Abstract
In mammals, the development of pluripotency and specification of primordial germ cells (PGCs) have been studied predominantly using mice as a model organism. However, divergences among mammalian species for such processes have begun to be recognized. Between humans and mice, pre-implantation development appears relatively similar, but the manner and morphology of post-implantation development are significantly different. Nevertheless, the embryogenesis just after implantation in primates, including the specification of PGCs, has been unexplored due to the difficulties in analyzing the embryos at relevant developmental stages. Here, we present a comprehensive single-cell transcriptome dataset of pre- and early post-implantation embryo cells, PGCs and embryonic stem cells (ESCs) of cynomolgus monkeys as a model of higher primates. The identities of each transcriptome were also validated rigorously by other way such as immunofluorescent analysis. The information reported here will serve as a foundation for our understanding of a wide range of processes in the developmental biology of primates, including humans.
Subject terms: RNA sequencing, Pluripotent stem cells, Germline development, Embryology
Background & Summary
For more than half a century, mice have been exploited as a representative model organism for mammalian development and physiology. The results acquired from these researches have greatly contributed to our understanding of such processes as well as diseased states. Nonetheless, it has been widely recognized that there are significant differences in development and physiology between mice and humans.
The epiblast (EPI) is made up of cells specified during the peri-implantation period of early embryogenesis and differentiates into three primary germ layers and the germ cell lineages; the EPI therefore bears the pluripotency. Even within such a transient period, the EPI cells show dynamic changes in pluripotency from a so-called naive to a primed state1, and both states in mice have been captured in vitro: the naïve state is replicated in ESCs/induced pluripotent stem cells (iPSCs), which have essentially the same status as the pre-implantation EPI in vivo2, while the primed state is replicated in epiblast stem cells (EpiSCs), which are derived from the post-implantation EPI and are homologs of the EPI of the gastrula 3,4,5. On the other hand, although human ESCs/iPSCs have been established, their characteristics—including morphology, culture requirements, and the molecular networks underlying the pluripotent state—have been considered to more closely resemble those of mouse EpiSCs. The underlying mechanisms of these differences have remained unresolved, in part due to the lack of in vivo analyses of human/primate early post-implantation development.
The PGCs emerge from the posterior EPI at the onset of gastrulation in mice6. The specification process of mouse PGCs has been extensively studied and was reconstructed in vitro7. Remarkably, the resultant cells (PGC-like cells: PGCLCs) have the ability to produce functional gametes, and the dynamics of PGCLC specification is highly similar to that in vivo. In humans, PGCLCs have also been induced from human ESCs/iPSCs8,9, but it has still remained unclear whether their properties recapitulate those in vivo, again due to the lack of in vivo information.
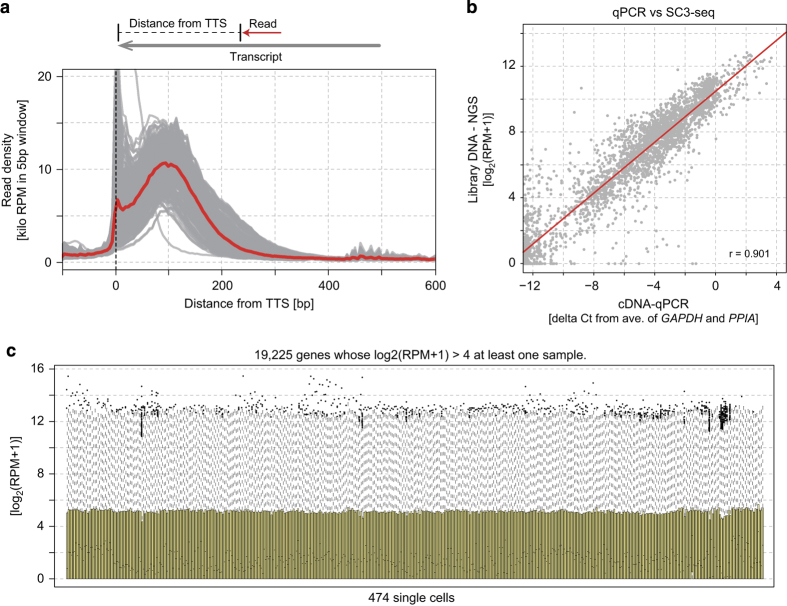
Here, we present a comprehensive transcriptome dataset at the single-cell level from pre- and post-implantation embryo cells, PGCs and ESCs of cynomolgus monkeys, one of the primates amenable to experiment and most closely related to humans. We employed the single-cell mRNA 3′ end sequencing (SC3-seq), which was designed to enrich the reads of the 3′ end of transcripts and enables highly quantitative and effective analysis10. We successfully amplified a total of 1,241 single-cell cDNAs and generated 474 transcriptomes (Table 1 (available online only)). The qualities of the transcriptomes and the representations of gene expression profile were validated by qPCR. The sample annotations were defined comprehensively by comparing the expression of key genes in transcriptome data with that obtained through the histological analysis such as immunofluorescent analysis and/or in situ hybridization11,12. Thus, the dataset in this Data Descriptor defined the first comprehensive molecular dynamics of primate early development, including early post-implantation embryogenesis, and will provide a foundation for future studies of primate development.
Table 1. Metadata and mapping statistics of SC3-seq analysis.
| GSM ID | GSM name | Cluster in Fgi3 | order in cluster Fig3 | Embryonic Day/ passage number | cells picked from | Embryo name | sex | GSE ID | Total reads | SC3-seq mapped reads | Mapped_others | Control_RNA | low_quality_adaptor_polyA | Unmapped |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GSM1932351 | E16_MS09T43 | Post_paTE | 1 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 6,128,858 | 2,774,597 | 1,297,685 | 30,256 | 1,289,211 | 737,109 |
| GSM1932349 | E14_MS13T86 | Post_paTE | 2 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 6,429,515 | 1,271,723 | 351,757 | 39,719 | 4,249,933 | 516,383 |
| GSM1932347 | E14_MS09T11 | Post_paTE | 3 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 4,456,818 | 1,061,281 | 1,264,024 | 39,322 | 1,401,801 | 690,390 |
| GSM1932350 | E16_MS09T42 | Post_paTE | 4 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 6,187,993 | 1,708,675 | 1,319,009 | 28,864 | 2,382,646 | 748,799 |
| GSM1932345 | E14_MS09T07 | Post_paTE | 5 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 5,692,791 | 2,230,595 | 1,137,592 | 28,407 | 1,594,158 | 702,039 |
| GSM1932348 | E14_MS13T81 | Post_paTE | 6 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,371,581 | 2,182,547 | 852,205 | 15,379 | 1,665,453 | 655,997 |
| GSM1932343 | E14_MS09T04 | Post_paTE | 7 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 5,434,597 | 2,156,596 | 1,112,574 | 23,223 | 1,468,406 | 673,798 |
| GSM1932344 | E14_MS09T06 | Post_paTE | 8 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 4,864,284 | 1,551,471 | 902,202 | 20,353 | 1,776,915 | 613,343 |
| GSM1932346 | E14_MS09T10 | Post_paTE | 9 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 5,696,608 | 2,089,772 | 918,302 | 19,568 | 2,000,647 | 668,319 |
| GSM1932341 | E14_MS09T01 | Post_paTE | 10 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 6,214,950 | 1,955,043 | 1,536,175 | 45,151 | 1,762,696 | 915,885 |
| GSM1932342 | E14_MS09T02 | Post_paTE | 11 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 5,879,574 | 2,345,386 | 1,338,822 | 18,193 | 1,333,109 | 844,064 |
| GSM1932533 | E08_MS02T34 | PreL_TE | 12 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 6,049,861 | 2,871,747 | 1,875,055 | 2,272 | 729,346 | 571,441 |
| GSM1932532 | E08_MS02T33 | PreL_TE | 13 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,060,125 | 2,390,667 | 1,419,805 | 1,296 | 752,289 | 496,068 |
| GSM1932527 | E08_MS02T11 | PreL_TE | 14 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 8,423,512 | 4,499,577 | 1,807,662 | 1,951 | 1,334,781 | 779,541 |
| GSM1932524 | E08_MS02T04 | PreL_TE | 15 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,224,475 | 2,745,820 | 1,234,798 | 1,390 | 766,219 | 476,248 |
| GSM1932528 | E08_MS02T19 | PreL_TE | 16 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,595,531 | 2,740,479 | 1,476,266 | 1,284 | 834,387 | 543,115 |
| GSM1932531 | E08_MS02T27 | PreL_TE | 17 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,887,381 | 3,081,912 | 1,317,162 | 1,181 | 919,368 | 567,758 |
| GSM1932529 | E08_MS02T24 | PreL_TE | 18 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 6,071,248 | 3,053,414 | 1,341,552 | 1,242 | 1,064,737 | 610,303 |
| GSM1932530 | E08_MS02T25 | PreL_TE | 19 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,670,112 | 2,850,725 | 1,377,720 | 973 | 888,091 | 552,603 |
| GSM1932525 | E08_MS02T07 | PreL_TE | 20 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,778,802 | 2,758,330 | 1,556,026 | 1,821 | 888,837 | 573,788 |
| GSM1932526 | E08_MS02T10 | PreL_TE | 21 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 7,316,886 | 3,642,213 | 1,799,007 | 2,012 | 1,150,717 | 722,937 |
| GSM1932514 | E07_MS03T55 | PreL_TE | 22 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 3,725,478 | 1,864,037 | 764,322 | 696 | 753,139 | 343,284 |
| GSM1932518 | E07_MS03T63 | PreL_TE | 23 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 6,513,999 | 2,476,661 | 1,071,889 | 1,384 | 2,302,184 | 661,881 |
| GSM1932560 | E09_MS03T77 | PreL_TE | 24 | E09 | whole embryo | 120725ICSIday9 | male | GSE74767 | 4,140,253 | 1,685,077 | 1,115,324 | 1,424 | 790,681 | 547,747 |
| GSM1932558 | E09_MS03T75 | PreL_TE | 25 | E09 | whole embryo | 120725ICSIday9 | male | GSE74767 | 4,557,502 | 2,211,118 | 727,286 | 1,337 | 1,111,516 | 506,245 |
| GSM1932559 | E09_MS03T76 | PreL_TE | 26 | E09 | whole embryo | 120725ICSIday9 | male | GSE74767 | 3,390,166 | 1,764,821 | 660,928 | 952 | 593,318 | 370,147 |
| GSM1932562 | E09_MS03T86 | PreL_TE | 27 | E09 | whole embryo | 120725ICSIday9 | male | GSE74767 | 5,438,653 | 2,421,176 | 1,136,347 | 620 | 1,225,112 | 655,398 |
| GSM1932561 | E09_MS03T84 | PreL_TE | 28 | E09 | whole embryo | 120725ICSIday9 | male | GSE74767 | 6,109,506 | 2,680,667 | 1,139,307 | 907 | 1,546,768 | 741,857 |
| GSM1932515 | E07_MS03T58 | PreL_TE | 29 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 4,678,468 | 2,160,704 | 1,101,343 | 373 | 952,714 | 463,334 |
| GSM1932517 | E07_MS03T60 | PreL_TE | 30 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 6,956,412 | 2,726,988 | 1,786,233 | 632 | 1,727,298 | 715,261 |
| GSM1932522 | E07_MS08T45 | PreL_TE | 31 | E07 | whole embryo | 130319ICSI#1day7 | male | GSE74767 | 5,740,510 | 2,232,954 | 1,483,724 | 2,228 | 1,475,049 | 546,555 |
| GSM1932521 | E07_MS08T44 | PreL_TE | 32 | E07 | whole embryo | 130319ICSI#1day7 | male | GSE74767 | 4,781,234 | 2,108,623 | 902,393 | 1,499 | 1,321,849 | 446,870 |
| GSM1932519 | E07_MS03T67 | PreL_TE | 33 | E07 | whole embryo | 130319ICSI#1day7 | male | GSE74767 | 4,151,770 | 1,584,429 | 946,058 | 666 | 1,216,838 | 403,779 |
| GSM1932520 | E07_MS03T68 | PreL_TE | 34 | E07 | whole embryo | 130319ICSI#1day7 | male | GSE74767 | 4,716,991 | 2,087,621 | 902,820 | 1,142 | 1,271,443 | 453,965 |
| GSM1932523 | E07_MS08T46 | PreL_TE | 35 | E07 | whole embryo | 130319ICSI#1day7 | male | GSE74767 | 4,890,708 | 2,248,541 | 822,944 | 1,319 | 1,431,515 | 386,389 |
| GSM1932540 | E08_MS02T69 | PreL_TE | 36 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,924,513 | 1,996,844 | 712,726 | 2,113 | 802,376 | 410,454 |
| GSM1932545 | E08_MS02T75 | PreL_TE | 37 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 4,022,342 | 1,975,185 | 905,004 | 2,377 | 704,341 | 435,435 |
| GSM1932542 | E08_MS02T72 | PreL_TE | 38 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,946,813 | 1,548,626 | 1,175,149 | 694 | 748,307 | 474,037 |
| GSM1932537 | E08_MS02T62 | PreL_TE | 39 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,569,030 | 1,682,482 | 703,577 | 1,377 | 713,905 | 467,689 |
| GSM1932544 | E08_MS02T74 | PreL_TE | 40 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 4,197,156 | 2,277,155 | 750,534 | 1,742 | 734,496 | 433,229 |
| GSM1932546 | E08_MS02T78 | PreL_TE | 41 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,321,507 | 1,630,392 | 725,159 | 936 | 600,705 | 364,315 |
| GSM1932548 | E08_MS02T82 | PreL_TE | 42 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,708,067 | 1,832,230 | 742,293 | 1,174 | 734,749 | 397,621 |
| GSM1932549 | E08_MS02T83 | PreL_TE | 43 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,339,408 | 1,811,022 | 606,102 | 727 | 588,348 | 333,209 |
| GSM1932536 | E08_MS02T61 | PreL_TE | 44 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,166,483 | 1,772,765 | 568,943 | 1,146 | 472,607 | 351,022 |
| GSM1932538 | E08_MS02T64 | PreL_TE | 45 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,876,847 | 2,034,822 | 634,940 | 1,007 | 737,731 | 468,347 |
| GSM1932539 | E08_MS02T65 | PreL_TE | 46 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 4,026,786 | 2,178,907 | 759,252 | 1,170 | 695,027 | 392,430 |
| GSM1932547 | E08_MS02T79 | PreL_TE | 47 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,433,160 | 1,861,980 | 604,684 | 767 | 622,869 | 342,860 |
| GSM1932556 | E08_MS02T95 | PreL_TE | 48 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,784,209 | 2,057,143 | 648,876 | 1,243 | 643,604 | 433,343 |
| GSM1932555 | E08_MS02T94 | PreL_TE | 49 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,620,267 | 2,008,743 | 601,331 | 635 | 629,385 | 380,173 |
| GSM1932557 | E08_MS02T96 | PreL_TE | 50 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 1,435,755 | 876,743 | 219,031 | 452 | 195,689 | 143,840 |
| GSM1932534 | E08_MS02T58 | PreL_TE | 51 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,538,090 | 1,978,308 | 566,088 | 1,469 | 611,554 | 380,671 |
| GSM1932550 | E08_MS02T86 | PreL_TE | 52 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,581,807 | 1,891,030 | 531,717 | 439 | 695,572 | 463,049 |
| GSM1932541 | E08_MS02T70 | PreL_TE | 53 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,448,892 | 1,883,198 | 627,628 | 930 | 596,782 | 340,354 |
| GSM1932543 | E08_MS02T73 | PreL_TE | 54 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,143,604 | 1,664,583 | 575,809 | 1,134 | 583,512 | 318,566 |
