Abstract
Ribosomal frameshifting is a rare but ubiquitous process that is being studied extensively. Meanwhile, frameshifting motifs without any secondary mRNA structures were identified but rarely studied experimentally. We report unambiguous observation of highly efficient “–1” and “–2” frameshiftings on a GA7G slippery mRNA without the downstream secondary structure, using force-induced remnant magnetization spectroscopy combined with unique probing schemes. The result represents the first experimental evidence of multiple frameshifting steps. It is also one of the rare reports of the “–2” frameshifting. Our assay removed the ambiguity of transcriptional slippage involvement in other frameshifting assays. Two significant insights for the frameshifting mechanism were revealed. First, EF-G·GTP is indispensable to frameshifting. Although EFG·GDPCP has been shown to prompt translocation before, we found that it could not induce frameshifting. This implies that the GTP hydrolysis is responsible for the codon–anticodon re-pairing in frameshifting, which corroborates our previous mechanical force measurement of EF-G·GTP. Second, translation in all three reading frames of the slippery sequence can be induced by the corresponding in-frame aminoacyl tRNAs. Although A-site tRNA is known to affect the partition between “0” and “–1” frameshifting, it has not been reported that all three reading frames can be translated by their corresponding tRNAs. The in vitro results were confirmed by toe-printing assay and protein sequencing.
Introduction
Frameshifting is a process in which the ribosome decodes mRNA in an alternative grouping of consecutive nucleotide triplets.1 Random frameshiftings are translational errors that often encounter stop codons shortly afterward, whereas programmed frameshiftings decode overlapping genes and regulate both mRNA stabilities and protein expression levels.2 Despite the varying motifs for “–1” and “+1” frameshiftings, cis-acting mRNA elements generally induce this process in the thermodynamically favored direction.2,3 Frameshifting is mostly studied in viral mRNAs for its association with infectiousness, although it occurs in both prokaryotic and eukaryotic cellular mRNAs. The putative “–1” frameshifting motif includes a slippery sequence in the form of “X XXY YYZ” (the blanks define the “0” reading frame), a downstream secondary structure, and a spacer between the two elements; less often upstream Shine–Dalgarno (SD) sequences can replace the downstream secondary structures.4 However, bioinformatics analysis identified frameshifted open reading frames (ORFs) that were not associated with proximal secondary structures.5,6 These ORFs were attributed to transcriptional slippage7,8 or trans-acting protein factor.9 Similar motifs were also identified in bacteria.4 However, frameshiftings without mRNA secondary structures are rarely experimentally studied.
The single nucleotide (nt) difference between the three reading frames makes it difficult to directly and precisely resolve them. The conventional dual luciferase assay measures the ratio of the proteins translated in the “0” and “–1” reading frames inside the cell.10 It cannot rule out the roles of transcription slippage and trans-acting factors as mentioned above.5,8 In addition, dual luciferase assay or mass spectrometry cannot distinguish different frameshifting pathways, such as multiple frameshifting steps and sizes that lead to the same peptides.11 Recently, single molecule and fast kinetic fluorescence signals have been tracked to deduce the ribosome reading frame, but the actual ribosome position was not directly probed.12,13 Optical trap cannot identify the frameshifting positions because of the intrinsic ribosome fluctuation on the slippery site.11 The toe-printing assay usually exhibits multiple-bands even for homologous ribosome complexes,14 making it difficult to quantify mixtures of frameshifting products unless a single frameshifting product dominates.15,16
Here, we report a new assay of using systematically designed DNA probes labeled with magnetic beads to precisely reveal the ribosome positions on mRNA with single nt resolution. This assay consists of force-induced remnant magnetization spectroscopy (FIRMS) that we invented and two novel probing schemes that are first reported here. The position of the ribosome was determined by precisely identifying the mRNA nucleotides adjacent to the ribosome entry site, which is 11–13 nucleotides away from the first nucleotide of P-site codon.17−19 The FIRMS measures the dissociation forces of nucleic acid duplexes formed with the mRNA and DNA probes with high resolution. Using this assay, we tracked three consecutive translocation steps to unambiguously identify nine possible ribosome positions on the mRNA under in vitro conditions. High-yield ribosomal “–1” and “–2” frameshiftings were revealed on a short slippery mRNA without a secondary structure, which was confirmed by the conventional toe-printing assay and in vitro mRNA-translations. Mechanistic studies were carried out by modifying the mRNA motif, introducing a secondary structure, and varying other experimental conditions.
Results and Discussion
Translocation Probing Strategies
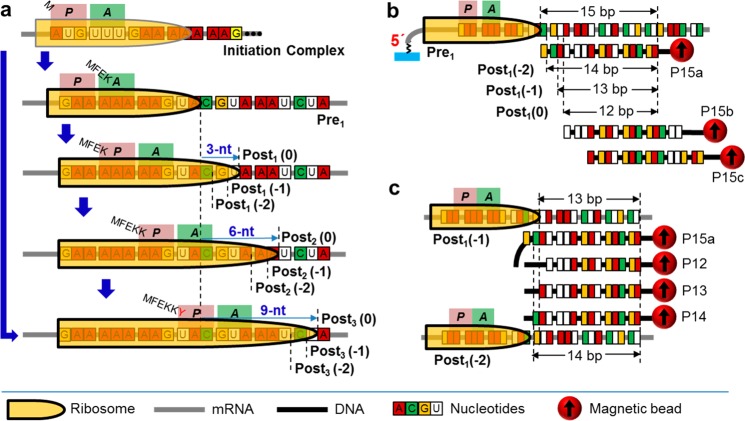
Figure 1a displays the ribosome complexes studied in this work that tracked the ribosome movements over the slippery sequence “GAA AAA AAG” (GA7G), from “AAA” at the A-site to “AAG” at the E-site. The overall displacement is 9 nt. The pretranslocation-complex-1 (Pre1) carried tRNAGlu and MFEK-tRNALys at the peptidyl-tRNA-binding site (P-site) and aminoacyl-tRNA-binding site (A-site), respectively. The mRNA sequence starting from the P-site to downstream was “GAA AAA AAG″. Then the EF-G·GTP complex was added to promote the first translocation step to form the post-translocation-complex-1 (Post1), which potentially possesses all three reading frames, “0”, “–1”, and “–2” (denoted as Post1(0), Post1(−1) and Post1(−2), respectively). The second translocation step proceeded by adding EF-G·GTP and Lys-tRNALys ternary complex, to form Post2 complexes that also potentially contained all three reading frames of Post2(0), Post2(−1) and Post2(−2). The Post3 complexes were generated in one-pot from the ribosome initiation complex with EF-G·GTP, total tRNAs, and only the corresponding set of amino acids for each specific frame. In addition, Post3(0) was also prepared from Post2 in the presence of the “0” frame substrate (Tyr-tRNATyr ternary complex) and EF-G·GTP.
