Abstract
The presence of heritable differences among cancer cells within a tumor, called intra-tumor genetic heterogeneity, has long been suspected of playing a role in poor responses to therapy. Research over the past decade has documented the existence of such heterogeneity within tumors of individual patients and documented its potential clinical significance. The research methods for identifying this heterogeneity were not, however, readily adaptable to widespread clinical application. After a brief review of this background, we describe the development of a measure of intra-tumor genetic heterogeneity, based on whole-exome sequencing of individual tumor samples, that could be applied to biopsy specimens in a clinical setting. This measure has now been used in head and neck squamous cell carcinoma (HNSCC) to document, for the first time, a relation of high intra-tumor genetic heterogeneity to shorter overall survival in a large, multi-institutional study. The implications of heterogeneity for research and clinical care thus now need to be addressed.
Keywords: Head and neck squamous cell carcinoma, Intra-tumor genetic heterogeneity, Next-generation sequencing, Targeted therapy
Introduction
The genetic progression model of head and neck squamous cell carcinoma (HNSCC),1 now twenty years old, assumed an essentially linear progression from normal mucosa through dysplasia and carcinoma in situ to invasive cancer. That work also, however, recognized that neighboring cells in or near a tumor could differ genetically. The authors noted that their work provided “a possible mechanism for field cancerization,” in that “early genetic events may be shared by cells in a local anatomic area, which are apparently derived from a common clone.” This model provided a reasonable explanation for multiple primary tumors arising from the same anatomic area in a patient, as “subsequent genetic events in various subclones produce different phenotypic alterations.”
A related type of genetic diversity, among the cancer cells within an already invasive tumor, has recently been found to be of great importance in HNSCC and other types of cancer. Here we review how such intra-tumor genetic heterogeneity has been found to be important in tumor development and responses to therapy. We then discuss some clinical implications of this heterogeneity, and a way to evaluate this heterogeneity that can be translated from research to the clinic.
Historical background
By intra-tumor genetic heterogeneity we mean the presence of differences in genomic DNA sequences among cancer cells of an individual tumor, which can be passed down to progeny as the cancer cells divide. These differences can include point mutations, copy-number alterations (CNA), and intra- or inter-chromosomal translocations.
The idea that cancer cells within a tumor might differ genetically is not new. Concepts of intra-tumor heterogeneity and its implications for therapy date back at least to the 1970s, as some results with early mouse models of cancer were best explained if there were cells having different sensitivities to cytotoxic therapy or different tendencies to metastasize in the same tumor.2, 3, 4 Around that time, low DNA replication fidelity in tumors was identified as a potential mechanism for genetic differences among cancer cells.5 Until a few years ago, however, little attention was paid to such potential differences among cancer cells within a tumor.
The frequent failures of targeted therapy recently brought renewed attention to intra-tumor heterogeneity. A drug designed to target the particular oncogene driving a tumor was expected to provide a magic bullet that would kill all the cancer cells depending on it.6 Although dramatic initial responses to such drugs could be obtained, these were almost always followed by relapse and tumor regrowth.7 Intra-tumor heterogeneity, as proposed decades previously, provided a reasonable explanation for such initial success followed by treatment failure: therapy selecting for pre-existing subclones that had mutated in ways that happened to provide resistance.
This lack of detailed attention to intra-tumor heterogeneity until a few years ago may in part have been due to a necessary focus on fundamental cellular and molecular mechanisms of cancer, before dealing with what might have originally been considered second-order issues like differences within a tumor. Perhaps more important, however, there were few useful tools for examining such heterogeneity until a few years ago.
Sequencing methods and intra-tumor heterogeneity
The most direct way to examine intra-tumor genetic heterogeneity is to compare DNA sequences among regions of a tumor. Until about 10 years ago, however, detailed genomic DNA analysis was based on Sanger sequencing, which had difficulty detecting even known variants present in 10%–20% of a sample's DNA,8 let alone quantifying unknown variants at that level. In practice this posed a significant problem for analyzing mutations present in tumor subclones. For example, say that a heterozygous mutation is present in a subclone representing 30% of the cancer cells. Then even if there were no normal cells within the tumor sample, the mutation (involving 15% of cancer-cell DNA) would already be down near the Sanger-sequencing detection limit. With many tumor samples containing relatively large amounts of normal cells and their normal DNA (called DNA “impurity” in the jargon of those who study tumor mutations), it would have been difficult to adapt Sanger sequencing to quantitative analysis of intra-tumor genetic heterogeneity.
