Abstract
Genome-wide association studies (GWASs) have revealed numerous loci for areal bone mineral density (aBMD). We completed the first GWAS meta-analysis (n = 15,275) of lumbar spine volumetric BMD (vBMD) measured by quantitative computed tomography (QCT), allowing for examination of the trabecular bone compartment. SNPs that were significantly associated with vBMD were also examined in two GWAS meta-analyses to determine associations with morphometric vertebral fracture (n = 21,701) and clinical vertebral fracture (n = 5893). Expression quantitative trait locus (eQTL) analyses of iliac crest biopsies were performed in 84 postmenopausal women, and murine osteoblast expression of genes implicated by eQTL or by proximity to vBMD-associated SNPs was examined. We identified significant vBMD associations with five loci, including: 1p36.12, containing WNT4 and ZBTB40; 8q24, containing TNFRSF11B; and 13q14, containing AKAP11 and TNFSF11. Two loci (5p13 and 1p36.12) also contained associations with radiographic and clinical vertebral fracture, respectively. In 5p13, rs2468531 (minor allele frequency [MAF] = 3%) was associated with higher vBMD (β = 0.22, p = 1.9 × 10−8) and decreased risk of radiographic vertebral fracture (odds ratio [OR] = 0.75; false discovery rate [FDR] p = 0.01). In 1p36.12, rs12742784 (MAF = 21%) was associated with higher vBMD (β = 0.09, p = 1.2 × 10−10) and decreased risk of clinical vertebral fracture (OR = 0.82; FDR p = 7.4 × 10−4). Both SNPs are noncoding and were associated with increased mRNA expression levels in human bone biopsies: rs2468531 with SLC1A3 (β = 0.28, FDR p = 0.01, involved in glutamate signaling and osteogenic response to mechanical loading) and rs12742784 with EPHB2 (β = 0.12, FDR p = 1.7 × 10−3, functions in bone-related ephrin signaling). Both genes are expressed in murine osteoblasts. This is the first study to linkSLC1A3 and EPHB2 to clinically relevant vertebral osteoporosis phenotypes. These results may help elucidate vertebral bone biology and novel approaches to reducing vertebral fracture incidence. © 2016 American Society for Bone and Mineral Research.
Keywords: Bone QCT/μCT, Analysis/Quantitation of Bone, Osteoporosis, Diseases and Disorders of/Related to Bone, General Population Studies, Epidemiology, Human Association Studies, Genetic Research, Fracture Risk Assessment
Introduction
Vertebral osteoporosis and fracture are substantial sources of pain, height loss, and mobility limitation in older adults.(1) Not only has the burden of these conditions increased with the aging of populations, but the age-specific incidence of vertebral fracture has risen or remained steady even as the incidence of hip and other osteoporotic fractures has declined.(2–4) Risk of fracture correlates with lower vertebral bone mineral density (BMD)(5) that declines with age, leaving the bone susceptible to compression and deformation even in the absence of traumatic force.
Both vertebral BMD and fracture are heritable,(6) and multiple genetic loci have been associated with BMD as determined by dual-energy X-ray absorptiometry (DXA) through large-scale genomewide association study (GWAS) meta-analyses.(7,8) Far fewer loci—either from BMD candidate genes or from GWAS— have been linked to fracture risk, even in large samples.(9,10) This is likely due to several factors, including the multifactorial causation of fracture, heterogeneity of fracture risk by skeletal sites, and the inability of DXA BMD to completely define bone strength and fracture risk.
Vertebral bone is unique in that its source of strength comes to a large extent from the trabecular compartment, with relatively little cortical area compared to other load-bearing skeletal sites. Quantitative computed tomography (QCT) of the vertebrae allows for volumetric BMD (vBMD) measures of the trabecular compartment specifically, which are more strongly associated with vertebral fracture than areal BMD (aBMD) measures.(11) aBMD measured by DXA is confounded by bone size. In contrast with DXA, CT-based measurements of the spine allow for exclusion of vertebral elements and artifacts, including osteophytic or extraskeletal calcification that is common in older adults.(12,13) For these reasons, we undertook a GWAS meta-analysis of QCT-based vBMD in the lumbar spine in older men and women, and we evaluated the resulting associations in separate meta-analyses of vertebral fracture.
Subjects and Methods
Study design and participants
Cohort design and characteristics are described in Supplemental Table 1.
Discovery studies
Six cohorts of men and women of European descent with CT imaging of the L2or L3 spine were included in a discovery metaanalysis of trabecular vBMD (n = 12,287). Discovery cohorts included the Age Gene/Environment Susceptibility-Reykjavik Study (AGES-Reykjavik)(14); Framingham Osteoporosis Study(6,15); Family Heart Study; Health Aging and Body Composition (Health ABC); Multi-Ethnic Study of Atherosclerosis (MESA)(16);and Osteoporotic Fractures in Men (MrOS).(17) Except for Health ABC, the cohorts also had an integral vBMD (n = 11,080) phenotype at the same lumbar vertebra. All participants were adults, and the mean age in each cohort ranged from 52 to 76 years (Supplemental Table 1).
Replication studies
Replication of trabecular vBMD associations were done in silico in the Diabetes Heart Study (DHS, n = 967) and with de novo genotyping on selected SNPs in an additional sample from the AGES-Reykjavik cohort (n = 2020).
Fracture studies
Two of the discovery cohorts (Framingham and MrOS) and five additional cohorts (MrOS-Sweden, Rotterdam I-III, and Study of Osteoporotic Fractures [SOF]) contributed to the evaluation of trabecular vBMD-associated loci for their association with radiographic vertebral fracture (n = 21,701; 20% fracture cases).
Lumbar spine CT phenotypes
CT scans of the spine provided vBMD phenotypes at either L2 or L3 (Fig. 1). In cohorts with vBMD measured at adjacent levels, correlations were high (r = 0.89 to 0.93). CT scanners and software used in each cohort are listed in Supplemental Table 1. In MESA, CT data were analyzed using Image Analysis QCT 3D PLUS software (Image Analysis, Columbia, Kentucky), and the remaining cohorts were analyzed using software developed by one author (TL).(18) In order to ensure regions of interest were defined consistently across cohorts, two authors (TL and MB) evaluated phenotype definitions. The tissue density of the analyzed volume calibrated to units of equivalent concentration of hydroxyapatite in g/cm3 yielded the BMD values. The following two BMD phenotypes were analyzed:
Fig. 1.
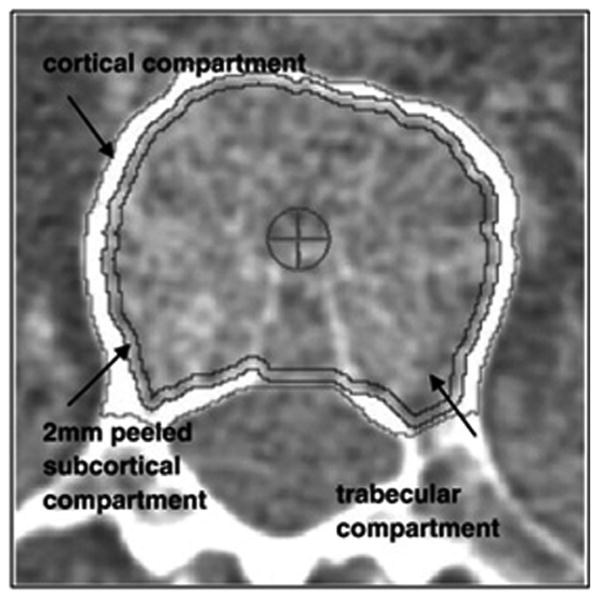
Cross-sectional view of the trabecular region of interest in the lumbar spine. Trabecular vBMD included this region only, whereas integral vBMD also included the cortical compartment. Both exclude the posterior elements that DXA measures of BMD incorporate, thus allowing CT measures to more precisely capture BMD of the vertebral body itself. Reprinted with permission from Elsevier from: Engelke K, Mastmeyer A, Bousson V, Fuerst T, Laredo J-D, Kalender WA. Reanalysis precision of 3D quantitative computed tomography (QCT) of the spine. Bone, 2009:44(4):566–72.
Trabecular vBMD is the average density in g/cm3 of all the voxels contained within the boundary of the trabecular region. This measure was calculated from a single slice of the region encompassing most of the trabecular bone in the vertebral body (Fig. 1).
