In the study herein, we examine the usefulness of germline BCL2 single-nucleotide polymorphisms (SNP) in identifying low- and high-risk patients. Our initial study identified 2 constitutional BCL2 SNPs significantly associated with survival of DLBCL patients treated with R-CHOP. We independently examined the impact of both SNPs in an independent replication set and the association with survival for 1 of the SNPs, rs7226979, was confirmed.
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoid neoplasm and accounts for about 30–40% of non-Hodgkin lymphomas (NHL).1 The prognosis of patients with DLBCL has improved for all ages and both sexes in recent years.2 One of the reasons for this improvement in survival is the addition of rituximab to conventional CHOP (cyclophosphamide, doxorubicin, vincristine and prednisone) chemotherapy.3 From March 1, 2001, the British Columbia (BC) Cancer Agency implemented this approach by recommending the combination of CHOP and rituximab (R-CHOP) for all newly diagnosed patients with DLBCL, regardless of age, in the province of BC.4
Subjects were patients diagnosed with DLBCL who consented and participated in the BC population-based case-control NHL study previously described.5 In summary, subjects were patients with NHL without evidence of HIV infection or prior organ transplantation, aged 20–79 at the time of diagnosis, diagnosed between March 2000 and February 2004.
BCL2 SNP genotyping results were selected from a larger GoldenGate assay (Illumina, San Diego, CA, USA). The procedures for the selection of SNPs and rigorous quality control of samples and genotypes have been described.5
Herein we restricted analysis to the 217 germline DNA samples that were derived from patients with DLBCL (primary mediastinal large B-cell lymphoma (PMBCL) cases were excluded), and examined the effect of genetic polymorphisms in the BCL2 gene. We analyzed data for 9 SNPs, including rs1462129, rs2850761, rs7236090, rs4456611, rs17759659, rs1026825, rs7226979, rs1801018 and rs7230970. Regarding the outcome, diagnostic and treatment information for patients was obtained from the BC Cancer Agency (BCCA) Lymphoid Cancer Database. Information obtained included the Eastern Cooperative Oncology Group (ECOG) performance status,6 serum lactate dehydrogenase (LDH), size of the largest mass of tumor (cm), Ann Arbor stage and the International Prognostic Index score (IPI). Treatment was categorized based on whether patients received primary CHOP (93 patients), or R-CHOP given with curative intent (124 patients), with or without radiation. For each patient at least 6 years of follow up was available.
A group of 168 DLBCL patients without evidence of HIV infection or prior organ transplantation, aged 20–80 years, diagnosed between 2004 and 2012, who received R-CHOP treatment, were selected as an independent replication group. Selection was based on the availability of constitutional DNA from peripheral blood only. All patients in the replication set were followed until 4 June 2015 and had at least 30 months of follow up.
Overall survival (OS) and progression-free survival (PFS) were the outcomes examined. Survival curves were estimated using the Kaplan-Meier method. Cox proportional hazards regression was used to investigate the effects of genetic factors on outcome. Hazard ratios for genetic variation were adjusted by age, sex, ECOG performance status, LDH, size of the largest mass of tumor (cm), Ann Arbor stage/symptom, IPI, ethnicity, and body mass index (BMI). The criterion used to identify confounders in the regression models was a 5% change in the hazard ratio (HR) Each SNP was examined for additive, dominant, recessive and co-dominant models. To account for multiple testing, we used the Bonferroni correction approach. The interaction between SNPs and treatment protocols (CHOP vs. R-CHOP) was examined using the addition of interaction terms in the Cox proportional analysis model. The effect of SNPs with a significant interaction with treatment was calculated separately for CHOP and R-CHOP treatment.
