Abstract
Nucleosomes at the promoters of genes regulate the accessibility of the transcription machinery to DNA, and function as a basic layer in the complex regulation of gene expression. Our understanding of the role of the nucleosome's spontaneous, thermally driven position changes in modulating expression is lacking. This is the result of the paucity of experimental data on these dynamics, at high‐resolution, and for DNA sequences that belong to real, transcribed genes. We have developed an assay that uses partial, reversible unzipping of nucleosomes with optical tweezers to repeatedly probe a nucleosome's position over time. Using the nucleosomes at the promoters of two model genes, Cga and Lhb, we show that the mobility of nucleosomes is modulated by the sequence of DNA and by the use of alternative histone variants, and describe how the mobility can affect transcription, at the initiation and elongation phases.
Keywords: nucleosomes, mobility, transcription, single molecule biophysics, optical tweezers
Introduction
The genetic information encoded in DNA provides instructions for how an organism develops and functions, with consequences throughout its life span. However, DNA does not exist in the cell in isolation, but in the form of chromatin, a complex structure of DNA and positively charged histone proteins, whose basic packaging unit is the nucleosome: a structure composed of ∼147 base pairs of DNA wrapped around two copies of histone proteins H2A, H2B, H3, and H4. Packaging of DNA into chromatin protects it from various hazards and reduces dramatically the volume it occupies, but also makes it largely inaccessible to the gene‐expression machinery; hence, to allow DNA transcription, this compaction must be disrupted in a controlled manner.
The structure of promoter chromatin is shaped by the combined effects of “local” features, such as the sequence of DNA, competition with transcription factors (TFs) and the incorporation of histone variants and post‐translational modifications, and other “long range” features such as enhancers and chromatin remodelers. However, we hypothesize that the physical properties of promoter nucleosomes (which are modulated by all the above factors) are the ones that ultimately dictate the ability of TFs to bind, and of RNA polymerase (RNAP) to transcribe, in order to achieve the expression level required for a specific gene, at a specific time. This motivates us to characterize, in vitro, nucleosomes that mimic those in vivo, and to explore the possible mechanisms that relate their physical properties to the transcriptional outcome.
Here, we describe the use of single‐molecule chromatin “unzipping” with optical tweezers, to shed light on the role that two specific factors (the sequence of DNA and the presence of the histone variant H2A.Z) play in modulating an important but experimentally understudied nucleosomal property: their spontaneous repositioning, or mobility, on the DNA. Recent experimental findings1 are discussed in the context of their possible implications for the regulation of two critical phases in transcription: initiation and early elongation.
The Dynamic Nature of Nucleosomes
While considerable efforts have been invested in elucidating how the mean position of nucleosomes on DNA is determined,2, 3, 4 even if still an unsettled matter,5 it is also clear that their dynamics play a crucial role in their function as regulators of expression. This is true not only for the long scale movements induced by ATP‐consuming chromatin remodelers, but also for smaller, thermally driven, positional and conformational changes. Among these, nucleosome “breathing”, i.e. the spontaneous wrapping/unwrapping of DNA at one end of the nucleosome [Fig. 1(A)], has been shown to play an important role in transcriptional initiation. By monitoring the ability of restriction enzymes to cut DNA at nucleosome‐protected sites, it was shown6 that TFs reach their binding site by exploiting these spontaneous, thermally driven conformational changes that momentarily expose their binding sites. These initial studies were followed by single‐molecule FRET experiments that directly detected the breathing fluctuations and their role in modulating TF accessibility.7, 8, 9, 10, 11, 12, 13, 14 Remarkably, nucleosomal breathing has also been shown to govern the ability of RNAP to transcribe through a nucleosome: Upon encountering the nucleosomal barrier, RNA often backtracks allowing the octamer to regain full contact with the DNA.15 Recovery from the backtracked state requires diffusion of RNAP on the DNA back into alignment of its active site with the 3'‐end of the transcript.16 However, the newly formed octamer‐DNA contacts prevent the realignment. Hodges et al. showed17 that recovery from the backtracked state can only take place concomitantly with a spontaneous breathing fluctuation of the nucleosome, making breathing also an important factor in transcriptional elongation.
Figure 1.
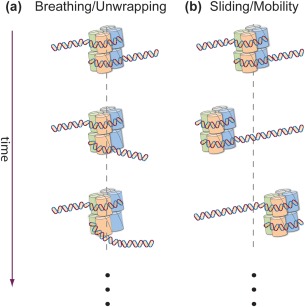
Thermally driven conformational dynamics of the nucleosome. (a) “Breathing”, spontaneous wrapping/unwrapping of DNA at one end of the nucleosome. (b) “Sliding”, spontaneous repositioning of the histone octamer as a whole relative to the DNA.
As opposed to breathing, spontaneous repositioning of the nucleosome, where the histone octamer as a whole moves relative to the DNA [often termed “sliding” or mobility; Fig. 1(B)] has not been experimentally characterized to much detail. Early studies that described sliding or repositioning of nucleosomes studied 200–400 bp DNA fragments which harbored nucleosomes, and were based on the differences in electrophoretic mobility of the complex as a function of the position of the octamer on the DNA.18, 19, 20 In a different approach, chemically modified histone proteins capable of inducing a nick in the DNA were used.21 These studies described sliding as a slow, thermally activated process: nucleosomes were shown to reposition on their templates at time scales of hours, if incubated at 37°C, but not at 5°C. While these are important findings, and stimulated the development of detailed models to characterize the potential physical mechanism behind this repositioning,22, 23, 24 the experiments are limited in a number of aspects. First, they were generally done with artificial, high affinity positioning sequences, such as Widom's “601” sequence,25 which can not only significantly reduce the magnitude of the mobility but can also introduce specific features such as the existence of a 10 bp periodicity in the repositioning.26 Next, they lack the resolution to detect small, bp‐scale movements, thus requiring very long incubation times. Finally, the thermally induced motion is a stochastic process, and many of its feature can be masked by the use of bulk methods, which suffer from the intrinsic averaging over an unsynchronized population. Hence, the ability to probe the mobility of nucleosomes at the single molecule level, with bp‐scale resolution, and on natural, biologically relevant sequences, is of great interest.