Integral vBMD is the average density in g/cm3 of all the voxels contained within the periosteal edge of the vertebral body, excluding all of the posterior elements (Fig. 1).
Vertebral fracture phenotypes
Two separate fracture GWAS meta-analyses were conducted on non-overlapping population samples. Radiographic vertebral fracture is often asymptomatic and was evaluated by imaging in cohort studies of older men and women (described below in Radiographic vertebral fracture). On the other hand, clinical vertebral fracture was defined by signs and symptoms such as back pain, height loss, and kyphosis and was confirmed by imaging.
Radiographic vertebral fracture
These studies included prevalent and incident fracture evaluated from T4 to L4. Prevalent radiographic vertebral fracture was defined for each cohort as described in Supplemental Table 1. Prevalent vertebral fracture was defined by Genant's scale(19) or by using vertebral body height ratios >3 SD,(20,21) evaluated by visual semiquantitative readings or quantitative assessments with review by a radiologist. In three cohorts (MrOS,(22) Rotterdam I and II), incident radiographic vertebral fracture was also included in the case definition. Between 15% and 22% of each cohort were cases.
Clinical vertebral fracture
Nine clinic-based or population-based case-control or cohort studies were included in the GWAS of this phenotype. All participants were postmenopausal women age 45 years or older from Europe or Australia. Cases were matched with controls from the same geographical region or same ethnic background.(23)
Genotyping and imputation
Genomewide genotyping was followed by imputation of up to 2.7 million non-genotyped SNPs using the HapMap II reference panels. Details of each cohort's genotyping and imputation methods are provided in Supplemental Table 2. Replication genotyping for 13 SNPs in the AGES cohort was done using TaqMan at LGC Genomics (Hoddesdon, Hertfordshire, UK, www.lgcgroup.com/genomics).
Statistical analysis
Association analyses were performed in each study first and then meta-analyses were applied to combine results from each study. Within each study, a Z-score transformation of BMD phenotypes was applied. A general linear regression model with the additive genetic effect was applied to test for association between each BMD phenotype (Z-score) and each autosomal SNP. In the regression model, we adjusted for age, age2, weight (kg), and principal components for ancestral genetic background and study specific covariates (such as study site). In mixed-sex cohorts, sex was included as an adjustment variable in the regression model. Sex-stratified results were also provided. In the Framingham Osteoporosis Study and the Family Heart Study, a linear mixed effects model with within-family correlations as a random effect was used to account for relatedness among participants. In the Diabetes Heart Study, a random effects model was implemented using Sequential Oligogenic Linkage Analysis Routines (SOLAR) version 6.3.4 (Texas Biomedical Research Institute, San Antonio, TX, USA) as described.(24,25) The kinship coefficient matrix used was verified by the genotypic data via the software KING. The fixed-effect inverse-variance meta-analyses were performed by two authors (YH and CN) independently. Meta-analysis results were filtered by sample size (less than one-half of the total sample size), number of studies with available results (fewer than three cohort studies), overall MAF (<1%) and the heterogeneity test across studies (I2 ≥ 50 or p values for heterogeneity χ2 test < 5 × 10−5). Genomewide significance (GWS) level was defined as p < 5 × 10−8, and suggestive significance level was defined as p < 5 × 10−6 after adjusting for genomic control λGC value within each cohort and after meta-analysis. Double genomic control was applied to the integral vBMD GWAS after observing an elevated λGC value. Q-Q and Manhattan plots were generated in R 3.2.2 (R Project for Statistical Computing; https://www.r-project.org/) using the plyrand qqman packages (Supplemental Figs. 1 and 2). For GWS and suggestive SNPs, effect sizes were evaluated for heterogeneity between sexes using fixed-effect inverse-variance meta-analysis.
Conditional associations
To identify whether SNPs that were associated with trabecular vBMD or integral vBMD were independent from (not in linkage disequilibrium [LD] with) SNPs that were reported to be associated with DXA BMD at the lumbar spine,(8) we performed a conditional analysis using the Genomewide Complex Trait Analysis (GCTA) tool package.(26,27) For GWS and suggestive loci, we conditioned on reported BMD GWAS SNPs (associated with lumbar spine BMD measured by DXA). We selected SNPs for replication genotyping that were associated with CT BMD (trabecular vBMD or integral vBMD) independently from SNPs associated with DXA BMD based on conditional analyses. After genotype quality control, 13 SNPs were available for inclusion in meta-analyses of discovery and replication.
Association with radiographic vertebral fracture
The most significant SNP at each GWS or suggestive trabecular vBMD locus (n = 12) was evaluated for its association with radiographic vertebral fracture using logistic regression, with results combined across cohorts using fixed-effect inverse-variance meta-analysis. In each cohort, adjustments for age, height, and weight were performed. In mixed-sex cohorts, sex adjustment was also included. In addition, results for each of the 12 SNPs were examined in an independent meta-analysis of clinical vertebral fracture (n = 5893(23)). False discovery rate (FDR) p values were calculated, and the direction of effect relative to that for trabecular vBMD was used to evaluate consistency across phenotypes (eg, an odds ratio [OR] >1 for fracture is consistent with a negative β for trabecular vBMD).
Expression quantitative trait locus analysis
We conducted cis-expression quantitative trait locus (cis-eQTL) analysis within a 2-Mb flanking region (1 Mb upstream and 1 Mb downstream) of each of the top SNPs to evaluate whether they influence transcript levels of genes in human pelvic crest bone biopsies(28) and human primary osteoblasts.(29) Expression experiments in human whole-bone biopsies and human primary osteoblasts were conducted in different study samples. For details of genotyping and microarray expression profiling, see Supplemental Methods. For eQTL analyses, a linear regression model with the additive genetic effect was used. We adjusted for age, weight, cigarette smoking, and genetic ancestry in the regression model. Locus-wide statistical significance was defined as FDR Q-values <0.05 in each locus.
Primary murine osteoblasts
Gene expression profiles of candidate genes near genomewide-associated SNPs were examined in primary mouse osteoblasts undergoing differentiation. These data have been described in detail(30) and are freely available from the Gene Expression Omnibus (GSE54461). For details, see Supplemental Methods.
In silico annotation and enrichment analyses
Because all GWS and suggestive SNPs were noncoding (either intronic and intergenic SNPs), as were all SNPs in high LD with them, we annotated potential regulatory functions of those SNPs based on experimental epigenetic evidence including chromatin states, DNAse hypersensitive sites, histone modifications, phylogenetic conservation, altered regulatory motifs for transcription factor binding sites in human tissues, primary cells, and cell lines from the ENCODE Project and the Roadmap Epigenomics Project.(31,32) This was done by searching the HaploReg4 web browser.(33) Position weight matrices (PWMs) of a motif sequence were scored for instances that passed a threshold of p<4 × 10−7. Only instances in which a motif in the sequence passed the threshold of a PWM in either the reference or the alternate genomic sequence with variable nucleotide(s) (thus changing the PWM score) were considered. The enhancer and promoter states were obtained by ChromHMM(34) and were visualized using the WashU Epigenome Browser (Washington University, St. Louis, MO, USA; http://epigenomegateway.wustl.edu/).
To evaluate whether top associated SNPs were enriched with regulatory elements in specific tissues (such as bone relevant tissues, including primary osteoblast, bone marrow-derived stem cell, or mesenchymal progenitor cells available in the ENCODE and the Roadmaps Epigenomics Project), a hyper-geometric test was performed and permutation was used to estimate enrichment p values. Because our imputation was based on the reference panel that is not generated by whole-genome sequencing (international HapMap Project Phase II reference panel), we expected that our top associated SNPs may predominantly serve only as surrogate markers that are in high LD with the un-genotyped and un-imputed functional SNPs that are responsible for GWAS signals. Therefore, we extended our in silico functional annotation and enrichment analysis to those common SNPs (MAF > 1% based on 1000 Genomes Project Phase I version 3 CEU references) that are in high LD (r2 > 0.8 based on 1000 Genomes Project Phase I version 3 CEU references) with the most significant SNP in each locus.
