Abstract
Background
Pesticides cause oxidative stress to plants and their residues persist in plant parts, which are a major concern for the environment as well as human health. Brassinosteroids (BRs) are known to protect plants from abiotic stress conditions including pesticide toxicity. The present study demonstrated the effects of seed-soaking with 24-epibrassinolide (EBR) on physiological responses of 10-day old Brassica juncea seedlings grown under imidacloprid (IMI) toxicity.
Results
In the seedlings raised from EBR-treated seeds and grown under IMI toxicity, the contents of hydrogen peroxide (H2O2) and superoxide anion (O. 2 −) were decreased, accompanied by enhanced activities of superoxide dismutase (SOD), catalase (CAT), glutathione reductase (GR), glutathione-S-transferase (GST), guaiacol peroxidase (POD) and the content of glutathione (GSH). As compared to controls, the gene expressions of SOD, CAT, GR, POD, NADH (NADH-ubiquinone oxidoreductase), CXE (carboxylesterase), GSH-S (glutathione synthase), GSH-T (glutathione transporter-1), P450 (cytochrome P450 monooxygenase) and GST1-3,5-6 were enhanced in the seedlings raised from EBR-treated seeds and grown in IMI supplemented substratum. However, expression of RBO (respiratory burst oxidase, the gene responsible for H2O2 production) was decreased in seedlings raised from EBR treated seeds and grown under IMI toxicity. Further, the EBR seed treatment decreased IMI residues by more than 38% in B. juncea seedlings.
Conclusions
The present study revealed that EBR seed soaking can efficiently reduce oxidative stress and IMI residues by modulating the gene expression of B. juncea under IMI stress. In conclusion, exogenous EBR application can protect plants from pesticide phytotoxicity.
Keywords: Brassica juncea, Imidacloprid toxicity, Brassinosteroids, Pesticide residues
Background
Plants are susceptible to attack by various insect pests like termites, soil bugs, aphids and leaf hoppers. Pesticides are widely utilized to control these insect pests, but these pesticides cause aerial pollution upon spraying due to their volatile nature [1]. Imidacloprid (IMI) is a neonicotinoid insecticide, which is systemic in action and applied via soil/seed treatment to protect plants from soil and aerial insect pests, without causing any aerial pesticide pollution [2]. Application of pesticides causes oxidative stress to plants by generating reactive oxygen species [3, 4]. Pesticides persist in the parts of plants in form of pesticide residues [4, 5] which cause a major threat to humans as well as pollinators, including honeybees [6]. Due to their persistence in form of residues, pesticides also enter the ecosystem via the food chain, hence cause serious threat to ecosystems and the environment [1]. Plants are able to degrade pesticides into soluble and less toxic metabolites through a three-phased enzyme-mediated degradation process [7, 8]. The first phase involves enzymes like cytochrome P450 monooxygenases (P450), peroxidases and carboxylesterases, which are involved in the activation of pesticides. In the 2nd phase, glutathione-S-transferase (GST) and UDP-glycosyltransferase help in the conjugation of activated pesticides with glutathione (GSH) and glucose, resulting in the formation of less toxic and more soluble metabolites. These metabolites are finally stored in vacuoles or in the apoplast in the 3rd phase of pesticide detoxification. Brassinosteroids (BRs), a class of plant hormones, are well known to protect plants from abiotic stresses including that caused by the pesticides [9–12]. BRs also decrease oxidative stress in plants caused by pesticides, accompanied by enhanced activities of antioxidative enzymes [3] and a decrease of pesticide residues in plants [5, 6]. Exogenous application of BRs have been reported to enhance the expression of genes encoding P450, GST, superoxide dismutase (SOD), ascorbate peroxidase (APOX), catalase (CAT), glutathione reductase (GR) and GSH synthase, resulting in pesticide detoxification [3, 5]. Pesticide application causes oxidative stress to plants by producing reactive oxygen species (ROS), and persists in the form of pesticide residues. Antioxidative and pesticide detoxification enzymes help in scavenging ROS and reduction of pesticide residues. Keeping in mind the roles of BRs in boosting the antioxidative defence system of plants under pesticide toxicity, the present work was carried out to understand the 24-epibrassinolide (EBR)-regulated pesticide detoxification mechanism in Brassica juncea seedlings. In earlier studies, researchers have mostly applied BRs via foliar mode, but the present study was undertaken to access the effects of seed-soaking with EBR on oxidative stress and IMI residues in 10-day old B. juncea seedlings grown under IMI toxicity.
Methods
Plant germination
Seeds of B. juncea L. variety RLC-1 were soaked in 0 or 100 nM EBR L−1 for 8 h. IMI concentrations (0, 150, 200 and 250 mg IMI L−1) were prepared by dissolving IMI in distilled water. 3 mL of IMI solution were poured into each Petri-plate lined with Whatman#1 filter-paper. The EBR-soaked seeds of B. juncea were germinated in Petri-plates containing IMI solutions and kept in a seed germinator (25 ± 0.5 °C temperature, 16 h photoperiod, and 175 μmol m−2 s−1 light intensity). Seedlings were harvested after 10 days of sowing and analysed for contents of ROS and GSH, the activities of antioxidative enzymes, expression of genes and IMI residues. All the experiments were performed in triplicates. Each replicate consisted of one Petri-plate, and 10 seedlings were randomly selected from it.
Estimation of reactive oxygen species
Superoxide anions (O.2−)
The superoxide anion content was estimated according to Wu et al. [13]. One g of plant tissue was homogenized in 6 mL of phosphate buffer (65 mM, pH = 7.8) containing 1% of polyvinylpyrrolidone. The homogenate was centrifuged at 5000 × g for 15 min at 4 °C. To 0.5 mL of supernatant, 0.5 mL of phosphate buffer (65 mM, pH = 7.8) and 0.1 mL of hydroxylamine hydrochloride (10 mM) were added. The mixture was incubated at 25 °C for 30 min. After incubation, 1 mL of 3-aminobenzenesulphonic acid (58 mM) and 1 mL of 1-naphthylamine (7 mM) were added to the mixture, followed by an incubation at 25 °C for 20 min. The absorbance was measured at 530 nm. To calculate the superoxide content, a standard curve of sodium nitrite was used and content was expressed as μmol g−1 FW of seedlings.