The usefulness of DNA sequencing for analyzing heterogeneity was dramatically enhanced by the adoption of next-generation sequencing (NGS) for large-scale tumor analysis. Besides dramatically reducing costs, NGS allows better quantitation of individual mutations. In contrast to the analog readout from Sanger sequencing, NGS analyzes each (typically amplified) piece of DNA separately.9 Thus the fraction of DNA bearing a particular mutation is simply the fraction of DNA pieces covering a particular locus that show the mutation, the “mutant-allele fraction” (MAF). Increasing the sequencing depth, the number of DNA pieces analyzed that cover the desired portion of the genome, thus can allow detection of MAF down to levels of a few percent.10 NGS can be combined with prior enrichment for the protein-coding regions of the genome, called exome sequencing, or with whole-genome amplification to allow sequencing down to individual cells.
Modern research measurements of heterogeneity
The work of Maley et al11 on the progression of Barrett's epithelium to invasive esophageal adenocarcinoma may mark the beginning of the recent resurgence of research on the clinical importance of genetic heterogeneity in cancer. Those authors compared several genetic markers (LOH and microsatellite shifts at multiple genomic loci, and mutations in TP53 and CDKN2A) among flow-purified fractions of cells from multiple sites within individual patients' Barrett's epithelia. Based on these results, they determined differences in subclonal composition among the sites of each patient's samples. Patients whose epithelia showed the most clonal diversity were also the most likely to progress to adenocarcinoma.
Genetic heterogeneity has been examined in several types of tumors. For example, in breast cancer, differences among cells were determined even down to the single cell level, with CNA patterns analyzed by fluorescent in-situ hybridization (FISH) of defined genomic regions12 or by whole-genome sequencing of single cells.13 In addition to identifying genetically distinct subclones within primary tumors, it was shown that a single clone of an aneuploid cell could seed a liver metastasis.13 Single-cell sequencing also found genetic diversity in the form of “pseudodiploid” cells with nonrecurring CNA patterns, presumably not forming distinct clones.13 FISH was used to examine CNAs of particular receptor tyrosine kinase (RTK) genes among glioblastoma cells.14 Distressingly for hopes of targeted therapies, neighboring cells within a tumor could differ in terms of which RTK showed amplification. These types of studies clearly documented the existence of intra-tumor genetic heterogeneity and supported the idea that heterogeneity might be clinically important.
The work of Gerlinger et al15 on renal cell carcinoma (RCC) demonstrated the power of NGS to elucidate clinical implications of intra-tumor genetic heterogeneity. They sampled and directly sequenced multiple samples from four RCCs. The authors could thus reconstruct subclonal progressions of mutations that led to the dramatic differences they found among separate portions of individual tumors. Different tumor-suppressor genes could be mutated in different regions of the same tumor, suggesting convergent functional evolution. A mutation leading to constitutively active mTOR was present in some portions of a tumor while missing from others. Associated CNA analysis also documented intra-tumor genetic heterogeneity, and a prognostic signature based on mRNA expression of 110 genes could show either good prognosis or poor prognosis, depending on the tumor region examined.
These and other studies of intra-tumor heterogeneity over the past several years16 had clear clinical implications, particularly for modern targeted therapy. As noted above, work four decades ago had suggested that resistant subclones within a tumor might account for failure of therapy, a possibility recognized even in the early days of targeted therapy.6 Even for therapy targeted against a presumed driver of all cells of a tumor, if a subclone lacked the targeted mutation or had acquired a different mutation that conferred resistance, then even a dramatic initial response to targeted therapy (representing the bulk of the tumor) could be negated by the outgrowth of persistent disease.
Despite these research advances and their clinical implications, the methods used in these studies were difficult to translate to clinical practice. Generally applicable methods like NGS required multiple sampling and multiple analyses that would not be practical in routine clinical application. Although techniques like FISH could be used to evaluate anatomic heterogeneity of defined genomic regions, such methods required pre-identification of genomic regions of interest. For translation to the clinic, there needed to be a generally applicable method for evaluating intra-tumor genetic heterogeneity that could be applied to the initial tumor biopsy specimen.
Mutant-allele tumor heterogeneity (MATH) and HNSCC
The first reports of large-scale exome NGS in HNSCC17, 18 illustrated the lack of attention to intra-tumor genetic heterogeneity as recently as 5 years ago. They also, however, provided the information needed to develop a general way to measure that heterogeneity.