| GSM1932535 | E08_MS02T60 | PreL_TE | 55 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,515,803 | 1,705,807 | 751,137 | 1,606 | 624,196 | 433,057 |
| GSM1932551 | E08_MS02T87 | PreL_TE | 56 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,391,374 | 1,794,959 | 653,114 | 941 | 551,402 | 390,958 |
| GSM1932553 | E08_MS02T90 | PreL_TE | 57 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,930,689 | 1,974,787 | 792,200 | 409 | 680,532 | 482,761 |
| GSM1932552 | E08_MS02T88 | PreL_TE | 58 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,312,599 | 1,599,652 | 605,333 | 540 | 701,419 | 405,655 |
| GSM1932554 | E08_MS02T91 | PreL_TE | 59 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 2,856,778 | 1,504,603 | 567,425 | 952 | 455,171 | 328,627 |
| GSM1932516 | E07_MS03T59 | PreL_TE | 60 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 3,548,957 | 1,432,388 | 703,706 | 1,092 | 1,048,476 | 363,295 |
| GSM1932564 | E09_MS03T90 | PreL_TE | 61 | E09 | whole embryo | 121212ICSI#2day9 | female | GSE74767 | 5,709,106 | 2,489,234 | 1,016,452 | 1,081 | 1,658,458 | 543,881 |
| GSM1932563 | E09_MS03T87 | PreL_TE | 62 | E09 | whole embryo | 121212ICSI#2day9 | female | GSE74767 | 4,017,390 | 1,754,220 | 721,675 | 450 | 1,153,287 | 387,758 |
| GSM1932565 | E09_MS03T94 | PreL_TE | 63 | E09 | whole embryo | 121212ICSI#2day9 | female | GSE74767 | 5,352,204 | 2,142,251 | 929,021 | 1,406 | 1,768,607 | 510,919 |
| GSM1932278 | E08_MS02T23 | Hypoblast | 64 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 6,253,800 | 3,311,662 | 1,150,934 | 2,685 | 1,136,944 | 651,575 |
| GSM1932265 | E07_MS03T72 | Hypoblast | 65 | E07 | whole embryo | 130319ICSI#1day7 | male | GSE74767 | 4,116,751 | 1,472,479 | 1,003,075 | 2,241 | 1,233,305 | 405,651 |
| GSM1932270 | E08_MS02T12 | Hypoblast | 66 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 8,297,727 | 2,819,205 | 3,729,548 | 3,238 | 912,304 | 833,432 |
| GSM1932275 | E08_MS02T20 | Hypoblast | 67 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 7,579,424 | 2,832,280 | 2,976,128 | 2,346 | 986,177 | 782,493 |
| GSM1932304 | E09_MS03T78 | Hypoblast | 68 | E09 | whole embryo | 120725ICSIday9 | male | GSE74767 | 6,213,540 | 2,610,118 | 1,362,017 | 3,049 | 1,458,223 | 780,133 |
| GSM1932258 | E07_MS03T54 | Hypoblast | 69 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 1,672,175 | 679,947 | 330,500 | 424 | 487,044 | 174,260 |
| GSM1932264 | E07_MS03T71 | Hypoblast | 70 | E07 | whole embryo | 130319ICSI#1day7 | male | GSE74767 | 4,374,061 | 1,858,315 | 1,004,343 | 1,253 | 1,108,676 | 401,474 |
| GSM1932283 | E08_MS02T35 | Hypoblast | 71 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,559,037 | 2,957,509 | 1,240,210 | 2,896 | 824,688 | 533,734 |
| GSM1932309 | E09_MS03T93 | Hypoblast | 72 | E09 | whole embryo | 121212ICSI#2day9 | female | GSE74767 | 4,047,218 | 1,727,535 | 611,741 | 1,057 | 1,318,679 | 388,206 |
| GSM1932301 | E08_MS02T80 | Hypoblast | 73 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 2,928,563 | 1,546,076 | 574,730 | 1,521 | 518,755 | 287,481 |
| GSM1932294 | E08_MS02T56 | Hypoblast | 74 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 3,703,150 | 1,801,845 | 679,234 | 752 | 817,974 | 403,345 |
| GSM1932307 | E09_MS03T88 | Hypoblast | 75 | E09 | whole embryo | 121212ICSI#2day9 | female | GSE74767 | 4,510,417 | 1,802,845 | 887,587 | 269 | 1,351,497 | 468,219 |
| GSM1932271 | E08_MS02T13 | Hypoblast | 76 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,456,909 | 2,489,880 | 1,597,628 | 2,722 | 805,899 | 560,780 |
| GSM1932273 | E08_MS02T15 | Hypoblast | 77 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 4,900,508 | 2,317,010 | 1,309,031 | 2,093 | 774,212 | 498,162 |
| GSM1932289 | E08_MS02T46 | Hypoblast | 78 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 2,740,751 | 1,443,739 | 271,601 | 2,550 | 760,299 | 262,562 |
| GSM1932297 | E08_MS02T67 | Hypoblast | 79 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,038,262 | 1,317,612 | 555,726 | 800 | 822,919 | 341,205 |
| GSM1932299 | E08_MS02T71 | Hypoblast | 80 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 4,334,836 | 2,296,508 | 826,140 | 2,254 | 776,926 | 433,008 |
| GSM1932296 | E08_MS02T66 | Hypoblast | 81 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,606,216 | 1,761,448 | 743,823 | 1,830 | 688,599 | 410,516 |
| GSM1932295 | E08_MS02T63 | Hypoblast | 82 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,825,054 | 1,888,277 | 675,624 | 1,339 | 803,707 | 456,107 |
| GSM1932298 | E08_MS02T68 | Hypoblast | 83 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 4,698,998 | 2,448,369 | 927,116 | 1,086 | 830,291 | 492,136 |
| GSM1932300 | E08_MS02T77 | Hypoblast | 84 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,503,384 | 1,762,556 | 787,325 | 1,375 | 589,409 | 362,719 |
| GSM1932292 | E08_MS02T53 | Hypoblast | 85 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 3,064,629 | 1,288,864 | 984,174 | 802 | 455,389 | 335,400 |
| GSM1932286 | E08_MS02T40 | Hypoblast | 86 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 2,999,552 | 1,314,476 | 543,353 | 684 | 813,435 | 327,604 |
| GSM1932266 | E07_MS08T43 | Hypoblast | 87 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 5,884,380 | 2,274,787 | 1,269,340 | 2,020 | 1,710,448 | 627,785 |
| GSM1932261 | E07_MS03T61 | Hypoblast | 88 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 5,050,434 | 2,679,033 | 830,810 | 1,108 | 1,063,682 | 475,801 |
| GSM1932262 | E07_MS03T62 | Hypoblast | 89 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 3,737,959 | 1,253,448 | 454,121 | 696 | 1,641,288 | 388,406 |
| GSM1932257 | E07_MS03T53 | Hypoblast | 90 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 3,247,062 | 1,380,594 | 561,520 | 978 | 985,829 | 318,141 |
| GSM1932260 | E07_MS03T57 | Hypoblast | 91 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 4,228,060 | 1,649,137 | 735,846 | 837 | 1,416,930 | 425,310 |
| GSM1932263 | E07_MS03T64 | Hypoblast | 92 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 5,658,536 | 2,448,635 | 869,028 | 4,642 | 1,783,411 | 552,820 |
| GSM1932293 | E08_MS02T54 | Hypoblast | 93 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 2,121,073 | 1,021,847 | 521,055 | 440 | 360,424 | 217,307 |
| GSM1932305 | E09_MS03T80 | Hypoblast | 94 | E09 | whole embryo | 120725ICSIday9 | male | GSE74767 | 5,386,009 | 2,755,211 | 938,497 | 1,999 | 1,122,425 | 567,877 |
| GSM1932306 | E09_MS03T81 | Hypoblast | 95 | E09 | whole embryo | 120725ICSIday9 | male | GSE74767 | 6,122,695 | 2,982,307 | 1,167,681 | 1,772 | 1,295,494 | 675,441 |
| GSM1932290 | E08_MS02T49 | Hypoblast | 96 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 2,382,124 | 1,168,237 | 433,819 | 603 | 518,283 | 261,182 |
| GSM1932259 | E07_MS03T56 | Hypoblast | 97 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 5,436,682 | 2,065,629 | 723,122 | 1,238 | 2,102,206 | 544,487 |
| GSM1932302 | E08_MS02T81 | Hypoblast | 98 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,168,404 | 1,534,046 | 587,887 | 759 | 702,026 | 343,686 |
| GSM1932288 | E08_MS02T42 | Hypoblast | 99 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 3,794,210 | 1,757,710 | 635,730 | 641 | 1,004,093 | 396,036 |
| GSM1932284 | E08_MS02T37 | Hypoblast | 100 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 3,543,589 | 1,583,178 | 726,407 | 480 | 862,831 | 370,693 |
| GSM1932291 | E08_MS02T50 | Hypoblast | 101 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 4,422,671 | 2,243,836 | 900,940 | 379 | 812,274 | 465,242 |
| GSM1932303 | E08_MS03T02 | Hypoblast | 102 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 4,924,109 | 1,686,073 | 995,253 | 1,376 | 1,750,956 | 490,451 |
| GSM1932285 | E08_MS02T39 | Hypoblast | 103 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 2,492,058 | 1,125,935 | 504,315 | 525 | 595,034 | 266,249 |
| GSM1932287 | E08_MS02T41 | Hypoblast | 104 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 2,784,628 | 1,064,290 | 416,610 | 762 | 981,362 | 321,604 |
| GSM1932308 | E09_MS03T89 | Hypoblast | 105 | E09 | whole embryo | 121212ICSI#2day9 | female | GSE74767 | 4,556,388 | 1,787,184 | 627,169 | 835 | 1,675,217 | 465,983 |
| GSM1932310 | E09_MS03T95 | Hypoblast | 106 | E09 | whole embryo | 121212ICSI#2day9 | female | GSE74767 | 3,987,985 | 1,618,676 | 529,743 | 1,452 | 1,445,851 | 392,263 |
| GSM1932282 | E08_MS02T32 | Hypoblast | 107 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,385,527 | 2,641,107 | 1,362,869 | 3,915 | 824,154 | 553,482 |
| GSM1932277 | E08_MS02T22 | Hypoblast | 108 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 7,441,517 | 3,341,675 | 2,087,441 | 3,203 | 1,237,182 | 772,016 |
| GSM1932280 | E08_MS02T28 | Hypoblast | 109 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,746,208 | 2,867,676 | 1,414,059 | 1,479 | 897,868 | 565,126 |
| GSM1932281 | E08_MS02T29 | Hypoblast | 110 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,122,182 | 2,500,056 | 1,341,557 | 1,863 | 788,259 | 490,447 |
| GSM1932269 | E08_MS02T09 | Hypoblast | 111 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,603,782 | 2,774,200 | 1,514,425 | 2,196 | 777,795 | 535,166 |
| GSM1932267 | E08_MS02T03 | Hypoblast | 112 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,466,255 | 2,874,759 | 1,207,637 | 2,313 | 863,322 | 518,224 |
| GSM1932279 | E08_MS02T26 | Hypoblast | 113 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,794,104 | 3,066,464 | 1,333,685 | 2,278 | 857,213 | 534,464 |
| GSM1932268 | E08_MS02T08 | Hypoblast | 114 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,182,804 | 2,283,877 | 1,437,874 | 1,996 | 901,736 | 557,321 |
| GSM1932276 | E08_MS02T21 | Hypoblast | 115 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,453,815 | 3,079,438 | 1,009,470 | 1,872 | 852,175 | 510,860 |
| GSM1932272 | E08_MS02T14 | Hypoblast | 116 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 7,097,619 | 3,522,049 | 1,840,343 | 1,871 | 1,039,334 | 694,022 |
| GSM1932274 | E08_MS02T18 | Hypoblast | 117 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 8,340,331 | 4,630,817 | 1,490,805 | 2,366 | 1,382,157 | 834,186 |
| GSM1932500 | E06_MS03T30 | PreE_TE | 118 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 6,944,413 | 2,754,186 | 1,623,766 | 568 | 1,723,852 | 842,041 |
| GSM1932328 | E06_MS03T38 | PreE_TE | 119 | E06 | whole embryo | 130306ICSI#7day6 | female | GSE74767 | 5,491,733 | 1,859,417 | 1,334,132 | 370 | 1,530,297 | 767,517 |
| GSM1932501 | E06_MS68T13 | PreE_TE | 120 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 4,122,463 | 1,641,710 | 928,557 | 879 | 1,104,197 | 447,120 |
| GSM1932496 | E06_MS03T24 | PreE_TE | 121 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 6,827,318 | 2,640,660 | 1,514,237 | 1,089 | 1,889,713 | 781,619 |
| GSM1932503 | E06_MS68T16 | PreE_TE | 122 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 2,893,961 | 1,093,097 | 510,406 | 603 | 1,017,745 | 272,110 |
| GSM1932493 | E06_MS03T19 | PreE_TE | 123 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 3,330,196 | 1,282,712 | 704,022 | 580 | 971,107 | 371,775 |
| GSM1932498 | E06_MS03T27 | PreE_TE | 124 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 5,420,210 | 2,280,662 | 1,152,813 | 625 | 1,349,682 | 636,428 |
| GSM1932497 | E06_MS03T25 | PreE_TE | 125 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 4,714,281 | 1,974,147 | 1,015,941 | 694 | 1,166,128 | 557,371 |
| GSM1932494 | E06_MS03T21 | PreE_TE | 126 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 4,913,293 | 2,089,458 | 970,368 | 685 | 1,316,400 | 536,382 |
| GSM1932491 | E06_MS03T16 | PreE_TE | 127 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 6,277,089 | 2,479,631 | 1,251,603 | 1,074 | 1,796,953 | 747,828 |
| GSM1932492 | E06_MS03T17 | PreE_TE | 128 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 4,979,264 | 2,199,305 | 953,015 | 539 | 1,272,417 | 553,988 |
| GSM1932502 | E06_MS68T15 | PreE_TE | 129 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 2,897,604 | 1,080,308 | 741,179 | 621 | 735,034 | 340,462 |
| GSM1932495 | E06_MS03T22 | PreE_TE | 130 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 4,651,187 | 1,704,550 | 996,999 | 509 | 1,420,828 | 528,301 |
| GSM1932499 | E06_MS03T29 | PreE_TE | 131 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 5,978,956 | 2,839,558 | 1,019,870 | 736 | 1,544,425 | 574,367 |
| GSM1932510 | E07_MS03T46 | PreE_TE | 132 | E07 | whole embryo | 121128ICSI#1day7 | male | GSE74767 | 4,582,757 | 1,027,348 | 1,450,613 | 1,842 | 1,257,947 | 845,007 |
| GSM1932508 | E07_MS03T44 | PreE_TE | 133 | E07 | whole embryo | 121128ICSI#1day7 | male | GSE74767 | 5,353,604 | 1,334,901 | 2,017,416 | 926 | 1,277,041 | 723,320 |
| GSM1932507 | E07_MS03T43 | PreE_TE | 134 | E07 | whole embryo | 121128ICSI#1day7 | male | GSE74767 | 6,033,396 | 2,176,475 | 1,358,292 | 770 | 1,644,763 | 853,096 |
| GSM1932506 | E07_MS03T42 | PreE_TE | 135 | E07 | whole embryo | 121128ICSI#1day7 | male | GSE74767 | 4,791,070 | 2,076,983 | 875,287 | 1,258 | 1,292,582 | 544,960 |
| GSM1932513 | E07_MS03T49 | PreE_TE | 136 | E07 | whole embryo | 121128ICSI#1day7 | male | GSE74767 | 4,747,124 | 1,973,432 | 1,245,732 | 1,678 | 927,008 | 599,274 |
| GSM1932509 | E07_MS03T45 | PreE_TE | 137 | E07 | whole embryo | 121128ICSI#1day7 | male | GSE74767 | 4,843,458 | 1,933,285 | 1,120,766 | 683 | 1,203,336 | 585,388 |
| GSM1932511 | E07_MS03T47 | PreE_TE | 138 | E07 | whole embryo | 121128ICSI#1day7 | male | GSE74767 | 5,774,869 | 2,221,827 | 1,240,890 | 700 | 1,492,525 | 818,927 |
| GSM1932505 | E07_MS03T41 | PreE_TE | 139 | E07 | whole embryo | 121128ICSI#1day7 | male | GSE74767 | 5,950,583 | 2,343,854 | 1,517,510 | 700 | 1,396,080 | 692,439 |