Hydrogen peroxide (H2O2)
H2O2 was evaluated using method given by Patterson et al. [14]. Plant tissue (0.5 g) was crushed in 1 mL of acetone, followed by centrifugation at 5000 × g for 15 min at 4 °C. To the supernatant, 20 μL of 20% titanium chloride in concentrated HCl were added. Then 200 μL of ammonia solution (17 M) were added, followed by repeated washing of the precipitate with acetone. Washed precipitates were dissolved in 1.5 mL of H2SO4 (2 N). Absorbance was read at 410 nm. The content of hydrogen peroxide was calculated from a standard curve of H2O2 and was expressed as μmol g−1 FW of seedlings.
Estimation of antioxidative enzymes and glutathione content
Superoxide dismutase (SOD)
SOD activity was estimated according to Kono [15] with minor modifications. One g of plant tissue was homogenized in 3 mL of sodium carbonate buffer, followed by centrifugation at 12,000 × g at 4 °C for 20 min. Supernatant was used as sample for further analysis. The reaction mixture consisted of 1630 μL of sodium carbonate buffer (pH = 10.2), 500 μL of nitroblue tetrazolium (24 μM), 100 μL of EDTA (0.1 mM), 100 μL of hydroxylamine hydrochloride (1 mM), 100 μL of Triton-X-100 (0.03%) and 70 μL of sample. The absorbance was measured at 560 nm.
Catalase (CAT)
CAT activity was estimated according to Aebi [16] with slight modifications. 3 mL of 100 mM potassium phosphate buffer (PPB) with pH 7.0 were used for homogenization of 1 g of fresh seedlings. The homogenate was then centrifuged at 2000 × g for 20 min at 4 °C, and the supernatant was used to estimate the CAT activity. In a cuvette, the reaction mixture consisted of 1500 μL of PPB (pH = 7.0, 50 mM), 930 μL of hydrogen peroxide (15 mM) and 70 μL of sample. The absorbance was measured at 240 nm.
Guaiacol peroxidase (POD)
The activity of POD was determined using method given by Putter [17]. One g of seedlings was homogenized in 3 mL of PPB (100 mM, pH = 7.0) buffer, followed by centrifugation at 12,000 × g at 4 °C for 20 min. The supernatant was used as sample for further analysis. The absorbance was recorded at 436 nm by preparing a reaction mixture containing 70 μL sample with 2350 μL PPB (50 mM, pH 7.0), 50 μL guaiacol solution (20 mM) and 30 μL H2O2 (12 mM).
Glutathione reductase (GR)
GR activity was determined according to Carlberg and Mannervik [18]. One g of fresh seedlings was homogenized in 3 mL of PPB (100 mM, pH = 7.0) buffer followed by centrifugation at 12,000 × g at 4 °C for 20 min. The supernatant was used as plant sample to determine GR activity. The reaction mixture contained 1530 μL PPB (50 mM, pH 7.0), 300 μL each of ethylenediaminetetraacetate (EDTA) (3.0 mM), NADPH (0.1 mM), oxidized glutathione (1.0 M), and 70 μL sample. The absorbance was recorded at 340 nm.
Glutathione-S-transferase (GST)
The glutathione-S-transferase activity was quantified based on Habig and Jacoby [19]. One g of fresh seedlings was homogenized in 3 mL of PPB (100 mM, pH = 7.5) buffer and centrifuged at 12,000 × g at 4 °C for 20 min. The supernatant was used as sample in reaction mixture. The reaction mixture contained 70 μL sample, 1930 μL PPB (50 mM, pH 7.5), and 250 μL each of reduced glutathione (10 mM) and 1-chloro-2,4-dinitrobenzene (10 mM). The absorbance was measured at 340 nm.
Glutathione (GSH)
The glutathione content was determined according to the scheme given by Sedlak and Lindsay [20]. Fresh plant tissue (1 g) was homogenized in 3 mL of Tris buffer (50 mM, pH 10.0) containing 1 mM EDTA. The homogenate was then subjected to centrifugation at 12,000 × g for 15 min, and the supernatant from the plant extract was used to estimate the GSH content. To the 100 μL of plant extract, 1 mL of Tris buffer, 50 μL of Ellman’s reagent and 4 mL of absolute methanol were added, and kept at room temperature for 15 min and then subjected to centrifugation at 3000 × g for 15 min. The absorbance of the supernatant was measured noted at 412 nm.
Gene expression analysis
Total RNA was extracted from whole seedlings using Trizol method according to the manufacturer’s instructions (Invitrogen). Total RNA was used for reverse transcription with an RNA to cDNA kit (Invitrogen), containing MuLV as reverse transcriptase, dNTP’s mix, random octamers and oligo (dT)16. Gene-specific primers (Table 1) were designed according to the mRNA sequence from Genbank and EMBL database, and the actin gene was used as an internal control due to its high expression stability in the vegetative stage of plants. Quantitative real time PCR (qRT-PCR) was performed using the StepOne™ real time detection system (Applied Biosystems) and Power SYBR® Green PCR Master Mix (three biological and technical replicates). A melting curve was generated at the end of the each PCR cycle, which verified that a single product was amplified, using the software provided along with the PCR system. The mRNA quantification was based on the method of Livak and Schmittgen [21]. ΔCt values were obtained by subtracting the threshold value (Ct) of the internal control (actin) from that of the gene of interest. ΔΔCt values were obtained by subtracting the Ct values of the untreated control sample from the ΔCt value. The fold-changes in the expression levels relative to the untreated samples were expressed as 2-ΔΔCt.
Table 1.