Even as large-scale genomic sequencing of tumors began about a decade ago, the focus generally remained on what mutations were identified within a tumor rather than on whether all cancer cells had the same mutations. Although the use of NGS rather than Sanger sequencing allowed analysis of subclonal mutations in a tumor, research interest was primarily on identifying “driver” mutations accounting for transformations of a normal cell into the cancer cells of a tumor, rather than on “passenger” mutations that had occurred before invasive cancer had developed or that emerged as a tumor progressed.19 Thus subclonal mutations were of secondary interest.
Neither of the first two papers on large-scale exome sequencing of HNSCC, appearing back-to-back in Science in 2011,17, 18 noted the possibility of subclonal mutations. Both simply used the number of mutations found per megabase of DNA analyzed as a measure of mutation rate, regardless of the fraction of tumor DNA that contained each mutation. One of those papers17 did not even provide data on mutant-allele fraction (MAF) values that might help distinguish subclonal mutations.
The second paper on HNSCC exome sequencing, however, provided the information needed to develop a simple, useful measure of intra-tumor genetic heterogeneity. For the somatic mutations they identified, Stransky et al18 provided (in supplemental data) the number of DNA sequence reads that showed the reference DNA sequence and the number that showed the mutated sequence. We realized that this might be sufficient to let us assess the heterogeneity within each tumor.
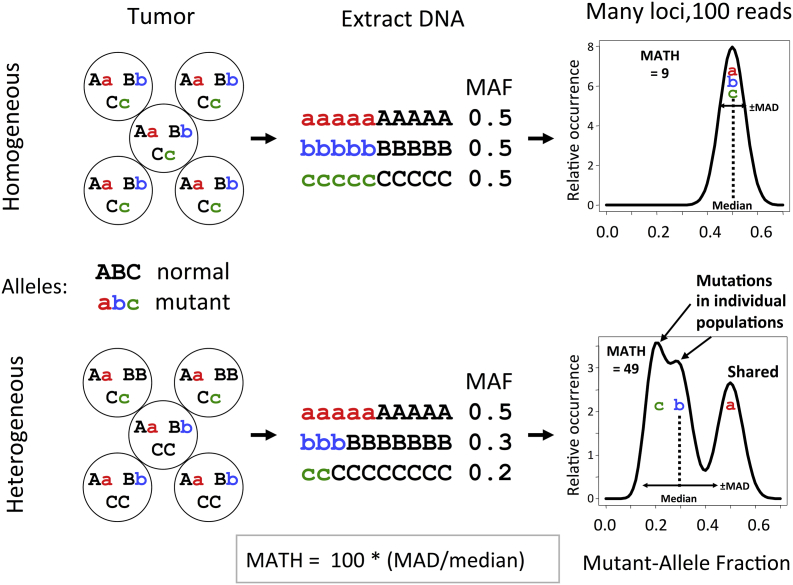
Our basic idea was simple, illustrated in Fig. 1. We reasoned that mutations restricted to subclones of a tumor would tend to appear at lower MAF values than would mutations shared among all cancer cells of a tumor. Thus tumors with higher genetic heterogeneity might be expected to have a wider range of MAF values among their mutated loci than would tumors with low heterogeneity, even in a single sample of a tumor. To minimize contributions from loci with MAF values affected massively by CNA, we used a robust measure of the width of the distribution, the median absolute deviation.
Fig. 1.
The idea behind mutant-allele tumor heterogeneity (MATH) measurement. Heterozygous mutations in a tumor in which all cells share the same mutations (top) will show a narrow distribution of mutant-allele fractions (MAF) after DNA is extracted and subjected to next-generation sequencing (left to right). The width of the distribution of MAF will represent random sampling of mutant and normal alleles, as illustrated hypothetically for 100 total reads at each locus (right). Cells whose combination of shared and unique mutations defines a genetically heterogeneous tumor (bottom) will show a wider distribution of MAF values and a lower median MAF value among mutated loci. MATH is the percentage ratio of the distribution width (the median absolute deviation, MAD) to the median MAF value. Adapted from Mroz and Rocco,20 with permission.
We also recognized, however, that we would have to correct for differences among tumor samples in the “impurity” of normal cell DNA diluting cancer-cell DNA. We reasoned that most somatic mutations in a tumor were likely to be heterozygous “passenger” mutations that had not undergone selection, and thus should have typical MAF values on the order of 0.5 among the cancer cells within a tumor. A lower median MAF value would suggest increasing “impurity” from normal DNA. Thus normalizing raw MAF values by the median MAF value among mutated loci for each tumor should provide a first-order correction for DNA “impurity,” allowing more reliable comparisons of heterogeneity among tumors.