In total, the clinical and outcome information of 217 cases of DLBCL were available for analysis. These included 201 diagnosed as DLBCL (93%), and 16 diagnosed with a composite histology of follicular lymphoma (FL) and DLBCL, treated as DLBCL (7.0%). Of these cases, 188 had quality approved genotyping data for rs1462129, rs2850761, rs7236090, rs1026825, rs7226979 and rs7230970; whereas 164 cases had quality approved genotyping data for rs4456611, rs17759659 and rs1801018. Survival analysis including all cases (i.e., regardless of treatment) did not reveal any significant associations between OS and PFS with any of the BCL2 SNPs. However, there were significant interactions between 2 BCL2 SNPs (rs7226979 and rs4456611) and R-CHOP treatment. Stratified analysis showed associations between rs7226979 (N=105) and rs4456611 (N=93) and OS that were evident only in R-CHOP treated patients (P<0.01). Of these 2 SNPs only rs7226979 showed a statistically significant association with PFS (P<0.05) (Figure 1A, Table 1A).
Figure 1.
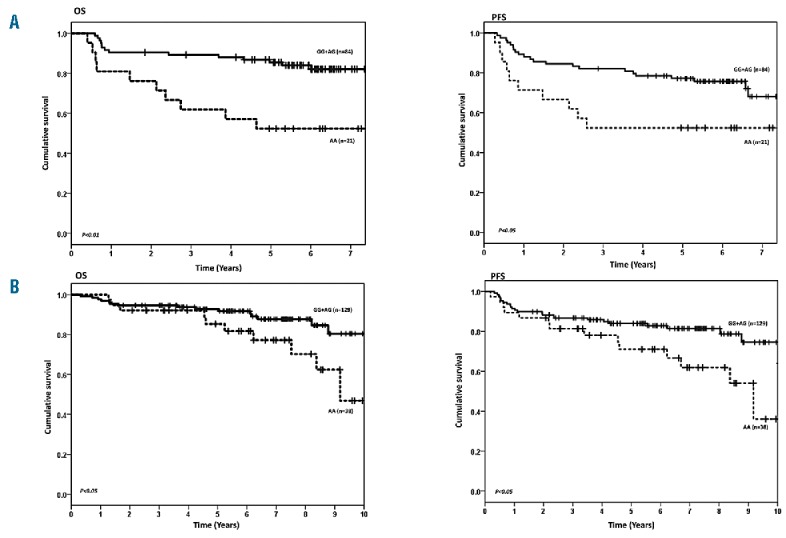
Survival of DLBCL treated with Primary R-CHOP by BCL2 SNP rs7226979. A) Test set. B) Replication set. OS: overall survival; PFS: progression-free survival.
Table 1.
Hazard ratios (HR) and 95% confidence intervals (CI) estimated for the association between BCL2 genotypes and survival of DLBCL patients treated with R-CHOP in A) the test set, and B) the replication set.
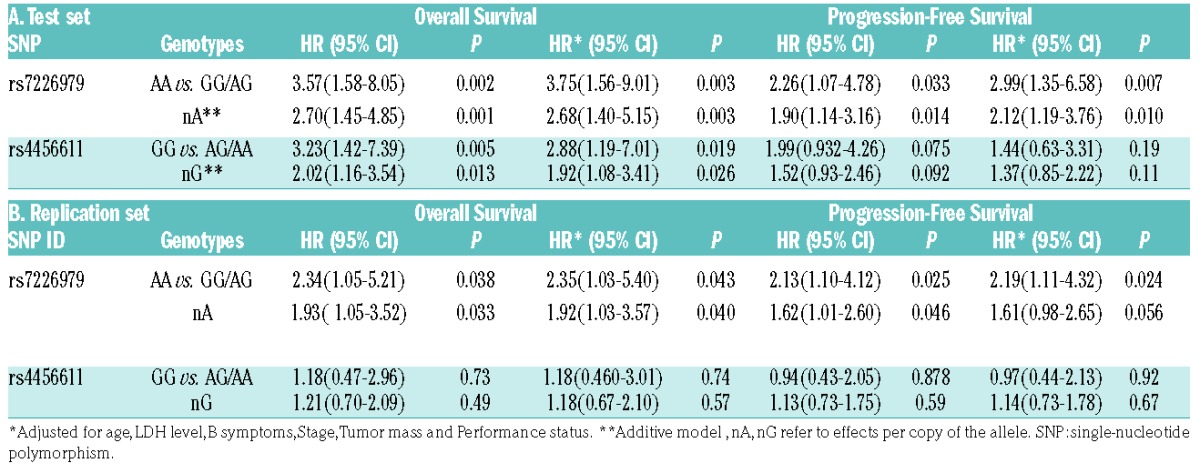
Patients receiving R-CHOP who had the AA genotype at rs7226979 had approximately a 4 times higher risk of death compared to patients with a G allele (P<0.01), which remained the same after adjustment (P<0.01). The same pattern was observed for PFS where carriers of AA had twice the risk of an event compared to other genotypes (P<0.05), and 3 times the risk of an event (P<0.01) after adjusting for other variables (Table 1A). For rs4456611, R-CHOP patients with the GG genotype had over a 3 times higher risk of death compared to patients with an A allele both with and without adjustment (P<0.01), which was attenuated slightly after adjustment for clinical factors (P=0.03). No statistically significant associations were observed between rs4456611 genotypes and PFS before and after adjustments.