Results
BMD and fracture associations
Five loci had genome-wide significant (GWS, p < 5 ×10−8) associations with trabecular vBMD in the meta-analysis of discovery (six cohort studies) and replication cohorts (two cohort studies, combined n = 15,275). These included four loci marked by common SNPs (MAF ≥ 5%) located in 1p36.12 (near WNT4, ZBTB40), 1p43 (GREM2), 8q24 (TNFRSF11B), and 13q14 (TNFSF11; Table 1). All except for GREM2 have been previously associated with lumbar spine aBMD. In addition, in the 5p13 locus, which has not previously been linked to lumbar spine BMD, the most significantly associated SNP with trabecular vBMD was rs2468531 (MAF = 3%). All GWS SNPs were intronic or intergenic (Supplemental Fig. 3). There was no significant heterogeneity of effect sizes between sexes for these SNPs (Supplemental Fig. 4). All GWS SNPs associated with trabecular vBMD were also strongly associated with integral vBMD and had similar effect sizes, although not all of the trabecular vBMD GWAS SNPs achieved genome-wide significance for integral vBMD, perhaps because of the smaller sample size with integral vBMD measurements (Supplemental Table 3). An additional six loci had common SNPs with suggestive-significant associations (5 × 10−8 < p < 5 × 10−6) with trabecular vBMD, and only one of these (6q25) reached a suggestive significance level for integral vBMD (Supplemental Table 3). An additional nine loci had SNPs that were suggestively associated with integral vBMD, though only two were suggestive after double GC adjustment (Supplemental Table 3). Among them, the most significantly associated SNP, rs3786178 (MAF = 2%) near CTIF on 18q21.2, had a relatively strong association (standardized β = 0.19, p = 1.86 × 10−6).
Table 1. Summary of Associations With Trabecular vBMD.
| Locus | Position (build 36) | SNP | Nearest gene(s) (distance in kb)a | A1/A2 | A1 frequency | Beta for A1 | SE | Heterogeneity I2 | p | |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Discovery (n = 12,287) | Combined (n = 15,275)b | |||||||||
| Combined p < 5 × 10−8 for trabecular vBMD | ||||||||||
| 1p36.12 | 22444772 | rs12755933 | WNT4 (103) | C/G | 0.27 | −0.084 | 0.013 | 0 | 2.66 ×10−10 | 9.80 ×10−11 |
| 1p36.12 | 22554953 | rs12742784 | ZBTB40 (96) | C/T | 0.21 | 0.094 | 0.015 | 0 | 1.18 × 10−10 | 1.05 ×10−10 |
| 1q43 | 238659259 | rs9661787 | FMN2 (0)-GREM2 (60) | C/G | 0.81 | −0.098 | 0.015 | 0 | 1.26 × 10−10 | 1.29 ×10−11 |
| 5p13 | 36516441 | rs2468531 | SLC1A3 (126)-RANBP3L (179) | G/C | 0.03 | 0.221 | 0.037 | 0 | 1.47 × 10−9 | 1.91 × 10−8 |
| 8q24 | 120045437 | rs1485303 | TNFRSF11B (12) | A/G | 0.44 | −0.057 | 0.012 | 14.6 | 7.35 × 10−7 | 4.63 × 10−9 |
| 13q14 | 41862203 | rs17457561 | AKAP11 (67) (TNFSF11 [700]) | A/G | 0.25 | −0.095 | 0.014 | 0 | 1.95 × 10−12 | 1.12 × 10−11 |
| Combined 5 × 10−8 ≤ p < 5 × 10−6 for trabecular vBMD | ||||||||||
| 2p21 | 54735125 | rs2941584 | SPTBN1 (0) | C/T | 0.32 | −0.060 | 0.012 | 33.5 | 1.26 × 10−6 | 1.29 ×10−7 |
| 6q25 | 151897286 | rs10872673 | C6orf97 (40) | G/T | 0.60 | 0.065 | 0.012 | 0 | 5.92 × 10−8 | 8.22 ×10−7 |
| 12p13.3 | 2378832 | rs7301013 | CACNA1C(298) | A/G | 0.83 | −0.076 | 0.016 | 39.4 | 1.45 × 10−6 | 4.90 × 10−7 |
| 12p12.1 | 28183168 | rs12813778 | CCDC91 (118)-PTHLH (167) | A/G | 0.23 | −0.068 | 0.014 | 32.9 | 1.16 × 10−6 | 2.35 ×10−6 |
| 12q21.3 | 88861177 | rs4842697 | ATP2B1 (287) | G/T | 0.57 | −0.057 | 0.012 | 22.4 | 9.09 × 10−7 | 2.60 × 10−6 |
| 16p13.3 | 315783 | rs9921222 | AXIN1 (27) | C/T | 0.47 | −0.059 | 0.012 | 0 | 6.09 × 10−7 | 4.90 × 10−7 |
vBMD = volumetric BMD.
Locus zoom plots are in Supplemental Fig. 3.
Observations were added by new genotyping in AGES (n = 2021) and/or by in silico replication in DHS (n = 967). The SNP association was considered to be replicated if the addition of the replication sample to the meta-analysis did not result in a p >5 × 10−8.
Among SNPs associated with trabecular vBMD or integral vBMD (listed in Table 1) with p < 5 × 10−6, only rs2468531 (5p13, near SLC1A3) was found to be associated with radiographic vertebral fracture after correction for multiple testing (OR = 0.75 per minor allele, FDR p = 0.01, Table 2, Fig. 2A, B). The same SNP was nominally associated with clinical vertebral fracture (OR = 0.66, = 0.01; Alonso and colleagues, unpublished data; but FDR p = 0.07). Among SNPs associated with trabecular vBMD (listed in Table 1), the GWAS SNP rs12742784 near ZBTB40 was associated with clinical vertebral fracture (OR = 0.82, FDR p = 7.4 × 10−4). Although SNPs at other loci (GREM2, C6orf97, ATP2B1) were nominally significantly associated with vertebral fracture, no other ORs were nearly as strong as for rs2468531 or rs12742784 (Table 2).
Table 2. Associations With Vertebral Fracture for SNPs With Combined p < 5 × 10−6 for Trabecular vBMD.
| Locus | SNP | Nearest gene(s) | Morphometry vertebral fracture (n = 21,701) | Clinical vertebral fracture (n = 5893) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| A1 frequency | OR | P | FDR p | Heterogeneity I2 | A1 frequency | OR | p | FDR p | Heterogeneity I2 | |||
| Combined p < 5 × 10−8 for trabecular vBMD | ||||||||||||
| 1p36.12 | rs12755933 | WNT4 | 0.26 | 0.99 | 0.680 | 0.816 | 0 | 0.27 | 1.02 | 0.707 | 0.943 | 9.8 |
| 1 p36.12 | rs12742784 | ZBTB40 | 0.20 | 0.99 | 0.798 | 0.826 | 10.2 | 0.20 | 0.82 | 6.2 × 10−5 | 7.4 ×10−4 | 5.4 |
| 1q43 | rs9661787 | FMN2-GREM2 | 0.82 | 1.06 | 0.102 | 0.306 | 48.7 | 0.82 | 1.12 | 0.055 | 0.220 | 0.1 |
| 5p13 | rs2468531 | SLC1A3-RANBP3L | 0.03 | 0.75 | 0.001 | 0.012 | 0 | 0.03 | 0.66 | 0.011 | 0.066 | 5.3 |
| 8q24 | rs1485303 | TNFRSF11B | 0.43 | 1.03 | 0.298 | 0.596 | 0 | 0.44 | 0.98 | 0.690 | 0.943 | 37.7 |
| 13q14 | rs17457561 | AKAP11 | 0.24 | 1.02 | 0.516 | 0.788 | 0 | 0.27 | 0.95 | 0.343 | 0.686 | 0.0 |
| Combined 5 × 10−8 < p < 5 × 10−6 for trabecular vBMD | ||||||||||||
| 2p21 | rs2941584 | SPTBN1 | 0.31 | 1.03 | 0.215 | 0.516 | 0 | 0.33 | 1.08 | 0.096 | 0.230 | 18.2 |
| 6q25 | rs10872673 | C6orf97 | 0.59 | 0.94 | 0.012 | 0.072 | 0 | 0.59 | 1.02 | 0.608 | 0.943 | 0.0 |
| 12p13.3 | rs7301013 | CACNA1C | 0.84 | 1.01 | 0.826 | 0.826 | 0 | 0.85 | 1.02 | 0.792 | 0.950 | 0.0 |
| 12p12.1 | rs12813778 | CCDC91-PTHLH | 0.23 | 0.98 | 0.525 | 0.788 | 75.7 | 0.24 | 1.10 | 0.081 | 0.230 | 0.0 |
| 12q21.3 | rs4842697 | ATP2B1 | 0.59 | 1.05 | 0.038 | 0.152 | 47.0 | 0.59 | 1.00 | 0.948 | 0.981 | 0.0 |
| 16p13.3 | rs9921222 | AXIN1 | 0.47 | 1.01 | 0.647 | 0.816 | 0 | 0.47 | 1.00 | 0.981 | 0.981 | 27.8 |
Bold values are statistically significant (FDR p < 0.05).
vBMD = volumetric BMD; OR = odds ratio, FDR = false-discovery rate.