Thus we decided to express the width of the distribution of MAF values as a percentage of the tumor's median MAF value. We called the resulting measure “mutant-allele tumor heterogeneity” (MATH).
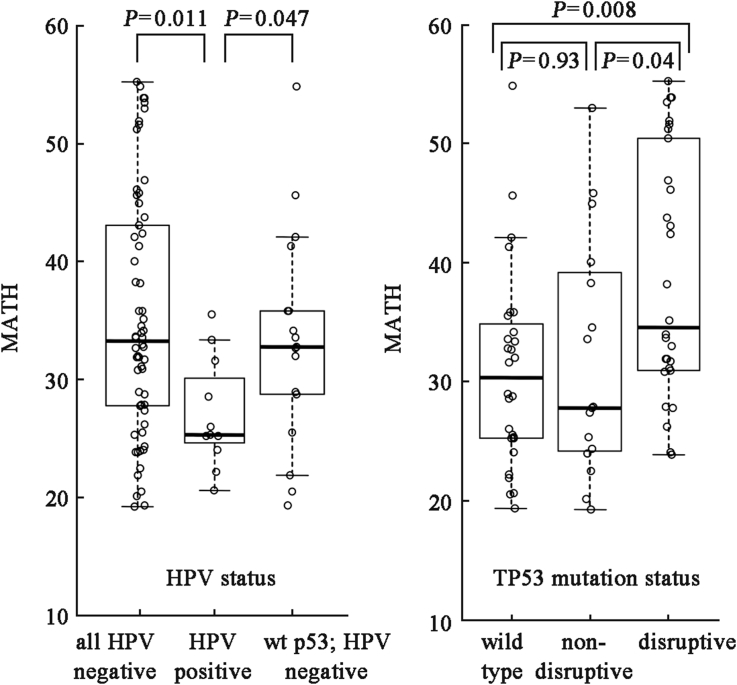
From the limited clinical data initially provided for 74 HNSCC cases by Stransky et al,18 we quickly realized that MATH was associated with poor-prognosis characteristics of HNSCC, as might be expected of a useful measure of intra-tumor genetic heterogeneity (Fig. 2). MATH was lower in tumors positive for human papillomavirus (HPV), with their typically better clinical outcomes, than in HPV-negative tumors, even if the HPV-negative tumors had wild-type TP53. MATH was higher in tumors that bore disruptiveTP53 mutations, with their reportedly worse clinical outcomes,21 than in tumors with wild-type TP53 or with non-disruptive mutations. In the Stransky et al18 cases, MATH increased with pack-years of smoking, a known poor prognostic marker, when TP53 mutation status was taken into account. Thus this very simple measure of heterogeneity, which could be obtained from a single sample of a tumor, showed considerable promise from the outset.20
Fig. 2.
Relation of MATH values to HPV status and TP53 mutation status in HNSCC. Box-and-whisker plots with individual tumor MATH values shown for HNSCC sequenced by Stransky et al.18 Adapted from Mroz and Rocco,20 with permission.
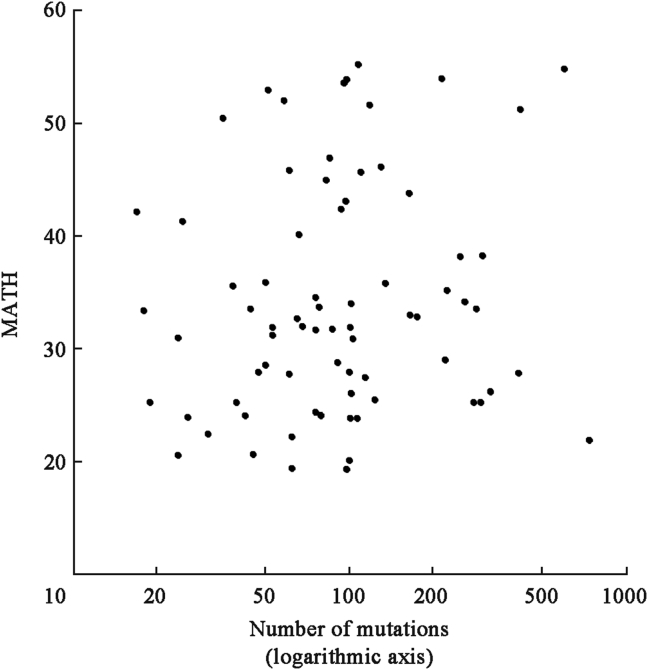
Notably, MATH values represented something other than simply the mutation rates within the tumors. To the initial surprise of some colleagues, who thought that MATH might simply represent mutation rate, the MATH value was independent of the number of mutations within a tumor (Fig. 3), which is proportional to the usual measure of mutation rate in exome-sequencing studies (mutations per megabase of targeted genomic DNA). MATH evidently was capturing, as intended, differences of mutations among cancer cells of a tumor.