Force‐Unzipping of Reconstituted Nucleosomes Reveals Their Mobility on DNA
Previous studies have shown that force‐spectroscopy experiments using single‐molecule manipulation techniques, such as magnetic tweezers or optical tweezers, can be used to probe the mechanical properties of both single nucleosomes and nucleosome arrays.27, 28 In particular, it was shown that that by force‐unwinding the DNA it is possible to generate a detailed histone–DNA interaction energy landscape.29 In our experiments, we use a DNA template that includes a reconstituted nucleosome, attached to dsDNA molecular “handles” harboring two tags [Fig. 2(A)]: a biotin tag, which binds to a streptavidin‐coated microscopic bead, and a digoxygenin tag, which binds to an anti‐digoxygenin coated bead. Using a dual‐trap optical tweezers,30 the beads are held in separate traps and tension is applied to the construct by moving one of them with a piezo‐controlled mirror with nanometer resolution. As the beads are separated [Fig. 2(B)], the force on the construct increases, stretching its handles, until a value is reached (F∼17 pN) at which the base‐pairing in the central DNA/chromatin region is mechanically disrupted, as indicated by a series of extension increases concomitant with force drops [Fig. 2(C)]. In the presence of a nucleosome, histone‐DNA interactions need to be disrupted in order to “unzip” the DNA, resulting in a higher rupture force (25–30 pN).
Figure 2.
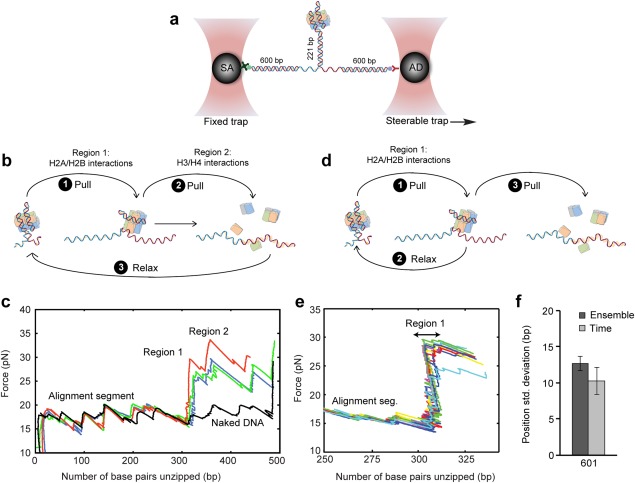
Force‐unzipping of in vitro reconstituted nucleosomes reveals their stability, position and mobility. (a) Reconstituted nucleosomes are connected to dsDNA molecular handles, which are attached to polystyrene beads trapped in two separate optical traps. One of the traps is moved to stretch the tethered construct. (b) Experimental procedure for probing interactions inside the nucleosome: pulling the DNA leads to unzipping and disruption of region 1 (off‐dyad), which correspond to interactions of DNA with H2A/H2B dimers, and region 2 (dyad), which corresponds to interactions with the H3/H4 tetramer. (c) Typical results for irreversible pulling and disruption of the first and second regions of interactions in 3 different 601 nucleosomes (green, blue and red). Irreversible interaction of region 2 leads to complete removal of histone proteins from DNA as shown after relaxing the DNA (black, only one experiment shown for clarity). (d) Experimental scheme for repetitive disruption of region 1 which reveals histone octamer movements on a single DNA molecule. (e) Typical results for reversible probing a nucleosome. Shown is data for a single nucleosome nucleosome reconstituted on 601 DNA. (f) The dispersion in position measured in an ensemble of irreversibly unzipped nucleosomes, is equivalent to the mean dispersion measured for single nucleosomes, interrogated reversibly with 30s intervals.
It has been shown, 29, 31, 32 that the disassembly pattern of nucleosomes under unzipping [Fig. 2(B)] is related to the strength of the histone–DNA interactions. Interactions with the H3/H4 tetramer result in a large rip at the nucleosome's dyad, and interactions of the DNA with H2A/H2B dimers give rise to additional regions of interaction located ∼±40 bp with respect to the dyad. Destabilizing the central H3/H4 interactions leads to disassembly of the nucleosome, so in most experiments only two interaction regions are observed [Fig. 2(C)]. These experiments can provide information on the stability of a nucleosome (e.g. the average force required to disrupt its dyad), and its position (e.g. the position at which the force crosses a certain threshold, or the position of the dyad interaction).
We have recently demonstrated1 that disruption of the first strong interaction region (about 40 bp before the dyad) is a reversible process: if the force is relaxed after the disruption, the interaction forms again. This observation allows us to subject nucleosomes to multiple cycles of unzipping of the H2A/H2B region followed by force relaxation to allow re‐zipping [Fig. 2(D)]. Since the repetitive, partial unzipping of a single nucleosome [Fig. 2(E)] can be used to probe the position of a single nucleosome several times as a function of time, this approach allows us to characterize the movement, or mobility of the nucleosome on DNA, which we quantify by the root‐mean‐square (RMS) position of the nucleosome over time [Fig. 2(F)]. Interestingly, when an interval of 30 s is used in between successive probing of a single nucleosome, the mean mobility (i.e. averaged over a number of identical experiments) is similar to the dispersion in position in a set of irreversible unzipping experiments where every nucleosome is probed once [Fig. 2(F)]. In other words, the dispersion over time is similar to the dispersion observed in a “snapshot” of the ensemble, indicating that the positional dispersion in the ensemble experiments [Fig. 2(C)] reflects, also, the nucleosome's mobility.
It is interesting to note that although there is a large difference in timescales for the observed repositioning between our experiments (30 s) and the early reports of nucleosome sliding (hours), the results are in fact consistent. This stems from the properties of thermal diffusion, which indicate that the typical time it takes to reach a certain distance scales as , where is the diffusion constant of the nucleosome on the DNA. From the early experiments, which used strong positioning sequences and monitored repositioning on a 200–400 bp DNA molecule, the diffusion constant of the nucleosome on the DNA was estimated as . Hence, for a 30 s time we expect , consistent with the 12.7+/–1 bp we measure for 601 DNA.