Fig. 2.
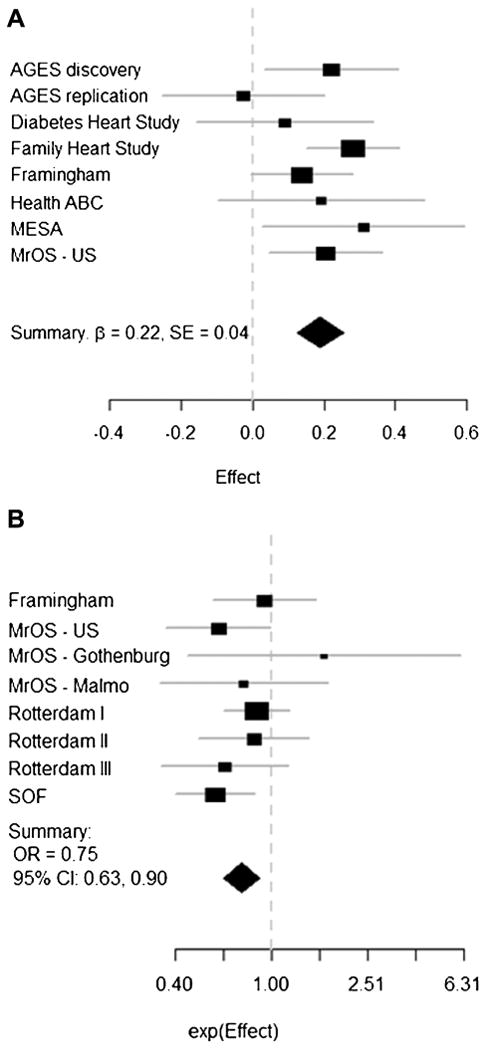
Beta coefficients and 95% CI for the additive effect of rs2468531 (SLC1A3) on trabecular vBMD (A) and ORs and 95% CI for the association with morphometric vertebral fracture (B). Detailed study-specific results are provided in Supplemental Table 7.
cis-eQTLs in human bone biopsies
The cis-eQTL results of the whole-bone biopsies are shown in Table 3. For GWS SNPs, we found two significant eQTLs after multiple testing correction. First, SNP rs12742784 in the 1p36.12 locus was associated with increased expression of EPHB2, about 355 kb downstream (β = 0.12, FDR p = 1.72 × 10−3), but was only marginally associated (p = 0.08) with ZBTB40, the nearest gene in this GWAS locus (about 96 kb downstream). The other significant cis-eQTL finding was for SNP rs2468531 in the 5p13 locus, which was associated with increased expression of SLC1A3, 126kb downstream from the associated SNP (β = 0.28, FDR p = 0.01). A suggestive SNP, rs2941584, was associated with EML6 expression in the 2p21 locus (FDR p = 0.04); and a suggestive SNP rs7301013 with WNT5B expression in the 12p13.3 locus (FDR p = 0.01). In most cases, the most significantly associated cis-eQTL gene in each locus was not the gene nearest to the top associated SNP, a phenomenon also observed in other studies.(35) CCDC91 was filtered out because of low signal values in the microarray.
Table 3. Summary of Associations With Gene Expression in Human Bone Biopsies.
| Locus | Position (build 36) | SNP | Region | Nearest gene(s) (distance in kb) | Association with the nearest genes | The most significant cis-eQTL | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| Beta | p | Gene | Beta | p | FDR p | Distance (kb) | |||||
| Combined p < 5 × 1 10−8 for trabecular vBMD | |||||||||||
| 1p36.12 | 22444772 | rs12755933 | Intergenic | WNT4 (103) | 0.030 | 0.37 | EPHB2 | −0.062 | 0.02 | 0.34 | 465 |
| 1p36.12 | 22554953 | rs 12742784 | Intergenic | ZBTB40 (96) | 0.105 | 0.08 | EPHB2 | 0.120 | 3.49 ×10−5 | 1.72 × 10−3 | 355 |
| 1q43 | 238659259 | rs9661787 | Intronic | FMN2 (0) | 0.027 | 0.27 | GREM2 | −0.049 | 0.10 | 0.61 | 60 |
| 5p13 | 36516441 | rs2468531 | Intergenic | SLC1A3 (126) | 0.279 | 4.36 × 10−3 | SLC1A3 | 0.279 | 4.36 × 10−3 | 0.01 | 126 |
| Intergenic | RANBP3L (179) | −0.107 | 0.33 | ||||||||
| 8q24 | 120045437 | rs1485303 | Intergenic | TNFRSF1 IB (12) | −0.019 | 0.61 | NOV | −0.136 | 0.04 | 0.53 | 452 |
| 13q14 | 41862203 | rs17457561 | Intergenic Intergenic | AKAP11 (67) TNFSF11 (700) | −0.068 0.035 | 0.03 0.28 | AKAP11 | −0.068 | 0.03 | 0.32 | 67 |
| Combined 5 × 10−8 < p < 5 × 10−6 for trabecular vBMD | |||||||||||
| 2p21 | 54735125 | rs2941584 | Intronic | SPTBN1 | 0.039 | 0.20 | EML6 | 0.096 | 3.33 × 10−3 | 0.04 | 69 |
| 6q25 | 151897286 | rs10872673 | Intronic Intergenic | C6orf97 (CCDC170) ESR1 (160) | −0.003 0.040 | 0.92 0.07 | MTHFD1L | −0.090 | 0.03 | 0.28 | 433 |
| 12p13.3 | 2378832 | rs7301013 | Intronic | CACNA1C | 0.056 | 0.17 | WNT5B | 0.280 | 1.81 × 10−4 | 0.01 | 752 |
| 12p12.1 | 28183168 | rs12813778 | Intergenic | CCDC91 (118) | n.a. | n.a. 0.41 | STK38L | −0.068 | 0.03 | 0.15 | 813 |
| Intergenic | PTHLH (167) | −0.018 | |||||||||
| 12q21.3 | 88861177 | rs4842697 | Intergenic | ATP2B1 (287) | −0.093 | 0.02 | ATP2B1 | −0.093 | 0.02 | 0.13 | 287 |
| 16p13.3 | 315783 | rs9921222 | Intronic | AXIN1 | −0.033 | 0.27 | POLR3K | 0.102 | 2.57 × 10−3 | 0.07 | 279 |
Bold values are statistically significant (FDR p < 0.05).
eQTL = expression quantitative trait locus; FDR = false-discovery rate; vBMD = volumetric BMD; n.a. = filtered out because of low signal values in the microarray.
Expression in mouse osteoblasts
Ten of the 11 genes located near a trabecular vBMD locus or implicated through eQTL analysis were expressed during murine osteoblastogenesis, and each presented with a unique level and/or pattern of expression. EphB2 maintained a consistent level of expression over time, whereas Slc1a3 expression rose rapidly through the period of rapid cell proliferation and reached a plateau coincident with a phase associated with increased expression of extracellular matrix genes (∼day 8 to day 10; Supplemental Fig. 5). Of the two genes near 1q43, Grem2 expression was extremely high during the period of cell culture associated with rapid growth and cell division. Expression of this gene decreased and entered a steady-state plateau at about day 8 postdifferentiation. Like Grem2, Fmn2 showed a pattern of decreasing expression during osteoblastogenesis; however, expression of this gene was very low at all points examined. Both Akap11 and Zbtb40 showed constant expression during osteoblast maturation (Supplemental Fig. 5).
In silico annotation of noncoding SNPs and of SNPs in LD with the top associated SNPs
Evaluation of whether the top associated SNPs (from Table 1) were enriched with regulatory elements in specific tissues showed the most significant enrichment for the cortex-derived primary cultured neurospheres (E053) with enrichment p value = 9.56 × 10−3. We observed three SNPs (out of 12) overlapping with regulatory elements in the cortex neurospheres (ENCODE, Supplemental Tables 4 and 5), compared to the 0.5 expected SNPs overlapping with regulatory elements in the same cell type if we randomly select 12 SNPs with the same MAF and LD pattern across the whole genome. After multiple testing corrections by FDR, the enrichment in the cortex neurospheres became nonsignificant.