Fig. 3.
MATH values of HNSCC are not closely related to the number of mutations, a standard measure of mutation rate. Shown for HNSCC sequenced by Stransky et al.18 From Mroz and Rocco,20 used with permission.
High tumor MATH values and shorter overall survival in HNSCC
These promising initial findings prompted us to test whether tumor MATH values were related to individual clinical outcomes, not just to poor-outcome classes of HNSCC. We worked with colleagues who had been involved in the Stransky et al18 study to obtain and analyze more detailed clinical data for those patients, including overall survival.
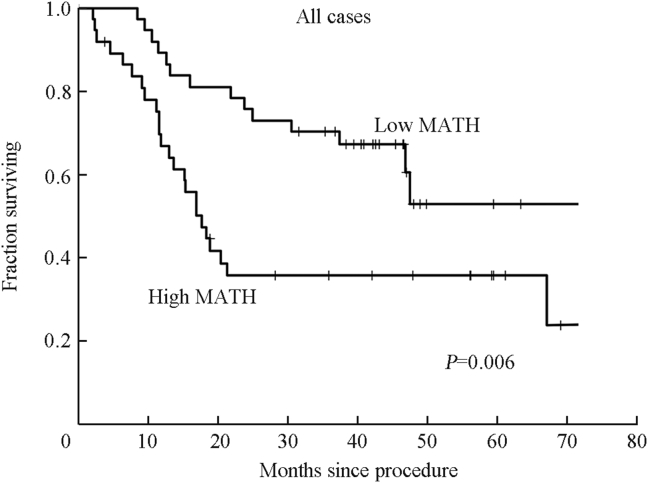
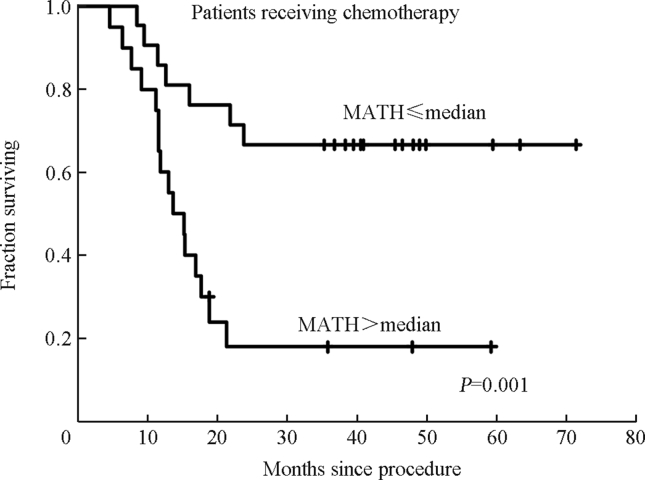
Increasing MATH was significantly related to shorter overall survival among those 74 patients, both as an individual variable and when other prognostic variables were taken into account.22 For plotting survival curves we chose a MATH cutoff of 32, equal to the median of these 74 tumor MATH values. On that basis, patients with high-MATH tumors had twice the hazard of death as those with low-MATH tumors (Fig. 4). For those who received systemic chemotherapy the high/low-MATH hazard ratio of 4 was even larger (Fig. 5).
Fig. 4.
Relation of MATH to overall survival in the 74 patients whose HNSCC had been sequenced by Stransky et al.18 Kaplan–Meier curves from Mroz et al,22 used with permission.
Fig. 5.
Relation of MATH to overall survival in patients whose HNSCC had been sequenced by Stransky et al18 and who had received systemic chemotherapy as part of treatment. Kaplan–Meier curves from Mroz et al,22 used with permission.
In order to generalize this finding of a relation of MATH to outcome, we needed to validate the finding in an independent data set. The Stransky et al18 data came from a single institution, the University of Pittsburgh, and were relatively limited in numbers (74 patients, 39 deaths) even though at the time it was the largest single group of HNSCC patients whose tumors had been subjected to exome NGS.