The TSS Regions of the Two LH Genes Harbor Mobile Nucleosomes
Although the 601 sequence is widely used in many in vitro studies, studying the role that spontaneous nucleosome repositioning plays in modulating gene expression cannot be based on DNA positioning sequences. These artificial sequences, selected for their high affinity for the formation of nucleosomes in an in vitro reconstitution assay,25 harbor nucleosomes that are not necessarily typical in their dynamics, as compared to nucleosomes found in natural sequences. Hence, it is important to look at real sequences derived from real genes. In our work, we use the promoters of Cga and Lhb, the genes that encode for the two subunits of the Luteinizing Hormone (LH), a glycoprotein secreted by the anterior pituitary that controls reproductive function. Although Cga and Lhb are both expressed in the pituitary gonadotropes under similar hormonal control, the α subunit comprises also a part of other hormones; thus, their basal levels of expression differ, with the Cga gene being expressed at much higher levels. We have recently shown1 that these large differences in expression pattern are the result, at least in part, of their distinct promoter chromatin structure [Fig. 3(A)], making these two genes a convenient model to study the interplay between chromatin structure and transcriptional outcome. Hence, we assemble nucleosomes mimicking the TSS and +1 nucleosomes at the proximal promoter sequences of Cga and Lhb [Fig. 3(A)], using mouse histones that were expressed in E. coli, and probe them by force‐unzipping.
Figure 3.
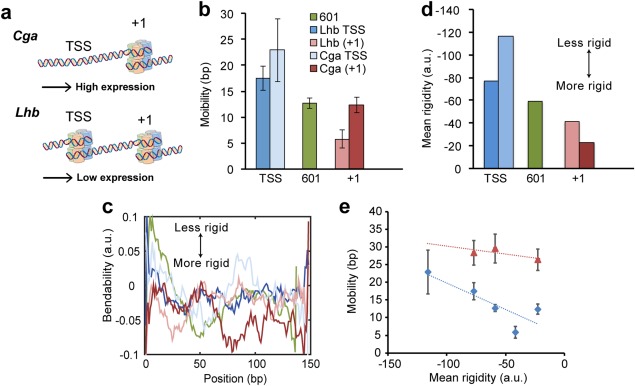
DNA sequence affects the mobility of promoter nucleosomes. (a) Promoter nucleosomes of Cga and Lhb in gonadotropes are remodeled in different ways, tailored to achieve a moderate expression of Lhb and a much higher expression of Cga. A +1 nucleosome is positioned on both genes, while the nucleosome on the TSS of Cga is depleted compared to Lhb. (b) The mobility of TSS nucleosomes in both Cga and Lhb, is significantly higher than the mobility of the corresponding +1 nucleosomes. (P = 0.025 for Lhb, P = 0.024 for Cga; two‐sample Ansari‐Bradley test). (c) Bendability of the underlying DNA sequences, smoothed with a 30 bp running window. (d) Mean rigidity, defined as the inverse of the sequence‐averaged bendability for each construct. (e) Rigidity and mobility are correlated for canonical nucleosomes (blue diamonds; r = 0.84, P = 0.05), but not for H2A.Z containing nucleosomes (red triangles).
When irreversibly unzipped, nucleosomes reconstituted on these gene sequences exhibit similar disassembly patterns as the ones reconstituted on the 601 positioning sequence, with two prominent regions of strong interaction.1 Moreover, although there is a small but significant difference in mean breaking force of Region 2 between the synthetic high‐affinity 601 DNA and the gene sequences, there are no significant differences among the latter (Table 1). In contrast, significant differences in the mobility of the nucleosome are observed, with both TSS nucleosomes exhibiting a higher mobility as compared with their respective +1 nucleosomes [P = 0.025 for Lhb, P = 0.024 for Cga; two‐sample Ansari‐Bradley test; Fig. 3(B)]. While the +1 nucleosomes exhibit a mobility comparable to that of the 601 sequence (or even smaller for Lhb), the TSS nucleosomes are much more mobile.
Table 1.
Parameters Measured for the Nucleosomes Reconstituted with Canonical Histones
| Region 1 | Region 2 | Mobility (bp) | |||
|---|---|---|---|---|---|
| Sequence | Force (pN) | Position (bp) | Force (pN) | Position (bp) | |
| 601 | 28.1 ± 0.3 | 327.7 ± 10.6 | 31.4 ± 0.3 | 368.7 ± 1.4 | 12.7 ± 1.0 |
| Lhb TSS | 27.8 ± 0.7 | −77.9 ± 18.6 | 30.5 ± 0.5 | −43.4 ± 3.3 | 17.5 ± 2.4 |
| Lhb +1 | 26.9 ± 0.7 | 82.6 ± 10.8 | 28.9 ± 1.0 | 117.0 ± 2.2 | 5.8 ± 1.7 |
| Cga TSS | 26.9 ± 1.2 | −104.2 ± 20.5 | 28.7 ± 0.9 | −64.4 ± 8.1 | 22.9 ± 6.1 |
| Cga +1 | 28.6 ± 0.5 | 87.3 ± 10.3 | 29.2 ± 0.5 | 118.5 ± 2.1 | 12.4 ± 1.5 |
Force: the mean force required to break the interaction at Region 1 or 2. Position: location of the interactions. For the genes' nucleosomes, relative to the TSS. For 601, relative to the beginning of the alignment sequence. Mobility: root‐mean‐square of the position of Region 1. For all parameters, mean +/– SE are shown.
How is the nucleosome's mobility affected by the sequence? The underlying sequence of DNA can affect the properties of nucleosomes by the formation of specific DNA‐histone interactions and by the sequence‐dependent mechanical properties of DNA.33 Previous works have shown that the sequence has an important effect on the positioning, structure, and stability of nucleosomes33, 34, 35 and, recently, it was demonstrated that the sequence can also affect the dynamics of the nucleosome's local conformational transitions.36 However, not much has been studied about the effect of sequence on the spontaneous sliding of nucleosomes.