Primers used for quantitative real time polymerase chain reaction (qRT-PCR)
| Gene name | Primer sequence |
|---|---|
| actin | Forward primer 5′ CTTGCACCTAGCAGCATGAA 3′ Reverse primer 5′ GGACAATGGATGGACCTGAC 3′ |
| SOD | Forward primer 5′GGTTTCCATGTCCATGCTCT 3′ Reverse primer 5′ATTGTGAAGGTGGCAGTTCC 3′ |
| CAT | Forward primer 5′ TCAGCTGCCAGTTAATGCAC 3′ Reverse primer 5′ GACAGCAGGTGGAGTTGGAT 3′ |
| GR | Forward primer 5′ AAGGCAAAAGAAGGTGCTGA 3′ Reverse primer 5′ AGTTCCCTTGCTGGTCTTCA 3′ |
| RBOH | Forward primer 5′ACGGGGTGTGATAGAGATGC 3′ Reverse primer 5′TTTTTCCAGTTGGGTCTTGC 3′ |
| NADH | Forward primer 5′CTCGGCCTTTCTCAACAGAC 3′ Reverse primer 5′CATTTCCCAAGTTTCCCAGA 3′ |
| CXE | Forward primer 5′ GGCGCTAACATGACTCATCA 3′ Reverse primer 5′ CTCCCAGAGTTGAGCGATTC 3′ |
| GSH-S | Forward primer 5′ CCCATCTTCAACGAGTTGGT 3′ Reverse primer 5′ GTGCAAACCCAAACGAATCT 3′ |
| GSH-T | Forward primer 5′GCTGGTCACAGGAACCATCT 3′ Reverse primer 5′CTACTTCAGTGCCCCACCAT 3′ |
| GST-1 | Forward primer 5′CGTCGTCGAAGAAGAAGAGG 3′ Reverse primer 5′TTTTTGGTGGGAGTTCCAAG 3′ |
| GST-2 | Forward primer 5′AGACCAAGCCGTTGTTGAAG 3′ Reverse primer 5′TTTTTGGTGGGAGTTCCAAG 3′ |
| GST-3 | Forward primer 5′TACGAGGCTAGGCTCAAGGA 3′ Reverse primer 5′AGCCACCCACTCGTTAACAC 3′ |
| GST-4 | Forward primer 5′CAAGGAACCAACCTTCTCCA 3′ Reverse primer 5′TGGTCAGTGGTCAAGCCATA 3′ |
| GST-5 | Forward primer 5′AGTGGCTGCAAAGCTTGTTT 3′ Reverse primer 5′TGTGGTGAAGATCGGTCAAA 3′ |
| GST-6 | Forward primer 5′GCCGAAGAGGAGGCTAAGTT 3′ Reverse primer 5′TCGGTGAAGAGCTTCTTGGT 3′ |
| POD | Forward primer 5′ TTCGAACGGAAAAAGATGCT 3′ Reverse primer 5′ AACCCTCCATGAAGGACCTC 3′ |
| P450 | Forward primer 5′ CATTTGTTCTCACCCACACG 3′ Reverse primer 5′ CACAACCGAGTTCGTGAATG 3′ |
Analysis of IMI residues
AOAC, the official method 2007.01 [22] was followed to prepare seedling extracts for IMI residue analysis. One g of seedlings was crushed in 1 mL 1% acetic acid in acetonitrile, and 200 mg of anhydrous MgSO4 and 50 mg sodium acetate were added. The mixture was shaken for 2 min, followed by centrifugation at 1500 × g for 5 min. 0.5 mL of the upper layer was taken, and 75 mg anhydrous MgSO4 along with 25 mg of primary secondary amine (PSA) sorbent were added. The mixture was again centrifuged, followed by filtration of the upper layer with 0.22 μ filters and stored at 4 °C until analysis.
GC-MS analysis
GC-MS (QP2010 Plus, Shimadzu, Kyoto, Japan) was used to analyse the seedling extracts for IMI residues. Helium was used as carrier gas, the initial oven column temperature was set at 50 °C, followed by increasing it to 125 °C at 25 °C min−1 and finally increased to 300 °C at 10 °C min−1 (hold for 15 min). The sample injector temperature was set to 250 °C, the injection mode was split, column flow rate was 1.70 mL min−1, the analytical column used was DB-5 ms. The ion source and interface temperatures were set to 200 °C and 280 °C respectively. The volume of the sample injected was 8 μL.
Statistical analysis
The results were analysed using Two-way ANOVA, Tukey’s HSD and multiple linear regression analysis (MLR), using self-coded software (MS-Excel 2010) and artificial neural networks (ANN), using Statistica-12. In ANN model, contents of oxidative stress markers and GSH, activities of antioxidative enzymes, relative expression of genes and IMI residues (targets) were regressed against the concentration of applied IMI and EBR (inputs), using 3 neurons, 1 hidden layer, tanh function from input to neurons, and from neurons to output.
Results
Effects of EBR on ROS
Imidacloprid (IMI) application was observed to increase the oxidative stress in B. juncea seedlings by enhancing the contents of H2O2 and O. 2 −. However, seed soaking with 100 nM EBR resulted in decreasing the contents of these reactive oxygen species (ROS) in the seedlings grown in IMI supplemented Petri-plates (Table 2). Statistical analysis of data using two-way ANOVA and Tukey’s HSD revealed that there was a significant difference in ROS contents in the seedlings of B. juncea plants. Multiple linear regression (MLR) analysis revealed that the partial regressions between the concentrations of EBR used for seed soaking and the contents of H2O2 and O. 2 − generated were regressed negatively, whereas IMI regressed positively on the generation of these ROS. This indicated the role of EBR in decreasing the ROS generation (Table 2).
Table 2.