The Cancer Genome Atlas (TCGA) provided such an independent data set. This multi-institutional, international collaboration supported by the US National Institutes of Health set out to obtain detailed molecular information, including tumor-specific genomic mutations, for hundreds of tumors from each of more than 20 types of cancer, along with corresponding patient clinical data.23 Molecular and clinical data on HNSCC were available to the public even before the initial publication of molecular analyses by TCGA.24
We were careful to treat these TCGA data as a validation data set for our initial findings on MATH and outcome. As the Stransky et al18 data included no MAF values below 0.075, we restricted MATH calculations in the TCGA data to genomic loci with MAF values at or above that cutoff, and used identical methods for calculating MATH values. When distinguishing high-heterogeneity from low-heterogeneity tumors, we also used the identical MATH value cutoff of 32, rather than optimize to the TCGA data set.
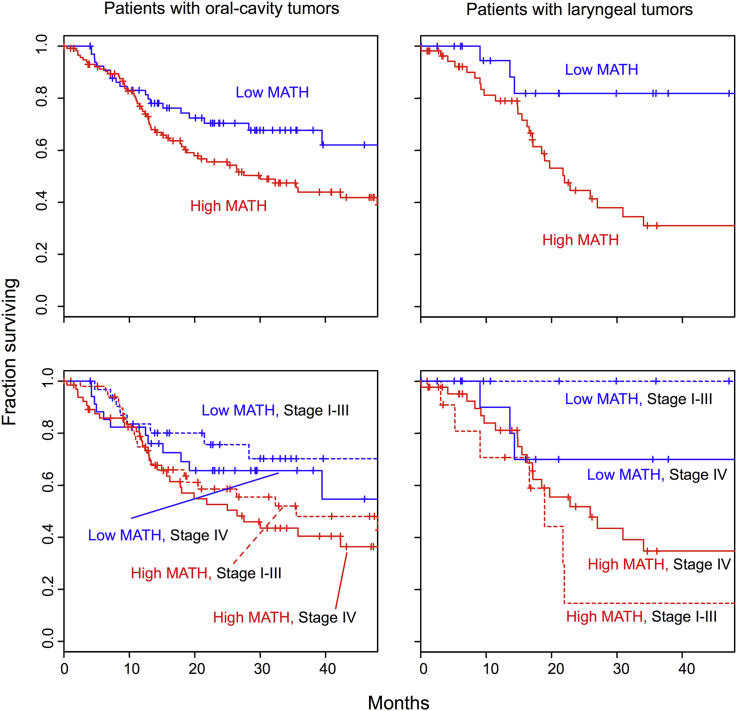
With these TCGA data on HNSCC we thus established a relation of high intra-tumor heterogeneity to worse outcome in a large, multi-institutional study, for the first time to our knowledge in any type of cancer.25 We validated both the overall relation of MATH to HNSCC outcome and a particularly strong relation among patients who received chemoradiation. With this larger data set (305 patients, 131 deaths) we also could demonstrate that high MATH was related to worse outcome even when HPV and TP53 mutation status, tumor grade, smoking history, T and N classifications, age, gender, and TNM stage were taken into account in a multi-variable survival model. Within both oral-cavity and laryngeal subsites, MATH values significantly distinguished outcome classes within TNM stage classes (Fig. 6). Although these data were not obtained from a prospectively designed study, the combination of cases from multiple institutions provided a patient population that was reasonably representative of surgically treated patients in the US having stage II, III, and IV HNSCC. Thus there is very strong evidence that high intra-tumor genetic heterogeneity is associated with worse clinical outcome in HNSCC. The predictions of clinical significance of intra-tumor heterogeneity based on pioneering basic science studies nearly 4 decades ago have been validated, at least for HNSCC.
Fig. 6.
MATH values (low, blue; high, red) distinguish outcomes in patients with oral-cavity (left) or laryngeal (right) tumors (top), even when TNM stage is taken into account (bottom). Kaplan–Meier of cases analyzed by The Cancer Genome Atlas. From Mroz et al,25 figure freely available under the Creative Commons Attribution license.