It is important to note that it is highly unlikely that nucleosome repositioning will take place in a single step, as this would involve the large energy cost (>75 kBT) of disrupting all the histone–DNA interactions.26 Hence, two mechanisms have been proposed for the sliding of nucleosomes, and both involve the diffusive propagation of defects along the nucleosome interaction points. In the first mechanism these are DNA loops, preferentially 10 bp long22, 23 in order for them to be twist‐free, while in the second these are twist defects, which carry either a missing or an additional single bp.24 If a defect created at one side of the nucleosome is able to propagate all the way to the other side, the nucleosome will effectively be translocated by the size of the propagating defect. Since the time it takes for a defect to form is much longer than its propagation time,22, 23 the movement of the nucleosome can be modeled as a 1D random walk, with a diffusion constant that depends on the rate of defects creation. Interestingly, these mechanisms offer possible scenarios to incorporate the effect of sequence on the mobility. In the DNA looping mechanism, DNA sequences with higher bendability lower the free energy of the looped DNA,33 increasing the rate of defects creation and inducing faster nucleosome translocation. In the twist defect model, specific DNA sequences that are more readily twistable will lower the free energy cost of the overwound or underwound structures, thus allowing for faster nucleosome translocation.33
To shed light on the mechanism governing the distinct degrees of mobility we observe for our nucleosomes, we calculated the bendability, , of all our constructs, using bendability parameters for overlapping trinucleotides37 [Fig. 3(C)]. Figure 3(D,E) show that there is a remarkable correlation between the mean rigidity of the sequence (which we define as and the measured mobility (r = 0.84, P = 0.05), lending support for a sequence‐dependent modulation of the nucleosome's diffusion constant via modulation of a defect formation rate.
The LH Gene Promoters are Enriched with Mobile, H2A.Z‐Containing Nucleosomes
We recently demonstrated that the promoters of Cga and Lhb are both enriched with the histone variant H2A.Z, but with different localization patterns1: while the widely expressed Cga (which contains a nucleosome depleted region, NDR, at the TSS) is enriched at the +1 nucleosome, the Lhb promoter is enriched at the TSS nucleosome [Fig. 4(A)]. H2A.Z is an evolutionarily conserved and essential variant of the canonical H2A histone, which has been shown to have roles in development, differentiation, T‐cell activation, and more.38 H2A.Z also seems to play an important role in transcriptional regulation, as it is enriched at the promoters of both active and silent genes.39 Moreover, nucleosomes containing H2A.Z, in addition to the variant H3.3, are present in regions that were previously considered to be depleted of nucleosomes in active promoters, enhancers, and insulators.40 However, the role of H2A.Z in stabilizing or destabilizing the nucleosome, its repressive or activating role in transcription41 and, more generally, the mechanism by which it influences gene expression, are still matters of debate.
Figure 4.
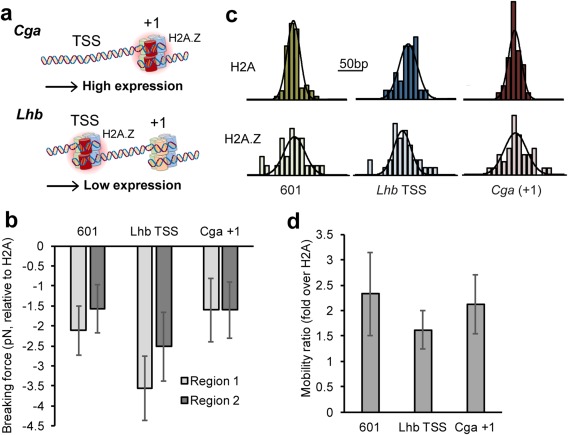
Incorporation of H2A.Z increases the mobility of promoter nucleosomes. (a) Promoter nucleosomes of Cga and Lhb in gonadotropes are incorporated with histone variant H2A.Z in distinct +1 and TSS positions, respectively. (b) H2A.Z containing nucleosomes exhibit significantly lower breaking forces than their canonical counterparts, in both dyad and off‐dyad interactions. (Region 1: P = 0.0008, 0.004, and 0.001; Region 2: P = 0.002, 0.04, and 0.02; for Lhb TSS, Cga +1 and 601, respectively; two‐sample Kolmogorov‐Smirnov test). (c) H2A.Z broadens the dispersion in the position of an ensemble of nucleosomes. (d) The mobility of H2A.Z‐containing nucleosomes is higher than the canonical nucleosomes, for all sequences probed.
To characterize the role that H2A.Z incorporation plays in our model genes, we reconstituted nucleosomes using H2A.Z, together with canonical H2B, H3, and H4, on the sequences of Lhb TSS and Cga +1. Unzipping experiments with H2A.Z‐containing nucleosomes reveals two regions of strong histone–DNA interactions, as observed in experiments with canonical nucleosomes. However, the mean breaking force is significantly reduced, for both Region 1 and Region 2 [mean reduction ∼2 pN; Fig. 4(B), Table 2]. Interestingly, nucleosomes reconstituted with H2A.Z on the 601 sequence showed a similar reduction in breaking forces. (Region 1: P = 0.0008, 0.004, and 0.001; Region 2: P = 0.002, 0.04, and = 0.02, for Lhb TSS, Cga +1 and 601, respectively; two‐sample Kolmogorov‐Smirnov test).
Table 2.
Parameters Measured for the Nucleosomes Reconstituted with H2A.Z
| Region 1 | Region 2 | Mobility (bp) | |||
|---|---|---|---|---|---|
| Sequence | Force (pN) | Position (bp) | Force (pN) | Position (bp)* | |
| 601 | 26.0 ± 0.5 | 329.7 ± 24.3 | 29.9 ± 0.7 | 365.0 ± 5.7 | 29.6 ± 4.1 |
| Lhb TSS | 24.3 ± 0.4 | −87.2 ± 22.8 | 28.0 ± 0.7 | −53.1 ± 4.9 | 28.4 ± 3.5 |
| Cga +1 | 25.9 ± 0.6 | 84.8 ± 28.2 | 27.6 ± 0.5 | 121.5 ± 4.3 | 26.3 ± 3.1 |
All parameters as detailed for Table I.
