ABSTRACT
Transforming growth factor β1 (TGF-β1) is a master cytokine in many biological processes, including tissue homeostasis, epithelial-to-mesenchymal transition, and wound repair. Here, we report that four and a half LIM-only protein 2 (FHL2) is a critical regulator of TGF-β1 expression. Devoid of a DNA-binding domain, FHL2 is a transcriptional cofactor that plays the role of coactivator or corepressor, depending on the cell and promoter contexts. We detected association of FHL2 with the TGF-β1 promoter, which showed higher activity in Fhl2−/− cells than in wild-type (WT) cells in a reporter assay. Overexpression of FHL2 abrogates the activation of the TGF-β1 promoter, whereas the upregulation of TGF-β1 gene transcription correlates with reduced occupancy of FHL2 on the promoter. Moreover, ablation of FHL2 facilitates recruitment of RNA polymerase II on the TGF-β1 promoter, suggesting that FHL2 may be involved in chromatin remodeling in the control of TGF-β1 gene transcription. Enhanced expression of TGF-β1 mRNA and cytokine was evidenced in the livers of Fhl2−/− mice. We tested the in vivo impact of Fhl2 loss on hepatic fibrogenesis that involves TGF-β1 activation. Fhl2−/− mice developed more severe fibrosis than their WT counterparts. These results demonstrate the repressive function of FHL2 on TGF-β1 expression and contribute to the understanding of the TGF-β-mediated fibrogenic response.
KEYWORDS: FHL2, TGF-β1, transcription regulation, fibrosis
INTRODUCTION
The transforming growth factor β (TGF-β) family is a multifunctional regulator of cell differentiation, immunity, morphogenesis, tissue homeostasis, and repair (1). TGF-β can be released from immune cells, epithelial cells, endothelial cells, and fibroblasts. Binding of TGF-β to their receptors activates Smad transcription factors that mediate transcription of TGF-β target genes. Among the three mammalian TGF-β isoforms that have distinct biological activities, TGF-β1 is the strongest inducer of fibrosis. In response to injury, TGF-β1 is critical for the activation of fibrogenic myofibroblasts, which express α smooth muscle actin (α-SMA) and secrete extracellular matrix proteins, mostly fibronectin and collagen types I and III. The TGF-β1 promoter contains AP1 and Sp1 transcription factor regulatory elements and responds positively to the TGF-β1 cytokine and the Ras oncoprotein (2–4).
Four and a half LIM-only protein 2 (FHL2) is involved in the regulation of various signaling pathways through protein-protein interaction. Thanks to its unique structure composed exclusively of four and a half LIM domains (for lin 11, isl-1, and mec-3), FHL2 has been identified in diverse protein complexes of multiple signaling pathways. In the cytoplasm, FHL2 interacts with integrins and focal adhesion kinases and plays a role in transmission of extracellular matrix (ECM)/integrin receptor-mediated signals (5–7). FHL2 binds to tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) and mediates the NF-κB signaling pathway (8). In the nucleus, FHL2 interacts with a diverse group of DNA-binding factors to control a broad range of transcription programs, including β-catenin/TCF, AP1, and androgen receptor (9–12).
Accumulating evidence indicates that the effects of FHL2 in the cell are different, even opposite, depending on the cell type and environment. In osteoclasts, FHL2 inhibits TRAF6-induced NF-κB activation (8), whereas FHL2 potently enhances NF-κB activity in epithelial cells (13). Similarly, interaction of FHL2 with β-catenin in myoblasts results in inhibition of β-catenin-mediated activation of a TCF/LEF-dependent reporter gene (10), whereas the same reporter was strongly activated by FHL2 in epithelial cancer cell lines (12). In vivo experience showed that injection of FHL2 into Xenopus embryos suppressed β-catenin-induced axis duplication (10), while enhanced expression of FHL2 in hepatocytes promoted liver tumorigenesis by activation of Wnt/β-catenin signaling (14).
FHL2 is intimately linked to the TGF-β signaling pathway at different levels. FHL2 acts as a coactivator of Smad proteins, including Smad2, Smad3, and Smad4, and plays a role in stabilizing the E3 ligase Arkadia to enhance TGF-β signaling (15, 16). Moreover, FHL2 is a target of TGF-β, responding positively to TGF-β stimulation in a Smad4-independent manner (17, 18). In human melanoma and colon cancer, enhanced expression of FHL2 is correlated with an increase of TGF-β, which predicts invasion, metastasis, and poor survival (18–20).
Here, we report a new aspect of the relationship between FHL2 and TGF-β in which deficiency of FHL2 upregulates TGF-β1 expression. In Fhl2−/− mice, enhanced TGF-β1 expression results in severe fibrosis in experimental models of liver fibrogenesis.
RESULTS
Deficiency of FHL2 increases TGF-β1 expression.
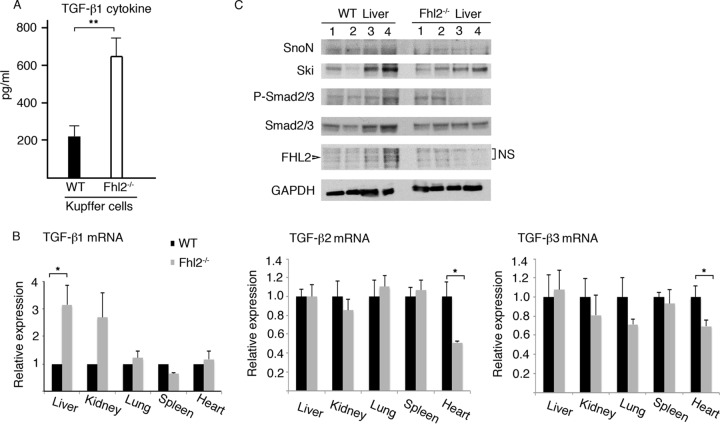
Previous studies have pointed to an important role of FHL2, in cooperation with several components of the TGF-β signaling pathway, in enhancing TGF-β signaling (15, 16). To investigate the effects of FHL2 on the expression of TGF-β, we examined TGF-β1 cytokine production in liver-resident macrophage Kupffer cells derived from Fhl2−/− animals, because Kupffer cells are the major source of TGF-β1, which is the most important member of the TGF-β family in the liver (21). We previously reported that FHL2 is expressed in Kupffer cells (13). We isolated Kupffer cells from WT and Fhl2−/− animals and cultured them in vitro for 24 h. The active form of the TGF-β1 cytokine in the supernatant was measured by enzyme-linked immunosorbent assay (ELISA). Remarkably, Fhl2−/− Kupffer cells expressed significantly higher levels of TGF-β1 than WT cells (Fig. 1A). To assess if the upregulation of TGF-β1 expression is at the transcriptional level and to have a broad view of the effect of FHL2 deficiency on the expression of three TGF-β isoforms, we extracted RNA from different tissues, including liver, kidney, lung, spleen, and heart, derived from Fhl2−/− and WT mice and examined TGF-β1, TGF-β2, and TGF-β3 mRNAs by quantitative PCR (qPCR). As shown in Fig. 1B, transcription of the TGF-β1 gene was significantly upregulated in the liver but not in the other organs from Fhl2−/− mice. Expression of TGF-β2 and TGF-β3 showed no difference in all the organs analyzed except the heart, where the mRNA levels of TGF-β2 and TGF-β3 were decreased in Fhl2−/− mice. We then examined whether FHL2 deficiency could have an impact on the expression of TGF-β targets in the liver, where FHL2 is weakly expressed in hepatocytes (14). TGF-β induces phosphorylation of Smad2/3 and degradation of the transcription repressors Ski and SnoN (22). Protein lysates prepared from WT and Fhl2−/− livers were analyzed for expression of Smad2/3, SnoN, and Ski and phosphorylation of Smad2/3. Besides individual heterogeneity, no significant difference was observed for P-Smad2/3, SnoN, and Ski between WT and Fhl2−/− mice (Fig. 1C).