| GSM1932504 | E07_MS03T40 | PreE_TE | 140 | E07 | whole embryo | 121128ICSI#1day7 | male | GSE74767 | 4,800,241 | 2,176,400 | 1,040,298 | 867 | 1,025,341 | 557,335 |
| GSM1932512 | E07_MS03T48 | PreE_TE | 141 | E07 | whole embryo | 121128ICSI#1day7 | male | GSE74767 | 5,530,186 | 2,309,733 | 1,096,898 | 900 | 1,387,157 | 735,498 |
| GSM1932334 | E06_MS68T06 | ICM | 142 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 2,942,999 | 1,033,265 | 654,289 | 214 | 1,024,403 | 230,828 |
| GSM1932330 | E06_MS68T02 | ICM | 143 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 3,662,545 | 1,260,599 | 854,612 | 112 | 1,264,635 | 282,587 |
| GSM1932336 | E06_MS68T08 | ICM | 144 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 1,718,888 | 688,659 | 381,073 | 135 | 507,758 | 141,263 |
| GSM1932340 | E06_MS68T12 | ICM | 145 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 5,716,490 | 2,156,704 | 1,214,981 | 223 | 1,852,795 | 491,787 |
| GSM1932338 | E06_MS68T10 | ICM | 146 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 3,036,423 | 1,127,865 | 496,022 | 106 | 1,172,828 | 239,602 |
| GSM1932311 | E06_MS03T03 | ICM | 147 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 4,310,758 | 1,732,807 | 933,992 | 90 | 1,214,988 | 428,881 |
| GSM1932331 | E06_MS68T03 | ICM | 148 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 2,995,292 | 1,236,380 | 587,957 | 106 | 939,372 | 231,477 |
| GSM1932332 | E06_MS68T04 | ICM | 149 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 3,590,300 | 1,303,501 | 674,612 | 120 | 1,352,639 | 259,428 |
| GSM1932327 | E06_MS03T34 | ICM | 150 | E06 | whole embryo | 130306ICSI#7day6 | female | GSE74767 | 3,667,894 | 1,784,545 | 774,838 | 349 | 716,505 | 391,657 |
| GSM1932329 | E06_MS68T01 | ICM | 151 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 3,035,025 | 1,077,323 | 859,981 | 199 | 834,737 | 262,785 |
| GSM1932339 | E06_MS68T11 | ICM | 152 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 2,261,084 | 947,479 | 494,808 | 67 | 633,725 | 185,005 |
| GSM1932314 | E06_MS03T06 | ICM | 153 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 5,871,201 | 2,615,551 | 1,173,228 | 121 | 1,520,325 | 561,976 |
| GSM1932333 | E06_MS68T05 | ICM | 154 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 3,128,282 | 1,180,113 | 915,981 | 93 | 757,868 | 274,227 |
| GSM1932337 | E06_MS68T09 | ICM | 155 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 2,904,099 | 934,305 | 630,137 | 189 | 1,121,586 | 217,882 |
| GSM1932321 | E06_MS03T14 | ICM | 156 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 8,533,262 | 3,753,526 | 1,959,436 | 409 | 2,024,870 | 795,021 |
| GSM1932318 | E06_MS03T11 | ICM | 157 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 5,235,517 | 1,923,956 | 1,385,484 | 165 | 1,401,869 | 524,043 |
| GSM1932335 | E06_MS68T07 | ICM | 158 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 2,175,030 | 791,535 | 470,022 | 93 | 734,063 | 179,317 |
| GSM1932319 | E06_MS03T12 | ICM | 159 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 4,737,642 | 1,935,172 | 1,217,903 | 389 | 1,107,964 | 476,214 |
| GSM1932320 | E06_MS03T13 | ICM | 160 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 6,715,658 | 2,623,120 | 1,568,355 | 217 | 1,843,399 | 680,567 |
| GSM1932312 | E06_MS03T04 | ICM | 161 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 4,626,086 | 1,596,212 | 1,343,358 | 263 | 1,214,874 | 471,379 |
| GSM1932313 | E06_MS03T05 | ICM | 162 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 5,933,397 | 2,005,358 | 1,431,224 | 247 | 1,874,416 | 622,152 |
| GSM1932316 | E06_MS03T08 | ICM | 163 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 4,894,876 | 1,677,663 | 1,430,825 | 171 | 1,274,381 | 511,836 |
| GSM1932315 | E06_MS03T07 | ICM | 164 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 5,905,135 | 2,179,900 | 1,646,276 | 188 | 1,474,665 | 604,106 |
| GSM1932317 | E06_MS03T10 | ICM | 165 | E06 | whole embryo | 130206ICSI#1day6 | male | GSE74767 | 5,598,031 | 2,064,974 | 1,490,856 | 115 | 1,463,758 | 578,328 |
| GSM1932326 | E06_MS03T28 | ICM | 166 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 5,825,270 | 1,864,102 | 1,781,052 | 553 | 1,395,959 | 783,604 |
| GSM1932325 | E06_MS03T26 | ICM | 167 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 7,305,783 | 2,903,429 | 1,666,231 | 1,755 | 1,939,730 | 794,638 |
| GSM1932324 | E06_MS03T23 | ICM | 168 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 5,453,926 | 1,952,589 | 1,494,436 | 741 | 1,324,192 | 681,968 |
| GSM1932322 | E06_MS03T15 | ICM | 169 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 7,115,508 | 1,852,598 | 2,569,191 | 615 | 1,637,754 | 1,055,350 |
| GSM1932323 | E06_MS03T20 | ICM | 170 | E06 | whole embryo | 130227ICSI#1day6 | female | GSE74767 | 3,383,309 | 1,414,237 | 691,234 | 647 | 912,830 | 364,361 |
| GSM1932470 | E08_MS02T36 | Pre_EPI | 171 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,030,935 | 2,503,909 | 1,361,791 | 2,032 | 706,360 | 456,843 |
| GSM1932464 | E08_MS02T05 | Pre_EPI | 172 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 6,383,922 | 3,136,083 | 1,587,943 | 2,845 | 1,031,883 | 625,168 |
| GSM1932466 | E08_MS02T16 | Pre_EPI | 173 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 6,462,396 | 3,215,522 | 1,511,766 | 1,791 | 1,073,194 | 660,123 |
| GSM1932467 | E08_MS02T17 | Pre_EPI | 174 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 6,095,474 | 2,924,677 | 1,593,399 | 1,447 | 972,598 | 603,353 |
| GSM1932469 | E08_MS02T31 | Pre_EPI | 175 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 5,540,759 | 2,865,042 | 1,229,822 | 1,746 | 909,487 | 534,662 |
| GSM1932468 | E08_MS02T30 | Pre_EPI | 176 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 6,222,423 | 3,052,728 | 1,554,247 | 1,748 | 986,756 | 626,944 |
| GSM1932463 | E08_MS02T02 | Pre_EPI | 177 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 6,997,596 | 3,521,486 | 1,715,673 | 2,519 | 1,085,482 | 672,436 |
| GSM1932465 | E08_MS02T06 | Pre_EPI | 178 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 8,210,584 | 3,855,825 | 2,311,515 | 1,033 | 1,244,790 | 797,421 |
| GSM1932487 | E09_MS03T82 | Pre_EPI | 179 | E09 | whole embryo | 120725ICSIday9 | male | GSE74767 | 5,911,096 | 2,630,317 | 1,144,709 | 3,174 | 1,454,901 | 677,995 |
| GSM1932489 | E09_MS03T85 | Pre_EPI | 180 | E09 | whole embryo | 120725ICSIday9 | male | GSE74767 | 5,756,563 | 2,151,618 | 1,595,354 | 1,394 | 1,264,180 | 744,017 |
| GSM1932481 | E08_MS02T85 | Pre_EPI | 181 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,226,087 | 1,652,615 | 614,776 | 756 | 622,276 | 335,664 |
| GSM1932483 | E08_MS02T92 | Pre_EPI | 182 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,446,447 | 1,870,723 | 583,028 | 775 | 621,742 | 370,179 |
| GSM1932480 | E08_MS02T59 | Pre_EPI | 183 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,975,653 | 2,175,367 | 667,993 | 1,021 | 689,611 | 441,661 |
| GSM1932484 | E08_MS02T93 | Pre_EPI | 184 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 3,290,342 | 1,760,659 | 577,301 | 742 | 594,363 | 357,277 |
| GSM1932488 | E09_MS03T83 | Pre_EPI | 185 | E09 | whole embryo | 120725ICSIday9 | male | GSE74767 | 6,247,104 | 2,850,546 | 1,164,757 | 1,409 | 1,506,375 | 724,017 |
| GSM1932486 | E09_MS03T79 | Pre_EPI | 186 | E09 | whole embryo | 120725ICSIday9 | male | GSE74767 | 4,696,577 | 2,037,773 | 980,869 | 1,115 | 1,122,284 | 554,536 |
| GSM1932471 | E08_MS02T38 | Pre_EPI | 187 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 3,953,616 | 1,820,660 | 785,167 | 735 | 922,793 | 424,261 |
| GSM1932482 | E08_MS02T89 | Pre_EPI | 188 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 4,143,227 | 2,091,560 | 846,679 | 477 | 709,076 | 495,435 |
| GSM1932485 | E08_MS03T01 | Pre_EPI | 189 | E08 | whole embryo | 130220ICSI#11day8 | male | GSE74767 | 5,560,690 | 2,130,831 | 1,435,800 | 916 | 1,316,457 | 676,686 |
| GSM1932459 | E07_MS03T70 | Pre_EPI | 190 | E07 | whole embryo | 130319ICSI#1day7 | male | GSE74767 | 5,017,381 | 1,908,365 | 1,113,926 | 1,317 | 1,487,919 | 505,854 |
| GSM1932462 | E08_MS02T01 | Pre_EPI | 191 | E08 | whole embryo | 121107ICSI13_day8 | male | GSE74767 | 6,961,614 | 2,885,495 | 2,328,845 | 2,925 | 1,027,889 | 716,460 |
| GSM1932490 | E09_MS03T91 | Pre_EPI | 192 | E09 | whole embryo | 121212ICSI#2day9 | female | GSE74767 | 3,746,140 | 1,502,322 | 753,474 | 876 | 1,092,474 | 396,994 |
| GSM1932457 | E07_MS03T52 | Pre_EPI | 193 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 3,913,000 | 1,296,786 | 582,916 | 640 | 1,601,122 | 431,536 |
| GSM1932458 | E07_MS03T65 | Pre_EPI | 194 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 3,326,435 | 1,206,204 | 477,638 | 707 | 1,312,096 | 329,790 |
| GSM1932474 | E08_MS02T45 | Pre_EPI | 195 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 2,633,093 | 809,519 | 807,506 | 1,038 | 699,159 | 315,871 |
| GSM1932476 | E08_MS02T48 | Pre_EPI | 196 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 4,618,458 | 2,267,311 | 855,484 | 1,393 | 1,036,888 | 457,382 |
| GSM1932478 | E08_MS02T55 | Pre_EPI | 197 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 2,702,684 | 1,369,998 | 520,748 | 843 | 534,669 | 276,426 |
| GSM1932460 | E07_MS08T41 | Pre_EPI | 198 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 4,487,291 | 1,685,809 | 905,915 | 758 | 1,431,994 | 462,815 |
| GSM1932472 | E08_MS02T43 | Pre_EPI | 199 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 3,663,627 | 1,150,153 | 1,098,507 | 822 | 986,098 | 428,047 |
| GSM1932475 | E08_MS02T47 | Pre_EPI | 200 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 3,624,214 | 1,455,132 | 641,544 | 779 | 1,139,473 | 387,286 |
| GSM1932461 | E07_MS08T42 | Pre_EPI | 201 | E07 | whole embryo | 121205ICSI#1day7 | male | GSE74767 | 4,966,321 | 1,898,568 | 786,661 | 693 | 1,794,445 | 485,954 |
| GSM1932473 | E08_MS02T44 | Pre_EPI | 202 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 3,663,855 | 1,572,060 | 749,161 | 561 | 977,792 | 364,281 |
| GSM1932477 | E08_MS02T52 | Pre_EPI | 203 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 3,431,679 | 1,681,422 | 587,613 | 878 | 787,337 | 374,429 |
| GSM1932479 | E08_MS02T57 | Pre_EPI | 204 | E08 | whole embryo | 121212ICSI#11day8 | male | GSE74767 | 3,634,685 | 1,821,962 | 709,497 | 913 | 708,169 | 394,144 |
| GSM1932383 | E14_MS08T52 | PostE_EPI | 205 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 5,018,860 | 2,310,794 | 849,358 | 41,034 | 1,223,490 | 594,184 |
| GSM1932385 | E14_MS08T54 | PostE_EPI | 206 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 3,654,049 | 1,639,829 | 687,972 | 28,356 | 900,880 | 397,012 |
| GSM1932390 | E14_MS10T62 | PostE_EPI | 207 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 3,267,201 | 1,313,405 | 573,627 | 18,403 | 970,348 | 391,418 |
| GSM1932380 | E14_MS08T49 | PostE_EPI | 208 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 4,131,755 | 2,061,105 | 660,675 | 38,247 | 938,004 | 433,724 |
| GSM1932382 | E14_MS08T51 | PostE_EPI | 209 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 4,649,292 | 1,929,765 | 944,233 | 29,120 | 1,241,423 | 504,751 |
| GSM1932391 | E14_MS10T63 | PostE_EPI | 210 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 5,357,391 | 1,646,750 | 1,206,422 | 33,881 | 1,752,267 | 718,071 |
| GSM1932389 | E14_MS10T61 | PostE_EPI | 211 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 3,452,452 | 1,470,294 | 543,136 | 14,227 | 1,075,532 | 349,263 |
| GSM1932379 | E14_MS08T48 | PostE_EPI | 212 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 5,380,776 | 2,294,964 | 1,131,044 | 22,573 | 1,313,719 | 618,476 |
| GSM1932386 | E14_MS08T55 | PostE_EPI | 213 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 3,504,639 | 1,512,957 | 645,824 | 23,157 | 910,255 | 412,446 |
| GSM1932384 | E14_MS08T53 | PostE_EPI | 214 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 4,143,768 | 2,084,074 | 848,006 | 12,178 | 737,024 | 462,486 |
| GSM1932387 | E14_MS08T56 | PostE_EPI | 215 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 4,370,587 | 1,327,520 | 784,876 | 20,863 | 1,643,860 | 593,468 |
| GSM1932381 | E14_MS08T50 | PostE_EPI | 216 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 5,381,220 | 2,042,304 | 786,798 | 30,413 | 1,899,262 | 622,443 |
| GSM1932388 | E14_MS10T60 | PostE_EPI | 217 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 3,406,261 | 1,130,970 | 398,835 | 31,279 | 1,493,031 | 352,146 |
| GSM1932352 | E13_MS10T34 | PostE_EPI | 218 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 3,631,183 | 1,310,399 | 574,706 | 25,917 | 1,339,825 | 380,336 |
| GSM1932372 | E13_MS61T52 | PostE_EPI | 219 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,888,568 | 2,168,287 | 853,587 | 42,138 | 1,281,081 | 543,475 |
| GSM1932356 | E13_MS10T39 | PostE_EPI | 220 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 3,381,261 | 1,205,866 | 498,956 | 21,784 | 1,280,474 | 374,181 |
| GSM1932374 | E13_MS61T55 | PostE_EPI | 221 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,153,188 | 2,039,724 | 640,879 | 28,133 | 1,040,898 | 403,554 |
| GSM1932366 | E13_MS10T58 | PostE_EPI | 222 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 3,661,460 | 1,244,055 | 479,841 | 21,986 | 1,522,610 | 392,968 |
| GSM1932361 | E13_MS10T52 | PostE_EPI | 223 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 6,969,917 | 2,243,634 | 828,708 | 36,783 | 3,079,261 | 781,531 |
| GSM1932355 | E13_MS10T37 | PostE_EPI | 224 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,732,417 | 1,692,455 | 631,569 | 19,068 | 1,872,668 | 516,657 |