Figure 1.
Schemes of the ribosome complexes and the FIRMS assay. (a) Ribosome complexes. Starting from the initiation complex, the pretranslocation complex was produced, followed by three consecutive steps of translocation going through the GA7G motif. (b) The FIRMS scheme of using different magnetically labeled DNAs for probing different translocation step. In each step, the formation of 12-, 13-, and 14-bp DNA-mRNA duplexes indicate normal translocation, “–1” frameshifting, and “–2” frameshifting, respectively. (c) Scheme of using probe DNAs with a single nt difference to confirm the reading frame.
Figure 1b,c shows two probing schemes using multiple magnetically labeled DNAs. For each scheme, FIRMS was used to determine the dissociation forces of the resulting DNA-mRNA duplexes by measuring the magnetic signal as a function of centrifugal force; the magnetic signal will show a decrease when the duplexes dissociate because of the removal of the associated magnetic beads (Figure S1). Specifically, in Figure 1b, three DNA oligomers were designed to have 3-nt shift in between, so that each one will probe one of the three translocation steps. In Figure 1c, to improve the precision of frameshifting assignments, a series of probing DNAs with1-nt difference in between and aligned at their 5′-termini were used to probe the same translocation step (details in the Supporting Information). Therefore, the reading frames can be precisely determined from the DNA–mRNA binding patterns, and multiple frameshiftings can be unambiguously assigned.
High-Yield Frameshiftings on the GA7G Motif without a Secondary Structure
In the first translocation step, we observed 55% “–1” and 45% “–2” frameshiftings but no “0” frame translocation. This observation was confirmed with two probing schemes and extensive control sequences.
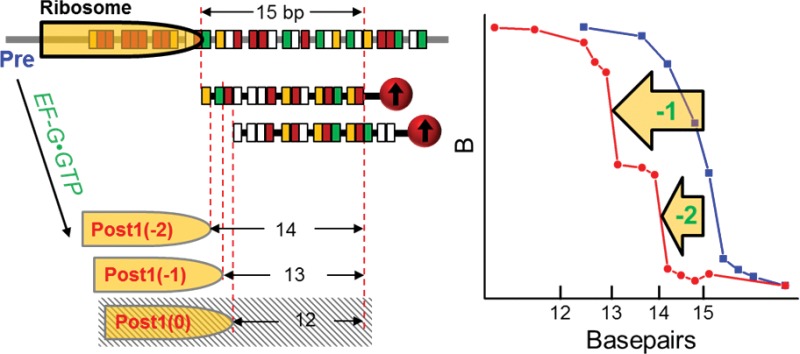
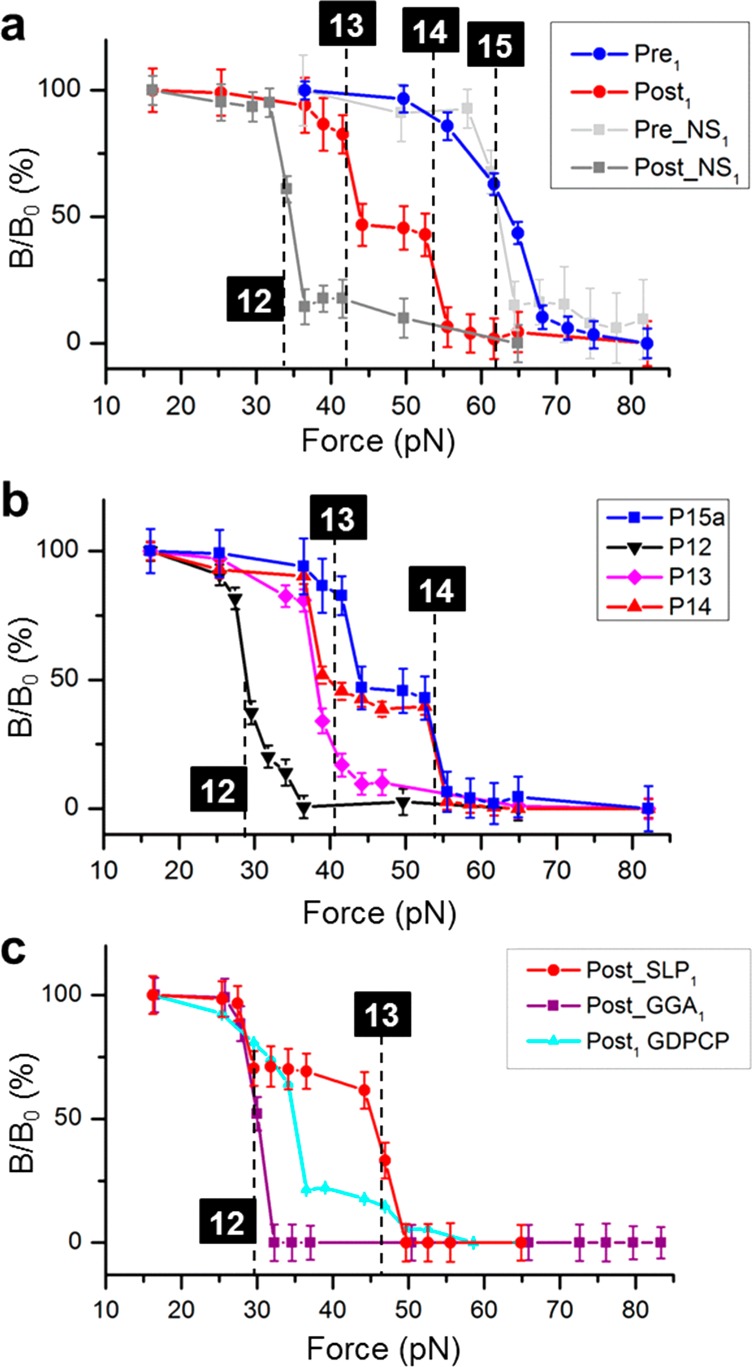
Figure 2 shows the results of the first translocation step. The dissociation of the DNA–mRNA duplexes is indicated by a sharp decrease in the magnetic signal. A calibration curve of dissociation force versus bp for a series of DNA–mRNA duplexes has been obtained (Figure S2). Using probe P15a, Pre1 complex exhibited 15-bp binding force (Figure 2a, blue trace). Post1 yielded two binding forces of 13- and 14-bp, respectively (Figure 2a, red trace). No 12-bp binding force was observed. This result indicates both “–1” and “–2” frameshiftings for the GA7G motif but no normal translocation. When the slippery motif was replaced by a nonslippery (NS) “GAA AGU AAG”, normal translocation occurred for its first translocation product, Post_NS1 (Figure 2a, dark gray trace). This was indicated by the binding force for 12-bp. For comparison, its pretranslocation complex, Pre_NS1, yielded binding force of 15-bp (Figure 2a, light gray trace).