FIG 1.
Loss of FHL2 enhances TGF-β1 expression in the liver. (A) TGF-β1 cytokine in Kupffer cells isolated from WT and Fhl2−/− animals, as evaluated by ELISA. **, P < 0.005. The data are the means and standard deviations (SD) obtained from five animals in each genotype. (B) Analysis of TGF-β1, TGF-β2, and TGF-β3 mRNAs by real-time reverse transcription (RT)-PCR in the indicated tissues from WT and Fhl2−/− mice. The values for the WT tissues were arbitrarily set as 1. The averages and SD values for three independent experiments are shown. *, P < 0.05. (C) Analysis of expression of SnoN, Ski, and Smad2/3 and phosphorylation of Smad2/3 in the livers of four WT and four Fhl2−/− animals by immunoblotting. NS, nonspecific.
FHL2 is present in transcriptional complexes on the TGF-β1 promoter.
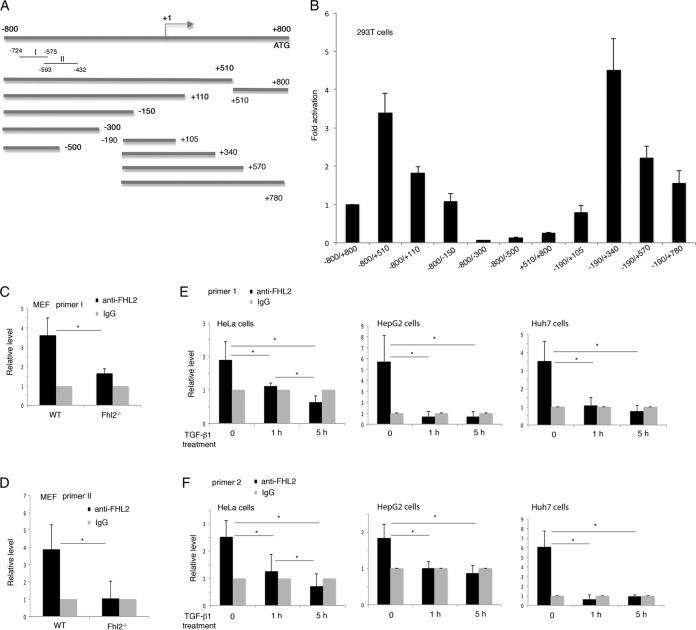
We focused the study on the liver, where FHL2 deletion appeared to create a condition that specifically promotes TGF-β1 expression at the transcriptional level. To explore how FHL2 is involved in TGF-β1 regulation, we first synthesized the genomic fragment comprising 800 bp upstream and 800 bp downstream of the transcription start site of the mouse TGF-β1 gene and constructed a series of deletion fragments in the firefly luciferase reporter vector for fine mapping of the region containing promoter activity (Fig. 2A). The fusion fragments were transiently transfected into 293T cells, and luciferase activities were determined after normalization to the expression of a control reporter. As shown in Fig. 2B, the fragment −190 to +340 displayed the highest level of luciferase activity, which is in agreement with a previous report (2). Potential binding sites of numerous transcription factors were identified in the sequence encompassing −800 to +800 of the TGF-β1 gene using the program PROMO (23; see File S1 in the supplemental material). Notably, sequences recognized by the AP-1 complex components c-Jun and c-Fos are localized in multiple regions of the promoter. AP1 binding sites were also identified in the human TGF-β1 promoter (24).
FIG 2.
FHL2 is associated with the TGF-β1 promoter. (A) Schematic representation of murine TGF-β1 promoter constructs in the luciferase reporter assay. (B) Reporter assay in 293T cells. The activity of the −800/+800 reporter was arbitrarily set at 1. The data are presented as mean induction in luciferase activity plus SD from duplicate samples. The results shown are representative of those from more than three independent assays. (C and D) Association of FHL2 with the murine TGF-β1 promoter. WT and Fhl2−/− MEFs were analyzed by ChIP-qPCR using chromatin immunoprecipitated by anti-FHL2 antibody or IgG with the primers I (C) and II (D), which are indicated in panel A. (E and F) Association of FHL2 with the human TGF-β1 promoter. HeLa, HepG2, and Huh7 cells were treated with SB431542 to inhibit the effects of endogenous TGF-β, followed by induction for 1 h and 5 h with human TGF-β1 (2 ng/ml), before being subjected to ChIP-qPCR with two independent primers (primer 1 [E] and primer 2 [F]) specific to the human TGF-β1 promoter. *, P < 0.05. The data presented are the means and SD obtained from three independent experiments.
To determine whether the TGF-β1 gene is a direct transcriptional target of FHL2, we carried out chromatin immunoprecipitation (ChIP)-qPCR assays to examine the potential association of FHL2 with the TGF-β1 promoter. Lacking a DNA-binding domain, FHL2 could be part of transcription regulatory complexes linking to the promoter through transcription factors. For the ChIP assay, wild-type (WT) MEFs were cross-linked with formaldehyde, and protein-DNA complexes were immunoprecipitated with anti-FHL2 antibody or control IgG. Fhl2−/− mouse embryonic fibroblasts (MEFs) were used as negative-control cells. Analysis by qPCR of immunoprecipitated chromatin with two pairs of primers (Fig. 2A) (see Materials and Methods) revealed the association of FHL2 with the TGF-β1 promoter (Fig. 2C and D). To prove that FHL2 is also linked to the human TGF-β1 promoter, which shares 66% nucleotide identify with the murine TGF-β1 promoter (2), we performed ChIP-qPCR in HeLa cells and two hepatic cell lines, HepG2 and Huh7. Using chromatin immunoprecipitated by FHL2 antibody as the template, quantitative-PCR analysis with two pairs of primers specific to the human TGF-β1 promoter successfully amplified the promoter region (Fig. 2E and F). Interestingly, when cells were treated with the TGF-β1 cytokine, which is known to induce TGF-β1 gene transcription (4), FHL2 occupancy on the promoter of TGF-β1 was significantly reduced (Fig. 2E and F), suggesting that FHL2 exerts a repressive effect on the promoter of TGF-β1. These results indicate that the TGF-β1 promoter is a direct target of FHL2.
FHL2 negatively regulates the TGF-β1 promoter.