Effect of EBR seed-soaking on the contents of stress markers and glutathione along with activities of antioxidative enzymes in Brassica juncea seedlings grown in IMI supplemented substratum (Mean ± SD, Two-way ANOVA, Tukey’s HSD, multiple linear regression)
| Treatments | H2O2 content (μmol g−1 FW) | O. 2 − content (μmol g−1 FW) | SOD activity (Units mg−1 protein) | CAT activity (μmol min−1 mg−1 protein) | POD activity (μmol min−1 mg−1 protein) | GR activity (μmol min−1 mg−1 protein) | GST activity (μmol min−1 mg−1 protein) | GSH content (mg g−1 FW) | |
| IMI (mg L−1) | EBR (nM) | ||||||||
| 0 | 0 | 12.70 ± 1.27 | 0.97 ± 0.15 | 29.27 ± 3.29 | 5.68 ± 0.74 | 35.10 ± 2.88 | 5.91 ± 0.24 | 26.48 ± 1.06 | 0.51 ± 0.02 |
| 0 | 100 | 11.48 ± 1.87 | 0.88 ± 0.12 | 38.85 ± 4.95 | 7.57 ± 0.83 | 52.94 ± 4.60 | 8.51 ± 0.59 | 34.20 ± 2.33 | 0.75 ± 0.05 |
| 150 | 0 | 15.22 ± 1.53 | 1.64 ± 0.29 | 36.25 ± 5.03 | 6.71 ± 0.98 | 44.14 ± 5.19 | 7.83 ± 0.60 | 28.99 ± 1.52 | 0.56 ± 0.03 |
| 150 | 100 | 10.11 ± 0.35 | 1.02 ± 0.20 | 53.98 ± 6.24 | 9.93 ± 1.14 | 71.46 ± 4.85 | 10.81 ± 0.66 | 37.47 ± 2.51 | 0.69 ± 0.03 |
| 200 | 0 | 17.41 ± 3.17 | 2.56 ± 0.52 | 32.85 ± 3.52 | 5.19 ± 0.85 | 68.23 ± 8.52 | 7.91 ± 0.41 | 31.25 ± 1.25 | 0.50 ± 0.04 |
| 200 | 100 | 10.94 ± 1.61 | 1.64 ± 0.17 | 50.72 ± 6.23 | 8.94 ± 0.50 | 94.40 ± 10.41 | 9.66 ± 1.20 | 38.67 ± 2.13 | 0.72 ± 0.07 |
| 250 | 0 | 21.70 ± 0.47 | 3.14 ± 0.60 | 27.98 ± 5.22 | 3.50 ± 0.74 | 43.93 ± 3.71 | 5.88 ± 0.64 | 25.71 ± 1.01 | 0.39 ± 0.06 |
| 250 | 100 | 14.81 ± 0.86 | 1.75 ± 0.29 | 42.67 ± 3.43 | 6.47 ± 0.78 | 74.28 ± 8.45 | 7.11 ± 1.03 | 32.89 ± 0.98 | 0.62 ± 0.04 |
| FIMI
FEBR FIMI×EBR HSD (p < 0.05) |
17.53*** 54.84*** 3.77* 4.59 |
25.53*** 30.32*** 3.98* 0.95 |
6.99** 56.66*** 0.95 13.77 |
16.14*** 74.60*** 1.32 2.31 |
33.01*** 89.59*** 0.99 18.60 |
19.73*** 51.55*** 1.77 2.06 |
14.00*** 122.45*** 0.16 4.81 |
10.79*** 128.84*** 2.03 0.08 |
|
| Multiple linear regression equation | β-regression coefficients | Multiple correlation coefficient | Significant at p < | ||||||
| βIMI | βEBR | ||||||||
| H2O2 content = 13.6710 + 0.0205 X1–0.0490 X 2 | 0.4994 | –0.6379 | 0.8102 | 0.001 | |||||
| O. 2 − content = 1.1777 + 0.0060 X1–0.0080 X 2 | 0.7037 | –0.4741 | 0.8485 | 0.001 | |||||
| SOD activity = 29.388 + 0.0146 X1 + 0.1497 X 2 | 0.1403 | 0.7662 | 0.7790 | 0.001 | |||||
| CAT activity = 5.8784–0.0041 X1 + 0.0296 X 2 | –0.1852 | 0.7222 | 0.7465 | 0.001 | |||||
| POD activity = 33.3930 + 0.0963 X1 + 0.2542 X 2 | 0.4660 | 0.6569 | 0.8054 | 0.001 | |||||
| GR activity = 6.8544 + 0.0002 X1 + 0.0214 X 2 | 0.0110 | 0.6247 | 0.6249 | 0.01 | |||||
| GST activity = 27.562 + 0.0036 X1 + 0.0770 X 2 | 0.0725 | 0.8226 | 0.8258 | 0.001 | |||||
| GSH content = 0.5507–0.0004 X1 + 0.0021 X 2 | –0.3077 | 0.8384 | 0.8931 | 0.001 | |||||
X1 = IMI (mg L−1), X 2 = EBR (nM), *, ** and *** = significant at p < 0.05, p < 0.01 and p < 0.001 respectively
Effects of EBR on Glutathione content and activities of antioxidative enzymes
The content of GSH as well as the activities of antioxidative enzymes like SOD, CAT, POD, GR and GST were enhanced in the seedlings raised from EBR-treated seeds and grown in Petri-plates supplemented with IMI solutions (Table 2). Significant differences in SOD, CAT, POD, GR and GST activities and GSH content were observed after analysing the data, using two-way ANOVA and Tukey’s HSD. MLR analysis and positive βEBR regression coefficients also showed that EBR application increases the GSH content and activities of antioxidative enzymes (Table 2).
Effects of EBR on gene expression
We observed that seed soaking with EBR significantly modulated the gene expression in B. juncea seedlings grown under IMI toxicity (Fig. 1). The fold-change expression of the RBO gene was decreased by 37.65% in seedlings raised from EBR-treated seed, and grown under IMI toxicity, as compared to seedlings raised from untreated seeds and grown under IMI toxicity. On similar comparison of B. juncea seedlings, an increase in the expression of SOD (39.42%), CAT (78.82%), GR (23.24%), POD (31.51%), GST1 (64.04%), GST2 (90.51%), GST3 (157.64%), GST5 (203.39%), GST6 (154.76%), GSH-S (56.52%), GSH-T (32.0%), CXE (99.28%), NADH (18.18%) and P450 (152.78%) was noticed (Fig. 1). We observed a significant difference in the expression of genes encoding enzymes involved in pesticide detoxification enzymes in B. juncea seedlings after analysing the data using two-way ANOVA and Tukey’s HSD. Positive βEBR values obtained from the MLR analysis revealed that EBR seed soaking up-regulated the expression of all the genes (encoding enzymes involved in pesticide detoxification enzymes), except GST-4 (Table 3).