Two common nonsynonymous SNPs were found to be in LD with the top associated SNPs. SNP rs2073618 (N3K) (discovery p value = 1.41 × 10−6) in TNFRSF11B is in high LD (r2 = 0.88) with rs1485303, and SNP rs35737760 (D859E) in CACNA1E is in high LD with SNP rs7301013 that was suggestive for integral vBMD. However, both variants were predicted not to affect protein function due to the corresponding amino acid substitution based on the conservation-based SIFT (Sorting Intolerant From Tolerant) package (http://sift.jcvi.org/) and were predicted to be benign by the PolyPhen-2 (Polymorphism Phenotyping v2) package (http://genetics.bwh.harvard.edu/pph2/). All common SNPs in high LD with top associated SNPs in all other loci were either intronic or intergenic. Twelve of the 21 loci with GWS or suggestive trabecular vBMD or integral vBMD associations had SNPs in LD that were located in either predicted enhancer or promoter regions in one of the three bone-relevant cell types: osteoblast primary cells, mesenchymal stem cell-derived chondrocyte-cultured cells, and bone marrow-derived mesenchymal cultured cells (E129, E049 and E026, Supplemental Table 5). These included intronic or intergenic SNPs in the GWS trabecular vBMD loci near GREM2, TNFRSF11B, and TNFSF11, as well as the 5p13 locus. Of note, the GREM2 locus (1q43) has quiescent annotations across an abundance of cell types except for two SNPs, rs1414660 (r2 for LD with top vBMD SNP = 0.87; discovery p value = 2.01 × 10−10) and rs9659023 (not analyzed), which are both located in an enhancer region that is specifically active in bone marrow-derived mesenchymal cells, osteoblasts, and other mesenchymal cells, suggesting a cell-type-specific gene regulation in those bone-relevant cells. The rs1414660 regulatory sequence is under evolutionary constraint, as estimated by GERP (Genomic Evolutionary Rate Profiling) and SiPhy (SIte-specific PHYlogenetic analysis) (http://www.broadinstitute.org/mammals/haploreg/detail_v4.1.php?query=&id=rs1414660). Furthermore, the rs1414660 intronic single nucleotide change was predicted to perturb conserved regulatory motifs for bone-relevant transcription factors, such as CEBPa, CEBPb and others (Supplemental Table 6). The 5p13 locus is quiescent except for a region surrounding the indel rs148073475 (http://www.broadinstitute.org/mammals/haploreg/detail_v4.1.php?query=&id=rs148073475). To evaluate whether these 12 SNPs in LD with variants are specifically located in enhancer regulatory regions for three bone-relevant cells, we tested for enrichment in regulatory regions in tissues other than bone-relevant tissues. We performed hypergeometric tests in 127 cell types and tissues from the Roadmap Epigenomics Project but did not find significant enrichment, suggesting that these selected SNPs are enriched only in bone-specific gene regulatory regions.
Discussion
Through these meta-analyses of vertebral trabecular vBMD and fracture, we identified two loci associated with lumbar spine BMD, vertebral fracture, and human bone expression of genes that were also expressed in mouse osteoblasts. One is a novel BMD locus, at 5p13, and the minor alleles of the most significantly associated SNPs had a lower frequency (3%), stronger positive associations with trabecular vBMD (β = 0.22), and lower risk of fracture (OR = 0.75) than did other variants associated with BMD or fracture in this study. Cis-eQTL analyses in human bone and mouse osteoblast expression studies suggest that the SLC1A3 gene, which is ∼126kb upstream of SNP rs2468531, may be the gene in this locus involved in bone modeling/remodeling. Common variants in another locus, 1p36.12, were significantly associated with trabecular vBMD, fracture (clinical vertebral OR: 0.82, FDR p = 7.4×10−4) and expression of EPHB2, providing an explanation for the associations with BMD phenotypes consistently reported in this region.(8,36–38) This is the first study to link EPHB2 expression to vertebral phenotypes in humans. The finding that these loci are associated with both vertebral trabecular BMD and vertebral fracture reinforces their clinical importance. Our expression studies suggest a regulatory function for the causal variants underlying these SNP associations.
SLC1A3
Studies have reported that the SLC1A3 gene is involved in glutamate signaling necessary for osteogenic response to mechanical loading.(39) SLC1A3 (aka GLAST or EAAT1) is a glutamate transporter expressed in human as well as rat and mouse osteoblasts and osteocytes, discovered through its downregulation in osteocytes in response to osteogenic mechanical loading.(40) SLC1A3 has been found to be expressed in most human tissues/cells, especially in brain and neurons. Although a Slc1a3 mouse knockout showed no differences in multiple femoral bone phenotypes through age 6 months,(41) there has been no examination of either vertebral bone phenotypes or of aged mice in such a knockout. These are important distinctions, because Slc1a3 knockout effects on another phenotype have been shown to depend on advanced age,(42) and axial and appendicular skeletal regions are known to have distinct patterns of development and regulation over the course of development (eg, Courtland and colleagues,(43) Sabsovich and colleagues,(44) and Zanotti and Canalis(45)).
SLC1A3 associations have not been reported to be genome-wide significantly associated with aBMD in any previous study. In the most recent lumbar spine aBMD GWAS meta-analysis, the most significant SNP in this locus was rs2468531 (MAF = 4.8%, β = 0.077, p = 7.4 × 10−3).(30) That aBMD GWAS meta-analysis had much larger sample size and included many of the participating studies in our CT GWAS meta-analysis. It is unclear whether the difference in phenotype or underlying population differences contributed to this discrepancy. Previously reported cell and animal model evidence for SLC1A3 suggests its biological function in bone physiology may interact with mechanical loading and aging; therefore, it may be fruitful to evaluate such interactions with potentially causative SLC1A3 SNPs in future studies.
EPHB2
Our findings suggest that regulation of the Ephb2 gene contributes to the previously reported associations with BMD and fracture at the 1p36.12 locus. ZBTB40 has been consistently linked to both spine and hip BMD as well as fracture risk,(8,36–38) but has an unknown role in bone development or maintenance. ZBTB40 was expressed in osteoblasts in our study, but the associated (intergenic) SNP was unrelated to any regulatory element in ENCODE. This gene is often reported concurrently with WNT4 associations with BMD; however, LD patterns and conditional analyses support the existence of two separate signals in this region of chromosome 1p36.12.(8) In addition, cis-eQTL analysis did not find significant associations between GWAS SNPs and ZBTB40 and WNT4 gene expression in whole bone. On the other hand, a cis-eQTL in this locus was found for the EPHB2 gene. The EPHB2 gene encodes a member of the Eph receptor family of receptor tyrosine kinase transmembrane glycoproteins. Several in vitro and animal studies have found that Ephb2 is involved in bone development,(46) homeostasis,(47) and fracture repair,(48) as well as skeletal response to PTH(49) and IGF.(50) This is the first study to link genetic variation in humans to EPHB2 expression and clinically important vertebral phenotypes.
GREM2
One additional locus associated with trabecular vBMD, an intronic SNP, rs9661787, in FMN2 and near GREM2, has not previously been reported for lumbar spine BMD by DXA. It has, however, been linked to trabecular BMD of the distal tibia, as measured by peripheral quantitative CT (pQCT), and was demonstrated to affect trabecular number and thickness as evaluated by HRpQCT.(51) In the current analyses, the FMN2/ GREM2 SNP was also associated with integral vBMD with a similar effect size, perhaps because of the sizeable contribution of trabecular vBMD in the overall measure of integral vBMD by CT of the lumbar spine. The homologous region in mice (174.8 Mb on chromosome 1) has consistently been associated with BMD phenotypes, including vertebral BMD.(52) In addition, our GWAS SNP rs9661787 in the FMN2/GREM2 locus is in high LD (r2 = 0.88) with a GWAS SNP (rs9287237) identified previously for a pQCT GWAS.(51) SNP rs9287237 had similar effect sizes to our top SNP for trabecular vBMD and integral vBMD in this study.