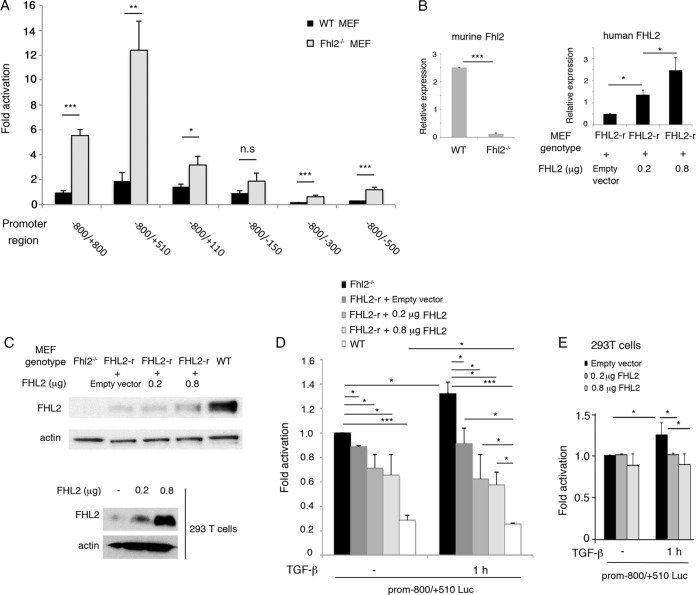
The upregulated expression of TGF-β1 in Fhl2-deficient cells implied a repressor effect of FHL2 on the TGF-β1 promoter. To test this, we carried out a TGF-β1 luciferase reporter assay in WT or Fhl2−/− MEFs. Remarkably, the activity of all the reporters except −800/−150 was significantly increased in Fhl2−/− MEFs compared to WT MEFs (Fig. 3A). To further link the suppressive effect to FHL2, we restored FHL2 expression in Fhl2−/− MEFs with human FHL2 (FHL2-r) (Fig. 3B and C). We tested the activity of the −800/+510 reporter in Fhl2−/−, FHL2-r, and WT MEFs treated with the TGF-β cytokine. As shown in Fig. 3D, the reporter activity was inversely correlated with the levels of FHL2 expression in Fhl2−/−, FHL2-r, and WT cells in the presence and absence of the TGF-β cytokine, indicating that the repressive effect of FHL2 on the TGF-β1 promoter is dose dependent. However, this dose dependency of FHL2 was not linear, as transfection of 0.2 μg and 0.8 μg of plasmid expressing FHL2 in FHL2-r cells (Fig. 3B and C) did not further repress the promoter activity (Fig. 3D). On the other hand, the promoter activity in FHL2-r cells was not significantly augmented by TGF-β cytokine treatment compared to untreated FHL2-r cells, while it was even higher in untreated WT MEFs than in treated WT MEFs (Fig. 3D). The TGF-β cytokine acts on diverse regulatory elements of the TGF-β1 promoter, of which FHL2 is a part. The TGF-β cytokine can inhibit FHL2 association with the TGF-β1 promoter, thus relieving the repressive function of FHL2 (Fig. 2E and F). The ultimate activity of the TGF-β1 promoter under the stimulation of the TGF-β cytokine, however, depends on the sum of the actions of the diverse regulatory elements, to which FHL2 makes a contribution.
FIG 3.
FHL2 is a negative regulator of the TGF-β1 promoter. (A) Luciferase reporter assay in WT and Fhl2−/− MEFs. The activity of the −800/+800 reporter in WT MEFs was arbitrarily set at 1. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; n.s., nonsignificant. (B) Analysis of FHL2 gene transcription by qPCR. Primers to detect murine Fhl2 were used in WT and Fhl2−/− MEFs. To restore FHL2 expression in Fhl2−/− MEFs, human FHL2 was stably expressed in Fhl2−/− MEFs (FHL2-r). Transient transfection of 0.2 μg or 0.8 μg of human FHL2 was carried out in FHL2-r cells. Human FHL2 primers were used to analyze mRNA in FHL2-r-related cells. (C) Analysis of FHL2 protein in MEFs and 293T cells with anti-FHL2 antibody reactive to both human and mouse FHL2 proteins. (D) The TGF-β1 promoter is negatively regulated by FHL2. The −800/+510 reporter was transfected into MEFs expressing different doses of FHL2. The cells were treated with 10 μM SB431542 to inhibit the effects of endogenous TGF-β, followed by induction for 1 h with human TGF-β1 (2 ng/ml), before being subjected to luciferase assay. (E) Reporter assay in 293T cells. 293T cells were cotransfected with 0.2 μg or 0.8 μg of plasmid expressing FHL2 and the murine TGF-β1 −800/+510 promoter reporter; 36 h later, the cells were treated with 10 μM SB431542, followed by induction for 1 h with human TGF-β1, before being subjected to luciferase assay. The data presented are the means and SD obtained from three independent experiments.
Next, we examined the effects of FHL2 on the TGF-β1 promoter in the 293T cell line. When 293T cells were subjected to treatment with the TGF-β cytokine, the activity of the −800/+510 reporter was modestly but significantly activated (Fig. 3E). This activation was blunted by overexpression of FHL2 (Fig. 3C and E). Overexpression of human FHL2 did not have an effect on the promoter activity without TGF-β1 stimulation (Fig. 3E), suggesting that under steady-state conditions, endogenous FHL2 may not be the limiting factor. Taken together, these results indicate that FHL2 is a negative regulator of the TGF-β1 promoter in both murine and human cells.
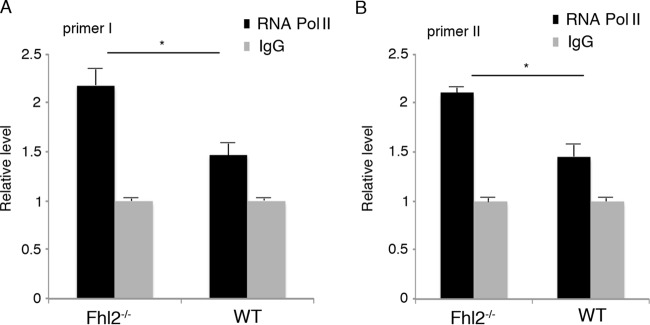
To further decipher the mechanisms of the negative regulation of the TGF-β1 promoter by FHL2, we used WT and Fhl2−/− MEFs to carry out ChIP-qPCR assays with antibody against RNA polymerase II (Pol II), which transcribes all the protein-coding genes. The primers I and II were employed for qPCR to amplify the endogenous TGF-β1 promoter, using chromatin immunoprecipitated with anti-Pol II antibody as the template (Fig. 2A). The recruitment of Pol II to the TGF-β1 promoter was significantly increased in Fhl2−/− MEFs in comparison to WT cells (Fig. 4). These data suggest that loss of FHL2 may alter the chromatin architecture to allow Pol II to gain access to the TGF-β1 promoter.
FIG 4.
Loss of FHL2 enhances the recruitment of RNA Pol II. Chromatin immunoprecipitated with anti-Pol II antibody or IgG was amplified with either primer I (A) or primer II (B), which are indicated in Fig. 2A. The value of amplification in chromatin immunoprecipitated with IgG was arbitrarily set at 1. *, P < 0.05. The data presented are the means and SD obtained from three independent experiments.
Fhl2−/− mice show increased susceptibility to fibrogenesis in the liver.