Fig. 1.
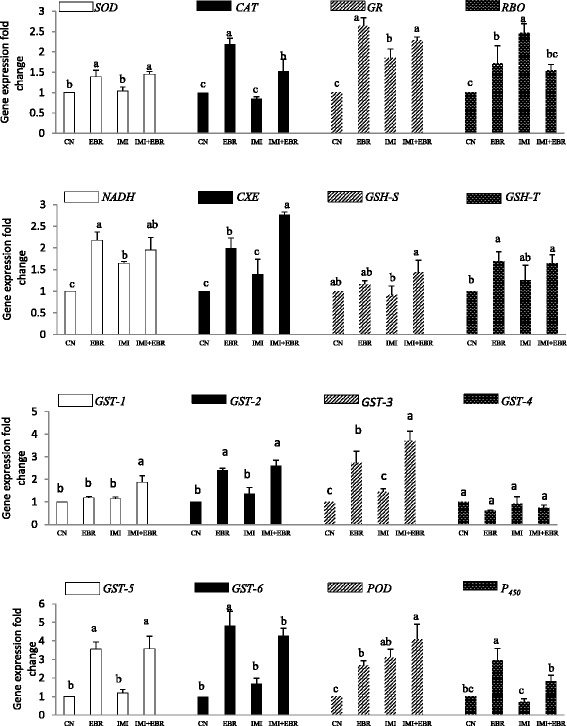
Effect of EBR seed-soaking on the relative expression of genes involved in IMI stress amelioration in Brassica juncea seedlings. Data are shown as the mean ± standard deviation (three biological replicates), bars with the same letters indicate no significant difference at p < 0.05 (comparison among treatments of same gene). HSD values for each gene have been mentioned in Table 3. (EBR concentration = 100 nM and IMI concentration = 200 mg IMI L−1)
Table 3.
Two-way ANOVA and multiple linear regression analysis (MLR) of the relative gene expression in Brassica juncea seedlings raised from EBR-soaked seeds and grown in IMI supplemented substratum
| Gene name | F-ratios | HSD (p < 0.05) | ||
| FIMI | FEBR | FIMI×EBR | ||
| SOD | 0.78 | 48.57*** | 0.01 | 0.26 |
| CAT | 16.66** | 87.07*** | 6.89* | 0.45 |
| GR | 7.05* | 124.34*** | 42.83*** | 0.41 |
| RBOH | 19.06** | 0.46 | 31.10*** | 0.66 |
| NADH | 4.16 | 51.72*** | 18.29** | 0.46 |
| CXE | 21.64** | 88.35*** | 2.26 | 0.56 |
| GSH-S | 0.93 | 10.57* | 2.86 | 0.46 |
| GSH-T | 0.59 | 17.10** | 1.14 | 0.60 |
| GST-1 | 21.14** | 26.40*** | 8.68* | 0.40 |
| GST-2 | 6.86* | 143.74*** | 0.52 | 0.49 |
| GST-3 | 12.82** | 99.63*** | 1.85 | 0.90 |
| GST-4 | 0.09 | 8.38* | 1.07 | 0.43 |
| GST-5 | 0.19 | 110.10*** | 0.11 | 1.06 |
| GST-6 | 0.059 | 143.70*** | 5.28 | 1.21 |
| POD | 40.71*** | 23.44** | 1.66 | 1.24 |
| P450 | 11.03* | 51.62*** | 4.06 | 0.96 |
| MLR equation | β-regression coefficient | Multiple correlation coefficient | Significant at p < | |
| βIMI | βEBR | |||
| SOD expression = 0.9960 + 0.0003 X1 + 0.0041 X 2 | 0.1169 | 0.9201 | 0.9274 | 0.001 |
| CAT expression = 1.1311–0.0020 X1 + 0.0093 X 2 | – 0.3748 | 0.8566 | 0.9351 | 0.001 |
| GR expression = 1.3034 + 0.0012 X1 + 0.0103 X 2 | 0.1968 | 0.8260 | 0.8491 | 0.001 |
| RBOH expression = 1.4136 + 0.0032 X1–0.0010 X 2 | 0.5702 | – 0.0892 | 0.5771 | 0.10 |
| NADH expression = 1.2212 + 0.0011 X1 + 0.0074 X 2 | 0.2250 | 0.7933 | 0.8246 | 0.01 |
| CXE expression = 0.9051 + 0.0029 X1 + 0.0118 X 2 | 0.4242 | 0.8571 | 0.9563 | 0.001 |
| GSH-S expression = 0.9113 + 0.0005 X1 + 0.0034 X 2 | 0.2040 | 0.6875 | 0.7170 | 0.02 |
| GSH-T expression = 1.0713 + 0.0005 X1 + 0.0055 X 2 | 0.1483 | 0.7983 | 0.8120 | 0.01 |
| GST-1 expression = 0.8681 + 0.0021 X1 + 0.0046 X 2 | 0.5737 | 0.6411 | 0.8604 | 0.001 |
| GST-2 expression = 1.0400 + 0.0015 X1 + 0.0133 X 2 | 0.2077 | 0.9504 | 0.9728 | 0.001 |
| GST-3 expression = 0.8635 + 0.0036 X1 + 0.0200 X 2 | 0.3238 | 0.9025 | 0.9588 | 0.001 |
| GST-4 expression = 0.9497 + 0.0002 X1–0.0030 X 2 | 0.0747 | – 0.6909 | 0.6949 | 0.02 |
| GST-5 expression = 1.0393 + 0.0005 X1 + 0.0248 X 2 | 0.0401 | 0.9643 | 0.9651 | 0.001 |
| GST-6 expression = 1.3087 + 0.0003 X1 + 0.0322 X 2 | 0.0195 | 0.9565 | 0.9567 | 0.001 |
| POD expression = 1.1777 + 0.0088 X1 + 0.0134 X 2 | 0.7426 | 0.5635 | 0.9322 | 0.001 |
| P450 expression = 1.2155–0.0036 X1 + 0.0154 X 2 | – 0.3842 | 0.8312 | 0.9157 | 0.001 |
X1 = IMI (mg L−1), X 2 = EBR (nM), *, ** and *** = significant at p < 0.05, p < 0.01 and p < 0.001 respectively
The Effect of EBR on IMI residues
IMI residues were reduced by 38% in the seedlings raised from EBR seed soaking and germinated in IMI-treated Petri-plates (Fig. 2). The analysis of data using two-way ANOVA and Tukey’s HSD showed a significant difference for IMI residues in B. juncea seedlings raised from untreated seeds and EBR treated seeds and grown under IMI toxicity. A negative βEBR regression coefficient for IMI residues also revealed that seed soaking with EBR results in decreasing the IMI residues (Table 4).