Our data also indicate that, for all sequences probed, H2A.Z incorporation results in a significant, ∼2‐fold increase in the nucleosomes' mobility [Fig. 4(D)]. Since the resulting mobility is similar for the different sequences tested, including intrinsically low‐mobility sequences such as 601 and Cga +1 [Fig. 2(F)], it seems that both sequence and histone identity determine the mobility of nucleosomes, but H2A.Z is a stronger determinant. These single‐molecule results are also consistent with previous gel‐based reports of increased sliding by H2A.Z containing nucleosome on DNA positioning sequences.42
How does the incorporation of H2A.Z increase the nucleosome's mobility? Although the sequence homology between H2A.Z and H2A is only ∼60%, the structure of the H2A.Z‐containing nucleosome is highly similar to that of the canonical ones.43 There are some differences, however, in particular in domain L1, important for the interactions between the two H2A/H2B dimers, and the C‐terminal docking domain, responsible for their interaction with the H3/H4 tetramer. The loss of hydrogen bonds between H2A.Z and H3/H4 is expected to weaken the interactions between H2A.Z/H2B and H3/H4, and therefore may be the source of the decreased breaking force of the H2A.Z nucleosomes, but are not likely to affect the nucleosomes mobility. Since residues at the C‐terminus of H2A make stable hydrogen bonds with the DNA, we believe that the increased mobility we observe here is the result of the absence of these bonds in H2A.Z‐containing nucleosomes. Notably, truncation of H2A C‐terminal domain has been reported to increase the thermal mobility of nucleosomes.44
Interestingly, there were indications in previous studies that H2A.Z not only increases the sliding of nucleosomes, but may also bias the repositioning towards different sites as compared to the position of canonical nucleosomes.45 This led to the suggestion that the effect of H2A.Z on nucleosome positioning, that is a possible mean repositioning to a different location, may be functionally important.46 Our results indicate that reconstitution with H2A.Z results in a shift of ∼ 10 bp upstream in the mean position of the Lhb TSS nucleosome (P = 0.02; two‐sample Kolmogorov‐Smirnov test; Tables 1, 2, II). No significant changes in the mean position of nucleosomes on the Cga +1, or 601 sequence were observed, indicating that such mechanism may be sequence dependent.
Of note, the single‐molecule experiments described above were performed with recombinant histone proteins, which lack post‐translational modifications, in order to address the effects of sequence and histone‐variant usage in a well‐controlled experiment. However, one may expect that the mobility will be affected also by the presence of specific modifications. For example, H3K56ac on the entry‐exit region was found to affect nucleosome breathing, and H3K122ac on the dyad was found to affect nucleosome stability.47 It will be interesting to elucidate the effect of these modifications on the mobility of nucleosomes, as well as the effect of other modifications on other histones such as H2A and H2A.Z.
Finally, it has been shown in vitro that nucleosome arrays containing H2A.Z are resistant to condensation.48, 49 It was later suggested that these special biophysical properties of H2A.Z are exploited by the cell as a mean of controlling the spread of chromatin silencing, by forming nucleosomes that are refractory to the propagation of Sir2/3‐induced deacetylation.50 Single‐molecule measurements of the mobility of H2A.Z nucleosomes, and the effect of changes in histone H4 acetylation on it, will perhaps enable clarifying the molecular mechanism responsible for this effect.
Mobile Nucleosomes Can Modulate Transcription Initiation
The dynamic equilibrium model of Polach and Widom6 postulates that binding of TFs to sites that are buried inside the nucleosome is modulated by the nucleosome's thermally driven spontaneous breathing. Breathing fluctuations are fast,8 and thus TFs bind to a buried site with an apparent dissociation constant , where is the dissociation constant on naked DNA, and quantifies the accessibility of the binding site, that is the probability for a breathing fluctuation that exposes it. Since the free‐energy cost for such a fluctuation depends on the amount of DNA that needs to unwrap to expose the site, is a sensitive function of the distance of the binding site from the nucleosome's dyad, ranging from ∼10−2 to 10−1 for sites at the edges of the nucleosome to ∼10−4–10−5 for sites near to the dyad.6, 8, 51
How should the mobility of a nucleosome be incorporated in such a model? Typical unwrapping and rewrapping rates are fast (∼4 s−1 and ∼20 to ∼90 s−1, respectively8) as compared to the typical rate of repositioning; thus, we can postulate a simple model in which breathing fluctuations are always in equilibrium for the instantaneous position of the nucleosome. In this case, we can assume that, on average, TF binding will be determined by a time‐averaged accessibility, , which now includes both breathing and mobility [Fig. 5(A)].
Figure 5.
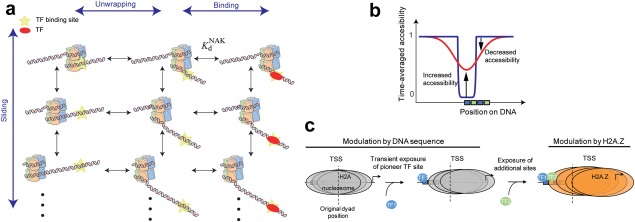
Model for the effect of nucleosome mobility on transcription initiation. (a) Schematic representation of the effect of nucleosome mobility on the accessibility of TF binding sites. (b) The mobility of the nucleosome can be modeled by an effective, time‐averaged accessibility. (c) TSS sequences support formation of inherently mobile nucleosomes, which as a consequence of repositioning facilitate binding of pioneer TFs, restricting nucleosome movement upstream. Such factors can recruit chromatin remodelers which incorporate H2A.Z and further facilitate binding of additional transcription factors.