Figure 2.
Probing the three reading frames of the first translocation step. (a) FIRMS profiles of Pre1 and Post1 for the GA7G motif, in comparison with those for a nonslippery (NS) motif. (b) Confirmation of the “–1” and “–2” frameshiftings for GA7G using a series of NDA probes. (c) FIRMS profiles for the stem loop (SLP), the GGA motifs, and GA7G motif promoted by EF-G·GDPCP, showing different translocation behaviors.
The unusual “–1” and “–2” frameshiftings of the GA7G motif were confirmed in Figure 2b, using the probing scheme depicted in Figure 1c. The P12, P13, and P14 exhibited binding forces for Post1 of 12-, 13-, and a combination of 13- and 14-bp, respectively. This indicated that the 13-bp DNA-mRNA duplex was limited by the ribosome front. The 13-/14-bp combination persisted when using P15a, but no 15-bp duplex appeared. This result again indicated that the ribosome front limited the duplexes to be 13 and 14 bps. Together, the two results conclusively determined the exact ribosome positions. Similarly, the exact ribosome front in Pre1 was confirmed to form exactly 15-bp duplex with P15a and a longer DNA probe (Figure S3). Therefore, our assay unambiguously revealed that only Post1(−1) (∼55%) and Post1(−2) were present after the first round of translocation.
To elucidate the unusual frameshifting mechanism, three approaches were used (Figure 2c). The first experiment was to reveal the role of GTP. When EF-G·GTP was replaced by its nonhydrolyzable analogue, EF-G·GDPCP, which also promotes translocation,20 mostly normal translocation occurred to produce Post1(0) of 32 pN binding force (cyan trace). The 12-bp complex was the major product, different from those in the EF-G·GTP experiments. These results indicate that GTP energy is indispensable to frameshifting.
The second experiment was to change the slippery sequence. We modified the GA7G sequence to GGA6G (denoted as GGA for simplicity). The post complex (Post_GGA1) formed only 12-bp duplexes with P15a, indicating only normal translocation (Figure 2c, purple trace). This result agrees with the literature,12,21 showing the critical role of the P-site codon–anticodon interaction in stimulating frameshifting. It also means the SD-sequence was probably too far to play a significant role (13 nt away from AAA). Similarly, no significant frameshifting (less than ∼10%, our current detection limit) was detected in the following two translocation steps to form Post_GGA2 and Post_GGA3 (Figure S4a,b). Even in the presence of the downstream aminoacyl tRNA of the “–1” reading frame, no Post_GGA3(−1) was induced. This result is consistent with the literature that A AAA AAG needs a downstream secondary structure to cause frameshifting.4,22 In addition, because the frameshifting has occurred on the GAA AAA sequence, we studied the mRNA that replaced the following AAG to CGC. As shown in Figure S4c,d, no frameshifting occurred for two translocation steps that led to Post_CGC1 and Post_CGC2. This result suggests that GAA AAA alone is insufficient to induce the frameshifting. Again, this result agrees with the literature.4
The third experiment was to determine the frameshifting with the authentic dnaX stem loop structure.22 The post complex (Post_SLP) formed both 12- and 13-bp duplexes with SLP15a, respectively (Figure 2c, red trace). The assignments were confirmed using the scheme in Figure 1c (Figure S3). The “–2” frameshifting was absent. Instead, Post_SLP1(−1) was the major product at 63 ± 12%, and the remaining was Post_SLP1(0). This result agrees with the literature that showed approximately 70% frameshifting efficiency.12,21,22 The Post_SLP3 was studied, and the frameshifting yield was preserved (Figure S4e,f).
Toe-Printing Assay and Protein Expressions Confirmed the “–1” and “–2” Frameshiftings on the GA7G Motif
The FIRMS results were confirmed with conventional biochemical assays. First, the ribosome toe-printing was conducted on an mRNA with GA7G motif implemented.23 The ribosomes were paused after synthesizing the “MFEKK” peptide. Two control mRNAs, one with a downstream stem-loop after GA7G and the other with “GAA AGU AAG” in place of GA7G, were assayed side-by-side. The sequences were named “GA7G” (Figure 3, Lane 3), “SLP” (Lane 2) and “AGU” (Lane 4), respectively. The standard protocol was followed, except that Cy5-labeled primers were used instead of 32P-labeled primers. Given the weak processivity of reverse transcriptase, toe-printing patterns are always present with discrete multibands because of enzyme drop-off. Therefore, this assay has limitations in quantifying frameshifting efficiencies. Regardless, the nonrandom multibands patterns supported the frameshifting processes. In the nonframeshifting sequence (Lane 4), the ribosome carrying MFESK was 16-nt away from the P-site codon “AAG”, generating a 47 nt cDNA. Meanwhile, the “GA7G” sequence exhibited both “–1” and “–2” frameshifted bands near 47-nt. In the presence of the stem loop, only “–1” frameshifting was observed. The pattern for “GA7G” was more diffuse because of the more branches of frameshifting pathways, similar to other reports.24
Figure 3.