SNP rs9287237 was robustly associated with GREM2 expression in human osteoblasts.(51) Each additional T allele of rs9287237 was associated with decreased expression of GREM2 in human osteoblasts, increased trabecular BMD, and decreased fracture risk.(51) In our eQTL analysis, we also found SNP rs9287237 was associated with lower GREM2 expression in human whole-bone biopsies, though the association was not statistically significant. GREM2 (aka PRDC and Gremlin2) is an extracellular antagonist of bone morphogenetic proteins (BMPs), and loss of this factor allows for an increase in osteoblastic differentiation.(53–55) We observed a marked decrease in expression of Grem2 as these cells transitioned from a committed but immature cell to a fully mature osteoblast in mouse. This suggests that Grem2 is an inhibitor of osteoblast maturation and/or function and that expression of this gene must be reduced to allow this process to occur. Our observation that GWAS SNP rs9661787 was associated with lower GREM2 expression (Table 3)and associated with higher trabecular vBMD (Table 1, G allele of rs9661787) supports previous findings that Grem2 inhibited osteoblast maturation and/or function. Available data suggest that this gene is not expressed or is only marginally expressed in mouse osteoclasts (BioGPS; Scripps Research Institute [TSRI], La Jolla, CA, USA; http://biogps.org). Unlike for Grem2, expression of Fmn2 was barely detectable in osteoblasts. This low/lack of expression of this gene in bone has been corroborated in other studies wherein whole-bone, osteoblasts, osteoclast, and osteoblast-like cell lines were examined (http://biogps.org). Together these data suggest that GREM2 is the more likely candidate at this locus.
Four other loci had common SNPs that were genome-wide significantly associated with trabecular vBMD (near WNT4, ZBTB40, TNFRSF11B, and TNFSF11), although effect sizes were much smaller than for the 5p13 locus. These loci have been consistently reported to be associated with lumbar spine BMD by DXA(8,29,30) and fracture.(10) The roles of WNT4, TNFRSF11B, and TNFSF11 are well described in WNT-signaling and RANK/ RANKL/osteoprotegerin (OPG) pathways, which are central to bone metabolism.(56) Of particular relevance to our phenotype of spine BMD in older age, Wnt4 has been demonstrated to prevent bone loss and one of its common causes, inflammation, by inhibiting NF-κB in macrophages and osteoclast precursors.(57) The SNP, however, is predicted not to be constrained by SIFT and is benign by PolyPhen-2.
Phenotypic considerations
vBMD measured by CT has advantages over DXA BMD for evaluating genetic regulation specific to three-dimensional (3D) structure and mineral density. CT measures allow for the removal of bone size from the phenotype, whereas DXA BMD is a two-dimensional (2D)-measured combination of size and density. Furthermore, DXA BMD of the lumbar spine includes the vertebral body and posterior elements, as well as other regions that are predisposed to degenerative changes. vBMD measures, including trabecular and integral BMD, were more strongly associated with prevalent vertebral fracture than was spine aBMD in a case-control study nested in the Framingham Osteoporosis Study.(11) Therefore, vBMD may be a more relevant phenotype for vertebral fracture risk. Although candidate gene analyses have discovered distinct associations with, eg, vBMD and vertebral cross-sectional area,(58) ours is the first GWAS of volumetric phenotypes in a multicohort meta-analysis and the first to link vBMD SNP associations to fracture and gene expression.
Limitations
Limitations of the study include a relatively small number (15,275) of participants with vBMD phenotypes available. As a polygenic and complex trait, BMD requires very large sample sizes to detect the effect sizes often observed for common SNPs. Of 49 loci associated with lumbar spine BMD in previous large GWAS meta-analyses,(8,30) we identified only nine in our spine vBMD traits. Whether this lack of confirmation of previously identified spine BMD associations is due to true phenotypic differences in association or simply lower sample size in the current study is unknown. However, four of six trabecular vBMD GWAS loci found in the current study have been reported by previous BMD GWAS meta-analysis; and four of six trabecular vBMD-suggestive GWAS loci have also been reported by a previous DXA-derived BMD GWAS meta-analysis, suggesting limited statistical power with relatively smaller sample size may play a major role in this lack of confirmation. Further investigation of the involvement of those GWAS loci unique to vBMD will improve our understanding of the molecular regulation of bone.
Moderate effect size for most of the common SNPs may also contribute to the limited statistical power of the current study. The majority of common variants previously reported are closer to zero (|standardized β| ≤ 0.03) than the range of effect sizes we observed for GWS associations with vBMD (0.057 ≤ |standardized β| ≤ 0.098), implying lack of power in this study to identify common SNPs with smaller effect sizes. Similarly, the study was not designed to detect less common (1% < MAF < 5%) or rare variants (MAF < 1%), and only two variants with 2%< MAF < 5% (SLC1A3 and CTIF) were found to be associated with trabecular vBMD or integral vBMD. These had effect sizes approximately three times as large as the more common variants and were similar in effect size to that reported for a low-frequency variant in EN1 for vertebral BMD,(30) again underscoring the value of studying less common variants for skeletal phenotypes. There are undoubtedly other genetic contributors to BMD that will require studies powered to detect moderate effect sizes in low-frequency or rare variants.
Limitations regarding heterogeneous radiographic vertebral fracture definitions have been described(9) and may have limited our power for observing fracture associations among the vertebral-BMD-associated loci. However, we observed similar prevalence of fracture across the cohorts that were included (15% to 22%), indicating that differences in case definitions may result in ascertainment of similar cases in these studies. Three trabecular vBMD loci (near ZBTB40, FMN2/GREM2, and SLC1A3) were associated more strongly with clinical vertebral fracture than with radiographic vertebral fracture, indicating a potential for clinical fracture to give a stronger association signal than radiographic fracture for these variants.
Several additional limitations of this study include limited statistical powerto detect SNPs with moderate eQTL; imputation based on HapMap II reference panel rather than whole-genome sequencing–based 1000G reference panel, providing limited coverage of genetic variants across genomes; and limited bone-relevant tissues in existing databases for the in silico annotations of associated loci. In addition, pinpointing causal variants and functionally validating them in cellular or animal models remains to be done.
Despite these limitations, we identified novel vertebral BMD and fracture associations related to SLC1A3 and EPHB2 expression. We also confirmed the importance of GREM2 and several genes involved in WNT-signaling for trabecular BMD and vertebral fracture in large cohorts of older adults. Our study reinforces the benefit of large-scale GWAS of more refined and clinically relevant skeletal phenotypes and the need for continued evaluation of noncoding genomic variants with potential regulatory function.
Supplementary Material
Acknowledgments
Age Gene/Environment Susceptibility-Reykjavik Study (AGES-Reykjavik): The Age, Gene/Environment Susceptibility Reykjavik Study was funded by NIH contract N01-AG-12100, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). The Fracture Registry is funded by the Icelandic Heart Association.
Diabetes Heart Study: The authors thank the other investigators, the staff, and the participants of the DHS study for their valuable contributions. This study was supported by the National Institutes of Health through R01 HL67348 and R01 HL092301 (to DWB), R01 AR48797 (to JJC), and F31 AG044879 (to LMR).
Family Heart Study: This study was supported by grants from the National Heart, Lung, and Blood Institute (U01 HL067893, U01 HL067894, U01 HL067896, U01 HL067897, U01 HL056567, U01 HL056568, and U01 HL067901).
Framingham: The study was funded by grants from the US National Institute for Arthritis, Musculoskeletal and Skin Diseases and National Institute on Aging (R01 AR 41398 and R01 AR061162 to DPK; R01 AR 050066 to DK). The Framingham Heart Study of the National Heart, Lung, and Blood Institute of the National Institutes of Health and Boston University School of Medicine were supported by the National Heart, Lung, and Blood Institute's Framingham Heart Study (N01-HC-25195) and its contract with Affymetrix, Inc. for genotyping services (N02-HL-6-4278). Analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center.
Health, Aging, and Body Composition (Health ABC): This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. This research was supported by the U.S. National Institute of Aging (NIA) contracts N01AG62101, N01AG62103, and N01AG62106. The genomewide association study was funded by NIA grant 1R01AG032098 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. We thank the participants of the Health, Aging, and Body Composition Study.
Multi-Ethnic Study of Atherosclerosis (MESA): MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079, UL1-TR-000040, and DK063491. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, CA, USA) and the Broad Institute of Harvard and MIT (Boston, MA, USA) using the Affymetrix Genome-Wide Human SNP Array 6.0. Support for the Bone Mineral Density dataset was provided by grant HL072403.