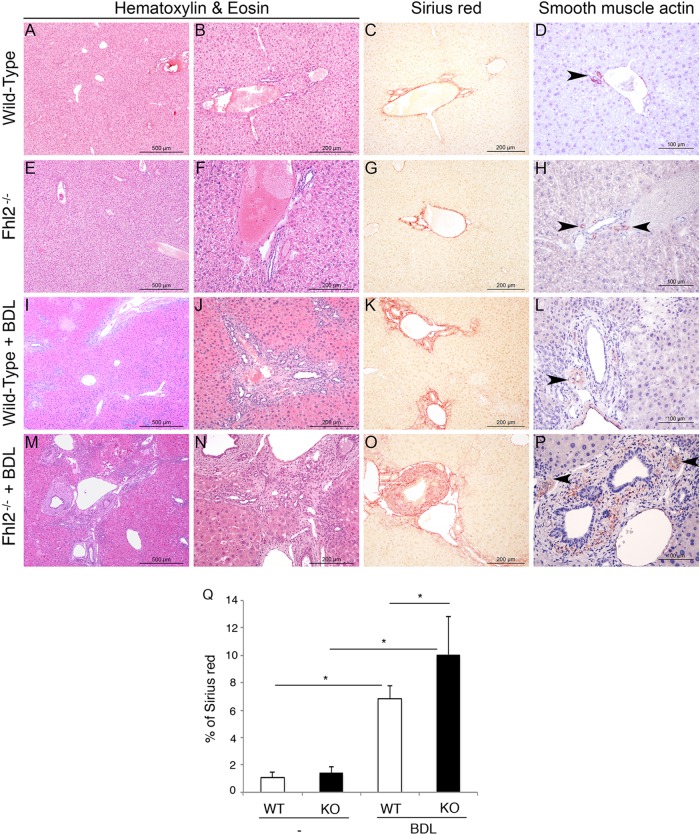
TGF-β1 is considered the most powerful mediator of fibrogenesis. The high expression level of TGF-β1 in Fhl2−/− liver suggested that Fhl2−/− mice might be profibrogenic. We subjected Fhl2−/− and WT mice to a bile duct ligation (BDL) operation, which is a widely used model for liver fibrosis through cholestatic liver injury (25). We sacrificed the mice 15 days after BDL. Livers derived from BDL-treated mice lacking FHL2 developed severe fibrosis compared to those of WT animals, showing wider fibrotic scars and substantially more scar branching, as assessed by staining with hematoxylin-eosin (H&E) and Sirius red, which directly marks the extracellular matrix deposited by activated hepatic stellate cells (HSCs) (Fig. 5A to C, E to G, I to K, M to O, and Q). Moreover, FHL2 knockout (KO) livers harbored a large increase in activated HSCs, as determined by α-SMA staining (Fig. 5D, H, L, and P). We further confirmed our findings in a second well-established model of liver fibrosis induced by thioacetamide (TAA) administration. Fhl2−/− mice showed increased fibrosis after 20 weeks of TAA treatment (data not shown). These results are in agreement with a previous report showing enhanced fibrosis in Fhl2−/− mice using a carbon tetrachloride (CCl4)-induced model of liver fibrosis (26).
FIG 5.
Enhanced hepatic fibrosis in Fhl2−/− mice. Fhl2−/− mice (n = 7) and WT mice (n = 6) underwent BDL. H&E staining of liver parenchyma was performed with control WT and Fhl2−/− mice (A, B, E, and F) or mice subjected to BDL (I, J, M, and N). Hepatic fibrosis was assessed by Sirius red staining (C, G, K, and O). Quantification is shown in panel Q. Expression of α-SMA was determined by immunohistochemistry (D, H, L, and P). The data presented are the means and SD obtained from at least five mice in each group. *, P < 0.05.
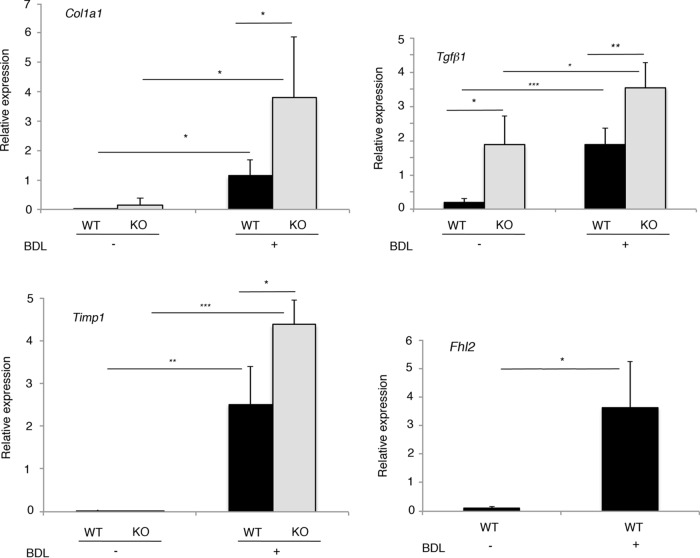
To investigate the molecular characteristics of fibrosis developed in Fhl2−/− liver, we examined by qRT-PCR expression of fibrogenesis markers, including alpha-1 type I collagen, tissue inhibitor of metalloproteinases 1 (TIMP1), and TGF-β1 encoded, respectively, by the Col1a1, Timp1, and Tgfb1 genes, in the livers of Fhl2−/− and WT mice 15 days after BDL or in control mice. This analysis revealed that expression of Col1a1 and Timp1 was activated after BDL in both Fhl2−/− and WT mice (Fig. 6). Moreover, the levels of Col1a1 and Timp1 activation following BDL were significantly higher in Fhl2−/− liver than in WT liver, correlating with the aggravated fibrosis in Fhl2−/− mice (Fig. 6).
FIG 6.
Expression of Col1a1, Tgfb1, TIMP1, and Fhl2 mRNAs in the livers of WT and Fhl2−/− mice after BDL. Hepatic levels of Col1a1, Tgfb1, TIMP1, and Fhl2 mRNAs were examined by qPCR in control WT and Fhl2−/− mice and those operated on. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. The data presented are the means and SD obtained from at least five mice in each group.
In accordance with the results shown in Fig. 1B, the mRNA level of TGF-β1 in the liver was higher in Fhl2−/− mice than in WT mice under steady-state conditions (Fig. 6). Transcription of the TGF-β1 gene was significantly enhanced in fibrotic liver compared to control liver in both Fhl2−/− and WT mice (Fig. 6). Moreover, livers from Fhl2−/− animals displayed higher levels of TGF-β1 mRNA following BDL treatment than those of their WT counterparts (Fig. 6), demonstrating that fibrogenic signaling was enhanced in the livers from Fhl2−/− animals. Of note, FHL2 expression was upregulated in fibrotic livers of WT mice (Fig. 6), which was correlated with previous observations in human liver fibrosis (14, 26). Therefore, ablation of FHL2 leads to an increased level of TGF-β1 and enhanced susceptibility to fibrogenesis.
FHL2 deficiency has no effect on renal fibrosis.