Interestingly, one could expect a priori that a symmetric movement of the nucleosome, such as that which we expect for diffusional reposition, will produce no net effect on the exposure of a site, as the distance from the dyad to the binding site will at some times be increased at by the mobility and at others it will be decreased. However, the exponential dependence of on breaks the symmetry, creating a net effect. It is also worth noting that the modulation by the mobility can have both a repressing as well as a facilitating effect: For example, if a binding site is outside the nucleosome, but in its vicinity, . In this case, the mobility can only have a repressing effect, as the mobile nucleosome is now able to momentarily cover the binding site, resulting in . Alternatively, if the binding site is at the dyad, has its minimal possible value, hence repositioning can only increase the exposure, that is . In general, sites that are closer to the dyad than a critical value will have their accessibility increased, while those that are further away than this value will see a decrease in accessibility. Taken together, this makes the modulation of the mobility a powerful and versatile tool that provides a way to moderately adjust TF binding, as opposed to the more radical effect of eviction of the nucleosome.
Of note, one can imagine a scenario where the mobility, as conveyed by the sequence of DNA and by the presence of H2A.Z can act sequentially to modulate expression: Mobile nucleosomes at the TSS may allow for a basal level of recruitment of pioneer TFs, which are responsible then for the recruitment of chromatin remodelers that direct the incorporation of H2A.Z. The resulting higher‐mobility nucleosome allows then recruitment of additional TFs [Fig. 5(C)].
Mobile Nucleosomes Can Facilitate RNAP Elongation
Nucleosomes present a hurdle for transcriptional elongation52, 53 and induce RNAP pausing. This is particularly true for the +1 nucleosome, which creates a ∼3 times higher barrier than downstream nucleosomes.54 Overcoming this obstacle is therefore critical for promoter escape during the early elongation phase of transcription. When RNAP encounters the nucleosomal barrier, ∼8–13 bp within the +1 nucleosome,54 it backtracks allowing the octamer to regain full contact with the DNA.15 The re‐formed contacts between the octamer and DNA prevent the recovery of RNAP from its backtracked state by diffusing back into alignment of its active site with the 3'‐end of the transcript. With no energy input, RNAP is not able to actively disrupt the nucleosome in order to reach alignment. Hence, the recovery is a passive process, where RNAP must exploit spontaneous breathing fluctuations in the nucleosome.17, 55 However, if nucleosomes are mobile, it is reasonable to expect that the recovery will be affected not only by the breathing dynamics of a nucleosome at a fixed position, but also by the repositioning of the nucleosome as a whole (Fig. 6).
Figure 6.
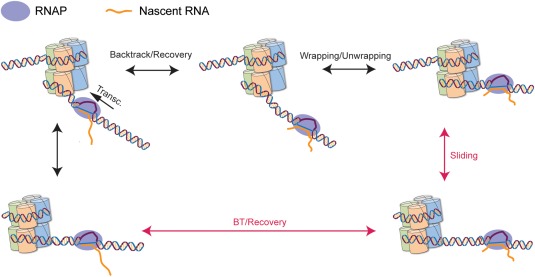
Schematic representation of the effect of nucleosome mobility on RNAP elongation. Recovery from the backtracked state is governed by the combined effect of nucleosome movement and breathing. Movement of the nucleosome provides an “alternative pathway” for backtrack recovery (red).
Backtrack recovery on naked DNA can be modeled as a first‐passage problem, in which the enzyme freely diffuses up to the first random encounter with the 3'‐end of the nascent RNA. It has been shown,56 that this results in a distribution for the backtrack recovery time given by , where and are the intrinsic forward and backward diffusional stepping rates of the polymerase, and is the modified Bessel function of the first kind.56 In the presence of a nucleosome, since RNAP cannot actively displace the nucleosome, the pause durations follow the same distribution, but with a modified forward stepping17 given by , where here is the probability for the nucleosome to be in the unwrapped state (i.e. in a fluctuation large enough that RNAP can move forward). As described in the previous section, to incorporate the effect of nucleosome mobility we can replace by . Since RNAP, and possibly the nascent RNA, may act as barriers for the movement of the nucleosome, effectively biasing its diffusion downstream on the DNA, we expect > . Hence, a mobile nucleosome may facilitate backtrack recovery, shortening the recovery time and increasing the efficiency of promoter escape. There are some caveats to this model: First, it is possible that the movement of the nucleosome will also affect the movement of RNAP, biasing the polymerase towards further backtracking. Moreover, it is not clear whether there is a separation of time‐scales between RNAP backtracking and nucleosome mobility that can justify the rapid‐equilibrium treatment above. A detailed model of the coupled kinetics of polymerase and nucleosome, or simulations, for example, by Markov state models and molecular dynamics,57 will be required to further characterize their interplay. Nevertheless, experimental data supports a facilitating effect for the dynamics of nucleosomes on the elongation of RNAP. First, it has been shown that the nucleosomal barrier is relieved by ISW2, an ATP‐dependent chromatin remodeler, which translocates the nucleosome over a short distance.53 Moreover, sin mutations, which do not significantly alter the structure of nucleosomes but increase their mobility, have been shown to rescue defects in SWI/SNF action,42 while deletion of H2A.Z in S. cerevisiae strongly increased the need for SWI/SNF.58 Finally, the efficiency of in vitro transcription on a template that contains an H2A.Z nucleosome is significantly higher than the efficiency through a canonical nucleosome.1
Summary
The structure and dynamics of promoter chromatin are shaped in a gene‐specific and cell‐specific way by the synergistic action of numerous factors, including the sequence of DNA, the identity of the histone proteins, post‐translational modifications, chromatin remodelers, distal enhancers and more. However, all these effects eventually converge into a structure whose biophysical properties then control the rate and fate of transcription. Thus, a mechanistic understanding of transcriptional regulation requires characterizing the interplay between the above‐mentioned factors and the resulting biophysical properties of the nucleosomes, such as position, stability, and mobility.