Osteoporotic Fractures in Men (MrOS)-USA: The Osteoporotic Fractures in The Osteoporotic Fractures in Men (MrOS) Study is supported by National Institutes of Health funding. The following institutes provide support: the National Institute on Aging (NIA), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Center for Advancing Translational Sciences (NCATS), and NIH Roadmap for Medical Research under the following grant numbers: U01 AG027810, U01 AG042124, U01 AG042139, U01 AG042140, U01 AG042143, U01 AG042145, U01 AG042168, U01 AR066160, and UL1 TR000128. The National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) provides funding for the MrOS ancillary study ‘GWAS in MrOS and SOF’ under the grant number RC2 AR058973. CMN is supported by NIAMS K01 AR062655.
Osteoporotic Fractures in Men (MrOS)-Sweden: The MrOS Sweden study is supported by the Swedish Research Council, the Swedish Foundation for Strategic Research, the ALF/LUA research grant from the Sahlgrenska University Hospital, the Lundberg Foundation, the Torsten and Ragnar Soderberg's Foundation and the Novo Nordisk Foundation.
Rotterdam Studies: The Rotterdam Study is funded by Erasmus Medical Centre and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European
Commission (DG XII), and the Municipality of Rotterdam. We are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists. The generation and management of GWAS genotype data for the Rotterdam Study (RS I, RS II, RS III) was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands. The GWAS datasets are supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA), project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera, and Marjolein Peters, MSc, and Carolina Medina-Gomez, MSc, for their help in creating the GWAS database, and Karol Estrada, PhD, Yurii Aulchenko, PhD, and Carolina Medina-Gomez, MSc, for the creation and analysis of imputed data.
Study of Osteoporotic Fractures: The Study of Osteoporotic Fractures (SOF) is supported by National Institutes of Health funding. The National Institute on Aging (NIA) provides support under the following grant numbers: R01 AG005407, R01 AR35582, R01 AR35583, R01 AR35584, R01 AG005394, R01 AG027574, and R01 AG027576.
Expression studies (C. L. Ackert-Bicknell): NIH/NIAMS AR060234 and AR060981.
Expression studies (S. Reppe and K. Gautvik): This work was supported by the 6th EU framework program to “OSTEOGENE” [LSHM-CT-2003-502941]; the South-Eastern Norway Regional Health Authority; and Oslo University Hospital, Ullevaal.
Authors' roles: Overseeing: CO, DB, DK, DM, DPK, ESO, FR, JC, KMG, MAA, MHC, MK, RHM, SC, SHR, SR, TBH, VG, and YHH; Genotyping: AJC, CLAB, CMN, CO, DB, DPK, ESO, FR, JMZ, LR, and MKK; Phenotyping: CO, DM, DPK, EHO, EJS, ESO, FR, GT, JC, JJ, JMZ, KMG, KS, MAA, MLB, MB, MHC, MKK, SR, TCR, TL, and VG; Analysis: AVS, CL, CLAB, CMN, CP, CTL, CW, EL, JCL, JJ, KMG, LAC, LO, LV, MAI, MAN, MC, NA, NP, PS, SR, WCC, and YHH; Writing group: CLAB, CMN, CO, CTL, DPK, ESO, FR, TCR, YHH.
JL is an employee of GNS Healthcare. ESO has received grant support from Merck and has a consulting relationship with Merck. DPK has grants from Merck Sharp & Dohme and Policy Analysis, Inc/Amgen, and receives royalties from Wolters Kluwer for UpToDate, and from Springer for book publishing; he serves on a Scientific Advisory Board for Merck Sharp & Dohme.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Disclosures: These relationships are not related to the article and are not expected to result in conflicts. All other authors state that they have nothing to disclose.
References
- 1.Kendler DL, Bauer DC, Davison KS, et al. Vertebral fractures: clinical importance and management. Am J Med. 2015;129(2):221, e1–e10. doi: 10.1016/j.amjmed.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 2.Oudshoorn C, Hartholt KA, Zillikens MC, et al. Emergency department visits due to vertebral fractures in the Netherlands, 1986-2008: steep increase in the oldest old, strong association with falls. Injury. 2012;43(4):458–61. doi: 10.1016/j.injury.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Leslie WD, Sadatsafavi M, Lix LM, et al. Secular decreases in fracture rates 1986-2006 for Manitoba, Canada: a population-based analysis. Osteoporos Int. 2011;22(7):2137–43. doi: 10.1007/s00198-010-1470-4. [DOI] [PubMed] [Google Scholar]
- 4.Amin S, Achenbach SJ, Atkinson EJ, Khosla S, Melton LJ., 3rd Trends in fracture incidence: a population-based study over 20 years. J Bone Miner Res. 2014;29(3):581–9. doi: 10.1002/jbmr.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perilli E, Briggs AM, Kantor S, et al. Failure strength of human vertebrae: prediction using bone mineral density measured by DXA and bone volume by micro-CT. Bone. 2012;50(6):1416–25. doi: 10.1016/j.bone.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Liu CT, Karasik D, Zhou Y, et al. Heritability of prevalent vertebral fracture and volumetric bone mineral density and geometry at the lumbar spine in three generations of the Framingham study. J Bone Miner Res. 2012;27(4):954–8. doi: 10.1002/jbmr.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richards JB, Zheng HF, Spector TD. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nat Rev Genet. 2012;13(8):576–88. doi: 10.1038/nrg3228. [DOI] [PubMed] [Google Scholar]
- 8.Estrada K, Styrkarsdottir U, Evangelou E, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oei L, Estrada K, Duncan EL, et al. Genome-wide association study for radiographic vertebral fractures: a potential role for the 16q24 BMD locus. Bone. 2014;59:20–7. [PMC free article] [PubMed] [Google Scholar]
- 10.Richards JB, Kavvoura FK, Rivadeneira F, et al. Genetic Factors for Osteoporosis Consortium. Collaborative meta-analysis: associations of 150 candidate genes with osteoporosis and osteoporotic fracture. Ann Intern Med. 2009;151(8):528–37. doi: 10.7326/0003-4819-151-8-200910200-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson DE, Demissie S, Allaire BT, et al. The associations between QCT-based vertebral bone measurements and prevalent vertebral fractures depend on the spinal locations of both bone measurement and fracture. Osteoporos Int. 2014;25(2):559–66. doi: 10.1007/s00198-013-2452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orwoll ES, Oviatt SK, Mann T. The impact of osteophytic and vascular calcifications on vertebral mineral density measurements in men. J Clin Endocrinol Metab. 1990;70(4):1202–7. doi: 10.1210/jcem-70-4-1202. [DOI] [PubMed] [Google Scholar]
- 13.Li N, Li XM, Xu L, Sun WJ, Cheng XG, Tian W. Comparison of QCT and DXA: osteoporosis detection rates in postmenopausal women. Int J Endocrinol. 2013;2013:895474. doi: 10.1155/2013/895474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris TB, Launer LJ, Eiriksdottir G, et al. Age, Gene/Environment Susceptibility-Reykjavik Study: multidisciplinary applied phenomics. Am J Epidemiol. 2007;165(9):1076–87. doi: 10.1093/aje/kwk115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samelson EJ, Christiansen BA, Demissie S, et al. QCT measures of bone strength at the thoracic and lumbar spine: the Framingham Study. J Bone Miner Res. 2012;27(3):654–63. doi: 10.1002/jbmr.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budoff MJ, Hamirani YS, Gao YL, et al. Measurement of thoracic bone mineral density with quantitative CT. Radiology. 2010;257(2):434–40. doi: 10.1148/radiol.10100132. [DOI] [PubMed] [Google Scholar]
- 17.Orwoll E, Blank JB, Barrett-Connor E, et al. Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study—a large observational study of the determinants of fracture in older men. Contemp Clin Trials. 2005;26(5):569–85. doi: 10.1016/j.cct.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Lang TF, Li J, Harris ST, Genant HK. Assessment of vertebral bone mineral density using volumetric quantitative CT. J Comput Assist Tomogr. 1999;23(1):130–7. doi: 10.1097/00004728-199901000-00027. [DOI] [PubMed] [Google Scholar]
- 19.Genant HK, Wu CY, van Kuijk C, Nevitt MC. Vertebral fracture assessment using a semiquantitative technique. J Bone Miner Res. 1993;8(9):1137–48. doi: 10.1002/jbmr.5650080915. [DOI] [PubMed] [Google Scholar]