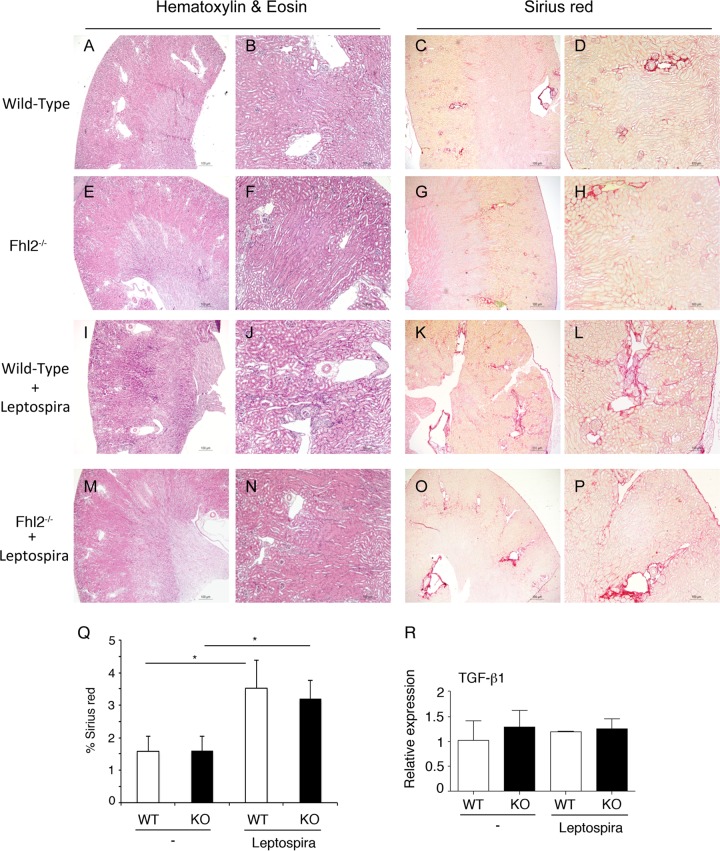
A previous report showed that Fhl2−/− mice developed stronger fibrotic alterations in a bleomycin-induced model of lung fibrosis following an unresolved inflammation (27). To test if FHL2 deficiency had a broad effect on fibrogenesis, we chose to examine fibrogenesis in the kidney, as FHL2 plays roles in the development of kidney disease (28). We used the Leptospira bacterial infection model of renal fibrosis (29) and infected age- and sex-matched WT and Fhl2−/− mice with equal numbers of Leptospira bacteria expressing luciferase. All the mice were successfully infected in the kidney, as attested by in vivo imaging (data not shown). After 21 days, the kidneys were examined for fibrosis using Sirius red staining and expression of TGF-β1. As shown in Fig. 7A to P, Leptospira bacteria induced fibrosis in the kidney in both WT and Fhl2−/− mice, but no significant difference in the accumulation of fibrotic tissue was observed between WT and Fhl2−/− animals (Fig. 7Q). Analysis of TGF-β1 expression by qPCR showed no change in mRNA after bacterial infection between WT and Fhl2−/− mice (Fig. 7R). These results attest to the tissue-dependent features in TGF-β1 activation and fibrogenesis associated with FHL2 deficiency.
FIG 7.
No effect of FHL2 deficiency on renal fibrosis. Kidneys were derived from naive or Leptospira-infected WT and Fhl2−/− mice. Light microscopy of kidneys stained with H&E (A, B, E, F, I, J, M, and N) and collagen deposition stained with Sirius red (C, D, G, H, K, L, O, and P) is shown. Original magnifications, ×40 (A, C, E, G, I, K, M, and O) and ×100 (B, D, F, H, J, L, N, and P). (Q) Sirius red quantification. (R) TGF-β1 mRNA was analyzed by qPCR in kidneys from naive or Leptospira-infected WT and Fhl2−/− mice. The data presented are the means and SD obtained from five mice in each group. *, P < 0.05.
DISCUSSION
Given the importance of TGF-β1 in many biological processes, it is vital to understand the mechanisms controlling its expression. In this report, we show that FHL2 acts as an important negative regulator of TGF-β1 gene transcription. The action of FHL2 in inhibiting TGF-β1 gene transcription is likely to involve both direct and indirect mechanisms.
Previous studies demonstrated that the effects of FHL2 on transcription can be positive or negative depending on the gene target and the cellular context (10, 30). Associated with the TGF-β1 promoter, FHL2 plays the role of corepressor in the transcription of the TGF-β1 gene. We show that overexpression of FHL2 inhibits the activity of the TGF-β1 promoter, whereas ablation of FHL2 increases TGF-β1 gene transcription. The inhibition of the TGF-β1 promoter by FHL2 is dose dependent, suggesting that the repressive activity could be intrinsic to FHL2 or FHL2-associated factors with repressive functions. The transcription factors with which FHL2 interacts on the TGF-β1 promoter remain to be determined. Our search of transcription factor binding sites on the TGF-β1 promoter has identified multiple responsive elements recognized by Fos and Jun, which are known FHL2-associated transcription factors (11). FHL2 could act through Jun and Fos to inhibit the transcription of the TGF-β1 gene. In the face of TGF-β1 cytokine treatment, which activates the TGF-β1 promoter, transient overexpression of FHL2 can successfully abrogate the activation, correlating with the repressive effects of FHL2 on TGF-β1 gene transcription. Under physiological conditions, TGF-β1-induced activation of the TGF-β1 promoter depends on a multitude of signaling pathways through the binding of signal-responsive activators and repressors, including FHL2, which also positively responds to the TGF-β1 cytokine (17, 18). The feedback loop of FHL2 and TGF-β1 may constitute one of the complex transcriptional regulation networks that determine the level and the outcome of TGF-β1 in cells. It is not clear how TGF-β1 induces hepatic fibrosis in Fhl2−/− mice. Our observations that expression of TGF-β targets Ski, SnoN, and phosphorylation of Smad2/3 were not altered in the liver deficient for FHL2, suggesting that the canonical TGF-β signaling may not be involved in this process. It remains to be determined if the profibrotic action of TGF-β1 in Fhl2−/− liver operates through noncanonical TGF-β signaling.
Our finding that loss of FHL2 resulted in increased recruitment of RNA polymerase II on the TGF-β1 promoter implies a novel role of FHL2 in regulation of gene transcription. The question of how FHL2 is involved in the dynamic process of genome packaging to allow or restrict transcriptional regulator access to the regulatory sequences remains to be addressed. Composed of multiple LIM domains for protein-protein interaction, FHL2 binds with several chromatin modifiers, including the histone acetyltransferase CBP/p300 (31), the histone ubiquitin ligase Brca1 (32), and the histone demethylase LSD1 (33), which actively participate in covalently modifying nucleosomes. It is possible that FHL2 could collaborate with these enzymes to maintain a repressed architecture by virtue of the restricted chromatin environment for transcription regulators. Ablation of FHL2 may tailor the way in which the chromatin is packaged to increase the access of RNA polymerase II to the TGF-β1 promoter.
Correlating with enhanced expression of TGF-β1, the fibrotic response in Fhl2−/− mice was dramatic in the liver. A previous report showed aggravated liver fibrosis after carbon tetrachloride injection in Fhl2−/− mice, but the authors failed to detect increased expression of TGF-β1 mRNA under steady-state conditions, as reported here (26). The reason for this difference is unclear but may reflect different sensitivities in the detection of TGF-β1 gene transcription. However, Huss et al. revealed a higher level of TGF-β1 in fibrotic tissues from Fhl2−/− mice than in those from WT mice, consistent with our findings that activation of TGF-β1 gene transcription is stronger in the absence of FHL2. A previous report pointed to a higher fibrosis score in the lungs of Fhl2−/− mice following bleomycin administration (27). It showed that loss of FHL2 resulted in increased expression of the proinflammatory matrix protein tenascin C and downregulation of the macrophage-activating C-type lectin receptor DC-SIGN. This unresolved inflammation in Fhl2−/− mice was the major driver of increased pulmonary fibrosis (27). In all organ systems, the wound repair process is initiated by inflammation, followed by proliferation and remodeling. It is thus highly likely that FHL2 interferes with distinct processes in lung and liver fibrosis. Our observations that loss of FHL2 had no drastic effect on renal fibrosis further underscore the tissue specificity of FHL2 actions.