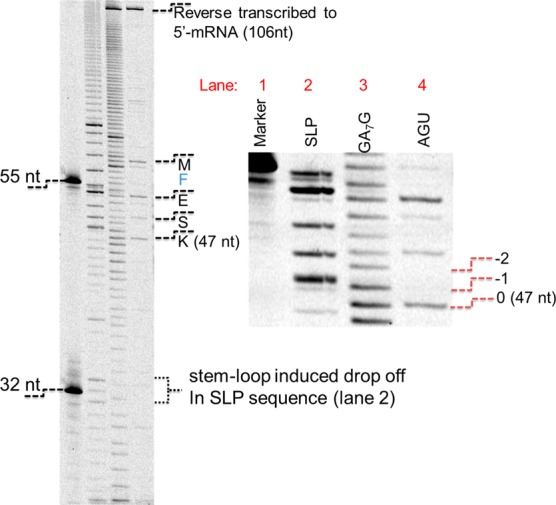
Toe-printing assays verified the frameshifting. The toe-printing assays of the cDNAs that were reverse transcribed with Cy5-labeled primer. Lane 1: markers of 32 and 55 nt in lengths; Lanes 2, 3, and 4: toe-printing of SLP, GA7G, and AGU sequences, respectively. The right panel was a close-up view that was obtained by averaging four repeated scans. In Lane 4, the distinct bands were consistent with the decoding of K, S, E, and M, respectively. The bands near 106-nt at the top of the plots were the cDNAs reverse transcribed to the 5′-of the mRNAs.
Second, the GA7G motif was tested with recombinant protein expression in the E. coli cells. The “GA7G” motif without the downstream stem loop was incorporated into three constructs that were inserted in the pET20b (+) vectors. The constructs were shown in Table S1. The 8.5 kDa protein sequence was modified from a shorter peptide sequence of ribosomal protein L27.25 Proteins were approximately 8.5, 6.5, and 4.6 kDa (Table S2). These constructs were expressed and purified via the Ni-NTA columns (Supporting Information). The constructs I and II generated the 8.5 kDa proteins with similar yields, via “0” and “–1” translocation processes, respectively (Figure S5a). The proteins were identified by N-terminal Edman sequencing (Figures S5b,c). The time-course of the IPTG induced protein synthesis was monitored with SDS-PAGE (Figure S6). Conversely, we did not isolate the similar protein in construct III, probably due to plasmid instability or protease digestion. However, the “–2” frameshifting protein was successfully isolated when the 28.8 kDa mCherry protein sequence was placed in the “–2” reading frame of construct II (Figure S7). However, no protein bands for the 6.5 or 4.6 kDa were observed. The 6.5 kDa protein sequence was further implemented in the same vector without the slippery site, and it was not isolated. Therefore, the proteins in the other two frames were not stable. Because the proteins decoded in the other two frames were fixed, we could not design a sequence which decodes for three stable proteins in all three reading frames simultaneously.
Although we cannot directly estimate the frameshifting efficiencies because not all of the proteins in the three reading frames were expressed simultaneously, the preparation protocol was exactly the same (Supporting Information) and 500–1000 pmol of the 8.5 kDa protein (for construct I and II) or mCherry protein was obtained, suggesting the similar partition in all three reading frames.
Third, the “GA7G” motif was tested in the PURExpress kit with mRNAs instead of DNAs. As shown in Table S3, four mRNA constructs were synthesized by in vitro transcription, which incorporated with the mCherry protein in the 0, −1, −2, and 0 (without slippery site) reading frames, respectively. Construct IV and V were the positive control and background, respectively. The proteins were synthesized for 2 h and fluorescence were measured. The measurements for experiments I–III were normalized with experiment IV after subtracting background from experiment V. The relative yields for the “0”, “–1”, and “–2” frameshiftings were then calculated to be 34%, 35%, and 31%, respectively. These results were consistent with the FIRMS observations and agreed with the relative yields deduced from the recombinant protein synthesis results. Although in vitro transcribed mRNA still could not rule out transcriptional slippage, the very high yield of the frameshifting efficiencies compared to the 1–2% yield of the transcriptional slippages7,8 strongly favored the ribosome slippage in our observations.
Post2 Complexes Maintain the Same Reading Frames as in Post1 but Can Form Post3(0) with the Next “0” Frame Substrate
After confirming the FIRMS results with conventional biochemical means, we tracked the second translation step with P15b by incubating Post1 with Lys-tRNALys ternary complex and EF-G·GTP. Therefore, 12-, 13-, and 14-bp still, respectively, refer to the “0”, “–1”, and “–2” reading frames (Figure 1b). The result showed only the “–1” and “–2” products (Figure 4a). The overlay of the traces for Post1 and Post2 showed that these two traces were almost identical (Figure S8), implying that frameshifting may occur only at the translocation of the “AAA” codon, and normal translocation proceeds from Post1 to Post2. However, we cannot rule out the possibility of a second frameshifting step that result in the same distribution of “–1” and “–2” frameshiftings, as indicated by the toe-printing experiments. Nevertheless, a second frameshifting step was indeed observed to form Post3(0) when the Post2 complexes were incubated with the next “0” frame substrate Tyr-tRNATyr ternary complex (decodes “UAC”) and EF-G·GTP. This complex was probe with P15c. Figure 4b shows the existence of 12-bp duplexes at 27 pN, corresponding to Post3(0) (red trace). Both Post3(−1) and Post3(−2) were absent. The 15-bp binding force was due to residual Post2 complex in which the ribosome front did not reach the probe. The “+1” or “+2” frameshifting to restore the “0” reading frame is probably via the “hungry codon” mechanism26 to form Pre3(0), which exhibited only 15 bp binding force in the absence of EF-G·GTP (green trace).
Figure 4.