- 20.Black DM, Cummings SR, Stone K, Hudes E, Palermo L, Steiger P. A new approach to defining normal vertebral dimensions. J Bone Miner Res. 1991;6(8):883–92. doi: 10.1002/jbmr.5650060814. [DOI] [PubMed] [Google Scholar]
- 21.Black DM, Palermo L, Nevitt MC, et al. Comparison of methods for defining prevalent vertebral deformities: the Study of Osteoporotic Fractures. J Bone Miner Res. 1995;10(6):890–902. doi: 10.1002/jbmr.5650100610. [DOI] [PubMed] [Google Scholar]
- 22.Cawthon PM, Haslam J, Fullman R, et al. Osteoporotic Fractures in Men Research Group. Methods and reliability of radiographic vertebral fracture detection in older men: the osteoporotic fractures in men study. Bone. 2014;67:152–5. doi: 10.1016/j.bone.2014.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alonso N, The Clinical Vertebral Fracture Consortium. Uitterlinden A, Rivadeneira F, Ralston SH. Identification of a novel locus on 2q13 of large effect size which predisposes to clinical vertebral fractures independently of BMD: the GEFOS Consortium. Bone Abstracts. 2016;5:P241. [Google Scholar]
- 24.Lange LA, Burdon K, Langefeld CD, et al. Heritability and expression of C-reactive protein in type 2 diabetes in the Diabetes Heart Study. Ann Hum Genet. 2006;70(Pt 6):717–25. doi: 10.1111/j.1469-1809.2006.00280.x. [DOI] [PubMed] [Google Scholar]
- 25.Hsu FC, Lenchik L, Nicklas BJ, et al. Heritability of body composition measured by DXA in the diabetes heart study. Obes Res. 2005;13(2):312–9. doi: 10.1038/oby.2005.42. [DOI] [PubMed] [Google Scholar]
- 26.Yang B, Sun H, Wang H. The downstream effects of vitamin D in spermatozoa needs further study. Hum Reprod. 2010;25(8):2152–3. doi: 10.1093/humrep/deq114. author reply 2153. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reppe S, Refvem H, Gautvik VT, et al. Eight genes are highly associated with BMD variation in postmenopausal Caucasian women. Bone. 2010;46(3):604–12. doi: 10.1016/j.bone.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Hsu YH, Zillikens MC, Wilson SG, et al. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility loci for osteoporosis-related traits. PLoS Genet. 2010;6(6):e1000977. doi: 10.1371/journal.pgen.1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng HF, Forgetta V, Hsu YH, et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526(7571):112–7. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) project. Science. 2004;306(5696):636–40. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 32.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9(3):215–6. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reppe S, Wang Y, Thompson WK, et al. GEFOS Consortium. Genetic sharing with cardiovascular disease risk factors and diabetes reveals novel bone mineral density loci. PLoS One. 2015;10(12):e0144531. doi: 10.1371/journal.pone.0144531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duncan EL, Danoy P, Kemp JP, et al. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011;7(4):e1001372. doi: 10.1371/journal.pgen.1001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karasik D, Myers RH, Hannan MT, et al. Mapping of quantitative ultrasound of the calcaneus bone to chromosome 1 by genome-wide linkage analysis. Osteoporos Int. 2002;13(10):796–802. doi: 10.1007/s001980200110. [DOI] [PubMed] [Google Scholar]
- 38.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, et al. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358(22):2355–65. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 39.Brakspear KS, Mason DJ. Glutamate signaling in bone. Front Endocrinol (Lausanne) 2012;3:97. doi: 10.3389/fendo.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mason DJ, Suva LJ, Genever PG, et al. Mechanically regulated expression of a neural glutamate transporter in bone: a role for excitatory amino acids as osteotropic agents? Bone. 1997;20(3):199–205. doi: 10.1016/s8756-3282(96)00386-9. [DOI] [PubMed] [Google Scholar]
- 41.Gray C, Marie H, Arora M, et al. Glutamate does not play a major role in controlling bone growth. J Bone Miner Res. 2001;16(4):742–9. doi: 10.1359/jbmr.2001.16.4.742. [DOI] [PubMed] [Google Scholar]
- 42.Schraven SP, Franz C, Ruttiger L, et al. Altered phenotype of the vestibular organ in GLAST-1 null mice. J Assoc Res Otolaryngol. 2012;13(3):323–33. doi: 10.1007/s10162-011-0311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Courtland HW, Elis S, Wu Y, et al. Serum IGF-1 affects skeletal acquisition in a temporal and compartment-specific manner. PLoS One. 2011;6(3):e14762. doi: 10.1371/journal.pone.0014762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabsovich I, Clark JD, Liao G, et al. Bone microstructure and its associated genetic variability in 12 inbred mouse strains: microCT study and in silico genome scan. Bone. 2008;42(2):439–51. doi: 10.1016/j.bone.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zanotti S, Canalis E. Activation of Nfatc2 in osteoblasts causes osteopenia. J Cell Physiol. 2015;230(7):1689–95. doi: 10.1002/jcp.24928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raft S, Coate TM, Kelley MW, Crenshaw EB, 3rd, Wu DK. Pou3f4-mediated regulation of ephrin-b2 controls temporal bone development in the mouse. PLoS One. 2014;9(10):e109043. doi: 10.1371/journal.pone.0109043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao C, Irie N, Takada Y, et al. Bidirectional ephrinB2-EphB4 signaling controls bone homeostasis. Cell Metab. 2006;4(2):111–21. doi: 10.1016/j.cmet.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 48.Zhu L, Dissanayaka WL, Green DW, Zhang C. Stimulation of EphB2/ ephrin-B1 signalling by tumour necrosis factor alpha in human dental pulp stem cells. Cell Prolif. 2015;48(2):231–8. doi: 10.1111/cpr.12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takyar FM, Tonna S, Ho PW, et al. EphrinB2/EphB4 inhibition in the osteoblast lineage modifies the anabolic response to parathyroid hormone. J Bone Miner Res. 2013;28(4):912–25. doi: 10.1002/jbmr.1820. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Menendez A, Fong C, ElAlieh HZ, Chang W, Bikle DD. Ephrin B2/EphB4 mediates the actions of IGF-I signaling in regulating endochondral bone formation. J Bone Miner Res. 2014;29(8):1900–13. doi: 10.1002/jbmr.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paternoster L, Lorentzon M, Lehtimaki T, et al. Genetic determinants of trabecular and cortical volumetric bone mineral densities and bone microstructure. PLoS Genet. 2013;9(2):e1003247. doi: 10.1371/journal.pgen.1003247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ackert-Bicknell CL, Karasik D, Li Q, et al. Mouse BMD quantitative trait loci show improved concordance with human genome-wide association loci when recalculated on a new, common mouse genetic map. J Bone Miner Res. 2010;25(8):1808–20. doi: 10.1002/jbmr.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ideno H, Takanabe R, Shimada A, et al. Protein related to DAN and cerberus (PRDC) inhibits osteoblastic differentiation and its suppression promotes osteogenesis in vitro. Exp Cell Res. 2009;315(3):474–84. doi: 10.1016/j.yexcr.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 54.Im J, Kim H, Kim S, Jho EH. Wnt/beta-catenin signaling regulates expression of PRDC, an antagonist of the BMP-4 signaling pathway. Biochem Biophys Res Commun. 2007;354(1):296–301. doi: 10.1016/j.bbrc.2006.12.205. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki D, Yamada A, Aizawa R, et al. BMP2 differentially regulates the expression of Gremlin1 and Gremlin2, the negative regulators of BMP function, during osteoblast differentiation. Calcif Tissue Int. 2012;91(1):88–96. doi: 10.1007/s00223-012-9614-5. [DOI] [PubMed] [Google Scholar]
- 56.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. 2013;19(2):179–92. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
- 57.Yu B, Chang J, Liu Y, et al. Wnt4 signaling prevents skeletal aging and inflammation by inhibiting nuclear factor-kappaB. Nat Med. 2014;20(9):1009–17. doi: 10.1038/nm.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zmuda JM, Yerges-Armstrong LM, Moffett SP, et al. Osteoporotic Fractures in Men Study Group. Genetic analysis of vertebral trabecular bone density and cross-sectional area in older men. Osteoporos Int. 2011;22(4):1079–90. doi: 10.1007/s00198-010-1296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


