) On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc 125:14859–14866. [DOI] [PubMed] [Google Scholar]
- 50. Choi UB, Zhao ML, Zhang YX, Lai Y, Brunger AT (2016) Complexin induces a conformational change at the membrane‐proximal C‐terminal end of the SNARE complex. Elife 5:e16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brunger AT (2005) Structure and function of SNARE and SNARE‐interacting proteins. Q Rev Biophys 38:1–47. [DOI] [PubMed] [Google Scholar]
- 52. Zorman S, Rebane AA, Ma L, Yang GC, Molski MA, Coleman J, Pincet F, Rothman JE, Zhang YL (2014) Common intermediates and kinetics, but different energetics, in the assembly of SNARE proteins. Elife 3:e03348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ma L, Rebane AA, Yang G, Xi Z, Kang Y, Gao Y, Zhang YL (2015) Munc18‐1‐regulated stage‐wise SNARE assembly underlying synaptic exocytosis. Elife 4:e09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ma L, Kang Y, Jiao J, Rebane AA, Cha HK, Xi Z, Qu H, Zhang YL (2016) alpha‐SNAP enhances SNARE zippering by stabilizing the SNARE four‐helix bundle. Cell Rep 15:531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang XM, Rebane AA, Ma L, Li F, Jiao J, Qu H, Pincet F, Rothman JE, Zhang YL (2016) Stability, folding dynamics, and long‐range conformational transition of the synaptic t‐SNARE complex. Proc Natl Acad Sci USA 113:E8031–E8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jiao JY, Rebane AA, Ma L, Zhang YL (2017) Single‐molecule protein folding experiments using high‐resolution optical tweezers. Methods Mol Biol 1486:357–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moffitt JR, Chemla YR, Smith SB, Bustamante C (2008) Recent advances in optical tweezers. Annu Rev Biochem 77:205–228. [DOI] [PubMed] [Google Scholar]
- 58. Zhang XM, Ma L, Zhang YL (2013) High‐resolution optical tweezers for single‐molecule manipulation. Yale J Biol Med 86:367–383. [PMC free article] [PubMed] [Google Scholar]
- 59. Gittes F, Schmidt CF (1998) Interference model for back‐focal‐plane displacement detection in optical tweezers. Optics Lett 23:7–9. [DOI] [PubMed] [Google Scholar]
- 60. Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM (2005) Direct observation of base‐pair stepping by RNA polymerase. Nature 438:460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moffitt JR, Chemla YR, Izhaky D, Bustamante C (2006) Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc Natl Acad Sci USA 103:9006–9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sirinakis G, Clapier CR, Gao Y, Viswanathanc R, Cairns BR, Zhang YL (2011) The RSC chromatin remodeling ATPase translocates DNA with high force and small step size. EMBO J 30:2364–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Neupane K, Foster DAN, Dee DR, Yu H, Wang F, Woodside MT (2016) Direct observation of transition paths during the folding of proteins and nucleic acids. Science 352:239–242. [DOI] [PubMed] [Google Scholar]
- 64. Bustamante C, Chemla YR, Forde NR, Izhaky D (2004) Mechanical processes in biochemistry. Annu Rev Biochem 73:705–748. [DOI] [PubMed] [Google Scholar]
- 65. Liphardt J, Onoa B, Smith SB, Tinoco I, Bustamante C (2001) Reversible unfolding of single RNA molecules by mechanical force. Science 292:733–737. [DOI] [PubMed] [Google Scholar]
- 66. Zhang YL, Jiao J, Rebane AA (2016) Hidden Markov modeling with detailed balance and its application to single protein folding. Biophys J 111:2110–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rebane AA, Ma L, Zhang YL (2016) Structure‐based derivation of protein folding intermediates and energies from optical tweezers. Biophys J 110:441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cecconi C, Shank EA, Bustamante C, Marqusee S (2005) Direct observation of the three‐state folding of a single protein molecule. Science 309:2057–2060. [DOI] [PubMed] [Google Scholar]
- 69. Cecconi C, Shank EA, Dahlquist FW, Marqusee S, Bustamante C (2008) Protein‐DNA chimeras for single molecule mechanical folding studies with the optical tweezers. Eur Biophys J 37:729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gao Y, Sirinakis G, Zhang YL (2011) Highly anisotropic stability and folding kinetics of a single coiled coil protein under mechanical tension. J Am Chem Soc 133:12749–12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Marko JF, Siggia ED (1995) Stretching DNA. Macromolecules 28:8759–8770. [Google Scholar]
- 72. Smith CL, Cui Y, Bustamante C (1996) Overstretching B‐DNA: the elastic response of individual double‐stranded and single‐stranded DNA molecules. Science 271:795–799. [DOI] [PubMed] [Google Scholar]