| GSM1932357 | E13_MS10T40 | PostE_EPI | 225 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,253,097 | 1,240,030 | 523,745 | 17,892 | 2,009,047 | 462,383 |
| GSM1932377 | E13_MS61T58 | PostE_EPI | 226 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 5,046,660 | 2,538,779 | 817,034 | 29,195 | 1,163,910 | 497,742 |
| GSM1932367 | E13_MS10T59 | PostE_EPI | 227 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 5,327,403 | 2,036,297 | 688,892 | 21,098 | 1,953,234 | 627,882 |
| GSM1932358 | E13_MS10T43 | PostE_EPI | 228 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,064,616 | 1,109,244 | 479,640 | 13,115 | 1,991,049 | 471,568 |
| GSM1932364 | E13_MS10T56 | PostE_EPI | 229 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 5,580,776 | 2,294,746 | 723,015 | 25,072 | 1,895,902 | 642,041 |
| GSM1932368 | E13_MS13T92 | PostE_EPI | 230 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,886,895 | 1,833,952 | 887,046 | 29,651 | 1,518,995 | 617,251 |
| GSM1932370 | E13_MS61T47 | PostE_EPI | 231 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 7,680,274 | 3,126,791 | 1,302,423 | 57,941 | 2,345,974 | 847,145 |
| GSM1932376 | E13_MS61T57 | PostE_EPI | 232 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 5,109,115 | 2,058,620 | 720,968 | 31,317 | 1,740,321 | 557,889 |
| GSM1932360 | E13_MS10T51 | PostE_EPI | 233 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 3,820,780 | 1,379,623 | 543,875 | 25,280 | 1,455,043 | 416,959 |
| GSM1932354 | E13_MS10T36 | PostE_EPI | 234 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,611,068 | 1,704,528 | 793,149 | 33,585 | 1,531,220 | 548,586 |
| GSM1932365 | E13_MS10T57 | PostE_EPI | 235 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 5,477,540 | 1,720,551 | 664,069 | 24,273 | 2,454,522 | 614,125 |
| GSM1932371 | E13_MS61T48 | PostE_EPI | 236 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 6,600,195 | 2,670,987 | 1,224,134 | 62,909 | 1,811,115 | 831,050 |
| GSM1932375 | E13_MS61T56 | PostE_EPI | 237 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,705,431 | 2,206,137 | 820,721 | 44,683 | 1,131,729 | 502,161 |
| GSM1932362 | E13_MS10T54 | PostE_EPI | 238 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 2,960,331 | 916,683 | 326,728 | 14,808 | 1,477,832 | 224,280 |
| GSM1932378 | E13_MS61T59 | PostE_EPI | 239 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,971,556 | 1,948,851 | 751,203 | 37,700 | 1,665,787 | 568,015 |
| GSM1932369 | E13_MS61T46 | PostE_EPI | 240 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 5,407,134 | 2,239,901 | 888,867 | 56,532 | 1,728,923 | 492,911 |
| GSM1932373 | E13_MS61T53 | PostE_EPI | 241 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 5,134,799 | 1,175,688 | 504,949 | 31,199 | 3,016,832 | 406,131 |
| GSM1932400 | E14_MS61T67 | PostE_EPI | 242 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 6,640,905 | 2,492,993 | 1,224,128 | 50,531 | 2,244,773 | 628,480 |
| GSM1932393 | E14_MS13T76 | PostE_EPI | 243 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,173,819 | 2,230,982 | 782,925 | 27,513 | 1,625,416 | 506,983 |
| GSM1932398 | E14_MS61T64 | PostE_EPI | 244 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 4,674,366 | 2,172,897 | 676,874 | 16,119 | 1,325,248 | 483,228 |
| GSM1932397 | E14_MS61T63 | PostE_EPI | 245 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 4,841,732 | 2,501,774 | 778,259 | 14,980 | 1,034,844 | 511,875 |
| GSM1932406 | E14_MS61T73 | PostE_EPI | 246 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 7,329,080 | 3,760,116 | 1,056,217 | 29,739 | 1,749,026 | 733,982 |
| GSM1932395 | E14_MS61T60 | PostE_EPI | 247 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,131,260 | 2,793,393 | 805,815 | 16,180 | 1,003,958 | 511,914 |
| GSM1932403 | E14_MS61T70 | PostE_EPI | 248 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,648,592 | 2,933,835 | 805,367 | 20,691 | 1,357,164 | 531,535 |
| GSM1932399 | E14_MS61T66 | PostE_EPI | 249 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 4,160,408 | 1,765,840 | 627,387 | 17,503 | 1,332,085 | 417,593 |
| GSM1932405 | E14_MS61T72 | PostE_EPI | 250 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 4,732,250 | 2,002,833 | 608,464 | 17,101 | 1,695,533 | 408,319 |
| GSM1932392 | E14_MS13T74 | PostE_EPI | 251 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,908,852 | 2,812,008 | 888,540 | 31,119 | 1,520,633 | 656,552 |
| GSM1932396 | E14_MS61T61 | PostE_EPI | 252 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 4,009,583 | 2,025,315 | 639,702 | 20,422 | 958,211 | 365,933 |
| GSM1932402 | E14_MS61T69 | PostE_EPI | 253 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,224,800 | 2,543,266 | 833,951 | 26,693 | 1,296,474 | 524,416 |
| GSM1932401 | E14_MS61T68 | PostE_EPI | 254 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,762,852 | 1,890,605 | 730,323 | 26,877 | 2,584,245 | 530,802 |
| GSM1932394 | E14_MS13T78 | PostE_EPI | 255 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 7,576,817 | 2,359,981 | 962,007 | 22,321 | 3,421,115 | 811,393 |
| GSM1932404 | E14_MS61T71 | PostE_EPI | 256 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,875,217 | 1,932,537 | 711,004 | 28,833 | 2,764,747 | 438,096 |
| GSM1932213 | E13_MS61T45 | Gast1 | 257 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 6,260,409 | 2,796,039 | 887,162 | 68,989 | 1,968,511 | 539,708 |
| GSM1932214 | E13_MS61T54 | Gast1 | 258 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,067,745 | 1,735,228 | 591,561 | 42,163 | 1,371,322 | 327,471 |
| GSM1932222 | E14_MS13T87 | Gast1 | 259 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,590,610 | 1,321,734 | 434,019 | 28,733 | 3,389,510 | 416,614 |
| GSM1932218 | E14_MS13T80 | Gast1 | 260 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 6,766,479 | 2,020,602 | 724,000 | 36,047 | 3,386,343 | 599,487 |
| GSM1932224 | E14_MS61T65 | Gast1 | 261 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 4,349,151 | 1,584,381 | 645,839 | 15,429 | 1,695,124 | 408,378 |
| GSM1932215 | E14_MS13T75 | Gast1 | 262 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,166,036 | 2,176,984 | 765,430 | 23,296 | 1,636,849 | 563,477 |
| GSM1932223 | E14_MS13T90 | Gast1 | 263 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,972,781 | 2,701,180 | 735,771 | 24,314 | 1,894,707 | 616,809 |
| GSM1932359 | E13_MS10T50 | Gast1 | 264 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,724,340 | 1,712,771 | 629,937 | 24,881 | 1,852,043 | 504,708 |
| GSM1932219 | E14_MS13T82 | Gast1 | 265 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 6,554,677 | 2,366,708 | 943,723 | 25,893 | 2,497,611 | 720,742 |
| GSM1932221 | E14_MS13T85 | Gast1 | 266 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,839,819 | 2,117,626 | 543,131 | 18,100 | 2,628,398 | 532,564 |
| GSM1932217 | E14_MS13T79 | Gast1 | 267 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,852,908 | 2,454,754 | 865,336 | 11,660 | 1,820,675 | 700,483 |
| GSM1932353 | E13_MS10T35 | Gast1 | 268 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,200,696 | 1,636,155 | 615,928 | 19,349 | 1,458,497 | 470,767 |
| GSM1932363 | E13_MS10T55 | Gast1 | 269 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 3,697,179 | 1,663,284 | 575,593 | 15,983 | 1,057,883 | 384,436 |
| GSM1932216 | E14_MS13T77 | Gast1 | 270 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,432,765 | 1,961,603 | 579,443 | 12,731 | 2,364,402 | 514,586 |
| GSM1932220 | E14_MS13T83 | Gast1 | 271 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 7,148,276 | 3,241,380 | 1,036,210 | 22,005 | 2,031,849 | 816,832 |
| GSM1932419 | E16_MS08T77 | PostL_EPI | 272 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 3,017,911 | 1,284,591 | 453,790 | 23,777 | 959,105 | 296,648 |
| GSM1932420 | E16_MS09T21 | PostL_EPI | 273 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 6,972,044 | 2,169,463 | 815,086 | 49,484 | 3,265,932 | 672,079 |
| GSM1932226 | E16_MS08T58 | PostL_EPI | 274 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 2,742,497 | 1,454,138 | 412,393 | 21,077 | 586,321 | 268,568 |
| GSM1932415 | E16_MS08T69 | PostL_EPI | 275 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 3,576,622 | 1,175,118 | 384,620 | 19,214 | 1,621,421 | 376,249 |
| GSM1932225 | E16_MS08T57 | PostL_EPI | 276 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 3,309,054 | 1,324,410 | 535,931 | 20,825 | 1,078,643 | 349,245 |
| GSM1932227 | E16_MS08T59 | PostL_EPI | 277 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 3,421,176 | 1,130,905 | 322,322 | 13,764 | 1,634,648 | 319,537 |
| GSM1932228 | E16_MS09T27 | PostL_EPI | 278 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 6,987,897 | 3,190,969 | 857,587 | 34,323 | 2,236,018 | 669,000 |
| GSM1932417 | E16_MS08T74 | PostL_EPI | 279 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 3,271,328 | 1,122,744 | 339,100 | 23,159 | 1,470,549 | 315,776 |
| GSM1932414 | E16_MS08T68 | PostL_EPI | 280 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 3,155,768 | 1,297,779 | 376,185 | 19,005 | 1,134,414 | 328,385 |
| GSM1932408 | E16_MS08T61 | PostL_EPI | 281 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 3,521,447 | 1,089,438 | 306,755 | 14,118 | 1,831,976 | 279,160 |
| GSM1932411 | E16_MS08T64 | PostL_EPI | 282 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 4,119,502 | 1,491,255 | 377,891 | 19,562 | 1,811,996 | 418,798 |
| GSM1932423 | E16_MS09T41 | PostL_EPI | 283 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 4,835,205 | 2,365,878 | 675,277 | 22,820 | 1,295,202 | 476,028 |
| GSM1932409 | E16_MS08T62 | PostL_EPI | 284 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 4,383,791 | 1,783,961 | 586,778 | 16,350 | 1,549,661 | 447,041 |
| GSM1932412 | E16_MS08T65 | PostL_EPI | 285 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 4,660,737 | 2,301,288 | 754,680 | 20,951 | 1,098,067 | 485,751 |
| GSM1932416 | E16_MS08T71 | PostL_EPI | 286 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 3,872,999 | 1,407,729 | 452,015 | 19,419 | 1,610,000 | 383,836 |
| GSM1932421 | E16_MS09T28 | PostL_EPI | 287 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 5,712,754 | 1,553,723 | 520,191 | 18,892 | 3,074,141 | 545,807 |
| GSM1932410 | E16_MS08T63 | PostL_EPI | 288 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 4,274,353 | 2,069,039 | 621,291 | 25,384 | 1,101,209 | 457,430 |
| GSM1932413 | E16_MS08T66 | PostL_EPI | 289 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 3,962,474 | 1,750,334 | 500,422 | 19,317 | 1,287,892 | 404,509 |
| GSM1932422 | E16_MS09T36 | PostL_EPI | 290 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 6,134,019 | 2,550,707 | 884,771 | 57,696 | 2,083,739 | 557,106 |
| GSM1932407 | E16_MS08T60 | PostL_EPI | 291 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 4,836,244 | 1,823,887 | 499,567 | 41,495 | 2,026,415 | 444,880 |
| GSM1932418 | E16_MS08T75 | PostL_EPI | 292 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 2,493,261 | 1,156,986 | 419,735 | 13,778 | 656,578 | 246,184 |
| GSM1932432 | E17_MS10T84 | PostL_EPI | 293 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,023,290 | 1,750,881 | 632,621 | 10,883 | 1,199,525 | 429,380 |
| GSM1932442 | E17_MS11T49 | PostL_EPI | 294 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 6,332,238 | 2,812,972 | 955,759 | 16,352 | 1,922,568 | 624,587 |
| GSM1932443 | E17_MS11T78 | PostL_EPI | 295 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 7,752,671 | 3,068,219 | 1,036,405 | 29,444 | 2,838,819 | 779,784 |
| GSM1932447 | E17_MS11T84 | PostL_EPI | 296 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 5,943,922 | 2,536,880 | 959,643 | 19,565 | 1,758,741 | 669,093 |
| GSM1932424 | E17_MS10T66 | PostL_EPI | 297 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,123,632 | 1,717,130 | 616,864 | 5,734 | 1,309,485 | 474,419 |
| GSM1932229 | E17_MS11T77 | PostL_EPI | 298 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 5,779,221 | 2,515,040 | 978,086 | 15,745 | 1,712,081 | 558,269 |
| GSM1932230 | E17_MS11T82 | PostL_EPI | 299 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 5,760,027 | 2,498,382 | 843,698 | 11,423 | 1,818,088 | 588,436 |
| GSM1932429 | E17_MS10T77 | PostL_EPI | 300 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,023,569 | 2,003,710 | 657,280 | 9,679 | 951,445 | 401,455 |
| GSM1932431 | E17_MS10T82 | PostL_EPI | 301 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,761,433 | 1,853,833 | 580,120 | 8,003 | 1,876,707 | 442,770 |
| GSM1932454 | E17_MS61T42 | PostL_EPI | 302 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 5,839,848 | 2,869,059 | 854,783 | 11,588 | 1,431,495 | 672,923 |
| GSM1932455 | E17_MS61T43 | PostL_EPI | 303 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,802,883 | 1,528,440 | 569,801 | 9,119 | 2,211,970 | 483,553 |
| GSM1932452 | E17_MS61T40 | PostL_EPI | 304 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 7,816,409 | 4,013,305 | 1,614,145 | 16,708 | 1,365,709 | 806,542 |
| GSM1932426 | E17_MS10T71 | PostL_EPI | 305 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 3,960,544 | 1,680,169 | 536,347 | 6,610 | 1,351,425 | 385,993 |
| GSM1932435 | E17_MS10T87 | PostL_EPI | 306 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,062,660 | 1,878,167 | 634,085 | 6,605 | 1,143,165 | 400,638 |
| GSM1932440 | E17_MS10T94 | PostL_EPI | 307 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 3,835,690 | 1,868,758 | 598,493 | 7,673 | 966,778 | 393,988 |
| GSM1932439 | E17_MS10T93 | PostL_EPI | 308 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,368,840 | 1,868,871 | 703,941 | 6,928 | 1,361,031 | 428,069 |