We have developed a novel assay that allows us to probe the mobility of nucleosomes, at the single molecule level and with high resolution, and have used this assay to probe the mobility of nucleosomes that mimic, in position and composition, those found at the promoter of two model genes, Cga and Lhb. Our findings indicate that the sequences of these promoters results in nucleosomes of high mobility on their TSS regions. Since the mobility of the nucleosomes correlates with the bendability of the underlying DNA, our results lend support for a model of nucleosome mobility by the diffusive propagation of a structural defect. In addition, we have shown that the selective incorporation of the histone variant H2A.Z, as we observe at the Lhb TSS and Cga +1 nucleosomes, results in a large increase in their mobility. A similar increase in mobility for the 601 positioning sequence suggests that H2A.Z is a very strong determinant of the nucleosome's mobility, able to overcome the small degrees of mobility dictated by certain sequences. Notably, the mobility measured here for H2A.Z nucleosomes is similar to the dispersion reported previously following the action of SWI/SNF remodelers,31 stressing the potential of the mobility as a modulator of gene expression.
Selective incorporation of mobile nucleosomes at different regions of the DNA has the potential to modulate the distinct stages of transcription. A mobile TSS nucleosome, as dictated by its sequence or the incorporation of H2A.Z, is expected to affect TF binding and therefore the initiation of transcription. Interestingly, since a higher mobility can increase the time‐averaged exposure of some TF binding sites, while reducing it for others, this simple model can have implications for the controversy on the repressing/activating effect of H2A.Z.46 Mobile nucleosomes after the TSS, and in particular the +1, will modulate the elongation phase, by facilitating recovery of RNAP from a backtracked state, thus increasing the rate of promoter escape.
Author Contributions
S.R., P.M. and A.K. designed the research; S.R., A.B. and L.P. performed the experiments. O.M. and A.K. designed and built the optical tweezers setup. S.R. and A.K. analyzed the data. A.K wrote the paper.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This research was supported in part by the Russell Berrie Nanotechnology Institute through funding to AK and PM.
References
- 1. Rudnizky S, Bavly A, Malik O, Pnueli L, Melamed P, Kaplan A (2016) H2A.Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes. Nat Commun 7:12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaplan N, Moore IK, Fondufe‐Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. (2009) The DNA‐encoded nucleosome organization of a eukaryotic genome. Nature 458:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K (2010) Evidence against a genomic code for nucleosome positioning. Reply to “Nucleosome sequence preferences influence in vivo nucleosome organization”. Nat Struct Mol Biol 17:920–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Struhl K, Segal E (2013) Determinants of nucleosome positioning. Nat Struct Mol Biol 20:267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takasuka TE, Stein A (2010) Direct measurements of the nucleosome‐forming preferences of periodic DNA motifs challenge established models. Nucleic Acids Res 38:5672–5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Polach KJ, Widom J (1995) Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol 254:130–149. [DOI] [PubMed] [Google Scholar]
- 7. Li G, Widom J (2004) Nucleosomes facilitate their own invasion. Nat Struct Mol Biol 11:763–769. [DOI] [PubMed] [Google Scholar]
- 8. Li G, Levitus M, Bustamante C, Widom J (2005) Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol 12:46–53. [DOI] [PubMed] [Google Scholar]
- 9. Tomschik M, Zheng H, Van Holde K, Zlatanova J, Leuba SH (2005) Fast, long‐range, reversible conformational fluctuations in nucleosomes revealed by single‐pair fluorescence resonance energy transfer. Proc Natl Acad Sci USA 102:3278–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kelbauskas L, Chan N, Bash R, Yodh J, Woodbury N, Lohr D (2007) Sequence‐dependent nucleosome structure and stability variations detected by Förster resonance energy transfer. Biochemistry 46:2239–2248. [DOI] [PubMed] [Google Scholar]
- 11. Kelbauskas L, Sun J, Woodbury N, Lohr D (2008) Nucleosomal stability and dynamics vary significantly when viewed by internal versus terminal labels. Biochemistry 47:9627–9635. [DOI] [PubMed] [Google Scholar]
- 12. Gansen A, To'th K, Schwarz N, Langowski J (2009) Structural variability of nucleosomes detected by single‐pair Förster resonance energy transfer: histone acetylation, sequence variation, and salt effects. J Phys Chem B 113:2604–2613. [DOI] [PubMed] [Google Scholar]
- 13. Gansen A, Valeri A, Hauger F, Felekyan S, Kalinin S, Toth K, Langowski J, Seidel CAM (2009) Nucleosome disassembly intermediates characterized by single‐molecule FRET. Proc Natl Acad Sci USA 106:15308–15313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koopmans WJA, Buning R, Schmidt T, Van Noort J (2009) spFRET using alternating excitation and FCS reveals progressive DNA unwrapping in nucleosomes. Biophys J 97:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaykalova DA, Kulaeva OI, Volokh O, Shaytan AK, Hsieh F‐K, Kirpichnikov MP, Sokolova OS, Studitsky VM (2015) Structural analysis of nucleosomal barrier to transcription. Proc Natl Acad Sci USA 112:E5787–E5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Galburt EA, Grill SW, Wiedmann A, Lubkowska L, Choy J, Nogales E, Kashlev M, Bustamante C (2007) Backtracking determines the force sensitivity of RNAP II in a factor‐dependent manner. Nature 446:820–823. [DOI] [PubMed] [Google Scholar]
- 17. Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C (2009) Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325:626–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pennings S, Meersseman G, Bradbury EM (1991) Mobility of positioned nucleosomes on 5 S rDNA. J Mol Biol 220:101–110. [DOI] [PubMed] [Google Scholar]
- 19. Meersseman G, Pennings S, Bradbury EM (1992) Mobile nucleosomes—a general behavior. embo J 1:2951–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pennings S, Meersseman G, Bradbury EM (1994) Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc Natl Acad Sci USA 91:10275–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Flaus A, Richmond TJ (1998) Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J Mol Biol 275:427–441. [DOI] [PubMed] [Google Scholar]