- 73. Min D, Kim K, Hyeon C, Cho YH, Shin YK, Yoon TY (2013) Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a force‐generating mechanism. Nat Commun 4:1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Heo P, et al. (2016) A chemical controller of SNARE‐driven membrane fusion that primes vesicles for Ca2+‐triggered millisecond exocytosis. J Am Chem Soc 138:4512–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Diao J, et al. (2012) Synaptic proteins promote calcium‐triggered fast transition from point contact to full fusion. Elife 1:e00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bharat TA, Malsam J, Hagen WJ, Scheutzow A, Sollner TH, Briggs JA (2014) SNARE and regulatory proteins induce local membrane protrusions to prime docked vesicles for fast calcium‐triggered fusion. EMBO Rep 15:308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Melia TJ, Weber T, McNew JA, Fisher LE, Johnston RJ, Parlati F, Mahal LK, Sollner TH, Rothman JE (2002) Regulation of membrane fusion by the membrane‐proximal coil of the t‐SNARE during zippering of SNAREpins. J Cell Biol 158:929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sudhof TC (2013) Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron 80:675–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Xi ZQ, Gao Y, Sirinakis G, Guo HL, Zhang YL (2012) Direct observation of helix staggering, sliding, and coiled coil misfolding. Proc Natl Acad Sci USA 109:5711–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Harbury PB, Zhang T, Kim PS, Alber T (1993) A switch between 2‐stranded, 3‐stranded and 4‐stranded coiled coils in GCN4 leucine‐zipper mutants. Science 262:1401–1407. [DOI] [PubMed] [Google Scholar]
- 81. Woodside MT, Behnke‐Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM (2006) Nanomechanical measurements of the sequence‐dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci USA 103:6190–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. van den Bogaart G, Holt MG, Bunt G, Riedel D, Wouters FS, Jahn R (2010) One SNARE complex is sufficient for membrane fusion. Nat Struct Mol Biol 17:358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shi L, Shen QT, Kiel A, Wang J, Wang HW, Melia TJ, Rothman JE, Pincet F (2012) SNARE proteins: one to fuse and three to keep the nascent fusion pore open. Science 335:1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jiao JY, Rebane AA, Ma L, Gao Y, Zhang YL (2015) Kinetically coupled folding of a single HIV‐1 glycoprotein 41 complex in viral membrane fusion and inhibition. Proc Natl Acad Sci USA 112:E2855–E2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Corbett MA, et al. (2011) A mutation in the Golgi Qb‐SNARE gene GOSR2 causes progressive myoclonus epilepsy with early ataxia. Am J Hum Genet 88:657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mohrmann R, de Wit H, Verhage M, Neher E, Sorensen JB (2010) Fast vesicle fusion in living cells requires at least three SNARE complexes. Science 330:502–505. [DOI] [PubMed] [Google Scholar]
- 87. Lagow RD, Bao H, Cohen EN, Daniels RW, Zuzek A, Williams WH, Macleod GT, Sutton RB, Zhang B (2007) Modification of a hydrophobic layer by a point mutation in syntaxin 1A regulates the rate of synaptic vesicle fusion. PLoS Biol 5:800–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ossig R, Schmitt HD, de Groot B, Riedel D, Keranen S, Ronne H, Grubmuller H, Jahn R (2000) Exocytosis requires asymmetry in the central layer of the SNARE complex. EMBO J 19:6000–6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yu H, Rathore SS, Shen C, Liu Y, Ouyang Y, Stowell MH, Shen J (2015) Reconstituting intracellular vesicle fusion reactions: the essential role of macromolecular crowding. J Am Chem Soc 137:12873–12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Stein A, Weber G, Wahl MC, Jahn R (2009) Helical extension of the neuronal SNARE complex into the membrane. Nature 460:525–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. McNew JA, Weber T, Parlati F, Johnston RJ, Melia TJ, Sollner TH, Rothman JE (2000) Close is not enough: SNARE‐dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J Cell Biol 150:105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhou P, Bacaj T, Yang X, Pang ZP, Sudhof TC (2013) Lipid‐anchored snares lacking transmembrane regions fully support membrane fusion during neurotransmitter release. Neuron 80:470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Han X, Wang CT, Bai JH, Chapman ER, Jackson MB (2004) Transmembrane segments of syntaxin line the fusion pore of Ca2+‐triggered exocytosis. Science 304:289–292. [DOI] [PubMed] [Google Scholar]