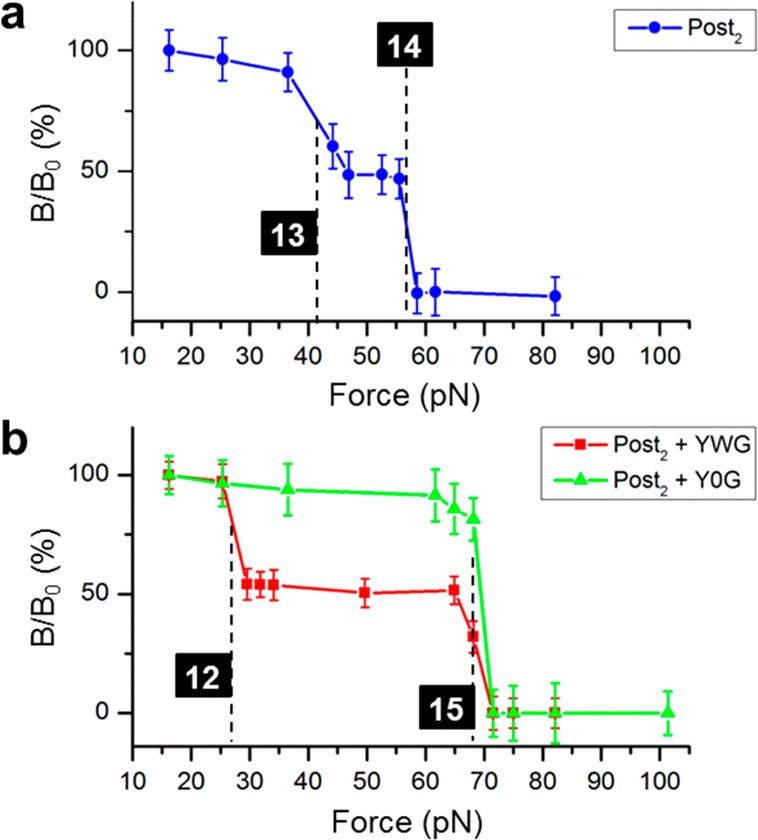
Products of the second- and third-step translocations. (a) FIRMS profile showing the formation of Post2(−1) and Post2(−2), indicated by the 13-bp dissociation at 42 pN and the 14-bp dissociation at 57 pN, respectively. (b) FIRMS profiles showing the formation of Post3(0) via Post2 only in the presence of EF-G·GTP, indicated by the 12-bp duplex. YWG: mix of Y-tRNAtyr, Tu·GTP, and EF-G·GTP. Y0G: mix of Y-tRNAtyr and Tu·GTP only, without EF-G·GTP.
Post3 Complexes in All Three Reading Frames Are Formed in the Presence of the In-Frame Aminoacyl tRNAs
To explain the lack of the “0” frame product in Post1 and Post2, the Post3 complexes were prepared in one-pot from the initiation complex, in which the ribosome had completed the slippery sequence. Under these conditions, Post3(0) was formed, which suggested that the “0” frame translocation may be favored kinetically in the presence of the in-frame aminoacyl tRNAs, without pausing on the slippery site. In addition, when aminoacyl tRNAs for the other reading frames were provided exclusively, the ribosome was biased to the corresponding frame efficiently, indicating the powerful decoding roles of tRNAs.
The initiation complex was incubated with total tRNA and one set of amino acids to form the Post3 complexes in the three frames separately: Phe, Glu, Lys, and Tyr for “0” frame; Phe, Glu, Lys, and Val for “–1” frame; Phe, Glu, Lys, and Ser for “–2” frame. Using probe P15c, the FIRMS results were expected to contain two transitions for each complex: the Post3 of one specific reading frame and the stalled Post2 of the other two frames. Post3(0), Post3(−1), and Post3(−2) will form 12-, 13-, and 14-bp duplexes with P15c, respectively (Figure 5a). All Post2 complexes will form 15-bp duplexes only. Figure 5b shows that under each condition, approximately 50% ribosome formed Post3 complex of the specific reading frame, and the remaining was Post2. The high-yield formation of Post3(−1) and Post3 (−2) demonstrated that the frameshiftings of Post1 on GA7G motif are intrinsic, not due to in vitro artificial pausing, which could induce “–1” and “+1” frameshiftings.26 However, it is possible that the frameshifting yields in Figure 2 were higher in our experiments than in the cell because of the pausing and more complicated factors in the cell.
Figure 5.
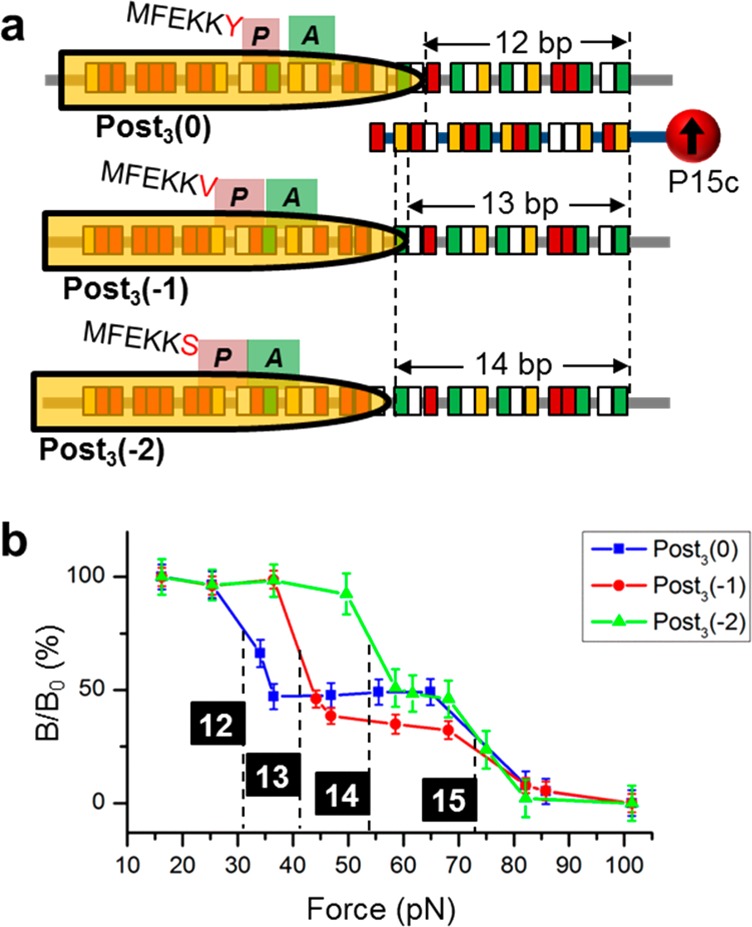
Frameshifting products after three continuous translocation steps from the initial complex. (a) Detection scheme of the three reading frames using P15c. (b) FIRMS profiles. Post3 in all three reading frames were formed, indicated by the 12-bp duplex for Post3(0), 13-bp for Post3(−1), and 14-bp for Post3(−2), respectively.