The effects of FHL2 loss on fibrosis stress the medical relevance of the repressive activity of FHL2 on the TGF-β1 promoter. However, we and others previously reported that FHL2 was upregulated in human liver fibrotic tissues and that a higher level of FHL2 was associated with advanced fibrosis (14, 26). This correlates with our finding of FHL2 activation in liver fibrotic tissues of WT mice (Fig. 6). In line with this observation, in human melanoma and colon cancer, enhanced expression of FHL2 is correlated with an increase of TGF-β in tumors, with invasion, metastasis, and poor survival prognosis (18–20). This multifaceted nature of the relationship between FHL2 and TGF-β1 further supports the notion that the cellular context and the expression level of each protein dictate the process and the outcome of the complex signal transduction. Our data described here contribute to unveiling new facets of TGF-β regulation and should facilitate additional investigations aimed at further exploring TGF-β biology and dysfunctions.
MATERIALS AND METHODS
Animals and fibrosis induction.
Fhl2−/− mice on a Black Swiss–129-SV/J background (34) were backcrossed into C57BL/6J mice over more than eight generations. All the animals were housed in a pathogen-free environment at the Institut Pasteur animal facility and received humane care according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals.
Animal experiments were carried out according to the European Commission directives for animal experimentation (decree 2001-131; Journal Officiel, 6 February 2001). BDL was performed on 6 WT and 7 Fhl2−/− mice (8 to 12 weeks old) by certified researchers. The mice were sacrificed 15 days postoperation. The livers were removed and either fixed in 4% paraformaldehyde (PFA) for staining or frozen for RNA extraction. Bioluminescent Leptospira interrogans bacteria (107) were used to infect five WT and five Fhl2−/− 8-week-old mice by intraperitoneal (i.p.) injection in 200 μl of phosphate-buffered saline (PBS) as described previously (29). The mice were sacrificed 21 days postinfection. These experiments were repeated twice. Kidneys from infected and naive mice were removed and fixed in 4% PFA for staining. The experimental procedures were approved by the Institut Pasteur ethics committee (number 2013-0034).
RNA analysis.
Total RNA was extracted from liver, lung, heart, spleen, and kidney from 10 Fhl2−/− and 10 WT mice using TRIzol (Invitrogen) after tissue homogenization. One microgram of RNA was reverse transcribed using random primers (Promega), and cDNA was generated using RevertAid H Minus reverse transcriptase (Thermo Fisher Scientific). The relative levels of mRNAs were determined by real-time PCR using Fast Start Universal SYBR green (Roche). The mean copy number of each gene from a triplicate determination was normalized to the mean copy number of hypoxanthine-guanine phosphoribosyltransferase (HPRT). The following primers were used for real-time PCR: human FHL2, 5′-GGCTGAGAACTGTGTCTTCC-3′ and 5′-ACAAGTCCTTGCAGTCACAG-3′; murine FHL2, 5′-GTCCTACAAGGATCGGCACT-3′ and 5′-ACAGGTGAAGCAGGTCTCGT-3′; HPRT, 5′-ATGCCGAGGATTTGGAAAAA-3′ and 5′-ACAATGTGATGGCCTCCCA-3′; murine TGF-β1, 5′-GCGTATCAGTGGGGGTCA-3′ and 5′-GTCAGACATTCGGGAAGCAG-3′; murine TGF-β2, 5′-GCAGATCCTGAGCAAGCTG-3′ and 5′-GTAGGGTCTGTAGAAAGTGG-3′; murine TGF-β3, 5′-AGCGCAGACACAACCCATAG-3′ and 5′-ACACGACTTCACCACCATGT-3′; murine Col1a1, 5′-GACCAGGAGGACCAGGAAGT-3′ and 5′-GAAACCCGAGGTATGCTTGA-3′; and murine Timp1, 5′-GTCACTCTCCAGTTTGCAAG-3′ and 5′-GACCACCTTATACCAGCGTT-3′.
Cytokine assay.
Kupffer cells were isolated from WT and Fhl2−/− animals as described previously (35), and 106 cells were plated in RPMI medium supplemented with 10% fetal calf serum (FCS) and 1% penicillin-streptomycin for 24 h. The active form of TGF-β1 was measured in supernatants by ELISA (R&D Systems, Minneapolis, MN) as specified by the manufacturer.
Cell culture, reagents, and reporter assay.
293T cells, HeLa cells, MEFs, and HepG2 and Huh7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS). The Fhl2−/− MEFs were described previously (36). The human FHL2 gene was cloned in the retroviral pBabe-puromycin vector. Phoenix ecotropic virus packaging cells were transfected with either pBabe-empty or pBabe-hFHL2, and virus-containing supernatant was collected. To restore FHL2 expression in Fhl2−/− MEFs, Fhl2−/− MEFs were transduced with virus-containing supernatant supplemented with 8 μg/ml Polybrene. Cells were selected with 4 μg/ml puromycin.
The 1.6-kb fragment encompassing 800 bp upstream and 800 bp downstream of the transcription initiation site of the murine TGF-β1 gene was synthesized by Eurofins MWG Operon. For reporter assays, 3 × 105 Fhl2−/− WT MEFs or 293T cells were plated in six-well plates and transfected using Lipofectamine 2000 (Invitrogen) with luciferase reporter containing the murine TGF-β1 promoter (0.05 μg). Luciferase activity was determined 48 h later using the Dual-Glo luciferase assay system (Promega). A Renilla plasmid under the control of thymidine kinase was used as the internal control, and the total DNA amount in each transfection was kept constant by adding empty-vector plasmid. All experiments were performed in duplicate and repeated at least three times.
To test the effects of FHL2 on the TGF-β1 promoter, MEFs and 293T cells were cotransfected with 0.2 μg or 0.8 μg of plasmid expressing human FHL2 or empty vector and the murine TGF-β1 −800/+510 promoter reporter. Thirty-six hours later, the cells were treated with 10 μM SB431542 (Sigma) for 16 h to inhibit the effects of endogenous TGF-β (37), followed by induction for 1 h with human TGF-β1 (2 ng/ml; PeproTech), and then collected for luciferase activity assay.
Putative transcription factor binding sites were searched in sequences 800 bp upstream and 800 bp downstream of the TGF-β1 gene transcription start site, using PROMO (23) with a dissimilarity of 10%. Transcription factor recognition sites with a length shorter than 5 nucleotides were not taken into account.
ChIP assay.
ChIP was performed as described previously (36). To investigate the association of FHL2 with the murine or human TGF-β1 promoter, MEFs or HeLa, HepG2, or Huh7 cells were used for ChIP assays with anti-FHL2 (MBL; c-76204). The HeLa, HepG2, or Huh7 cells were treated with 10 μM SB431542 for 16 h, followed by induction for 1 h or 5 h with human TGF-β1 (2 ng/ml), and then were subjected to ChIP-qPCR assay. Because the Tgfb1 promoter region contains highly GC-rich sequences, only 2 pairs of primers among 10 pairs of primers tested were able to amplify the murine TGF-β1 promoter by quantitative PCR (Fig. 2A). Their sequences are as follows: I, 5′-ATGGAGTGGAGTGTTGAGGG-3′ and 5′-CTTGCAGTCCATGGCATAGG-3′, and II, 5′-CTATGCCATGGACTGCAAGG-3′ and 5′-GAGGCACCTTACCCATGAGA-3′. Two pairs of primers were used to amplify the human TGF-β1 promoter by quantitative PCR: 1, 5′-GGCAGTTGGCGAGAACAGT-3′ and 5′-CTGGGGTCAGCTCTGACAGT-3′, and 2, 5′-TGGGAGGTGCTCAGTAAAGG-3′ and 5′-ACCCAGAACGGAAGGAGAGT-3′. To examine the effect of FHL2 on chromatin accessibility, Fhl2−/− and WT MEFs were used for ChIP assays with anti-RNA polymerase II antibodies (Santa Cruz Biotechnology; sc-9001X). Immunoprecipitated chromatin was amplified with the primers I and II by qPCR.