| GSM1932441 | E17_MS11T46 | PostL_EPI | 309 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,819,748 | 2,325,097 | 687,338 | 8,723 | 1,351,647 | 446,943 |
| GSM1932425 | E17_MS10T68 | PostL_EPI | 310 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,314,460 | 1,988,346 | 699,701 | 7,463 | 1,168,035 | 450,915 |
| GSM1932445 | E17_MS11T80 | PostL_EPI | 311 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 6,360,692 | 2,770,899 | 1,028,820 | 10,546 | 1,923,227 | 627,200 |
| GSM1932438 | E17_MS10T92 | PostL_EPI | 312 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,056,315 | 1,738,857 | 658,106 | 5,791 | 1,207,803 | 445,758 |
| GSM1932450 | E17_MS61T38 | PostL_EPI | 313 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 6,802,869 | 3,662,405 | 1,232,000 | 14,467 | 1,193,026 | 700,971 |
| GSM1932451 | E17_MS61T39 | PostL_EPI | 314 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,388,044 | 2,345,632 | 780,819 | 9,864 | 854,664 | 397,065 |
| GSM1932453 | E17_MS61T41 | PostL_EPI | 315 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 6,199,367 | 2,871,477 | 1,024,328 | 6,945 | 1,637,822 | 658,795 |
| GSM1932456 | E17_MS61T62 | PostL_EPI | 316 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 5,438,048 | 2,796,011 | 1,041,016 | 9,925 | 987,067 | 604,029 |
| GSM1932437 | E17_MS10T91 | PostL_EPI | 317 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 3,281,102 | 1,523,138 | 547,329 | 4,038 | 881,090 | 325,507 |
| GSM1932446 | E17_MS11T81 | PostL_EPI | 318 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 7,434,085 | 2,926,883 | 857,048 | 19,107 | 2,891,178 | 739,869 |
| GSM1932436 | E17_MS10T88 | PostL_EPI | 319 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 9,291,965 | 3,534,721 | 1,299,099 | 11,996 | 3,438,825 | 1,007,324 |
| GSM1932444 | E17_MS11T79 | PostL_EPI | 320 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 7,011,810 | 3,495,850 | 1,077,672 | 7,063 | 1,692,890 | 738,335 |
| GSM1932428 | E17_MS10T73 | PostL_EPI | 321 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,991,604 | 2,405,558 | 754,428 | 3,101 | 1,275,170 | 553,347 |
| GSM1932448 | E17_MS13T95 | PostL_EPI | 322 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 2,818,753 | 1,484,505 | 522,896 | 1,438 | 501,125 | 308,789 |
| GSM1932434 | E17_MS10T86 | PostL_EPI | 323 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 3,919,946 | 1,161,278 | 775,011 | 5,914 | 1,502,712 | 475,031 |
| GSM1932427 | E17_MS10T72 | PostL_EPI | 324 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,853,357 | 2,041,471 | 665,068 | 10,098 | 1,640,388 | 496,332 |
| GSM1932433 | E17_MS10T85 | PostL_EPI | 325 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,018,000 | 1,408,426 | 650,193 | 7,385 | 1,531,369 | 420,627 |
| GSM1932430 | E17_MS10T81 | PostL_EPI | 326 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 10,177,704 | 4,585,826 | 1,764,589 | 19,875 | 2,650,876 | 1,156,538 |
| GSM1932449 | E17_MS61T37 | PostL_EPI | 327 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 5,771,399 | 2,997,376 | 1,103,821 | 10,191 | 1,047,149 | 612,862 |
| GSM1932150 | C6FF_MS11T85 | cyESC | 328 | p35 | on feeder | CMK6 | male | GSE74767 | 5,685,879 | 1,394,685 | 762,044 | 72,737 | 2,997,581 | 458,832 |
| GSM1932157 | C6FF_MS11T94 | cyESC | 329 | p35 | on feeder | CMK6 | male | GSE74767 | 5,399,985 | 1,666,197 | 750,437 | 58,148 | 2,474,841 | 450,362 |
| GSM1932151 | C6FF_MS11T86 | cyESC | 330 | p35 | on feeder | CMK6 | male | GSE74767 | 4,718,245 | 1,290,189 | 983,718 | 30,524 | 1,965,411 | 448,403 |
| GSM1932154 | C6FF_MS11T89 | cyESC | 331 | p35 | on feeder | CMK6 | male | GSE74767 | 7,617,392 | 2,282,589 | 1,209,734 | 52,433 | 3,221,062 | 851,574 |
| GSM1932155 | C6FF_MS11T91 | cyESC | 332 | p35 | on feeder | CMK6 | male | GSE74767 | 6,165,768 | 2,344,540 | 871,538 | 45,552 | 2,363,629 | 540,509 |
| GSM1932153 | C6FF_MS11T88 | cyESC | 333 | p35 | on feeder | CMK6 | male | GSE74767 | 6,568,211 | 2,610,226 | 1,110,630 | 46,406 | 2,161,056 | 639,893 |
| GSM1932152 | C6FF_MS11T87 | cyESC | 334 | p35 | on feeder | CMK6 | male | GSE74767 | 9,385,186 | 3,951,387 | 1,527,775 | 55,172 | 2,993,424 | 857,428 |
| GSM1932156 | C6FF_MS11T92 | cyESC | 335 | p35 | on feeder | CMK6 | male | GSE74767 | 7,049,544 | 2,571,496 | 955,412 | 41,141 | 2,813,948 | 667,547 |
| GSM1932158 | C6FF_MS11T95 | cyESC | 336 | p35 | on feeder | CMK6 | male | GSE74767 | 5,765,027 | 2,174,233 | 820,136 | 39,835 | 2,258,408 | 472,415 |
| GSM1932166 | C6oF_MS05T60 | cyESC | 337 | p32 | on feeder | CMK6 | male | GSE74767 | 5,602,944 | 2,038,658 | 853,198 | 81,768 | 1,999,860 | 629,460 |
| GSM1932170 | C6oF_MS05T65 | cyESC | 338 | p32 | on feeder | CMK6 | male | GSE74767 | 4,338,016 | 1,872,461 | 732,407 | 73,732 | 1,150,358 | 509,058 |
| GSM1932161 | C6oF_MS05T55 | cyESC | 339 | p32 | on feeder | CMK6 | male | GSE74767 | 4,573,153 | 1,803,673 | 808,437 | 74,413 | 1,357,596 | 529,034 |
| GSM1932164 | C6oF_MS05T58 | cyESC | 340 | p33 | on feeder | CMK6 | male | GSE74767 | 5,260,014 | 2,402,288 | 1,088,409 | 57,867 | 1,095,462 | 615,988 |
| GSM1932160 | C6oF_MS05T54 | cyESC | 341 | p32 | on feeder | CMK6 | male | GSE74767 | 4,252,513 | 1,868,236 | 747,226 | 58,557 | 1,097,275 | 481,219 |
| GSM1932171 | C6oF_MS05T66 | cyESC | 342 | p32 | on feeder | CMK6 | male | GSE74767 | 6,482,149 | 2,543,430 | 1,177,311 | 77,141 | 1,862,423 | 821,844 |
| GSM1932165 | C6oF_MS05T59 | cyESC | 343 | p33 | on feeder | CMK6 | male | GSE74767 | 4,931,267 | 1,972,305 | 790,650 | 47,680 | 1,459,006 | 661,626 |
| GSM1932163 | C6oF_MS05T57 | cyESC | 344 | p33 | on feeder | CMK6 | male | GSE74767 | 5,926,478 | 2,280,339 | 855,457 | 56,093 | 2,067,006 | 667,583 |
| GSM1932159 | C6oF_MS05T53 | cyESC | 345 | p32 | on feeder | CMK6 | male | GSE74767 | 4,946,669 | 1,916,811 | 1,362,942 | 50,225 | 1,011,756 | 604,935 |
| GSM1932162 | C6oF_MS05T56 | cyESC | 346 | p33 | on feeder | CMK6 | male | GSE74767 | 5,493,273 | 2,518,346 | 932,134 | 54,525 | 1,281,251 | 707,017 |
| GSM1932168 | C6oF_MS05T63 | cyESC | 347 | p32 | on feeder | CMK6 | male | GSE74767 | 7,969,936 | 3,146,701 | 1,089,650 | 139,992 | 2,679,399 | 914,194 |
| GSM1932172 | C6oF_MS05T67 | cyESC | 348 | p32 | on feeder | CMK6 | male | GSE74767 | 7,307,147 | 3,357,337 | 984,953 | 128,640 | 2,021,391 | 814,826 |
| GSM1932167 | C6oF_MS05T61 | cyESC | 349 | p32 | on feeder | CMK6 | male | GSE74767 | 6,181,340 | 2,867,046 | 957,189 | 97,759 | 1,639,481 | 619,865 |
| GSM1932169 | C6oF_MS05T64 | cyESC | 350 | p32 | on feeder | CMK6 | male | GSE74767 | 6,002,302 | 2,547,962 | 856,777 | 83,920 | 1,801,952 | 711,691 |
| GSM1932180 | CMK9_MS05T75 | cyESC | 351 | p30 | on feeder | CMK9 | female | GSE74767 | 7,016,731 | 2,694,784 | 1,161,606 | 120,177 | 2,233,176 | 806,988 |
| GSM1932174 | CMK9_MS05T69 | cyESC | 352 | p25 | on feeder | CMK9 | female | GSE74767 | 5,994,987 | 2,640,013 | 774,385 | 85,814 | 1,822,376 | 672,399 |
| GSM1932177 | CMK9_MS05T72 | cyESC | 353 | p30 | on feeder | CMK9 | female | GSE74767 | 5,917,510 | 2,083,645 | 785,929 | 89,618 | 2,321,327 | 636,991 |
| GSM1932178 | CMK9_MS05T73 | cyESC | 354 | p30 | on feeder | CMK9 | female | GSE74767 | 6,484,221 | 2,576,534 | 1,059,246 | 85,767 | 2,016,955 | 745,719 |
| GSM1932179 | CMK9_MS05T74 | cyESC | 355 | p30 | on feeder | CMK9 | female | GSE74767 | 6,377,212 | 1,744,318 | 712,750 | 60,254 | 3,203,996 | 655,894 |
| GSM1932173 | CMK9_MS05T68 | cyESC | 356 | p25 | on feeder | CMK9 | female | GSE74767 | 6,015,452 | 2,546,174 | 719,918 | 59,132 | 2,005,387 | 684,841 |
| GSM1932175 | CMK9_MS05T70 | cyESC | 357 | p25 | on feeder | CMK9 | female | GSE74767 | 6,520,471 | 3,392,703 | 915,233 | 48,455 | 1,379,355 | 784,725 |
| GSM1932176 | CMK9_MS05T71 | cyESC | 358 | p30 | on feeder | CMK9 | female | GSE74767 | 6,099,778 | 2,032,247 | 1,008,912 | 39,444 | 2,313,087 | 706,088 |
| GSM1932243 | E17_MS11T48 | Gast2a | 359 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 5,806,294 | 2,678,662 | 846,001 | 12,627 | 1,636,045 | 632,959 |
| GSM1932238 | E17_MS10T70 | Gast2a | 360 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 2,294,680 | 895,882 | 307,165 | 8,389 | 853,773 | 229,471 |
| GSM1932239 | E17_MS10T75 | Gast2a | 361 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 3,944,616 | 1,858,387 | 640,854 | 3,601 | 1,040,613 | 401,161 |
| GSM1932233 | E16_MS09T30 | Gast2a | 362 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 8,574,550 | 2,828,497 | 803,937 | 24,489 | 4,195,868 | 721,759 |
| GSM1932241 | E17_MS10T89 | Gast2a | 363 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 3,919,592 | 1,754,548 | 673,529 | 4,666 | 1,064,154 | 422,695 |
| GSM1978223 | E16_MS09T61 | Gast2a | 364 | E16 | posterior third of embryonic disk | 140708ICSIday16 | female | GSE76267 | 4,049,199 | 2,282,575 | 853,571 | 267 | 560,632 | 352,154 |
| GSM1932235 | E17_MS10T65 | Gast2a | 365 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,440,931 | 2,268,581 | 538,336 | 6,382 | 1,210,161 | 417,471 |
| GSM1932237 | E17_MS10T69 | Gast2a | 366 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,433,787 | 1,199,634 | 448,072 | 3,640 | 2,367,667 | 414,774 |
| GSM1932231 | E16_MS08T72 | Gast2a | 367 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 3,659,156 | 627,889 | 165,971 | 9,682 | 2,619,601 | 236,013 |
| GSM1978241 | E17_MS11T51 | Gast2a | 368 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE76267 | 5,841,383 | 2,885,799 | 986,986 | 10,887 | 1,364,571 | 593,140 |
| GSM1932232 | E16_MS08T73 | Gast2a | 369 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 3,337,171 | 961,445 | 293,657 | 17,746 | 1,748,821 | 315,502 |
| GSM1932240 | E17_MS10T76 | Gast2a | 370 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 3,057,128 | 1,377,378 | 463,617 | 8,849 | 930,996 | 276,288 |
| GSM1932242 | E17_MS11T47 | Gast2a | 371 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 6,081,119 | 3,015,809 | 909,886 | 12,009 | 1,543,543 | 599,872 |
| GSM1932234 | E17_MS10T64 | Gast2a | 372 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,683,382 | 1,799,438 | 610,713 | 9,817 | 1,755,512 | 507,902 |
| GSM1932236 | E17_MS10T67 | Gast2a | 373 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 3,571,880 | 1,451,245 | 431,905 | 8,664 | 1,376,458 | 303,608 |
| GSM1932252 | E17_MS10T95 | Gast2b | 374 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 3,682,906 | 790,702 | 251,159 | 3,396 | 2,378,270 | 259,379 |
| GSM1932255 | E17_MS13T94 | Gast2b | 375 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 5,297,975 | 2,463,188 | 942,433 | 8,642 | 1,216,573 | 667,139 |
| GSM1932245 | E16_MS08T76 | Gast2b | 376 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 2,299,055 | 604,446 | 193,625 | 10,199 | 1,272,468 | 218,317 |
| GSM1932246 | E16_MS09T22 | Gast2b | 377 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 4,897,138 | 1,742,463 | 461,643 | 31,755 | 2,208,400 | 452,877 |
| GSM1932256 | E17_MS13T96 | Gast2b | 378 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 3,365,476 | 1,423,386 | 524,005 | 10,309 | 1,060,600 | 347,176 |
| GSM1932248 | E16_MS09T25 | Gast2b | 379 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 5,437,975 | 1,113,448 | 370,742 | 22,315 | 3,496,380 | 435,090 |
| GSM1932251 | E17_MS10T83 | Gast2b | 380 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,855,016 | 1,984,269 | 658,425 | 9,846 | 1,706,078 | 496,398 |
| GSM1932250 | E17_MS10T78 | Gast2b | 381 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 4,682,344 | 2,164,705 | 656,983 | 7,979 | 1,341,594 | 511,083 |
| GSM1932244 | E16_MS08T67 | Gast2b | 382 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 4,073,298 | 1,422,526 | 361,201 | 14,969 | 1,827,696 | 446,906 |
| GSM1932247 | E16_MS09T24 | Gast2b | 383 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 5,581,486 | 2,005,020 | 522,470 | 22,318 | 2,546,170 | 485,508 |
| GSM1932249 | E16_MS09T33 | Gast2b | 384 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 8,922,904 | 2,781,866 | 861,468 | 46,039 | 4,401,670 | 831,861 |
| GSM1932253 | E17_MS10T96 | Gast2b | 385 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 1,652,495 | 753,658 | 308,308 | 3,416 | 411,239 | 175,874 |
| GSM1932254 | E17_MS11T83 | Gast2b | 386 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 6,352,155 | 2,446,879 | 959,800 | 13,730 | 2,301,042 | 630,704 |
| GSM1978213 | E20_MS08T83 | Gast2b | 387 | E20 | around body stalk | 140401ICSIday20 | male | GSE76267 | 2,533,340 | 1,222,210 | 462,118 | 17,427 | 546,024 | 285,561 |
| GSM1978211 | E20_MS08T81 | Gast2b | 388 | E20 | around body stalk | 140401ICSIday20 | male | GSE76267 | 3,415,819 | 1,614,001 | 579,720 | 19,755 | 844,059 | 358,284 |
| GSM1978209 | E20_MS08T78 | Gast2b | 389 | E20 | around body stalk | 140401ICSIday20 | male | GSE76267 | 2,877,068 | 1,491,499 | 558,264 | 2,960 | 515,863 | 308,482 |
| GSM1978212 | E20_MS08T82 | Gast2b | 390 | E20 | around body stalk | 140401ICSIday20 | male | GSE76267 | 2,913,013 | 1,373,265 | 415,812 | 11,837 | 817,774 | 294,325 |
| GSM1978210 | E20_MS08T79 | Gast2b | 391 | E20 | around body stalk | 140401ICSIday20 | male | GSE76267 | 3,711,400 | 1,396,069 | 459,471 | 35,709 | 1,472,680 | 347,471 |
| GSM1978214 | E20_MS08T84 | Gast2b | 392 | E20 | around body stalk | 140401ICSIday20 | male | GSE76267 | 3,830,874 | 1,522,648 | 522,212 | 32,553 | 1,362,770 | 390,691 |
| GSM1978215 | E20_MS08T85 | Gast2b | 393 | E20 | around body stalk | 140401ICSIday20 | male | GSE76267 | 3,347,144 | 1,107,501 | 455,724 | 32,267 | 1,459,582 | 292,070 |