What MATH is measuring
The explanation above focused on genetically distinct subclones within a tumor as the justification for the MATH algorithm and for a relation of high MATH to worse outcome. Nevertheless, in our first report we noted that the MATH algorithm might represent additional phenomena contributing to worse outcome.20 In particular, the MATH algorithm for identifying intra-tumor heterogeneity would pick up diversity in MAF values among mutated loci arising not only from subclones but also from a diversity of CNA values among genomic loci, even in a monoclonal tumor. It also could pick up the type of “pseudodiploid” CNA, not necessarily associated with defined subclones, reported in breast cancer by Navin et al.13
There is some evidence that CNA makes an important contribution to tumor MATH values and their relation to outcome. A pan-cancer analysis of TCGA tumors identified two major classes of tumors based on patterns of genomic alterations. One, class M, was predominantly characterized by somatic point mutations while the other, class C, showed high levels of CNA instead.26 Many of the TCGA HNSCC had been classified in that paper, so we evaluated the relations of these tumor classes to MATH and to outcome. Class C tumors, with high CNA, showed significantly higher MATH values than did class M tumors, and class C tumors were associated with shorter overall survival (although the association of class C with outcome lost significance when MATH and HPV status were taken into account).25 So a contribution of CNA both to high MATH values and to worse outcome seems likely.
Whether MATH is primarily measuring diversity of point mutations or of CNA, it evidently is capturing some aspect of a tumor that is related to worse outcome. The idea that MATH is measuring tumor subclones that may have pre-existing resistance to systemic therapy, as suggested 40 years ago as an explanation for therapy resistance, is only one way that high MATH scores might be related to poor outcome. Another possibility is that MATH provides a measure of the evolutionary age of the tumor, with an older tumor more likely to have developed an occult metastasis at time of presentation. Consistent with that possibility, patients with high-heterogeneity HNSCC were more likely to show spread of disease to loco-regional nodes.25 Alternatively, high MATH might represent a potential to develop non-lethal point mutations or CNA continually, consistent with the “mutator phenotype” proposed by Loeb.27 In that circumstance, cytotoxic therapy might lead to the appearance and selection of resistance mutations even if they were not present prior to therapy.
As these different processes may have different therapeutic implications it is crucial to learn which are at work. To understand better what MATH is capturing at a cellular and molecular level, we are working with colleagues who have developed methods for teasing out the contributions of normal DNA, overall changes in DNA ploidy, more localized CNA, and subclonal somatic point mutations to the observed MAF values in a tumor.28
Some clinical implications of intra-tumor genetic heterogeneity
Routine clinical application of intra-tumor heterogeneity assessment must await validation of relations between heterogeneity and outcome in samples from prospectively designed clinical studies. Nevertheless, given the compelling evidence from many types of cancer of the clinical importance of intra-tumor heterogeneity, beginning the discussion about its clinical implications is clearly warranted now.
As noted above, one most obvious implication of intra-tumor heterogeneity is the limit that it may place on modern “targeted” therapy. The idea that a tumor is driven by a particular deranged cellular signaling pathway or even is “addicted to” a particular mutation in such a pathway formed the basis for developing drugs that target such pathways or mutations. Even if the target itself is a founder event and is present in all cancer cells of a tumor, intra-tumor heterogeneity may mean that some subclone of cells already harbors additional mutations that will prevent cell death when the target is inhibited. Heterogeneity could even mean that the target was lost from a subclone in later stages of tumor development, as mutations in other proteins replaced its originally necessary function. Either explanation is certainly consistent with typical clinical responses of tumors to targeted therapy, where initial dramatic reductions in tumor volume are typically followed by relapse in a few months. At the least, trials of targeted therapies might best be stratified by intra-tumor heterogeneity to evaluate these possibilities.
A second implication is that measurement of intra-tumor heterogeneity may help inform treatment choices, not just provide prognostic information. Results to date in HNSCC suggest that the relation of MATH to outcome may be most striking when standard systemic therapy is part of initial treatment,22, 25 consistent either with selection of pre-existing resistance mutations or with a tendency to develop mutations that may lead to resistance in high-heterogeneity tumors. If MATH is highly related to systemic therapy outcome but not so strongly to surgical outcome, high-heterogeneity tumors might preferentially be treated with surgery if there is a choice.
This implication of intra-tumor heterogeneity may be particularly important in HNSCC, in which there often is a choice between surgical and organ-preserving cytotoxic therapies for primary treatment. With recent advances in surgical technologies, both surgery and organ-preserving chemoradiation can be considered even for previously poorly accessible anatomic sites like the oropharynx.29 If future studies show that high intra-tumor genetic heterogeneity is strongly related to poor outcome following chemoradiation but not following surgical therapy, then assessment of heterogeneity may help inform this fundamental treatment choice. Similarly, the relation of high primary tumor heterogeneity to nodal metastasis noted above might help inform the choice of whether to perform a neck dissection in oral-cavity cancer in the clinical N0 setting.