Immunohistochemistry and immunoblotting.
Immunohistochemistry (IHC) was performed on PFA-fixed paraffin-embedded sections as previously described (14). Briefly, tissues were dewaxed in xylene and unmasked in a citric acid solution. After blocking with normal horse serum, sections were incubated with primary antibody against α-SMA (Sigma; A5228). Endogenous peroxidase activity was blocked by incubating the sections with 3% hydrogen peroxide. The sections were then incubated with secondary antibodies (Vector Laboratories) for 30 min. The peroxidase reaction was developed using a DAB substrate kit (SK-4100; Vector Laboratories). A combination of hematoxylin and eosin was used for counterstaining. For Sirius red staining, after deparaffination and rehydration, tissue sections were labeled for 30 min with a solution of 0.1% (wt/vol) Sirius red in saturated picric acid. For fibrosis quantification, a Sirius red/fast green collagen-staining kit (Amsbio; 9046) was used.
To test the effects of FHL2 on the downstream effectors of TGF-β signaling and FHL2 expression in cells, protein lysates were prepared from liver tissues and MEFs and 293T cells as described previously (13). Protein extract was subjected to electrophoresis on NuPAGE 4 to 12% Bis-Tris gels (Invitrogen; NP0323BOX) in NuPAGE MOPS (morpholinepropanesulfonic acid) SDS running buffer (Thermo Fisher Scientific; NP000102). The following antibodies were used for immunoblotting analysis: Smad2/3 (Cell Signaling; 8685S), phosphorylated Smad2/3 (Cell Signaling; 8828S), Ski (Santa Cruz; sc-33693), SnoN (Santa Cruz; sc-9141), FHL2 and actin (Sigma; MA1-744), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Cell Signaling; 2118S).
Statistical analysis.
Statistical analysis was performed by Mann-Whitney U test.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sophie Lotersztajn and Fatima Teixeira-Clerc for insightful discussions. We are grateful to Jean-Marc Panaud for technical help.
This work was funded in part by the French Ligue contre le Cancer, Comité de Paris, the Fondation ARC pour la Recherche sur le Cancer (ARC), and the Institut National du Cancer (INCA). J.D. was supported by the French Ministère de l'Enseignement Supérieur et de la Recherche, Y.N. was supported by the Cancéropôle Ile-de-France, and T.X. was supported by Total Foundation.
We dedicate this paper to Minou Adib-Conquy.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00636-16.
REFERENCES
- 1.Massague J. 2012. TGFbeta signalling in context. Nat Rev Mol Cell Biol 13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geiser AG, Kim SJ, Roberts AB, Sporn MB. 1991. Characterization of the mouse transforming growth factor-beta 1 promoter and activation by the Ha-ras oncogene. Mol Cell Biol 11:84–92. doi: 10.1128/MCB.11.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim SJ, Glick A, Sporn MB, Roberts AB. 1989. Characterization of the promoter region of the human transforming growth factor-beta 1 gene. J Biol Chem 264:402–408. [PubMed] [Google Scholar]
- 4.Van Obberghen-Schilling E, Roche NS, Flanders KC, Sporn MB, Roberts AB. 1988. Transforming growth factor beta 1 positively regulates its own expression in normal and transformed cells. J Biol Chem 263:7741–7746. [PubMed] [Google Scholar]
- 5.Samson T, Smyth N, Janetzky S, Wendler O, Muller JM, Schule R, von der Mark H, von der Mark K, Wixler V. 2004. The LIM-only proteins FHL2 and FHL3 interact with alpha- and beta-subunits of the muscle alpha7beta1 integrin receptor. J Biol Chem 279:28641–28652. doi: 10.1074/jbc.M312894200. [DOI] [PubMed] [Google Scholar]
- 6.Park J, Will C, Martin B, Gullotti L, Friedrichs N, Buettner R, Schneider H, Ludwig S, Wixler V. 2008. Deficiency in the LIM-only protein FHL2 impairs assembly of extracellular matrix proteins. FASEB J 22:2508–2520. doi: 10.1096/fj.07-095521. [DOI] [PubMed] [Google Scholar]
- 7.Wixler V, Geerts D, Laplantine E, Westhoff D, Smyth N, Aumailley M, Sonnenberg A, Paulsson M. 2000. The LIM-only protein DRAL/FHL2 binds to the cytoplasmic domain of several alpha and beta integrin chains and is recruited to adhesion complexes. J Biol Chem 275:33669–33678. doi: 10.1074/jbc.M002519200. [DOI] [PubMed] [Google Scholar]
- 8.Bai S, Kitaura H, Zhao H, Chen J, Muller JM, Schule R, Darnay B, Novack DV, Ross FP, Teitelbaum SL. 2005. FHL2 inhibits the activated osteoclast in a TRAF6-dependent manner. J Clin Invest 115:2742–2751. doi: 10.1172/JCI24921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller JM, Isele U, Metzger E, Rempel A, Moser M, Pscherer A, Breyer T, Holubarsch C, Buettner R, Schule R. 2000. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J 19:359–369. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin B, Schneider R, Janetzky S, Waibler Z, Pandur P, Kuhl M, Behrens J, von der Mark K, Starzinski-Powitz A, Wixler V. 2002. The LIM-only protein FHL2 interacts with beta-catenin and promotes differentiation of mouse myoblasts. J Cell Biol 159:113–122. doi: 10.1083/jcb.200202075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morlon A, Sassone-Corsi P. 2003. The LIM-only protein FHL2 is a serum-inducible transcriptional coactivator of AP-1. Proc Natl Acad Sci U S A 100:3977–3982. doi: 10.1073/pnas.0735923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei Y, Renard CA, Labalette C, Wu Y, Levy L, Neuveut C, Prieur X, Flajolet M, Prigent S, Buendia MA. 2003. Identification of the LIM protein FHL2 as a coactivator of beta-catenin. J Biol Chem 278:5188–5194. doi: 10.1074/jbc.M207216200. [DOI] [PubMed] [Google Scholar]
- 13.Dahan J, Nouet Y, Jouvion G, Levillayer F, Adib-Conquy M, Cassard-Doulcier AM, Tebbi A, Blanc F, Remy L, Chen J, Cairo S, Werts C, Si-Tahar M, Tordjmann T, Buendia MA, Wei Y. 2013. LIM-only protein FHL2 activates NF-kappaB signaling in the control of liver regeneration and hepatocarcinogenesis. Mol Cell Biol 33:3299–3308. doi: 10.1128/MCB.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nouet Y, Dahan J, Labalette C, Levillayer F, Julien B, Jouvion G, Cairo S, Vives FL, Ribeiro A, Huerre M, Colnot S, Perret C, Nhieu JT, Tordjmann T, Buendia MA, Wei Y. 2012. The four and a half LIM-only protein 2 regulates liver homeostasis and contributes to carcinogenesis. J Hepatol 57:1029–1036. doi: 10.1016/j.jhep.2012.06.035. [DOI] [PubMed] [Google Scholar]