| GSM1932570 | E17_MS13T91 | VEYE | 394 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE74767 | 5,370,495 | 2,364,228 | 760,318 | 27,171 | 1,646,584 | 572,194 |
| GSM1932569 | E16_MS09T31 | VEYE | 395 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 6,719,326 | 1,904,855 | 497,391 | 61,908 | 3,651,103 | 604,069 |
| GSM1932567 | E14_MS09T18 | VEYE | 396 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 8,900,878 | 2,724,546 | 1,025,611 | 81,392 | 4,094,701 | 974,628 |
| GSM1932566 | E13_MS13T65 | VEYE | 397 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 6,550,968 | 2,745,730 | 866,410 | 74,838 | 2,183,023 | 680,967 |
| GSM1932568 | E16_MS09T29 | VEYE | 398 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 4,699,044 | 1,326,601 | 497,129 | 27,374 | 2,413,441 | 434,499 |
| GSM1932196 | E13_MS61T51 | EXMC | 399 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 2,380,449 | 649,422 | 505,261 | 35,654 | 935,396 | 254,716 |
| GSM1932193 | E13_MS61T44 | EXMC | 400 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,992,419 | 1,777,702 | 1,014,748 | 61,755 | 1,511,700 | 626,514 |
| GSM1932192 | E13_MS13T73 | EXMC | 401 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,968,605 | 1,565,778 | 570,780 | 31,702 | 2,089,133 | 711,212 |
| GSM1932194 | E13_MS61T49 | EXMC | 402 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,514,622 | 1,726,413 | 587,602 | 39,365 | 1,791,584 | 369,658 |
| GSM1932188 | E13_MS13T69 | EXMC | 403 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 7,418,081 | 3,187,534 | 981,406 | 59,464 | 2,389,629 | 800,048 |
| GSM1932189 | E13_MS13T70 | EXMC | 404 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 5,351,896 | 2,036,025 | 612,586 | 41,610 | 2,112,910 | 548,765 |
| GSM1932187 | E13_MS13T68 | EXMC | 405 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,728,036 | 1,696,836 | 539,997 | 37,436 | 1,972,157 | 481,610 |
| GSM1932195 | E13_MS61T50 | EXMC | 406 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 5,113,147 | 2,568,357 | 856,786 | 42,108 | 1,124,353 | 521,543 |
| GSM1932183 | E13_MS10T48 | EXMC | 407 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 5,003,608 | 1,667,614 | 505,878 | 28,894 | 2,299,225 | 501,997 |
| GSM1932186 | E13_MS13T67 | EXMC | 408 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 7,259,032 | 2,935,365 | 947,510 | 53,442 | 2,528,829 | 793,886 |
| GSM1932191 | E13_MS13T72 | EXMC | 409 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 3,647,420 | 1,351,719 | 436,631 | 23,525 | 1,477,061 | 358,484 |
| GSM1932182 | E13_MS10T46 | EXMC | 410 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 3,462,825 | 1,558,079 | 482,451 | 22,280 | 1,042,687 | 357,328 |
| GSM1932181 | E13_MS10T45 | EXMC | 411 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 4,215,974 | 1,682,418 | 602,849 | 17,471 | 1,430,047 | 483,189 |
| GSM1932184 | E13_MS10T49 | EXMC | 412 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 3,890,228 | 1,699,838 | 480,106 | 19,696 | 1,319,844 | 370,744 |
| GSM1932185 | E13_MS13T66 | EXMC | 413 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 6,730,740 | 2,073,784 | 639,340 | 32,378 | 3,356,625 | 628,613 |
| GSM1932190 | E13_MS13T71 | EXMC | 414 | E13 | around embryonic disk | 140812ICSId13A | male | GSE74767 | 5,010,347 | 1,782,709 | 514,103 | 30,293 | 2,213,392 | 469,850 |
| GSM1932201 | E14_MS09T12 | EXMC | 415 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 4,499,258 | 1,396,689 | 673,212 | 47,127 | 1,909,433 | 472,797 |
| GSM1932198 | E14_MS09T05 | EXMC | 416 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 4,712,170 | 1,972,492 | 726,892 | 32,778 | 1,421,622 | 558,386 |
| GSM1932202 | E14_MS09T13 | EXMC | 417 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 4,887,552 | 1,994,152 | 611,182 | 30,203 | 1,732,975 | 519,040 |
| GSM1932197 | E14_MS09T03 | EXMC | 418 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 6,179,641 | 2,674,674 | 922,508 | 38,436 | 1,809,175 | 734,848 |
| GSM1932199 | E14_MS09T08 | EXMC | 419 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 6,284,532 | 2,799,760 | 1,035,924 | 38,507 | 1,673,776 | 736,565 |
| GSM1932200 | E14_MS09T09 | EXMC | 420 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 5,235,294 | 1,435,777 | 1,072,389 | 30,040 | 1,885,009 | 812,079 |
| GSM1932212 | E16_MS09T40 | EXMC | 421 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 6,122,799 | 2,420,950 | 678,658 | 55,194 | 2,425,850 | 542,147 |
| GSM1932208 | E14_MS09T20 | EXMC | 422 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 5,951,877 | 1,828,462 | 1,514,995 | 71,181 | 1,703,653 | 833,586 |
| GSM1932211 | E16_MS09T35 | EXMC | 423 | E16 | around embryonic disk | 140401ICSIday16 | male | GSE74767 | 5,783,250 | 1,645,154 | 595,237 | 37,661 | 3,030,070 | 475,128 |
| GSM1932210 | E14_MS13T89 | EXMC | 424 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,107,578 | 1,751,425 | 561,929 | 30,353 | 2,284,863 | 479,008 |
| GSM1932203 | E14_MS09T14 | EXMC | 425 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 8,424,537 | 3,525,377 | 1,339,289 | 64,959 | 2,522,369 | 972,543 |
| GSM1932205 | E14_MS09T16 | EXMC | 426 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 6,405,722 | 2,397,545 | 975,445 | 46,620 | 2,132,229 | 853,883 |
| GSM1932207 | E14_MS09T19 | EXMC | 427 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 4,994,805 | 1,689,785 | 873,867 | 25,957 | 1,794,598 | 610,598 |
| GSM1932209 | E14_MS13T88 | EXMC | 428 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE74767 | 5,374,005 | 2,084,772 | 649,894 | 20,540 | 2,102,412 | 516,387 |
| GSM1932204 | E14_MS09T15 | EXMC | 429 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 7,495,611 | 3,087,547 | 1,692,548 | 42,352 | 1,720,382 | 952,782 |
| GSM1932206 | E14_MS09T17 | EXMC | 430 | E14 | around embryonic disk | 140401ICSIday14 | male | GSE74767 | 6,623,690 | 2,835,967 | 1,161,044 | 43,281 | 1,855,882 | 727,516 |
| GSM1978240 | E17_MS11T50 | ePGC | 431 | E17 | around embryonic disk | 140722ICSIday17A | male | GSE76267 | 6,484,754 | 2,759,696 | 1,025,123 | 32,728 | 2,042,132 | 625,075 |
| GSM1978227 | E20_MS09T80 | ePGC | 432 | E20 | around body stalk | 140401ICSIday20 | male | GSE76267 | 20,379,189 | 2,768,207 | 1,134,960 | 55,288 | 15,228,572 | 1,192,162 |
| GSM1978242 | E14_MS13T84 | ePGC | 433 | E14 | around embryonic disk | 141014ICSIday14A | male | GSE76267 | 6,451,769 | 2,371,638 | 989,233 | 28,464 | 2,312,769 | 749,665 |
| GSM1978229 | E13_MS10T41 | ePGC | 434 | E13 | around embryonic disk | 140812ICSId13A | male | GSE76267 | 6,195,292 | 1,832,200 | 745,359 | 53,311 | 2,891,781 | 672,641 |
| GSM1978228 | E13_MS10T38 | ePGC | 435 | E13 | around embryonic disk | 140812ICSId13A | male | GSE76267 | 4,544,428 | 1,348,416 | 474,886 | 34,187 | 2,300,868 | 386,071 |
| GSM1978230 | E13_MS10T42 | ePGC | 436 | E13 | around embryonic disk | 140812ICSId13A | male | GSE76267 | 5,397,010 | 1,793,362 | 669,596 | 34,896 | 2,292,925 | 606,231 |
| GSM1978221 | E16_MS09T59 | ePGC | 437 | E16 | posterior third of embryonic disk | 140708ICSIday16 | female | GSE76267 | 4,725,321 | 1,976,399 | 912,879 | 7,891 | 1,380,225 | 447,927 |
| GSM1978219 | E16_MS09T57 | ePGC | 438 | E16 | posterior third of embryonic disk | 140708ICSIday16 | female | GSE76267 | 6,592,996 | 2,471,100 | 1,046,508 | 10,372 | 2,502,009 | 563,007 |
| GSM1978220 | E16_MS09T58 | ePGC | 439 | E16 | posterior third of embryonic disk | 140708ICSIday16 | female | GSE76267 | 5,846,395 | 2,445,922 | 1,061,205 | 9,531 | 1,788,066 | 541,671 |
| GSM1978216 | E16_MS09T54 | ePGC | 440 | E16 | posterior third of embryonic disk | 140708ICSIday16 | female | GSE76267 | 6,440,845 | 2,768,662 | 1,250,856 | 11,287 | 1,828,087 | 581,953 |
| GSM1978218 | E16_MS09T56 | ePGC | 441 | E16 | posterior third of embryonic disk | 140708ICSIday16 | female | GSE76267 | 7,420,875 | 3,629,404 | 1,440,353 | 8,838 | 1,606,541 | 735,739 |
| GSM1978224 | E16_MS09T62 | ePGC | 442 | E17 | around body stalk | 140701ICSIday17 | female | GSE76267 | 6,593,092 | 3,223,754 | 1,231,392 | 11,764 | 1,493,770 | 632,412 |
| GSM1978225 | E16_MS09T63 | ePGC | 443 | E17 | around body stalk | 140701ICSIday17 | female | GSE76267 | 4,563,768 | 2,197,235 | 678,906 | 8,189 | 1,241,506 | 437,932 |
| GSM1978226 | E16_MS09T64 | ePGC | 444 | E17 | around body stalk | 140701ICSIday17 | female | GSE76267 | 4,933,267 | 2,292,021 | 797,954 | 8,037 | 1,357,713 | 477,542 |
| GSM1978217 | E16_MS09T55 | ePGC | 445 | E16 | posterior third of embryonic disk | 140708ICSIday16 | female | GSE76267 | 4,860,698 | 2,001,514 | 878,010 | 8,010 | 1,542,364 | 430,800 |
| GSM1978222 | E16_MS09T60 | ePGC | 446 | E16 | posterior third of embryonic disk | 140708ICSIday16 | female | GSE76267 | 5,728,320 | 2,418,845 | 1,280,759 | 10,686 | 1,480,333 | 537,697 |
| GSM1978231 | E36m_MS11T36 | lPGC | 447 | E36 | genital ridge | 140708ICSId36#60 | male | GSE76267 | 5,400,628 | 2,564,184 | 719,087 | 88,017 | 1,615,308 | 414,032 |
| GSM1978233 | E36m_MS11T39 | lPGC | 448 | E36 | genital ridge | 140708ICSId36#60 | male | GSE76267 | 4,484,534 | 2,101,368 | 659,813 | 53,644 | 1,351,214 | 318,495 |
| GSM1978234 | E36m_MS11T40 | lPGC | 449 | E36 | genital ridge | 140708ICSId36#60 | male | GSE76267 | 5,367,304 | 2,674,202 | 883,623 | 74,230 | 1,293,189 | 442,060 |
| GSM1978232 | E36m_MS11T37 | lPGC | 450 | E36 | genital ridge | 140708ICSId36#60 | male | GSE76267 | 4,398,119 | 838,423 | 233,628 | 24,041 | 3,044,209 | 257,818 |
| GSM1978236 | E36m_MS11T42 | lPGC | 451 | E36 | genital ridge | 140708ICSId36#60 | male | GSE76267 | 4,060,696 | 835,082 | 255,584 | 20,899 | 2,730,162 | 218,969 |
| GSM1978235 | E36m_MS11T41 | lPGC | 452 | E36 | genital ridge | 140708ICSId36#60 | male | GSE76267 | 6,465,443 | 3,042,654 | 1,113,433 | 59,106 | 1,693,668 | 556,582 |
| GSM1643205 | D51_MS04T45 | lPGC | 453 | E51 | genital ridge | 130522ICSI-PGCday51 | female | GSE67259 | 6,217,902 | 1,987,429 | 953,264 | 48,821 | 2,421,909 | 806,479 |
| GSM1978237 | E47m_MS11T43 | lPGC | 454 | E47 | genital ridge | 130522ICSI-PGCday47 | male | GSE76267 | 4,906,783 | 1,142,966 | 471,600 | 30,758 | 2,847,557 | 413,902 |
| GSM1643192 | D43_MS04T07 | lPGC | 455 | E43 | genital ridge | 130522ICSI-PGCday43 | female | GSE67259 | 5,257,575 | 1,388,708 | 600,162 | 59,621 | 2,738,634 | 470,450 |
| GSM1643189 | D43_MS04T03 | lPGC | 456 | E43 | genital ridge | 130522ICSI-PGCday43 | female | GSE67259 | 7,810,554 | 2,442,060 | 836,572 | 92,390 | 3,690,427 | 749,105 |
| GSM1643193 | D43_MS04T08 | lPGC | 457 | E43 | genital ridge | 130522ICSI-PGCday43 | female | GSE67259 | 5,315,584 | 2,054,875 | 818,982 | 50,805 | 1,852,925 | 537,997 |
| GSM1643188 | D43_MS04T02 | lPGC | 458 | E43 | genital ridge | 130522ICSI-PGCday43 | female | GSE67259 | 6,937,663 | 2,311,985 | 863,645 | 67,975 | 3,022,088 | 671,970 |
| GSM1643190 | D43_MS04T04 | lPGC | 459 | E43 | genital ridge | 130522ICSI-PGCday43 | female | GSE67259 | 7,382,718 | 2,568,659 | 1,245,873 | 62,589 | 2,833,619 | 671,978 |
| GSM1643195 | D50_MS04T13 | lPGC | 460 | E50 | genital ridge | 130523ICSI-PGCAday50 | female | GSE67259 | 5,032,528 | 658,565 | 286,527 | 57,956 | 3,612,581 | 416,899 |
| GSM1643201 | D51_MS04T12 | lPGC | 461 | E51 | genital ridge | 130522ICSI-PGCday51 | female | GSE67259 | 5,051,665 | 1,272,937 | 447,202 | 78,010 | 2,843,959 | 409,557 |
| GSM1643200 | D51_MS04T05 | lPGC | 462 | E51 | genital ridge | 130522ICSI-PGCday51 | female | GSE67259 | 5,158,521 | 1,146,301 | 429,323 | 76,297 | 3,065,474 | 441,126 |
| GSM1643203 | D51_MS04T32 | lPGC | 463 | E51 | genital ridge | 130522ICSI-PGCday51 | female | GSE67259 | 4,714,689 | 1,255,199 | 469,655 | 53,814 | 2,582,227 | 353,794 |
| GSM1978238 | E55m_MS11T44 | lPGC | 464 | E55 | genital ridge | 130522ICSI-PGCAday55 | male | GSE76267 | 4,192,986 | 1,006,194 | 281,403 | 49,950 | 2,546,091 | 309,348 |
| GSM1643204 | D51_MS04T44 | lPGC | 465 | E51 | genital ridge | 130522ICSI-PGCday51 | female | GSE67259 | 5,838,129 | 1,445,421 | 618,080 | 76,858 | 3,186,196 | 511,574 |
| GSM1643202 | D51_MS04T25 | lPGC | 466 | E51 | genital ridge | 130522ICSI-PGCday51 | female | GSE67259 | 4,819,220 | 1,697,101 | 740,520 | 60,610 | 1,831,995 | 488,994 |
| GSM1643206 | D51_MS04T51 | lPGC | 467 | E51 | genital ridge | 130522ICSI-PGCday51 | female | GSE67259 | 5,931,039 | 2,780,764 | 970,445 | 79,686 | 1,557,214 | 542,930 |
| GSM1978239 | E55m_MS11T45 | lPGC | 468 | E55 | genital ridge | 130522ICSI-PGCBday55 | male | GSE76267 | 5,041,062 | 1,427,413 | 450,922 | 49,535 | 2,744,972 | 368,220 |
| GSM1643199 | D50_MS04T17 | lPGC | 469 | E50 | genital ridge | 130523ICSI-PGCAday50 | female | GSE67259 | 5,167,957 | 913,813 | 306,932 | 49,705 | 3,442,011 | 455,496 |
| GSM1643196 | D50_MS04T14 | lPGC | 470 | E50 | genital ridge | 130523ICSI-PGCAday50 | female | GSE67259 | 8,528,249 | 1,656,845 | 524,818 | 77,031 | 5,716,794 | 552,761 |
| GSM1643197 | D50_MS04T15 | lPGC | 471 | E50 | genital ridge | 130523ICSI-PGCAday50 | female | GSE67259 | 5,334,342 | 985,333 | 496,368 | 31,348 | 3,319,044 | 502,249 |
| GSM1643198 | D50_MS04T16 | lPGC | 472 | E50 | genital ridge | 130523ICSI-PGCAday50 | female | GSE67259 | 8,113,939 | 1,566,367 | 485,739 | 55,837 | 5,306,171 | 699,825 |
| GSM1643191 | D43_MS04T06 | lPGC | 473 | E43 | genital ridge | 130522ICSI-PGCday43 | female | GSE67259 | 8,190,359 | 3,428,491 | 1,230,319 | 101,251 | 2,577,107 | 853,191 |
| GSM1643194 | D43_MS04T10 | lPGC | 474 | E43 | genital ridge | 130522ICSI-PGCday43 | female | GSE67259 | 19,071,477 | 4,732,262 | 1,885,120 | 213,117 | 10,580,768 | 1,660,210 |
Methods
The description of the method is extended from the related research manuscripts11,12.