Under an additive model, each A allele of rs7226979 increased the risk of death by 2.5 times, with or without adjustment (P<0.01). For rs4456611, G genotype increased the risk of death (P=0.01), however this association was not significant after applying Bonferroni correction. The possibility of having both SNPs in the same haplotype was also tested, and showed that the 2 SNPs are not in linkage disequilibrium (LD; r2=0.13).
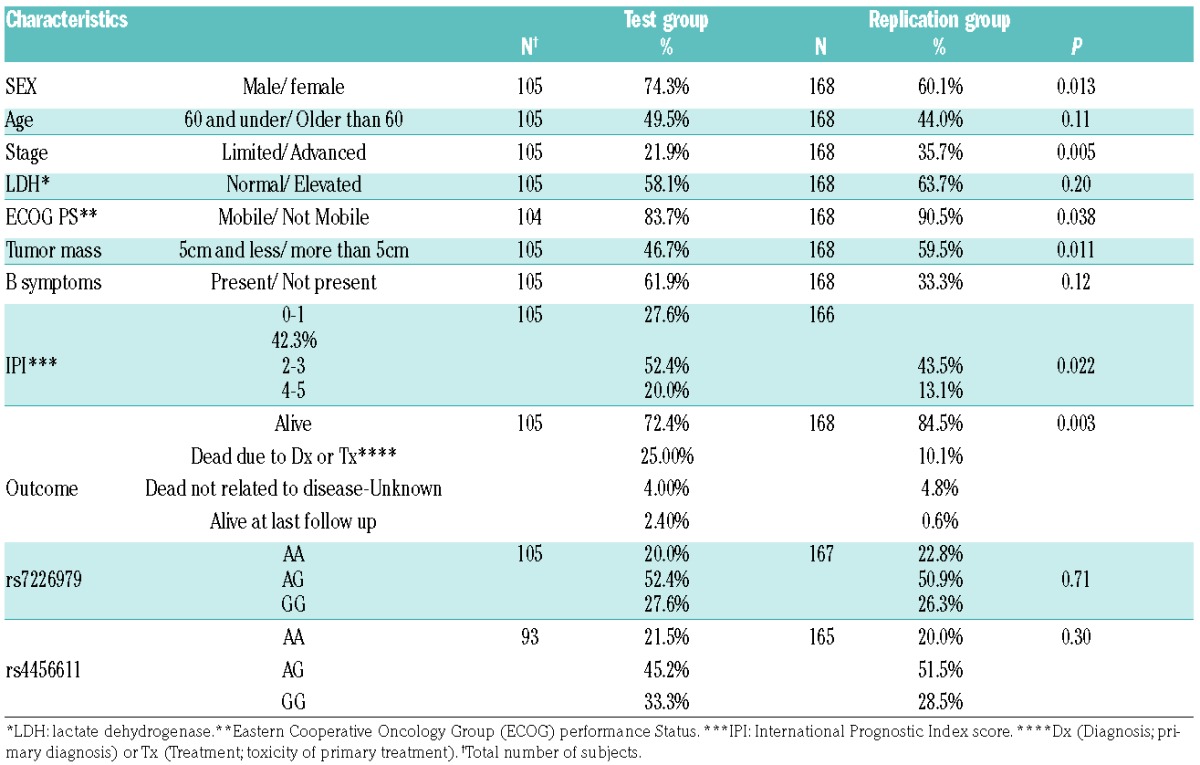
Subjects in the replication set were slightly more likely to be female, older than 60, and have a lower IPI (less advanced stage, better performance status, smaller tumor mass and fewer B symptoms) (Table 2). The results from the replication study are consistent with the association of rs7226979 with OS and PFS seen in the initial study; carriers of the AA genotype had more than double the chance of dying or progression compared to carriers of AG and GG genotypes, with and without adjustment for the same clinical factors (P<0.05). Under an additive model, every A genotype of rs7226979 significantly increased the risk of death 1.9 times, with or without adjustment (P<0.05). The replication study did not confirm the observed association between rs4456611 and OS and PFS. (Table 1B and Figure 1B)
Table 2.
Characteristics and clinical features of the test group† and the replication group.
BCL2 has a key role in DLBCL tumorigenesis; its involvement in lymphoma translocations was the basis for its discovery.7 This gene’s critical role is in the inhibition of apoptosis.8 BCL2 expression is also a strong predictive marker for DLBCL patients treated with R-CHOP.9
Host genetic variation is a promising tool for identifying susceptibility to carcinogenic exposure as well as patient prognosis and prediction of outcome. A recent genome-wide association study (GWAS) identified polymorphisms in patients with DLBCL initially treated with immunochemotherapy that are associated with event-free survival and OS.10 Several reasons may have contributed to the fact that our identified SNPs have not been discovered by previous GWAS. Of those, differences in population structure11 via the use of multicenter studies,10 differences in treatment modalities and the use of different genotyping assays could be responsible for missing BCL2 SNPs.