Fig. 2.
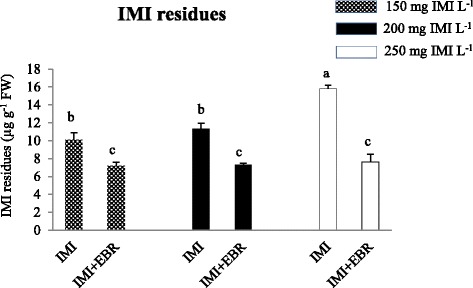
Effect of EBR seed-soaking on the IMI residues in B. juncea seedlings. Data are shown as the mean ± standard deviation (three biological replicates), bars with the same letters indicate no significant difference at p < 0.05. (EBR concentration = 100 nM)
Table 4.
Two-way ANOVA and multiple linear regression analysis (MLR) of the IMI residues in Brassica juncea seedlings raised from EBR-soaked seeds and grown in IMI supplemented substratum
| Parameter | F-ratios | HSD (p < 0.05) | ||
|---|---|---|---|---|
| FIMI | FEBR | FIMI×EBR | ||
| IMI residues | 44.21*** | 322.65*** | 33.31*** | 1.62 |
| MLR equation | β-regression coefficients | Multiple correlation coefficient | ||
| βIMI | βEBR | |||
| IMI residues = 6.2997 + 0.0305 X1 − 0.0500 X 2 | 0.4031 | –0.8117 | 0.9062*** | |
X1 = IMI (mg L−1), X 2 = EBR (nM), *** = significant at p < 0.001
Our data analysis using ANN also showed that experimental (target) and simulated (output) data were highly correlated, which indicates that ANN simulates the physiological studies carried out in the current experiment at a very high level of significance (Table 5).
Table 5.
Data showing correlation coefficients obtained from artificial neural networks model
| Parameter | Correlation coefficient | ||
|---|---|---|---|
| Train | Test | Validation | |
| SOD expression | 0.9255*** | 0.9990*** | 0.9178*** |
| CAT expression | 0.8995*** | 0.9681*** | 0.9940*** |
| POD expression | 0.9231*** | 0.9999*** | 0.9546*** |
| GST-1 expression | 0.9629*** | 0.9987*** | 0.9999*** |
| GST-2 expression | 0.9274*** | 0.7158*** | 0.9653*** |
| GST-3 expression | 0.9632*** | 0.9866*** | 0.9629*** |
| GST-4 expression | 0.9688*** | 0.9993*** | 0.9904*** |
| GST-5 expression | 0.7949*** | 0.4758** | 0.9498*** |
| GST-6 expression | 0.9696*** | 0.9988*** | 0.9976*** |
| GR expression | 0.9769*** | 0.9936*** | 0.9835*** |
| CXE expression | 0.9833*** | 0.9868*** | 0.9846*** |
| P450 expression | 0.9421*** | 0.9603*** | 0.9873*** |
| NADH expression | 0.9593*** | 0.9992*** | 0.9243*** |
| RBO expression | 0.9469*** | 0.9966*** | 0.9997*** |
| GSH-T expression | 0.9555*** | 0.9984*** | 0.9295*** |
| GSH-S expression | 0.8435*** | 0.9260*** | 0.9822*** |
| H2O2 content | 0.9053*** | 0.8859*** | 0.9317*** |
| O2 − content | 0.9017*** | 0.9784*** | 0.9379*** |
| GSH content | 0.8829*** | 0.8760*** | 0.9166*** |
| SOD activity | 0.8899*** | 0.9082*** | 0.9565*** |
| CAT activity | 0.9193*** | 0.9275*** | 0.9615*** |
| POD activity | 0.8475*** | 0.9081*** | 0.8491*** |
| GST activity | 0.9396*** | 0.9401*** | 0.9090*** |
| GR activity | 0.9235*** | 0.9269*** | 0.9573*** |
| IMI residues | 0.9890*** | 0.9809*** | 0.9766*** |
** and *** = significant at p < 0.01 and p < 0.001 respectively
Discussion
In the present investigation, the contents of ROS including and O. 2 − and H2O2 were noticed to increase with the application of IMI. A decrease in the contents of these ROS was noticed after EBR seed treatment. One of the reasons for the production of these ROS might be oxidative burst caused by abiotic stress conditions due to the disruption of the antioxidative defense system [23]. It has also been demonstrated that RBOH1 (Respiratory burst oxidase homologue1) is responsible for the production of H2O2 in plants under pesticide stress [4]. Moreover, in the current study, we also observed that the expression of the RBO gene (respiratory burst oxidase, a gene which is responsible for the production of H2O2) was up-regulated in B. juncea plants grown under IMI toxicity. It has been observed that the O2 .- content was decreased after seed application of EBR, and it might have occurred as a result of the conversion of O2 .- to H2O2 by superoxide dismutase (SOD), whose activity as well as gene expression (SOD) in the current experiment was also observed to be enhanced after the seed treatment of EBR. Additionally, the content of H2O2 was also noticed to decline with seed application of EBR, and the possible reason behind this could be the conversion of H2O2 to water and molecular oxygen by the antioxidative enzyme catalase (CAT). In the current study, the activity of the CAT enzyme and the expression of the CAT gene were also increased in seedlings raised from EBR-soaked seeds and grown in Petri-plates containing IMI. Moreover, the expression of RBO has also been observed to decrease with the EBR application, suggesting another reason for the significant reduction of H2O2. Studies carried out by Hayat et al. [24] and Fariduddin et al. [25] also showed that BRs play an important role in the scavenging of ROS in plants under environmental stress conditions.