- 22. Schiessel H, Widom J, Bruinsma RF, Gelbart WM (2001) Polymer reptation and nucleosome repositioning. Phys Rev Lett 86:4414–4417. [DOI] [PubMed] [Google Scholar]
- 23. Kulić IM, Schiessel H (2003) Nucleosome repositioning via loop formation. Biophys J 84:3197–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kulić IM, Schiessel H (2003) Chromatin dynamics: nucleosomes go mobile through twist defects. Phys Rev Lett 91:148103. [DOI] [PubMed] [Google Scholar]
- 25. Lowary PT, Widom J (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence‐directed nucleosome positioning. J Mol Biol 276:19–42. [DOI] [PubMed] [Google Scholar]
- 26. Eslami‐Mossallam B, Schiessel H, van Noort J (2016) Nucleosome dynamics: sequence matters. Adv Colloid Interf Sci 232:101–113. [DOI] [PubMed] [Google Scholar]
- 27. Killian JL, Li M, Sheinin MY, Wang MD (2012) Recent advances in single molecule studies of nucleosomes. Curr Opin Struct Biol 22:80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chien F‐T, Van Noort J (2009) 10 years of tension on chromatin: results from single molecule force spectroscopy. Curr Pharm Biotechnol 10:474–485. [DOI] [PubMed] [Google Scholar]
- 29. Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD (2009) High‐resolution dynamic mapping of histone‐DNA interactions in a nucleosome. Nat Struct Mol Biol 16:124–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moffitt JR, Chemla YR, Izhaky D, Bustamante C (2006) Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc Natl Acad Sci USA 103:9006–9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shundrovsky A, Smith CL, Lis JT, Peterson CL, Wang MD (2006) Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat Struct Mol Biol 13:549–554. [DOI] [PubMed] [Google Scholar]
- 32. Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K (2011) Structure and Scm3‐mediated assembly of budding yeast centromeric nucleosomes. Nat Commun 2:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Widom J (2001) Role of DNA sequence in nucleosome stability and dynamics. Q Rev Biophys 34:269–324. [DOI] [PubMed] [Google Scholar]
- 34. Chua EYD, Vasudevan D, Davey GE, Wu B, Davey CA (2012) The mechanics behind DNA sequence‐dependent properties of the nucleosome. Nucleic Acids Res 40:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tóth K, Bohm V, Sellmann C, Danner M, Hanne J, Berg M, Barz I, Gansen A, Langowski J (2013) Histone‐ and DNA sequence‐dependent stability of nucleosomes studied by single‐pair FRET. Cytometry A 83:839–846. [DOI] [PubMed] [Google Scholar]
- 36. Ngo TTM, Zhang Q, Zhou R, Yodh JG, Ha T (2015) Asymmetric unwrapping of nucleosomes under tension directed by DNA local flexibility. Cell 160:1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brukner I, Sánchez R, Suck D, Pongor S (1995) Trinucleotide models for DNA bending propensity: comparison of models based on DNaseI digestion and nucleosome packaging data. J Biomol Struct Dyn 13:309–317. [DOI] [PubMed] [Google Scholar]
- 38. Subramanian V, Fields PA, Boyer LA (2015) H2A.Z: a molecular rheostat for transcriptional control. F1000 Prime Rep 7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA (2008) H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135:649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G (2009) H3.3/H2A.Z double variant–containing nucleosomes mark ‘nucleosome‐free regions’ of active promoters and other reglatory regions. Nat Genet 41:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marques M, Laflamme L, Gervais AL, Gaudreau L (2010) Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics 5:267–272. [DOI] [PubMed] [Google Scholar]
- 42. Flaus A, Rencurel C, Ferreira H, Wiechens N, Owen‐Hughes T (2004) Sin mutations alter inherent nucleosome mobility. embo J 23:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suto RK, Clarkson MJ, Tremethick DJ, Luger K (2000) Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol 7:1121–1124. [DOI] [PubMed] [Google Scholar]
- 44. Vogler C, Huber C, Waldmann T, Ettig R, Braun L, Izzo A, Daujat S, Chassignet I, Lopez‐Contreras AJ, Fernandez‐Capetillo O, Dundr M, Rippe K, Langst G, Schneider R (2010) Histone H2A C‐terminus regulates chromatin dynamics, remodeling, and histone H1 binding. PLoS Genet 6:e1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL (2005) Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci USA 102:18385–18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zlatanova J, Thakar A (2008) H2A.Z: view from the top. Structure 16:166–179. [DOI] [PubMed] [Google Scholar]
- 47. Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen‐Hughes T, van Noort J, Rhodes D, Chin JW (2009) A method for genetically installing site‐specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell 36:153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausió J (2001) Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J Biol Chem 276:41945–41949. [DOI] [PubMed] [Google Scholar]
- 49. Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ (2002) The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol 9:172. [DOI] [PubMed] [Google Scholar]
- 50. Meneghini MD, Wu M, Madhani HD (2003) Conserved histone variant H2A.Z protects Euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725–736. [DOI] [PubMed] [Google Scholar]
- 51. Anderson J, Widom J (2000) Sequence and position‐dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol 296:979–987. [DOI] [PubMed] [Google Scholar]
- 52. Kornberg RD, Lorch Y (1999) Twenty‐five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285–294. [DOI] [PubMed] [Google Scholar]
- 53. Bondarenko VA, Steele LM, Ujvári A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. (2006) Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell 24:469–479. [DOI] [PubMed] [Google Scholar]
- 54. Weber CM, Ramachandran S, Henikoff S (2014) Nucleosomes are context‐specific, H2A.Z‐modulated barriers to RNA polymerase. Mol Cell 53:819–830. [DOI] [PubMed] [Google Scholar]
- 55. Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M (2005) Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell 18:97–108. [DOI] [PubMed] [Google Scholar]
- 56. Depken M, Galburt EA, Grill SW (2009) The origin of short transcriptional pauses. Biophys J 96:2189–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Silva D‐A, Weiss DR, Avila FP, Da L‐T, Levitt M, Wang D, Huang X (2014) Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc Natl Acad Sci USA 111:7665–7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Santisteban MS, Kalashnikova T, Smith MM (2000) Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell 103:411–422. [DOI] [PubMed] [Google Scholar]