Our study has several strengths: participants for the test and replication samples are patients from a single institution that provides cancer treatment to the entire province of BC. Our use of a candidate gene design addresses genetic pathways of known biological relevance, and is based on a prior hypothesis. This approach simplifies the interpretation of findings based on the biological plausibility of the BCL2 gene and minimizes the loss of study power due to correction for multiple tests.
A limitation of this study is that we had limited information on molecular heterogeneity of DLBCLs (i.e., germinal center vs. activated B-cell subtypes). Comparison of the test and replication sets revealed different clinical and outcome characteristics that reflect different recruitment approaches.
The exact mechanism through which BCL2 SNPs interact with R-CHOP is not clear. Both of the SNPs studied herein are located in the single intron of BCL2, and so are somewhat unlikely to be functional variants. Some studies reported associations of these SNPS with other biological outcomes and human disorders.12–15 Rs7226979 is significantly associated with the risk of non-obstructive azoospermia.12 BCL2 variant rs12457893 (which is in high LD with rs7226979, r2=0.98), is associated with acute kidney injury in septic shock13 and with the hyperdiploid phenotype of acute lymphoblastic leukemia (ALL).14 Another SNP (rs1944419) in high LD with rs7226979 is associated with neurobehavioral disturbance.15
Currently, most clinically applied predictive markers are based on tumor material; however, the discovery of host related prognostic and predictive factors is also important. Compared to genetic analysis of the tumor, the patient’s constitutional genetic profile is relatively easy to obtain, and can be assessed before treatment is started. Our result has the potential to be useful as a complementary tool to predict the outcome of patients treated with R-CHOP and enhance clinical decision making after confirmation by further studies.