As a result of oxidative stress caused by pesticide toxicity, the antioxidative defense system gets activated in order to efficiently scavenge the ROS and ultimately reduce the oxidative stress [3]. In the current study, the GSH content and the activities of enzymatic antioxidants including SOD, CAT, POD, GR and GST were observed to increase with IMI toxicity (except higher IMI concentration) as well as with the EBR seed application. SOD and CAT are involved in the conversion of harmful superoxide anions to non-toxic water and molecular oxygen. Additionally, it has also been reported that another pathway, the ascorbate-glutathione cycle, is involved in the detoxification of H2O2 [26]. Glutathione is involved in reduction of H2O2 into water by the ascorbate-glutathione cycle, which is also catalyzed by the GR enzyme [27]. In the ascorbate-glutathione cycle, GR also plays an important role in reduction of oxidative stress by maintaining the ratio of reduced and oxidized glutathione. In the current study, specific activity of GR was also observed to increase with the application of IMI, which suggests that GR is actively involved in detoxification of ROS generated as a result of IMI toxicity. Similar increase in GR activity was observed in Vigna radiata plants grown under chlorpyrifos toxicity [28]. EBR seed application in the current study further enhanced the activities of all these antioxidative enzymes under IMI toxicity. The alterations of the activities of these antioxidative enzymes might be due to the EBR modulated protein synthesis or altered enzyme kinetics [29, 30]. Moreover, in the current study, we noticed that gene expression of SOD, CAT, GR, POD and GST1-3,5-6 was also up-regulated in seedlings raised from EBR-treated seeds and grown under IMI toxicity. This suggests that the increase in the expression of the genes encoding these antioxidative enzymes might be one of the reasons for enhanced specific activities of antioxidative enzymes in B. juncea plants grown from EBR-treated seeds and grown under IMI stress.
In the current study, IMI residues were observed to decrease in seedlings raised from EBR-treated seeds and grown under IMI toxicity. As described in the introduction section, the three phased enzyme-mediated detoxification system is responsible for pesticide degradation in plants [7, 8]. In the current study, activities of POD and GST enzymes, which are involved in three phased pesticide detoxification system, were observed to increase with the IMI application as well as EBR seed treatment. Additionally, the gene expression of phase-1 enzyme viz., P450, POD, CXE and phase-2 enzyme GST along with the GSH-S and GSH-T was also observed to increase with EBR application under IMI toxicity. Oxidoreductase helps in pesticide detoxification [4] and the gene expression of NADH was also noticed to enhance after EBR seed application under IMI toxicity. Since the expression of genes (encoding enzymes involved in pesticide detoxification system) was modulated by EBR-seed soaking, this could be the possible reason for the reduction of IMI residues in B. juncea seedlings raised from EBR-treated seeds and grown under IMI toxicity.
Conclusions
The current study demonstrates that seed soaking with EBR enhances IMI detoxification and decreases oxidative stress in B. juncea seedlings through the up-regulation of SOD, CAT, GR, POD, NADH, CXE, GSH-S, GSH-T, P450 and GST1-3,5-6 genes involved in enzymatic pesticide detoxification accompanied by down-regulation of RBO gene. As compared to earlier studies that were based on foliar application of BRs, the important point in the current study is that a single application of EBR via seed treatment can efficiently activate the plant defence system against pesticide stress by modulation of gene expression.
Acknowledgements
The authors express their sincere gratitude to Department of Science and Technology, Govt. of India for providing INSPIRE fellowship to AS, and to the University Grants Commission, New Delhi for providing sophisticated instrumentation facilities. Thanks to Axios Review, two anonymous reviewers and their editor who provided valuable comments on an earlier version of this manuscript.
Funding
Not applicable.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Author’s contributions
AS, ST and VK performed the experiments. AS and AKT statistically analysed the data and wrote the manuscript. RB, AKT and AKK designed the experiment, supervised the work and critically checked the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- ANOVA
Analysis of variance
- AOAC
Association of official analytical chemists
- cDNA
Complimentary deoxyribonucleic acid
- EBR
24-epibrassionolide
- EMBL
European molecular biology laboratory
- FW
Fresh weight
- HSD
Honestly significant difference
- MLR
Multiple linear regression
- PCR
Polymerase chain reaction
- PSA
Primary secondary amine
- RNA
Ribonucleic acid
Contributor Information
Anket Sharma, Email: anketsharma@gmail.com.
Renu Bhardwaj, Email: renubhardwaj82@gmail.com.
References
- 1.Rajendran S. Environment and health aspects of pesticides use in Indian agriculture. In: Martin J, Bunch V, Suresh M, Kumaran TV, editors. Proceedings of the third international conference on environment and health. Department of Geography, University of Madras and Faculty of Environmental Studies, York University; 2003. p. 353–373.