- 15.Ding L, Wang Z, Yan J, Yang X, Liu A, Qiu W, Zhu J, Han J, Zhang H, Lin J, Cheng L, Qin X, Niu C, Yuan B, Wang X, Zhu C, Zhou Y, Li J, Song H, Huang C, Ye Q. 2009. Human four-and-a-half LIM family members suppress tumor cell growth through a TGF-beta-like signaling pathway. J Clin Invest 119:349–361. doi: 10.1172/JCI35930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia T, Levy L, Levillayer F, Jia B, Li G, Neuveut C, Buendia MA, Lan K, Wei Y. 2013. The four and a half LIM-only protein 2 (FHL2) activates transforming growth factor beta (TGF-beta) signaling by regulating ubiquitination of the E3 ligase Arkadia. J Biol Chem 288:1785–1794. doi: 10.1074/jbc.M112.439760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy L, Hill CS. 2005. Smad4 dependency defines two classes of transforming growth factor {beta} (TGF-{beta}) target genes and distinguishes TGF-{beta}-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol Cell Biol 25:8108–8125. doi: 10.1128/MCB.25.18.8108-8125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Jiang B, Guo Z, Sardet C, Zou B, Lam CS, Li J, He M, Lan HY, Pang R, Hung IF, Tan VP, Wang J, Wong BC. 2010. Four-and-a-half LIM protein 2 promotes invasive potential and epithelial-mesenchymal transition in colon cancer. Carcinogenesis 31:1220–1229. doi: 10.1093/carcin/bgq094. [DOI] [PubMed] [Google Scholar]
- 19.Gullotti L, Czerwitzki J, Kirfel J, Propping P, Rahner N, Steinke V, Kahl P, Engel C, Schule R, Buettner R, Friedrichs N. 2011. FHL2 expression in peritumoural fibroblasts correlates with lymphatic metastasis in sporadic but not in HNPCC-associated colon cancer. Lab Invest 91:1695–1705. doi: 10.1038/labinvest.2011.109. [DOI] [PubMed] [Google Scholar]
- 20.Westphal P, Mauch C, Florin A, Czerwitzki J, Olligschlager N, Wodtke C, Schule R, Buttner R, Friedrichs N. 2015. Enhanced FHL2 and TGF-beta1 expression is associated with invasive growth and poor survival in malignant melanomas. Am J Clin Pathol 143:248–256. doi: 10.1309/AJCPXEC6CIT2TXAF. [DOI] [PubMed] [Google Scholar]
- 21.Seki E, Brenner DA. 2015. Recent advancement of molecular mechanisms of liver fibrosis. J Hepatobiliary Pancreat Sci 22:512–518. doi: 10.1002/jhbp.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy L, Howell M, Das D, Harkin S, Episkopou V, Hill CS. 2007. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol Cell Biol 27:6068–6083. doi: 10.1128/MCB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. 2002. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 24.Birchenall-Roberts MC, Ruscetti FW, Kasper J, Lee HD, Friedman R, Geiser A, Sporn MB, Roberts AB, Kim SJ. 1990. Transcriptional regulation of the transforming growth factor beta 1 promoter by v-src gene products is mediated through the AP-1 complex. Mol Cell Biol 10:4978–4983. doi: 10.1128/MCB.10.9.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, Schwabe RF. 2007. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med 13:1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 26.Huss S, Stellmacher C, Goltz D, Khlistunova I, Adam AC, Trebicka J, Kirfel J, Buttner R, Weiskirchen R. 2013. Deficiency in four and one half LIM domain protein 2 (FHL2) aggravates liver fibrosis in mice. BMC Gastroenterol 13:8. doi: 10.1186/1471-230X-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alnajar A, Nordhoff C, Schied T, Chiquet-Ehrismann R, Loser K, Vogl T, Ludwig S, Wixler V. 2013. The LIM-only protein FHL2 attenuates lung inflammation during bleomycin-induced fibrosis. PLoS One 8:e81356. doi: 10.1371/journal.pone.0081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li SY, Huang PH, Tarng C, Lin TP, Yang WC, Chang YH, Yang AH, Lin CC, Yang MH, Chen JW, Schmid-Schonbein GW, Chien S, Chu PH, Lin SJ. 2015. Four-and-a-half LIM domains protein 2 is a coactivator of Wnt signaling in diabetic kidney disease. J Am Soc Nephrol 26:3072–3084. doi: 10.1681/ASN.2014100989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fanton d'Andon M, Quellard N, Fernandez B, Ratet G, Lacroix-Lamande S, Vandewalle A, Boneca IG, Goujon JM, Werts C. 2014. Leptospira interrogans induces fibrosis in the mouse kidney through Inos-dependent, TLR- and NLR-independent signaling pathways. PLoS Negl Trop Dis 8:e2664. doi: 10.1371/journal.pntd.0002664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei Y, Fabre M, Branchereau S, Gauthier F, Perilongo G, Buendia MA. 2000. Activation of beta-catenin in epithelial and mesenchymal hepatoblastomas. Oncogene 19:498–504. doi: 10.1038/sj.onc.1203356. [DOI] [PubMed] [Google Scholar]
- 31.Labalette C, Renard CA, Neuveut C, Buendia MA, Wei Y. 2004. Interaction and functional cooperation between the LIM protein FHL2, CBP/p300, and beta-catenin. Mol Cell Biol 24:10689–10702. doi: 10.1128/MCB.24.24.10689-10702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan J, Zhu J, Zhong H, Lu Q, Huang C, Ye Q. 2003. BRCA1 interacts with FHL2 and enhances FHL2 transactivation function. FEBS Lett 553:183–189. doi: 10.1016/S0014-5793(03)00978-5. [DOI] [PubMed] [Google Scholar]
- 33.Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J, Metzger E, Schule R, Buettner R. 2006. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res 66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 34.Chan KK, Tsui SK, Lee SM, Luk SC, Liew CC, Fung KP, Waye MM, Lee CY. 1998. Molecular cloning and characterization of FHL2, a novel LIM domain protein preferentially expressed in human heart. Gene 210:345–350. doi: 10.1016/S0378-1119(97)00644-6. [DOI] [PubMed] [Google Scholar]
- 35.Leroux A, Ferrere G, Godie V, Cailleux F, Renoud ML, Gaudin F, Naveau S, Prevot S, Makhzami S, Perlemuter G, Cassard-Doulcier AM. 2012. Toxic lipids stored by Kupffer cells correlates with their pro-inflammatory phenotype at an early stage of steatohepatitis. J Hepatol 57:141–149. doi: 10.1016/j.jhep.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 36.Labalette C, Nouet Y, Sobczak-Thepot J, Armengol C, Levillayer F, Gendron MC, Renard CA, Regnault B, Chen J, Buendia MA, Wei Y. 2008. The LIM-only protein FHL2 regulates cyclin D1 expression and cell proliferation. J Biol Chem 283:15201–15208. doi: 10.1074/jbc.M800708200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. 2002. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol 62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.