On the other hand, the absence of the “0” product in Post1 and Post2 may be because the prolonged pausing has weakened the kinetic advantage of normal translocation.13 To examine this hypothesis, under Post3(0) formation condition (blue trace of Figure 5b), the composition of the residual post2 complexes were studied with the P15b. Post3(0) would form 9-bp duplex with this probe, which is unstable to be detected by FIRMS. Figure S9 showed that Post2 complexes in all three reading frames were formed. The “0” frame ribosomal complexes at Post2(0)/Pre3(0) were the major products (indicated by the 12 bp binding force), while Post2(−1) and Post2(−2) complexes were also formed with significant percentages (indicated by the 13- and 14-bp binding forces, respectively). Note that Pre3(0) could form, but it would be indistinguishable from Post2(0). Therefore, Figure S9 showed that the ribosome preferred the “0” frame if it was not halted at the slippery site. Our study implies that the “0” frame product is either the kinetically favored product in the cell (Figures 5 and S7) or the accumulating outcome of multistep frameshiftings (Figure 4).
Both “–1” and “–2” Frameshiftings Were Observed on GA7G-Only Motif
We have unambiguously revealed the intrinsic frameshifting on GA7G motif without entangling with the other factors, such as the downstream aminoacyl tRNAs and stimulators. To our best knowledge, this is the first time that high-efficiency “–2” frameshifting was observed without a stimulator, a trans-acting factor, or transcriptional slippage, although the slipper sequence alone is known to induce “–1” frameshifting.4 Compared to the common 1–5% frameshifting efficiencies, the frameshifting percentages observed here are substantially higher. This is probably due to the lack of the stimulator because similar high efficiencies were reported elsewhere in the absence of stimulators.24,27,28
Extensive ribosome pausing over the poly(A) sequences was observed that could lead to translational frameshiftings.24 Meanwhile, “–1”, “–4”, and “+2” frameshiftings were observed simultaneously on a similar mRNA.11 A systematic bioinformatics and experimental analysis also indicated that the poly(A) sequence was slippery to induce “–1” frameshifting.4 In this report, we further found “–2” frameshifting occurred at comparable efficiency as “–1” frameshifting, probably due to the ambiguities of codon-anticodon re-pairing over the poly(A) motifs.
Frameshifting Occurs When “GAA AAA” Moves from “P- and A-” to “E- and P-” Sites
The exact codon may vary at which frameshifting can occur,13 and frameshifting can occur at multiple sites with multiple slipping-distances.11 The prevailing view is that frameshifting occurs during the “YYZ” translocation when the mRNA secondary structure clashes with the ribosome entry site. However, our results have shown that frameshifting occurs when “XXY” translocates, either with or without the secondary structure. It appeared that the location of the mRNA secondary structure did not matter. However, Figure 4b showed that Post2 complex, which lacked the 0-frame ribosomes, has been pulled into the 0-frame to form Post3(0) with the 0-frame Tyr-tRNATyr substrate. This result suggests that a second “+1” frameshifting step is possible on the YYZ codon. The multiple frameshifting sites agreed with the GC/LC-MS study.11 However, in the MS study, the fundamental slippery sequence has to be replaced (AAA AAG to AAC AAG); in our study, it was intact. Because the slippery site is the major motif under investigation, our method is more applicable. In addition, multiple frameshifting pathways were proposed,11,13,29 but multiple steps of frameshifting has not been reported before. Our method is unique in this regard because other methods cannot distinguish multiple steps from a single large step that generates the same peptide.
Finally, frameshifting at the “XXY” codon is not inconsistent with a single-molecule study showing that a noncanonical, ratcheted, and long-lived ribosome conformation emerged after decoding the “XXY” codon, although the ribosome movement on the mRNA was not directly determined in that study.12
tRNAs Can Define the Ribosome Translation Frame on the Slippery Site
We observed two tRNA effects in governing the frameshifting. The first one is the suppression of frameshifting when the P-site codon is changed from “GAA” to “GGA”, probably because of the stronger codon–anticodon interaction for an “A–G” than a “T–A” pair. The second one is the induction of the ribosome into any of the three reading frames with the corresponding set of substrates, showing the role of the A-site tRNA. These two observations suggest that frameshifting is the synergistic outcome of P-site tRNA re-pairing and A-site tRNA sampling, which corroborate a previous model.30 The A-site tRNA has been suggested to decode with only two nucleotides at a hungry codon, which can prompt frameshifting in both the “+1” and “–1” directions.26 In addition, changing the codon at the slippery site or its proximity has changed the frameshifting efficiencies.12,31 However, to our best knowledge, this report is the first time to show that all three reading frames can be translated by their in-frame tRNAs. Therefore, our results showed more prominent active role of tRNAs in guiding the ribosome into certain ORFs.