Experimental study design
The overall experimental design is illustrated in Fig. 1. For the generation of SC3-seq data, single cells were prepared from pre- and post-implantation embryos, genital ridges and ESCs. They were picked manually and the SC3-seq cDNAs were amplified. After the quality validation and the selection of cDNAs, the DNA libraries for massive parallel sequencers were constructed (see the results of prior qPCR analyses11,12). The sequence reads were acquired by SOLiD5500xl and were mapped onto the cynomolgus monkey genome, Macaca fascicularis 5.0 (MacFas5.0). Then, the reads count was converted into reads per million (RPM). Finally, we verified the global distributions of gene expression and obtained quality-validated transcriptomes. Generally, single-cell transcriptome analysis drops off the positional information. Therefore, the expression patterns of key genes were also examined by immunofluorescent analysis or in situ hybridization of their expression, and each transcriptome data point was annotated accurately by comparison with its histologically examined counterpart11,12.
Figure 1. Overview of the study design and work flow.
The overall experimental design and work flow. The isolated cells dissociated from each samples were picked up for the cDNA synthesis. Then the quality and key gene expression of the generated cDNAs were examined by qPCR and the selected samples were used for the library construction. The libraries were sequenced and the resultant reads were mapped onto the cynomolgus monkey genome. The global distribution of gene expression in each cell was validated. Finally, the annotations of cell clusters were defined by comparing the gene expression patterns of the transcriptome and histological analysis such as immunofluorescent analysis.
Animals
The experimental procedures were approved by the Animal Care and Use Committee of Shiga University of Medical Science, and the methods were carried out in accordance with the approved guidelines. The procedures in cynomolgus monkeys for housing, oocyte collection, intra- cytoplasmic sperm injection (ICSI), pre-implantation embryo culture, and transfer of pre- implantation embryos into foster mothers were performed as described previously with some modifications9, 11,12,13,14 . Briefly, monkeys were housed individually in appropriate cages, and the light cycle consisted of 12 h of artificial light from 8 AM to 8 PM. Temperature and humidity in the animal rooms were maintained at 25±2 °C and 50±5%, respectively. Each animal was fed 20 g kg‒1 of body weight of commercial pellet monkey chow (CMK-1; CLEA Japan Inc., Tokyo, Japan) in the morning, supplemented with 20–50 g of sweet potato in the afternoon. Water was available ad libitum.
For super-ovulation, ovarian stimulation with follicle-stimulating hormone (Gonapure; ASKA) was performed by embedding an implantable and programmable micro-fusion device (iPRECIO; Primetech Corporation) subcutaneously. The day when the ICSI was performed was designated as embryonic day 0 (E0). For embryo transfer, 4 to 5 two-cell to blastocyst-stage embryos were selected and transferred into an appropriate recipient female. For the detection of pregnancy of post-implantation embryos, implanted embryos were monitored by ultrasound scanning. For the collection of early post-implantation embryos (until around E20), the implanted uterus was surgically removed and bisected for the isolation of embryos. For the germ cell collection in gonads, the embryos were delivered by Caesarean section and bisected for the isolation of gonads. All the embryos used in this Data Descriptor are listed in Table 2 (available online only).
Table 2. Summary of embryos and ESCs where single cells were picked from.
| Embryo ID/Cell Line | sex | Embryonic Day/Passage number | cells picked from | cDNA constructed | good quality | SC3-seqed |
|---|---|---|---|---|---|---|
| 130206ICSI#1day6 | male | E06 | whole embryo | 47 | 45 | 23 |
| 130227ICSI#1day6 | female | E06 | whole embryo | 47 | 42 | 18 |
| 130306ICSI#7day6 | female | E06 | whole embryo | 39 | 26 | 2 |
| 121128ICSI#1day7 | male | E07 | whole embryo | 47 | 32 | 10 |
| 121205ICSI#1day7 | male | E07 | whole embryo | 47 | 35 | 17 |
| 130319ICSI#1day7 | male | E07 | whole embryo | 23 | 15 | 8 |
| 121107ICSI13_day8 | male | E08 | whole embryo | 59 | 53 | 36 |
| 121212ICSI#11day8 | male | E08 | whole embryo | 47 | 38 | 21 |
| 130220ICSI#11day8 | male | E08 | whole embryo | 47 | 47 | 38 |
| 120725ICSIday9 | male | E09 | whole embryo | 58 | 45 | 12 |
| 121212ICSI#2day9 | female | E09 | whole embryo | 47 | 37 | 8 |
| 140812ICSId13A | male | E13 | around embryonic disk | 60 | 58 | 49 |
| 140401ICSIday14 | male | E14 | around embryonic disk | 48 | 43 | 33 |
| 141014ICSIday14A | male | E14 | around embryonic disk | 60 | 58 | 30 |
| 140401ICSIday16 | male | E16 | around embryonic disk | 48 | 46 | 36 |
| 140708ICSIday16 | female | E16 | posterior third of embryonic disk | 96 | 94 | 8 |
| 140701ICSIday17 | female | E17 | around body stalk | 96 | 92 | 3 |
| 140722ICSIday17A | male | E17 | around embryonic disk | 96 | 94 | 55 |
| 140401ICSIday20 | male | E20 | around body stalk | 96 | 79 | 8 |
| 140708ICSId36#60 | male | E36 | genital ridge | 48 | 13 | 6 |
| 130522ICSI-PGCday43 | female | E43 | genital ridge | 58 | 55 | 7 |
| 130522ICSI-PGCday47 | male | E47 | genital ridge | 60 | 34 | 1 |
| 130523ICSI-PGCAday50 | female | E50 | genital ridge | 60 | 9 | 5 |
| 130522ICSI-PGCday51 | female | E51 | genital ridge | 60 | 47 | 7 |
| 130522ICSI-PGCAday55 | male | E55 | genital ridge | 30 | 17 | 1 |
| 130522ICSI-PGCBday55 | male | E55 | genital ridge | 30 | 16 | 1 |
| CMK9 | female | p25 | on feeder | 24 | 9 | 3 |
| CMK9 | female | p30 | on feeder | 21 | 14 | 5 |
| CMK6 | male | p32 | on feeder | 46 | 32 | 10 |
| CMK6 | male | p33 | on feeder | 24 | 13 | 4 |
| CMK6 | male | p35 | off feeder | 24 | 3 | 9 |
Cell culture
The cynomolgus ESCs [CMK6 (male) and CMK9 (female)] were gifts from H. Suemori15. For cultivation on feeders, they were cultured with conventional human ESC medium [DMEM/F12 (D6421; Sigma-Aldrich) supplemented with 20% (vol/vol) of KSR (10828-028; Thermo Fisher Scientific), 1 mM of sodium pyruvate (11360-070; Thermo Fisher Scientific), 2 mM of GlutaMax (35050-061; Thermo Fisher Scientific), 0.1 mM of non-essential amino acids (11143-050; Thermo Fisher Scientific), 0.1 mM of 2-mercaptoethanol (M3148; Sigma-Aldrich), 1,000 U ml‒1 of ESGRO mouse LIF (ESG1107; Millipore), and 4 ng ml‒1 of recombinant human bFGF (060-04543; Wako Pure Chemical Industries)] on mouse embryonic feeders (MEFs)16. For feeder-free cultivation, cynomolgus ESCs were cultured under the same condition as human iPSCs (StemFit; Ajinomoto) on recombinant LAMININ511 (iMatrix511, Nippi), as described previously17. All of the cell lines were tested for mycoplasma contamination by MycoAlert (LT07-118; Lonza Japan), according to the manufacturer’s instructions.
Single-cell preparation
For pre-implantation embryos, the zona pellucida was removed by acid tyrode solution treatment (T1788; Sigma-Aldrich). Then the whole embryo was incubated with 0.25% trypsin/PBS (T4799; Sigma-Aldrich) for around 10 min at 37 °C, then dissociated into single cells by repeated pipetting, and dispersed in 0.1 mg ml‒1 of PVA/PBS (P8136; Sigma-Aldrich) for preparation of single-cell cDNAs.
For early post-implantation embryos, the implantation site was dissected out from the uterus and the embryonic fragment containing the EPI, amnion, hypoblast, and yolk-sac endoderm was isolated manually. For isolation of PGCs, in several instances relatively posterior parts of the embryonic fragments were dissected. The embryo information and dissected positions are summarized in Table 1 (available online only). For the embryonic gonads, the genital ridges were dissected out from embryos manually. Then each fragment was incubated with 0.25% trypsin/PBS for around 10 min at 37 °C, dissociated into single cells by repeated pipetting, and dispersed in 0.1 mg ml‒1 of PVA/PBS.
For cynomolgus ESCs, the cells were first detached as clumps with CTK solution [0.25% trypsin (15090-046; Thermo Fisher Scientific), 0.1 mg ml‒1 of collagenase IV (17104-019; Thermo Fisher Scientific), and 1 mM of CaCl2 (06729-55; Nacalai Tesque)], incubated in TrypLE Select (12563029; Thermo Fisher Scientific) for around 10 min at 37 °C, and dispersed into single cells in 1% (vol/vol) KSR/PBS containing 10 μM of the ROCK inhibitor Y-27632 (257-00511; Wako Pure Chemical Industries)16. Cells under the feeder-free condition were directly incubated in TrypLE Select for around 5 min at 37 °C, and dispersed into single cells in 1% (vol/vol) KSR/PBS containing 10 μM of the ROCK inhibitor Y-27632.
Acquisition of SC3-seq data
The SC3-seq process consists of two steps: the synthesis and amplification of cDNAs from isolated single cells, and the construction of a DNA library for sequencing by the SOLiD5500xl sequencer. The first step, the synthesis and amplification of the single-cell cDNA, was performed essentially as described previously10. Briefly, cells were picked up manually into the lysis buffer containing tagged dT primer [V1dT(24)], and lysed by heating. Then the Reverse transcription buffer was added and the 800~1,500 nt cDNA strand was synthesized with short reaction time (5 min). The reaction mixture contained an excess amount of tagged dT primers, which would have interrupted proper amplification. Therefore, the remaining dT primers were digested by exonuclease. Next, in order to add the poly A tail at the end of the synthesized cDNA strand, a terminal deoxynucleotidyl transferase mixture was added into the reaction. Then the second cDNA strand with another tag was synthesized by using a V3-tagged dT primer [V3(dT)24]. Finally, the synthesized double-strand cDNAs were amplified by PCR using V1 and V3 sequences, and the cDNAs with V3 and V1 tags at the 5′ and 3′ sides of the mRNA were obtained.
Before the construction of the DNA library, the quality of the amplified cDNAs was evaluated by examining the Ct values of the qPCR of several endogenous genes and by examining the distribution of the lengths of cDNA fragments using a LabChip GX (CLS760672; Perkin Elmer) or Bioanalyzer 2,100 (5,067-4,626; Agilent Technologies) system. qPCR was performed using Power SYBR Green PCR Master Mix (4367659; Thermo Fisher Scientific) with a CFX384 real-time qPCR system (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. According to the pilot experiments, the samples whose Ct values of GAPDH were more than 21 tended to have poor quality (data not shown). Therefore, we considered the samples whose Ct values of GAPDH and PPIA were less than 19 and 20 as those with good quality. Then we chose appropriate samples from good quality cDNAs for library construction based on the combinations of the lineage-specific gene expression (Table 3 (available online only)). For the cells from pre-implantation embryos, NANOG, GATA4 and GATA6 were used for the marker of EPI, hypoblast and hypoblast/Trophectoderm. For the cells from post-implantation embryos, EPI cells were defined as POU5F1(+)/ NANOG(+)/ SOX2(+)/ PRDM14(+)/ T(−)/ GATA4(−), and gastrulating cells were POU5F1(+)/ NANOG(low)/ PRDM14(low)/ some of T, GATA4, GATA6(+). The extraembryonic cells such as visceral endoderm, yolk sac endoderm and extraembryonic mesenchyme were classified as POU5F1(low) and other lineage-specific genes (+). The early PGCs were identified as PRDM1(+)/ TFAP2C(+)/ SOX17(+)/SOX2(−). The late PGCs from embryonic gonads were identified as POU5F1(+)/ NANOG(+)/ TFAP2C(+)/ SOX2(−). Most of the primer sets were designed using Primer-Blast (NCBI) within a distance of 500 base pairs (bp) from the transcription termination sites (TTSs). The primer sets and oligo DNA sequences used in this Data Descriptor are given in Table 3 (available online only).
Table 3. Primer list.
| Gene | Primer Name | sense | anti-sense |
|---|---|---|---|
| BMP4 | mfBMP4-qPCR-s1/as1 | ATTCCAGTATCCCCAAAGCCTG | GATCTCAGCGACACCCACATC |
| CDX2 | mfCDX2-qPCR-s3/as3 | CTTCCCAGACCAGGAAAGGC | GGCCTGGAGTCCAATAACCA |
| CER1 | mfCER1-qPCR-s1/as1 | GTAATTTGCTTGGTTGCCTCCT | CCAGTGATTGTTCCTCTTCCCT |
| COL6A1 | mfCOL6A1-qPCR-s1/as1 | CTTTTCCCACCAATCCTCACCT | CCTTGGTTTCCCAAAACGAGAC |
| DDX4 | mmDDX4-qPCR-s1/as1 | CATTCCTGGCTTCAGCGGTA | GGGTTGGGAGCTTGTGAAGA |
| DNMT3A | mfDNMT3A_qPCR-s3/as3 | CTGCCAAAAAGGGGGCTAGA | CTGAAGACTCCGTACCCTGC |
| DNMT3L | mfDNMT3L-qPCR-s1/as1 | CGGAAGAAGAATTGTCCCTGC | GGGAGAAAGCAGTTCTTCACCA |
| DPPA3 | mfDPPA3-qPCR-s1/as1 | AGGGATCTGTGTCATTTGAATGTAT | GGCTTCTAAGACACATGGGAATACT |
| FN1 | mfFN1-qPCR-s1/as1 | TGTGCAAGTCTCTCTAATTGTTGA | TTAAAGCAGAAAGTATAAGGCTGTC |
| FOXA1 | mfFOXA1-qPCR-s1/as1 | AGCTGGATTTGAAAATGTGGTCC | CCCGTCTGGCTATACTAACACC |
| FOXA2 | mfFOXA2-qPCR-s1/as1 | GAGGGTCACACTTGATACCCCAC | CAGGTGCTTGAAGAAGCAGGAG |
| FOXH1 | mfFOXH1-qPCR-s1/as1 | GAAACACCAGGCAGTAACCTTG | AGTGTAGGAGAGACAGAGGCAT |
| GAPDH | mfGAPDH-qPCR-s2/as2 | AACAGGGTGGTGGACCTCAT | TTCCTCTTGTGCTCTCGCTG |
| GATA2 | mfGATA2-qPCR-s1/as1 | TTCTGTATGCGGTGATGGCCT | AGCGACTGCCCATCCATATT |
| GATA4 | mfGATA4-qPCR-s1/as1 | TGGCTATAGCAGAGAATACCTTTGAACCA | ACAGGTTTGTGGGTTAGGGAGGGT |
| GATA6 | mfGATA6-qPCR-s1/as1 | AGAAAGGCAATTTTCCTGAGAGA | ACATCCCATTTCTATATTTCCAATTGTT |
| IGF1 | mfIGF1-qPCR-s1/as1 | CCGCTGCTAAACACACCACAG | TAAGCTGCATGATATTTGAAAGGTTTTG |
| KRT18 | mmKRT18-qPCR-s1/as1 | CCAAGATCATGGCAGACATCCG | CTCTCCTCAATCTGCTGAGACC |
| LAMA5 | mfLAMA5-qPCR-s1/as1 | CGTCACCATGACTCGCTCTG | AGTGAGAGGAGTCACCTGCAA |
| MIXL1 | mfMIXL1-qPCR-s1/as1 | TTCAAAACACTCGAGGACTCCC | TGCTAAGGGTAACATTGGAGTGA |
| NANOG | mfNANOG-qPCR-s2/as2 | TGTTCCGGTTTCCATTATGCC | TAGGCTCCAACCATACTCCA |
| NANOS3 | mfNANOS3-qPCR-s1/as1 | ATTTCCAGGAAGACCCACCCTA | GATTTCAGCTGTTGCCTCGC |
| PDGFRA | mfPDGFRA-qPCR-s1/as1 | GCTGGCCTGAGAAACACAATTT | ACGTCCCTCTTCACAAAAATAGGA |
| POU5F1 | mfPOU5F1-qPCR-s1/as1 | GGGAGGAGCTAGGGAAAGAGAACCTA | CCCCCACCCGTTGTGTTCCCA |
| PPIA | mfPPIA-qPCR-s1/as1 | ACAGGTCCTGGCATCTTGTC | CTCAGTCTTGGCAGTGCAGA |
| PRDM1 | mfPRDM1-qPCR-s1/as1 | TTCCCAACTACTCGTTTGTTCTTTG | CATGTAAGAGGCAGAAAAAGGAAGG |
| PRDM14 | mfPRDM14-qPCR-s5/as5 | TGCCCTGTTGTTTTAGGACTGT | AACCAGCAGTTAAGGAAAGGCT |
| SOX15 | mfSOX15-qPCR-s1/as1 | CCTAACCCACCTCTAACTCTGG | TGTAGTCCAACAGGAGAAAGGG |
| SOX17 | mmSOX17-qPCR-s3/as3 | TGCATTGTCAAAAACCCTATTTCCA | CACCCGGGACAACATTTCTTTG |
| SOX2 | mfSOX2-qPCR-s1/as1 | AAGGTTTTTCCCCCTTTATTTTCCG | GATTCTCGGCAGACTGATTCAAATA |
| SOX7 | mfSOX7-qPCR-s1/as1 | ATGATTGACGGGTGTGCAGTC | TGACAACTGTTTCAAAGTATGTGTCG |
| T | mfBRACHYURY-qPCR-s2/as2 | TGCTGTCCCAAGTGGCTTAC | CTGGACCCTGGCAAACATCT |
| TCL1B | mfTCL1B-qPCR-s1/as1 | ATAGATCCAGTGCTGAGCCAGT | TCGTGAGCTGAAGAACTGAACA |
| TFAP2C | mmTFAP2C-qPCR-s2/as2 | TCGGAGATCAAGTCCTCTGG | CCTTTGAACACGGGGTTTAG |
| XIST | mfXIST-qPCR-s1/as1 | TGTGAACTGATGTGAAATGCAGA | CCAACTCCCCAGTTTGTTTCAA |
SC3-seq libraries of quality-checked cDNAs were constructed as described previously10. Briefly, the cDNAs were digested into 150–250 bp fragment by sonication (M210, Covaris). Then the damaged cDNA fragments were end-polished by T4 DNA polymerase (M203; NEB) and T4 polynucleotide kinase (M201; NEB). The cDNAs are now expected to be broken into three types of fragments; the ones with the V3-tag (5′ side of the original mRNA), the ones with the V1-tag (3′ side of the original mRNA), and the ones without any tag (internal part of the original mRNA). This fragment mixture was subjected to a one-cycle DNA polymerization step to allocate the internal adaptor sequence (the essential tag for the SOLiD sequencer) at the end of V1 tag. Then the P1-adaptor (another essential tag for the SOLiD sequencer) was added by ligation at the internal adaptor-free end. Finally, the index sequence and P2 tag (tag for the SOLiD sequencer) were added by PCR, and the library DNAs were obtained.