- 94. Chang CW, Chiang CW, Gaffaney JD, Chapman ER, Jackson MB (2016) Lipid‐anchored synaptobrevin provides little or no support for exocytosis or liposome fusion. J Biol Chem 291:2848–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ellena JF, Liang BY, Wiktor M, Stein A, Cafiso DS, Jahn R, Tamm LK (2009) Dynamic structure of lipid‐bound synaptobrevin suggests a nucleation‐propagation mechanism for trans‐SNARE complex formation. Proc Natl Acad Sci 106:20306–20311. USA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Borisovska M, et al. (2012) Membrane‐proximal tryptophans of synaptobrevin II stabilize priming of secretory vesicles. J Neurosci 32:15983–15997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kesavan J, Borisovska M, Bruns D (2007) v‐SNARE actions during Ca2+‐triggered exocytosis. Cell 131:351–363. [DOI] [PubMed] [Google Scholar]
- 98. Wu Z, Auclair SM, Bello O, Vennekate W, Dudzinski NR, Krishnakumar SS, Karatekin E (2016) Nanodisc‐cell fusion: control of fusion pore nucleation and lifetimes by SNARE protein transmembrane domains. Sci Rep 6:27287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Dhara M et al (2016) v‐SNARE transmembrane domains function as catalysts for vesicle fusion. Elife 5. [Article #]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Fang QH, Lindau M (2014) How could snare proteins open a fusion pore? Physiology 29:278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ngatchou AN, Kisler K, Fang QH, Walter AM, Zhao Y, Bruns D, Sorensen JB, Lindau M (2010) Role of the synaptobrevin C terminus in fusion pore formation. Proc Natl Acad Sci USA 107:18463–18468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bao H, Goldschen‐Ohm M, Jeggle P, Chanda B, Edwardson JM, Chapman ER (2016) Exocytotic fusion pores are composed of both lipids and proteins. Nat Struct Mol Biol 23:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Schwartz ML, Merz AJ (2009) Capture and release of partially zipped trans‐SNARE complexes on intact organelles. J Cell Biol 185:535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sudhof TC (2014) The molecular machinery of neurotransmitter release (Nobel lecture). Angew Chem Int Ed 53:12696–12717. [DOI] [PubMed] [Google Scholar]
- 105. Baker RW, Jeffrey PD, Zick M, Phillips BP, Wickner WT, Hughson FM (2015) A direct role for the Sec1/Munc18‐family protein Vps33 as a template for SNARE assembly. Science 349:1111–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Rizo J, Sudhof TC (2012) The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices‐guilty as charged? Annu Rev Cell Dev Biol 28:279–308. [DOI] [PubMed] [Google Scholar]
- 107. Hui EF, Johnson CP, Yao J, Dunning FM, Chapman ER (2009) Synaptotagmin‐mediated bending of the target membrane is a critical step in Ca2+‐regulated fusion. Cell 138:709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lin CC, Seikowski J, Perez‐Lara A, Jahn R, Hobartner C, Walla PJ (2014) Control of membrane gaps by synaptotagmin‐Ca2+ measured with a novel membrane distance ruler. Nat Commun 5.[PAGE #S]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Diao JJ, Cipriano DJ, Zhao ML, Zhang YX, Shah S, Padolina MS, Pfuetzner RA, Brunger AT (2013) Complexin‐1 enhances the on‐rate of vesicle docking via simultaneous SNARE and membrane interactions. J Am Chem Soc 135:15274–15277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Winter U, Chen X, Fasshauer D (2009) A conserved membrane attachment site in alpha‐SNAP facilitates N‐ethylmaleimide‐sensitive factor (NSF)‐driven SNARE complex disassembly. J Biol Chem 284:31817–31826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Park Y, Vennekate W, Yavuz H, Preobraschenski J, Hernandez JM, Riedel D, Walla PJ, Jahn R (2014) alpha‐SNAP interferes with the zippering of the snare protein membrane fusion machinery. J Biol Chem 289:16326–16335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lou XC, Shin YK (2016) SNARE zippering. Biosci Rep 36.[PAGE #S]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Shin J, Lou XC, Kweon DH, Shin YK (2014) Multiple conformations of a single SNAREpin between two nanodisc membranes reveal diverse pre‐fusion states. Biochem J 459:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Naumann C, Brumm T, Bayerl TM (1992) Phase‐transition behavior of single phosphatidylcholine bilayers on a solid spherical support studied by DSC, NMR and FT‐IR. Biophys J 63:1314–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Burre J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Sudhof TC (2010) αSynuclein promotes SNARE‐complex assembly in vivo and in vitro. Science 329:1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shen C, Rathore SS, Yu H, Gulbranson DR, Hua R, Zhang C, Schoppa NE, Shen J (2015) The trans‐SNARE‐regulating function of Munc18‐1 is essential to synaptic exocytosis. Nat Commun 6:8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Shen XM, Selcen D, Brengman J, Engel AG (2014) Mutant SNAP25B causes myasthenia, cortical hyperexcitability, ataxia, and intellectual disability. Neurology 83:2247–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]