GTP Energy Is Essential to Frameshifting
We have shown that translocation on the “GA7G-only” motif with EF-G·GDPCP generated normal translocation, on the contrary to the EF-G·GTP experiments; A-site substrate without EF-G cannot drive Post2 into Post3, on the contrary to when EF-G·GTP is present. Some recent kinetics studies have revealed transient translocation intermediates (67–280 ms lifetime) in which the mRNA has moved three nucleotides while the ribosome is in the process to form the canonical post-translocation configuration.32,33 However, it is unlikely that the lack of frameshifting with EF-G·GDPCP observed here is due to this intermediate state because of the very different time scale in this study. On the other hand, EF-G·GDPCP is competent in translocation at 0.5 s–1 turnover rate,33,34 which means that under our experimental conditions, most ribosomes were turned into the post-translocation configuration. Therefore, these results indicate that the GTP energy is essential to overcome the frameshifting reaction barrier, whereas without GTP translocation the process proceeds via alternative pathways. This conclusion is consistent with our previous report that an 89 pN mechanical force accompanies the GTP hydrolysis by EF-G.17 A Cryo-EM study has revealed significant tRNA deformation induced by EF-G, which also implies the involvement of mechanical force.35 An X-ray structural study observed the ribosome-EF-G complex in the midtranslocation, showing that while the P-site tRNA moved precisely along the 30S-head swiveling, the A-site tRNA moved 0.65 nm further to avoid clash with the EF-G domain IV.36 This structure implied that the EF-G exerted its force on the A-site tRNA. Then the mRNA moves accordingly via its interactions with the tRNAs. In fact, on the basis of the 89 pN force measurement, we have estimated the EF-G catalyzed translocation has a transition-state distance of approximately 0.5 nm,17 which agreed well with the 0.65 nm displacement in this structure. During frameshifting, the mechanical force exerted on the A-site tRNA can disrupt the weaker codon–anticodon interaction on the slippery site, giving the ribosome an opportunity to re-pair the tRNA–mRNA in a different frame. Therefore, we expect the power stroke on the slippery sites will be smaller because of the weaker force transmission from the tRNA to the mRNA, compared to normal translocation. We are currently testing this hypothesis.
Methods
The MRE600 ribosomes were purified according to the literature.37 The plasmids of His-tagged IF1, IF3, EF-Tu, EF-G were provided by Drs. Yale Goldman and Barry Cooperman at the University of Pennsylvania. The IF2 plasmid was provided by Dr. Rachel Green at the Johns Hopkins University. The total aminoacyl-tRNA synthetases were purified from the S100 extract of E. coli cells.38 The tRNAfMet, tRNAPhe, and tRNALys were purchased from Chemical Block or Sigma-Aldrich. The biotinylated mRNAs and DNA oligos were purchased from Integrated DNA Technologies. The sequence of the mRNA containing the GA7G motif was 5′-Bio-C AAC UGU UAA UUA AAU UAA AUU AAA AAG GAA AUA AAA AUG UUU GAA AAA AAG UAC GUA AAU CUA CUG CUG AAC UC-3′. Other sequences are provided in the Supporting Information.
The in vitro ensemble of ribosome complexes was similar to the previous procedure.25 The FIRMS measurements were similar to those in our recent reports.17,39 The mRNAs for toe-printings were transcribed and purified in vitro using the HiScribe T7 Quick High Yield RNA Synthesis Kit (NEB). Details are provided in the Supporting Information.
Acknowledgments
This work is supported by the National Institutes of Health (R01GM111452, to Y.W. and S.X) and the Welch Foundation (E-1721, to Y.W.). T.-W.T. and H.Y. acknowledge support by the chemical biology program at the University of Houston. S.X. acknowledges support by the Texas Center for Superconductivity (TcSUH). We thank D. Winant at the Stanford University PAN facility for his excellent Edman sequencing technique.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.7b00028.
Detailed experimental procedures, nine supplemental figures, and three supplemental tables (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Baranov P. V.; Gesteland R. F.; Atkins J. F. (2002) Recoding: translational bifurcations in gene expression. Gene 286, 187–201. 10.1016/S0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- Firth A. E.; Brierley I. (2012) Non-canonical translation in RNA viruses. J. Gen. Virol. 93, 1385–1409. 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamski F. M.; Donly B. C.; Tate W. P. (1993) Competition between frameshifting, termination and suppression at the frameshift site in the Escherichia coli release factor-2 mRNA. Nucleic Acids Res. 21, 5074–5078. 10.1093/nar/21.22.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V.; Prere M. F.; Canal I.; Firth A. E.; Atkins J. F.; Baranov P. V.; Fayet O. (2014) Analysis of tetra- and hepta-nucleotides motifs promoting −1 ribosomal frameshifting in Escherichia coli. Nucleic Acids Res. 42, 7210–7225. 10.1093/nar/gku386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y.; Treffers E. E.; Li Y.; Tas A.; Sun Z.; van der Meer Y.; de Ru A. H.; van Veelen P. A.; Atkins J. F.; Snijder E. J.; Firth A. E. (2012) Efficient −2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc. Natl. Acad. Sci. U. S. A. 109, E2920–2928. 10.1073/pnas.1211145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B. Y.; Miller W. A.; Atkins J. F.; Firth A. E. (2008) An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U. S. A. 105, 5897–5902. 10.1073/pnas.0800468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodamilans B.; Valli A.; Mingot A.; San Leon D.; Baulcombe D.; Lopez-Moya J. J.; Garcia J. A. (2015) RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the potyviridae family. J. Virol. 89, 6965–6967. 10.1128/JVI.00337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olspert A.; Chung B. Y.; Atkins J. F.; Carr J. P.; Firth A. E. (2015) Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep. 16, 995–1004. 10.15252/embr.201540509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Treffers E. E.; Napthine S.; Tas A.; Zhu L.; Sun Z.; Bell S.; Mark B. L.; van Veelen P. A.; van Hemert M. J.; Firth A. E.; Brierley I.; Snijder E. J.; Fang Y. (2014) Transactivation of programmed ribosomal frameshifting by a viral protein. Proc. Natl. Acad. Sci. U. S. A. 111, E2172–2181. 10.1073/pnas.1321930111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G.; Ingram J. A.; Kelly P. J.; Gesteland R. F.; Atkins J. F. (1998) A dual-luciferase reporter system for studying recoding signals. RNA 4, 479–486. [PMC free article] [PubMed] [Google Scholar]