The quality and quantity of the constructed libraries were evaluated by using a LabChip GX or Bioanalyzer 2,100 system, a Qubit dsDNA HS assay kit (Q32851; Thermo Fisher Scientific), and a SOLiD Library TaqMan Quantitation kit (4449639; Thermo Fisher Scientific). The clonal amplification of the libraries on beads by emulsion PCR was performed using a SOLiD EZ Bead System (4449639; Thermo Fisher Scientific) at the E120 scale according to the manufacturer's instruction. The resulting bead libraries were loaded onto flowchips and sequenced for 50 and 5 bp barcode plus Exact Call Chemistry (ECC) on a SOLiD 5500XL system (4449639; Thermo Fisher Scientific).
Note that the SC3-seq method has now been modified to be applicable to the illumina sequencers. The detailed protocol has been recently published18.
Reference sequences
The genome sequence for MacFas5.0 (mfa_ref_Macaca_fascicularis_5.0_X.fa (X: chr1 - chr20, chrX)) and transcript definition file (GFF3) for MacFas5.0 (ref_Macaca_fascicularis_5.0_top_level.gff3) were obtained from the NCBI ftp site. ERCC spike-in sequences were obtained from the Thermo Fisher Scientific website (https://tools.thermofisher.com/content/sfs/manuals/cms_095047.txt).
Modification of transcript definition file
The SC3-seq protocol enriches ~300 bp from the 3′-end of transcription termination sites (TTSs). Therefore, an inaccurate definition of a TTS, especially one that defines TTS as too far short of the appropriate site, could result in an absence of expression values, even if there are reads accumulated further downstream. To ensure that all possible transcript signals are covered, we extended all 3′-end of TTSs by up to 10 kb in the transcript definition file (GFF3 file, all ‘gene’, ‘transcript’ and ‘exon’ type category, Supplementary File 1) by applying the following rules according to the previous report10:
Identify genes sharing identical TTSs. Remove all these genes but one at each TTS position.
Extend TTS by 10 kb if no gene is found within the targeted area.
If the extended TTS hits a transcription start site (TSS) of a downstream gene (same strand), stop the extension at 1 bp upstream of the TSS.
If the extended TTS hits a TTS of a downstream gene (opposite strand), stop the extension at the mid of two TTS.
Remove all pseudogenes (marked as ‘pseudo=True’), tRNAs or ncRNAs.
Since the gene names in these MacFas5.0 annotations were not fully annotated like their human counterparts, we searched for genomic coordinates of the MacFas5.0 genes on the human genome, hg19, using the LiftOver tool of UCSC, and matched the MacFas5.0 gene name with the human one if the names were different12.
Read processing, mapping and conversion to expression values
All reads were pre-processed by cutadapt v1.11 with ‘-c -e 0.1 -q 20 -n 2 -O 1 -m 30’ options and ‘-a’ and ‘-g’ options for sequence CTCGAGGGCGCGCCGGATCCATATACGCCTTGGCCGTACAGCAG, and -a option for sequence A(20) to remove adaptor and poly-A sequences, and low quality bases19. Untrimmed and trimmed reads of 30 bp or longer were mapped onto the MacFas5.0 genome and ERCC spike-in RNA sequences with tophat v2.0.11/bowtie1.0.1 with ‘-bowtie1 -C -no-coverage search’’ options20. Mapped reads on the genome and ERCC in the bam file were separated and reads on the genome were processed with cufflinks v2.2.0 with ‘—compatible-hit-norm —no-length-correction—library-type fr-secondstrand’ options and MafFas5.0 reference gene annotation with extended TTSs21.
Data Records
The raw csfasta and QV.qual files were deposited in the Gene Expression Omnibus (GEO) database under acquisition numbers GSE67259, GSE76267 and GSE74767 (Data Citation 1, Data Citation 2 and Data Citation 3). The deposited data also contain the abundance of processed expression data, including the Entrez gene IDs and gene names. The sample information is summarized in Table 1 (available online only).
Technical Validation
Verification of the qualities of SC3-seq data
A total of 474 cDNAs from each single cells were sequenced to a depth of between 0.64–4.73 mega mapped reads (Table 1 (available online only)). According to the previous study10, 0.5 mega reads were sufficient for an accurate analysis by SC3-seq. The positions of mapped reads were significantly enriched at the very 3′ ends of mapped genes (Fig. 2a). The comparison of data between cDNAs and subsequent library DNAs indicated that the expression profiles were highly conserved during the experiments (Fig. 2b). Finally, in order to validate the quality of the SC3-seq libraries, we evaluated the profile of expression levels of all the expressed genes in at least one cell (Fig. 2c). Although the single-cell transcriptome is affected by both technical and biological variation, the expression values of the 75th percentile gene showed uniform distribution, indicating that all samples were successfully generated with appropriate qualities.
Figure 2. Verification of the qualities of SC3-seq data.
(a) The SC3-seq track [read density (kilo RPM) plotted against the read position from the annotated TTSs] of all cells (gray) and the average of all cells (red) as indicated above. (b) Scatter plot comparison of the gene expressions acquired by qPCR [delta Ct] and by NGS analysis [log2(RPM+1)]. The delta Ct values of qPCR analysis were calculated from mean values of GAPDH and PPIA. The regression line is shown in red, and the correlation coefficient (r) is indicated at the bottom right of the plot. (c) Distribution of gene expression levels of all expressed genes. All the expressed genes (19,225 genes) were defined as the genes whose log2(RPM+1) values were more than 4 in at least one sample among all 474 cells. The bars at the top, middle and bottom of the box indicate the 75th, 50th, and 25th percentile expression levels, and the top bar of the whisker indicates the expression levels encompassing the expression of 2 s.d. from the median of the genes, respectively.
Clustering and annotating of the cells
The cells were grouped by unsupervised hierarchical clustering (UHC) using the hclust function of R software with Pearson correlation distance and Ward’s method (ward.D2) (Fig. 3a), and t-SNE analysis using the Rtsne function in the Rtsne package with default parameters22 (Fig. 3b). Their annotations were defined by comparing the expression of key genes in transcriptome data with that obtained through the histological analysis11,12 and are summarized in Table 1 (available online only).
Figure 3. Clustering and annotation of the cells.
(a) Unsupervised hierarchical clustering (UHC) with all the expressed genes and a heat map of the levels of selected marker genes. The colored bars under the dendrogram indicate grouping (top) and embryonic days/cell type (bottom), respectively. Post-paTE, post-implantation parietal trophectoderm; PreL-TE, pre-implantation late TE; PreE-TE, pre-implantation early TE; ICM, inner cell mass; Pre-EPI, pre-implantation epiblast; PostE-EPI, post-implantation early epiblast; G1, gastrulating cells group 1; PostL-EPI, post-implantation late epiblast; G2a/G2b, gastrulating cells group 1/2; VE/YE, visceral endoderm/yolk sac endoderm; EXMC, extraembryonic mesenchyme; ePGC, early PGC; lPGC, late PGC. (b) Plot of two-dimensional t-SNE analysis with all the expressed genes. The color codes is as indicated.
In UHC analysis, all cells were classified into two large clusters: 6 clusters mainly from pre-implantation embryos and 10 clusters from post-implantation embryos, genital ridges and ESCs (Fig. 3a). In the left side of the dendrogrham in Fig. 3a, the green, blue and orange groups of the pre-implantation cluster consisted of cells from E7 to E9, and each of them expressed the key genes for the pre-implantation epiblast (Pre-EPI) (NANOG and SOX2), hypoblast (SOX17, GATA4 and GATA6) and late trophectoderm (PreL-TE) (TFAP2C, GATA3 and GATA6) according to the histological analysis12. On the other hand, the light gray and dark gray groups in the pre-implantation cluster consisted of the cells from E6 and E7 only, and while the light gray group expressed a high level of trophectoderm-related genes, the dark gray group did not. Consistent with this, in the t-SNE analysis, the cells in the light gray cluster were plotted close to the PreL-TE, while those in the dark gray cluster were located on the side of Pre-EPI and Hypoblast (Fig. 3b). Therefore we annotated them as pre-implantation early trophectoderm (PreE-TE) and inner cell mass (ICM) cells. The brown group, which was derived from post-implantation embryos but located close to PreL-TE in the pre-implantation cluster (Fig. 3a,b), had a similar gene expression pattern to PreL-TE (Fig. 3a), suggesting that the members of the brown group are the derivative of the PreL-TE. Therefore we annotated these cells as post-implantation parietal trophectoderm (Post-paTE) cells.
In the other large cluster in UHC dendrogram (Fig. 3a), the PGCs from genital ridges (late PGCs; lPGCs) were grouped into one cluster (red), and the cells in yellow clusters from the early post-implantation embryo (E13–20) were annotated as early PGCs (ePGCs) because they were clustered with lPGCs both in the UHC dendrogram and t-SNE plot, and also shared characteristic gene expression patterns with lPGCs(Fig. 3a,b). Next, the other clusters of post-implantation cells were classified into two groups according to their POU5F1 expression (Fig. 3a); POU5F1 is expressed in embryonic cells during the early post-implantation embryo stage12. The light and dark blue groups were annotated as extraembryonic mesenchyme (EXMC) and visceral endoderm/yolk sac endoderm (VE/YE) cells because they showed high and uniform expression of COL6A1 and FOXA1 (Fig. 3a), whose expression patterns were confirmed by histological analysis12. The remaining clusters were classified as EPI [post-implantation early epiblast, PostE-EPI (E13, 14); post-implantation late epiblast, PostL-EPI (E16,17)] or gastrulating cells (Gast1, 2a, 2b) due to the expression of the pluripotency-associated genes and differentiation-related genes. We could not provide a detailed explanation of the cell types of gastrulating cells due to the highly variable expression of the differentiation-related genes. Consistent with this, the cells in Gast2a and Gast2b were not separated clearly in the t-SNE analysis (Fig. 3b). Both the male and female ESCs (CMK6 and CMK9) were clustered close to PostL-EPI (Fig. 3a,b).
Additional Information
Tables 1, 2 and 3 are only available in the online version of this paper.
How to cite this article: Nakamura, T. et al. Single-cell transcriptome of early embryos and cultured embryonic stem cells of cynomolgus monkeys. Sci. Data 4:170067 doi: 10.1038/sdata.2017.67 (2017).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported in part by a Grant-in-Aid from MEXT and by JST-ERATO. We thank Y. Nagai, R. Kabata, N. Konishi, Y. Sakaguchi, M. Kasawaki, T. Sato, M. Kabata, T. Yamamoto, J. Matsushita, C. Iwatani, H. Tsuchiya, Y. Seita, S. Nakamura, and M. Matsutani for their technical assistance. We are grateful to H. Suemori for the provision of CMK6/9, to M. Ema for encouragement, and to the animal care staff at the Research Center for Animal Life Science, Shiga University of Medical Science for their assistance. This work was supported by JST ERATO Grant Numbers JPMJER1104, Japan.
Footnotes
The authors declare no competing financial interests.
Data Citations
References
- Nichols J. & Smith A. Naive and primed pluripotent states. Cell Stem Cell 4, 487–492 (2009). [DOI] [PubMed] [Google Scholar]
- Boroviak T., Loos R., Bertone P., Smith A. & Nichols J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 16, 516–528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons I. G. M. et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 (2007). [DOI] [PubMed] [Google Scholar]
- Kojima Y. et al. The Transcriptional and Functional Properties of Mouse Epiblast Stem Cells Resemble the Anterior Primitive Streak. Cell Stem Cell 14, 107–120 (2014). [DOI] [PubMed] [Google Scholar]
- Tesar P. J. et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 (2007). [DOI] [PubMed] [Google Scholar]
- Saitou M., Kagiwada S. & Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 139, 15–31 (2012). [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S. & Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532 (2011). [DOI] [PubMed] [Google Scholar]
- Irie N. et al. SOX17 is a critical specifier of human primordial germ cell fate. Cell 160, 253–268 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K. et al. Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Cell Stem Cell 17, 178–194 (2015). [DOI] [PubMed] [Google Scholar]
- Nakamura T. et al. SC3-seq: a method for highly parallel and quantitative measurement of single-cell gene expression. Nucleic Acids Res. 43, e60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K. et al. The Germ Cell Fate of Cynomolgus Monkeys Is Specified in the Nascent Amnion. Dev. Cell 39, 1–17 (2016). [DOI] [PubMed] [Google Scholar]
- Nakamura T. et al. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537, 57–62 (2016). [DOI] [PubMed] [Google Scholar]
- Seita Y. et al. Generation of transgenic cynomolgus monkeys that express green fluorescent protein throughout the whole body. Sci. Rep. 6, 24868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki J. et al. Vitrification and transfer of cynomolgus monkey (Macaca fascicularis) embryos fertilized by intracytoplasmic sperm injection. Theriogenology 76, 33–38 (2011). [DOI] [PubMed] [Google Scholar]
- Suemori H. et al. Establishment of embryonic stem cell lines from cynomolgus monkey blastocysts produced by IVF or ICSI. Dev. Dyn. 222, 273–279 (2001). [DOI] [PubMed] [Google Scholar]
- Watanabe K. et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25, 681–686 (2007). [DOI] [PubMed] [Google Scholar]
- Nakagawa M. et al. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci. Rep. 4, 3594 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikura Y. et al. In Vitro Derivation and Propagation of Spermatogonial Stem Cell Activity from Mouse Pluripotent Stem Cells. Cell reports 17, 2789–2804 (2016). [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10 (2011). [Google Scholar]
- Kim D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten L. J. P. & Hinton G. E. Visualizing high-dimensional data using t-sne. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.