A third implication is that the processes that lead to high heterogeneity might themselves represent novel targets for therapy. An analogy can be drawn to mutational variation in microorganisms: too little genetic diversity and the population may be at risk of a novel environmental threat, too much diversity and genetic catastrophe results. The population of cancer cells may similarly be balanced between homogeneity and excess diversity.30 If the processes that lead to that balance can be identified, it might be possible to drive the balance either way to favor destruction of cancer cells.
Intra-tumor genetic heterogeneity has now been documented in many types of cancer, and a large-scale association of intra-tumor genetic heterogeneity and cancer outcome as been established in HNSCC. It is thus time to consider intra-tumor heterogeneity and its implications explicitly as translational research and clinical practice progress.
Disclosure
The Massachusetts General Hospital has filed a patent application based on material discussed in this article, with EAM and JWR as inventors.
Acknowledgment
This work was supported in part by grant R01 DE022087 from the National Institute of Dental and Craniofacial Research.
Footnotes
Peer review under responsibility of Chinese Medical Association.
References
- 1.Califano J., van der Riet P., Westra W. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56:2488–2492. [PubMed] [Google Scholar]
- 2.Hakansson L., Trope C. On the presence within tumours of clones that differ in sensitivity to cytostatic drugs. Acta Pathol Microbiol Scand A. 1974;82:35–40. doi: 10.1111/j.1699-0463.1974.tb03825.x. [DOI] [PubMed] [Google Scholar]
- 3.Fidler I.J., Kripke M.L. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893–895. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 4.Dexter D.L., Kowalski H.M., Blazar B.A., Fligiel Z., Vogel R., Heppner G.H. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 1978;38:3174–3181. [PubMed] [Google Scholar]
- 5.Loeb L.A., Springgate C.F., Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 6.Sawyers C. Targeted cancer therapy. Nature. 2004;432:294–297. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 7.Huang M., Shen A., Ding J., Geng M. Molecularly targeted cancer therapy: some lessons from the past decade. Trends Pharmacol Sci. 2014;35:41–50. doi: 10.1016/j.tips.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Sestini R., Provenzano A., Bacci C., Orlando C., Genuardi M., Papi L. NF2 mutation screening by denaturing high-performance liquid chromatography and high-resolution melting analysis. Genet Test. 2008;12:311–318. doi: 10.1089/gte.2007.0096. [DOI] [PubMed] [Google Scholar]
- 9.Metzker M.L. Sequencing technologies – the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 10.Stead L.F., Sutton K.M., Taylor G.R., Quirke P., Rabbitts P. Accurately identifying low-allelic fraction variants in single samples with next-generation sequencing: applications in tumor subclone resolution. Hum Mutat. 2013;34:1432–1438. doi: 10.1002/humu.22365. [DOI] [PubMed] [Google Scholar]
- 11.Maley C.C., Galipeau P.C., Finley J.C. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 12.Park S.Y., Gonen M., Kim H.J., Michor F., Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navin N., Kendall J., Troge J. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snuderl M., Fazlollahi L., Le L.P. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Gerlinger M., Rowan A.J., Horswell S. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burrell R.A., McGranahan N., Bartek J., Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 17.Agrawal N., Frederick M.J., Pickering C.R. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stransky N., Egloff A.M., Tward A.D. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mroz E.A., Rocco J.W. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49:211–215. doi: 10.1016/j.oraloncology.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poeta M.L., Manola J., Goldwasser M.A. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–2561. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mroz E.A., Tward A.D., Pickering C.R., Myers J.N., Ferris R.L., Rocco J.W. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer. 2013;119:3034–3042. doi: 10.1002/cncr.28150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Cancer Genome Atlas Research Network, Weinstein J.N., Collisson E.A. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mroz E.A., Tward A.D., Hammon R.J., Ren Y., Rocco J.W. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12:e1001786. doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciriello G., Miller M.L., Aksoy B.A., Senbabaoglu Y., Schultz N., Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loeb L.A. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 28.Carter S.L., Cibulskis K., Helman E. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonilla-Velez J., Mroz E.A., Hammon R.J., Rocco J.W. Impact of human papillomavirus on oropharyngeal cancer biology and response to therapy: implications for treatment. Otolaryngol Clin North Am. 2013;46:521–543. doi: 10.1016/j.otc.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cahill D.P., Kinzler K.W., Vogelstein B., Lengauer C. Genetic instability and Darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–M60. [PubMed] [Google Scholar]