- Yan S.; Wen J. D.; Bustamante C.; Tinoco I. Jr. (2015) Ribosome Excursions during mRNA Translocation Mediate Broad Branching of Frameshift Pathways. Cell 160, 870–881. 10.1016/j.cell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Petrov A.; Johansson M.; Tsai A.; O’Leary S. E.; Puglisi J. D. (2014) Dynamic pathways of −1 translational frameshifting. Nature 512, 328–332. 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan N.; Katunin V. I.; Belardinelli R.; Peske F.; Rodnina M. V. (2014) Programmed −1 frameshifting by kinetic partitioning during impeded translocation. Cell 157, 1619–1631. 10.1016/j.cell.2014.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirokikh N. E.; Alkalaeva E. Z.; Vassilenko K. S.; Afonina Z. A.; Alekhina O. M.; Kisselev L. L.; Spirin A. S. (2010) Quantitative analysis of ribosome-mRNA complexes at different translation stages. Nucleic Acids Res. 38, e15. 10.1093/nar/gkp1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper H. B.; Masuda I.; Frenkel-Morgenstern M.; Hou Y. M. (2015) Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNA. Nat. Commun. 6, 7226. 10.1038/ncomms8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper H. B.; Masuda I.; Frenkel-Morgenstern M.; Hou Y. M. (2015) The UGG Isoacceptor of tRNAPro Is Naturally Prone to Frameshifts. Int. J. Mol. Sci. 16, 14866–14883. 10.3390/ijms160714866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L.; Li Y.; Tsai T. W.; Xu S.; Wang Y. (2013) Noninvasive measurement of the mechanical force generated by motor protein EF-G during ribosome translocation. Angew. Chem., Int. Ed. 52, 14041–14044. 10.1002/anie.201307419. [DOI] [PubMed] [Google Scholar]
- Qu X.; Wen J. D.; Lancaster L.; Noller H. F.; Bustamante C.; Tinoco I. Jr. (2011) The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 475, 118–121. 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S.; Hickerson R. P.; Noller H. F. (2005) mRNA helicase activity of the ribosome. Cell 120, 49–58. 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Katunin V. I.; Savelsbergh A.; Rodnina M. V.; Wintermeyer W. (2002) Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry 41, 12806–12812. 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- Kim H. K.; Liu F.; Fei J.; Bustamante C.; Gonzalez R. L. Jr.; Tinoco I. Jr. (2014) A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc. Natl. Acad. Sci. U. S. A. 111, 5538–5543. 10.1073/pnas.1403457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B.; Wills N. M.; Gesteland R. F.; Atkins J. F. (1994) rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J. Bacteriol. 176, 6842–6851. 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N.; Thum C.; Mankin A. S. (2008) Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell 30, 190–202. 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Koutmou K. S.; Schuller A. P.; Brunelle J. L.; Radhakrishnan A.; Djuranovic S.; Green R. (2015) Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife 4, e05534. 10.7554/eLife.05534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuntop M. E.; Ly C. T.; Wang Y. (2010) Single-molecule study of ribosome hierarchic dynamics at the peptidyl transferase center. Biophys. J. 99, 3002–3009. 10.1016/j.bpj.2010.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J.; Lindsley D. (1993) Ribosome frameshifting at hungry codons: sequence rules, directional specificity and possible relationship to mobile element behaviour. Biochem. Soc. Trans. 21, 817–821. 10.1042/bst0210817. [DOI] [PubMed] [Google Scholar]
- Belfield E. J.; Hughes R. K.; Tsesmetzis N.; Naldrett M. J.; Casey R. (2007) The gateway pDEST17 expression vector encodes a −1 ribosomal frameshifting sequence. Nucleic Acids Res. 35, 1322–1332. 10.1093/nar/gkm003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P. V.; Fayet O.; Hendrix R. W.; Atkins J. F. (2006) Recoding in bacteriophages and bacterial IS elements. Trends Genet. 22, 174–181. 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Liao P. Y.; Choi Y. S.; Dinman J. D.; Lee K. H. (2011) The many paths to frameshifting: kinetic modelling and analysis of the effects of different elongation steps on programmed −1 ribosomal frameshifting. Nucleic Acids Res. 39, 300–312. 10.1093/nar/gkq761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P. V.; Gesteland R. F.; Atkins J. F. (2004) P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA 10, 221–230. 10.1261/rna.5122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew S. F.; Crowe-McAuliffe C.; Graves R.; Cardno T. S.; McKinney C.; Poole E. S.; Tate W. P. (2015) The highly conserved codon following the slippery sequence supports −1 frameshift efficiency at the HIV-1 frameshift site. PLoS One 10, e0122176. 10.1371/journal.pone.0122176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman M. R.; Alejo J. L.; Altman R. B.; Blanchard S. C. (2016) Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat. Struct. Mol. Biol. 23, 333–341. 10.1038/nsmb.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belardinelli R.; Sharma H.; Caliskan N.; Cunha C. E.; Peske F.; Wintermeyer W.; Rodnina M. V. (2016) Choreography of molecular movements during ribosome progression along mRNA. Nat. Struct. Mol. Biol. 23, 342–348. 10.1038/nsmb.3193. [DOI] [PubMed] [Google Scholar]
- Rodnina M. V.; Savelsbergh A.; Katunin V. I.; Wintermeyer W. (1997) Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385, 37–41. 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Namy O.; Moran S. J.; Stuart D. I.; Gilbert R. J.; Brierley I. (2006) A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441, 244–247. 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Lancaster L.; Donohue J. P.; Noller H. F. (2014) How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science 345, 1188–1191. 10.1126/science.1255030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. J.; Green R. (2007) Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol. Cell 28, 458–467. 10.1016/j.molcel.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemkhadze K. S.; Odintsov V. B.; Semenkov Y. P.; Kirillov S. V. (1981) Quantitative study of the interaction of aminoacyl-tRNA with the a site of Escherichia coli ribosomes: equilibrium and kinetic parameters of binding in the absence of EF-Tu factor and GTP. FEBS Lett. 125, 10–14. 10.1016/0014-5793(81)80985-4. [DOI] [PubMed] [Google Scholar]
- De Silva L.; Yao L.; Wang Y.; Xu S. (2013) Well-defined and sequence-specific noncovalent binding forces of DNA. J. Phys. Chem. B 117, 7554–7558. 10.1021/jp403817b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.