- 2.Bonmatin JM, Marchand PA, Charvet R, Moineau I, Bengsch ER, et al. Quantification of imidacloprid uptake in maize crops. J Agric Food Chem. 2005;53:5336–41. doi: 10.1021/jf0479362. [DOI] [PubMed] [Google Scholar]
- 3.Sharma I, Bhardwaj R, Pati PK. Exogenous application of 28-homobrassinolide modulates the dynamics of salt and pesticides induced stress responses in an elite rice variety Pusa Basmati-1. J Plant Growth Regul. 2015;34:509–18. doi: 10.1007/s00344-015-9486-9. [DOI] [Google Scholar]
- 4.Zhou Y, Xia X, Yu G, Wang J, Wu J, et al. Brassinosteroids play a critical role in the regulation of pesticide metabolism in crop plants. Sci Rep. 2015;5:9018. doi: 10.1038/srep09018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia XJ, Zhang Y, Wu JX, Wang JT, Zhou YH, et al. Brassinosteroids promote metabolism of pesticides in cucumber. J Agric Food Chem. 2009;57:8406–13. doi: 10.1021/jf901915a. [DOI] [PubMed] [Google Scholar]
- 6.Cresswell JE. A meta-analysis of experiments testing the effects of a neonicotinoid insecticide (imidacloprid) on honey bees. Ecotoxicol. 2011;20:149–57. doi: 10.1007/s10646-010-0566-0. [DOI] [PubMed] [Google Scholar]
- 7.Coleman J, Blake-Kalff M, Davies E. Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation. Trends Plant Sci. 1997;2:144–51. doi: 10.1016/S1360-1385(97)01019-4. [DOI] [Google Scholar]
- 8.Cherian S, Oliveira MM. Transgenic plants in phytoremediation: recent advances and new possibilities. Environ Sci Technol. 2005;39:9377–90. doi: 10.1021/es051134l. [DOI] [PubMed] [Google Scholar]
- 9.Krishna P. Brassinosteroid-mediated stress responses. J Plant Growth Regul. 2003;22:289–97. doi: 10.1007/s00344-003-0058-z. [DOI] [PubMed] [Google Scholar]
- 10.Kanwar MK, Poonam, Bhardwaj R. Arsenic induced modulation of antioxidative defense system and brassinosteroids in Brassica juncea L. Ecotoxicol Environ Saf. 2015;115:119–25. doi: 10.1016/j.ecoenv.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 11.Sharma A, Kumar V, Singh R, Thukral AK, Bhardwaj R. Effect of seed pre-soaking with 24-epibrassinolide on growth and photosynthetic parameters of Brassica juncea L. in imidacloprid soil. Ecotoxicol Environ Saf. 2016;133:195–201. doi: 10.1016/j.ecoenv.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Sharma A, Kumar V, Thukral AK, Bhardwaj R. Epibrassinolide-imidacloprid interaction enhances non-enzymatic antioxidants in Brassica juncea L. Ind J Plant Physiol. 2016;21:70–5. doi: 10.1007/s40502-016-0203-x. [DOI] [Google Scholar]
- 13.Wu GL, Cui J, Tao L, Yang H. Fluroxypyr triggers oxidative damage by producing superoxide and hydrogen peroxide in rice (Oryza sativa) Ecotoxicol. 2010;19:124–32. doi: 10.1007/s10646-009-0396-0. [DOI] [PubMed] [Google Scholar]
- 14.Patterson BD, Mackae EA, Mackae I. Estimation of hydrogen peroxide in plants extracts using titanium (iv) Anal Biochem. 1984;139:487–92. doi: 10.1016/0003-2697(84)90039-3. [DOI] [PubMed] [Google Scholar]
- 15.Kono Y. Generation of superoxide radical during auto oxidation of hydroxylamine and an assay for superoxide dismutase. Arch Biochem Biophy. 1978;186:189–95. doi: 10.1016/0003-9861(78)90479-4. [DOI] [PubMed] [Google Scholar]
- 16.Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–6. doi: 10.1016/S0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 17.Putter J. Peroxidases. In: Bergmeyer HU, editor. Methods of enzymatic analysis. New York: Verlag Chemie-Academic Press; 1974. pp. 685–90. [Google Scholar]
- 18.Carlberg I, Mannervik B. Purification and characterization of the flavoenzyme glutathione reductase from rat liver. J Biol Chem. 1975;250:5475–80. [PubMed] [Google Scholar]
- 19.Habig WH, Jakoby WB. Glutathione S-transferases (rat and human) Methods Enzymol. 1981;77:218–31. doi: 10.1016/S0076-6879(81)77029-0. [DOI] [PubMed] [Google Scholar]
- 20.Sedlak J, Lindsay RHC. Estimation of total, protein bound and non protein sulfhydryl groups in tissue with Ellmann’s reagent. Anal Biochem. 1968;25:192–205. doi: 10.1016/0003-2697(68)90092-4. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.AOAC Official Method 2007.01. Pesticide residues in foods by acetonitrile extraction and partitioning with magnesium sulfate, gas chromatography/mass spectrometry and liquid chromatography/tandem mass spectrometry. AOAC Int. 2007.
- 23.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:405–10. doi: 10.1016/S1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 24.Hayat S, Khalique G, Wani AS, Alyemeni MN, Ahmad A. Protection of growth in response to 28-homobrassinolide under the stress of cadmium and salinity in wheat. Int J Biol Macromol. 2014;64:130–6. doi: 10.1016/j.ijbiomac.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Fariduddin Q, Yusuf M, Ahmad I, Ahmad M. Brassinosteroids and their role in response of plants to abiotic stresses. Biol Planta. 2014;58:9–17. doi: 10.1007/s10535-013-0374-5. [DOI] [Google Scholar]
- 26.Asada K, Takahash M. Production and scavenging of active oxygen in photosynthesis. In: Kyle DJ, Osmond CB, Arntzen CJ, editors. Photoinhibition. Amsterdam: Elsevier; 1987. pp. 227–87. [Google Scholar]
- 27.Chew O, Whelan J, Millar AH. Molecular definition of the Ascorbate-Glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem. 2003;47:46869–77. doi: 10.1074/jbc.M307525200. [DOI] [PubMed] [Google Scholar]
- 28.Parween T, Jan S. Mahmooduzzafar, Fatma T. Evaluation of oxidative stress in Vigna radiata L. in response to chlorpyrifos. Int J Environ Sci Technol. 2012;9:605–12. doi: 10.1007/s13762-012-0095-x. [DOI] [Google Scholar]
- 29.Lascano HR, Gomez LD, Casano LM, Trippi VS. Changes in glutathione reductase activity and protein content in wheat leaves and chloroplasts exposed to photooxidative stress. Plant Physiol Biochem. 1998;36:321–9. doi: 10.1016/S0981-9428(98)80046-6. [DOI] [Google Scholar]
- 30.Thao NP, Tran LS. Potentials toward genetic engineering of drought tolerant soybean. Crit Rev Biotechnol. 2012;32:349–62. doi: 10.3109/07388551.2011.643463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.