Supplementary Material
Acknowledgments
The authors thank Agnes Lai and Zenaida Abanto (Cancer Control Research, British Columbia Cancer Agency) for their contributions to this study. JMC is the Clinical Director of the British Columbia Cancer Agency Centre for Lymphoid Cancer and receives research funding support from the Terry Fox Research Institute, Genome Canada, Genome British Columbia, the Canadian Institutes for Health Research, and the British Columbia Cancer Foundation.
Footnotes
Funding: this work was supported by the National Cancer Institute of Canada (grant to RDG, JMC, and JJS), and the Canadian Institutes of Health Research (grant to RDG, JMC, and JJS).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Swerdlow SH, International Agency for Research on Cancer., World Health Organization. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon, France: International Agency for Research on Cancer, 2008. [Google Scholar]
- 2.Pulte D, Gondos A, Brenner H. Ongoing improvement in outcomes for patients diagnosed as having Non-Hodgkin lymphoma from the 1990s to the early 21st century. Arch Intern Med. 2008;168(5):469–476. [DOI] [PubMed] [Google Scholar]
- 3.Pfreundschuh M, Trumper L, Osterborg A, et al. CHOP-like chemotherapy plus rituximab versus CHOP-like chemotherapy alone in young patients with good-prognosis diffuse large-B-cell lymphoma: a randomised controlled trial by the MabThera International Trial (MInT) Group. Lancet Oncol. 2006;7(5):379–391. [DOI] [PubMed] [Google Scholar]
- 4.Sehn LH, Donaldson J, Chhanabhai M, et al. Introduction of combined CHOP plus rituximab therapy dramatically improved outcome of diffuse large B-cell lymphoma in British Columbia. J Clin Oncol. 2005;23(22):5027–5033. [DOI] [PubMed] [Google Scholar]
- 5.Schuetz JM, Daley D, Graham J, et al. Genetic variation in cell death genes and risk of non-Hodgkin lymphoma. PloS One. 2012; 7(2):e31560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oken MM, Creech RH, Tormey DC, et al. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol. 1982;5(6):649–655. [PubMed] [Google Scholar]
- 7.Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science. 1984;226(4678):1097–1099. [DOI] [PubMed] [Google Scholar]
- 8.Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335(6189):440–442. [DOI] [PubMed] [Google Scholar]
- 9.Iqbal J, Meyer PN, Smith LM, et al. BCL2 predicts survival in germinal center B-cell-like diffuse large B-cell lymphoma treated with CHOP-like therapy and rituximab. Clin Cancer Res. 2011;17(24): 7785–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghesquieres H, Slager SL, Jardin F, et al. Genome-wide association study of event-free survival in diffuse large B-cell lymphoma treated with immunochemotherapy. J Clin Oncol. 2015;33(33):3930–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11(5):356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin Y, Ji J, Du G, et al. Comprehensive pathway-based analysis identifies associations of BCL2, GNAO1 and CHD2 with non-obstructive azoospermia risk. Hum Reprod. 2014;29(4):860–866. [DOI] [PubMed] [Google Scholar]
- 13.Frank AJ, Sheu CC, Zhao Y, et al. BCL2 genetic variants are associated with acute kidney injury in septic shock. Crit Care Med. 2012; 40(7):2116–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lautner-Csorba O, Gezsi A, Semsei AF, et al. Candidate gene association study in pediatric acute lymphoblastic leukemia evaluated by Bayesian network based Bayesian multilevel analysis of relevance. BMC Med Genomics. 2012;5:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoh NZ, Wagner AK, Alexander SA, et al. BCL2 genotypes: functional and neurobehavioral outcomes after severe traumatic brain injury. J Neurotrauma. 2010;27(8):1413–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


